Summary
In the c-ring rotor of ATP synthases ions are shuttled across the membrane during ATP synthesis by a unique rotary mechanism. We investigated characteristics of the c-ring from the alkaliphile Bacillus pseudofirmus OF4 with respect to evolutionary adaptations to operate with protons at high environmental pH. The X-ray structures of the wild-type c13 ring at pH 9.0 and a ‘neutralophile-like’ mutant (P51A) at pH 4.4, at 2.4 and 2.8 Å resolution, respectively, reveal a dependency of the conformation and protonation state of the proton-binding glutamate (E54) on environmental hydrophobicity. Faster labeling kinetics with the inhibitor dicyclohexylcarbodiimide (DCCD) demonstrate a greater flexibility of E54 in the mutant due to reduced water occupancy within the H+-binding site. A second ‘neutralophile-like’ mutant (V21N) shows reduced growth at high pH, which is explained by restricted conformational freedom of the mutant's E54 carboxylate. The study directly connects subtle structural adaptations of the c-ring ion-binding site to in vivo effects of alkaliphile cell physiology.
Keywords: F1Fo-ATP-synthase, rotary ATPase, membrane protein complex, X-ray structure, bioenergetics, extremophiles, alkaliphiles
Introduction
F 1Fo-ATP synthases are highly conserved, membrane-embedded enzyme complexes that use a rotary-mechanism to drive the synthesis of the universal energy currency adenosine5’-triphosphate (ATP). The simplest version of the enzyme can be found in bacteria consisting of a soluble F1 (subunits α3β3γδε) and a membrane-embeddedFo (subunits ab2cn) domain. ATP is synthesized within three catalytic β-subunits (Abrahams et al., 1994, Boyer, 1993) using rotational energy that is converted from the transmembrane electrochemical gradient of ions (Na+ or H+) within the enzyme's Fo domain (Dimroth et al., 2006). The c-ring (or rotor ring), the rotating part of the Fo domain, is an oligomeric, membrane-embedded protein complex, consisting of a species-dependent, varying number (n) of c-subunit copies (cn ring with n=8-15) (Meier et al., 2011). With every 360° rotation (Noji et al., 1997) of the rotor ring, n ions are transferred across the membrane in Fo and three ATP are produced in F 1. This stoichiometry ratio (n/3) defines the so-called ion-to-ATP ratio (i) of the enzyme, which is a precisely adapted cell physiological parameter (Ferguson, 2010, Watt et al., 2010, Pogoryelov et al., 2012, Meier et al., 2011, Preiss et al., 2013). Ion translocation through the Fo complex involves both the Fo rotor and stator. In the so-called two channel model (Vik & Antonio, 1994, Junge et al., 1997), two pathways within the stator subunit are proposed to shuttle the ions to and from the c-ring ion-binding site at the middle of the hydrophobic membrane. In the ATP synthesis mode, the ions are first translocated through the periplasmic “half channel” (an as yet structurally undefined ion pathway) to bind and neutralize the ion-specific ion-binding sites on the c-ring, involving a conserved carboxylate. Once tightly bound in the so-called ion locked conformation (Meier et al., 2005, Pogoryelov et al., 2009, Preiss et al., 2010) they are able to be shuttled along the hydrophobic environment of the membrane alkyl chains without an increased Coulomb penalty (Junge et al., 1997). After an almost 360° rotation and reaching the stator from the opposite side, the ions finally can be released again when they face the second hydrophilic pathway within the stator a-subunit; this is referred to as the open conformation (Pogoryelov et al., 2010, Symersky et al., 2012, Mizutani et al., 2011).
Alkaliphilic bacteria living in high pH environments maintain an inverse ΔpH that reduces the net proton-motive force (pmf). Despite this challenging constellation, these bacteria use proton-dependent ATP synthases to drive the synthesis of ATP (Krulwich et al., 2011). To support robust growth at high environmental pH, alkaliphiles have developed a set of adaptative strategies (Hicks et al., 2010, Krulwich & Ito, 2013). While the effective, local pmf at the membrane surface may be larger than in the bulk and help promoting pmf-dependent ATP synthesis (Heberle et al., 1994, Mulkidjanian et al., 2005, Sanden et al., 2010), some of the adaptations have manifested directly as features of the alkaliphile ATP synthase itself. These include an enzyme-intrinsic blockage of F1-ATP hydrolysis (Hicks & Krulwich, 1990, Cook et al., 2003, Hoffmann & Dimroth, 1990) and a set of alkaliphile-specific sequence motifs in the transmembrane subunits a and c that were proposed to affect the function of the ATP synthase at alkaline pH. Experimental evidence confirmed that several of these amino acid motifs and residues facilitate ATP synthase function. For example, a specific lysine residue in the a-subunit was proposed to contribute importantly to proton capture by the ion-translocating Fo domain (Wang et al., 2004, McMillan et al., 2007, Fujisawa et al., 2010). Other amino acid motifs were shown to enhance the c-ring stoichiometry, to promote higher ion-to-ATP ratios (Matthies et al., 2009, Preiss et al., 2013). The high resolution crystal structure of the c-rotor ring of alkaliphilic Bacillus pseudofirmus OF4 c-ring revealed a slightly constricted cylinder with a central pore and a tridecameric (c13) stoichiometry (Preiss et al., 2010). A noteworthy difference between this c-ring and other rotor rings was its less pronounced hour-glass shape. The outer α-helices are more loosely packed and a rather extended c-ring diameter allowed assembly of a larger n per cn ring, which enhances i and hence supports ATP synthesis at a low pmf (Matthies et al., 2009, Preiss et al., 2010, Preiss et al., 2013). In addition to these structural characteristics, the B. pseudofirmus OF4 c13 ring was specifically shown to harbour a water molecule within each of the 13 ion-binding sites. Each of these water molecules is coordinated by four amino acid residues, including a conserved key glutamate (E54), and assists directly in the coordination of the translocated proton. However, rather than forming a hydronium ion (Boyer, 1988, von Ballmoos & Dimroth, 2007) by binding the proton stably, the water was shown to reside in H+- bonding distance to the conserved and protonated E54 (Leone et al., 2010)) and was hence proposed to enhance proton affinity at the ion-binding site of this enzyme from an alkaliphile (Preiss et al., 2010).
With this in mind, we hypothesized that these water molecules together with the previously reported alkaliphile-specific amino acid residues near E54 directly help to capture protons within the enzyme and hence support enzyme operation at a high environmental pH. Here, we investigated the subtle structural adaptations of this rotor ring and their effects on the ability of the ATP synthase to operate properly in cells growing at neutral and alkaline pH. We used high-resolution X-ray crystallography to explore structural features of the alkaliphilic wild-type (WT) vs. two distinct “neutralophile-like” mutant c-rings. The structural work was combined with a biochemical study using the ATP synthase inhibitor dicyclohexylcarbodiimide (DCCD), which made it possible to follow the kinetics of (de-)protonation events directly at the glutamates of the c-ring ion-binding site. Finally, we further complemented these results with growth experiments, which illustrate direct physiological effects of mutations that affect the ion-binding site of the c-ring.
