Abstract
The clinical course of infections with the hepatitis B virus (HBV) substantially varies between individuals, as a consequence of a complex interplay between viral, host, environmental and other factors. Due to the high genetic variability of HBV, the virus can be categorized into different HBV genotypes and subgenotypes, which considerably differ with respect to geographical distribution, transmission routes, disease progression, responses to antiviral therapy or vaccination, and clinical outcome measures such as cirrhosis or hepatocellular carcinoma. However, HBV (sub)genotyping has caused some controversies in the past due to misclassifications and incorrect interpretations of different genotyping methods. Thus, an accurate, holistic and dynamic classification system is essential. In this review article, we aimed at highlighting potential pitfalls in genetic and phylogenetic analyses of HBV and suggest novel terms for HBV classification. Analyzing full-length genome sequences when classifying genotypes and subgenotypes is the foremost prerequisite of this classification system. Careful attention must be paid to all aspects of phylogenetic analysis, such as bootstrapping values and meeting the necessary thresholds for (sub)genotyping. Quasi-subgenotype refers to subgenotypes that were incorrectly suggested to be novel. As many of these strains were misclassified due to genetic differences resulting from recombination, we propose the term “recombino-subgenotype”. Moreover, immigration is an important confounding facet of global HBV distribution and substantially changes the geographic pattern of HBV (sub)genotypes. We therefore suggest the term “immigro-subgenotype” to distinguish exotic (sub)genotypes from native ones. We are strongly convinced that applying these two proposed terms in HBV classification will help harmonize this rapidly progressing field and allow for improved prophylaxis, diagnosis and treatment.
Keywords: Hepatitis B virus, Hepatitis, Classification, Genotype, Subgenotype, Phylogenetic tree
Core tip: Hepatitis B virus (HBV) eradication could be achieved through three important points: (1) efficient universal vaccination; (2) accurate diagnostic assays; and (3) effective treatment of HBV carriers. Undoubtedly, these critical measures are not possible without fully understanding the genetics of the virus and being able to differentiate the isolates. In this review article we provide an update of HBV virology, focusing on classification and its impact on diagnosis, clinical outcomes, therapy, prophylaxis, evolution and epidemiology. Subsequently, the role of correct classification in describing HBV is highlighted, and misclassifications together with their causes are recounted. Finally, through the proposal of novel terms, HBV strains are reclassified.
POSSIBLE ERADICATION OF HEPATITIS B VIRUS; MULTI-FACTORIAL COMPLICATIONS
Undoubtedly, the World Health Organization’s (WHO) announcement in 1980 that smallpox virus was eradicated through vaccination represented one of the extraordinary human breakthroughs in the battle against infectious diseases[1,2]. Unfortunately, it seems that vaccination alone is not enough for hepatitis B virus (HBV) eradication.
HBV was discovered by Blumberg et al[3] (1925-2011) in 1965. Five years later, the first HBV vaccine and diagnostic blood test were developed[4]. The HBV vaccine is considered to be the first widely used vaccine against cancer and a chronic disease. In contrast to other vaccines, the clinical trial of this vaccine was short, and the vaccine was quickly approved by the United States Food and Drug Administration (FDA)[5]. Multiple studies have confirmed that the incidence of acute hepatitis, chronic liver disease as well as hepatocellular carcinoma (HCC) is decreased in the HBV-vaccinated population. Owing to the administration of more than one billion doses of HBV vaccines since 1982[6], the worldwide mortality rate of HBV has diminished significantly[7].
The WHO and other alliance organizations established the annual World Hepatitis Day in 2008. July 28th was selected for this particular day in honor of Prof. Blumberg’s birth date. The experts and healthcare organizations put their efforts into raising global awareness of viral hepatitis, especially about HBV and hepatitis C virus (HCV). The experts educate people all around the world about prevention, transmission, diagnosis and treatment against viral hepatitis infection. Undoubtedly, like smallpox eradication and its global preventative program, the World Hepatitis Day also moves up the knowledge of the global strengthening of preventive and control measures against viral hepatitis. Consequently, it is anticipated that increasing HBV vaccination coverage worldwide will definitely have positive impact on HBV eradication in the near future.
Despite advances that have resulted in several generations of HBV vaccines, a series of viral screening assays and effective treatment options, HBV is still considered a dangerous, life-threatening illness and a serious public health problem. The WHO estimates that at least two billion people (one fourth to one third of the world’s population) had been infected with the virus; 400 million people are infected chronically[8]. HBV-related diseases are currently ranked ninth on the global list for causes of mortality, and HBV is the fifth most important infectious agent, resulting in about one million deaths annually[9]. Due to the significant public health risk that HBV poses, it is important to compile comprehensive knowledge of both viral and host properties to enable elimination of HBV infection in the near future.
Host-related complications
The existence of a large reservoir of chronically infected HBV carrier patients hampers eradication of the virus[10]. The prevalence of chronic infection varies from region to region, such that different geographical parts of the world exhibit different sero-epidemiological patterns of HBV infection. The highest seroprevalence (8%) is found in Asia and the South Pacific region, which is considered a highly endemic region. In Sub-Saharan Africa, Alaska, the Mediterranean region and India, HBV seroprevalence is between 2% and 7%, which is considered an intermediate endemic range. In European countries and some parts of Central and South America, HBV seroprevalence is less than 2%, which is considered low[11].
Patients chronically infected with HBV have a greater than 100-fold chance of developing HCC compared to uninfected people[12]. From a global perspective, HBV is the leading cause of HCC and causes one million deaths annually[13]. Although the eradication of hepatitis B by means of universal vaccination seems technically achievable, this task is made difficult by the fact that hundreds of millions individuals are already chronically infected with HBV. The elimination of hepatitis B will only be successful when this group of chronically infected patients is cured naturally or through antiviral treatments. Only accurate and continuously improved diagnostic policies will identify the pool of carriers for appropriate treatment. To further complicate diagnosis, different types of chronicity like normal obvious infection (overt) or masked infection (occult) increase the complexity of the diagnostic algorithms[14,15]. Vaccinating patients in high-risk groups, which come in contact with infected people, would decrease the infectivity and risk of transmission to the healthy population and should therefore be considered a priority[4,6].
Virus related complexity
HBV is the prototypic species of a family of DNA viruses called Hepadnaviridae. The HBV genome is approximately 3200 bp long, circular, and consists of four genes and seven open reading frames (ORFs). HBV itself evolves inter- and intra-genetically in reservoirs. Since the reverse transcriptase enzyme lacks proofreading activity, the nucleotide substitution rate for HBV is higher than that of other DNA viruses[16]. During persistent, long-term HBV infection and under different selective pressures, variants of HBV can emerge. Some variants are able to evade diagnostic, prophylactic and therapeutic measures. This extraordinary genomic diversity, together with a high replication capacity, allows HBV to adapt to different hosts[17]. Based on evolutionary analysis, HBV has eight confirmed genotypes (named A to H) and two tentative genotypes (called I and J), and almost forty subgenotypes (Figure 1A)[18,19]. Phylogenetic and evolutionary analyses of complete genome sequences have classified HBV into these eight distinct genotypes; each has an intergroup nucleotide divergence greater than 7.5%[20]. “Subgenotypes” are subgroups within the same genotype that meet two particular criteria. First, they have a nucleotide divergence between 4% and 7.5% over the full-length genome[21], and secondly, there is high phylogenetic bootstrap support. “Clades” further divide subgenotypes, and have less than 4% nucleotide diversity based on the complete genome sequences[20]. Prior to molecular analyses (genotype, subgenotype and clades), classification of HBV strains was based on the immunological heterogeneity of HBsAg, which led to the categorization into different HBV serotypes (subtypes). The development of DNA sequencing revealed that amino acid changes in the major hydrophilic region (MHR) region of the HBsAg are responsible for this classification. This serotype-based classification is still used, and epidemiological studies describe associations between serologic subtypes and genotypes[20,22].
Figure 1.
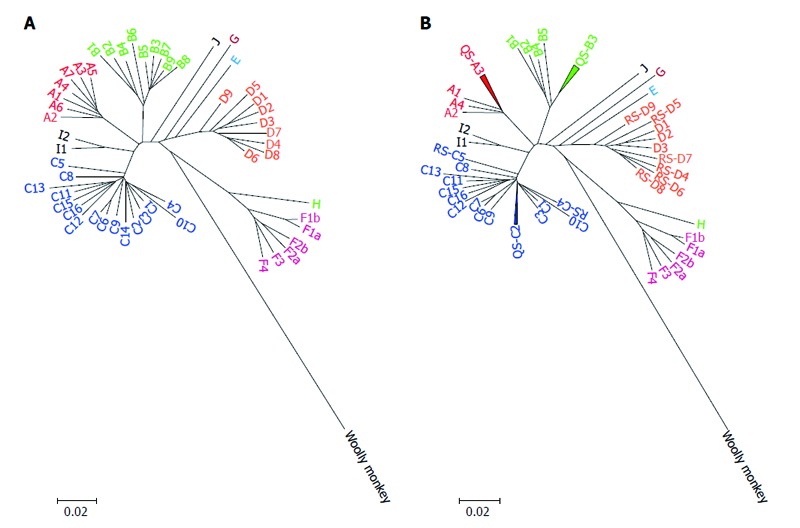
Neighbour-joining phylogenetic tree was conducted based on hepatitis B virus full-length genomes of all proposed genotypes and subgenotypes before (A) and after (B) reclassification. HBV genome sequence that used are listed below: JN182318: A1; HE576989: A2; AB194951: A3; AY934764: A4; FJ692613: A5; GQ331047: A6; FN545833: A7; AB642091: B1; FJ899779: B2; GQ924617: B3; GQ924626: B4; GQ924640: B5; JN792893: B6; GQ358137: B7; GQ358147: B8; GQ358149: B9; AB697490: C1; GQ358158: C2; DQ089801: C3; HM011493: C4; EU410080: C5; EU670263: C6; GU721029: C7; AP011106: C8; AP011108: C9; AB540583: C10; AB554019: C11; AB554025: C12; AB644280: C13; AB644284: C14; AB644286: C15; AB644287: C16; GU456636: D1; GQ477452: D2; EU594434: D3; GQ922003: D4; GQ205377: D5; KF170740: D6; FJ904442: D7; FN594770: D8; JN664942: D9; FN594748: E; FJ709464: F1b; DQ899146: F2b; AY090459: F1a; DQ899142: F2a; AB036920: F3; AF223965: F4; GU563556: G; AB516393: H; FJ023659: I1; FJ023664: I2 ; AB486012: J and AY226578: Woolly monkey as an out-group. HBV: Hepatitis B virus.
HBV CLASSIFICATION METHODS
Phylogenetic analysis of the nucleotide sequences of the whole HBV genome represents the most conclusive method for HBV genotyping[23]. This method is considered the “gold standard” approach for genotyping and subgenotyping, though it is relatively expensive and time-consuming. Fortunately, sequencing is becoming cheaper and faster, so it may serve as a common molecular method in the very near future even for clinical routine purposes. Phylogenetic analysis can also be performed on individual genes instead of the complete genome, in particular on the HBV envelope (S) gene[24]. The results obtained from the partial sequence (HBV S gene) may be useful for determining the HBV genotype, but it will not be appropriate for determining the HBV subgenotype.
Online methods to genotype HBV
Bioinformatics is currently used in many fields of science, and numerous bioinformatics software tools are available online. Genotyping of microorganisms has also become readily available, particularly for viruses, since they carry small genomes compared to eukaryotic organisms. One of the most popular online systems is a sequence similarity algorithm, such as BLAST, which is available through the National Center for Biotechnology Information (NCBI). BLAST analysis can be used to identify the sequences that are most similar to the sample by comparing the sample sequence to sequences archived in GenBank. NCBI has generated additional online tools specifically to determine the sequence of certain genotypes and subgenotypes of viruses. Thus, HBV and several other viruses can currently be genotyped using the NCBI web-based HBV genotyping tool[25]. To further facilitate HBV genotype determination, several experts in the field have introduced different online HBV genotyping tools, such as the hepatitis B virus database (HBVdb)[26], HBV STAR[27], BioAfrica-Oxford HBV Automated Subtyping Tool[28], HepSEQ Genotyper[29,30] and the jumping profile Hidden Markov Model (jpHMM)[31].
