ABSTRACT
Loop-mediated isothermal amplification (LAMP) combined with enzyme-linked immunosorbent assay (LAMP–ELISA) and with lateral flow dipstick (LAMP–LFD) are rapid, sensitive and specific methods for the visual detection of clinical pathogens. In this study, LAMP–ELISA and LAMP–LFD were developed for the visual detection of canine parvovirus (CPV). For LAMP, a set of four primers (biotin-labeled forward inner primers) was designed to specifically amplify a region of the VP2 gene of CPV. The optimum time and temperature for LAMP were 60 min and 65°C, respectively. The specific capture oligonucleotide probes, biotin-labeled CPV probe for LAMP–ELISA and fluorescein isothiocyanate-labeled CPV probe for LAMP–LFD were also designed for hybridization with LAMP amplicons on streptavidin-coated wells and LFD strips, respectively. For the comparison of detection sensitivity, conventional PCR and LAMP for CPV detection were also performed. The CPV detection limits by PCR, PCR–ELISA, LAMP, LAMP–ELISA and LAMP–LFD were 102, 102, 10−1, 10−1 and 10−1 TCID50/ml, respectively. In tests using artificially contaminated dog fecal samples, the samples with CPV inoculation levels of ≥1 TCID50/ml gave positive results by both LAMP–ELISA and LAMP–LFD. Our data indicated that both LAMP–ELISA and LAMP–LFD are promising as rapid, sensitive and specific methods for an efficient diagnosis of CPV infection.
Keywords: canine, ELISA, lateral flow dipstick, loop-mediated amplification, parvovirus
Canine parvovirus type 2 (CPV-2), a member of the Parvovirus genus of the family Parvoviridae, was first identified in 1978 [1, 14]. It has a small non-enveloped and icosahedral capsid containing single-stranded DNA. The CPV genome is approximately 5.2-kb [28] long and contains two open reading frames (ORFs). ORF1 encodes two non-structural proteins (NS1 and NS2) through alternative splicing of the transcribed viral mRNA, and ORF2 encodes two structural proteins (VP1 and VP2) [33]. The VP2 is the major capsid protein containing the antigenic determination sites to play an important role in determining CPV antigenic properties [17, 35]. CPV-2 is an epidemic enteric pathogen of dogs and causes acute gastroenteritis and lymphopenia mostly in puppies [26, 27]. A few years after CPV-2 outbreak, two new antigenic variants were characterized and termed as CPV-2a and CPV-2b to be the predominant type and were spread and distributed all over the world rapidly [28, 29]. In 2000, a new antigenic type of CPV was detected in Italy and rapidly spread to several countries [7].
It is difficult to diagnose CPV infection from the main clinical signs, such as vomiting and diarrhea, because these symptoms are common to other enteric diseases [12]. Some conventional methods used to detect CPV include electron microscopy, virus isolation [36], latex agglutination [3, 34], hemagglutination [19, 20, 36] and enzyme-linked immunosorbent assay (ELISA) [11, 15]. Many of these methods are effective and accurate in detecting viral infections in the laboratory. However, they are often laborious, time-consuming, expensive and/or lack specificity and sensitivity. With advances in molecular detection techniques, PCR [12, 37] and real-time PCR [8, 10, 14, 16] have been established for CPV diagnosis with a varying degree of sensitivity and specificity [9]. However, these techniques require skilled technicians and can only be performed in a diagnostic or commercial laboratory by employing specialized equipment not commonly available to veterinary clinics and impractical for use in the field. A rapid, accurate, sensitive, simple and economical on-site method is therefore needed for CPV detection, and one of the candidate methods is the technology of loop-mediated isothermal amplification (LAMP). The major advantages of LAMP comparing with conventional PCR are that (1) LAMP does not require a thermal cycler and can be performed simply with a heating block and/or water bath, (2) the reaction result of LAMP can be observed and justified by naked eyes, and (3) LAMP detection has a high sensitivity and can be completed within 1 hr under well experimental operation [21, 22, 24].
LAMP, first developed by Notomi et al. [24], is a powerful nucleic acid amplification technique that is sensitive and fast. It easily amplifies target sequences under isothermal conditions usually ranging from 60 to 65°C. In LAMP, specific primers are combined with Bst polymerase, which has strand displacement activity, to produce a large amount of amplified target DNA in <1 hr. The amplified product can be analyzed by gel electrophoresis and/or visual inspection of turbidity resulting from the formation of the magnesium pyrophosphate by-product [22]. Accordingly, LAMP technology has been widely used for the detection of different pathogens [21]. To date, LAMP has been developed to diagnose canine viruses, including canine distemper virus (CDV) [5], rabies virus [4], influenza virus [13] and parvovirus [6, 23, 30]. LAMP has been found to be promising as a sensitive and cost-effective method for CPV detection.
Recent studies have shown that LAMP combined with ELISA and with lateral flow dipstick (LFD) are promising for application to pathogen diagnosis by visual field testing because they are nearly instrument-free [2, 31, 32]. However, the application of these detection methods remains to be explored in veterinary clinics. The objective of this study was to appropriately develop and evaluate a detection system based on the application of LAMP in conjunction with ELISA and LFD for convenient visual detection of CPV with high sensitivity and specificity.
MATERIALS AND METHODS
CPV strain and genomic DNA preparation: The culture supernatant of CPV Strain C154 (CPV-2b) containing viral particles at 1 × 107 TCID50/ml was kindly provided by Dr. C. K. Chuang of the Agricultural Technology Research Institute, Taiwan. The CPV suspension was serially diluted tenfold (106–10−2 TCID50/ml), and the CPV genomic DNA was prepared from 100 µl of CPV supernatant using Viral Nucleic Acid Extraction Kit II (GeneDirex, Las Vegas, NV, U.S.A.), according to the manufacturer’s instructions. The extracted DNA was subjected to PCR and LAMP.
