Abstract
TCF-1 and LEF-1 are essential for early T cell development, but their roles beyond the CD4+CD8+ double positive (DP) stage are unknown. By specific ablation in DP thymocytes, we demonstrated that deficiency in TCF-1 and LEF-1 diminished CD4+ T cell output and redirected CD4+ T cells to a CD8+ T cell fate. The role of TCF-1 and LEF-1 in CD4-CD8 lineage choice was partly mediated by direct positive regulation of Th-POK. Furthermore, loss of TCF-1 and LEF-1 unexpectedly caused CD4 derepression in CD8+ lineage-committed T cells without affecting the expression of Runx factors. Instead, TCF-1 physically interacted with Runx3 to cooperatively silence the Cd4 gene. Thus, TCF-1 and LEF-1 adopt distinct genetic wiring to program CD4+ fate decision and establish CD8+ T cell identity.
CD4+ and CD8+ T cells, the essential mediators of cellular immune responses, are produced in the thymus following sequential maturation stages. Hematopoietic progenitors first seed the thymus and then make T cell lineage specification and commitment decisions at the CD4−CD8− double negative (DN) stage1, 2. While TCRβ recombination is completed at the CD25+CD44− DN3 stage, rearrangements at the TCRα locus occur after DN cells mature to CD4+CD8+ double positive (DP) thymocytes, followed by negative and positive selection. The positively selected DP thymocytes first give rise to CD4+CD8lo intermediate cells, which then differentiate into MHC class II-restricted CD4+ or MHC class I-restricted CD8+ single positive (SP) T cells, a decision known as CD4+ versus CD8+ lineage choice3.
The CD4+ versus CD8+ T cell lineage decision is influenced by the timing, intensity and duration of signals derived from TCR and cytokines3. A number of transcriptional factors intrinsically regulate this critical fate decision4, 5. Myb, GATA-3, Tox and Th-POK factors are specifically required for CD4+ T cell differentiation6, 7, 8, 9, and combined mutations of Runx1 and Runx3 completely abrogates CD8+ T cell production with limited effects on CD4+ T cell output10, 11. In terms of genetic interaction, Myb is required for induction of GATA-3 by TCR signals in DP thymocytes7. Upregulation of Th-POK is most evident in the CD4+8lo intermediates12 and depends on both Tox and GATA-36, 9. Th-POK is required to antagonize Runx3 activity and/or expression to promote CD4+ T cell lineage commitment11, and conversely, Runx3-mediated repression of Th-POK is critical for CD8+ T cell differentiation10, 12. Collectively, the Th-POK-Runx3 axis appears to be a critical convergence point in the CD4-CD8 lineage choice.
Once the decision to become either CD4+ or CD8+ SP thymocytes is made, lineage-inappropriate genes must be silenced in the committed T cells to ensure the distinct identity and functional divergence. Thus far, silencing of CD4+ T cell-specific genes, such as the CD4 coreceptor itself and the Th-POK transcription factor in CD8+ SP T cells is well characterized. Cd4 repression is mediated by a ~430 bp silencer sequence in its first intron13. Th-POK is encoded by Zbtb7b (called Thpok here for simplicity and consistency with the literature), and its repression in CD8+ T cells is regulated by a ~560 bp sequence upstream of the Thpok exon 1a10, 12. Both Cd4 and Thpok silencers contain consensus binding motifs for Runx factors, and combined mutations of Runx1 and Runx3 result in derepression of Cd4 and Thpok in CD8+ T cells10, 13.
TCF-1 and LEF-1 are members of the TCF-LEF family of transcription factors and are abundantly expressed in T cells14, 15. TCF-1 is induced by Notch activation and is essential for specification of hematopoietic progenitors to T cell lineage16, 17. TCF-1 and LEF-1 then act together to promote complete T lineage commitment, β-selection and maturation of DN thymocytes to the DP stage18, 19. In these early thymocytes, TCF-1 also restrains the expression of LEF-1, Id2 and key components in the Notch signaling pathway to prevent malignant transformation18, 20, 21. However, because germline deletion of TCF-1 and LEF-1 causes severe early T cell developmental block and embryonic lethality, respectively19, 22, their roles beyond the DP stage are unknown. In this study, we overcame these obstacles by conditionally ablating both TCF-1 and LEF-1 in DP thymocytes using CD4-Cre. Loss of TCF-1 and LEF-1 specifically impaired the differentiation of CD4+ SP T cells from the bipotent DP and CD4+8lo precursor cells and caused derepression of CD4 in committed CD8+ SP T cells. These findings broaden the spectra of TCF-1 and LEF-1-mediated regulatory activities in late stages of T cell development and reveal new insight into cell-fate decision mechanisms and establishment of cell identity.
Results
TCF-1 and LEF-1 are required for production of CD4+ T cells
To investigate a role for TCF-1 and LEF-1 in late stages of T cell development, we used CD4-Cre to conditionally inactivate both factors in DP thymocytes. Lef1-floxed mice have been previously established18. The Tcf7 gene (encoding TCF-1) was conditionally targeted by the International Knockout Mouse Consortium (IKMC, project 37596). Exon 4 of Tcf7 was flanked by two LoxP sites, and deletion of this exon resulted in a nonsense frame-shift mutation (Supplementary Fig. 1). Immunoblotting confirmed that CD4-Cre-mediated deletion was initiated in pre-select DP thymocytes and complete in the post-select DP cells, effectively eliminating all isoforms of both proteins (Fig. 1a).
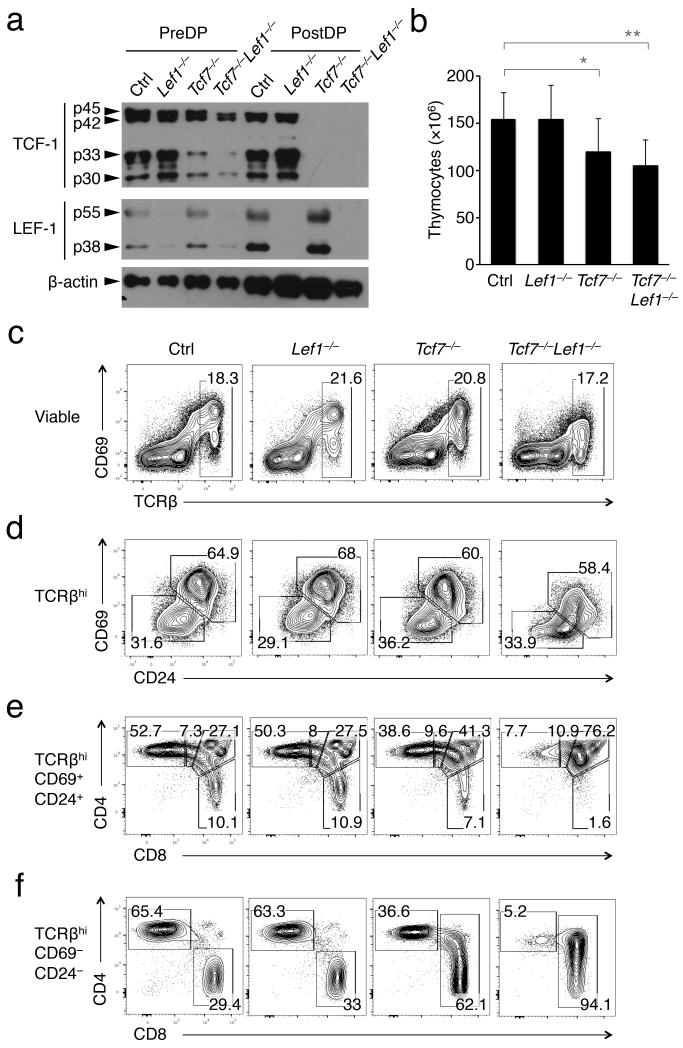
Figure 1. CD4-Cre-mediated deletion of TCF-1 or both TCF-1 and LEF-1 impairs CD4+ SP thymocyte development.
(a) Complete ablation of TCF-1 and LEF-1 proteins in post-select DP thymocytes. Thymocytes were surface-stained, FACS-sorted for TCRβlo-medCD69−CD4+CD8+ as pre-select DP (PreDP), and TCRβhiCD24+CD69+CD4+CD8+ as post-select DP (PostDP) thymocytes. Note that both TCF-1 and LEF-1 are expressed in multiple isoforms in thymocytes due to differential promoter usage and alternative splicing. Data are representative from 2 experiments. (b) Total thymic cellularity. Data are means ± s.d. from ≥ 5 independent experiments. *, p < 0.05; **, p < 0.01. (c) and (d) Analysis of post-select thymocytes. The TCRβhi thymocytes (c) are further fractionated into immature (CD24+CD69+) and mature (CD24− CD69−) subsets (d). The frequency of each subset is shown. (e) Loss of TCF-1 or both TCF-1 and LEF-1 diminishes the frequency of CD4+ SP thymocytes. The immature CD69+CD24+TCRβhi thymocytes were fractionated into DP, CD4+CD8lo, CD4+ SP and CD8+ SP subsets with their frequencies shown. (f) Loss of TCF-1 or both TCF-1 and LEF-1 causes derepression of the CD4 coreceptor in CD8+ lineage thymocytes. The mature CD69−CD24−TCRβhi thymocytes were separated into CD4+ and CD8+ subsets. The values are frequencies of CD4+ and CD8+ cells, with the latter including both CD8+CD4− and CD8+CD4+ cells that appear in Tcf7−/− and Tcf7−/−Lef1−/− mice. Data in c-f are representative of ≥ 5 independent experiments.
Due to the requirements of TCF-1 for T cell lineage specification, β-selection and thymocyte survival, germline deletion of TCF-1 results in diminished thymic cellularity to <5% of wild-type mice22. In contrast, CD4-Cre-mediated late deletion of TCF-1 or both TCF-1 and LEF-1 (called Tcf7−/− and Tcf7−/−Lef1−/− in this paper) only moderately reduced thymocyte counts, and CD4-Cre-Lef1fl/fl (Lef1−/−) mice showed similar thymic cellularity as littermate controls (Fig. 1b). Although the frequency of TCRβhi subset was not affected in Tcf7−/− and Tcf7−/−Lef1−/− thymi, the expression of CD69 was reduced on Tcf7−/−Lef1−/− TCRβhi thymocytes (Fig. 1c). The decreased CD69 expression was unlikely to be a result of diminished TCR signaling, because TCR-dependent upregulation of Gata3 and Tox was not affected in Tcf7−/−Lef1−/− post-select DP thymocytes (Supplementary Fig. 2). Downregulation of CD69 and CD24 marks intrathymic maturation of positively selected TCRβhi thymocytes. We found that maturation of CD24+CD69+ to CD24−CD69− thymocytes was not detectably perturbed in Tcf7−/− and Tcf7−/−Lef1−/− thymi (Fig. 1d). The CD24+CD69+TCRβhi subset contains post-selected DP thymocytes and CD4+8lo intermediates, which are immediate precursors to immature CD4+ or CD8+ SP thymocytes3. While T cell development was not apparently altered in Lef1−/− mice, we observed the accumulation of cells with a DP phenotype and the concomitant reduction in both CD4+ and CD8+ SP thymocytes in Tcf7−/− and Tcf7−/−Lef1−/− thymi (Fig. 1e). The CD24−CD69−TCRβhi subset contains mature SP thymocytes only11, and deletion of TCF-1 alone or with LEF-1 progressively diminished the frequency of CD4+ SP T cells (Fig. 1f), suggesting a requirement for TCF-1 and LEF-1 factors in effective production of CD4+ thymocytes.