French experts developed the HBVdb online genotyping system, allowing researchers not only to genotype HBV but also to create virus recombination and drug resistance profiles. A group from the United Kingdom designed the HBV STAR online tool to genotype HBV based on a statistically defined, position-specific scoring model. This tool is able to predict recombinant and non-human primate isolates. The HBV STAR tool also supports human immunodeficiency virus-1 (HIV-1) subtyping. Another group established a rapid high-throughput-genotyping system called BioAfrica-Oxford Automated Subtyping Tool, which is able to genotype and subtype several viruses, including HBV. The tool employs a phylogenetic approach to genotype viruses and uses bootscanning methods to detect recombination[30]. The HepSEQ Genotyper online tool is an international public health repository for hepatitis B developed by British scientists, which provides molecular, clinical and epidemiological information regarding HBV. HepSEQ Genotyper is capable of determining the HBV genotype and classifying clinically relevant mutations within the HBV genome such as vaccine escape, precore or antiviral-resistant mutations. This tool uses only HBV polymerase/surface genes for genotype computation and is therefore less accurate for detecting recombination. The jpHMM online tool was developed to subtype/genotype two viruses: HIV-1 and HBV. This tool uses a probabilistic approach to compare a sequence to a multiple alignment of a sequence family and can also be extended to detect recombination.
Molecular-based methods to genotype HBV
Several molecular approaches have been developed to characterize HBV genotypes, including polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP)[32,33], genotype-specific primers in a single or a multiplex-PCR set[34], oligonucleotide microarray chip (DNA Chip), restriction fragment mass polymorphism (RFMP), mass spectrometry (MS), PCR-invader assay, real-time PCR, hybridization strips as INNO-LiPA, reverse dot blot assay and sequencing[35].
Among them, PCR-RFLP is widely used to genotype HBV since it is simple and inexpensive. Nevertheless, there are many reports that indicate this technique is not always accurate and may often result in an ‘untypable’ genotype[36]. Genotype-specific primers in single and multiplex PCR sets are also utilized to distinguish different genotypes. This method is rapid and inexpensive and widely used for large population studies; however, any mutation in the genome may alter the result of the assay due to primer-DNA hybridization mismatching[37]. Recently, the DNA Chip method[38], invader assays[39], real-time PCR[40-42], hybridization and dot blot methods[43] have been employed for HBV genotype detection[38]. These techniques are highly sensitive; however, fidelity can be affected by any mutation within the HBV genome. Mass spectrometry[44] and RFMP[45] can also be used for genotyping of HBV. These methods can detect drug resistant variants, but fidelity is also affected by mutations, and the methods themselves are costly and require specialized equipment. Despite the emergence of many methods to genotype HBV, sequencing is still considered the “gold standard” method for genotyping, followed by phylogenetic and evolutionary analysis. This method is the most reliable, since it is able to determine the whole genome sequence of HBV.
Methods to determine HBV subtypes
Three decades ago, HBV subtyping was introduced as the first classification system for hepatitis B, in which categories were assigned based on the amino acid sequences at the surface antigen region[22,46]. This sero-molecular taxonomy based on the specific motifs of HBsAg led to 10 different sero-subtypes of HBV. Different HBV subtypes can be distinguished based on enzyme-linked immunosorbent assay (ELISA) or enzyme immunoassays (EIA) using specific monoclonal antibodies against pre-S2 and S regions[47]. Since sequencing of the HBV HBsAg region reveals subtype-specific motifs, serological subtyping (serotyping) of HBV can also be predicted by HBsAg amino acid sequence mapping[22].
The relationship between particular subgenotypes and subtypes has already been demonstrated[20,21]. Also, different impacts of HBV subtypes on results of serological assays for HBsAg have been demonstrated[48]. Little is known about the driving forces behind the divergence of HBV genotypes, subgenotypes and subtypes. We believe that they are the result of long-term adaptation to the host’s genetic background of certain human populations where they circulated. Subtypes distributed consistently with the pre-historic human migration[49]. Genotype, subgenotype and subtype-specific variability is stably transmitted within the host population and is considered to stay constant from the beginning of the infection in an individual[50].
Methods to determine HBV subgenotype
Although all aforementioned methods are able to detect HBV genotype, no molecular method has been introduced to determine HBV subgenotypes accurately. There are several studies that used PCR-based approaches to distinguish subgenotypes of HBV, however, they were designed to differentiate only limited subgenotypes. For instance, investigators introduced multiplex-PCR or semi-nested PCR assays to differentiate between HBV subgenotypes B1, B2, C1 and C2[51] or Ba and Bj[52]. Therefore, sequencing is still the only method that is able to determine subgenotypes of HBV accurately following careful phylogeny analysis. Based on our own experience in the field of HBV epidemiology, we would like to explain in more detail how the HBV subgenotype can be accurately determined.
Introducing “HBV subgenotyping guidelines”
To determine HBV subgenotype, several rules must be strictly applied: (1) Analyzing the full-length genome is the foremost prerequisite for determining the accurate subgenotype. This is necessary for the analysis of entire genes and ORFs at the nucleotide level. It also allows for accurate subgenotyping, even if mutations or recombinations occur in the particular strain of interest. In the past, using only the partial genome has led to several misclassifications of HBV subgenotypes. The preeminent example is the introduction of the subgenotypes A4 and A5 through partial genome analysis by Olinger et al[53] in 2006. Further analysis revealed that these strains should not be considered independent subgenotypes[54,55]. The partial genome is inadequate for HBV subgenotyping, because of the particular genomic structure in which four genes and seven proteins are arrayed in a concise genome. Furthermore, analysing partial genes instead of the full-length genome can sometimes lead to false epidemiological signals from distant geographical regions. For instance, Khedive et al[56] analysed 681 bp of isolated strains from HBV carriers in Iran and reported five strains with HBV subgenotypes D5 and D8, which is inconsistent with the known geographical distribution of subgenotypes. According to all epidemiological studies from Iran, D1 is the most frequent subgenotype of HBV, followed by a minor population of D2. Moreover, recent analysis showed that Iran is the most probable location of the common ancestor of subgenotype D1-D3[57]. Also, there is no accurate evidence of other subgenotypes of genotype D in Iran or its neighbouring countries. Thus, the reported identification of D5 and D8 is not geographically congruent with previous studies from Iran. Also, in other studies, like searching for the common source of HBV infection, full-length genome analysis provides strong evidence[10,58,59]; (2) Adherence to the ranges of intra-genotypic nucleotide divergence (more than 4.5% and less than 7.5%) that define distinct subgenotypes is the second most important factor for correctly identifying subgenotypes of HBV. Disregarding this rule in several cases, like with A5 and A7, has resulted in misclassification of strains[60,61]; (3) Bootstrap values greater than 75% are necessary to support the monophyletic tree to introduce a cluster as an independent subgenotype. There are several subgenotypes that have been introduced without considering this critical rule. Shi et al[62] misclassified the B3 subgenotype and Huy et al[63] misclassified C2 of HBV due to ignoring this criteria; (4) Recombinant strains should be excluded from any subgenotyping analysis as much as possible because they can disrupt the topology of a phylogenetic tree and falsely increase nucleotide divergence. For example, subgenotypes D8, D9, CD1 and CD2 have been misclassified[18,64-67]. The impact of recombination on HBV strain analysis will be discussed in more detail below; (5) To introduce novel subgenotypes, strains harbouring specific nucleotide and amino acid motifs should be identified. Some investigators have demonstrated this particular criteriaon for subgenotypes of A1, A3 and A6[55,68]; and (6) To avoid sampling bias, a minimum of three purported novel strains, together with all available subgenotype strains belonging to the same genotype, should be subjected to evolutionary and phylogeny analysis. Using random reference sequences, as opposed to selecting some particular reference sequence, is highly recommended for subgenotyping phylogeny analysis.
Since recombination is an inevitable part of HBV’s evolution, most subgenotypes that belong to a HBV genotype (such as genotype B) show a trace of recombination[69]. However, in several cases, the level of recombination is significant and the strain should not be classified as a subgenotype. This is discussed below in more detail. Table 1 presents examples of misclassified subgenotypes, their methodological errors, and the proposed correct subgenotype classification of HBV.
Table 1.
The misclassifications detected in hepatitis B virus subgenotyping
| Genotypes | Subgenotypes old classification | Reasons (R) for misclassification | Suggested resolution | New proposed subgenotypes reclassification | ||||
| A | A1, A2 | A1, A2 | ||||||
| A3, A4, A5 | R1 | R2 | R3 | R5 | Quasisubgenotype-A3 | QS-A3 | ||
| A6 | A4 | |||||||
| A7 | R2 | R3 | R5 | Quasisubgenotype-A3 | QS-A3 | |||
| B | B1, B2 | B1, B2 | ||||||
| B3, B5, B7, B8, B9 | R2 | R3 | Quasisubgenotype-B3 | QS-B3 | ||||
| B4 | B4 | |||||||
| B6 | B5 | |||||||
| C | C1 | C1 | ||||||
| C2 | R3 | Quasisubgenotype-C2 | QS-C2 | |||||
| C3 | C3 | |||||||
| C4 | R4 | Trace recombination | RS-C4 | |||||
| C5 | R4 | Trace recombination | RS-C5 | |||||
| C6-C13 | C6-C13 | |||||||
| C14 | R2 | Quasisubgenotype-C2 | QS-C2 | |||||
| C15, C16 | C15, C16 | |||||||
| C/D1, C/D2 | R4 | Inter-genotypic recombinant | Not considered as subgenotype | |||||
| D | D1, D2, D3 | R2 | Not decided yet | |||||
| D4, D5, D6, D7, D8, D9 | R4 | Recombino-subgenotype | RS-D4, RS-D5, RS-D6, RS-D7, RS-D8, RS-D9 | |||||
Number (R1-R5) indicates the reason of problems in classification. R1: Applying partial gene in introducing subgenotype; R2: < 4% nucleotide divergence; R3: Weak bootstrap value or no monophyletic cluster; R4: Recombination; R5: Bias in reference collection. QS: Quasi-subgenotype; RS: Recombino-subgenotype.
IMPORTANCE OF HBV CLASSIFICATION
Before passing away in 2011, the Nobel Prize winner Professor Blumberg emphasized the importance of eliminating HBV. He believed eradication could be achieved through three important points: (1) efficient universal vaccination; (2) accurate diagnostic assays; and (3) effective treatment of HBV carriers[4]. Undoubtedly, eradicating HBV is not possible without fully understanding the genetics of the virus and being able to differentiate the isolates. Thus, before delving into HBV classification and current methodological drawbacks, we first present a comprehensive overview of differences in HBV strains in terms of virological, epidemiological, clinical and evolutionary aspects.
Genotypic virological differences
It has been proposed that genotypic virological differences have evolved and adapted through long-term evolution, as well as through sequence insertions and deletions in the HBV genome. The HBV prototype strains comprises 3215 nucleotide base-pairs (bp), which are found in HBV genotypes B, C and F and H. The length of HBV strains varies from 3182 bp in the shortest genotype (genotype D) to 3248 bp in the longest one (genotype G)[17]. Particular genetic characteristics are present in some genotypes. For example, because of the existence of two stop codons in the core region of genotype G of HBV, this strain does not have the ability to secrete hepatitis B e antigen (or HBeAg)[70]. Also, the G1896A precore (PC) mutation, which also ablates HBeAg expression due to a stop codon in the precore region, is rare in genotypes A, F and H, while this mutation has the highest prevalence in genotype D[16,71,72]. Some reports have demonstrated the high prevalence of the PC mutation in non-Japanese subgenotypes of genotype B (B2-B5) compared to subgenotype B1[73]. Also, A1762T and G1764A/T basal core promoter (BCP) mutation, which is significantly associated with advanced liver diseases, is more prevalent in genotypes A and H of HBV[74,75]. Moreover, nucleotide variation and deletion in the pre-S region are particularly prevalent in distinct genotypes, such as genotype C in comparison with genotype B[76]. Studies revealed that diversity within the HBV genome is directly associated with cirrhosis as well as HCC incidence and outcome[77]. Moreover, heterogeneity and substitution rates are dissimilar in different HBV genotypes. Based on intra-genotype genetic diversity, some genotypes like genotypes A, B, C, D, and F have been classified into different subgenotypes, while genotypes of E, G and H do not contain enough heterogeneity to be subdivided into subgenotypes[69].
Genotype epidemiological distinctions
In addition to virological aspects, epidemiological studies revealed that HBV genotypes are associated with differences in geographical distribution and route of transmission.
Geographical distribution: Different genotypes of HBV show distinct geographical distribution patterns. For example, genotype A is dominant in Northwest Europe and North America. Also, some strains of genotype A have been found in the Philippines, Hong Kong and in some parts of Africa and Asia. Genotypes B and C are mainly prevalent in Southeast Asia and can be also found in the Pacific islands[21]. Genotype D is the most globally distributed genotype, and it can be found from Southern Europe and Africa to India. It can be also detected among intravenous drug users on all continents[78]. Genotype E is mainly dominant in West Africa[79]. Genotype F is found in South and Central America[19]. Genotype G was first discovered in France and the United States[80], and it has recently been reported in Belgium[81]. Genotype H has been described in America and Japan[82]. Recently, two controversial genotypes (I and J) have been proposed in South Asia[37,83], which will be discussed comprehensively later in Section 6 (recombination).