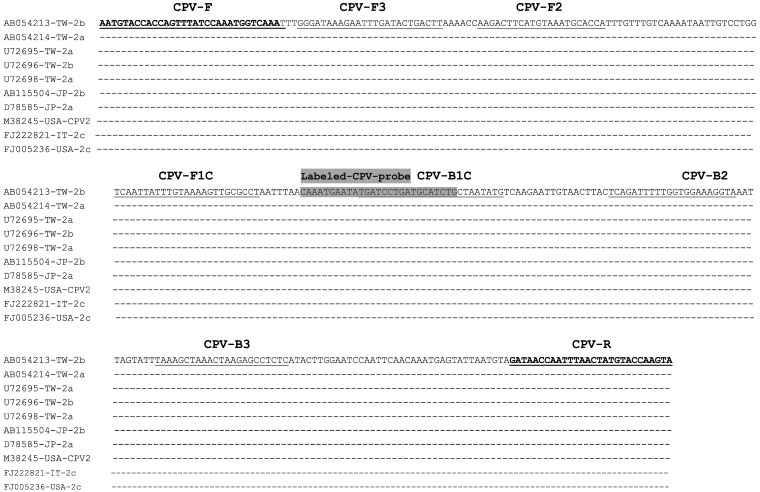
Design and synthesis of the primers and probe: According to the published VP2 gene sequences of CPV [38] in GenBank (accession numbers including AB054213, AB054214, U72695, U72696, U72698, AB115504, D78585, M38245, FJ222821 and FJ005236) (including type 2a, 2b and 2c), sequences were aligned using the software program DNASTAR (Madison, WI, U.S.A.). The conserved sequence within the VP2 gene with high homology was selected as the target for designing the LAMP primer set using the software PrimerExplorer V4 (http://primerexplorer.jp/elamp4.0.0/index.html). A set of four specific primers was synthesized by MissionBiotech Inc. (Taipei, Taiwan). The primer pair, CPV-F (5′-biotin labeled) and CPV-R, was used as the PCR primer to amplify a 319-bp fragment (Fig. 1).
Fig. 1.
Nucleotide sequences alignment of the VP2 gene from different CPV isolates including antigenic variants of type 2a, 2b and 2c. There were 5 (AB054213, AB054214, U72695, U72696 and U72698), 2 (AB115504 and D78585), 2 (M38245 and FJ005236) and 1 (FJ222821) CPV isolates from Taiwan, Japan, United State America and Italy, respectively. Partial sequences of the VP2 were aligned. The designed nucleic acid sequences of labeled-CPV-probe and primers were indicated, boxed and/or bolded.
The region selected as the internal capture probe was located inside the forward inner primer (FIP)/backward inner primer (BIP) LAMP-amplified fragment of VP2 and labeled with biotin at the 5′ end (MDBio Inc., Taipei, Taiwan). The optimum hybridization temperatures for capture probes [biotin-labeled probe for LAMP–ELISA or PCR–ELISA and fluorescein isothiocyanate (FITC)-labeled probe for LFD] were also evaluated.
PCR: PCR was performed in a 25-µl volume containing 1 µl of CPV template DNA; 4 µM of each primer; 200 µM each of dATP, dCTP, dGTP and dTTP (Promega, Madison, WI, U.S.A.); 5 µl of 5 × PCR buffer (100 mM Tris-HCl, 9 mM MgCl2, 110 mM NH4Cl, 110 mM KCl, 0.3% IGEPAL CA-630 and 0.25% Tween 20, pH 8.9); 0.5 U of OneTaq DNA polymerase (New England BioLabs, Ipswich, MA, U.S.A.); and H2O. The PCR conditions were as follows: denaturation at 95°C for 5 min, followed by 35 cycles of denaturation at 95°C for 30 sec, annealing at 58°C for 45 sec, extension at 72°C for 45 sec and final extension at 72°C for a further 10 min. The amplified PCR products were analyzed by electrophoresis on a 2% agarose gel containing 0.5 µg/ml ethidium bromide.
LAMP: LAMP was performed in a 20-µl volume containing 1.2 µM each of FIP and BIP; 0.3 µM each of the F3 and B3 primers; 10 µl of the 2 × reaction mixture (40 mM Tris-HCl, 20 mM KCl, 16 mM MgSO4, 20 mM (NH4)2SO4, 0.2% Tween 20, 1.6 M betaine and 2.8 mM dNTPs, pH 8.8); 1 µl template DNA; and 1 µl of Bst DNA polymerase (DNA Amplification Kit; Eiken Chemical Co., Tochigi, Japan). The reaction temperature was optimized by incubating the LAMP mixture at 59, 61, 63 or 65°C for 60 min. The reaction time was optimized by incubating the mixture for 15, 30, 45 and 60 min at a predetermined temperature (65°C). After heating at 80°C for 5 min to terminate the LAMP reaction, the LAMP products were analyzed by electrophoresis on a 2% agarose gel stained with ethidium bromide.
CPV detection by the LFD assay: A generic LFD strip (Milenia GenLine HybriDetect; Milenia Biotec GmbH, Gieβen, Germany) was used for the LFD assay. Under the test condition, a specific DNA probe was designed from the sequences between the F1P and B1P regions of LAMP amplicons (Table 1). According to the detection system established, the DNA probe was labeled with biotin or FITC at the 5′ end.
Table 1. The information of primers and probes used in this study for the sequence detection of VP2 gene of canine parvovirus.
| Primers and probes | Nucleotide position | Sequences (5’→3’) |
|---|---|---|
| Primer sets for LAMP | ||
| CPV-F3 | 1409–1433 | GGGATAAAGAATTTGATACTGACTT |
| CPV-B3 | 1627–1605 | GAGAGGCTCTTAGTTTAGCTTTA |
| Biotin-CPV-FIP (F1C-F2) | 1512–1488/1440–1461 | Biotin-AGGCGCAACTTTTACAAATAATTGAAAGACTTCATGTAAATGCACCA |
| CPV-BIP (B1C-B2) | 1530–1554/1595–1573 | TGATCCTGATGCATCTGCTAATATGTACCTTTCCACCAAAAATCTGA |
| Primer pair for PCR | ||
| Biotin-CPV-F | 1375–1398 | Biotin-AATGTACCACCAGTTTATCCAAAT |
| CPV-R | 1666–1693 | TACTTGGTACATAGTTAAATTGGTTATC |
| Probe for LAMP-ELISA | ||
| Biotin-CPV-probe | 1520–1546 | Biotin-CAAATGAATATGATCCTGATGCATCTG |
| Probe for LAMP-LFD | ||
| FITC-CPV-probe | 1520–1546 | FITC-CAAATGAATATGATCCTGATGCATCTG |
For this assay, 20 pmol of the FITC-labeled CPV probe was added to the biotin-labeled LAMP amplicons and hybridized at 58°C for 15 min. After hybridization, 8 µl of the reaction solution was mixed with 120 µl assay buffer, and the LFD strip was dipped into it for 5 min. The detection results were determined by observing the control and test lines on the LFD strips.