TCF-1 and LEF-1 contribute to Cd4 gene silencing in CD8+ T cells
A fraction of mature Tcf7−/− CD8+ SP thymocytes showed increased expression of the CD4 coreceptor, and this fraction was substantially increased in Tcf7−/−Lef1−/− CD8+ T cells (Fig. 1f). By dividing the Tcf7−/−Lef1−/− TCRβhi CD8+ cells into CD8+CD4− and CD8+CD4+ subsets (the latter is called CD8*4 hereafter to distinguish from true DP cells), we found that both subsets expressed CD8β protein (Fig. 2a). Although not a focus of this study, we noted that both CD8α and CD8β expression was moderately reduced in Tcf7−/−Lef1−/− TCRβhi CD8+ cells (Fig. 2a). We next measured the expression of genes that are characteristic of CD4+ or CD8+ SP T cells. Runx3 is expressed in both cell types, but a distal promoter of the Runx3 gene is exclusively utilized in CD8+ T cells, generating a Runx3d transcript23. On the other hand, Thpok is only expressed in CD4+ T cells. The CD4−CD8+ and CD8*4 subsets from Tcf7−/−Lef1−/− thymi both expressed Runx3d but not Thpok (Fig. 2b). In addition, total Runx3 and Prf1 (encoding perforin) were more highly expressed in naïve CD8+ than in CD4+ SP T cells, and this trend was preserved in CD8+CD4− and CD8*4 subsets in Tcf7−/−Lef1−/− animals (Fig. 2b). These data suggest that the CD8*4 cells from Tcf7−/−Lef1−/− mice are bona-fide cytotoxic CD8+ T cells with derepressed CD4 coreceptor, similar to Runx3- or Runx3d-deficient CD8+ T cells11, 13.
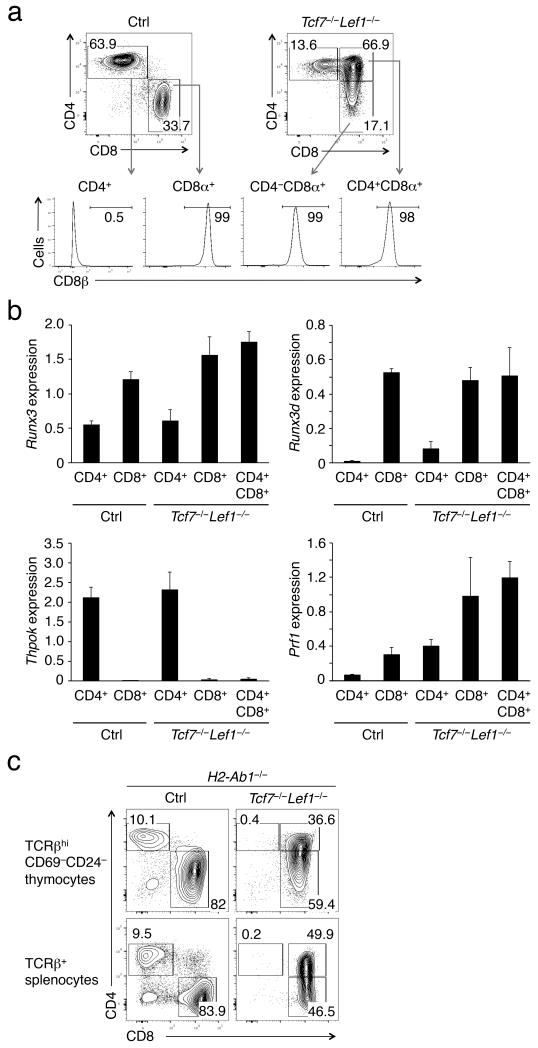
Figure 2. The CD4+CD8+ (CD8*4) mature thymocytes in Tcf7−/−Lef1−/− mice belong to the CD8+ lineage.
(a) The Tcf7−/−Lef1−/− CD8*4 mature thymocytes express CD8β. Mature TCRβhi thymocytes were analyzed for CD4, CD8α, and CD8β expression. (b) The Tcf7−/−Lef1−/− CD8*4 mature thymocytes express CD8+-characteristic genes. Shown is relative expression of each gene after normalization to Hprt1. Mature CD4+, CD8+, and CD8*4 thymocytes were sorted from Tcf7−/−Lef1−/− mice and littermate controls and analyzed for gene expression (n ≥ 3). (c) The Tcf7−/−Lef1−/− CD8*4 subset persists in the absence of MHC-II I-A and I-E molecules. Tcf7−/−Lef1−/− and control mice were crossed to an H2-Ab1−/− background, and the mature TCRβhi thymocytes were analyzed for CD4+ and CD8+ lineage distribution. Shown are representative data from 3 experiments.
To further substantiate this point, we crossed Tcf7−/−Lef1−/− and control mice to an H2-Ab1−/− background, in which CD4+ T cells were greatly diminished due to lack of MHC-II I-A and I-E molecules24. The MHC-I-selected T cells showed CD4 derepression in the absence of TCF-1 and LEF-1 (Fig. 2c), formally excluding the possibility that the CD8*4 T cells were CD4+ T cells with improper expression of CD8 coreceptors. These findings suggest a role of TCF-1 and LEF-1 in Cd4 gene silencing in CD8+ T cells.
TCF-1 and LEF-1 are critical for CD4+ T cell fate decision
Due to CD4 derepression in Tcf7−/−Lef1−/− CD8+ SP thymocytes, the true DP and the CD8*4 cells cannot be phenotypically distinguished in the immature CD24+CD69+ TCRβhi compartment. To accurately measure CD4+ and CD8+ SP T cell output, we therefore focused on the mature CD24−CD69− TCRβhi thymocytes, which do not contain DP cells in wild-type mice. Loss of TCF-1 alone caused a 40% reduction in mature CD4+ SP thymocytes, and deleting both TCF-1 and LEF-1 caused >80% reduction (Fig. 3a). Whereas TCF-1 deficiency did not have a significant impact on mature CD8+ SP thymocytes (counted as sum of CD8+CD4− and CD8*4 cells), loss of both factors substantially increased CD8+ SP T cell numbers (Fig. 3a). The ratio of mature CD4+ to CD8+ SP cells is approximately 2:1 in wild-type mice, but this ratio was reduced to 1:1 in Tcf7−/− mice and reversed to about 0.1:1 in Tcf7−/−Lef1−/− animals (Fig. 3b). The same phenotypic defects, including reduction in CD4+ SP T cell frequency and numbers and decreased CD4/CD8 ratio, persisted in the periphery (Supplementary Fig. 3a-c).
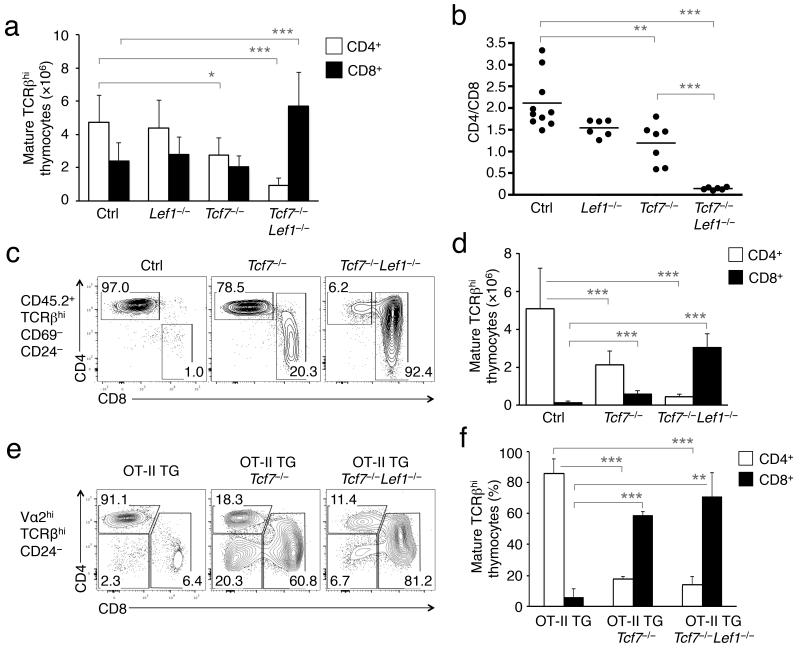
Figure 3. Deficiency in TCF-1 or both TCF-1 and LEF-1 redirects CD4+ T cells to the CD8+ lineage.
(a) Loss of TCF-1 or both TCF-1 and LEF-1 greatly diminishes CD4+ T cell output. Numbers of mature CD4+ and CD8+ SP thymocytes are shown as means ± s.d. (n ≥ 6). (b) Loss of TCF-1 or both factors reverses the CD4/CD8 ratio. Ratio of mature CD4+ to CD8+ cells was calculated from a. The horizontal line denotes mean value. *, p<0.05; **, p<0.01; ***, p<0.001. (c) MHC-II-selected thymocytes are redirected to CD8+ lineage in the absence of TCF-1 or both factors. BM cells from Tcf7−/−, Tcf7−/−Lef1−/−, or littermate controls were transplanted into lethally irradiated CD45.1+ congenic β2m−/− mice. Six weeks later, donor-derived (CD45.2+) mature CD69−CD24− TCRβhi thymocytes were analyzed for CD4+ and CD8+ lineage distribution. Representative contour plots (c) are from 4 independent experiments. Numbers of mature CD4+ and CD8+ thymocytes in the BM chimeras are shown in (d) as means ± s.d. (n ≥ 14). ***, p<0.001. (e) and (f) OT-II TCR transgenic T cells adopt a CD8+ fate in the absence of TCF-1 or both factors. The OT-II TG was crossed onto Tcf7−/−, Tcf7−/−Lef1−/−, or littermate controls. After gating on the Vα2+TCRβhi subset, mature CD24− thymocytes were analyzed for CD4+ and CD8+ lineage distribution. Representative contour plots (e) are shown (n ≥ 5 from 5 experiments). Frequencies of mature CD4+ and CD8+ OT-II thymocytes are summarized in (f). ***, p<0.001.
Germline deletion of TCF-1 reduces thymic cellularity by >95%, partly due to a critical requirement of TCF-1 for survival of early thymocytes25. By measuring active caspase-3 and caspase-7, we confirmed that early deletion of TCF-1 caused caspase activation in ~35% of post-select TCRβhi thymocytes (Supplementary Fig. 3d). However, the increase in caspase activation in TCRβhi thymocytes from Tcf7−/− or Tcf7−/− Lef1−/− mice was rather moderate (Supplementary Fig. 3e), indicating that CD4-Cre-mediated late deletion of TCF-1 and LEF-1 greatly alleviated the dependence of post-select thymocytes on TCF-1 and LEF-1 for survival. Importantly, caspase activation was similar between mature CD4+ and CD8+ SP thymocytes in Tcf7−/− or Tcf7−/−Lef1−/− animals (Supplementary Fig. 3f). These data suggest that TCF-1 and LEF-1 critically regulate CD4+ T cell fate decision, rather than preferentially promoting survival of CD4+ T cells. In addition, the residual CD4+ T cells in H2-Ab1−/− mice, which may have been selected on the H-2O MHC-II molecule26, were completely abrogated by loss of TCF-1 and LEF-1 (Fig. 2c), lending additional support for an essential role of TCF-1 and LEF-1 in promoting CD4+ lineage differentiation.