Results and discussion
2.4 Å resolution X-ray structure of the Bacillus pseudofirmus OF4 rotor ring at pH 9.0
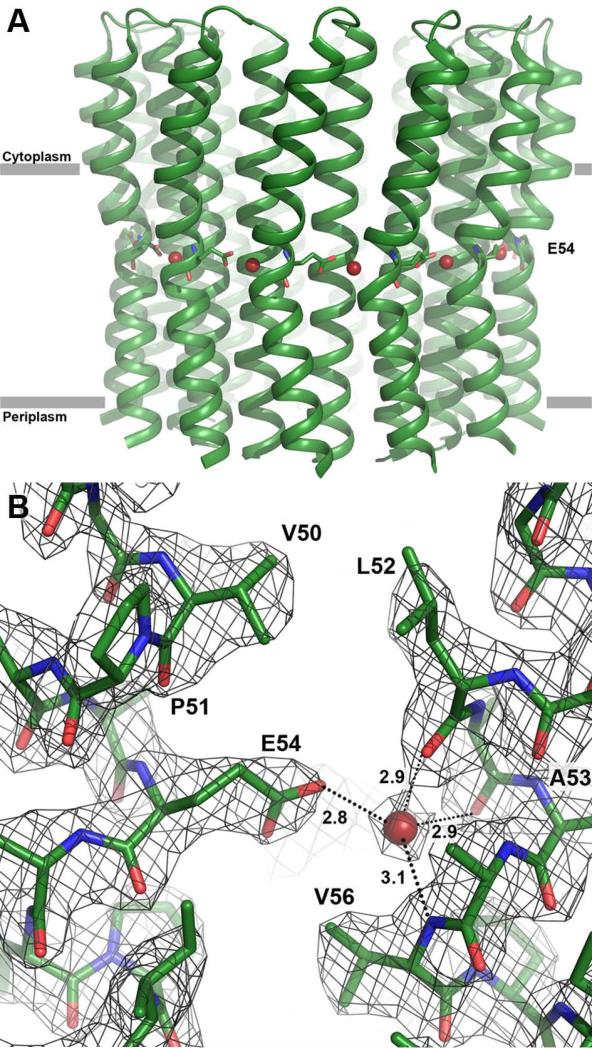
To ensure ATP synthase operation under alkaline conditions, the protons must be tightly bound during their rotation. In the B. pseudofirmus OF4 c13 ring structure, crystallized at pH 4.3, we observed water molecules within each binding site, which were proposed to support proton binding to this particular c-ring at high pH (Preiss et al., 2010). That work also showed that its ion-binding glutamate (E54) is de-protonated at very alkaline pH values, i.e. ≥pH 9.0 (Preiss et al., 2010). It therefore seemed likely that insights into the conformation of the deprotonated ATP synthase and information about the status of the previously observed ionbinding site water molecules could be obtained from a crystal structure of the WT B. pseudofirmus OF4 rotor at such a high pH. Crystals of purified WT c-subunit rotor rings were therefore grown at pH 9.0 and the structure was solved by X-ray crystallography at 2.4 Å (Table 1 and Fig. 1A). The structure showed the c-ring as a tridecameric assembly, forming a slightly constricted cylinder with a central pore. A comparison of the pH 4.3 (2x2v) and the new pH 9.0 c13 ring structure revealed that they were almost identical with an overall RMSD of only 0.179 (Cα backbone). Surprisingly, E54 was again found in a locked state (Fig. 1B), indicating its protonated state at pH 9.0. In addition, each ion-binding site still harboured a water molecule. These features are consistent with the structure of the c15 ring from Spirulina platensis, solved from pH 10.2-soaked crystals, showing an ion locked conformation with a protonated carboxylate, despite the high pH (Pogoryelov et al., 2010). The finding of protonated E54 residues in the pH 9 structure, however, seemed to challenge earlier biochemical investigations, which reported de-protonation of these residues at pH 9 (Preiss et al., 2010). This suggested that some aspect of the chemical environment in the crystals that differs from the environment in the earlier labeling experiments might affect the protonation state. A more detailed biochemical investigation of this observation was undertaken.
Table 1.
Data collection and refinement statistics.
| wild-type c13 ring at pH 9.0 | P51A mutant c13 ring at pH 4.4 | |
|---|---|---|
| Data collection | 1 crystal | 1 crystal |
| Space group | P21 | P21 |
| Cell dimensions | ||
| a, b, c (Å) | 73.8, 96.75, 118.33 | 73.94, 98.33, 121.93 |
| α, β, γ (°) | 90, 105.4, 90 | 90, 104.28, 90 |
| Resolution (Å) | 40 – 2.42 (2.5 – 2.42)* | 48.33 – 2.8 (2.9 – 2.8)* |
| R meas | 0.08 (0.89) | 0.125 (1.6) |
| R mergd-F | 0.17 (0.96) | 0.178(1.05) |
| I / σI | 12.3 (1.8) | 10.3 (1.8) |
| Completeness (%) | 99.3 (90.0) | 99.0 (98.6) |
| Redundancy | 4.5 (3.6) | 9.1(8.65) |
| Refinement | ||
| Resolution (Å) | 20 – 2.42 | 48.0 – 2.8 |
| No. reflections | 60880 | 41447 |
| Rwork / Rfree (%) | 19.49 / 22.83 | 23.51 / 28.78 |
| No. Atoms | 6745 | 6732 |
| Protein | 6327 | 6345 |
| Ligand/ion | 344 | 367 |
| Water | 74 | 20 |
| B-factors | 52.1 | 53.9 |
| Protein | 50.2 | 52.1 |
| Water | 50.6 | 43.7 |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.004 | 0.002 |
| Bond angles (°) | 0.793 | 0.573 |
Values in parentheses are for highest-resolution shell.
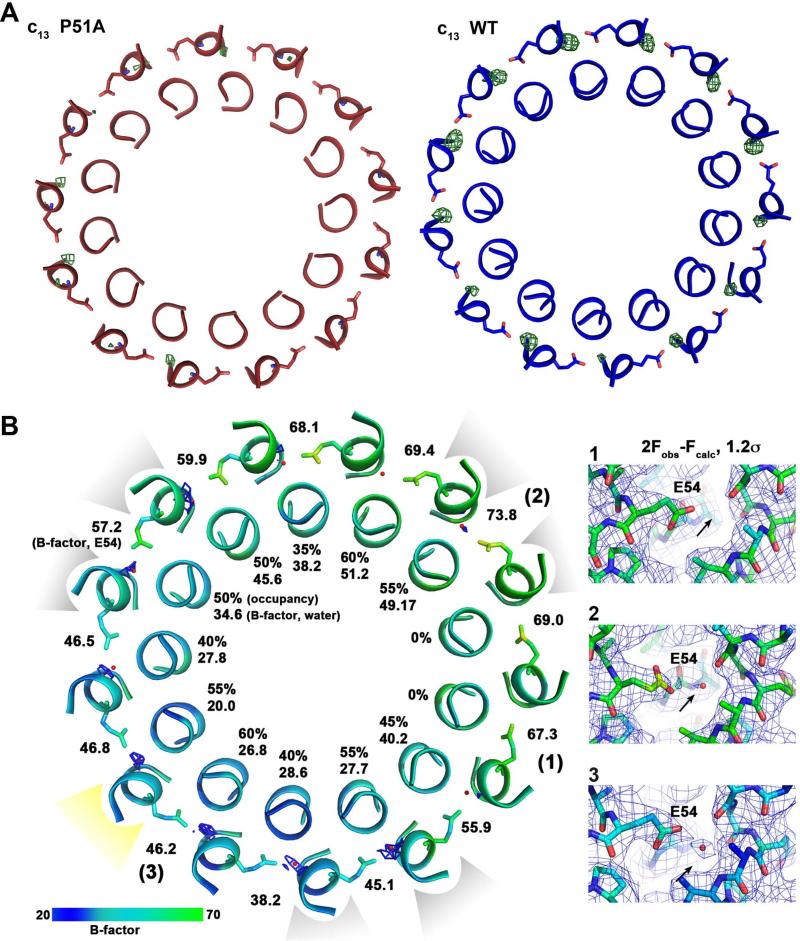
Figure 1. Structure at 2.4 Å resolution of the B. pseudofirmus OF4 c13 ring at pH 9.0.