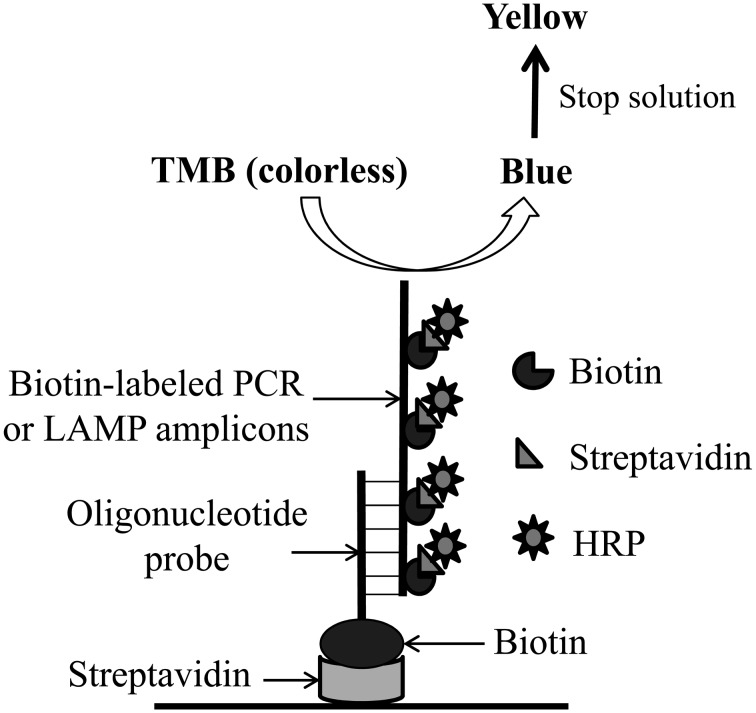
PCR and LAMP amplicon detection combined with ELISA: Detection using PCR–ELISA and LAMP–ELISA (Fig. 2) was conducted as described by Ravan et al. [32]. Each well of a 96-well microtiter plate (Nunc A/S, Roskilde, Denmark) was coated with 100 µl streptavidin (5 mg/ml; Sigma, St. Louis, MO, U.S.A.) in 10 mM phosphate-buffered saline (PBS, pH 7, Sigma) and refrigerated at 4°C overnight. The streptavidin-unbound sites were blocked with blocking solution [1% (w/v) bovine serum albumin in PBS] for 1 hr at room temperature. The plate was washed three times with PBST (PBS containing 0.05% Tween 20). Subsequently, each well received 100 µl of 2.5 µM biotin-labeled CPV probe diluted in PBST. The plates were incubated at 37°C for 1 hr. After washing three times with PBST, the plate was subjected to PCR–ELISA or LAMP–ELISA.
Fig. 2.
Schematic representation of PCR-ELISA and LAMP-ELISA assays. The biotin-labeled specific oligonucleotide probes are applied onto the surface of streptavidin-coated well and used to hybridize with biotin-labeled LAMP amplicons or biotin-labeled PCR products. After the nonspecific binding LAMP amplicons or biotin-labeled PCR products were removed, the conjugates of streptavidin- horseradish peroxidase (HRP) were added to perform the ELISA detection.
After amplification, 5 µl of the PCR or LAMP amplicons were diluted with 95 µl hybridization solution (50 mM phosphate buffer and 2 mM EDTA, pH 7.2) and denatured at 95°C for 5 min. After cooling on ice, the denatured biotin-labeled PCR or LAMP amplicons were added to the capture probe–streptavidin-coated well and incubated at 58°C for 1 hr. After washing three times with PBST, 100 µl of a 1:1,000 dilution of streptavidin–horseradish peroxide (PerkinElmer, Waltham, MA, U.S.A.) in PBS was added to each well, and the plates were incubated at 37°C for 45 min. The wells were then washed five times with PBST, and the 3,3′,5,5′-tetramethylbenzidine liquid substrate system for ELISA (100 µl, Sigma) was added to each well. After incubation in the dark at room temperature, the reaction was stopped by adding 50 µl of 2 M H2SO4. Absorbance was detected at 450 nm using the TECAN/Sunrise ELISA reader (Advance Biotechnology, Taipei, Taiwan).
Preparation of DNA from fecal samples artificially contaminated with CPV: Five grams of a CPV-free dog fecal sample in 50 ml of sterile PBS was completely mixed by vortexing and distributed into 50 vials. Each vial contained a 1-ml aliquot of the fecal sample. The aliquots were inoculated with 100 µl CPV suspension (103–10−1 TCID50/ml). The samples were centrifuged at 6,000 × g for 15 min, and the supernatant was collected. The supernatant was used for DNA extraction, and the samples were pretreated by rapid boiling and chilling as described previously [23]. Template DNA was prepared from the CPV-contaminated fecal samples using an Ultraclean Faecal DNA isolation kit (Mo Bio laboratories Inc., Carlsbad, CA, U.S.A.), according to the manufacturer’s instructions.
RESULTS
Establishment of the LAMP assay for CPV detection: Several Taiwan isolates and strains from Japan, Italy and USA including antigenic types 2a, 2b and 2c were selected for developing effective visual detection methods for different CPV strains [38]. Given that the sequence of the VP2 gene of CPV is frequently used as the target for CPV detection, the PCR/LAMP primers were designed on the basis of the alignment of VP2 gene sequences from the selected strains.
According to alignment analysis, a highly conserved sequence was selected as a suitable target for designing the PCR/LAMP primers (Fig. 1). A set of four primers capable of recognizing six distinct regions on the target sequence was designed: two outer primers (CPV-F3 and CPV-B3) and two inner primers (CPV-FIP and CPV-BIP). For ELISA and LFD analyses, the CPV-FIP primer was labeled with biotin at the 5′ end. Locations and sequences of the primers and specific probes for CPV detection are shown in Fig. 1 and Table 1, respectively.
