Abstract
Genome-wide association scans with high-throughput metabolic profiling provide unprecedented insights into how genetic variation influences metabolism and complex disease. Here we report the most comprehensive exploration of genetic loci influencing human metabolism to date, including 7,824 adult individuals from two European population studies. We report genome-wide significant associations at 145 metabolic loci and their biochemical connectivity regarding more than 400 metabolites in human blood. We extensively characterize the resulting in vivo blueprint of metabolism in human blood by integrating it with information regarding gene expression, heritability, overlap with known drug targets, previous association with complex disorders and inborn errors of metabolism. We further developed a database and web-based resources for data mining and results visualization. Our findings contribute to a greater understanding of the role of inherited variation in blood metabolic diversity, and identify potential new opportunities for pharmacologic development and disease understanding.
The discovery of mutations causing severe congenital metabolic disorders, or inborn errors of metabolism, has revolutionized our early understanding of how genes control biochemical reactions and metabolic pathways in the human body 1. Recent technological advances in metabolomics and genetics allowing the collection of high dimensional datasets in large population samples suggest that inborn errors of metabolism are only extreme cases of a wide spectrum of genetic variation in human metabolism, and that these loci are often at the basis of multifactorial traits and complex diseases. The so-called genetically influenced metabotypes (GIMs) identified to date have been shown to display larger effect sizes compared to most complex trait loci 2 and map preferentially in or near genes that encode enzymes, metabolite transporters and regulators of metabolism 3-13.
However, while the biomedical and pharmaceutical relevance of these associations may become clearer as focused gene-by-gene investigations are conducted, little is known about their system-wide interconnectivity, and how this knowledge can be translated into medical practice. A comprehensive blueprint of human metabolic pathways and the genes that regulate them would inform strategies for modifying deregulated metabolites in a rational and targeted manner, potentially using already existing drugs, as has been suggested for other GWAS findings 14. In this context, genetic associations provide powerful tools to identify genes that may be targeted to modulate metabolite levels.
Here we present the most comprehensive investigation of genetic influences on human metabolism to date, extending previous studies based on the same metabolomic platform 11,15. We applied powerful hypothesis-generating genome-wide scans to survey regions of the genome associated with a wide range of metabolic traits. The hundreds of associations and their metabolic context reported in this study identify a system-wide atlas of molecular readouts of activity for human genes measured in vivo. We not only provide information on hundreds of single genotype-metabolite associations, greatly expanding knowledge of metabolic intermediates of gene functions and disease, but we also generate a network including the majority of metabolites reliably measurable in blood using the Metabolon platform, allowing future exploration of perturbations caused by individual genetic variants across hundreds of metabolites at once. Finally, this information is based on experimental in-vivo data from a human population study. In this respect, this work contrasts the classical metabolic pathway maps that are obtained as composites of single in vitro biochemistry experiments. The novel loci empower future clinical and pharmacologic research in a number of key areas, spanning from a better understanding of genetic predisposition to disease, to the identification of potential novel biomarkers and drug surveillance tools, drug targets and the causal evaluation of environmental and modifiable influences on human traits and disease 16. Moreover, in order to maximize the downstream utility of the data for the broader scientific community, we make the atlas freely available through an extensive suite of web resources, including a database of detailed functional annotations and disease associations for each locus, and a network view of the data with linkage to genetic and metabolic web resources (see URLs). In the following we summarize the key results of our study.
Results and discussion
Eighty-four novel metabolic loci
The study sample included a total of 529 metabolites profiled using liquid-phase chromatography and gas chromatography separation coupled with tandem mass spectrometry in either plasma or serum from 7,824 adult individuals from two European population studies (Supplementary Table 1). Of these, the entire KORA dataset (N=1,768) and a small proportion of the TwinsUK dataset (1,052 individuals and 250 metabolites) have been described in two previous studies 11,15, with 5,002 TwinsUK individuals newly profiled in this study. Over half of the 529 metabolites (N=333, 63%) were chemically identified and could be assigned to eight broad metabolic groups (amino acid, carbohydrate, cofactors and vitamin, energy, lipid, nucleotide, peptide and xenobiotic metabolism) as described in the KEGG database 17. These groups could be further subdivided into 63 distinct biochemical pathways. Another 196 metabolites (37%) were classified as ‘unknown’, indicating that their chemical identity has not yet been conclusively established. Further information on the unknown metabolites, including measurement platform, retention time, m/z and fragmentation spectra can be found in Supplementary Table 2. Analyte overlap between unknowns and knowns was excluded using correlation analysis (Supplementary Table 3). After stringent quality controls, a subset of 486 metabolites was available for genetic analysis in both cohorts, including 309 known and 177 unknown metabolites.