Transmission route: Generally, HBV is transmitted through contaminated body fluids. Various routes of transmission include blood transfusions from infected patients, sexual intercourse, unsafe injections and mother-to-neonate transmission. Recently, tears, urine, saliva, bites and broken skin have also become accepted as probable modes of transmission[7]. Since HBV is able to survive on surfaces for up to seven days, direct transmission through contaminated surfaces to persons that have frequent contact with HBV carriers has been also reported[84]. To complicate matters, the transmission routes largely depend on the regional prevalence of chronic carriers of HBV-infected individuals[84]. Differences in the natural history of infection with different genotypes have also impacted the modes of HBV transmission[85]. The local prevalence of HBV with its geographical distribution, regional and social cultures, as well as taboos can help to draw an informative transmission pattern for each genotype[10]. In highly endemic areas (prevalence rate > 8%), such as South Asia where genotypes B and C are dominant, perinatal (mother-to-child) transmission is most common. It has been proposed that genotype C is primarily transmitted perinatally in this region, since the seroconversion of HBeAg to anti-HBe took longer in these patients, which is consistent with the fact that it takes longer for genotype C than for other genotypes[86].
In European countries with a low prevalence of HBV (prevalence rate < 2%), where genotype A and D are dominant, sexual transmission and unsafe injection practices are the main modes of HBV transmission. Nosocomial transmission and unsafe injection practices are considered responsible for more than 60% of HBV infections in Central Europe[87]. Though genotype E is transmitted by heterosexual relations in Africa, genotype A and particularly genotype G were isolated from men who have sex with men in Europe and Canada[81,88]. In West Asia and in the Middle East (where genotype D is dominant), the route of transmission and HBV seroprevalence depend on the region. For instance, Iran has a low HBV endemicity (around 2%), and intravenous drug injections, tattooing and phlebotomy are considered the major potential risk factors and transmission routes of HBV infection in the country. Furthermore, socioeconomic status, life style, occupation, and cultural attitude in different ethnic groups greatly impact the route of HBV transmission[89,90].
Genotypes’ impact on clinical outcomes
Increasing evidence suggests that HBV genotyping is important for determining HBV disease progression and designing appropriate antiviral treatment. Some genotypes are more associated with particular kinds of prognoses, such as acute forms of disease[88]. Several reports showed that genotype A evolves more rapidly in patients than genotype D does, which poses problems for treatment[16]. Also, patients infected with genotype C progressed to end stage liver disease earlier than those infected by genotype B[91]. Interestingly, it has been shown that patients infected with genotype F have higher mortality rates than those infected with genotype A or D[92]. In India, genotype D is associated with more severe liver complications than other genotypes[93]. In the United States, it has been shown that genotype D is an independent risk factor for fulminant hepatitis[94]. Interestingly, however, patients infected with genotype F have a higher rate of liver-related mortality than those infected with genotype D[95].
Genotype C and D generally tend to be related to more severe liver disease than genotype A and B and are more frequently associated with HCC[96]. In HBeAg-positive patients treated with standard interferon-alpha, the post-treatment HBeAg seroconversion and normalization of serum ALT levels are considerably better in those infected by genotype A and B than patients infected with genotypes C and D[97]. Furthermore, following pegylated interferon-alpha therapy, HBeAg seroconversion and a substantial decrease in serum titer of HBsAg was observed in genotypes A and B but not in patients infected with genotypes C and D[98-100]. In the case of antiviral therapy, it has been shown that genotype B is more frequently associated with lamivudine-resistant variants than genotype C. Likewise, in some studies, it has been observed that genotype A develops antiviral-resistant variants earlier than genotype D[74].
Genotypes’ impact on prophylaxis measures
Current HBV vaccines are recombinant peptides that cover a ‘‘super antigenic’’ and highly conserved motif of HBsAg. Protective serum titers of anti-HBs (greater than 10 IU/L) develop in 95%-99% of healthy infants, children[101,102] and young adults[103]. Many studies have demonstrated that HBV vaccination has dramatically decreased the HBV chronicity rates and HBV-related complications[7]. The current vaccine is derived from genotypes A and D of HBV[104]. Despite its success in most cases, there are several reports of vaccination failure due to genotype complications[105]. In one case, a German patient who was vaccinated and showed production of formally protective anti-HBs antibodies developed acute hepatitis B following infection with genotype F of HBV[104]. In a similar report from Europe, an Irish man was vaccinated, but developed infection by genotype F[106]. Both patients received HBV vaccines produced using the S gene of HBV genotypes A and D, which are the most common genotypes in Europe. Furthermore, in both reports, the HBV isolates did not carry typical vaccine escape mutations. Several investigators have described S gene variations isolated from vaccine failure cases[107,108]. The frequency of these mutations varies in different genotypes. Of note, some of these mutations are considered wild type motifs in another genotype[109]. Such instances of failure serve as reminders that our current vaccination program is imperfect. Future efforts should be directed towards developing vaccines that protect against all genotypes of HBV and also account for vaccine escape mutations.
Evolutionary differences of genotypes
Phylogenetic analysis has demonstrated that genotype H is closely related to genotype F, though there is enough inter-nucleotide divergence between these two particular genotypes to distinguish them as distinct genotypes. Phylogenetic analysis also revealed that these two genotypes (‘‘New World human genotypes’’) lie on the branch of the Woolly Monkey, which suggests cross-speciation between non-human and human genotypes of HBV[110]. This scenario was reiterated after evolutionary relationship analysis between genotypes D and E. Furthermore, they showed different evolutionary rates (number of substitution per site per year)[49].
SUBGENOTYPES ARE MORE DETAILED GENOTYPES
For some HBV genotypes, several subgroups can be easily defined as when the intra-genotypic nucleotide divergence stays between 4% and 7.5% over the full-length genome. According to conventions of identification, HBV subgenotypes are differentiated by numbers corresponding to the order of discovery; the numbers do not correspond to subgenotype evolution. For instance, D1-D4 were identified by Norder et al[21] earlier than D5[111]. However, D5 is the most ancient of all known subgenotypes for genotype D[112]. Also, it was noted that A6 (currently known as A4) is from a basal lineage that diverged earlier than the other African subgenotypes of genotype A[54,55]. Figure 2 illustrates the updated time line of HBV subgenotype identification. Uncovering the relationship between subgenotypes and subtypes of HBV has added significant value to molecular epidemiological studies of HBV[74].
Figure 2.

The time line of identification and designation of hepatitis B virus subgenotypes.
It should be noted that because of inappropriately applied methods, some subgenotypes have been incorrectly classified in the past. One of the most common mistakes is applying phylogenetic analysis over a partial genome sequence instead of the full-length genome. Experts in the field have attempted to correct the errors in numbering and misclassification (Figure 1B, Table 1), but inaccurate subgenotyping of HBV is continuously being reported[54,58,62,69,113-118].
HBV subgenotypes and geographical distribution
Subgenotypes reflect properties and distributions of genotypes. Figure 3 shows the geographical distribution of genotypes and subgenotypes of HBV together with the prevalence of HBsAg in different areas around the world. For instance, subgenotype A2 is dominant in Europe, A1 is prevalent in Asia and most of Africa, A3 is found in Cameroon and Gambia, A6 (currently named A4) and quasi-subgenotype A3 (which includes previously named A4 from Mali, A5 from Nigeria and A7 from Cameroon) have been isolated from other regions[69]. Subgenotype B1 was isolated from Japan, while B2, B3, B4, B5, B6, B7, B8 and B9 were isolated from Taiwan, Indonesia, Vietnam, Philippines, the Arctic region, Nusa Tenggara (a region from Eastern Indonesia) and Indonesia, respectively[119]. Subgenotype C1 was isolated from Taiwan, C2-C16 were isolated from China, Oceania, Australian Aborigines, Philippines, Papua-Indonesia and Nusa Tenggara[120]. Two recombinant mixed strains C/D1 and C/D2 (combination of HBV genotype C and D) were specifically reported in Tibet[65,121]. Although the genotype D of HBV is distributed globally, its subgenotypes are locally confined to certain geographical regions. For example, subgenotype D1 is restricted to Iran and its neighboring countries[116,122-126]. Subgenotype D2 is derived from East Europe and Russia, D3 was detected in Serbia, South Africa and Alaska, D4 was found in Oceania and Somalia, and D5-D9 were reported from India, Indonesia Tunisia and Nigeria[67]. Genotype F is also widely distributed: F1a is dominant in Costa Rica and Salvador, F1b in Argentina, Chile and Alaska, F2a in Venezuela and Brazil, F2b in Venezuela, F3 in Panama, Venezuela and Colombia and F4 is circulating in Brazil, Argentina and Bolivia[127].
Figure 3.

Geographical distribution of hepatitis B virus genotypes and subgenotypes in different regions of the world. Yellow arrows illustrate the directions of HBV subgenotype dispersal through immigration from highly and intermediate endemic countries to low HBV endemic areas (map of hepatitis B surface antigen (HBsAg) prevalence adapted from the website of the WHO). HBV: Hepatitis B virus; WHO: World Health Organization.
HBV subgenotypes and ethnic origin
It has been demonstrated that in some cases, such as for genotype B8, the distribution of HBV genotypes/subgenotypes is related to the ethnic origin[128], or B6 is considered confined to indigenous populations[129]. Interestingly, C4 can be detected in the Australian Aborigines but nowhere else, which suggests that C4 independently evolved from its ancestor in that region[130]. Strains isolated from indigenous populations, such as C3 and C5-C10 from Indonesia and B6 from the Canadian Arctic, proposed that HBV (sub)genotypes have evolved in different streams of human immigration[49].
HBV subgenotypes and clinical outcome
HBV subgenotypes also present differently in terms of clinical outcome. It is possible to uncover more details regarding the natural history of infection by comparing genotypes. For example, subgenotype A1, which is prevalent in West and South Africa, has a more severe clinical outcome compared to subgenotype A2. Patients infected with this subgenotype developed HCC at a young age in West and South Africa, whereas those who were infected with A2 and developed HCC in Europe were mainly older patients[131]. European patients infected with A2 had a mild clinical outcome and high chance of clearing HBsAg and losing HBV DNA[95]. Furthermore, in Europe more occult cases have been infected by A1 comparing with A2 or other (sub)genotypes[81]. Nevertheless, it is important to keep in mind that all these studies are hampered by the fact that it is difficult to adjust the studied patient cohorts for all potential confounding factors.
Subgenotype B6 is commonly associated with a mild clinical outcome in infected patients, while B1 can result in fulminant and acute hepatitis B infection. Patients infected with subgenotype B1 also developed advanced liver disease at an older age compared to patients infected with subgenotypes B2-B5[73,132]. In most studies in South Asia, genotypes B and C were compared. Though there is a paucity of data of comparing the clinical findings of different subgenotypes of genotype C, one study demonstrated an increased risk for HCC in patients infected by subgenotype C2 compared to C1 and genotype B subgenotypes[133]. Interestingly, precore mutations have not been observed in subgenotype F2, whereas subgenotype F1 frequently carries precore mutations[16]. In one large study, a higher percentage of patients who developed HCC had been infected by subgenotypes F2 or C2 compared to subgenotype A2, B6 or subgenotypes of genotype D[131].
CLASSIFICATION AND IMPORTANCE OF ACCURACY
In order to accurately investigate the impact of different (sub)genotypes of HBV on different aspects of infection from prophylaxis, diagnosis and therapy, it is crucial to agree to a holistic classification. Numerous HBV strains have been described through PCR and sequencing, however, many disregarded well-established HBV (sub)genotype definitions, which has resulted in several misclassifications. Subsequently, experts in the field agreed that the classification of HBV should be mended and rectified[54,69,113-115]. There are several reviews or commentary articles regarding the misclassification of HBV that describe the history of identification of different (sub)genotypes. However, the reclassification and renovation of this system has been considered less often. We, along with others, believe that recombination is the main cause of misclassifications and major evolutionary characteristics of HBV should be investigated to help identify strains that require additional analysis for proper classification[115]. Besides this factor, massive but gradual conversion of geographical distribution of HBV should be concomitantly investigated.