The view is from the membrane plane. E54 is shown as sticks; the red spheres indicate the water molecules in the ion-binding sites. (A) Overview of the c13 ring with cytoplasmic side on top in ribbon representation. (B) Close-up of the ion-binding site. E54 is protonated and in locked conformation, despite the high pH. The 2Fobs-Fcalc electron density map is shown as a grey mesh at 1.8σ.
pH-dependent modification of the B. pseudofirmus OF4 c13 ring with the inhibitor dicyclohexylcarbodiimide (DCCD)
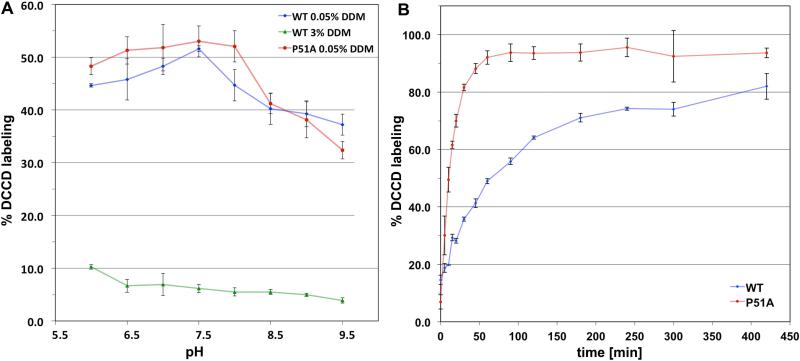
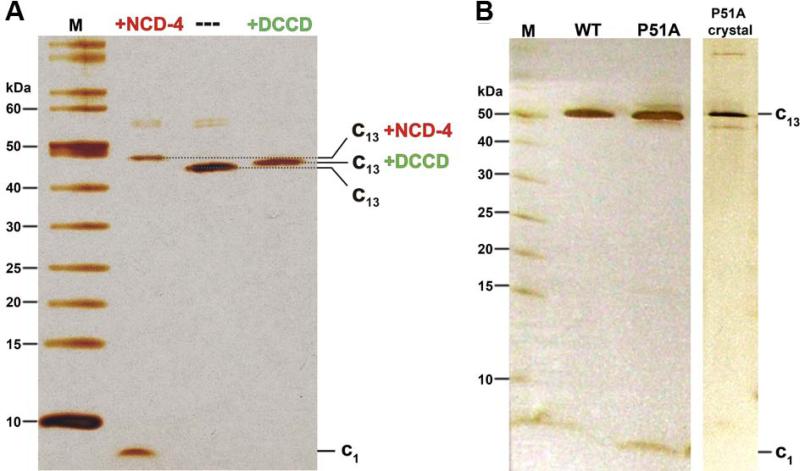
To investigate the issue of E54 protonation at pH 9, we studied the B. pseudofirmus OF4 cring using the ATP synthase inhibitor dicyclohexylcarbodiimide (DCCD) (Fig. 2). As previously reported for other c-rings (Meier et al., 2003, Pogoryelov et al., 2010), DCCD covalently reacts with protonated carboxylates (here E54); this is illustrated by a shift of the c ring bands on SDS-PAGE (Fig. 3A). Since the reaction is dependent on the protonation state and the conformation of the carboxylate, it can be used to specifically probe glutamate (or aspartate) residues and investigate an apparent pKa (pKapp) as well as its ability to switch from an ion-locked to an open state (Pogoryelov et al., 2010, Symersky et al., 2012). A pH-dependent labeling approach was carried out on the solubilized c13 ring at various detergent concentrations. As shown in Fig. 2A (blue trace), the highest DCCD labeling of the solubilized c13 ring in 0.05% dodecylmaltoside (DDM) was observed at pH 7.5 (52% of modified c1 after 90 min) with a sigmoid drop towards higher pH (37% of modified c1 after 90 min, pH 9.5). Due to non-specific reactions of DCCD, the labeling kinetics also dropped slightly towards lower pH (Pogoryelov et al., 2010, von Ballmoos & Dimroth, 2007). Thus, the reaction was pH-dependent with a pKa,app of ~8.0, and the de-protonation of E54 was most pronounced at alkaline pH values.
Figure 2. Effect of detergent concentration on pH-dependent modification and labeling kinetics of the isolated WT and P51A mutant B. pseudofirmus OF4 c13 ing with dicyclohexylcarbodiimide (DCCD).
(A) pH-dependent modification of E54 of the WT and P51A mutant with 375 μM DCCD at low and high dodecylmaltoside (DDM) detergent concentrations. At 0.05% DDM concentration the WT and the P51A c-ring mutant show a similar pH-dependent reaction profile, with fastest labeling at pH 7.5. In the presence of 3% DDM (same concentration used for 3D crystallization), DCCD modification was dramatically reduced, independent of the pH. DCCD reactions were carried out for 90 min for the WT and 15 min for P51A. (B) DCCD reaction kinetics of the isolated WT and P51A mutant c-ring. The labeling was performed at pH 7.5 using the same protein and DCCD concentrations as in (A), using 0.05% DDM. The P51A mutant c-ring showed much faster labeling kinetics compared with the WT c-ring. The standard deviation in both figures was calculated from at least three individual measurements.
Figure 3. SDS-PAGE of purified B. pseudofirmus OF4 c13 ring samples.
(A) SDS-PAGE before and after reaction with the ATP synthase inhibitor N,N’-dicyclohexylcarbodiimide (DCCD) and its fluorescent derivative N-cyclohexyl-N'-(4-(dimethylamino)naphthyl)carbodiimide (NCD-4). A sample of ~0.4 μg purified B. pseudofirmus OF4 c13 ring was incubated with 0.4 mM NCD-4 or 0.5 mM DCCD for 24 h at pH 8.0 and then analyzed on 11% SDS-PAGE. The 13 covalently modified c-subunits (all reacted with E54) resulted in an increased molecular mass of the c13 ring, which was visualized by a slower migration on the silver-stained SDS-PAGE (lanes: +NCD-4 and +DCCD), as compared to an untreated c-ring sample (lane: ---). (B) SDS-PAGE purified B. pseudofirmus OF4 WT c13 ring, P51A c-ring and P51A c-ring crystals. All samples appear at the same level, suggesting that they all have a c13 stoichiometry, as confirmed later by the crystal structure. All gels were performed using the Schägger system (Schägger & von Jagow, 1987) and stained with silver. Molecular weight markers (M) in kDa are indicated.
To understand why de-protonation of E54 can be monitored in labeling experiments, but was not observed in the crystal structure at pH 9.0, we changed the detergent concentration to that used in c-ring crystallization setup of the low pH structure, i.e., 3% (wt/vol) of DDM (Preiss et al., 2010) instead of 0.05%, and then conducted the same labeling experiments. As shown in Fig. 2A (green trace), the average labeling rate dropped dramatically and only a minor, residual pH-dependency of the DCCD modification reaction remained (~10% at pH 6 and <5% at pH 9.5 of modified c1 after 90 min). The observation indicates that in a more hydrophobic environment, such as 3% DDM, E54 is able to keep the glutamate in a stable, ion locked conformation. The result concurs with a DCCD labeling study performed with the proteoliposome-reconstituted S. platensis c15 ring, in which the ion locked conformation was observed and hardly reacted with DCCD at any of the pH values investigated (Pogoryelov et al., 2010). In summary, the probability of a conformational switch of E54 from locked to open and with it, de-protonation events at E54, are not primarily dependent on the bulk environmental pH, but are rather strongly affected by the hydrophobicity of the ion binding site's chemical micro-environment.
Modification of the “neutralophile-like” P51A c13ring mutant with the inhibitor DCCD
In the structure of the B. pseudofirmus OF4 c13 ring, proline-51 is located just one helix turn above E54 (Fig. 1B). It breaks the α-helical pattern and thereby provides optimal coordination distances for the translocated proton. In alkaliphilic Bacillus species, this proline is conserved whereas alanine is often found in neutralophilic bacteria (Fig. 4). A P51A mutant was previously reported to significantly reduce growth and ATP synthesis activity of B. pseudofirmus OF4 cells in a high alkaline environment (Wang et al., 2004). It is therefore considered as a sequence feature that is typical of alkaliphilic Bacillus species (Arechaga & Jones, 2002). Although earlier experiments had indicated that the P51A mutant has a severe growth deficit on non-fermentative carbon sources (i.e. malate) and not on fermentative glucose, molar growth yields on malate had not been determined in comparison to WT. Thus we here determined growth yields on limiting malate at both pH 7.5 and 10.5 in comparison with WT by methods described earlier (Preiss et al., 2013). As shown in Table 2, the mutant growth yield was not significantly reduced at pH 7.5. At pH 10.5, a significant reduction was observed in the molar growth yield of the mutant on malate, but it was not nearly as dramatic as the growth deficit of the same mutant strain on malate-replete growth media at pH 10.5. This may reflect the heightened importance of the efficiency of energy conserving mechanisms near the upper edge of the pH range, such that a small deficit has a large impact.