The primary genome-wide discovery analysis was carried out on approximately 2.1 million single nucleotide polymorphisms (SNPs) either directly genotyped, or imputed from the HapMap2 panel 18, and passing stringent quality control metrics (Online Methods, Supplementary Figure 1). From this initial discovery effort, 137 independent variants were significantly associated with metabolite concentrations at a stringent genome-wide cutoff of 1.03×10−10 (=5×10−8/486 metabolites; Supplementary Tables 4 and 5) 19. Subsequent discovery analysis on 98,346 pairwise metabolite ratios identified eight additional loci at a more stringent cutoff of 5.08×10−13 (=5×10−8/98,346 ratios; see the Supplementary Note for a discussion on interpretation of ratio). Overall, our study revealed 299 SNP-metabolite associations of genome-wide significance at 145 statistically independent SNPs (Supplementary Table 5, Figure 1 and Supplementary Figure 2). Of these, 84 loci have not been reported before, while three loci were reported while this work was in revision 20. Another 61 loci were identified in previous studies, thus suggesting validation across different platforms 2,10,11,15,21. A subset of the loci were also reported in smaller scale studies conducted in other tissues, suggesting that metabolite associations are reproducible across tissues and that blood metabolite loci may be for the most part representative of associations in other tissues. For instance, all five genome-wide significant loci detected in a study in urine in a discovery sample of 862 individuals 22 were also detected in blood in this study. As another example, the PYROXD2 locus was associated with an NMR metabolite in urine 23. The novel loci identify a rich catalog of novel metabolic associations, allowing linkage of genetic and disease associations with underlying molecular mechanisms and greatly enriching understanding of the genetic control of human metabolism. We systematically evaluated evidence for genes within a 1-Mb window centered on the sentinel SNPs to identify cases where the function of the gene matches the relevant metabolite (Online Methods). This effort identified a plausible or established biochemical link for 101 of the 145 loci, and involving 94 unique genes (reviewed in Supplementary Table 6). We henceforth refer to these 94 genes as ‘predicted causal’. For the remaining loci we annotated the gene nearest to the association peak.
Figure 1. Ideogram of metabolomic associations.
Chromosomal map illustrating the location of the 145 loci identified in this study. Locus label colors are indicative of metabolite pathway class for the strongest associated metabolite at each locus, and are carried through additional figures and in the corresponding Cytoscape file (see URLs). An interactive web version of this figure at the supporting online website (see URLs) provides an entry point to biologic and functional annotation for each locus.
Where a predicted causal gene was associated with a metabolite ratio, and both metabolites have been identified, we can characterize that ratio in terms of the underlying biochemistry. In eleven cases, the metabolite ratio seems to reflect the flux through a particular metabolic reaction that is influenced by the SNP (annotated as ‘activity’ in Supplementary Table 6). For instance, GOT2 (encoding mitochondrial glutamic-oxaloacetic transaminase 2) was associated with the ratio between phenyllactate (PLA) and phenylalanine. GOT2 catalyzes the conversion of phenylalanine to phenylpyruvate, which is then converted to phenyllactate 24. Another such example was MBOAT7, associated with the ratio of arachidonate (20:4n6) to 1-arachidonoylglycerophosphoinositol. MBOAT7 encodes a lysophosphatidylinositol acyltransferase that has specificity for arachidonoyl-CoA as an acyl donor 25. Arachidonate is readily converted to arachidonoyl-CoA.
In five cases both metabolites in a ratio were linked to a substrate or both linked to a product (for instance at ACE, SULT2A1, AKR1C4, ABP1 and THEM4). In these cases the effect of the genetic variant may be explained as causing one molecule to be consumed or acted on faster than the other (‘selectivity’).
In seven cases the levels of one metabolite ‘normalizing’ the statistical signal for the other may explain the ratio. For instance, PRODH encodes proline dehydrogenase, which catalyzes the first step in proline degradation 26. However, the p-value for the ratio of valine to proline was even stronger than the p-value for proline itself, suggesting that valine may be normalizing the proline concentration against the overall amino acid pool.