Deficiency in Th-POK or GATA-3 results in redirection of CD4+ T cells to the CD8+ lineage6, 8. In Tcf7−/−Lef1−/− mice, the reduction of CD4+ SP thymocytes was accompanied by an increase in CD8+ SP cells (Fig. 3a), indicative of lineage redirection. To further test this notion, we transplanted CD45.2+ bone marrow (BM) cells from Tcf7−/− and Tcf7−/−Lef1−/− mice into irradiated congenic CD45.1+β2m−/− recipients (Lef1−/− cells were not tested because loss of LEF-1 alone showed little impact). The β2m−/− mice are defective in MHC-I expression and thus have very few CD8+ SP T cells27. In β2m−/− hosts, mature TCRβhi thymocytes derived from BM cells of littermate controls were predominantly CD4+ (Fig. 3c,d). In contrast, Tcf7−/− BM cells gave rise to substantial amounts of mature CD8+ thymocytes, and mature thymocytes generated from Tcf7−/−Lef1−/− BM cells were predominantly CD8+ (Fig. 3c,d). These data indicate that MHC-II-selected thymocytes undergo a fate change from CD4+ to CD8+ T cells in the absence of TCF-1 or both TCF-1 and LEF-1. Consistent with the essential role of TCF-1 and LEF-1 in Cd4 silencing in CD8+ T cells (Fig. 2), the redirected CD8+ T cells lacking TCF-1 or both factors continued to exhibit derepression of the CD4 coreceptor (Fig. 3c).
In Tcf7−/−- or Tcf7−/−Lef1−/−-reconstituted β2m−/− BM chimeras, the redirected CD8+ T cells persisted in the periphery (Supplementary Fig. 4a,b). To determine if the redirected cells truly acquired CD8+ T cell identity and function or were CD4+ T cells with aberrant CD8 expression, we sorted the redirected CD8+ cells from the Tcf7−/−Lef1−/− BM chimeras into two subsets, CD8+CD4− and CD8*4. Gene expression analysis revealed that both subsets expressed CD8+ T cell-specific Runx3d and higher basal amounts of Prf1, but express little, if any, CD4+ T cell-specific Thpok transcripts (Supplementary Fig. 4c). We also activated the redirected CD8+ T cells in vitro. Similar to wild-type CD8+ T cells, the redirected Tcf7−/−Lef1−/− CD8+ T cells more proficiently produced granzyme B and interferon-γ, but less effectively produced IL-2 or induced CD40L than CD4+ T cells (Supplementary Fig. 4d). These observations further corroborate a complete fate change of MHC-II-selected T cells to cytotoxic CD8+ T cell lineage in the absence of TCF-1 and LEF-1.
We next investigated if deficiency in TCF-1 and LEF-1 changed the fate of T cells that express a fixed MHC-II-restricted TCR. To this end, we crossed the OT-II CD4+ TCR transgene (TG) with Tcf7−/−Lef1−/− mice. We detected the OT-II TCR by gating on TCRβhiVα2+ thymocytes, which are predominantly CD4+ T cells under TCF-1 and LEF-1-sufficient conditions (Fig. 3e,f). In contrast, deletion of TCF-1 or both TCF-1 and LEF-1 reduced the frequency of TCRβhiVα2+ mature CD4+ thymocytes with a concomitant increase in the CD8+ compartment (Fig. 3e,f). In these mice, derepression of CD4 was also observed in the redirected OT-II CD8+ T cells (Fig. 3e). Of note, in the presence of the OT-II transgene, CD4-Cre-mediated deletion of TCF-1 or both factors caused more severe reduction in total thymic cellularity (Supplementary Fig. 5a, compare with Fig. 1b). This is likely due to altered timing of target excision by CD4-Cre due to the TCR TG, because we observed early deletion of Tcf7 and Lef1 in DN thymocytes from OT-II TG Tcf7−/−Lef1−/− mice (Supplementary Fig. 5b). Nonetheless, mature OT-II+CD8+ thymocytes were more dominant over CD4+ SP thymocytes upon deletion of TCF-1 or both TCF-1 and LEF-1 (Supplementary Fig. 5c). Collectively, loss of TCF-1 and LEF-1 resulted in a fate change of MHC-II-selected cells to the CD8+ lineage, regardless if polyclonal TCRs or a fixed MHC-II-restricted TCR was expressed on the post-select DP thymocytes. These findings unambiguously reveal an essential role of TCF-1 and LEF-1 in directing the bipotent precursors to the CD4+ T cell lineage.
TCF-1 regulates balanced expression of Thpok and Runx3d
Several transcriptional factors have been characterized as intrinsic regulators of CD4-CD8 fate decision. Among these, Myb, GATA-3, Tox and Th-POK factors direct post-select DP cells to the CD4+ T cell lineage, and Runx factors ensure CD8+ T cell differentiation4, 5. We thus investigated if TCF-1 and LEF-1 regulate these key factors involved in lineage choice. Because double deficiency in TCF-1 and LEF-1 resulted in strong CD4 derepression in CD8+ SP T cells (Fig. 2), the resulting CD8*4 cells cannot be adequately separated from actual post-select DP thymocytes in Tcf7−/−Lef1−/− mice. To avoid misinterpretation of the data, we focused our gene expression analysis on Tcf7−/− thymocytes.
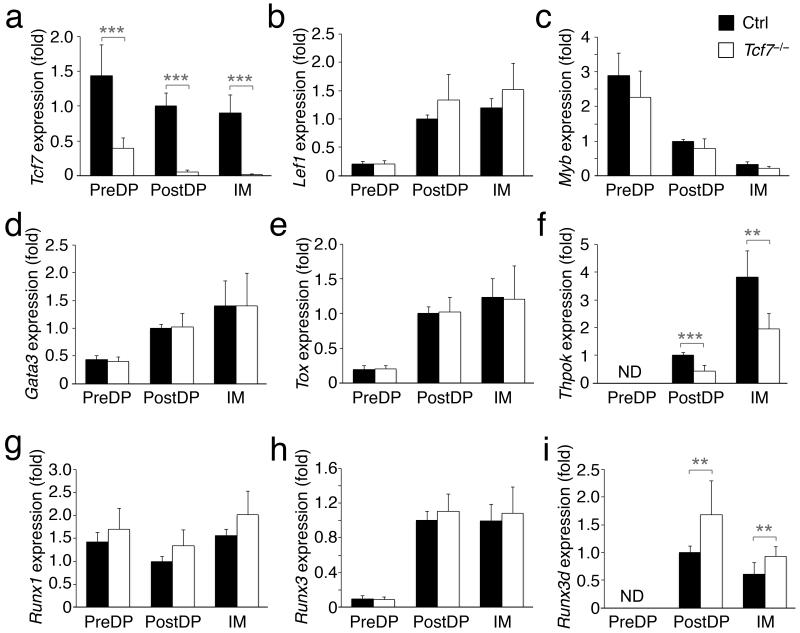
To discern kinetic changes of gene expression at distinct stages prior to complete lineage commitment, we purified pre-select DP (TCRβlo-med DP), post-select DP (TCRβhi DP) and CD4+8lo intermediates by cell sorting. Similar to immunoblotting results (Fig. 1a), CD4-Cre-mediated deletion of Tcf7 was more complete in post-select DP stage and beyond (Fig. 4a). While Tcf7 expression was relatively constant throughout these stages, Lef1 exhibited dynamic changes similar to Gata3 and Tox, as it was upregulated by positive selection signals and had a sustained high expression in CD4+8lo thymocytes (Fig. 4b,d,e)6, 9. The induction of Thpok was more potent at the CD4+8lo stage (Fig. 4f)12; in contrast, Myb expression was progressively downregulated in post-select DP and CD4+8lo cells (Fig. 4c). Deletion of TCF-1 did not affect the kinetic changes in Gata3, Tox and Myb expression (Fig. 4c-e); but substantially diminished the expression of Thpok in both post-select DP and CD4+8lo thymocytes (Fig. 4f). TCF-1 deficiency did not significantly alter the expression of Runx1 or total Runx3 (transcribed from both distal and proximal promoters) (Fig. 4g,h). Runx3 protein in CD8+ SP T cells is exclusively produced from the Runx3d transcript, and in fact, transcription from the Runx3 distal promoter is initiated at the post-select DP stage11. Significantly, specific deletion of Runx3d and complete ablation of Runx3 show remarkably similar effects on CD8+ lineage differentiation and Cd4 silencing in CD8+ T cells11, 23. Thus, Runx3d expression is more directly linked to the activity of Runx3 in lineage choice. Detection of Runx3d transcripts, using primers that were specifically complementary to cDNA transcribed from the distal promoter in quantitative RT-PCR, revealed that TCF-1 deficiency resulted in increased expression of Runx3d in both post-select DP and CD4+8lo cells (Fig. 4i). These gene expression analyses collectively suggest that TCF-1 and LEF-1 impinge on balanced expression of Thpok and Runx3d to regulate CD4-CD8 fate decision.
Figure 4. TCF-1 deficiency decreases Thpok but increases Runx3d expression in the bipotent precursors.
Thymocytes from Tcf7−/− and littermate controls were sorted for 3 subsets, pre-select DP (PreDP), post-select DP (PostDP), and CD4+8lo intermediate (IM). The expression of indicated genes was measured by quantitative RT-PCR. To demonstrate kinetic changes of each gene during these developmental stages, the expression of each gene in control post-select DP (after normalization to Hprt1) was arbitrarily set to 1, and the relative expression of this gene in all other control or Tcf7−/− subsets was then normalized accordingly and presented as fold changes. Data are means ± s.d. from 4 independent experiments (n ≥ 6). ND, not reliably detected. *, p<0.05; **, p<0.01; ***, p<0.001.
TCF-1 and LEF-1 act upstream of Th-POK to promote CD4+ T cell differentiation
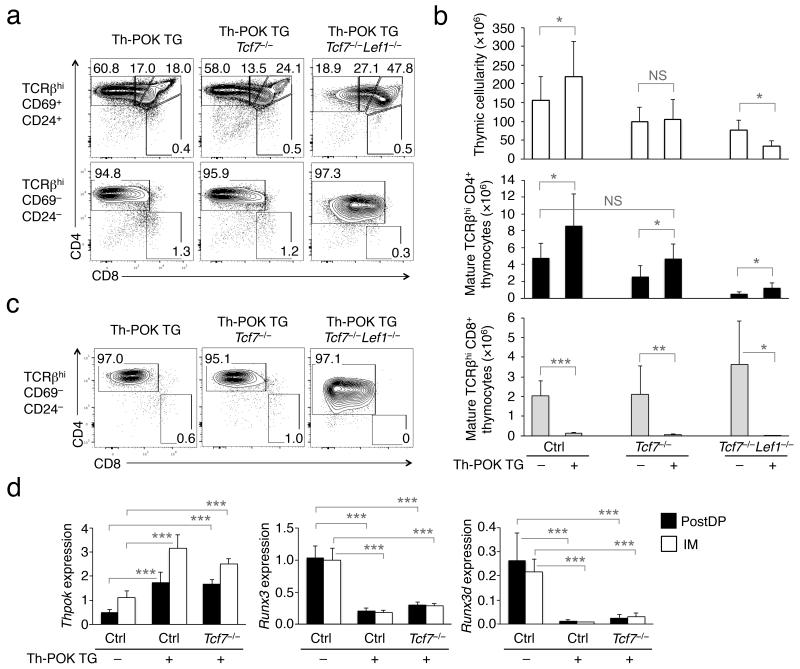
Because Th-POK and Runx3d mutually antagonize each other’s expression and/or activity11, 28, we next investigated which is the primary factor that acts downstream of TCF-1 and LEF-1. We crossed Tcf7−/− or Tcf7−/−Lef1−/− mice with a Th-POK TG driven by the human CD2 promoter. Consistent with previous reports6, 29, ectopic expression of Th-POK directed all post-select DP thymocytes to the CD4+ lineage, regardless of MHC restrictions. In Th-POK TG Tcf7−/− mice, Th-POK overexpression suppressed CD8+ T cell differentiation and increased the frequency of CD4+ T cells in immature and mature TCRβhi thymocytes (Fig. 5a). Importantly, Th-POK TG Tcf7−/− mice had similar numbers of mature CD4+ SP thymocytes as littermate controls without the TG (Fig. 5b), suggesting that ectopic Th-POK expression is sufficient to rescue CD4+ T cell differentiation defects caused by loss of TCF-1.