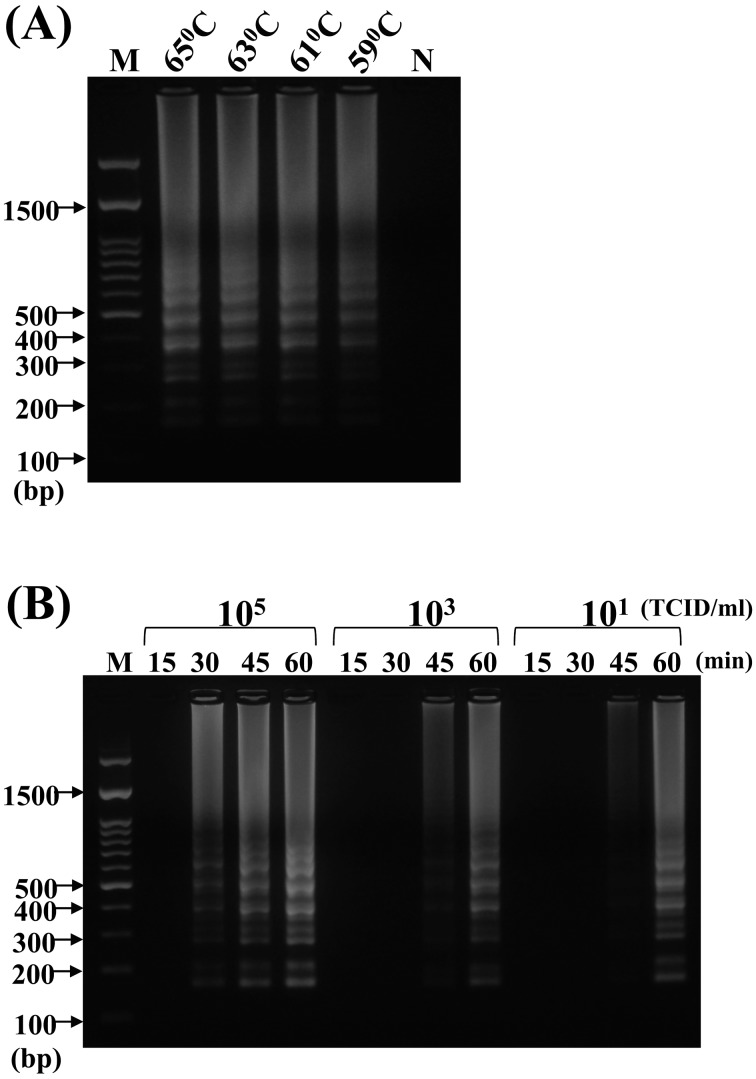
Optimization of the LAMP assay: When LAMP was performed to determine the optimal temperature and time of reaction, a ladder-like pattern of the LAMP products appeared on the 2% agarose gel at 59, 61, 63 and 65°C (Fig. 3A). A slight difference in band clarity was observed with increasing reaction temperature. We selected 65°C as the optimal working temperature for LAMP, because of the intense signal and specificity observed at the higher temperature. The LAMP product could be amplified as early as 30 min when the template DNA concentration was high (isolated from 105 TCID50/ml of CPV), whereas at a low template DNA concentration (10 TCID50/ml of CPV), the amplified LAMP was observed at 45 min at least (Fig. 3B). We selected the optimal reaction condition of 65°C for 60 min to ensure positive detection at a low template DNA concentration.
Fig. 3.
Optimization of loop-mediated isothermal amplification (LAMP) assay for the detection of VP2 sequence from CPV genomic DNA. (A) For the test of optimal temperature, reaction temperature of 65, 63, 61 and 59°C was tested using the genomic DNA extracted from 100 µl of CPV suspension (106 TCID50/ml) as template. (B) For the interactive test of reaction time and template DNA concentration, the CPV genomic DNAs were extracted from 105, 103 and 101 TCID50/ml, and then, each CPV genomic DNA was used in the LAMP reaction for 15, 30, 45 and 60 min. “N” indicated the negative control. “M” indicated the 100-bp ladder DNA marker, and the molecular of partial DNA ladders were also noted.
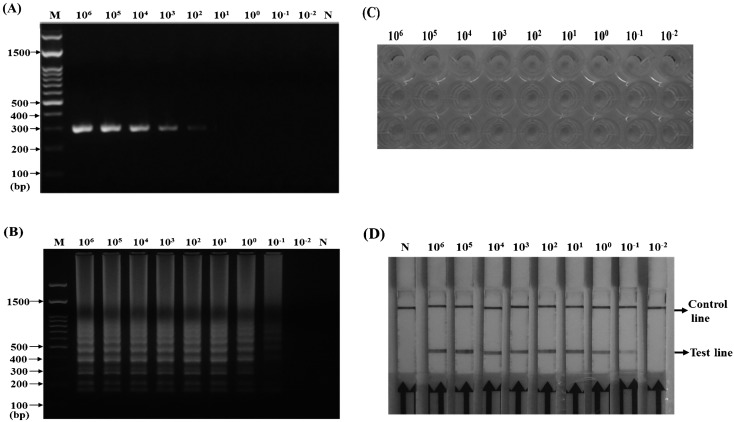
Sensitivity of PCR, LAMP and LAMP combined with the LFD assay: To determine the sensitivity of CPV detection by LAMP and LAMP combined with LFD, the CPV stock (107 TCID50/ml) was serially diluted tenfold, and the CPV genomic DNA was prepared from 100 µl of CPV supernatant. Each viral DNA (1 µl) was used as a template for conventional PCR or LAMP. In PCR, the respective dilutions were subjected to thermal cycling using the primer pair of biotin-labeled CPV-F and CPV-R (Table 1), which amplified a 319-bp fragment from the VP2 gene of CPV (Fig. 4A). In LAMP, a 20-µl reaction mixture was the same as for conventional LAMP; however, a 5′-biotinylated FIP was used to replace the FIP primer. The reaction was performed at 65°C for 60 min, and the products were analyzed by electrophoresis on a 2% agarose gel and the LFD assay. The results showed that PCR and LAMP could detect CPV at concentrations of 102 TCID50/ml and 10−1 TCID50/ml, respectively (Fig. 4A and 4B) and that LAMP–LFD was also able to detect CPV at concentrations as low as 10−1 TCID50/ml (Fig. 4C). Thus, LAMP and LAMP–LFD were both 1,000 times more sensitive than conventional PCR.
Fig. 4.
Sensitivity of PCR (A), loop-mediated isothermal amplification (LAMP) (B), LAMP combined with enzyme-linked immunosorbent assay (ELISA) (C) and LAMP combined with lateral flow dipstick (LFD) (D) assays for the detection of VP2 sequence from CPV genomic DNA. The templates of CPV genomic DNA were extracted from CPV suspension ranging from 106 to 10−2 TCID50/ml. “N” indicated the negative control, and “M” indicated the 100-bp DNA ladder marker.