A de novo atlas of human metabolic relationships in blood
We next generated a network view of genetic-metabolic interactions in the two cohorts by combining genetic and metabolite information. First, we connected metabolites with metabolites using Gaussian graphical models (GGMs). We have previously demonstrated that GGMs connect biochemically related metabolites 15,27, and can be applied to reconstruct metabolic pathways directly from the metabolomics data. Second, we connected metabolites with genetic loci based on our primary GWAS results, i.e. one link for each genome-wide significant association. For a more detailed description of the network generation process, see the Supplementary Note. To verify the stability of partial correlation values in both cohorts, we performed a bootstrapping-based subsampling approach. We observe generally low standard deviations, especially for high partial correlation values, indicating a high stability of the estimation. The resulting network (Figure 2) recapitulates relationships between 397 metabolites from 60 different pathways (249 known and 148 unknown) and 131 of the 145 genetic loci, and provides the first comprehensive and high-resolution reference map of human metabolic reactions and their genetic influences measured in a single in vivo “experiment” in blood.
Figure 2. A network view of genetic and metabolic associations.
The network view was built by combining genetic associations with a GGM network created from metabolite concentrations. A. Each node represents either a set of metabolites belonging to the same pathway (circular nodes) or a genetic locus (diamond-shaped nodes). An edge between a pathway and a locus was drawn if at least one metabolite showed a genome-wide significant association with the locus. A line between two pathway nodes was drawn if there was at least one connection in the underlying metabolite GGM network between two metabolites of the respective pathways (see Online Methods). Node color keys are the same as in Figure 1. Numbers within metabolite loci indicate pathway name as detailed in the legend. Numbers associated with each pathway name indicate the number of metabolites contained within each pathway node. B. Example of a network with full-detail resolution. The network is provided in digital form for interactive viewing at the supporting online website (see URLs). The fully annotated network is also available for download in Cytoscape format from the same website.
This in vivo reference map of metabolite relationships in blood complements existing knowledge of gene-metabolite relationships in specialized biochemical databases. It facilitates the visualization of genetic associations in the context of complex relationships between metabolites and SNPs, as in the case of the peptide sub-network of dipeptide and oligopeptide metabolites and peptide-related genes (Figure 2, box). To maximize the use of the data to the scientific community, we make the network available for download with extensive biochemical and biological annotation (URLs). We further make available a version of the network with basic annotation through a web browser, allowing rapid visualization and exploration of the data. Furthermore, an extensive database of genetic associations and their biologic, medical and pharmacologic annotation is available in Supplementary Table 6 and through the supporting online website (see URLs).
Allelic architecture of metabolic loci
To fully quantify the extent to which the metabolic loci capture metabolite variance, we carried out exhaustive characterizations of the allelic architecture of the novel loci. Metabolite heritabilities were estimated using the classical twin (ACE) model and the twin structure of the TwinsUK cohort (Figure 3, Supplementary Note). The contribution of metabolic loci to metabolite variance was high (median 6.9%, range 1-62%, Supplementary Note and Supplementary Table 7), with variants explaining greater than 20% heritability at approximately 10% of the metabolites and greater than 50% in four cases. This supports previous observations that variants explain on average greater proportions of trait variance for some metabolite classes compared to what is generally observed for complex traits 2, confirming the value of these intermediate molecular readouts for dissecting genetic contributions to complex traits with greater statistical power. We carried out local imputation with a denser haplotype reference map (1000 Genomes Project) for each of the 145 loci to explore the contribution of additional variants with lower minor allele frequency present in this panel and poorly represented by HapMap2 imputation. With the exception of the CYP3A cluster, the two imputation panels yielded lead SNPs with highly correlated frequency, p-values and explained variance at most loci (Supplementary Table 8, Supplementary Figure 3). We further explored the contribution of SNP-SNP interactions between the metabolic loci, defined as a departure from additive marginal effects (Online Methods). The analysis suggested that the effects of metabolic loci were predominantly additive, apart for the statistically significant interaction observed between NAT8 and PYROXD2 variants 23 (Figure 4, Supplementary Figure 4, Supplementary Table 9).