Figure 5. Ectopic expression of Th-POK rectifies defects in CD4+ T cell differentiation caused by loss of TCF-1.
(a) Th-POK TG represses CD8+ differentiation in the absence of TCF-1 or both TCF-1 and LEF-1. Th-POK TG was crossed onto Tcf7−/− or Tcf7−/−Lef1−/− mice. The immature and mature TCRβhi thymocytes were analyzed for CD4+ and CD8+ lineage distribution. Frequency of each subset is shown in the representative data from ≥ 3 experiments. (b) Cumulative data on the impact of Th-POK TG on the numbers of total, mature CD4+ and CD8+ SP thymocytes. Data are means ± s.d. from 5 experiments (n ≥ 6). *, p<0.05; **, p<0.01; ***, p<0.001; NS, not statistically significant. (c) Th-POK TG prevents CD4+ to CD8+ lineage redirection caused by deficiency in TCF-1 or both TCF-1 and LEF-1. BM cells from Th-POK TG, Th-POK TG Tcf7−/−, or Th-POK TG Tcf7−/−Lef1−/− were transplanted into irradiated CD45.1+ β2m−/− recipients. Six weeks later, CD45.2+ mature TCRβhi thymocytes were analyzed for CD4+ and CD8+ distribution. Representative data from ≥ 3 experiments are shown. (d) Ectopic expression of Th-POK represses Runx3d in the presence or absence of TCF-1. Post-select DP (PostDP) and CD4+8lo intermediate (IM) thymocytes were sorted and analyzed for gene expression. Shown is relative expression of each gene after normalization to Hprt1. Data are representative from 3 independent experiments. ***, p<0.001.
Although Tcf7−/−Lef1−/− mice had increased CD8+ output at the expense of CD4+ T cells, ectopic Th-POK expression repressed CD8+ T cell differentiation in Th-POK TG Tcf7−/−Lef1−/− mice (Fig. 5a,b). Consequently, mature TCRβhi thymocytes in Th-POK TG Tcf7−/−Lef1−/− mice were almost exclusively CD4+, and the frequency of CD4+ SP thymocytes in the immature TCRβhi subset was increased compared with Tcf7−/−Lef1−/− mice (compare Fig. 5a with Fig. 1e,f). In Th-POK TG Tcf7−/−Lef1−/− mice, the number of mature CD4+ SP thymocytes increased by about 50% over that in Tcf7−/−Lef1−/− animals, but remained substantially lower than those in littermate controls without the TG (Fig. 5b). These observations suggest that compared with loss of TCF-1 alone, double deficiency in TCF-1 and LEF-1 caused additional alterations, and as a result, the defective CD4+ T cell differentiation cannot be sufficiently reversed by overexpressing Th-POK. In line with this notion, the combination of Th-POK overexpression and TCF-1 and LEF-1 double deficiency may have more complex effects on late T cell development, causing further reduction in total thymocytes (Fig. 5b) and less aggregated expression of the CD4 coreceptor in the “rectified” mature CD4+ SP thymocytes (Fig. 5a).
We next examined the effect of ectopic Th-POK expression on CD4+ to CD8+ lineage redirection caused by loss of TCF-1 or both TCF-1 and LEF-1. We transplanted BM cells from Th-POK TG Tcf7−/− or Th-POK TG Tcf7−/−Lef1−/− mice into CD45.1+ β2m−/− mice. In the chimeric hosts, lineage redirection was completely blocked by the Th-POK TG in mature TCRβhi thymocytes (compare Fig. 5c with 3c). In line with this observation, ectopic Th-POK expression diminished Runx3 and Runx3d in TCF-1-sufficient post-select DP and CD4+8lo thymocytes, and more importantly, prevented Runx3d upregulation in Tcf7−/− cells (compare Fig. 5d with 4i). These data collectively suggest that Th-POK acts downstream of TCF-1 and LEF-1 in regulating the CD4-CD8 lineage choice.
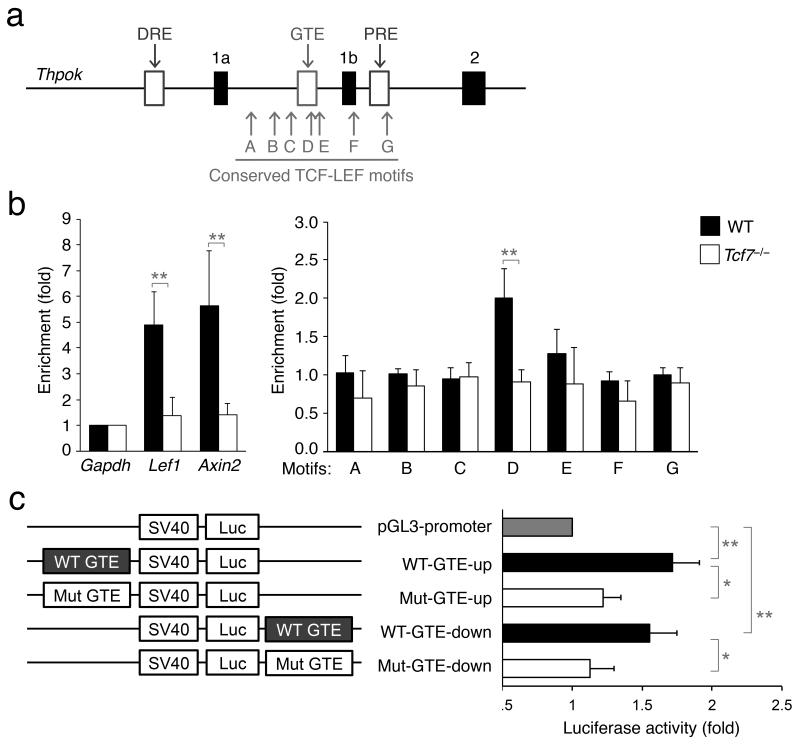
We then investigated if TCF-1 directly regulates Thpok gene expression. Previous studies have demonstrated that the −17 kb to +1 kb region flanking the first Thpok coding exon contains all the cis-elements required for its dynamic expression during thymocyte development12. Within this region, we found seven conserved TCF-LEF consensus binding sequences (T/A)CAAAG, designated as A-G, between exons 1a and 2 of the Thpok gene (Fig. 6a). We performed chromatin immunoprecipitation (ChIP) with a TCF-1 antibody30 or control IgG using sorted post-select DP and CD4+8lo thymocytes. We observed enriched binding of TCF-1 to the Lef1 and Axin2 gene segments (Fig. 6b), two known TCF-1 target genes. Among the seven conserved TCF-LEF motifs, TCF-1 bound specifically to motif D (Fig. 6b). TCF-1 binding to these genomic locations was abrogated in sorted post-select DP and CD4+8lo thymocytes from Tcf7−/− mice (Fig. 6b), indicating the binding specificity. It is important to note that motif D is located in a previously defined “general T lymphoid element” (GTE) which contributes to positive regulation of Thpok in T cells12. Analysis of data from a recent study that performed TCF-1 ChIP-Seq on whole thymocytes31 identified a strong TCF-1 binding peak at the Thpok GTE element (Supplementary Fig. 6a), consistent with our finding using the ChIP-PCR tiling assay.
Figure 6. TCF-1 acts through the GTE in the Thpok gene locus.
(a) Identification of conserved TCF-LEF motifs in the Thpok gene locus, as marked by arrows (A through G). Partial structure of the Thpok gene is shown, with filled bars denoting exons. Open bars denote the following cis-elements, DRE (distal regulatory element, also known as Thpok silencer), GTE, and PRE (proximal regulatory element). The sizes of exons and regulatory elements are not drawn to scale. (b) TCF-1 binds to the GTE in the Thpok locus. Post-select DP and CD4+8lo thymocytes were sorted together from WT or Tcf7−/− mice and used in ChIP with anti-TCF-1 or control IgG followed by quantitative PCR. Enrichment by TCF-1 antibody at each motif or locus was first normalized to IgG, and then normalized to that at the Gapdh locus. Two known TCF-1 target genes, Lef1 and Axin2, were detected as positive controls. Data are pooled results from ≥3 experiments. **, p<0.01. (c) Mutation of TCF-1 sites in the GTE abrogates its enhancer activity. The luciferase reporter constructs (shown on the left) were transfected into the EL-4 cells by electroporation, and 48 hrs later the luciferase activity was measured. Luciferase activity driven by the SV40 alone (pGL3 promoter) is arbitrarily set to 1, and that containing WT or mutant (Mut) GTE was normalized accordingly. Data are means ± s.d. from 2 experiments (n = 3). *, p<0.05; **, p<0.01.
The 473-bp GTE contains two sets of highly conserved CAAAG motifs (Supplementary Fig. 7a,b). To investigate the contribution of these potential TCF-1 sites to the enhancer activity of GTE, we cloned wild-type GTE either upstream or downstream of a luciferase reporter driven by an SV40 promoter. We then mutated both “CAAAG” motifs in the GTE to “accct” to generate mutant reporter constructs. Regardless of the location of insertion, inclusion of the wild-type GTE increased the reporter activity, and importantly, mutation of both TCF-1 sites almost completely abrogated the increase (Fig. 6c). This was observed in EL-4 thymoma as well as 293T cell lines (Supplementary Fig. 7c). Collectively, these findings support the notion that TCF-1 acts directly upstream of Th-POK in directing the bipotent DP or CD4+8lo precursors to the CD4+ T cell lineage.