Figure 4. Amino acid alignment of selected bacterial ATP synthase c-subunits.
The c-subunits are shown of alkaliphilic (*) and neutralophilic Bacillus species as well as from a selection of other well-characterized species. The residues targeted in this work are P51 and V21. A valine (V21 in B. pseudofirmus OF4, located in the c-ring ion-binding site) is conserved in alkaliphilic Bacillus species, while neutralophilic species have an asparagine or glutamine in this position. In the outer C-terminal σ-helix of alkaliphilic Bacillus species, the first proline (P51, B. pseudofirmus OF4) of the motif P51xxE54xxP57 (highlighted in red/grey) is located one helix turn above the ion-binding glutamate E54 (highlighted in red). Whereas P57 is present in several bacterial species, P51 exclusively exists in alkaliphilic Bacillus species.
Table 2.
Molar growth yields on limiting L-malate at pH 7.5 and 10.5 for WT and P51A.
| Strain | pH 7.5 | pH 10.5 |
|---|---|---|
| Wild-type | 40.1±0.81 | 39.9±1.6 |
| P51A | 36.2±2.9 | 32.9±1.9 |
mg dry weight (nmoles L-malate consumed)−1. The data are the average of 4-8 samples ± standard deviation.
To characterize the influence of the proline-51 residue on structure and function of the enzyme, we performed another pH-dependent DCCD modification study at low detergent concentration (0.05% DDM) using the isolated P51A mutant c13 ring (Fig. 2A, red trace). As with the WT, maximum labeling was observed at pH 7.5. The pH profile of the mutant was thus similar to the WT’s, with only a slightly steeper drop of the reaction rates towards higher pH values (Fig. 2A). Hence the pKa,app in this experiment did not change significantly with respect to the wild-type. Noticeably however, the P51A mutant c-ring showed much faster labeling kinetics than the WT ring at the identical low (0.05%) DDM concentration. While for the WT samples it took 60 min to get 50% of the c-subunits modified, the P51A mutant preparation showed the same yield of labeled c-subunits after 10 min (Fig. 2B). This significant difference in the reaction rate indicates that the protonated E54 in this mutant is more prone to react with DCCD than the WT. Since both pKa,app of the WT and the P51A mutant are similar but the reaction rates diverge, the accessibility of DCCD to the protonated E54 is apparently enhanced in the mutant. As noted above, the reaction rate is not only dependent on the protonation state of the carboxyl moiety but also on the conformational state of E54 and its ability to switch from locked to open (Pogoryelov et al., 2010). Therefore, the faster labeling rate together with its similar pKa,app is best explained by an E54 site, which can statistically undergo conformational switches more often in the mutant. Given this, we hypothesized that the reason for this observed difference lies in subtle structural changes of the mutant c13 ring itself. We therefore pursued an effort to determine the P51A c13 ring structure.
2.8 Å crystal structure of the B. pseudofirmus OF4 P51A mutant c13ring
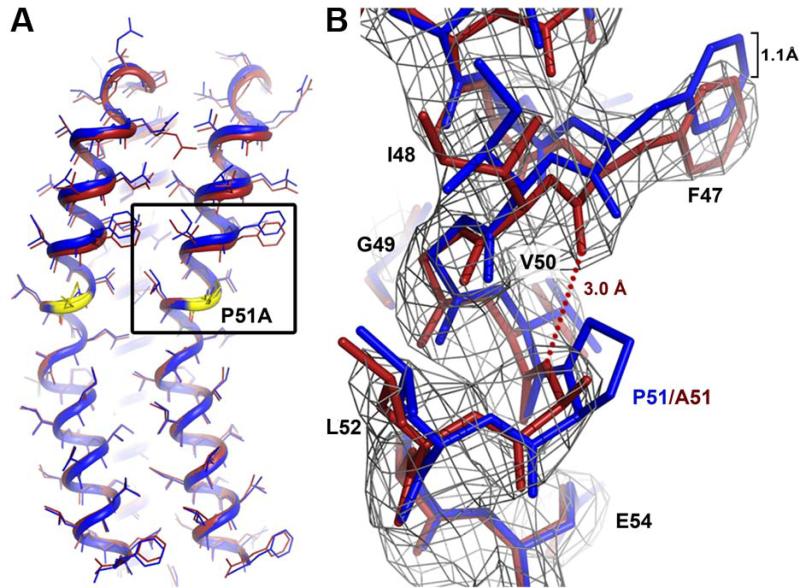
The purified P51A mutant c-ring from B. pseudofirmus OF4 shows an identical migration behaviour on SDS-PAGE as compared with the WT rotor ring. It suggests that their stoichiometries are identical (Fig. 3B) while excluding an earlier consideration of a stoichiometry change introduced by the mutation (Wang et al., 2004). We were therefore able to use the wild-type c13 model for molecular replacement to solve the P51A mutant structure by X-ray crystallography from crystals grown at pH 4.4 (Table 1). The final protein model was mostly identical to the wild-type c13 ring, with the ion-binding glutamate E54 in ion locked state (Fig. 5A). In the 2Fobs-Fcalc electron density map, the P51A mutation is directly visible (Fig. 5B) and fits to alanine (but not proline). The A51 backbone nitrogen now lies within hydrogen bonding distance with the F47 backbone carbonyl. It thereby restores the regular α-helix and, in consequence, shortens the outer C-terminal helix by 1.1 Å (Fig. 5B). Whereas the overall influence of the mutation on the shape of the c-ring remains small, a notable difference was revealed by the analysis of the water molecules at the ion-binding sites. Most of them either show no occupancy or a low occupancy in the mutant, as exemplified by two comparable Fobs-Fcalc maps of the mutant and WT, both with the datasets cut at the same 2.8 Å resolution (Fig. 6A).
Figure 5. Structure of the B. pseudofirmus OF4 P51A mutant c-ring ion-binding site at 2.8 Å.
(A) Overlay of two c-subunits from WT (blue) and P51A mutant (red). The position of the P51A mutation is highlighted in yellow. The overall RMSD of the two structures is 0.434 (calculated for the two Cα backbones). (B) Close up of the P51A mutation (area framed in (A)). The mutation causes the formation of a new hydrogen bond (red dots) within the α-helix backbone, which results in a slight down-shift (1.1 Å) of the amino acids preceding P51A, on the level of F47. The 2Fobs-Fcalc map is shown as grey mesh at 2σ.
Figure 6. Analysis of the water occupancies in the WT and P51A mutant B. pseudofirmus c13 ring ion-binding sites.
(A) The slate view is from the cytoplasm on the WT (blue) and P51A mutant (red) c13 ring at the level of the ion-binding sites. The helices are shown in ribbon representation, E54 as sticks. Omit map (Fobs-Fcalc, displayed at 3.5σ of the water molecules within the site is shown as green mesh. While in the WT c-ring all binding sites show strong densities, corresponding to 100% occupancy of the water molecules, just a few binding sites contain mostly weak densities for the waters, with lower occupancies in the P51A mutant. This indicates either absence or a higher flexibility of water molecules at these positions. (B) Slate view from cytoplasm on the P51A c-ring at level of the E54. The 2Fobs-Fcalc maps at 1.2σ show only partial water occupancy within the sites. The contact positions of crystallographic c-ring symmetry mates are indicated in light grey: three contacts at the cytoplasmic c-ring side (loop) and yellow: one contact at the periplasmic c-ring side (N- and C-termini). The water occupancies at c-subunits involved in crystal contacts are slightly higher. Water occupancies and B-factors are given for each individual water molecule. The B-factors for E54 side chains are also indicated. [1], [2] and [3]: Three examples for 2Fobs-Fcalc electron density maps shown at 1.2σ as grey mesh. Colors: A B-factor color scale from low (blue) to high (green) is indicated. The arrow points to the expected position of the water molecule.