RECOMBINATION AND ITS ETIOLOGY
Recombination in HBV is principally the result of the co-infection of a host with more than one strain from different (sub)genotypes. Different HBV strains can exchange their genetic material within the host cells. Recombination is favoured in particular geographical regions by three conditions: (1) two or more different HBV genotypes are circulating in the population; (2) the chronicity of HBV is high; and (3) public health level is low. Most recombinant strains have been reported from East Asia or Africa, where the prevalence of HBV infection is high and prophylaxis and control of infection is low. Due to the dense population, the chance of co-infection is boosted so risk of recombination is subsequently elevated. In contrast, HBV recombinant isolates have been reported rarely within typical European strains (such as subgenotypes A2, D2 and D3). This corroborates the necessity of efficient health control and prophylactic measures in order to decrease the risk of infection, co-infection and eventually recombination.
Currently, more than 30 recombinant strains have been described[126,134]. Sometimes recombination can occur between strains with high genetic homology, in which two different subgenotypes from a similar genotype are co-infected in a patient. Such recombination has been reported between HBV subgenotype B2 with B5 and between B1 with B6[128]. Also, there are some reports regarding recombination between two strains of different genotypes[65,66,121,135]. Although more than 60% of recombinant isolates have their breakpoint between nucleotides that span 1640-1900 (which encompasses the core region), recombinant strains with breakpoints in the S gene (350-500, 3150-100 or 650-830) have also been identified[134]. Markedly, the largest breakpoints have been detected among HBV isolates from Tibet, in which two different genotypes of C and D (subgenotypes C1 or C2 with D1 or D2) formed 50% of the recombination[135,136].
Recombination as a source of HBV misclassification
Recombination is an inevitable event in evolution and can cause errors in classification of HBV genotyping or subgenotyping[18,69,114,115]. In many cases, recombinant strains have been erroneously introduced as new genotypes or subgenotypes. For instance, Ghosh et al[137] introduced six HBV strains as a novel subgenotype of HBV named D9. In the phylogenetic tree, subgenotype D9 strains are grouped as a monophyletic and distinct cluster with a maximum bootstrap value. Not surprisingly, according to their analysis, these recombinant strains showed extraordinary nucleotide divergence from the other subgenotypes of genotype D (from minimum 5.2 ± 0.3 with D1 to 6.7 ± 0.4 with D7 and D8). This nucleotide divergence was odd, because it did not meet the threshold of genotype definition, yet it was much higher than normal nucleotide divergence within well-classified subgenotypes. More interestingly, there was a highly diverse pattern of recombination in these strains. Although all strains showed recombination with genotype E in the HBV core and pol regions, there was no identifiable recombination pattern among all strains. Indeed, different lengths of genotype D recombined with genotype E. Moreover, in some strains, some recombination with genotype C was observable. If PCR and sequencing had been perfectly carried out, subgenotype D9 strains should be classified as a D/E recombinant strain. Exactly the same scenario was suggested by another group for the D8 subgenotype[18]. Similar concerns have been raised regarding subgenotype D7, which was introduced as a novel subgenotype of genotype D. However, these strains harbored at least 900 nucleotides of genotype E in the backbone of this genotype D strain[138]. Likewise, subgenotype D4 may have recombined with D7 or D8, which were grouped in an exclusive cluster in the phylogeny tree[64]. The same concern was raised for other HBV subgenotypes, like genotype C, in which the recombinant strains C/D1 and C/D2 were suggested as new subgenotypes[65,66].
Proposing the term “recombino-subgenotype”
Owing to the availability of sequencing and free online phylogenetic software, novel subgenotypes of HBV are continually being suggested by researchers that disregard the fundamental definitions of HBV classification. To address the misclassification of HBV subgenotypes that lack enough nucleotide divergence and bootstrapping value to definitively be considered subgenotypes, we previously proposed the term of quasi-subgenotype[54]. This term has somewhat settled irregularities in subgenotyping of HBV and was respected by other studies[10,62,69,81,113,118,139-143]. To address the classification of recombinant strains, which are being introduced as independent subgenotypes, we propose to call the indefinitely recombinant strains as “recombino-subgenotype” rather than an independent subgenotype. According to our proposed definition, the “recombino-subgenotype” is a lineage that shows strong evidence of recombination and its nucleotide divergence, together with supportive bootstrap value, fall within the range necessary to define subgenotypes. Although these strains are recombinant, in a comprehensive phylogenetic tree (only based on full-length genome), “recombino-subgenotypes” are clustered around intra-genotype clusters and could have supporting bootstrap value. While this new term cannot prohibit the introduction of HBV recombinants as novel subgenotypes, it can clearly differentiate the pure subgenotypes from recombinant strains. Therefore, we would like to offer the term “recombino-subgenotype” to differentiate recombinant from non-recombinant (sub)genotypes. We sincerely hope that this term will help to remind scientists about recombination as a potential pitfall in HBV classification as well as allow a more accurate description of novel HBV isolates derived by recombination in HBV taxonomy.
IMMIGRATION: INTRODUCING THE TERM “IMMIGRO-SUBGENOTYPE”
While recombination is an important virological aspect to be considered in classification, the epidemiological profile of HBV is another valuable scope to study. Although ancient dispersal of HBV alone has made the HBV pool a dynamic viral population, recent trends of globalization and increasing human mobility are significantly speeding up HBV distribution and recombination between viral strains. Immigration and direct human contact are the two main causes of changes in geographic dispersal of HBV genotypes and subgenotypes; however, their profound impact has yet to be studied in more detail. People typically emigrate from countries with high HBV endemicity, which alters the geographical distribution of HBV (sub)genotypes around the world, notably in low sero-prevalence regions including Europe and North America.
Recently, novel subgenotypes have been reported from immigrants who are not living in their original home areas. For instance, HBV subgenotype A6 (currently named A4) strains have been isolated from African immigrants currently residing in Europe and North America[54,55,81,113,135]. It has been predicted that within the next few decades, HBV epidemiological patterns will be completely modified by waves of immigration. Thus, such “immigro-subgenotypes” may replace native strains in regions with high immigration. Their integration might alter the local prevalence of carriers, routes of transmission, and will have a great impact on prophylactic, diagnostic and therapeutic measures. In a recent study, Mitchell et al[144] showed that roughly 53800 HBV chronic carriers settled in the United States each year between 2004 and 2008 from countries of intermediate or high HBV endemicity (2%-31%). In all states of the United States, genotypes A, B, C and D have been identified in immigrants, who were born in HBV-endemic areas[145]. This is one of the clearest pieces of evidence highlighting the direct impact of immigration on the introduction of exotic genotypes to areas with low endemicity. Therefore, we would like to propose the term ”immigro-subgenotypes“ to differentiate native strains from imported stains.
Effect of “immigro-subgenotype” on clinical outcome
The genotype-specific history of HBV strains should be considered when studying imported strains[133]. In the HBV-endemic area, the main route of transmission is usually perinatal. It has been estimated that over 21% of worldwide HBV-related mortality is associated with perinatal transmission[146]. The risk of perinatal transmission is 100%, when the mother is HBeAg-positive and does not take any antiviral medications or HBV immunoglobulins (HBIg). The risk of chronicity of the infant will be 90%, if prophylactic countermeasures are not administrated directly after birth[147]. When the patient is HBeAg-positive, viral load is usually high, which further increases the risk of transmission. As a genotype-related characteristic in HBV endemic regions, the HBeAg test of mothers is positive in the years of childbearing, and just after four decades of life seroconversion might happen. This shows the infectivity potential of HBV carriers infected by (sub)genotypes circulating in endemic regions like East Asia, Africa, Alaska and East Europe[133]. In a recent investigation in Italy, the immigrant population (mostly from Eastern European countries) showed a high prevalence of HBeAg-positivity with a mean age of 31 years[148]. In another study, Dervicevic et al[149] showed the integration of exotic genotypes of HBV with HBeAg-positivity and high viral load among antenatal women in the United Kingdom. Dervicevic et al[149] emphasized the trend of changing epidemiological patterns of HBV in the United Kingdom, where an influx of immigration brings almost 6000 HBV carriers annually to this country. In numerous studies in Belgium, exotic (sub)genotypes of HBV have been identified and found to have integrated into the native population[55,59,81,135,150]. In Bolivia, the exchange of native HBV subgenotype F4 and exotic ones (subgenotypes B2 and C2) between Bolivian and Japanese immigrants was clearly demonstrated by phylogenetic analysis[151]. Interestingly, the exotic strains have different mutational patterns in different ORFs of HBV[123], which would have a different impact on the course of infection, therapeutic, diagnostic and prophylactic measures[152,153].
Naturally occurring mutations associated with drug resistance have been reported in native populations in Asia and Europe[154]. Additionally, in a study conducted by Bottecchia et al[155], a primary drug resistance mutation (rtM204V) was found in the course of treatment-naïve immigrants infected by (sub)genotype E and A3. In a recent study from our group, we found that exotic (sub)genotypes (A6) carried clinically important mutations, which could help the virus to escape from diagnostic assays or prophylactic measures[81,135]. Finally, it should be added that different disasters such as wars[156] or mass-casualties[157] can have direct and indirect impacts on the epidemiology of HBV. Since virological and clinical characteristics of HBV (sub)genotypes differ, it is crucial to monitor changes in epidemiological patterns of HBV infection as it relates to immigration[158].
CONCLUSION
In this review, we attempted to provide strong and up-to-date evidence about the impact of different (sub)genotypes on prophylaxis, diagnosis, clinical outcomes and treatment of HBV infection. Controlling HBV requires massive and unified efforts because modern human measures like vaccination and antiviral therapies have led to the rise of invasive strains, drug resistant, vaccine and diagnosis escape variants. Furthermore, immigration has changed the distribution of HBV and resulted in the emergence of exotic strains in destination territories. These strains, together with intra- and inter-(sub)genotypes recombination, complicate diagnosis, treatment and classification. Elimination of HBV infection requires concomitant vaccination, effective treatment and a vigorous diagnostic scheme. To organize all measures from prophylaxis to therapy, an accurate, holistic and dynamic classification system is essential. This system should be based on robust virological and epidemiological facts to cover all existing strains, and also have the capacity for newly identified strains in the future. Analyzing full-length genome sequences when classifying genotypes and subgenotypes is the foremost prerequisite of this classification system. Careful attention must be paid to all aspects of phylogenetic analysis, such as bootstrapping values and meeting the necessary thresholds for (sub)genotyping. Quasi-subgenotype refers to subgenotypes that were incorrectly suggested to be novel. As many of these strains were misclassified due to genetic differences resulting from recombination, we propose the term “recombino-subgenotype”. We also suggest to introduce the term “immigro-subgenotype” to distinguish exotic (sub)genotypes from native ones; immigration demonstrates a confounding facet of global HBV distribution. We are strongly convinced that applying these two proposed terms in HBV classification will help harmonize this field and allow for improved prophylaxis, diagnosis and treatment.