TCF-1 and LEF-1 do not depend on Runx3 in T cell lineage choice
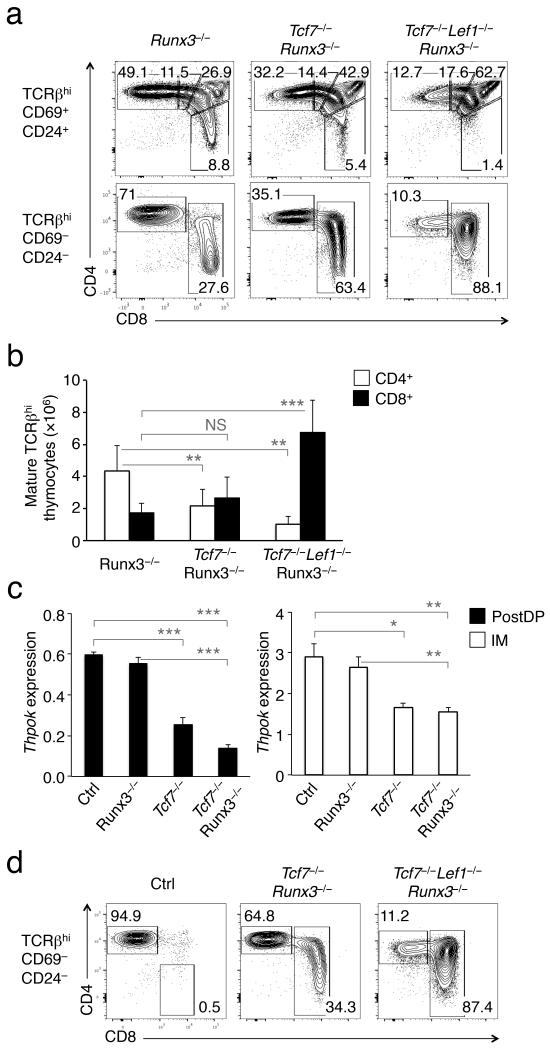
We showed above that TCF-1 positively regulated Thpok, which in turn repressed Runx3d expression. A parallel mechanism could be that TCF-1 directly represses Runx3d, which then negatively regulates Thpok. To test this, we ablated total Runx3 expression by crossing CD4-Cre-Runx3fl/fl (Runx3−/−) mice to Tcf7−/− and Tcf7−/−Lef1−/− strains. Runx3−/− mice have a moderate reduction of CD8+ SP T cells in the thymi and periphery23. However, Tcf7−/−Runx3−/− mice showed clearly reduced CD4+ T cell frequency and numbers in both immature and mature TCRβhi thymocytes compared with Runx3−/− mice (Fig. 7a,b). Moreover, Tcf7−/−Lef1−/−Runx3−/− mice exhibited more severe loss of mature CD4+ SP thymocytes, with CD8+ T cells remaining dominantly abundant, similar to Tcf7−/−Lef1−/− animals (compare Fig. 7a,b with Fig. 1e,f and 3a). Thus, Runx3 deficiency failed to rectify defective CD4+ lineage choice in Tcf7−/− or Tcf7−/−Lef1−/− mice. In line with this, we found that Thpok expression in post-select DP and CD4+8lo thymocytes from Runx3−/− mice was similar to littermate controls, and importantly, Tcf7−/−Runx3−/− post-select DP and CD4+8lo thymocytes expressed similar lower amounts of Thpok transcripts as Tcf7−/− cells (Fig. 7c).
Figure 7. Deletion of Runx3 does not rescue CD4+ differentiation defects caused by loss of TCF-1 and LEF-1.
(a) CD4-Cre-Runx3fl/fl (Runx3−/−) mice were crossed with Tcf7−/− and Tcf7−/−Lef1−/− strains to acquire Tcf7−/−Runx3−/− and Tcf7−/−Lef1−/−Runx3−/− animals. The immature and mature TCRβhi thymocytes were analyzed for CD4+ and CD8+ lineage distribution. Frequency of each subset is shown in the representative data from 4 experiments. (b) Cumulative data on the impact of Runx3 deletion on numbers of mature CD4+ and CD8+ SP thymocytes (n ≥ 5 from 4 experiments). (c) Deletion of Runx3 does not reverse the decrease in Thpok expression caused by TCF-1 deficiency. Post-select DP (PostDP) and CD4+8lo intermediate (IM) thymocytes were sorted and analyzed for Thpok gene expression. Shown is relative Thpok expression after normalization to Hprt1. *, p<0.05; **, p<0.01; ***, p<0.001. NS, not statistically significant. (d) Deletion of Runx3 does not prevent CD4+ to CD8+ lineage redirection caused by deficiency in TCF-1 or both TCF-1 and LEF-1. BM cells from Runx3−/−, Tcf7−/−Runx3−/− and Tcf7−/−Lef1−/−Runx3−/− mice were transplanted into irradiated CD45.1+ β2m−/− recipients. Six weeks later, CD45.2+ mature TCRβhi thymocytes were analyzed for CD4+ and CD8+ distribution. Data are representative from 3 experiments (n ≥ 6).
Overexpression of Runx3 increases CD8+ T cell output, but whether this is a result of lineage redirection remains controversial32. To determine if increased CD8+ T cell frequency and numbers in Tcf7−/−Lef1−/− mice results from aberrant upregulation of Runx3d and ensuing lineage redirection, we made β2m−/− BM chimeras with donor cells from Runx3−/−, Tcf7−/−Runx3−/− and Tcf7−/−Lef1−/−Runx3−/− mice. As expected, mature TCRβhi thymocytes derived from Runx3−/− BM cells were predominantly CD4+ in the β2m−/− recipients. However, mature TCRβhi thymocytes derived from Tcf7−/−Runx3−/− or Tcf7−/− Lef1−/−Runx3−/− BM cells exhibited CD4+ to CD8+ T cell lineage redirection, similar to those from Tcf7−/− or Tcf7−/−Lef1−/− BM cells (compare Fig. 7d with Fig. 3c). Therefore, regardless if TCF-1 and LEF-1 directly or indirectly suppress Runx3d expression, Runx3 proteins are not essential for TCF-1- and LEF-1-mediated regulation of CD4-CD8 lineage choice.
TCF-1 and LEF-1 cooperate with Runx3 in Cd4 gene silencing in CD8+ T cells
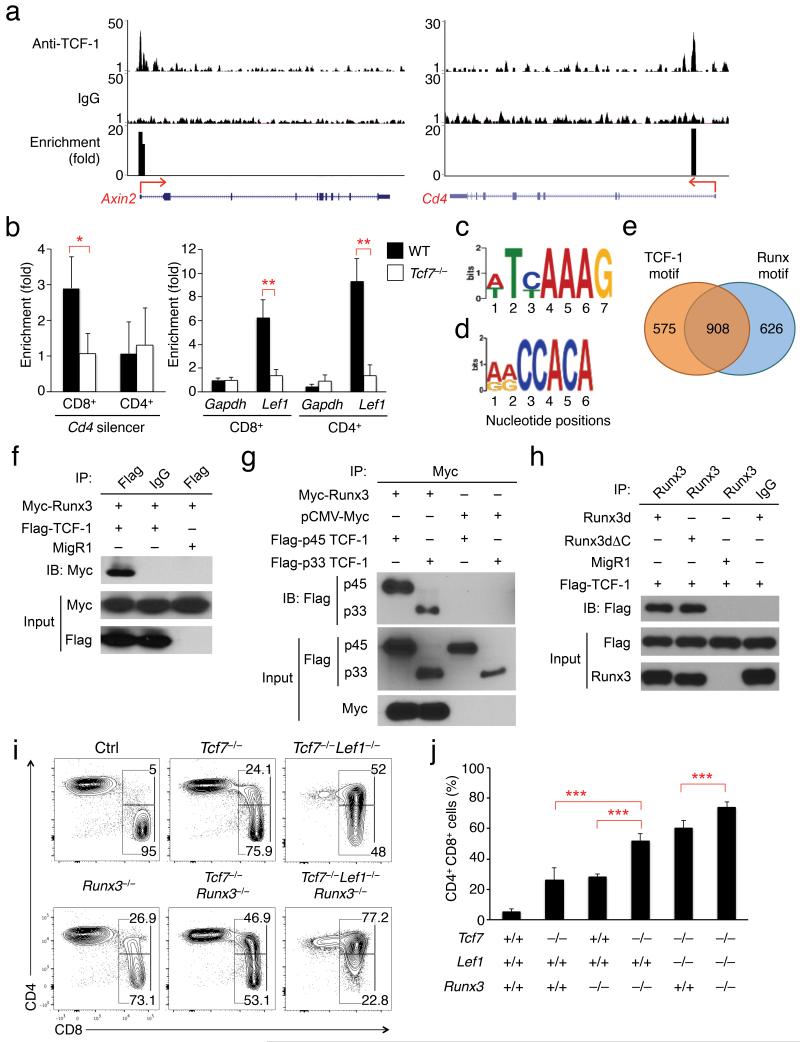
Detailed mapping of cis-elements in the Cd4 silencer has revealed that several sites, in addition to the Runx-binding motifs, contribute to stable repression of CD4 coreceptor in CD8+ T cells13, 33. However, the identity of the factors binding to these additional sites remains unknown. To better understand the role of TCF-1 and LEF-1 in this process, we performed ChIP-seq with anti-TCF-130 or control IgG in naïve splenic CD8+ T cells and used the model-based analysis for ChIP-Seq (MACS) algorithm34 to identify TCF-1 binding peaks. By stringent criteria of ≥ 4 fold enrichment, p < 10−5 and a false discovery rate (FDR) < 5%, we identified 2,827 high-confidence, strong TCF-1 binding peaks. Using more permissive criteria of p < 10−3, we located additional 6,577 weak TCF-1 peaks (Supplementary Fig. 8a). Genomic distribution analysis of all TCF-1 peaks revealed that only 6% were located in the promoter regions (−5k ~ +1 kb of transcription start sites) of known RefSeq genes (Supplementary Fig. 8b). Two histone modification marks, the activating H3K4me3 and repressive H3K27me3, were previously mapped by ChIP-seq in human naïve CD8+ T cells35. Peak overlap analysis showed that 15% of TCF-1 peaks in the promoter regions overlapped with H3K4me3, whereas the overlap with H3K27me3 was lower (Supplementary Fig. 8c). Among the strong TCF-1 binding peaks, we confirmed the direct association of TCF-1 with its known target genes including Axin2 and Lef1 (Fig. 8a and data not shown). One TCF-1 binding peak was found in the first intron of Cd4 at a location corresponding to the Cd4 silencer (Fig. 8a). Analysis of TCF-1 ChIP-Seq in whole thymocytes confirmed TCF-1 binding to the same location in the Cd4 gene (Supplementary Fig. 6b). In addition, a recent study reported Runx3 ChIP-Seq in CD8+ T cells and NK cells36. Collective analysis of these three sets of ChIP-seq data revealed that TCF-1 binding at the Cd4 intron 1 was perfectly aligned with the Runx3 binding peak (Supplementary Fig. 6b), suggesting co-occupancy of TCF-1 and Runx factors at the Cd4 silencer in CD8+ T cells. The ~430 bp Cd4 silencer indeed contains a perfect TCF-LEF motif “ACAAAG” in its 3′-terminus, with two known Runx motifs in its 5′ half. We validated enriched TCF-1 occupancy at the Cd4 silencer in wild-type CD8+ T cells, which was abrogated in Tcf7−/− CD8+ T cells (Fig. 8b). The Cd4 silencer was not occupied by TCF-1 in CD4+ T cells (Fig. 8b), suggesting lineage specificity. Consistent with this observation, Runx3 binding to the Cd4 silencer is specific to CD8+ T cells and not observed in NK cells36.
Figure 8. TCF-LEF and Runx factors cooperate in Cd4 silencing in CD8+ lineage T cells.