Figure 3. Heritability and variance explained.
We used the ACE model to partition the variance of each metabolite into narrow-sense heritability (orange), and common (purple) and unique environmental components. The proportion of heritability explained by all SNPs associated with a given metabolite at the genome-wide level is shown in red. The corresponding numeric values for heritability estimates are given in Supplementary Table 7.
Figure 4. Epistatic effects and Mendelian randomization analyses on eQTL loci.
(A) Interaction between NAT8 and PYROXD2 variants. Levels of the metabolite X-12093 are plotted as a function of genotypes at the two variants rs10469966 (NAT8) and rs4488133 (PYROXD2), showing a significant SNP*SNP interaction on metabolite levels. See also Supplementary Figure 4. (B) Regional plots illustrating overlap of association of SNPs with metabolites (red) and gene expression levels measured in fat (black), skin (blue) and LCLs (green) at locus 11. Associations p-values (–log10 scale) are plotted for a 1 Mb window surrounding the lead SNP rs6693388, and for associations of the SNP with linoleate (18:2n6)/ 5,8-tetradecadienoate (red), and with THEM4 expression in fat (black), skin (blue) and LCL (green) respectively. (C) Example of relationships tested by Mendelian randomization analysis at the same locus, where expression of gene THEM4 is shown to mediate the association between rs6693388 and the ratio linoleate (18:2n6) to 5,8-tetradecadienoate. All analyses were done in a subset of 484 unrelated TwinsUK participants with gene expression measured at the same time of visit of metabolomic measurements. The full results are in Supplementary Table 11.
Finally, we systematically compared the metabolite-associated SNPs against public repositories of variants (cis-eQTLs) affecting gene expression in liver 28, fat, skin and lymphoblastoid cell lines (LCLs) 29. For each lead metabolomic SNP, we first retrieved all SNPs with high linkage disequilibrium (r2≥0.8) in the 1000 Genomes Project pilot phase (CEU population). Each lead SNP and its proxies were then used as baits to search the two expression databases. A total of 57 lead SNPs identified ciseQTLs in at least one of four tissues searched under the nominal permutation p-value<0.001, defining a total of 101 SNP-gene pairs and 97 different genes (Supplementary Table 10). Of the 97 genes, 38 were predicted as causal based on our annotation, and 59 as non-causal. When compared to non-causal genes, causal genes showed 3.25-fold enrichment in the liver eQTL dataset (Fisher’s exact test p-value = 0.023, two-tailed), possibly reflecting the greater contribution that liver metabolism makes to blood metabolite levels. Furthermore, causal genes were enriched by 1.6-fold in fat compared to non-causal genes (Fisher’s exact test p-value = 0.038, two-tailed). No enrichment was seen in LCL and skin.
One major challenge of interpreting associations from GWAS is formulating and testing hypotheses on the causal effect of a SNP on an associated trait. Molecular QTL studies on metabolite or gene expression have greater statistical power compared to more complex traits like for instance HDL or LDL cholesterol levels 2. In this context, testing correlations between SNPs, metabolite concentrations and other molecular phenotypes (for instance epigenetic profiles or transcript levels) provides new opportunities to investigate metabolite pathways at the molecular level (see also Supplementary Information). We applied Mendelian randomization analysis to test the hypothesis that metabolite-associated SNPs affected metabolites through variation in transcript levels of corresponding causal genes. We focused on a subset of 32 predicted causal genes that had a matching eQTL in at least one MuTHER tissue and exploited a subset of 484 individuals of the MuTHER dataset where gene expression levels were measured at the time of metabolomic measurement (Online Methods). Such analysis revealed two loci significant at the Bonferroni-corrected p-value<1.5×10−3, namely THEM4 and CYP3A5. In both cases the allele associated with increased metabolite levels was associated with decreased gene expression in one or more tissues (Figure 4, Supplementary Table 11). This analysis provides support for the underlying causal variants having regulatory consequences for these two loci.