Footnotes
Supported by Mahmoud Reza Pourkarim is supported by a postdoctoral grant from the ''Fonds voor Wetenschappelijk Onderzoek Vlaanderen''
P- Reviewers: Kim K, Koch-Institute R S- Editor: Cui XM L- Editor: A E- Editor: Liu XM
References
- 1.Ladnyi ID, Breman JG. Smallpox eradication: progress and problems. Dev Biol Stand. 1978;41:281–290. [PubMed] [Google Scholar]
- 2.Kaiser J. Eradication. Pressure growing to set a date to destroy remaining smallpox stocks. Science. 2011;331:389. doi: 10.1126/science.331.6016.389. [DOI] [PubMed] [Google Scholar]
- 3.Blumberg BS, Gerstley BJ, Hungerford DA, London WT, Sutnick AI. A serum antigen (Australia antigen) in Down‘s syndrome, leukemia, and hepatitis. Ann Intern Med. 1967;66:924–931. doi: 10.7326/0003-4819-66-5-924. [DOI] [PubMed] [Google Scholar]
- 4.Blumberg BS. Primary and secondary prevention of liver cancer caused by HBV. Front Biosci (Schol Ed) 2010;2:756–763. doi: 10.2741/s98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blumberg BS. Baruch Blumberg--hepatitis B and beyond. Interviewed by Pam Das. Lancet Infect Dis. 2002;2:767–771. doi: 10.1016/s1473-3099(02)00458-9. [DOI] [PubMed] [Google Scholar]
- 6.Lavanchy D. Viral hepatitis: global goals for vaccination. J Clin Virol. 2012;55:296–302. doi: 10.1016/j.jcv.2012.08.022. [DOI] [PubMed] [Google Scholar]
- 7.Van Damme P, Zanetti AR, Shouval D, Van Herck K. Strategies for global prevention of hepatitis B virus infection. Adv Exp Med Biol. 2010;659:175–188. doi: 10.1007/978-1-4419-0981-7_14. [DOI] [PubMed] [Google Scholar]
- 8.Lavanchy D. Worldwide epidemiology of HBV infection, disease burden, and vaccine prevention. J Clin Virol. 2005;34 Suppl 1:S1–S3. doi: 10.1016/s1386-6532(05)00384-7. [DOI] [PubMed] [Google Scholar]
- 9.Szmaragd C, Balloux F. The population genomics of hepatitis B virus. Mol Ecol. 2007;16:4747–4758. doi: 10.1111/j.1365-294X.2007.03564.x. [DOI] [PubMed] [Google Scholar]
- 10.Pourkarim MR, Van Ranst M. Guidelines for the detection of a common source of hepatitis B virus infections. Hepat Mon. 2011;11:783–785. doi: 10.5812/kowsar.1735143X.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Valsamakis A. Molecular testing in the diagnosis and management of chronic hepatitis B. Clin Microbiol Rev. 2007;20:426–39, table of contents. doi: 10.1128/CMR.00009-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yuen MF, Tanaka Y, Fong DY, Fung J, Wong DK, Yuen JC, But DY, Chan AO, Wong BC, Mizokami M, et al. Independent risk factors and predictive score for the development of hepatocellular carcinoma in chronic hepatitis B. J Hepatol. 2009;50:80–88. doi: 10.1016/j.jhep.2008.07.023. [DOI] [PubMed] [Google Scholar]
- 13.Shen FC, Su IJ, Wu HC, Hsieh YH, Yao WJ, Young KC, Chang TC, Hsieh HC, Tsai HN, Huang W. A pre-S gene chip to detect pre-S deletions in hepatitis B virus large surface antigen as a predictive marker for hepatoma risk in chronic hepatitis B virus carriers. J Biomed Sci. 2009;16:84. doi: 10.1186/1423-0127-16-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alavian SM. Occult hepatitis B virus infection among hemodialysis patients. Hepat Mon. 2012;12:242–243. doi: 10.5812/hepatmon.869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alavian SM, Miri SM. Dilemma of HBsAg seroconversion in chronic hepatitis B infection: Dilemma of HBsAg in chronic HBV. Hepat Mon. 2011;11:67–68. [PMC free article] [PubMed] [Google Scholar]
- 16.Schaefer S. Hepatitis B virus: significance of genotypes. J Viral Hepat. 2005;12:111–124. doi: 10.1111/j.1365-2893.2005.00584.x. [DOI] [PubMed] [Google Scholar]
- 17.Schaefer S. Hepatitis B virus taxonomy and hepatitis B virus genotypes. World J Gastroenterol. 2007;13:14–21. doi: 10.3748/wjg.v13.i1.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yousif M, Kramvis A. Genotype D of hepatitis B virus and its subgenotypes: An update. Hepatol Res. 2013;43:355–364. doi: 10.1111/j.1872-034X.2012.01090.x. [DOI] [PubMed] [Google Scholar]
- 19.Santos AO, Alvarado-Mora MV, Botelho L, Vieira DS, Pinho JR, Carrilho FJ, Honda ER, Salcedo JM. Characterization of hepatitis B virus (HBV) genotypes in patients from Rondônia, Brazil. Virol J. 2010;7:315. doi: 10.1186/1743-422X-7-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kramvis A, Arakawa K, Yu MC, Nogueira R, Stram DO, Kew MC. Relationship of serological subtype, basic core promoter and precore mutations to genotypes/subgenotypes of hepatitis B virus. J Med Virol. 2008;80:27–46. doi: 10.1002/jmv.21049. [DOI] [PubMed] [Google Scholar]
- 21.Norder H, Couroucé AM, Coursaget P, Echevarria JM, Lee SD, Mushahwar IK, Robertson BH, Locarnini S, Magnius LO. Genetic diversity of hepatitis B virus strains derived worldwide: genotypes, subgenotypes, and HBsAg subtypes. Intervirology. 2004;47:289–309. doi: 10.1159/000080872. [DOI] [PubMed] [Google Scholar]
- 22.Purdy MA, Talekar G, Swenson P, Araujo A, Fields H. A new algorithm for deduction of hepatitis B surface antigen subtype determinants from the amino acid sequence. Intervirology. 2007;50:45–51. doi: 10.1159/000096312. [DOI] [PubMed] [Google Scholar]
- 23.Bartholomeusz A, Schaefer S. Hepatitis B virus genotypes: comparison of genotyping methods. Rev Med Virol. 2004;14:3–16. doi: 10.1002/rmv.400. [DOI] [PubMed] [Google Scholar]
- 24.Hardie DR, Williamson C. Analysis of the preS1 gene of hepatitis B virus (HBV) to define epidemiologically linked and un-linked infections in South Africa. Arch Virol. 1997;142:1829–1841. doi: 10.1007/s007050050200. [DOI] [PubMed] [Google Scholar]
- 25.Rozanov M, Plikat U, Chappey C, Kochergin A, Tatusova T. A web-based genotyping resource for viral sequences. Nucleic Acids Res. 2004;32:W654–W659. doi: 10.1093/nar/gkh419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hayer J, Jadeau F, Deléage G, Kay A, Zoulim F, Combet C. HBVdb: a knowledge database for Hepatitis B Virus. Nucleic Acids Res. 2013;41:D566–D570. doi: 10.1093/nar/gks1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Myers R, Clark C, Khan A, Kellam P, Tedder R. Genotyping Hepatitis B virus from whole- and sub-genomic fragments using position-specific scoring matrices in HBV STAR. J Gen Virol. 2006;87:1459–1464. doi: 10.1099/vir.0.81734-0. [DOI] [PubMed] [Google Scholar]
- 28.Alcantara LC, Cassol S, Libin P, Deforche K, Pybus OG, Van Ranst M, Galvão-Castro B, Vandamme AM, de Oliveira T. A standardized framework for accurate, high-throughput genotyping of recombinant and non-recombinant viral sequences. Nucleic Acids Res. 2009;37:W634–W642. doi: 10.1093/nar/gkp455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gnaneshan S, Ijaz S, Moran J, Ramsay M, Green J. HepSEQ: International Public Health Repository for Hepatitis B. Nucleic Acids Res. 2007;35:D367–D370. doi: 10.1093/nar/gkl874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shin-I T, Tanaka Y, Tateno Y, Mizokami M. Development and public release of a comprehensive hepatitis virus database. Hepatol Res. 2008;38:234–243. doi: 10.1111/j.1872-034X.2007.00262.x. [DOI] [PubMed] [Google Scholar]
- 31.Schultz AK, Bulla I, Abdou-Chekaraou M, Gordien E, Morgenstern B, Zoaulim F, Dény P, Stanke M. jpHMM: recombination analysis in viruses with circular genomes such as the hepatitis B virus. Nucleic Acids Res. 2012;40:W193–W198. doi: 10.1093/nar/gks414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lindh M, Gonzalez JE, Norkrans G, Horal P. Genotyping of hepatitis B virus by restriction pattern analysis of a pre-S amplicon. J Virol Methods. 1998;72:163–174. doi: 10.1016/s0166-0934(98)00026-3. [DOI] [PubMed] [Google Scholar]
- 33.Mizokami M, Nakano T, Orito E, Tanaka Y, Sakugawa H, Mukaide M, Robertson BH. Hepatitis B virus genotype assignment using restriction fragment length polymorphism patterns. FEBS Lett. 1999;450:66–71. doi: 10.1016/s0014-5793(99)00471-8. [DOI] [PubMed] [Google Scholar]
- 34.Kirschberg O, Schüttler C, Repp R, Schaefer S. A multiplex-PCR to identify hepatitis B virus--enotypes A-F. J Clin Virol. 2004;29:39–43. doi: 10.1016/s1386-6532(03)00084-2. [DOI] [PubMed] [Google Scholar]
- 35.Guirgis BS, Abbas RO, Azzazy HM. Hepatitis B virus genotyping: current methods and clinical implications. Int J Infect Dis. 2010;14:e941–e953. doi: 10.1016/j.ijid.2010.03.020. [DOI] [PubMed] [Google Scholar]
- 36.Amini-Bavil-Olyaee S, Tacke F, Alavian SM. HBV Subgenotypes D1, D2, D-del! Are ‘Old’ Genotyping Methods Interpreted Correctly? Hepat Mon. 2013;13:e13048. doi: 10.5812/hepatmon.13048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tatematsu K, Tanaka Y, Kurbanov F, Sugauchi F, Mano S, Maeshiro T, Nakayoshi T, Wakuta M, Miyakawa Y, Mizokami M. A genetic variant of hepatitis B virus divergent from known human and ape genotypes isolated from a Japanese patient and provisionally assigned to new genotype J. J Virol. 2009;83:10538–10547. doi: 10.1128/JVI.00462-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tang XR, Zhang JS, Zhao H, Gong YH, Wang YZ, Zhao JL. Detection of hepatitis B virus genotypes using oligonucleotide chip among hepatitis B virus carriers in Eastern China. World J Gastroenterol. 2007;13:1975–1979. doi: 10.3748/wjg.v13.i13.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tadokoro K, Kobayashi M, Yamaguchi T, Suzuki F, Miyauchi S, Egashira T, Kumada H. Classification of hepatitis B virus genotypes by the PCR-Invader method with genotype-specific probes. J Virol Methods. 2006;138:30–39. doi: 10.1016/j.jviromet.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 40.Payungporn S, Tangkijvanich P, Jantaradsamee P, Theamboonlers A, Poovorawan Y. Simultaneous quantitation and genotyping of hepatitis B virus by real-time PCR and melting curve analysis. J Virol Methods. 2004;120:131–140. doi: 10.1016/j.jviromet.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 41.Liu WC, Mizokami M, Buti M, Lindh M, Young KC, Sun KT, Chi YC, Li HH, Chang TT. Simultaneous quantification and genotyping of hepatitis B virus for genotypes A to G by real-time PCR and two-step melting curve analysis. J Clin Microbiol. 2006;44:4491–4497. doi: 10.1128/JCM.01375-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Amini-Bavil-Olyaee S, Pourkarim MR, Schaefer S, Mahboudi F, Van Ranst M, Adeli A, Trautwein C, Tacke F. Single-step real-time PCR to quantify hepatitis B virus and distinguish genotype D from non-D genotypes. J Viral Hepat. 2011;18:300–304. doi: 10.1111/j.1365-2893.2010.01308.x. [DOI] [PubMed] [Google Scholar]
- 43.Zhang R, Deng Y, Muller CP, Ou ZY, Ma L, Wang M, Li PQ, He YS. Determination of hepatitis B virus genotype by flow-through reverse dot blot. J Clin Virol. 2007;39:94–100. doi: 10.1016/j.jcv.2007.01.014. [DOI] [PubMed] [Google Scholar]
- 44.Ganova-Raeva LM, Dimitrova ZE, Campo DS, Khudyakov Y. Application of mass spectrometry to molecular surveillance of hepatitis B and C viral infections. Antivir Ther. 2012;17:1477–1482. doi: 10.3851/IMP2466. [DOI] [PubMed] [Google Scholar]