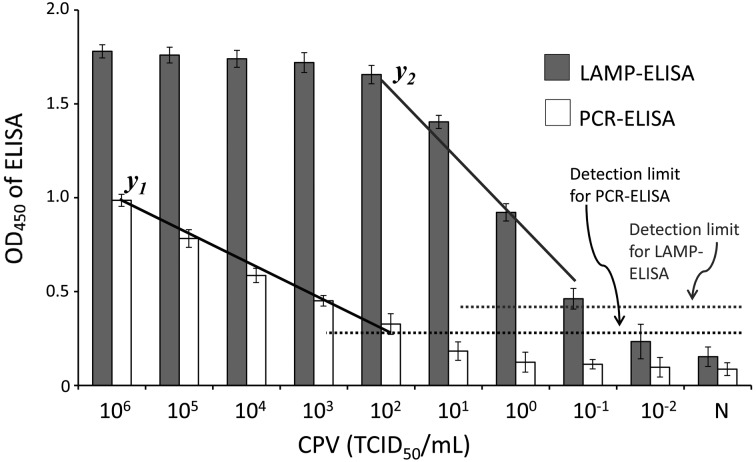
Sensitivity of PCR and LAMP combined with ELISA: The genomic DNAs extracted from serially tenfold diluted CPV suspensions were used as templates for biotin-labeled PCR and LAMP. As described in Materials and methods, the labeled PCR products and LAMP amplicons were analyzed by ELISA. The overall scheme of PCR–ELISA and LAMP–ELISA is shown in Fig. 2. The PCR–ELISA and LAMP–ELISA results were spectrophotometrically obtained using a microplate reader that provided an absorbance value corresponding to the amount of labeled PCR products or labeled LAMP amplicons attached to the surface of microtiter plate wells. In PCR–ELISA, a linear relationship (y1=−0.4064x + 2.127, R2=0.9833) was found at higher CPV titers, from 106 to 102 TCID50/ml. In LAMP–ELISA, CPV dilutions corresponding to 106–102 TCID50/ml gave a plateau absorbance value of OD450 ranging from 1.78 to 1.66, and a linear relationship (y2=−0.165x + 1.1216, R2=0.9869) was observed with CPV titers decreasing from 102 to 10−1 TCID50/ml (Fig. 5). It is indicated the detection limit of the developed PCR-ELISA and LAMP-ELISA was 102 and 10−1 TCID50/ml, respectively.
Fig. 5.
Semi-quantification and limitation of LAMP-ELISA and PCR-ELISA for the detection of VP2 sequence from CPV genomic DNA. Different titers of CPV (106 − 10−2 TCID50/ml) were applied to prepare the genomic DNA and then applied for PCR or LAMP amplification. The PCR products and LAMP amplicoms were used for the ELISA assays as the demonstration in Fig. 2. Each value was derived from three independent detections, and the error bars mean standard deviation (SD).
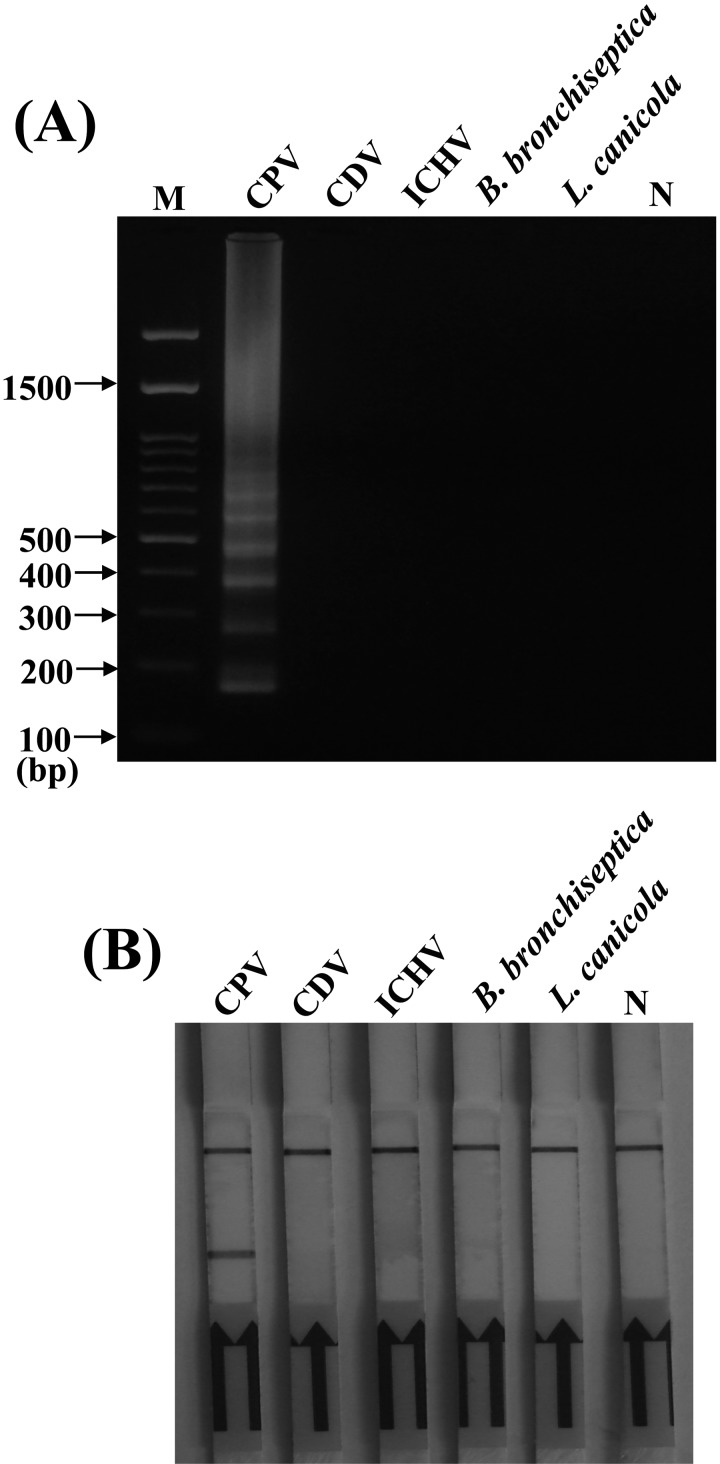
Specificity of LAMP detection by gel electrophoresis and LFD: In order to evaluate the specificity of established LAMP, potential cross-reactions were performed using DNA/RNA extracted from different pathogens including CDV, infectious canine hepatitis virus (ICHV), Leptospira canicola and Bordetella bronchiseptica. Biotin-labeled LAMP amplicons were analyzed by 2% agarose gel electrophoresis with ethidium bromide and by the LFD assay. As shown in Fig. 6A, cross-amplification tests using templates from CDV, ICHV, L. canicola and B. bronchiseptica showed that no amplicons were detected, whereas the reaction using the CPV template gave a positive result. The similar results were also observed in the LFD assay; the test band appeared only for CPV detection (Fig. 6B). These results indicated that the LAMP-based assay methods developed in this study were specific for CPV.