It has been shown previously that highly coordinated water molecules can become visible in F obs-Fcalc maps to 3.2 Å resolution (Minichino et al., 2003). So we hypothesized that the water molecules in the P51A mutant are either present in smaller numbers or are more flexible than they are in their WT positions. To validate these ideas, we tested the occupancies of the water molecules at each individual site in the mutant c-rings. The asymmetric unit of the P21 crystal contained one complete c-ring, giving a total of 13 individual c-subunit structures (no non-crystallographic symmetry averaging was applied), which we could take into the analysis (Fig. 6B). More specifically, we placed 13 water molecules in the present Fobs-Fcalc densities in the mutant structure and the calculated 2Fobs-Fcalc maps. At some sites we were able to generate significant 2Fobs-Fcalc maps, while in others no corresponding 2Fobs-Fcalc density could be generated. To determine the exact occupancy of the water we raised it stepwise until no positive Fobs-Fcalc density was visible any more (at 3.5σ). Finally, the occupancies of the questionable waters found in the 13 ion-binding sites varied from 60% to 0% (Fig. 6B). In sum, the occupancies of the mutant were significantly lower than those found in the corresponding WT settings, both for the pH 9.0 structure and the pH 4.3 structure (Preiss et al., 2010). It is also noteworthy that the occupancies were generally higher at those c-subunits that formed crystal contacts with other c-rings, while binding sites of c-subunits that were not involved in crystal contacts showed lowest occupancies (Fig. 6B). This effect could be caused by a higher flexibility of the individual c-subunits, resulting either in loss of the water molecule or a less defined position of the water molecules that would lead to weaker or no density. The observations suggest that the crystal contacts may influence the water occupancy at least modestly, perhaps by rigidifying the overall σ-helical construction.
Taken together, the structure of the “neutralophile-like” P51A mutant, with its apparent reduction in water occupancy, results in a higher conformational freedom of the glutamate side chain and allows the site to stochastically switch its conformation more often from locked to open. This change can account for the observed faster DCCD labeling rates and provides an underlying molecular reason, namely a subtle but important reduction of the water occupancy within the P51A c-ring ion-binding sites. Notably, this rather small difference on a molecular scale can be manifested also in vivo on the physiological scale of the bacterium's bioenergetics and growth under stringent alkaline pH conditions (Wang et al., 2004).
Growth studies of the B. pseudofirmus OF4 ‘neutralophile-like’ V21N c-ring mutant
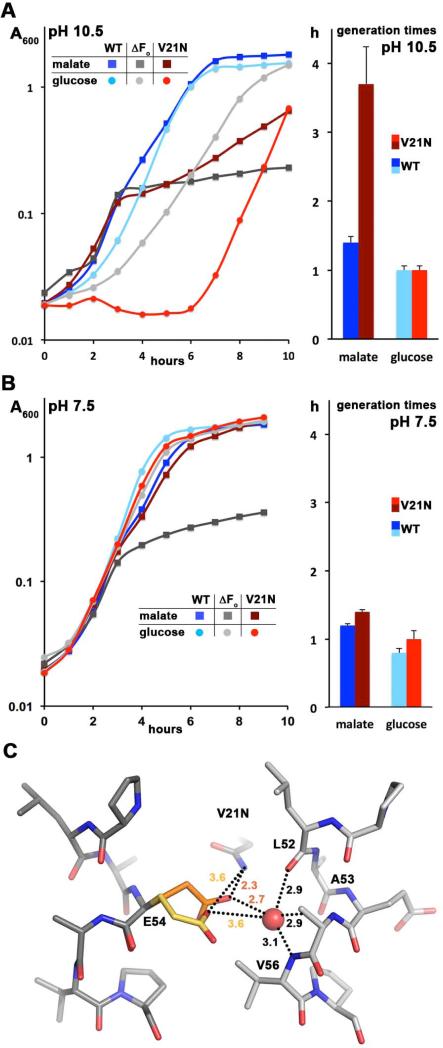
In the WT B. pseudofirmus OF4 c13 ring the E54 side chain has impressive rotamer freedom but still keeps an optimal hydrogen bonding distance with one of its carboxyl oxygens to the water molecule in the ion-binding site (Preiss et al., 2010). In other previously solved high-resolution c-ring structures (Meier et al., 2005, Pogoryelov et al., 2009, Symersky et al., 2012, Schulz et al., 2013) the glutamate is fixed in one position by a hydrogen bonding network involving a glutamine or glutamate residue from the inner N-terminal helix. The c-subunits from alkaliphilic Bacillus species contain a valine (V21 in B. pseudofirmus OF4) instead, making such an interaction impossible, while neutralophilic Bacillus species have an asparagine at that position (Fig. 4). We therefore considered the possibility that V21 represents another adaptation in alkaliphilic Bacillus species, to allow the rotameric flexibility of E54 and thereby provide an optimal distance to the water with highest possible proton affinity. To assess this hypothesis, we mutated V21 to asparagine, as is found in neutralophilic species (Preiss et al., 2010), and performed growth studies at pH 7.5 and 10.5, both on fermentable and non-fermentable media. We included a comparison of the growth of the V21N mutant with the WT and a ΔFo mutant, the latter being a control showing only fermentative growth (Fig. 7A, B). On the semi-defined malate or glucose medium at pH 7.5 (Fig. 7B), the V21N mutant grew at rates very similar to the WT. At pH 10.5, on the other hand, V21N exhibited a much more profound deficit using malate as the energy source (Fig. 7A). The generation time for V21N was 2.5 times slower than the WT, 3.7 hours compared to 1.4 hours. The V21N mutant also exhibited a pronounced lag growing on glucose at pH 10.5, although its growth rate was the same as the WT under these conditions (Fig. 7B, red trace. This resulted in a poorer growth profile than the negative control, the ΔFo mutant raising the possibility that asparagine at this position might lead to proton leakiness. Whereas modest leakiness might not greatly impair fermentative growth near neutral pH, it would be more problematic at high pH and/or during non-fermentative growth. The complex pattern of growth deficits of the V21N mutant made it impossible to assess a molar growth yield on malate, as has been conducted for several other mutants, including the P51A, in this study. The findings highlight the importance of V21 for growth at alkaline pH values, especially for non-fermentable substrates but even for glucose.
Figure 7. Growth phenotype of c-subunit V21N mutant at pH 7.5 and 10.5 and a model of its ion-binding site.
Growth curves for the WT, V21N c-ring mutant, and ΔFo mutant strains from B. pseudofirmus OF4 811M were performed at pH 10.5 (A) and 7.5 (B) on semi-defined fermentable and non-fermentable media for which the WT is the positive control and the ΔFo strain is the negative control. The V21N mutant strain showed a significant growth deficit at high pH, especially on non-fermentable malate medium. The marginal growth of the ΔFo strain on malate media is due to the small quantity of fermentable substrates in yeast extract. The growth curves and the calculated generation times are from 10 and 9 individual growth curves on malate at pH 10.5, for the WT and V21N, respectively, and the other growth curves and generation times are from 4 individual growth curves. (C) Ion-binding site model of the B. pseudofirmus OF4 V21N mutant c-ring. V21 was replaced in silico by asparagines and its conformation optimized using Scrwl (Krivov et al., 2009). Glutamate (E54) is shown in two rotameric conformations: orange, a c13 WT conformation in 2x2v (Preiss et al., 2010); yellow, ion locked conformation as observed in 1yce (I. tartaricus c11, (Meier et al., 2005)). In both cases a new hydrogen bond with N21 is formed (indicated by dashed lines), stabilizing the ion coordination network of the site in locked conformation.
Attempts to isolate the V21N c-ring from the B. pseudofirmus OF4 ATP synthase failed due to protein complex stability issues. As an alternative, we built a structural model of the V21N c-ring ion-binding site (Fig. 7C). In agreement with the mutants’ reduced growth on non-fermentable carbon-sources the model suggests that this mutant has a reduced rotamer freedom of E54 but still keeps optimal hydrogen-bonding distance between one of the oxygens from the proton-binding carboxylate and the water molecule (Preiss et al., 2010). In consequence, and similar to the effect of the P51A mutation, the capability of the ion-binding site to conformationally open and close is affected. The model hence provides a good rational basis for the experimentally-observed reduced growth at alkaline pH.