- 45.Han KH, Hong SP, Choi SH, Shin SK, Cho SW, Ahn SH, Hahn JS, Kim SO. Comparison of multiplex restriction fragment mass polymorphism and sequencing analyses for detecting entecavir resistance in chronic hepatitis B. Antivir Ther. 2011;16:77–87. doi: 10.3851/IMP1702. [DOI] [PubMed] [Google Scholar]
- 46.Kurbanov F, Tanaka Y, Mizokami M. Geographical and genetic diversity of the human hepatitis B virus. Hepatol Res. 2010;40:14–30. doi: 10.1111/j.1872-034X.2009.00601.x. [DOI] [PubMed] [Google Scholar]
- 47.Usuda S, Okamoto H, Iwanari H, Baba K, Tsuda F, Miyakawa Y, Mayumi M. Serological detection of hepatitis B virus genotypes by ELISA with monoclonal antibodies to type-specific epitopes in the preS2-region product. J Virol Methods. 1999;80:97–112. doi: 10.1016/s0166-0934(99)00039-7. [DOI] [PubMed] [Google Scholar]
- 48.Pourkarim MR, Sharifi Z, Soleimani A, Amini-Bavil-Olyaee S, Elsadek Fakhr A, Sijmons S, Vercauteren J, Karimi G, Lemey P, Maes P, et al. Evolutionary analysis of HBV “S” antigen genetic diversity in Iranian blood donors: a nationwide study. J Med Virol. 2014;86:144–155. doi: 10.1002/jmv.23798. [DOI] [PubMed] [Google Scholar]
- 49.Paraskevis D, Magiorkinis G, Magiorkinis E, Ho SY, Belshaw R, Allain JP, Hatzakis A. Dating the origin and dispersal of hepatitis B virus infection in humans and primates. Hepatology. 2013;57:908–916. doi: 10.1002/hep.26079. [DOI] [PubMed] [Google Scholar]
- 50.Günther S. Genetic variation in HBV infection: genotypes and mutants. J Clin Virol. 2006;36 Suppl 1:S3–S11. doi: 10.1016/s1386-6532(06)80002-8. [DOI] [PubMed] [Google Scholar]
- 51.Chen J, Yin J, Tan X, Zhang H, Zhang H, Chen B, Chang W, Schaefer S, Cao G. Improved multiplex-PCR to identify hepatitis B virus genotypes A-F and subgenotypes B1, B2, C1 and C2. J Clin Virol. 2007;38:238–243. doi: 10.1016/j.jcv.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 52.Wang J, Gao JW, Li J, Zhuang H, Wang J, Li YJ, Jin H. [Establishment of a semi-nested PCR for identifying the sub-genotypes (Ba and Bj) of hepatitis B virus of genotype B] Zhonghua Liuxingbingxue Zazhi. 2008;29:177–180. [PubMed] [Google Scholar]
- 53.Olinger CM, Venard V, Njayou M, Oyefolu AO, Maïga I, Kemp AJ, Omilabu SA, le Faou A, Muller CP. Phylogenetic analysis of the precore/core gene of hepatitis B virus genotypes E and A in West Africa: new subtypes, mixed infections and recombinations. J Gen Virol. 2006;87:1163–1173. doi: 10.1099/vir.0.81614-0. [DOI] [PubMed] [Google Scholar]
- 54.Pourkarim MR, Amini-Bavil-Olyaee S, Lemey P, Maes P, Van Ranst M. Are hepatitis B virus “subgenotypes” defined accurately? J Clin Virol. 2010;47:356–360. doi: 10.1016/j.jcv.2010.01.015. [DOI] [PubMed] [Google Scholar]
- 55.Pourkarim MR, Lemey P, Amini-Bavil-Olyaee S, Maes P, Van Ranst M. Novel hepatitis B virus subgenotype A6 in African-Belgian patients. J Clin Virol. 2010;47:93–96. doi: 10.1016/j.jcv.2009.09.032. [DOI] [PubMed] [Google Scholar]
- 56.Khedive A, Norouzi M, Ramezani F, Karimzadeh H, Alavian SM, Malekzadeh R, Montazeri G, Nejatizadeh A, Ziaee M, Abedi F, et al. Hepatitis B virus surface protein mutations clustered mainly in CTL immune epitopes in chronic carriers: results of an Iranian nationwide study. J Viral Hepat. 2013;20:494–501. doi: 10.1111/jvh.12045. [DOI] [PubMed] [Google Scholar]
- 57.Zehender G, Ebranati E, Gabanelli E, Shkjezi R, Lai A, Sorrentino C, Lo Presti A, Basho M, Bruno R, Tanzi E, et al. Spatial and temporal dynamics of hepatitis B virus D genotype in Europe and the Mediterranean Basin. PLoS One. 2012;7:e37198. doi: 10.1371/journal.pone.0037198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Amini-Bavil-Olyaee S, Maes P, Van Ranst M, Pourkarim MR. Providing strong evidence of nosocomial outbreak of hepatitis B virus infection. J Hosp Infect. 2012;80:269–270; author reply 270-272. doi: 10.1016/j.jhin.2011.10.017. [DOI] [PubMed] [Google Scholar]
- 59.Pourkarim MR, Verbeeck J, Rahman M, Amini-Bavil-Olyaee S, Forier AM, Lemey P, Maes P, Van Ranst M. Phylogenetic analysis of hepatitis B virus full-length genomes reveals evidence for a large nosocomial outbreak in Belgium. J Clin Virol. 2009;46:61–68. doi: 10.1016/j.jcv.2009.06.015. [DOI] [PubMed] [Google Scholar]
- 60.Andernach IE, Nolte C, Pape JW, Muller CP. Slave trade and hepatitis B virus genotypes and subgenotypes in Haiti and Africa. Emerg Infect Dis. 2009;15:1222–1228. doi: 10.3201/eid1508.081642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hübschen JM, Mbah PO, Forbi JC, Otegbayo JA, Olinger CM, Charpentier E, Muller CP. Detection of a new subgenotype of hepatitis B virus genotype A in Cameroon but not in neighbouring Nigeria. Clin Microbiol Infect. 2011;17:88–94. doi: 10.1111/j.1469-0691.2010.03205.x. [DOI] [PubMed] [Google Scholar]
- 62.Shi W, Zhu C, Zheng W, Carr MJ, Higgins DG, Zhang Z. Subgenotype reclassification of genotype B hepatitis B virus. BMC Gastroenterol. 2012;12:116. doi: 10.1186/1471-230X-12-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huy TT, Ushijima H, Quang VX, Win KM, Luengrojanakul P, Kikuchi K, Sata T, Abe K. Genotype C of hepatitis B virus can be classified into at least two subgroups. J Gen Virol. 2004;85:283–292. doi: 10.1099/vir.0.19633-0. [DOI] [PubMed] [Google Scholar]
- 64.Ghosh S, Banerjee P, Deny P, Mondal RK, Nandi M, Roychoudhury A, Das K, Banerjee S, Santra A, Zoulim F, et al. New HBV subgenotype D9, a novel D/C recombinant, identified in patients with chronic HBeAg-negative infection in Eastern India. J Viral Hepat. 2013;20:209–218. doi: 10.1111/j.1365-2893.2012.01655.x. [DOI] [PubMed] [Google Scholar]
- 65.Wang Z, Liu Z, Zeng G, Wen S, Qi Y, Ma S, Naoumov NV, Hou J. A new intertype recombinant between genotypes C and D of hepatitis B virus identified in China. J Gen Virol. 2005;86:985–990. doi: 10.1099/vir.0.80771-0. [DOI] [PubMed] [Google Scholar]
- 66.Wang Z, Hou J, Zeng G, Wen S, Tanaka Y, Cheng J, Kurbanov F, Wang L, Jiang J, Naoumov NV, et al. Distribution and characteristics of hepatitis B virus genotype C subgenotypes in China. J Viral Hepat. 2007;14:426–434. doi: 10.1111/j.1365-2893.2006.00813.x. [DOI] [PubMed] [Google Scholar]
- 67.Abdou Chekaraou M, Brichler S, Mansour W, Le Gal F, Garba A, Dény P, Gordien E. A novel hepatitis B virus (HBV) subgenotype D (D8) strain, resulting from recombination between genotypes D and E, is circulating in Niger along with HBV/E strains. J Gen Virol. 2010;91:1609–1620. doi: 10.1099/vir.0.018127-0. [DOI] [PubMed] [Google Scholar]
- 68.Kurbanov F, Tanaka Y, Fujiwara K, Sugauchi F, Mbanya D, Zekeng L, Ndembi N, Ngansop C, Kaptue L, Miura T, et al. A new subtype (subgenotype) Ac (A3) of hepatitis B virus and recombination between genotypes A and E in Cameroon. J Gen Virol. 2005;86:2047–2056. doi: 10.1099/vir.0.80922-0. [DOI] [PubMed] [Google Scholar]
- 69.Shi W, Zhang Z, Ling C, Zheng W, Zhu C, Carr MJ, Higgins DG. Hepatitis B virus subgenotyping: history, effects of recombination, misclassifications, and corrections. Infect Genet Evol. 2013;16:355–361. doi: 10.1016/j.meegid.2013.03.021. [DOI] [PubMed] [Google Scholar]
- 70.Zaaijer HL, Boot HJ, van Swieten P, Koppelman MH, Cuypers HT. HBsAg-negative mono-infection with hepatitis B virus genotype G. J Viral Hepat. 2011;18:815–819. doi: 10.1111/j.1365-2893.2010.01397.x. [DOI] [PubMed] [Google Scholar]
- 71.Amini-Bavil-Olyaee S, Alavian SM, Adeli A, Sarrami-Forooshani R, Sabahi F, Sabouri E, Tavangar HR, Azizi M, Mahboudi F. Hepatitis B virus genotyping, core promoter, and precore/core mutations among Afghan patients infected with hepatitis B: a preliminary report. J Med Virol. 2006;78:358–364. doi: 10.1002/jmv.20547. [DOI] [PubMed] [Google Scholar]
- 72.Amini-Bavil-Olyaee S, Sarrami-Forooshani R, Adeli A, Sabahi F, Abachi M, Azizi M, Mahboudi F. Complete genomic sequence and phylogenetic relatedness of hepatitis B virus isolates from Iran. J Med Virol. 2005;76:318–326. doi: 10.1002/jmv.20362. [DOI] [PubMed] [Google Scholar]
- 73.Sugauchi F, Orito E, Ichida T, Kato H, Sakugawa H, Kakumu S, Ishida T, Chutaputti A, Lai CL, Gish RG, et al. Epidemiologic and virologic characteristics of hepatitis B virus genotype B having the recombination with genotype C. Gastroenterology. 2003;124:925–932. doi: 10.1053/gast.2003.50140. [DOI] [PubMed] [Google Scholar]
- 74.Kramvis A, Kew MC. Relationship of genotypes of hepatitis B virus to mutations, disease progression and response to antiviral therapy. J Viral Hepat. 2005;12:456–464. doi: 10.1111/j.1365-2893.2005.00624.x. [DOI] [PubMed] [Google Scholar]
- 75.Amini-Bavil-Olyaee S, Herbers U, Luedde T, Trautwein C, Tacke F. Impact of hepatitis B e antigen-suppressing mutations on the replication efficiency of entecavir-resistant hepatitis B virus strains. J Viral Hepat. 2011;18:804–814. doi: 10.1111/j.1365-2893.2010.01378.x. [DOI] [PubMed] [Google Scholar]
- 76.Sugauchi F, Ohno T, Orito E, Sakugawa H, Ichida T, Komatsu M, Kuramitsu T, Ueda R, Miyakawa Y, Mizokami M. Influence of hepatitis B virus genotypes on the development of preS deletions and advanced liver disease. J Med Virol. 2003;70:537–544. doi: 10.1002/jmv.10428. [DOI] [PubMed] [Google Scholar]
- 77.Bahramali G, Sadeghizadeh M, Amini-Bavil-Olyaee S, Alavian SM, Behzad-Behbahani A, Adeli A, Aghasadeghi MR, Amini S, Mahboudi F. Clinical, virologic and phylogenetic features of hepatitis B infection in Iranian patients. World J Gastroenterol. 2008;14:5448–5453. doi: 10.3748/wjg.14.5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kidd-Ljunggren K, Myhre E, Bläckberg J. Clinical and serological variation between patients infected with different Hepatitis B virus genotypes. J Clin Microbiol. 2004;42:5837–5841. doi: 10.1128/JCM.42.12.5837-5841.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Odemuyiwa SO, Mulders MN, Oyedele OI, Ola SO, Odaibo GN, Olaleye DO, Muller CP. Phylogenetic analysis of new hepatitis B virus isolates from Nigeria supports endemicity of genotype E in West Africa. J Med Virol. 2001;65:463–469. [PubMed] [Google Scholar]
- 80.Stuyver L, De Gendt S, Van Geyt C, Zoulim F, Fried M, Schinazi RF, Rossau R. A new genotype of hepatitis B virus: complete genome and phylogenetic relatedness. J Gen Virol. 2000;81:67–74. doi: 10.1099/0022-1317-81-1-67. [DOI] [PubMed] [Google Scholar]
- 81.Pourkarim MR, Lemey P, Amini-Bavil-Olyaee S, Houspie L, Verbeeck J, Rahman M, Maes P, Vanwijngaerden E, Nevens F, Van Ranst M. Molecular characterization of hepatitis B virus strains circulating in Belgian patients co-infected with HIV and HBV: overt and occult infection. J Med Virol. 2011;83:1876–1884. doi: 10.1002/jmv.22174. [DOI] [PubMed] [Google Scholar]