(a) Genome-wide mapping of TCF-1 occupancy reveals its direct association with the Cd4 silencer. Splenic CD8+ T cells were used in ChIP followed by high throughput sequencing, and ChIP-Seq track wiggle files were uploaded to the UCSC genome browser for visualization of enriched TCF-1 binding peaks. Top two tracks are sequencing reads from ChIP with anti-TCF-1 and IgG, respectively. The bottom track shows fold enrichment of TCF-1 binding peaks. All tracks are shown for the Axin2 and Cd4 gene loci, with their transcription orientations marked with arrows. (b) Binding of TCF-1 to the Cd4 silencer is specific to CD8+ T cells. CD8+ or CD4+ SP thymocytes were sorted from WT or Tcf7−/− mice and used in ChIP with anti-TCF-1 or control IgG. Enrichment at the Gapdh and Lef1 loci was measured as negative and positive controls, respectively. Data are pooled results from at least 3 independent experiments. *, p<0.05; **, p<0.01. (c) TCF-LEF motif and (d) Runx motif enriched in the TCF-1 ChIP-seq peaks. (e) Distribution of TCF-LEF and Runx motifs in the TCF-1 binding peaks is summarized in a Venn diagram. (f) Runx3 is coimmunoprecipitated with TCF-1. Myc-tagged Runx3 and Flag-tagged full-length TCF-1 were overexpressed in 293T cells, and the lysates were immunoprecipitated with anti-Flag or control IgG followed by immunoblotting with anti-Myc. Lysate input without immunoprecipitation was blotted to detect the expressed proteins. (g) Both p45 and p33 TCF-1 isoforms are coimmunoprecipitated with Runx3. Myc-tagged Runx3 was overexpressed together with Flag-tagged full length (p45) or p33 TCF-1 isoform. The lysates were immunoprecipitated with anti-Myc and then immunoblotted with anti-Flag. As negative controls, pCMV-Myc was transfected in place of Myc-Runx3. (h) TCF-1 is coimmunoprecipitated with full length Runx3d or Runx3d lacking the VWRPY motif (Runx3ΔC). Runx3d or Runx3ΔC were overexpressed together with Flag-tagged full length TCF-1 in 293T cells. The cell lysates were immunoprecipitated with anti-Runx and then blotted with anti-Flag. Data in (f)-(h) are representative of at least 3 independent experiments. (i) and (j) TCF-1 and LEF-1 cooperate with Runx3 in Cd4 gene silencing. Mature TCRβhi thymocytes were analyzed in the compound knockout mice. Representative contour plots are shown in (i), and the values are percentages of CD8+CD4− and CD8*4 subsets within the CD8+ population (to avoid the influence of varied CD4+ frequency). Cumulative data on the frequency of the CD8*4 subset are in (j). n ≥ 5. ***, p < 0.001.
De novo motif discovery analysis of the strong TCF-1 binding peaks recovered the known consensus TCF-LEF motif in 52.5% and the Runx motif in 54.3 % of the peaks (Fig. 8c,d). Overall, 908 peaks (32% of the total) contained both TCF-LEF and Runx motifs (Fig. 8e). Motif analysis of the 6,577 weak TCF-1 binding peaks revealed a similar trend, with TCF-LEF and Runx motifs found in 37.3% and 41.4 % of the peaks, respectively, and 1,257 peaks containing both motifs (Supplementary Fig. 8d,e,f). By applying our stringent peak calling setting to the Runx3 ChIP-seq data in CD8+ T cells36, we identified 4,785 Runx3 binding peaks, and 1,270 of these overlapped with strong TCF-1 binding peaks. These high-throughput data suggest that TCF-LEF and Runx factors may have a broadly cooperative role in gene regulation. It has been shown that Runx3 interacts with TCF-4 and attenuated TCF-4-β-catenin signaling during intestinal tumorigenesis37. To further substantiate this point, we overexpressed Myc-tagged Runx3 with Flag-tagged full-length p45 TCF-1 isoform in 293T cells and immunoprecipitated the cell lysates with anti-Flag or control IgG. Immunoblotting with anti-Myc revealed that TCF-1 was co-immunoprecipitated with Runx3 (Fig. 8f). In the reciprocal experiment, immunoprecipitation with anti-Myc detected Flag-tagged p45 TCF-1 (Fig. 8g). The p33 isoform of TCF-1, which is truncated on the N-terminus and lacks the β-catenin-binding domain, was also coimmunoprecipitated with Runx3 (Fig. 8g). Runx3 recruits the Groucho-TLE corepressors through its last five amino acids, the “VWPRY” motif, in the C-terminus38. Specific deletion of this sequence in mouse germline causes CD4 derepression in CD8+ T cells38, similar to the complete deletion of Runx3, indicating an essential role for Runx3-mediated recruitment of Groucho-TLE corepressors in Cd4 silencing. Because TCF-LEF factors interact with Groucho-TLE14, the interaction between Runx3 and TCF-1 may be bridged by these corepressor proteins. To determine if Runx3 can directly interact with TCF-1, we generated a C-terminus-truncated version of Runx3d (Runx3dΔC) that specifically lacked the VWRPY motif. The Runx3dΔC mutant did coimmunoprecipitate with TCF-1 (Fig. 8h), suggesting that direct physical interaction between TCF-1 and Runx3 can occur independent of the Groucho-TLE corepressors.
To further delineate the TCF-Runx cooperation in vivo, we examined CD4 derepression in CD8+ T cells. In Tcf7−/− or Runx3−/− mice, a small portion of mature CD8+ thymocytes showed CD4 derepression; however, in Tcf7−/−Runx3−/− mice, a much larger portion of CD8+ T cells expressed CD4 (Fig. 8i, j). Tcf7−/−Lef1−/− mice showed stronger CD4 derepression in CD8+ T cells than Tcf7−/− mice, because of the functional redundancy between TCF-1 and LEF-1 (Fig. 2 and Fig. 8i), while Tcf7−/−Lef1−/−Runx3−/− mice showed a further increase in the frequency of CD8*4 cells within the mature TCRβhi CD8+ thymocytes, with cells in the CD8+CD4− gate showing a strong shift toward increased CD4 expression (Fig. 8i, j), analogous to what is observed in Runx3−/−Runx1+/- or Runx3−/− Runx1+/Δ446 mice10, 39. These observations demonstrate a functional synergy between TCF-LEF and Runx factors in achieving stable Cd4 silencing in CD8+ T cells.
Discussion
TCF-1 and LEF-1 have well documented roles in early T cell development. By conditional targeting of both factors, our studies reveal their roles in late developmental stages, CD4+ vs. CD8+ lineage choice and establishing CD8+ T cell identity. Lineage specification and commitment involve activation of lineage-appropriate genes and inactivation of lineage-inappropriate genes40. Before lineage commitment, the DP precursors are likely biased toward CD4+ specification5, because the post-select DP thymocytes lacking both Th-POK and Runx complex adopt a CD4+ T cell fate11. In addition to Myb, Tox and GATA-3, we identified TCF-1 and LEF-1 as independent factors in promoting CD4+ lineage specification. Whereas Myb is downregulated, Tox and GATA-3 are upregulated by positive selection signals. In contrast, TCF-1 and LEF-1 expression is induced in early DN stages, with TCF-1 abundantly expressed thereafter and LEF-1 exhibiting further induction in post-select DP thymocytes. Thus, TCF-1 and LEF-1 may act as a constant “inner drive” toward the CD4+ T cell lineage.
TCF-1 contributes to CD4+ T cell lineage commitment by direct positive regulation of Thpok. Thpok expression was diminished but not completely abrogated by TCF-1 deficiency. Hypomorphic Th-POK expression is known to cause redirection of CD4+ to CD8+ T cell lineage11, indicating that a threshold of Th-POK expression is required for complete commitment to the CD4+ lineage to occur. The diminished expression of Thpok in TCF-1-deficient post-select DP and CD4+8lo thymocytes was sufficient to reduce CD4+ T cell output and cause CD8+ lineage redirection when tested on a β2m−/− background or with an MHC-II-restricted TCR. Although significant CD4 derepression in Tcf7−/−Lef1−/− CD8+ T cells precluded us from decisive gene expression analysis in post-select DP cells lacking both TCF-1 and LEF-1, we did observe stronger CD4+ to CD8+ lineage redirection in all models tested, suggesting that loss of TCF-1 and LEF-1 causes a more severe reduction in Thpok expression.
TCF-1 deficiency also caused increased expression of Runx3d in the bipotent post-select DP and CD4+8lo thymocytes. However, this increase is most likely secondary to decreased Thpok expression, because a Th-POK transgene suppressed Runx3d expression in both wild-type and Tcf7−/− cells. In addition, whereas ectopic Th-POK expression restored CD4+ T cell output in Tcf7−/− mice, Runx3 deletion failed to do so. On the other hand, in mature TCRβhi thymocytes from Tcf7−/−Lef1−/− mice, the Th-POK transgene was able to suppress CD8+ T cell differentiation and lineage redirection in β2m−/− chimeric hosts, but was inefficient in restoring CD4+ T cell numbers. This resembles impaired generation of CD4+ SP thymocytes caused by CD4-Cre-mediated inactivation of GATA-3, which cannot be rectified by the Th-POK transgene6. Therefore, loss of both TCF-1 and LEF-1 may have perturbed expression of other critical genes that promote CD4+ T cell differentiation, in addition to Th-POK.
In spite of the intrinsic bias toward CD4+ lineage specification, the Th-POK expression and/or activity is opposed by Runx3d to ensure the MHC-I signal-selected DP thymocytes to commit to the CD8+ T cell lineage. It is well accepted that potent and persistent TCR signaling promotes CD4+ lineage choice. Recent studies further demonstrate that intrathymic cytokine signaling, rather than TCR, promotes CD8+ lineage choice41, 42. Indeed, IL-7 is shown to induce the expression of Runx3d, which in turn activates the Thpok silencer41. It is important to note that IL-7-derived signals inhibit the expression of TCF-1 and LEF-143. Given our new findings that TCF-1 and LEF-1 positively regulate Th-POK, IL-7 signaling may annihilate Th-POK expression via multiple mechanisms, including repressing its positive regulators (such as TCF-1 and LEF-1) in addition to inducing its negative regulator, Runx3d.
Once a lineage decision is made, lineage-inappropriate genes must be silenced to ensure that the cell identity is inheritably maintained. Runx factors play an important role in Cd4 gene silencing in CD8+ lineage-committed cells4, 44. Our results reveal a critical contribution of TCF-1 and LEF-1 to Cd4 silencing. TCF-1 and Runx3 physically interacted with each other and exhibited strong cooperativity in silencing the Cd4 gene in CD8+ T cells. Although it remains to be elucidated if TCF-1 is recruited to the Cd4 silencer directly by the DNA element(s) or indirectly by Runx factors, Runx and TCF-LEF factors are both essential components of a protein complex that occupies the Cd4 silencer. Compelling evidence indicates that epigenetic mechanisms are involved in inheritable Cd4 silencing in CD8+ T cells45. Identification of TCF-LEF factors in CD4 repression thus expands the contact surface of the Cd4 silencing complex for recruiting histone modification enzymes. Beyond Cd4 gene silencing, the cooperativity between TCF-LEF and Runx factors might be essential for positive regulatory functions in other gene regulatory contexts, or extend to different cell types such as hematopoietic stem cells, as previously suggested46.
While TCF-1 and LEF-1 are required for Cd4 silencing in CD8+ T cells, early deletion of these factors did not cause aberrant expression of CD4 in DN thymocytes18. In contrast, loss of Ikaros results in the opposite effect, causing CD4 derepression in DN thymocytes but not in CD8+ T cells47. Among Runx factors, Runx1 has a more dominant role in CD4 repression in DN cells, but Runx3 is more potent for Cd4 silencing in CD8+ T cells. Thus, the protein complex at the Cd4 silencer appears to undergo dynamic component changes as thymocytes progress through different developmental stages. TCF-1 and LEF-1 are both expressed in multiple isoforms, with the full-length isoform having the capacity to interact with β-catenin. The short isoforms are found to have suppressive and/or dominant negative functions16, 21, and in fact, p33 TCF-1 interacts with Runx3. It would be interesting to determine if there is a division of labor between the full-length and short isoforms of TCF-1 and LEF-1 in Cd4 silencing in CD8+ T cells, and by extension, in promoting Thpok expression in lineage choice.