Biological relevance
The novel associations span the large majority of the metabolic pathways explored, indicating widespread genetic influences on the human metabolome. We could assign a known biochemical function to approximately two-thirds of the overall associations (101 out of the 145 loci, Supplementary Table 6). One third (N=34) of the novel loci involved amino acids, and a similar number (N=33) were with intermediates of lipid metabolism, including notably sterols, carnitines and intermediates of inositol and fatty acid metabolism. The remaining novel associations were across a wide range of metabolic classes and functions, including importantly pathways with a central role in cellular metabolism and energy, intermediates of purine and pyrimidine metabolism, glucose homeostasis and vitamins and cofactor levels among others. In cases where a metabolite is of unknown identity, the metabolic function of the associated gene provides an hypothesis on its identity, as described earlier 15.
The current characterization of hundreds of loci embedded in their metabolic context further allows exploration of complex systems to a significantly greater depth and breadth compared to previous studies 2,10,11,15,21. For instance, 12 novel associations were within phenylalanine, tyrosine and tryptophan metabolism pathways, implicated in key brain functions through dopamine and serotonin biosynthesis. Among them, two common variants in TDO2 (encoding tryptophan 2,3-dioxygenase) and IDO1 (encoding indoleamine 2,3-dioxygenase 1) were associated with tryptophan and 4-hydroxytryptophan (X-12100) respectively, two intermediates of synthesis of the neurotransmitter serotonin. Several loci mapped to transporters, including association of plasma tyrosine and tryptophan levels with SLC16A10 (encoding a T-type amino acid transporter 1 (TAT1) for tryptophan, tyrosine and phenylalanine 30), associations between plasma kynurenine levels and SLC7A5 (encoding LAT1), which mediates cellular exchange of tryptophan and the tryptophan metabolite kynurenine 31, and many others. It will be important to characterize whether these associations provide reliable molecular readouts for the function of these genes in the brain, to ascertain the value of accessible tissues such as blood to dissect genetic influences on systems that are not readily accessible in vivo. As shown with this example, all associations in this study can now be used in a similar manner to explore other disease-relevant connections. Finally, we note that metabolomics analysis in blood reflects a cumulative readout of processes that occur in different metabolically active tissues, including uptake, release, production, and disposal of biochemicals from individual organs. Thus whether one of the genetic variants that we report here has an effect in a specific tissue will depend on whether the respective protein is actually expressed and active in that tissue. It is possible, and even likely, that analyses in individual tissues will show even more distinct metabolic profiles and stronger genetic associations than those observed here. A greater characterization of patterns of tissue specificity for metabolite levels and associations will be necessary to address this question.
Disease and pharmacological relevance
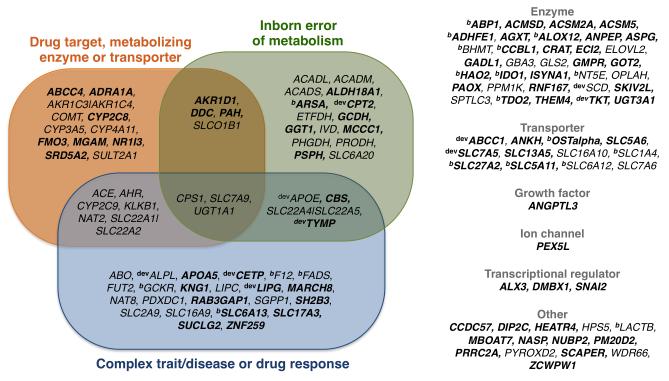
Integration of metabolic associations with complex traits and disease empowers understanding of molecular underpinnings of disease, as shown in the case of the bradykinin/kininogen/kinin system and cardiovascular disease (Supplementary Figure 5 and Supplementary Information). We searched the NHGRI GWAS Catalog (July 2013) for cases where the sentinel SNP either matched a complex trait or disease variant or was in high linkage disequilibrium with one (defined as r2≥0.8, Online Methods). A total of 41 metabolite-associated SNPs (Supplementary Table 4) and 14 out of 84 novel, matched SNPs previously associated with complex disease or drug response endpoints (Figure 5, Supplementary Table 4). We further searched the 1-Mb intervals centered on the associated SNP for genes causative for inborn errors of metabolism, identifying a significant enrichment (26 cases, hypergeometric test P-value = 5.9×10−16; Supplementary Table 12). As recently postulated by Mootha and Hirschhorn 32, this suggests that several genes (including CPS1 UGT1A1, CBS and SLC22A4-5 and others) harbor genetic variants with effects ranging from loss-of-function alleles in metabolic disorders all the way to common polymorphisms with moderate phenotypic consequences in multifactorial complex diseases and finally quantitative variation in the ‘normal’ range. One such example was the CPS1 locus, encoding a mitochondrial carbamoyl phosphate synthetase, an enzyme catalyzing the generation of carbamoyl phosphate from ammonia and bicarbonate. Mutations in CPS1 cause carbamyl phosphate synthetase deficiency, an autosomal recessive disorder characterized by congenital hyperammonemia and defective citrulline synthesis. A common variant in CPS1 was previously associated with increased risk of chronic kidney disease 33. The same variant was associated with glycine levels in this and previous studies 10,34,35. Glycine is interconverted to ammonia via the glycine cleavage complex 36, and provides a molecular intermediate trait for the disease.