- 82.Arauz-Ruiz P, Norder H, Robertson BH, Magnius LO. Genotype H: a new Amerindian genotype of hepatitis B virus revealed in Central America. J Gen Virol. 2002;83:2059–2073. doi: 10.1099/0022-1317-83-8-2059. [DOI] [PubMed] [Google Scholar]
- 83.Tran TT, Trinh TN, Abe K. New complex recombinant genotype of hepatitis B virus identified in Vietnam. J Virol. 2008;82:5657–5663. doi: 10.1128/JVI.02556-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shepard CW, Simard EP, Finelli L, Fiore AE, Bell BP. Hepatitis B virus infection: epidemiology and vaccination. Epidemiol Rev. 2006;28:112–125. doi: 10.1093/epirev/mxj009. [DOI] [PubMed] [Google Scholar]
- 85.Allain JP. Epidemiology of Hepatitis B virus and genotype. J Clin Virol. 2006;36 Suppl 1:S12–S17. doi: 10.1016/s1386-6532(06)80003-x. [DOI] [PubMed] [Google Scholar]
- 86.Livingston SE, Simonetti JP, Bulkow LR, Homan CE, Snowball MM, Cagle HH, Negus SE, McMahon BJ. Clearance of hepatitis B e antigen in patients with chronic hepatitis B and genotypes A, B, C, D, and F. Gastroenterology. 2007;133:1452–1457. doi: 10.1053/j.gastro.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 87.Custer B, Sullivan SD, Hazlet TK, Iloeje U, Veenstra DL, Kowdley KV. Global epidemiology of hepatitis B virus. J Clin Gastroenterol. 2004;38:S158–S168. doi: 10.1097/00004836-200411003-00008. [DOI] [PubMed] [Google Scholar]
- 88.Tamada Y, Yatsuhashi H, Masaki N, Nakamuta M, Mita E, Komatsu T, Watanabe Y, Muro T, Shimada M, Hijioka T, et al. Hepatitis B virus strains of subgenotype A2 with an identical sequence spreading rapidly from the capital region to all over Japan in patients with acute hepatitis B. Gut. 2012;61:765–773. doi: 10.1136/gutjnl-2011-300832. [DOI] [PubMed] [Google Scholar]
- 89.Smolle E, Zöhrer E, Bettermann K, Haybaeck J. Viral hepatitis induces hepatocellular cancer: what can we learn from epidemiology comparing iran and worldwide findings? Hepat Mon. 2012;12:e7879. doi: 10.5812/hepatmon.7879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zidan A, Scheuerlein H, Schüle S, Settmacher U, Rauchfuss F. Epidemiological pattern of hepatitis B and hepatitis C as etiological agents for hepatocellular carcinoma in iran and worldwide. Hepat Mon. 2012;12:e6894. doi: 10.5812/hepatmon.6894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sumi H, Yokosuka O, Seki N, Arai M, Imazeki F, Kurihara T, Kanda T, Fukai K, Kato M, Saisho H. Influence of hepatitis B virus genotypes on the progression of chronic type B liver disease. Hepatology. 2003;37:19–26. doi: 10.1053/jhep.2003.50036. [DOI] [PubMed] [Google Scholar]
- 92.Gomes-Gouvêa MS, Soares MC, Bensabath G, de Carvalho-Mello IM, Brito EM, Souza OS, Queiroz AT, Carrilho FJ, Pinho JR. Hepatitis B virus and hepatitis delta virus genotypes in outbreaks of fulminant hepatitis (Labrea black fever) in the western Brazilian Amazon region. J Gen Virol. 2009;90:2638–2643. doi: 10.1099/vir.0.013615-0. [DOI] [PubMed] [Google Scholar]
- 93.Thakur V, Guptan RC, Kazim SN, Malhotra V, Sarin SK. Profile, spectrum and significance of HBV genotypes in chronic liver disease patients in the Indian subcontinent. J Gastroenterol Hepatol. 2002;17:165–170. [Google Scholar]
- 94.Wai CT, Fontana RJ, Polson J, Hussain M, Shakil AO, Han SH, Davern TJ, Lee WM, Lok AS. Clinical outcome and virological characteristics of hepatitis B-related acute liver failure in the United States. J Viral Hepat. 2005;12:192–198. doi: 10.1111/j.1365-2893.2005.00581.x. [DOI] [PubMed] [Google Scholar]
- 95.Sánchez-Tapias JM, Costa J, Mas A, Bruguera M, Rodés J. Influence of hepatitis B virus genotype on the long-term outcome of chronic hepatitis B in western patients. Gastroenterology. 2002;123:1848–1856. doi: 10.1053/gast.2002.37041. [DOI] [PubMed] [Google Scholar]
- 96.Shi YH. Correlation between hepatitis B virus genotypes and clinical outcomes. Jpn J Infect Dis. 2012;65:476–482. doi: 10.7883/yoken.65.476. [DOI] [PubMed] [Google Scholar]
- 97.Erhardt A, Blondin D, Hauck K, Sagir A, Kohnle T, Heintges T, Häussinger D. Response to interferon alfa is hepatitis B virus genotype dependent: genotype A is more sensitive to interferon than genotype D. Gut. 2005;54:1009–1013. doi: 10.1136/gut.2004.060327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Buster EH, Flink HJ, Cakaloglu Y, Simon K, Trojan J, Tabak F, So TM, Feinman SV, Mach T, Akarca US, Schutten M, Tielemans W, van Vuuren AJ, Hansen BE, Janssen HL. Sustained HBeAg and HBsAg loss after long-term follow-up of HBeAg-positive patients treated with peginterferon alpha-2b. Gastroenterology. 2008;135:459–467. doi: 10.1053/j.gastro.2008.05.031. [DOI] [PubMed] [Google Scholar]
- 99.Sonneveld MJ, Rijckborst V, Cakaloglu Y, Simon K, Heathcote EJ, Tabak F, Mach T, Boucher CA, Hansen BE, Zeuzem S, et al. Durable hepatitis B surface antigen decline in hepatitis B e antigen-positive chronic hepatitis B patients treated with pegylated interferon-α2b: relation to response and HBV genotype. Antivir Ther. 2012;17:9–17. doi: 10.3851/IMP1887. [DOI] [PubMed] [Google Scholar]
- 100.Janssen HL, van Zonneveld M, Senturk H, Zeuzem S, Akarca US, Cakaloglu Y, Simon C, So TM, Gerken G, de Man RA, Niesters HG, Zondervan P, Hansen B, Schalm SW. Pegylated interferon alfa-2b alone or in combination with lamivudine for HBeAg-positive chronic hepatitis B: a randomised trial. Lancet. 2005;365:123–129. doi: 10.1016/S0140-6736(05)17701-0. [DOI] [PubMed] [Google Scholar]
- 101.Chen JH, Yu YS, Liu HH, Chen XH, Xi M, Zang GQ, Tang ZH. Ubiquitin conjugation of hepatitis B virus core antigen DNA vaccine leads to enhanced cell-mediated immune response in BALB/c mice. Hepat Mon. 2011;11:620–628. doi: 10.5812/kowsar.1735143X.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Tosun S, Deveci S, Kaplan Y, Kasirga E. Should a booster dose be administered in children after mass immunization for hepatitis B? Hepat Mon. 2011;11:440–444. [PMC free article] [PubMed] [Google Scholar]
- 103.Grosso G, Mistretta A, Marventano S, Ferranti R, Mauro L, Cunsolo R, Proietti L, Malaguarnera M. Long-term persistence of seroprotection by hepatitis B vaccination in healthcare workers of southern Italy. Hepat Mon. 2012;12:e6025. doi: 10.5812/hepatmon.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tacke F, Amini-Bavil-Olyaee S, Heim A, Luedde T, Manns MP, Trautwein C. Acute hepatitis B virus infection by genotype F despite successful vaccination in an immune-competent German patient. J Clin Virol. 2007;38:353–357. doi: 10.1016/j.jcv.2006.12.024. [DOI] [PubMed] [Google Scholar]
- 105.Cassidy A, Mossman S, Olivieri A, De Ridder M, Leroux-Roels G. Hepatitis B vaccine effectiveness in the face of global HBV genotype diversity. Expert Rev Vaccines. 2011;10:1709–1715. doi: 10.1586/erv.11.151. [DOI] [PubMed] [Google Scholar]
- 106.O'Halloran JA, De Gascun CF, Dunford L, Carr MJ, Connell J, Howard R, Hall WW, Lambert JS. Hepatitis B virus vaccine failure resulting in chronic hepatitis B infection. J Clin Virol. 2011;52:151–154. doi: 10.1016/j.jcv.2011.06.020. [DOI] [PubMed] [Google Scholar]
- 107.Hsu HY, Chang MH, Liaw SH, Ni YH, Chen HL. Changes of hepatitis B surface antigen variants in carrier children before and after universal vaccination in Taiwan. Hepatology. 1999;30:1312–1317. doi: 10.1002/hep.510300511. [DOI] [PubMed] [Google Scholar]
- 108.Ngui SL, O’Connell S, Eglin RP, Heptonstall J, Teo CG. Low detection rate and maternal provenance of hepatitis B virus S gene mutants in cases of failed postnatal immunoprophylaxis in England and Wales. J Infect Dis. 1997;176:1360–1365. doi: 10.1086/514133. [DOI] [PubMed] [Google Scholar]
- 109.Olinger CM, Weber B, Otegbayo JA, Ammerlaan W, van der Taelem-Brulé N, Muller CP. Hepatitis B virus genotype E surface antigen detection with different immunoassays and diagnostic impact of mutations in the preS/S gene. Med Microbiol Immunol. 2007;196:247–252. doi: 10.1007/s00430-007-0050-5. [DOI] [PubMed] [Google Scholar]
- 110.Kay A, Zoulim F. Hepatitis B virus genetic variability and evolution. Virus Res. 2007;127:164–176. doi: 10.1016/j.virusres.2007.02.021. [DOI] [PubMed] [Google Scholar]
- 111.Banerjee A, Kurbanov F, Datta S, Chandra PK, Tanaka Y, Mizokami M, Chakravarty R. Phylogenetic relatedness and genetic diversity of hepatitis B virus isolates in Eastern India. J Med Virol. 2006;78:1164–1174. doi: 10.1002/jmv.20677. [DOI] [PubMed] [Google Scholar]
- 112.Ghosh S, Banerjee P, RoyChoudhury A, Sarkar S, Ghosh A, Santra A, Banerjee S, Das K, Dwibedi B, Kar SK, et al. Unique hepatitis B virus subgenotype in a primitive tribal community in eastern India. J Clin Microbiol. 2010;48:4063–4071. doi: 10.1128/JCM.01174-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Pourkarim MR, Amini-Bavil-Olyaee S, Lemey P, Maes P, Van Ranst M. HBV subgenotype misclassification expands quasi-subgenotype A3. Clin Microbiol Infect. 2011;17:947–949. doi: 10.1111/j.1469-0691.2010.03374.x. [DOI] [PubMed] [Google Scholar]
- 114.Schaefer S, Magnius L, Norder H. Under construction: classification of hepatitis B virus genotypes and subgenotypes. Intervirology. 2009;52:323–325. doi: 10.1159/000242353. [DOI] [PubMed] [Google Scholar]
- 115.Kurbanov F, Tanaka Y, Kramvis A, Simmonds P, Mizokami M. When should “I” consider a new hepatitis B virus genotype? J Virol. 2008;82:8241–8242. doi: 10.1128/JVI.00793-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Mumtaz K, Hamid S, Ahmed S, Moatter T, Mushtaq S, Khan A, Mizokami M, Jafri W. A study of genotypes, mutants and nucleotide sequence of hepatitis B virus in Pakistan: HBV genotypes in pakistan. Hepat Mon. 2011;11:14–18. [PMC free article] [PubMed] [Google Scholar]
- 117.Shi W, Carr MJ, Dunford L, Zhu C, Hall WW, Higgins DG. Identification of novel inter-genotypic recombinants of human hepatitis B viruses by large-scale phylogenetic analysis. Virology. 2012;427:51–59. doi: 10.1016/j.virol.2012.01.030. [DOI] [PubMed] [Google Scholar]
- 118.Shi W, Zhu C, Zheng W, Zheng W, Ling C, Carr MJ, Higgins DG, Zhang Z. Subgenotyping of genotype C hepatitis B virus: correcting misclassifications and identifying a novel subgenotype. PLoS One. 2012;7:e47271. doi: 10.1371/journal.pone.0047271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Mulyanto SN, Surayah K, Tsuda F, Ichiyama K, Takahashi M, Okamoto H. A nationwide molecular epidemiological study on hepatitis B virus in Indonesia: identification of two novel subgenotypes, B8 and C7. Arch Virol. 2009;154:1047–1059. doi: 10.1007/s00705-009-0406-9. [DOI] [PubMed] [Google Scholar]
- 120.Mulyanto P, Depamede SN, Wahyono A, Jirintai S, Nagashima S, Takahashi M, Nishizawa T, Okamoto H. Identification of four novel subgenotypes (C13-C16) and two inter-genotypic recombinants (C12/G and C13/B3) of hepatitis B virus in Papua province, Indonesia. Virus Res. 2012;163:129–140. doi: 10.1016/j.virusres.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 121.Cui C, Shi J, Hui L, Xi H, Zhuoma G. The dominant hepatitis B virus genotype identified in Tibet is a C/D hybrid. J Gen Virol. 2002;83:2773–2777. doi: 10.1099/0022-1317-83-11-2773. [DOI] [PubMed] [Google Scholar]