Conclusions
Ion-binding sites in ATP synthase rotor rings are finely-tuned networks involving a variety of coordinating amino acid residues. In this work we showed that the sites in alkaliphilic Bacillus species are structurally and functionally adapted to ensure ion-binding and release even at enhanced pH levels. Two residues, P51 and V21, which are conserved in c-subunits of these species, influence the site's coordination network and promote the presence of a water molecule. P51 ensures a coordination geometry that is optimal to stably coordinate the water molecule and thereby bind the proton with high affinity. This coordination geometry is further ensured by the presence of V21, which allows rotamer freedom of the glutamate's carboxylate. Therefore, both residues are indirectly involved in proton coordination; it influences the capability of B. pseudofirmus OF4 to synthesize ATP via oxidative phosphorylation at high pH. Moreover, a general feature of the ATP synthase ion-binding glutamate was addressed on a structural and functional level, namely its ability to remain in an ion locked, protonated conformation, despite the high bulk pH. These features are critical for Fo motor function and non-fermentative growth at high pH and thus contribute to the alkaliphile's ability to cope with its bioenergetic challenges. The data shown in this work do not alter or influence the overall mechanism of ion translocation of the B. pseudofirmus OF4 ATP synthase, outlined in the introduction and in an earlier study that specifically addresses this topic in alkaliphiles (Preiss et al., 2010). However, the new findings highlight how the structure and biochemistry of the c-ring's ion-binding site is directly connected with growth and survival at stringent, high environmental pH and therefore how this enzyme is specifically adapted to the cell bioenergetics and physiology of alkaliphiles.
EXPERIMENTAL PROCEDURES
Cloning, protein expression and ATP synthase purification
Mutagenesis of the chromosomal c-subunit in Bacillus pseudofirmus OF4 811M was carried out by previously published methods (Wang et al., 2004, Liu et al., 2009). The F1Fo-ATP synthase was purified from a strain of B. pseudofirmus OF4 811M harbouring a six histidine tag after the N-terminal methionine in the β-subunit as detailed in (Fujisawa et al., 2010).
Purification of B. pseudofirmus OF4 c-rings
The c-rings (wild-type and P51A mutant) were purified using the method as described (Preiss et al., 2010). Briefly, purified B. pseudofirmus OF4 ATP synthase (3 mg/ml) was solubilized using 1% (wt/vol) N-lauroylsarcosine at 45°C for 10 min and all proteins except c13b2δ were precipitated by 62.5% (NH4)2SO4. After dialysis, the sample was again solubilized using 1.5% (wt/vol) Fos-12 at 45°C for 10 min and then loaded on a 5-35% (wt/vol) sucrose gradient. The c-ring was finally purified by hydroxyapatite, concentrated using polyethylene glycol (PEG) 35’000 and stored at 4°C.
Modification of the B. pseudofirmus OF4 c-ring with dicyclohexylcarbodiimide (DCCD) and MALDI-MS analysis of the labeled products
The purified c-ring was dialysed for 48 h at 4°C against volatile buffers: either 20 mM cacodylate/trimethylamine/NH3 (for pH 6.0-8.5) or 20 mM ammonium carbonate/NH3 (pH 9.0-9.5) using Spectrapor 1 membranes (MWCO 6'000-8'000, Spectrum Labs). Next, the samples were diluted to 0.1 mg/ml protein in either 0.05 or 3% (wt/vol) n-dodecyl-β-D-maltopyranoside (DDM) in the above buffers for the DCCD labeling experiments. To start the labeling reaction, DCCD (21 mM in 60% EtOH) was added to a final concentration of 0.375 mM to each sample. Aliquots were taken after 90 min (WT) or 15 min (P51A) for pH-depended labelings, or as indicated in the kinetics (Fig. 2). To stop the reaction 1 μl reaction setup was mixed with 1 μl matrix (dihydroxyacetophenone, saturated solution in 75% ethanol with 20 mM ammonium citrate) and spotted directly onto a ground-steel MALDI target in duplicates. MALDI mass spectra were acquired in the mass range of 5-20 kDa on a Bruker Autoflex III Smartbeam MALDI-TOF mass spectrometer using optimized ionization, ion optics and detector settings. Spectra were recalibrated using the near-neighbour method with a calibrant mixture (Bruker Protein Calibration standard 1, Bruker Daltonics, Bremen).
All spectra were evaluated and recalibrated using the software Bruker FlexAnalysis 3.3 (Build 75). After background substraction (TopHat), smoothing (Savitzky-Golay, width 1 m/z, 3 cycles) and peak picking (Centroid, s/n 5) the intensities of the c-monomer and the DCCD-labeled species were used for calculating the efficiency of DCCD binding. The “labeling efficiency” (Fig. 2) was calculated as the intensity ratio of the DCCD-bound species to the sum of the labeled and unlabeled species. All values were calculated from 3–4 experimental replications and technical MALDI measurement duplicates.
Determination of n-dodecyl-β-D-maltopyranoside concentration
The maltoside concentration was quantified in a colorimetric assay by determination of the sugar moiety with phenol and sulphuric acid (Urbani & Warne, 2005). Briefly, a 25 μl sample was mixed with 25 μl 5% (wt/vol) phenol. After the addition of 150 μl sulphuric acid the sample was incubated at 80°C for 30 min. Thereafter, the absorption of the samples at 492 nm was measured at room temperature. The detergent concentration was calculated using a standard curve determined from 0.01% to 0.1% n-dodecyl-β-D-maltopyranoside.
Crystallization of the B. pseudofirmus OF4 c13 ring, wild-type and P51A mutant
3D crystals were grown using the hanging drops vapour diffusion method. 0.5 μl WT c13 ring (2.5 mg/ml) was supplied with 0.5% n-tridecyl-β-D-maltopyranoside and mixed with 0.25 μl crystallization mixture containing 0.1 M Tris/HCl pH 9.0, 20% (vol/vol) PEG 400. For P51A mutant crystals 0.5 μl protein (3 mg/ml) was supplied with 0.4% n-undecyl-β-D-maltopyranoside and mixed with 0.25 µl crystallization mixture containing 0.1 M NaOAc pH 4.4, 18% PEG 400. Rod-shaped crystals of the wild-type and the P51A mutant grew within 24 h. Before flash freezing in liquid nitrogen, crystals were soaked in cryogenic solution containing 30% PEG 400, 0.1 M Tris/HCl pH 9.0, 0.5% DDM or 30% PEG 400, 0.1M NaOAc pH 4.4, 0.5% DDM respectively.
Data collection, structure determination and refinement
Data to 2.4 Å and 2.8 Å were collected at the ID23.2 beamline of the European Synchrotron Radiation Facility (ESRF, Grenoble, France) and Max-Planck/Novartis beamline X10SA (PXII) of the Swiss Light Source (SLS, Villigen, Switzerland), respectively. The data were processed using the XDS package (Kabsch, 1993); phases were determined by molecular replacement using Phaser from the CCP4 package (McCoy, 2007) using the B. pseudofirmus OF4 c13 ring structure (2x2v) as a search model. To remove model bias, the area around the ion-binding site was removed from the model for initial phasing and manually build into the resulting densities afterwards. Iterative cycles of refinement and model building were performed using phenix.refine (Zwart et al., 2008) and Coot (Emsley & Cowtan, 2004) without any non-crystallographic symmetry operation applied to visualize details within the individual copies of the ring. In both cases the refinement resulted in electron density maps, in which all amino acids were unambiguously traceable. The Ramachandran plots showed no outliers in any of the two structures. The structure figures were drawn with Pymol (DeLano, 2002). The atomic coordinates were deposited in the Protein Data Bank (PDB) with accession codes 4cbk and 4cbj for the wild-type c13 ring at pH 9.0 and the P51A mutant c13 ring at pH 4.4, respectively.