Figure 5. Medical and pharmacological relevance of metabolomic associations.
Genes reported in Supplementary Table 4 were classified based on their overlap with inborn errors of metabolism, or for being targets, metabolizing enzymes or transporters of FDA-approved drugs. Variants were annotated based on their overlap with complex trait and disease loci. Novel associations reported for the first time in this study are highlighted in bold. The symbols dev and b identify genes associated with compounds in active stages of drug development (preclinical, Phase I-III, pre-registration to registration) and bioactive drug-like compounds respectively. Full details on locus annotation are provided in Supplementary Tables 6, 13 and 14 and in the full version available on the supporting online website (see URLs).
The extension of clinical chemistry analysis to a richer set of metabolites in this study, and the underlying linkage to genetic differences, further present important novel opportunities to identify variants of possible pharmacogenomic relevance and new pharmacologic targets. Among the 132 unique genes associated with the 145 metabolic loci, 56 (42%) identify genes of potential interest for pharmacological development. Of them, 24 (18%) were drug targets (10 genes), drug-metabolizing enzymes (11) or transporters (3) for FDA- and/or EMA-approved drugs. For instance, 45 different FDA-approved drugs with a broad range of indications were linked with the ten drug targets alone (Supplementary Table 13). A further 11 genes were drug targets with compounds in early to late stages of development (from pre-clinical to Phase I, II and III and to registration) and 24 genes were linked to drugs that are either suspended, discontinued or have no reported development activity (Supplementary Table 14). Thus, overall 21 of the 132 (16%) reported genes were either established or promising drug targets. This indicates that druggable targets are highly enriched within metabolomic loci (2.8-fold enrichment, Chi-squared test P-value=1.19×10−5) when compared to the estimated druggable fraction of human genes (1,089 of 19,258 human genes, or 5.6% 14). Finally, 21 additional genes are targets, drug metabolizing enzymes or transporters for bioactive drug-like compounds (Supplementary Table 6). In these and other cases, the metabolic associations and linkage to disease status identifies possible new therapeutic targets concordant with biomarkers for clear establishment of efficacy, and may inform patient stratification based on genetic profiling. Furthermore the associated metabolites identify potential biomarkers that are readily detectable in accessible samples, for example urine or plasma, and that may improve evaluation of disease or efficacy of new medicines while accounting for individual genetic background. Finally, this catalogue of associations provides important new opportunities for drug repositioning or the identification of novel indications for existing drugs, thus potentially unlocking some of the original investment.
Conclusions
In summary, we carried out the most comprehensive analysis of genetic influences on human blood metabolites to date. Our observations suggest widespread genetic control over a large range of different pathways and functions, and support the notion of human metabolism as a complex continuum governed by genetic effects of variable intensity, complex regulatory influences and non-genetic effects. Our results advance knowledge in a number of areas of biomedical and pharmacogenetic interest, generating nearly one hundred novel hypotheses of SNP-metabolite and disease correlates and identifying a large catalog of novel potential biomarkers as well as associations to drug targets, transporters and metabolic enzymes. Lastly, the network provides a comprehensive in vivo reference map of genetic influences on blood metabolites in a healthy human population sample, linking genetic variants to hundreds of biochemical relationships and pathways. The data and results generated by this study are made freely available through a web database and a downloadable network to facilitate the necessary functional and clinical exploration of these novel hypotheses.