- 122.Ataei B, Yazdani MR, Kalantari H, Yaran M, Nokhodian Z, Javadi AA, Babak A, Adibi P. Hepatitis D virus infection in Isfahan, central Iran: Prevalence and risk factors among chronic HBV infection cases. Hepat Mon. 2011;11:269–272. [PMC free article] [PubMed] [Google Scholar]
- 123.Ayari R, Lakhoua-Gorgi Y, Bouslama L, Safar I, Kchouk FH, Aouadi H, Jendoubi-Ayed S, Najjar T, Ayed K, Abdallah TB. Investigation of DNA sequence in the Basal core promoter, precore, and core regions of hepatitis B virus from Tunisia shows a shift in genotype prevalence. Hepat Mon. 2012;12:e6191. doi: 10.5812/hepatmon.6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Khanizadeh S, Ravanshad M, Mohebbi SR, Naghoosi H, Abrahim Tahaei M, Mousavi Nasab SD, Romani S, Azimzadeh P, Sanati A, Zali MR. Polymorphisms within the Promoter Region of the Gamma Interferon (IFN-γ) Receptor1 Gene are Associated with the Susceptibility to Chronic HBV Infection in an Iranian Population. Hepat Mon. 2012;12:e7283. doi: 10.5812/hepatmon.7283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Tahaei SM, Mohebbi SR, Azimzadeh P, Vahedi M, Almasi S, Romani S, Sharifian A, Derakhshan F, Zali MR. Frequency of HIV and HCV Co-Infections in Chronic HBV Patients Referred to Taleghani Hospital, Tehran, Iran from 2006 to 2010. Hepat Mon. 2011;11:993–996. doi: 10.5812/kowsar.1735143X.740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sayan M, Dogan C. Hepatitis B virus genotype G infection in a Turkish patient undergoing hemodialysis therapy. Hepat Mon. 2012;12:118–121. doi: 10.5812/hepatmon.826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Alvarado-Mora MV, Pinho JR. Distribution of HBV genotypes in Latin America. Antivir Ther. 2013;18:459–465. doi: 10.3851/IMP2599. [DOI] [PubMed] [Google Scholar]
- 128.Sakamoto T, Tanaka Y, Simonetti J, Osiowy C, Borresen ML, Koch A, Kurbanov F, Sugiyama M, Minuk GY, McMahon BJ, et al. Classification of hepatitis B virus genotype B into 2 major types based on characterization of a novel subgenotype in Arctic indigenous populations. J Infect Dis. 2007;196:1487–1492. doi: 10.1086/523111. [DOI] [PubMed] [Google Scholar]
- 129.Thedja MD, Muljono DH, Nurainy N, Sukowati CH, Verhoef J, Marzuki S. Ethnogeographical structure of hepatitis B virus genotype distribution in Indonesia and discovery of a new subgenotype, B9. Arch Virol. 2011;156:855–868. doi: 10.1007/s00705-011-0926-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Davies J, Littlejohn M, Locarnini SA, Whiting S, Hajkowicz K, Cowie BC, Bowden DS, Tong SY, Davis JS. Molecular epidemiology of hepatitis B in the Indigenous people of northern Australia. J Gastroenterol Hepatol. 2013;28:1234–1241. doi: 10.1111/jgh.12177. [DOI] [PubMed] [Google Scholar]
- 131.Livingston SE, Simonetti JP, McMahon BJ, Bulkow LR, Hurlburt KJ, Homan CE, Snowball MM, Cagle HH, Williams JL, Chulanov VP. Hepatitis B virus genotypes in Alaska Native people with hepatocellular carcinoma: preponderance of genotype F. J Infect Dis. 2007;195:5–11. doi: 10.1086/509894. [DOI] [PubMed] [Google Scholar]
- 132.Ozasa A, Tanaka Y, Orito E, Sugiyama M, Kang JH, Hige S, Kuramitsu T, Suzuki K, Tanaka E, Okada S, et al. Influence of genotypes and precore mutations on fulminant or chronic outcome of acute hepatitis B virus infection. Hepatology. 2006;44:326–334. doi: 10.1002/hep.21249. [DOI] [PubMed] [Google Scholar]
- 133.McMahon BJ. The influence of hepatitis B virus genotype and subgenotype on the natural history of chronic hepatitis B. Hepatol Int. 2009;3:334–342. doi: 10.1007/s12072-008-9112-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Simmonds P, Midgley S. Recombination in the genesis and evolution of hepatitis B virus genotypes. J Virol. 2005;79:15467–15476. doi: 10.1128/JVI.79.24.15467-15476.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Pourkarim MR, Amini-Bavil-Olyaee S, Verbeeck J, Lemey P, Zeller M, Rahman M, Maes P, Nevens F, Van Ranst M. Molecular evolutionary analysis and mutational pattern of full-length genomes of hepatitis B virus isolated from Belgian patients with different clinical manifestations. J Med Virol. 2010;82:379–389. doi: 10.1002/jmv.21726. [DOI] [PubMed] [Google Scholar]
- 136.Zhou B, Xiao L, Wang Z, Chang ET, Chen J, Hou J. Geographical and ethnic distribution of the HBV C/D recombinant on the Qinghai-Tibet Plateau. PLoS One. 2011;6:e18708. doi: 10.1371/journal.pone.0018708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ghosh S, Mondal RK, Banerjee P, Nandi M, Sarkar S, Das K, Santra A, Banerjee S, Chowdhury A, Datta S. Tracking the naturally occurring mutations across the full-length genome of hepatitis B virus of genotype D in different phases of chronic e-antigen-negative infection. Clin Microbiol Infect. 2012;18:E412–E418. doi: 10.1111/j.1469-0691.2012.03975.x. [DOI] [PubMed] [Google Scholar]
- 138.Meldal BH, Moula NM, Barnes IH, Boukef K, Allain JP. A novel hepatitis B virus subgenotype, D7, in Tunisian blood donors. J Gen Virol. 2009;90:1622–1628. doi: 10.1099/vir.0.009738-0. [DOI] [PubMed] [Google Scholar]
- 139.Teshale EH, Ramachandran S, Xia GL, Roberts H, Groeger J, Barry V, Hu DJ, Holmberg SD, Holtzman D, Ward JW, et al. Genotypic distribution of hepatitis B virus (HBV) among acute cases of HBV infection, selected United States counties, 1999-2005. Clin Infect Dis. 2011;53:751–756. doi: 10.1093/cid/cir495. [DOI] [PubMed] [Google Scholar]
- 140.Mulyanto SN, Wahyono A, Jirintai S, Takahashi M, Okamoto H. Analysis of the full-length genomes of novel hepatitis B virus subgenotypes C11 and C12 in Papua, Indonesia. J Med Virol. 2011;83:54–64. doi: 10.1002/jmv.21931. [DOI] [PubMed] [Google Scholar]
- 141.Foupouapouognigni Y, Mba SA, Betsem à Betsem E, Rousset D, Froment A, Gessain A, Njouom R. Hepatitis B and C virus infections in the three Pygmy groups in Cameroon. J Clin Microbiol. 2011;49:737–740. doi: 10.1128/JCM.01475-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Stanojević B, Osiowy C, Schaefer S, Bojović K, Blagojević J, Nešić M, Yamashita S, Stamenković G. Molecular characterization and phylogenetic analysis of full-genome HBV subgenotype D3 sequences from Serbia. Infect Genet Evol. 2011;11:1475–1480. doi: 10.1016/j.meegid.2011.05.004. [DOI] [PubMed] [Google Scholar]
- 143.Ma MR, Ha XQ, Ling H, Wang ML, Zhang FX, Zhang SD, Li G, Yan W. The characteristics of the synonymous codon usage in hepatitis B virus and the effects of host on the virus in codon usage pattern. Virol J. 2011;8:544. doi: 10.1186/1743-422X-8-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Mitchell T, Armstrong GL, Hu DJ, Wasley A, Painter JA. The increasing burden of imported chronic hepatitis B--United States, 1974-2008. PLoS One. 2011;6:e27717. doi: 10.1371/journal.pone.0027717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zacharakis GH, Koskinas J, Kotsiou S, Papoutselis M, Tzara F, Vafeiadis N, Archimandritis AJ, Papoutselis K. Natural history of chronic HBV infection: a cohort study with up to 12 years follow-up in North Greece (part of the Interreg I-II/EC-project) J Med Virol. 2005;77:173–179. doi: 10.1002/jmv.20434. [DOI] [PubMed] [Google Scholar]
- 146.Lankarani KB. The necessity of booster vaccination after neonatal hepatitis B vaccination. Hepat Mon. 2011;11:419–421. [PMC free article] [PubMed] [Google Scholar]
- 147.Alavian SM, Fallahian F, Lankarani KB. The changing epidemiology of viral hepatitis B in Iran. J Gastrointestin Liver Dis. 2007;16:403–406. [PubMed] [Google Scholar]
- 148.Contini C, Badia L, Cultrera R, Grilli A, De Togni A. Epidemiological, clinical and laboratory features of chronic hepatitis B infection in a cohort of immigrant and Italian patients from Ferrara, Italy. Ann Hepatol. 2012;11:862–869. [PubMed] [Google Scholar]
- 149.Dervisevic S, Ijaz S, Chaudry S, Tedder RS. Non-A hepatitis B virus genotypes in antenatal clinics, United Kingdom. Emerg Infect Dis. 2007;13:1689–1693. doi: 10.3201/eid1311.070578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Micalessi MI, De Cock L, Vranckx R. Hepatitis B virus (HBV) genotyping in Belgian patients with chronic HBV infection. Clin Microbiol Infect. 2005;11:499–501. doi: 10.1111/j.1469-0691.2005.01144.x. [DOI] [PubMed] [Google Scholar]
- 151.Khan A, Tanaka Y, Saito H, Ebinuma H, Sekiguchi H, Iwama H, Wakabayashi G, Kamiya T, Kurbanov F, Elkady A, et al. Transmission of hepatitis B virus (HBV) genotypes among Japanese immigrants and natives in Bolivia. Virus Res. 2008;132:174–180. doi: 10.1016/j.virusres.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 152.Arababadi MK, Hassanshahi G, Pourfathollah AA, Zarandi ER, Kennedy D. Post-transfusion occult hepatitis B (OBI): a global challenge for blood recipients and health authorities. Hepat Mon. 2011;11:714–718. doi: 10.5812/kowsar.1735143X.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Amini-Bavil-Olyaee S, Vucur M, Luedde T, Trautwein C, Tacke F. Differential impact of immune escape mutations G145R and P120T on the replication of lamivudine-resistant hepatitis B virus e antigen-positive and -negative strains. J Virol. 2010;84:1026–1033. doi: 10.1128/JVI.01796-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Mahabadi M, Norouzi M, Alavian SM, Samimirad K, Azad TM, Saberfar E, Mahmoodi M, Ramezani F, Karimzadeh H, Malekzadeh R, et al. Drug-related mutational patterns in hepatitis B virus (HBV) reverse transcriptase proteins from Iranian treatment-naïve chronic HBV patients. Hepat Mon. 2013;13:e6712. doi: 10.5812/hepatmon.6712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Bottecchia M, Madejón A, Puente S, García-Samaniego J, Rivas P, Herrero D, Soriano V. Detection of hepatitis B virus genotype A3 and primary drug resistance mutations in African immigrants with chronic hepatitis B in Spain. J Antimicrob Chemother. 2011;66:641–644. doi: 10.1093/jac/dkq484. [DOI] [PubMed] [Google Scholar]
- 156.Reuben A. The thin red line. Hepatology. 2002;36:770–773. doi: 10.1002/hep.510360341. [DOI] [PubMed] [Google Scholar]
- 157.Italiano CM, Speers DJ, Chidlow GR, Dowse GK, Robertson AG, Flexman JP. Hepatitis B outbreak following a mass-casualty incident, Australia. J Infect Dis. 2011;204:400–407. doi: 10.1093/infdis/jir269. [DOI] [PubMed] [Google Scholar]
- 158.Fung SK, Wong FS, Wong DK, Hussain MT, Lok AS. Hepatitis B virus genotypes, precore and core promoter variants among predominantly Asian patients with chronic HBV infection in a Canadian center. Liver Int. 2006;26:796–804. doi: 10.1111/j.1478-3231.2006.01297.x. [DOI] [PubMed] [Google Scholar]