Stepwise analysis of the water occupancy at the ion-binding site
Generally, highly coordinated water molecules can become visible in Fobs-Fcalc maps to at least 3.2 Å (Minichino et al., 2003). To analyse the occupancy of water molecules at the individual ion-binding sites of the P51A mutant c13 ring structure (2.8 Å resolution) and compare it with the corresponding occupancies of the wild-type structure (2x2v, data truncated at 2.8 Å), we proceeded as follows. 1) all water molecules in the mutant structure were removed as an initial step to create a corresponding omit map (Fobs-Fcalc). 2) water molecules were placed into the densities of the omit map, as far as visible at 3.5σ. 3) After a refinement cycle with phenix.refine (Zwart et al., 2008), with the automatic occupancy-refinement option turned off, the placed water molecules were inspected for Fobs-Fcalc maps. 4) For those waters showing positive density in the Fobs-Fcalc map, indicating that too few atoms were placed, the occupancy of the water molecule was enhanced until no positive density was visible any more, indicating that the given occupancy is the most probable one. This procedure was done in iterative steps of Coot (Emsley & Cowtan, 2004) and phenix.refine (Zwart et al., 2008) for each site. Fig. 6B shows the final result of this analysis.
Generation of the B. pseudofirmus OF4 V21N mutant c-ring model
The mutation V21N was introduced into the c13 wild-type structure (2x2v) using Coot (Emsley & Cowtan, 2004). Based on rotamer libraries the software Scrwl4 (Krivov et al., 2009) was used to calculate the most probable side chain orientation.
Growth experiments
Growth media for B. pseudofirmus OF4 811M were buffered at either pH 7.5 with 0.1 M 3-(N-morpholino)propanesulfonic acid (MOPS) or at pH 10.5 with 0.1 M NaHCO3/Na2CO3. The carbon sources were glucose or D,L-malate at 50 mM and the media also contained 1 mM potassium phosphate, 0.1% yeast extract and 1% trace metal solution (STS) (Wang et al., 2004) as well as MgSO4 (5 mM at pH 7.5 and 0.1 mM at pH 10.5). Strains were pre-grown in pH 7.5 glucose-containing medium overnight. A volume of cells equivalent to 5 O.D. 600 nm were pelleted, washed with a pH 7.5 buffer (50 mM MOPS, 1 mM potassium phosphate, 0.1 mM MgSO4, pH 7.5 with NaOH), suspended in 1 ml of the same buffer and 0.2 ml of this suspension was used to inoculate 50 ml of medium in a 250 ml flask prewarmed to 37°C. Flasks were shaken at 225 rpm at 37°C and hourly readings at 600 nm were taken. Where appropriate, samples were diluted to read between 0.2 and 0.4 in the spectrophotometer (Shimadzu UV-1601). Molar growth yields on limiting malate (5 mM L-malate) at pH 7.5 and 10.5 were determined as in (Preiss et al., 2013).
Acknowledgements
The authors thank José Faraldo-Gómez for his helpful comments on the manuscript. We also thank the staffs of the Swiss-Light-Source (SLS) in Villigen and the European Synchrotron Radiation Facility (ESRF) in Grenoble for their technical support at the beamlines. Funding: This work was supported by the Collaborative Research Center (SFB) 807 of the German Research Foundation (DFG), the Cluster of Excellence ’Macromolecular Complexes‘ (CEFMC) at the Goethe University Frankfurt (Project EXC 115), the ESFRI-Instruct initiative by the European Union and research grant R01GM28454 from the National Institute of General Medical Sciences (TAK).
Abbreviations
- ATP
adenosine-5’-triphosphate
- SDS-PAGE
sodium dodecylsulfate-polyacrylamide gel electrophoresis
- DCCD
N,N'-dicyclohexylcarbodiimide
- NCD-4
N-cyclohexyl-N'-(4-(dimethylamino)naphthyl)carbodiimide
- pmf
proton-motive force
References
- Abrahams JP, Leslie AGW, Lutter R, Walker JE. Structure at 2.8 Å resolution of F1-ATPase from bovine heart mitochondria. Nature. 1994;370:621–628. doi: 10.1038/370621a0. [DOI] [PubMed] [Google Scholar]
- Arechaga I, Jones PC. The rotor in the membrane of the ATP synthase and relatives. FEBS Lett. 2002;494:1–5. doi: 10.1016/s0014-5793(01)02300-6. [DOI] [PubMed] [Google Scholar]
- Boyer PD. Bioenergetic coupling to protonmotive force: should we be considering hydronium ion coordination and not group protonation? Trends Biochem. Sci. 1988;13:5–7. doi: 10.1016/0968-0004(88)90005-9. [DOI] [PubMed] [Google Scholar]
- Boyer PD. The binding change mechanism for ATP synthase - some probabilities and possibilities. Biochim. Biophys. Acta. 1993;1140:215–250. doi: 10.1016/0005-2728(93)90063-l. [DOI] [PubMed] [Google Scholar]
- Cook GM, Keis S, Morgan HW, von Ballmoos C, Matthey U, Kaim G, Dimroth P. Purification and biochemical characterization of the F1Fo-ATP synthase from thermoalkaliphilic Bacillus sp. strain TA2.A1. J. Bacteriol. 2003;185:4442–4449. doi: 10.1128/JB.185.15.4442-4449.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL. The Pymol Molecular Graphics System. DeLano Scientific; San Carlos, CA: 2002. [Google Scholar]
- Dimroth P, von Ballmoos C, Meier T. Catalytic and mechanical cycles in F-ATP synthases. Fourth in the Cycles Review Series. EMBO Rep. 2006;7:276–282. doi: 10.1038/sj.embor.7400646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Ferguson SJ. ATP synthase: From sequence to ring size to the P/O ratio. Proc. Natl. Acad. Sci. U.S.A. 2010;107:16755–16756. doi: 10.1073/pnas.1012260107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisawa M, Fackelmayer OJ, Liu J, Krulwich TA, Hicks DB. The ATP synthase a-subunit of extreme alkaliphiles is a distinct variant: mutations in the critical alkaliphile-specific residue Lys-180 and other residues that support alkaliphile oxidative phosphorylation. J. Biol. Chem. 2010;285:32105–32115. doi: 10.1074/jbc.M110.165084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heberle J, Riesle J, Thiedemann G, Oesterhelt D, Dencher NA. Proton migration along the membrane surface and retarded surface to bulk transfer. Nature. 1994;370:379–382. doi: 10.1038/370379a0. [DOI] [PubMed] [Google Scholar]
- Hicks DB, Krulwich TA. Purification and reconstitution of the F1F0-ATP synthase from alkaliphilic Bacillus firmus OF4. Evidence that the enzyme translocates H+ but not Na+. J Biol Chem. 1990;265:20547–20554. [PubMed] [Google Scholar]
- Hicks DB, Liu J, Fujisawa M, Krulwich TA. F1F0-ATP synthases of alkaliphilic bacteria: lessons from their adaptations. Biochim. Biophys. Acta. 2010;1797:1362–1377. doi: 10.1016/j.bbabio.2010.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann A, Dimroth P. The ATPase of Bacillus alcalophilus. Purification and properties of the enzyme. Eur. J. Biochem. 1990;194:423–430. doi: 10.1111/j.1432-1033.1990.tb15635.x. [DOI] [PubMed] [Google Scholar]
- Junge W, Lill H, Engelbrecht S. ATP synthase: an electrochemical transducer with rotatory mechanics. Trends in biochemical sciences. 1997;22:420–423. doi: 10.1016/s0968-0004(97)01129-8. [DOI] [PubMed] [Google Scholar]
- Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Cryst. 1993;26:795–800. [Google Scholar]
- Krivov GG, Shapovalov MV, Dunbrack RL., Jr. Improved prediction of protein side-chain conformations with SCWRL4. Proteins. 2009;77:778–795. doi: 10.1002/prot.22488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krulwich TA, Ito M. Prokaryotic alkaliphiles. In: Rosenberg E, DeLong EF, Thompson F, Lory S, Stackebrandt E, editors. The Prokaryotes. Vol. 2. Springer; New York: 2013. pp. 441–469. [Google Scholar]