Online Methods
Study Samples
The TwinsUK cohort is an adult twin British registry composed of mostly women aged 18 to 102. These twins were recruited from the general UK population through national media campaigns and shown to have similar disease-related and lifestyle characteristics to population-based singletons in the same age group 37. The samples used in this study are 93% female in the age range 17 to 85 (mean 53 years). The Cooperative Health Research in the Region of Augsburg (KORA) study is a series of independent population-based epidemiological surveys and follow-up studies of participants living in the region of Augsburg, Southern Germany 38 The present study includes data of the follow-up study KORA F4 (2006–2008) of the KORA S4 survey (1999/2000). Individuals were aged between 32 to 77 years (mean 61 years) and had equal numbers of males and females (Supplementary Table 1). All participants in both TwinsUK and KORA have given written informed consent and the local ethics committees, Guy’s and St. Thomas’ Hospital Ethics Committee for TwinsUK, and Bayerische Landesärztekammer for KORA, approved the studies.
Data analysis
Primary genome-wide association analysis for metabolites
Genome-wide association
The primary association testing was carried out at each SNP (in the HapMap2 based imputed genotype dataset) for all 486 metabolite concentrations present in both TwinsUK and KORA datasets after QC steps. Linear regression models (assuming an additive genetic model) were used. Age and sex were included as covariates in both cohorts. Additionally, batch effect (described above in Metabolomics data acquisition and pre-processing) was added to the model only in TwinsUK, since the 1,768 KORA F4 individuals were processed in a single batch. In TwinsUK, associations were carried out using the MERLIN software 39. Briefly, MERLIN is based on the variance-component regression model and provides two family-based association tests under multivariate normality assumptions: a likelihood ratio test and a score test. In contrast to recently developed linear-mixed-model-based programs that estimate individual relationships from genotypes, MERLIN takes pedigree information from direct input of family pedigrees and reported twin status (monozygotic or dizygotic). Association tests were run on best guess genotypes (where genotypes were called if they had a posterior probability >0.9) using the computationally more efficient score test. In KORA, linear models were fit for unrelated individuals using the software QUICKTEST 40. Briefly, QUICKTEST is based on maximum likelihood estimation and assesses association tests for unrelated individuals under the linear model framework with the assumption of normal mixture model for the error distribution. QUICKTEST can deal with uncertain genotypes (for example, imputed genotypes with uncertain scores) and non-normal trait (for example, traits with skewness and heavy tails). Association tests were carried out on allelic dosages (0-2) accounting for uncertainty in imputed genotypes. For the 145 most associated SNP-metabolite pairs, associations were recalculated in TwinsUK using allelic dosage to ensure that association results were not affected by the genotype modeling. The resulting association P-values were virtually identical to those obtained from MERLIN (R2=0.99), demonstrating that there was no bias associated with the use of best-guess genotypes in TwinsUK and allelic dosages in KORA.
Meta-analysis
Association results in TwinsUK and KORA were combined using inverse variance meta-analysis based on effect size estimates and standard errors adjusting for genomic control. Heterogeneity in each association between datasets was tested using Cochran’s Q test (which is equivalent to McNemar test here as the number of datasets is two). All above analyses were carried out using the METAL software 41. To control for false positive error rates deriving from the large number of SNPs tested, a conservative Bonferroni adjusted p-value of p=1.03×10−10 (=5×10−8/486) was applied for declaring genome-wide significance for the SNP-metabolite associations. SNPs with low imputation quality (info<0.4), low minor allele frequency (<0.01), significant heterogeneity of effects between the two cohorts (defined as heterogeneity p<0.001 and p-value in either cohort being ≥0.001=0.05/486) or present in only one cohort were removed after meta-analysis. A lead SNP at each locus was selected to be the SNP with the lowest p-value against any trait at that locus. For each metabolite, all associations passing this cutoff were assigned to independent loci by iteratively allocating the lead SNP with the lowest p-value and SNPs within 500kb away from it into the same locus. These assignments were further revised as described later to estimate the effective number of independent loci.