Fig. 6.
Specificity of loop-mediated isothermal amplification (LAMP) (A) combined with lateral flow dipstick (LFD) (B) for the CPV detection. CPV genomic DNA, canine distemper virus (CDV) RNA, canine hepatitis virus (ICHV) genomic DNA, B. bronchiseptica genomic DNA and L. canicola genomic DNA were applied in the LAMP detections. “N” indicated the negative control. “M” indicated the DNA marker.
CPV detection in artificially contaminated dog fecal samples: CPV was artificially inoculated into dog fecal samples and subjected to CPV detection by PCR–ELISA, LAMP–ELISA and LAMP–LFD. According to the results shown in Table 2, the fecal samples with CPV inoculated at ≥102 TCID50/ml gave positive results by PCR–ELISA, and both LAMP–ELISA and LAMP–LFD provided positive results at ≥1 TCID50/ml of CPV. Importantly, the positive signals produced by all of the detection methods including PCR–ELISA, LAMP–ELISA and LAMP–LFD could be easily read with the naked eye.
Table 2. Sensitivity of PCR-ELISA, LAMP-ELISA and LAMP-LFD assays for the CPV detection in artificially contaminated fecal sample of dog.
| Methods | PBS (negative control) |
CPV inoculation
(TCID50/ml) d) |
||||
|---|---|---|---|---|---|---|
| 10–1 | 1 | 101 | 102 | 103 | ||
| PCR-ELISAa) | − | − | − | − | + | + |
| LAMP-ELISAb) | − | − | + | + | + | + |
| LAMP-LFDc) | − | − | + | + | + | + |
a) Positive was determined by the value of OD450>0.261 determined by a spectrophotometry (as shown in Fig. 6). b) Positive was determined by the value of OD450>0.447 determined by a spectrophotometry (as shown in Fig. 6). c) Positive was determined by yielding test band on LFD strip. d) Each CPV inoculation detected by different assays was performed from three independent fecal samples and got the same detection result.
DISCUSSION
Several detection methods have been developed to detect CPV proteins and nucleic acids, and many of these tests are effective and accurate in detecting the viral infection in laboratory. However, they require expensive equipment and are often laborious and time-consuming. Early and rapid diagnosis is necessary so that CPV-infected dogs can be isolated to prevent the spread of the disease and to administer supportive treatment for reducing morbidity and mortality. Therefore, a novel nucleic acid amplification method, termed LAMP, which amplifies specific DNA sequences under isothermal conditions within a few hours, was developed as a simple, rapid, specific and cost-effective alternative [24]. LAMP is an excellent technology for the detection of nucleic acids present at very low levels in biological and environmental samples with its remarkable sensitivity. Therefore, LAMP could be applied to gene analysis and study of genetic traits and mostly to detect etiological cause of infections.
In the present study, the CPV detection limit by PCR, PCR-ELISA, LAMP, LAMP–ELISA and LAMP–LFD was 102, 102, 10−1, 10−1 and 10−1 TCID50/ml, respectively. The results indicated that the sensitivity of LAMP–ELISA and LAMP–LFD for CPV detection is higher than that of conventional molecular methods. The results could be easily visualized with the naked eye. In addition to reducing assay time and elevating the sensitivity, combination of LAMP with ELISA or with LFD confirmed amplicon identity by hybridization and eliminates the need to handle, such as ethidium bromide.
Our results confirmed the previous report of CPV detection from CPV-suspected fecal samples by LAMP having a detection limit of 10−1 TCID50/ml [6] and to show that LAMP is more sensitive than PCR-based tests. We have demonstrated this consistently in the present study. Our results also showed that LAMP–ELISA was more sensitive than PCR–ELISA; a positive signal was detected at 102 TCID50/ml by PCR–ELISA, while LAMP–ELISA provided a positive signal at 10−1 TCID50/ml. Furthermore, the performance of LAMP and PCR diagnostic systems has been extensively compared by several groups. According to the report of Cho et al. [6], the detection rates of CPV-suspected fecal samples by LAMP and PCR were 80% and 74%, respectively. Mukhopadhyay et al. [23] also compared the detection rates of CPV by LAMP and PCR from clinical samples to be 74.28% and 57.85%. In general, LAMP has been found to have sensitivity similar or superior to that of PCR [18, 25, 32]. We have demonstrated this consistency of the 12 clinical samples tested and 10 (83%) and 8 (67%) were detected positive for CPV by LAMP and PCR, respectively (data not shown). The positive results of CPV detection could be presented fully when LAMP combined with ELISA and LFD. However, immunization with modified-live CPV vaccine may result in shedding of the virus for a period of 3 to 14 days post vaccination. Therefore, it is possible that there is a positive result that can be produced by a recent CPV vaccination with our developed LAMP–ELISA and LAMP–LFD assays.
In the present report, we described the development of LAMP–ELISA and LAMP–LFD diagnostic systems with an assay time of <3 hr for CPV levels of clinical concern with a pretty high sensitivity. We have known the most commercial test kits for CPV diagnosis are fabricated by the immunochromatographic assay or ELISA technology. The detection by the kits can be completed within mins, but the detection limit of the kits is approximate 103 TCID50/ml of CPV in canine feces. The purpose of our study was to develop a highly sensitive assay used to detect the canine CPV as possible as early after the virus infection. Early detection is a key in the control of virus transmission among dogs. Although the overall detection time is about 3 hr including sample preparation and detection, the test procedure of the developed LAMP–ELISA and LAMP–LFD assays is easy to be performed and can be used in field test. Our system comprised the amplification of a part of VP2 sequences that is unique to CPV, followed by hybridization to a specific probe for exact identification of CPV. Our results confirmed the results of recent reports that indicated LAMP–ELISA or LAMP–LFD to be highly-sensitive methods that can be easily applied for the visual detection of clinical pathogens [2, 32]. The high sensitivity of LAMP–ELISA and LAMP–LFD may allow the identification of dogs shedding CPV at low titers in their feces, helping veterinarians to adopt adequate measures of prophylaxis to prevent CPV infection.