Although ablation of the Runx complex causes derepression of both Cd4 and Thpok in CD8+ T cells10, loss of TCF-1 and LEF-1 specifically derepressed Cd4, but not Thpok. Consistent with this observation, ChIP-seq of TCF-1 in CD8+ T cells revealed no direct association of TCF-1 with the Thpok locus, and the previously defined Thpok silencer (DRE) does not contain a TCF-LEF consensus motif. Thus, the TCF-LEF and Runx cooperativity is highly gene context-dependent. In addition, regulation of Thpok by TCF-1 at the GTE was restricted to the bipotent precursors, implying that the GTE may not be accessible to TCF-1 in CD8+ lineage-committed T cells.
In summary, our studies demonstrate a role switch for TCF-1 and LEF-1 in late stages of T cells development. They promote CD4+ T cell fate decision in DP and CD4+8lo thymocytes by positively regulating Th-POK without directly involving Runx3, In CD8+ lineage-committed T cells; however, TCF-1 and LEF-1 cooperate with Runx3 to repress the lineage-inappropriate Cd4 gene. These new findings reveal that the same transcriptional regulator contributes to fate decision and establishment of cell identity through distinct genetic and molecular wiring.
ONLINE METHODS
Animals
The Tcf7-targeted mice were obtained from Institut Clinique de la Souris, France, part of the IKMC. Following rederivation at the animal use facility, University of Iowa, the mice were crossed with Rosa26-Flippase knock-in mice (Jackson Laboratory) to delete the LacZ-Neo cassette flanked by the Frt sites (Supplementary Fig. 1), converting the targeted allele into Tcf7-floxed allele (Tcf7fl/+). The Lef1fl/fl mice were previously described18, Runx3fl/fl mice were from the Jackson Laboratory, β2m−/− and H2-Ab1−/− mice from Taconic, Th-POK TG mice were provided by R. Bosselut29. All animals were analyzed at 5-10 weeks of age, and both genders included without randomization or “blinding”. All the BM chimeras were analyzed within 6-10 weeks after the BM transplantation. For all mouse phenotypic analysis, at least 5 animals of each genotype were analyzed in at least 3 independent experiments. All mouse experiments were performed under protocols approved by the Institutional Animal Use and Care Committee of the University of Iowa.
Flow cytometry
Single cell suspension was prepared from thymus and spleen and surface-stained as previously described18. All fluorochrome-conjugated antibodies were from eBiosciences or BD Biosciences. The antibodies and their clone numbers are CD4 (RM4-5), CD8α (53-6.7), CD8β (H35-17.2); TCRβ (H57-597), CD24 (M1/69), CD69 (H1.2F3), CD45.2 (104), B220 (RA3-6B2), Gr-1 (RB6-C5), NK1.1 (PK136), TER-119 (TER-119), γδTCR (GL3), Vα2 TCR (B20.1), IFN-γ (XMG1.2), IL-2 (JES6-5H4), and Streptavidin (eBiosciences Cat. No. 48-4317-82). Granzyme B (GB11) and control mouse IgG1 (Cat. No. MG104) were from Life Technologies. Data were collected on FACSVerse (BD Biosciences) and analyzed with FlowJo software (Version X, TreeStar).
Generation of BM chimeras
β2m−/− mice were crossed with the B6.SJL strain to acquire homozygous expression of CD45.1 and used as recipients. Whole BM cells were isolated from various donors, and 2 × 106 cells were transplanted into irradiated recipients via tail vein injection.
Luciferase assays
The 473-bp WT GTE was PCR-amplified, and mutant GTE was synthesized (GenScript) with proper flanking enzyme sites. These segments were cloned into the pGL3 promoter vector (Promega), via the KpnI and NheI sites upstream of the SV40 promoter or via the BamHI and SalI sites downstream of the luciferase gene. The reporter constructs were transfected into EL-4 cells by electroporation (GenePulser Xcell, BioRad) or 293T cells using Lipofectamine 2000 (Life Technologies) following standard protocols48. pRL-TK, which expresses Renilla luciferase driven by a thymidine kinase promoter, was cotransfected as an internal control. Forty-eight hrs later, cell lysates were extracted and analyzed for luciferase activity with the Dual-Luciferase Reporter Assay System (Promega).
Gene expression assay
Target cell populations were sorted from thymocytes or splenocytes, RNA extraction, reverse-transcription, and quantitative PCR were performed as described18. The primer sequences are in Supplementary Table 1.
Chromatin immunoprecipitation (ChIP)
Post-select DP and CD4+8lo thymocytes, mature CD8+ or CD4+ SP thymocytes, or splenic CD8+ T cells were sorted from either wild-type C57BL/6 or Tcf7−/− mice. The cells were cross-linked with 1% formaldehyde in medium for 5 minutes, processed using truChIP Chromatin Shearing Reagent Kit (Covaris), and sonicated for 5 minutes on Covaris S2 ultrasonicator. The sheared chromatin was immunoprecipitated with anti-TCF-130 or control IgG and washed as previously described. The immunoprecipitated DNA segments were used for library construction or PCR quantification. For calculation of enriched TCF-1 binding in a given cell type in ChIP-PCR experiments, each TCF-1 ChIP sample was first normalized to corresponding IgG ChIP sample, and the signal at a target region was then normalized to that at the Hprt1 or Gapdh promoter region. The primers for assessing enriched TCF-1 binding are listed in Supplementary Table 1.
ChIP-seq and data analysis
DNA segments from ChIP were end-repaired and ligated to indexed Illumina adaptors followed by low-cycle PCR. The resulting libraries were sequenced with the Illumina Hiseq-2000 platform. Sequencing reads were mapped to the mouse genome (mm9) using Bowtie v.0.12.5. The mapping statistics are summarized in the following table:
| Sample | Total numbers of reads |
Numbers of mapped reads |
|---|---|---|
| Tcf1 ChIP | 24,390,268 | 23,262,097 (95.4%) |
| IgG ChIP | 48,752,154 | 46,601,382 (95.6%) |
MACS34 was used for peak calling with two sets of cutoffs. The first set used ≥ 4 fold enrichment, p-value < 10−5, and FDR < 5% and thus identified strong TCF-1 binding peaks. The second set used the same fold enrichment and FDR cutoffs but p < 10−3, which identified additional weak TCF-1 binding peaks. The identified TCF-1 peaks were analyzed with MEME-ChIP v.4.9.0 to define consensus DNA binding motifs49. Peak sequences were padded with 200 bp genomic sequences on both sides for motif scanning. Patser v.3b was used to scan each TCF-1 peak region for the co-occurrence of TCF-1 and Runx binding sites50.
Immunoprecipitation and immunoblotting
The Tcf7 and Runx3d coding sequence was amplified from CD8+ T cell cDNA and cloned in Mig-R1 retroviral vector, and a 3×Flag tag was placed in-frame on the N-terminus of TCF-1. The expression plasmids were transfected into 293T cells using Lipofectamine 2000 (Life Technologies), and 48 hours later, cell lysates were extracted and incubated overnight with 2 μg of anti-Runx3 (R&D Systems, clone 527327), anti-Flag (Sigma-Aldrich, clone M2), or mouse IgG, followed by 2-hr incubation with Dynabeads Protein G (Life Technologies). After proper washing, the immunoprecipitated samples were analyzed by immunoblotting with anti-Myc (Cell Signaling Technologies, clone 71D10), Flag or Runx3 antibodies.
For assessing TCF-1 and LEF-1 deletion efficiency, pre- and post-select DP thymocytes (5 ×105 each) were sorted, and the lysates were probed with TCF-1 or LEF-1 antibodies (Cell Signaling Technology, clone C46C7 and C18A7, respectively), or anti-β-actin (Santa Cruz Biotechnology, clone I-19) as an equal loading control.
Statistical analysis
Data sets were analyzed with the Student’s t-test with a two-tailed distribution assuming equal sample variance.
Supplementary Material
ACKNOWLEDGEMENTS
We thank R. Bosselut (NCI, NIH) for providing the Th-POK TG mice, S.-C. Bae (Chungbuh National University, South Korea) for the Myc-tagged Runx3 expression plasmid. We also thank B. J. Fowlkes (NIAID) for her insightful input and discussion, Y. Wakabayashi and Y. Luo (NHLBI) for performing high throughput sequencing and data processing, T. Zhao for animal husbandry. We appreciate support from the Flow Cytometry Core facility (J. Fishbaugh, H. Vignes and G. Rasmussen) for cell sorting and Radiation Core facility (A. Kalen) for mouse irradiation. This study is supported by grants from the American Cancer Society (RSG-11-161-01-MPC to H.-H.X.) and the NIH (HL095540 and AI105351 to H.-H.X., HG006130 to K.T.). F.C.S. is a recipient of T32 pre-doctoral training grant (AI007485, NIAID, NIH). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
Database accession numbers
The TCF-1 ChIP-Seq data are available in the Gene Expression Omnibus (GEO) database under the accession number GSE 52070 (including GSM1258235 and GSM1258236).