- Krulwich TA, Sachs G, Padan E. Molecular aspects of bacterial pH sensing and homeostasis. Nat. Rev. Microbiol. 2011;9:330–343. doi: 10.1038/nrmicro2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone V, Krah A, Faraldo-Gómez JD. On the question of hydronium binding to ATP-synthase membrane rotors. Biophys. J. 2010;99:L53–55. doi: 10.1016/j.bpj.2010.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Fujisawa M, Hicks DB, Krulwich TA. Characterization of the functionally critical AXAXAXA and PXXEXXP motifs of the ATP synthase c-subunit from an alkaliphilic Bacillus. J. Biol. Chem. 2009;284:8714–8725. doi: 10.1074/jbc.M808738200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthies D, Preiss L, Klyszejko AL, Muller DJ, Cook GM, Vonck J, Meier T. The c13 ring from a thermoalkaliphilic ATP synthase reveals an extended diameter due to a special structural region. Journal of molecular biology. 2009;388:611–618. doi: 10.1016/j.jmb.2009.03.052. [DOI] [PubMed] [Google Scholar]
- McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr. D Biol. Crystallogr. 2007;63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan DGG, Keis S, Dimroth P, Cook GM. A specific adaptation in the a subunit of thermoalkaliphilic F1Fo-ATP synthase enables ATP synthesis at high pH but not at neutral pH values. J. Biol. Chem. 2007;282:17395–17404. doi: 10.1074/jbc.M611709200. [DOI] [PubMed] [Google Scholar]
- Meier T, Faraldo-Gómez JD, Börsch M. ATP synthase, a paradigmatic molecular machine. Molecular Machines in Biology Cambridge University Press; 2011. pp. 208–238. [Google Scholar]
- Meier T, Matthey U, von Ballmoos C, Vonck J, Krug von Nidda T, Kühlbrandt W, Dimroth P. Evidence for structural integrity in the undecameric c-rings isolated from sodium ATP synthases. Journal of molecular biology. 2003;325:389–397. doi: 10.1016/s0022-2836(02)01204-4. [DOI] [PubMed] [Google Scholar]
- Meier T, Polzer P, Diederichs K, Welte W, Dimroth P. Structure of the rotor ring of F-Type Na+-ATPase from Ilyobacter tartaricus. Science. 2005;308:659–662. doi: 10.1126/science.1111199. [DOI] [PubMed] [Google Scholar]
- Minichino A, Habash J, Raftery J, Helliwell JR. The properties of (2Fo - Fc) and (Fo - Fc) electron-density maps at medium-to-high resolutions. Acta Crystallogr. D Biol. Crystallogr. 2003;59:843–849. doi: 10.1107/s0907444903004219. [DOI] [PubMed] [Google Scholar]
- Mizutani K, Yamamoto M, Suzuki K, Yamato I, Kakinuma Y, Shirouzu M, Walker JE, Yokoyama S, Iwata S, Murata T. Structure of the rotor ring modified with N,N’-dicyclohexylcarbodiimide of the Na+-transporting vacuolar ATPase. Proc. Natl. Acad. Sci. U.S.A. 2011;108:13474–13479. doi: 10.1073/pnas.1103287108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulkidjanian AY, Cherepanov DA, Heberle J, Junge W. Proton transfer dynamics at membrane/water interface and mechanism of biological energy conversion. Biochemistry (Mosc) 2005;70:251–256. doi: 10.1007/s10541-005-0108-1. [DOI] [PubMed] [Google Scholar]
- Noji H, Yasuda R, Yoshida M, Kinosita K., Jr. Direct observation of the rotation of F1-ATPase. Nature. 1997;386:299–302. doi: 10.1038/386299a0. [DOI] [PubMed] [Google Scholar]
- Pogoryelov D, Klyszejko AL, Krasnoselska G, Heller E-M, Leone V, Langer JD, Vonck J, Muller DJ, Faraldo-Gómez JD, Meier T. Engineering rotor ring stoichiometries in the ATP synthase. Proc. Natl. Acad. Sci. U.S.A. 2012;109:E1599–1608. doi: 10.1073/pnas.1120027109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogoryelov D, Krah A, Langer JD, Yildiz Ö, Faraldo-Gomez JD, Meier T. Microscopic rotary mechanism of ion translocation in the Fo complex of ATP synthases. Nat. Chem. Biol. 2010;6:891–899. doi: 10.1038/nchembio.457. [DOI] [PubMed] [Google Scholar]
- Pogoryelov D, Yildiz Ö, Faraldo-Gómez JD, Meier T. High-resolution structure of the rotor ring of a proton-dependent ATP synthase. Nat. Struct. Mol. Biol. 2009;16:1068–1073. doi: 10.1038/nsmb.1678. [DOI] [PubMed] [Google Scholar]
- Preiss L, Klyszejko AL, Hicks DB, Liu J, Fackelmayer OJ, Yildiz Ö, Krulwich TA, Meier T. The c-ring stoichiometry of ATP synthase is adapted to cell physiological requirements of alkaliphilic Bacilllus pseudofirmus OF4. Proc. Natl. Acad. Sci. U.S.A. 2013;110:7874–7879. doi: 10.1073/pnas.1303333110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preiss L, Yildiz Ö, Hicks DB, Krulwich TA, Meier T. A new type of proton coordination in an F1Fo-ATP synthase rotor ring. PLoS Biol. 2010;8:e000443. doi: 10.1371/journal.pbio.1000443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanden T, Salomonsson L, Brzezinski P, Widengren J. Surface-coupled proton exchange of a membrane-bound proton acceptor. Proc. Natl. Acad. Sci. U.S.A. 2010;107:4129–4134. doi: 10.1073/pnas.0908671107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schägger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- Schulz S, Iglesias-Cans M, Krah A, Yildiz Ö, Leone V, Matthies D, Cook GM, Faraldo-Gómez JD, Meier T. A new type of Na+-driven ATP synthase membrane rotor with a two-carboxylate ion-coupling motif. PLoS Biology. 2013;11:e1001596. doi: 10.1371/journal.pbio.1001596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symersky J, Pagadala V, Osowski D, Krah A, Meier T, Faraldo-Gómez JD, Mueller DM. Structure of the c10 ring of the yeast mitochondrial ATP synthase in the open conformation. Nat. Struct. Mol. Biol. 2012;19:485–491. S481. doi: 10.1038/nsmb.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbani A, Warne T. A colorimetric determination for glycosidic and bile salt-based detergents: applications in membrane protein research. Anal. Biochem. 2005;336:117–124. doi: 10.1016/j.ab.2004.09.040. [DOI] [PubMed] [Google Scholar]
- Vik SB, Antonio BJ. A mechanism of proton translocation by F1F0 ATP synthases suggested by double mutants of the a subunit. J Biol Chem. 1994;269:30364–30369. [PubMed] [Google Scholar]
- von Ballmoos C, Dimroth P. Two distinct proton binding sites in the ATP synthase family. Biochemistry. 2007;46:11800–11809. doi: 10.1021/bi701083v. [DOI] [PubMed] [Google Scholar]
- Wang Z, Hicks DB, Guffanti AA, Baldwin K, Krulwich TA. Replacement of amino acid sequence features of a- and c-subunits of ATP synthases of alkaliphilic Bacillus with the Bacillus consensus sequence results in defective oxidative phosphorylation and nonfermentative growth at pH 10.5. J. Biol. Chem. 2004;279:26546–26554. doi: 10.1074/jbc.M401206200. [DOI] [PubMed] [Google Scholar]
- Watt IN, Montgomery MG, Runswick MJ, Leslie AG, Walker JE. Bioenergetic cost of making an adenosine triphosphate molecule in animal mitochondria. Proc. Natl. Acad. Sci. U.S.A. 2010;107:16823–16827. doi: 10.1073/pnas.1011099107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwart PH, Afonine PV, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, McKee E, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Storoni LC, Terwilliger TC, Adams PD. Automated structure solution with the PHENIX suite. Methods Mol. Biol. 2008;426:419–435. doi: 10.1007/978-1-60327-058-8_28. [DOI] [PubMed] [Google Scholar]