Our data also showed that the CPV detection limit for artificially contaminated fecal samples using LAMP–ELISA and LAMP–LFD was ≥1 TCID50/ml. This sensitivity was lower than the results shown in Figs. 4, 5 and Table 2. We suggest that the lower sensitivity of CPV detection in fecal samples than in PBS may be related to loss of the virus during isolation of CPV from the fecal samples. It may also be related to the presence of intestinal cells and bacteria along with CPV in the prepared DNA, which might have reduced the efficiency of LAMP in amplifying the target CPV DNA from the samples. However, our results indicated that both LAMP–ELISA and LAMP–LFD developed in the present study are applicable to CPV detection in naturally contaminated fecal samples.
In conclusion, when LAMP was combined with ELISA or with LFD, the detection signal of LAMP amplicons could be spectrophotometrically obtained and easily read with the naked eye and without agarose gel electrophoresis. The assays could be completed within 3 hr. To our knowledge, this is the first report employing LAMP combined with ELISA or LFD for detection of CPV. These results indicated that a simple and cost-effective LAMP-based technique can be developed into a rapid and reliable molecular diagnostic method with potential for routine use in the clinical detection of CPV and other veterinary clinical pathogens.
ACKNOWLEDGMENTS
This study was supported by the grant of 102-EC-17-A-02-04-0454 from the Ministry of Economic Affairs of Executive Yuan, Taiwan, R.O.C. The authors gratefully thank Dr. Chin-Kai Chuang for kindly providing CPV.
REFERENCES
- 1.Appel M. J., Scott F. W., Carmichael L. E.1979. Isolation and immunization studies of a canine parco-like virus from dogs with haemorrhagic enteritis. Vet. Rec. 105: 156–159. doi: 10.1136/vr.105.8.156 [DOI] [PubMed] [Google Scholar]
- 2.Arunrut N., Seetang-Nun Y., Phromjai J., Panphut W., Kiatpathomchai W.2011. Rapid and sensitive detection of Laem-Singh virus by reverse transcription loop-mediated isothermal amplification combined with a lateral flow dipstick. J. Virol. Methods 177: 71–74. doi: 10.1016/j.jviromet.2011.06.020 [DOI] [PubMed] [Google Scholar]
- 3.Bodeus M., Cambiaso C., Surleraux M., Burtonboy G.1988. A latex agglutination test for the detection of canine parvovirus and corresponding antibodies. J. Virol. Methods 19: 1–12. doi: 10.1016/0166-0934(88)90002-X [DOI] [PubMed] [Google Scholar]
- 4.Boldbaatar B., Inoue S., Sugiura N., Noguchi A., Orbina J. R., Demetria C., Miranda M. E., Yamada A.2009. Rapid detection of rabies virus by reverse transcription loop-mediated isothermal amplification. Jpn. J. Infect. Dis. 62: 187–191 [PubMed] [Google Scholar]
- 5.Cho H. S., Park N. Y.2005. Detection of canine distemper virus in blood samples by reverse transcription loop-mediated isothermal amplification. J. Vet. Med. B. Infect. Dis. Vet. Public Health 52: 410–413. doi: 10.1111/j.1439-0450.2005.00886.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cho H. S., Kang J. I., Park N. Y.2006. Detection of canine parvovirus in fecal samples using loop-mediated isothermal amplification. J. Vet. Diagn. Invest. 18: 81–84. doi: 10.1177/104063870601800111 [DOI] [PubMed] [Google Scholar]
- 7.Decaro N., Buonavoglia C.2012. Canine parvovirus-A review of epidemiological and diagnostic aspects, with emphasis on type 2c. Vet. Microbiol. 155: 1–12. doi: 10.1016/j.vetmic.2011.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Decaro N., Elia G., Martella V., Desario C., Campolo M., Di Trani L., Tarsitano E., Tempesta M., Buonavoglia C.2005. A real-time PCR assay for rapid detection and quantitation of canine parvovirus type 2 in the feces of dogs. Vet. Microbiol. 105: 19–28. doi: 10.1016/j.vetmic.2004.09.018 [DOI] [PubMed] [Google Scholar]
- 9.Desario C., Decaro N., Campolo M., Cavalli A., Cirone F., Elia G., Martella V., Lorusso E., Camero M., Buonavoglia C.2005. Canine parvovirus infection: Which diagnostic test for virus? J. Virol. Methods 126: 179–185. doi: 10.1016/j.jviromet.2005.02.006 [DOI] [PubMed] [Google Scholar]
- 10.Elia G., Cavalli A., Desario C., Lorusso E., Lucente M. S., Decaro N., Martella V., Buonavoglia C.2007. Detection of infectious canine parvovirus type 2 by mRNA real-time RT-PCR. J. Virol. Methods 146: 202–208. doi: 10.1016/j.jviromet.2007.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elia G., Desario C., Pezzoni G., Camero M., Brocchi E., Decaro N., Martella V., Buonavoglia C.2012. Recombinant ELISA using baculovirus-expressed VP2 for detection of antibodies against canine parvovirus. J. Virol. Methods 184: 98–102. doi: 10.1016/j.jviromet.2012.05.025 [DOI] [PubMed] [Google Scholar]
- 12.Hirasawa T., Kaneshige T., Mikazuki K.1994. Sensitive detection of canine parvovirus DNA by the nested polymerase chain reaction. Vet. Microbiol. 41: 135–145. doi: 10.1016/0378-1135(94)90143-0 [DOI] [PubMed] [Google Scholar]
- 13.Ito M., Watanabe M., Nakagawa N., Ihara T., Okuno Y.2006. Rapid detection and typing of influenza A and B by loop-mediated isothermal amplification: comparison with immunochromatography and virus isolation. J. Virol. Methods 135: 272–275. doi: 10.1016/j.jviromet.2006.03.003 [DOI] [PubMed] [Google Scholar]
- 14.Kelly W. R.1978. An enteric disease of dogs resembling feline panleulopenia. Aust. Vet. J. 54: 593. doi: 10.1111/j.1751-0813.1978.tb02426.x [DOI] [PubMed] [Google Scholar]