AUTHOR CONTRIBUTIONS F.C.S. and S.Y. performed experiments and analyzed the data. X.Z. and B.Z. did the coIP experiments. B.H. and W.Y. analyzed the ChIP-seq data under the supervision of K.T. and J.Z. H.K. provided the TCF-1 antibody. H.H.X designed and supervised the study. H.H.X wrote the paper with F.C.S. and S.Y.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Rothenberg EV, Moore JE, Yui MA. Launching the T-cell-lineage developmental programme. Nat Rev Immunol. 2008;8(1):9–21. doi: 10.1038/nri2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang Q, Jeremiah Bell J, Bhandoola A. T-cell lineage determination. Immunol Rev. 2010;238(1):12–22. doi: 10.1111/j.1600-065X.2010.00956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Singer A, Adoro S, Park JH. Lineage fate and intense debate: myths, models and mechanisms of CD4-versus CD8-lineage choice. Nat Rev Immunol. 2008;8(10):788–801. doi: 10.1038/nri2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collins A, Littman DR, Taniuchi I. RUNX proteins in transcription factor networks that regulate T-cell lineage choice. Nat Rev Immunol. 2009;9(2):106–115. doi: 10.1038/nri2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang L, Bosselut R. CD4-CD8 lineage differentiation: Thpok-ing into the nucleus. J Immunol. 2009;183(5):2903–2910. doi: 10.4049/jimmunol.0901041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang L, Wildt KF, Zhu J, Zhang X, Feigenbaum L, Tessarollo L, et al. Distinct functions for the transcription factors GATA-3 and ThPOK during intrathymic differentiation of CD4(+) T cells. Nat Immunol. 2008;9(10):1122–1130. doi: 10.1038/ni.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maurice D, Hooper J, Lang G, Weston K. c-Myb regulates lineage choice in developing thymocytes via its target gene Gata3. EMBO J. 2007;26(15):3629–3640. doi: 10.1038/sj.emboj.7601801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He X, He X, Dave VP, Zhang Y, Hua X, Nicolas E, et al. The zinc finger transcription factor Th-POK regulates CD4 versus CD8 T-cell lineage commitment. Nature. 2005;433(7028):826–833. doi: 10.1038/nature03338. [DOI] [PubMed] [Google Scholar]
- 9.Aliahmad P, Kaye J. Development of all CD4 T lineages requires nuclear factor TOX. J Exp Med. 2008;205(1):245–256. doi: 10.1084/jem.20071944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Setoguchi R, Tachibana M, Naoe Y, Muroi S, Akiyama K, Tezuka C, et al. Repression of the transcription factor Th-POK by Runx complexes in cytotoxic T cell development. Science. 2008;319(5864):822–825. doi: 10.1126/science.1151844. [DOI] [PubMed] [Google Scholar]
- 11.Egawa T, Littman DR. ThPOK acts late in specification of the helper T cell lineage and suppresses Runx-mediated commitment to the cytotoxic T cell lineage. Nat Immunol. 2008;9(10):1131–1139. doi: 10.1038/ni.1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He X, Park K, Wang H, Zhang Y, Hua X, Li Y, et al. CD4-CD8 lineage commitment is regulated by a silencer element at the ThPOK transcription-factor locus. Immunity. 2008;28(3):346–358. doi: 10.1016/j.immuni.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 13.Taniuchi I, Osato M, Egawa T, Sunshine MJ, Bae SC, Komori T, et al. Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell. 2002;111(5):621–633. doi: 10.1016/s0092-8674(02)01111-x. [DOI] [PubMed] [Google Scholar]
- 14.Staal FJ, Luis TC, Tiemessen MM. WNT signalling in the immune system: WNT is spreading its wings. Nat Rev Immunol. 2008;8(8):581–593. doi: 10.1038/nri2360. [DOI] [PubMed] [Google Scholar]
- 15.Xue HH, Zhao DM. Regulation of mature T cell responses by the Wnt signaling pathway. Ann N Y Acad Sci. 2012;1247:16–33. doi: 10.1111/j.1749-6632.2011.06302.x. [DOI] [PubMed] [Google Scholar]
- 16.Weber BN, Chi AW, Chavez A, Yashiro-Ohtani Y, Yang Q, Shestova O, et al. A critical role for TCF-1 in T-lineage specification and differentiation. Nature. 2011;476(7358):63–68. doi: 10.1038/nature10279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Germar K, Dose M, Konstantinou T, Zhang J, Wang H, Lobry C, et al. T-cell factor 1 is a gatekeeper for T-cell specification in response to Notch signaling. Proc Natl Acad Sci U S A. 2011;108(50):20060–20065. doi: 10.1073/pnas.1110230108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu S, Zhou X, Steinke FC, Liu C, Chen SC, Zagorodna O, et al. The TCF-1 and LEF-1 Transcription Factors Have Cooperative and Opposing Roles in T Cell Development and Malignancy. Immunity. 2012;37(5):813–826. doi: 10.1016/j.immuni.2012.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okamura RM, Sigvardsson M, Galceran J, Verbeek S, Clevers H, Grosschedl R. Redundant regulation of T cell differentiation and TCRalpha gene expression by the transcription factors LEF-1 and TCF-1. Immunity. 1998;8(1):11–20. doi: 10.1016/s1074-7613(00)80454-9. [DOI] [PubMed] [Google Scholar]
- 20.Yu S, Xue HH. TCF-1 mediates repression of Notch pathway in T lineage-committed early thymocytes. Blood. 2013;121(19):4008–4009. doi: 10.1182/blood-2013-01-477349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tiemessen MM, Baert MR, Schonewille T, Brugman MH, Famili F, Salvatori DC, et al. The nuclear effector of wnt-signaling, tcf1, functions as a T-cell-specific tumor suppressor for development of lymphomas. PLoS Biol. 2012;10(11):e1001430. doi: 10.1371/journal.pbio.1001430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Verbeek S, Izon D, Hofhuis F, Robanus-Maandag E, te Riele H, van de Wetering M, et al. An HMG-box-containing T-cell factor required for thymocyte differentiation. Nature. 1995;374(6517):70–74. doi: 10.1038/374070a0. [DOI] [PubMed] [Google Scholar]
- 23.Egawa T, Tillman RE, Naoe Y, Taniuchi I, Littman DR. The role of the Runx transcription factors in thymocyte differentiation and in homeostasis of naive T cells. J Exp Med. 2007;204(8):1945–1957. doi: 10.1084/jem.20070133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grusby MJ, Johnson RS, Papaioannou VE, Glimcher LH. Depletion of CD4+ T cells in major histocompatibility complex class II-deficient mice. Science. 1991;253(5026):1417–1420. doi: 10.1126/science.1910207. [DOI] [PubMed] [Google Scholar]
- 25.Goux D, Coudert JD, Maurice D, Scarpellino L, Jeannet G, Piccolo S, et al. Cooperating pre-T-cell receptor and TCF-1-dependent signals ensure thymocyte survival. Blood. 2005;106(5):1726–1733. doi: 10.1182/blood-2005-01-0337. [DOI] [PubMed] [Google Scholar]
- 26.Karlsson L, Surh CD, Sprent J, Peterson PA. A novel class II MHC molecule with unusual tissue distribution. Nature. 1991;351(6326):485–488. doi: 10.1038/351485a0. [DOI] [PubMed] [Google Scholar]
- 27.Zijlstra M, Bix M, Simister NE, Loring JM, Raulet DH, Jaenisch R. Beta 2-microglobulin deficient mice lack CD4-8+ cytolytic T cells. Nature. 1990;344(6268):742–746. doi: 10.1038/344742a0. [DOI] [PubMed] [Google Scholar]
- 28.Muroi S, Naoe Y, Miyamoto C, Akiyama K, Ikawa T, Masuda K, et al. Cascading suppression of transcriptional silencers by ThPOK seals helper T cell fate. Nat Immunol. 2008;9(10):1113–1121. doi: 10.1038/ni.1650. [DOI] [PubMed] [Google Scholar]
- 29.Sun G, Liu X, Mercado P, Jenkinson SR, Kypriotou M, Feigenbaum L, et al. The zinc finger protein cKrox directs CD4 lineage differentiation during intrathymic T cell positive selection. Nat Immunol. 2005;6(4):373–381. doi: 10.1038/ni1183. [DOI] [PubMed] [Google Scholar]
- 30.Hattori N, Kawamoto H, Fujimoto S, Kuno K, Katsura Y. Involvement of transcription factors TCF-1 and GATA-3 in the initiation of the earliest step of T cell development in the thymus. J Exp Med. 1996;184(3):1137–1147. doi: 10.1084/jem.184.3.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li L, Zhang JA, Dose M, Kueh HY, Mosadeghi R, Gounari F, et al. A far downstream enhancer for murine Bcl11b controls its T-cell specific expression. Blood. 2013;122(6):902–911. doi: 10.1182/blood-2012-08-447839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kohu K, Sato T, Ohno S, Hayashi K, Uchino R, Abe N, et al. Overexpression of the Runx3 transcription factor increases the proportion of mature thymocytes of the CD8 single-positive lineage. J Immunol. 2005;174(5):2627–2636. doi: 10.4049/jimmunol.174.5.2627. [DOI] [PubMed] [Google Scholar]
- 33.Taniuchi I, Sunshine MJ, Festenstein R, Littman DR. Evidence for distinct CD4 silencer functions at different stages of thymocyte differentiation. Molecular cell. 2002;10(5):1083–1096. doi: 10.1016/s1097-2765(02)00735-9. [DOI] [PubMed] [Google Scholar]
- 34.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, et al. Model-based analysis of ChIP-Seq (MACS) Genome biology. 2008;9(9):R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Araki K, Turner AP, Shaffer VO, Gangappa S, Keller SA, Bachmann MF, et al. mTOR regulates memory CD8 T-cell differentiation. Nature. 2009;460(7251):108–112. doi: 10.1038/nature08155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lotem J, Levanon D, Negreanu V, Leshkowitz D, Friedlander G, Groner Y. Runx3-mediated transcriptional program in cytotoxic lymphocytes. PLoS ONE. 2013;8(11):e80467. doi: 10.1371/journal.pone.0080467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ito K, Lim AC, Salto-Tellez M, Motoda L, Osato M, Chuang LS, et al. RUNX3 attenuates beta-catenin/T cell factors in intestinal tumorigenesis. Cancer cell. 2008;14(3):226–237. doi: 10.1016/j.ccr.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 38.Yarmus M, Woolf E, Bernstein Y, Fainaru O, Negreanu V, Levanon D, et al. Groucho/transducin-like Enhancer-of-split (TLE)-dependent and -independent transcriptional regulation by Runx3. Proc Natl Acad Sci U S A. 2006;103(19):7384–7389. doi: 10.1073/pnas.0602470103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Woolf E, Xiao C, Fainaru O, Lotem J, Rosen D, Negreanu V, et al. Runx3 and Runx1 are required for CD8 T cell development during thymopoiesis. Proc Natl Acad Sci U S A. 2003;100(13):7731–7736. doi: 10.1073/pnas.1232420100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rothenberg EV. Decision by committee: new light on the CD4/CD8-lineage choice. Immunol Cell Biol. 2009;87(2):109–112. doi: 10.1038/icb.2008.100. [DOI] [PubMed] [Google Scholar]
- 41.Park JH, Adoro S, Guinter T, Erman B, Alag AS, Catalfamo M, et al. Signaling by intrathymic cytokines, not T cell antigen receptors, specifies CD8 lineage choice and promotes the differentiation of cytotoxic-lineage T cells. Nat Immunol. 2010;11(3):257–264. doi: 10.1038/ni.1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McCaughtry TM, Etzensperger R, Alag A, Tai X, Kurtulus S, Park JH, et al. Conditional deletion of cytokine receptor chains reveals that IL-7 and IL-15 specify CD8 cytotoxic lineage fate in the thymus. J Exp Med. 2012;209(12):2263–2276. doi: 10.1084/jem.20121505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu Q, Erman B, Park JH, Feigenbaum L, Singer A. IL-7 receptor signals inhibit expression of transcription factors TCF-1, LEF-1, and RORgammat: impact on thymocyte development. J Exp Med. 2004;200(6):797–803. doi: 10.1084/jem.20032183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Taniuchi I, Ellmeier W. Transcriptional and epigenetic regulation of CD4/CD8 lineage choice. Adv Immunol. 2011;110:71–110. doi: 10.1016/B978-0-12-387663-8.00003-X. [DOI] [PubMed] [Google Scholar]
- 45.Zou YR, Sunshine MJ, Taniuchi I, Hatam F, Killeen N, Littman DR. Epigenetic silencing of CD4 in T cells committed to the cytotoxic lineage. Nature genetics. 2001;29(3):332–336. doi: 10.1038/ng750. [DOI] [PubMed] [Google Scholar]
- 46.Wu JQ, Seay M, Schulz VP, Hariharan M, Tuck D, Lian J, et al. Tcf7 is an important regulator of the switch of self-renewal and differentiation in a multipotential hematopoietic cell line. PLoS genetics. 2012;8(3):e1002565. doi: 10.1371/journal.pgen.1002565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Naito T, Gomez-Del Arco P, Williams CJ, Georgopoulos K. Antagonistic interactions between Ikaros and the chromatin remodeler Mi-2beta determine silencer activity and Cd4 gene expression. Immunity. 2007;27(5):723–734. doi: 10.1016/j.immuni.2007.09.008. [DOI] [PubMed] [Google Scholar]
Additional references associated with Methods
- 48.Xue HH, Bollenbacher J, Rovella V, Tripuraneni R, Du YB, Liu CY, et al. GA binding protein regulates interleukin 7 receptor alpha-chain gene expression in T cells. Nat Immunol. 2004;5(10):1036–1044. doi: 10.1038/ni1117. [DOI] [PubMed] [Google Scholar]
- 49.Machanick P, Bailey TL. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics. 2011;27(12):1696–1697. doi: 10.1093/bioinformatics/btr189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hertz GZ, Stormo GD. Identifying DNA and protein patterns with statistically significant alignments of multiple sequences. Bioinformatics. 1999;15(7-8):563–577. doi: 10.1093/bioinformatics/15.7.563. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.