- 15.Kumar M., Nandi S., Chidri S.2010a. Development of a polyclonal antibody-based AC-ELISA and its comparison with PCR for diagnosis of canine parvovirus infection. Virol. Sin. 25: 352–360. doi: 10.1007/s12250-010-3132-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumar M., Nandi S.2010b. Development of a SYBR Green based real-time PCR assay for detection and quantitation of canine parvovirus in faecal samples. J. Virol. Methods 169: 198–201. doi: 10.1016/j.jviromet.2010.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Langeveld J. P. M., Casal J. I., Vela C., Dalsgaard K., Smale S. H., Puijk W. C., Meloen R. H.1993. B-cell epitopes of canine parvovirus: distribution on the primary structure and exposure on the viral surface. J. Virol. 67: 765–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li X., Zhang S., Zhang H., Zhang L., Tao H., Yu J., Zheng W., Liu C., Lü D., Xiang R., Liu Y.2009. A loop-mediated isothermal amplification method targets the phoP gene for the detection of Salmonella in food samples. Int. J. Food Microbiol. 133: 252–258. doi: 10.1016/j.ijfoodmicro.2009.05.027 [DOI] [PubMed] [Google Scholar]
- 19.Mathys A., Mueller R., Pedersen N. C., Theilen G. H.1983. Comparison of hemagglutination and competitive enzyme-linked immunosorbent assay procedures for detecting canine parvovirus in feces. Am. J. Vet. Res. 44: 152–154 [PubMed] [Google Scholar]
- 20.Mochizuki M., San Gabriel M. C., Nakatani H., Yoshida M., Harasawa R.1993. Comparison of polymerase chain reaction with virus isolation and haemagglutination assays for the detection of canine parvoviruses in faecal specimens. Res. Vet. Sci. 55: 60–63. doi: 10.1016/0034-5288(93)90035-E [DOI] [PubMed] [Google Scholar]
- 21.Mori Y., Kanda H., Notomi T.2013. Loop-mediated isothermal amplification (LAMP): recent progress in research and development. J. Infect. Chemother. 19: 404–411. doi: 10.1007/s10156-013-0590-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mori Y., Nagamine K., Tomita N., Notomi T.2001. Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem. Biophys. Res. Commun. 289: 150–154. doi: 10.1006/bbrc.2001.5921 [DOI] [PubMed] [Google Scholar]
- 23.Mukhopadhyay H. K., Amsaveni S., Matta S. L., Antony P. X., Thanislass J., Pillai R. M.2012. Development and evaluation of loop-mediated isothermal amplification assay for rapid and sensitive detection of canine parvovirus DNA directly in fecal specimens. Lett. Appl. Microbiol. 55: 202–209. doi: 10.1111/j.1472-765X.2012.03284.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N., Hase T.2000. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 28: E63. doi: 10.1093/nar/28.12.e63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okamura M., Ohba Y., Kikuchi S., Suzuki A., Tachizaki H., Takehara K., Ikedo M., Kojima T., Nakamura M.2008. Loop-mediated isothermal amplification for the rapid, sensitive, and specific detection of the O9 group of Salmonella in chickens. Vet. Microbiol. 132: 197–204. doi: 10.1016/j.vetmic.2008.04.029 [DOI] [PubMed] [Google Scholar]
- 26.Parrish C. R.1990. Emergence, natural history, and variation of canine, mink, and parvoviruses. Adv. Virus Res. 38: 403–450. doi: 10.1016/S0065-3527(08)60867-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parrish C. R.1999. Host range relationships and the evolution of canine parvovirus. Vet. Microbiol. 69: 29–40. doi: 10.1016/S0378-1135(99)00084-X [DOI] [PubMed] [Google Scholar]
- 28.Parrish C. R., Aquadro C. F., Strassheim M. L.1991. Rapid antigenic-type replacement and DNA sequence evolution of canine parvovirus. J. Virol. 65: 6544–6552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parrish C. R., O’Connell P. H., Evermann J. F., Carrmichael L. E.1985. Natural variation of canine parvovirus. Science 230: 1046–1048. doi: 10.1126/science.4059921 [DOI] [PubMed] [Google Scholar]
- 30.Parthiban M., Kurunchi C., Divya K., Kumanan K., Bargavi D. S.2012. A rapid and highly reliable field-based LAMP assay of canine parvovirus. Acta Virol. 56: 71–74. doi: 10.4149/av_2012_01_71 [DOI] [PubMed] [Google Scholar]
- 31.Prompamorn P., Sithigorngu P., Rukpratanporn S., Longyant S., Sridulyakul P., Chaivisuthangkura P.2011. The development of loop-mediated isothermal amplification combined with lateral flow dipstick for detection of Vibrio parahaemolyticus. Lett. Appl. Microbiol. 52: 344–351. doi: 10.1111/j.1472-765X.2011.03007.x [DOI] [PubMed] [Google Scholar]
- 32.Ravan H., Yazdanparast R.2012. Development and evaluation of a loop-mediated isothermal amplification method in conjunction with an enzyme-linked immunosorbent assay for specific detection of Salmonella serogroup D. Anal. Chim. Acta 733: 64–70. doi: 10.1016/j.aca.2012.04.034 [DOI] [PubMed] [Google Scholar]
- 33.Reed A. P., Jones E. V., Miller T. J.1988. Nucleotide sequence and genome organization of canine parvovirus. J. Virol. 62: 266–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanekata T., Sugimoto T., Ueda S., Tsubokura M., Yamane Y., Senda M.1996. Latex agglutination test for canine parvovirus. Aust. Vet. J. 73: 215–217. doi: 10.1111/j.1751-0813.1996.tb10038.x [DOI] [PubMed] [Google Scholar]
- 35.Strassheim M. L., Gruenberg A., Veijalainen P., Sgro J. Y., Parrish C. R.1994. Two dominant neutralizing antigenic determinants of canine parvovirus are found on the threefold spike of the virus capsid. Virology 198: 175–184. doi: 10.1006/viro.1994.1020 [DOI] [PubMed] [Google Scholar]
- 36.Teramoto Y. A., Mildbrand M. M., Carlson J., Collins J. K., Winston S.1984. Comparison of enzyme-linked immunosorbent assay, DNA hybridization, hemagglutination, and electron microscopy for detection of canine parvovirus infections. J. Clin. Microbiol. 20: 373–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uwatoko K., Sunairi M., Nakajima M., Yamaura K.1995. Rapid method utilizing the polymerase chain reaction for detection of canine parvovirus in feces of diarrheic dogs. Vet. Microbiol. 43: 315–323. doi: 10.1016/0378-1135(94)00102-3 [DOI] [PubMed] [Google Scholar]
- 38.Wang H. C., Chen W. D., Lin S. L., Chan J. P. W., Wong M. L.2005. Phylogenetic analysis of canine parvovirus VP2 gene in Taiwan. Virus Genes 31: 171–174. doi: 10.1007/s11262-005-1791-0 [DOI] [PubMed] [Google Scholar]