Genome-wide analysis of metabolite/metabolite ratios
A second discovery step was carried out by testing genome-wide associations on all 98,346 (=444*443/2) pairwise ratios of two metabolite concentrations present in both cohorts after QC steps, following the principle described previously 11. Xenobiotic metabolites were excluded due to the low average call rates per individual. Due to the high number of traits and computation time and costs, for metabolite ratios genome-wide analyses were carried out in the TwinsUK study alone, followed by replication of significant hits (Bonferroni corrected p<5.08×10−13=5×10−8/98,346) in KORA F4. SNPs with low MAF (<1%) and info (<0.4) were removed from analysis, as in the association analysis on single metabolites. From the analysis in TwinsUK, a total of 430 loci survived Bonferroni correction and only the top association for each locus (i.e. the metabolite ratio and the SNP pair with the lowest p-value) was carried forward for replication in KORA. Both discovery and replication results were meta-analysed using the inverse variance model, and the combined result was filtered again on Bonferroni adjusted p-value of p<5.08×10−13 and heterogeneity (p≥0.001). Further notes on the interpretation and reporting of ratios are provided in the Supplementary Note.
Refinement of independence at metabolite loci
A series of additional analyses were carried out to further refine the identity of single metabolite- and metabolite ratio-associated loci. These analyses were aimed at identifying (i) loci where secondary, independent association signals map to less than 500kb from each other, or (ii) non-independent signals spanning >500kb owing to long-range linkage disequilibrium (LD). As a first step, the lead SNPs from analysis of metabolite concentrations and ratios were pooled. Regional association plots were created for each locus and trait, and were visually inspected to identify possible independent signals within each locus, or to detect possible signals spanning two consecutive loci. For these loci where putative independent SNPs were identified (defined as the two SNPs having an r2≤0.5 in the 1000 Genomes CEU population), conditional analyses were used to assess statistical independence between two loci. Specifically, we tested reciprocally association with each SNP while adding the second SNP as a term in the linear model. A Bonferroni correction for the number of pairs tested was applied to declare significance for the conditional test p-value. In case of non-independence, the best SNP-metabolite pair with the lowest association p-value was reported at each locus.
Total heritability estimates
The genetic and environmental influences on 503 single metabolites present in TwinsUK after QC steps were inferred by two common methods. First, the correlations of monozygotic and dizygotic twin pairs under the ACE model (which models trait variance as a function of additive genetics, common environment and unique environment and/or error effects) were compared, and narrow-sense heritabilities were inferred from the proportion of the total variance explained by estimated additive genetic effect. To estimate parameters of ACE model, a maximum likelihood method was applied under multivariate normality assumptions using the OpenMx software 42 adjusting for age, sex and batch effects (Supplementary Table 7). Metabolites measured in fewer than 30 twin pairs (either monozygotic or dizygotic) were excluded from calculations for heritability estimation.
Known heritability estimates under additive models
Next, the known heritability was estimated for 170 single metabolites associated with genetic variants in our GWAS. The explained genetic variance of each metabolite was estimated by multiple regression analysis including all associated SNPs under additive genetic models, after adjusting for covariates (age and sex in KORA; age, sex and experimental batch effect in TwinsUK). Only unrelated twin individuals were used for the analysis to avoid biases due to familial correlation. The known heritability was defined as the ratio of total variance of the metabolite to the variance explained by the multiple regression models including all SNPs significantly associated with the metabolite. This known heritability was then compared with the total heritability inferred above (Supplementary Table 7).
Imputation using denser haplotype maps from the 1000 Genomes Project
Genetic variants associated with metabolite concentrations below the genome-wide significance cutoff may contain true signals contributing to metabolite heritability. To survey more comprehensively genetic variation at the 145 metabolic loci, the associations at each locus were recalculated following imputation using a denser reference set (1000 Genomes Project, 1KGP) 43. Genome-wide genotypes were imputed using the 1KGP multi-population panel (March 2012 release for TwinsUK; June 2011 release for KORA), and associations with metabolites were tested as described before within a 1Mb window centered on each lead SNP. The analyses were carried out in TwinsUK and KORA separately and the results were combined by inverse variance meta-analysis following the procedure used in the HapMap2-based GWAS. Then, minor allele frequency, association p-value and the variances explained by two models (Y=α+β×SNP1KG+ε and Y=α+β×SNPHM2+ε) were compared between the most associated SNPs identified in the HapMap2- and 1GK-based imputations (respectively SNPHM2 and SNP1KG). Finally, the explained genetic variance of each metabolite was estimated by multiple regression analysis including all associated SNPs1KG under additive genetic model, and compared with the total heritability and with the known heritability estimated above using the original lead SNPs (referred as SNPsHM2 above). Overall, at 80% of the loci the 1KG-based imputation yielded a different variant compared to the HM2 imputation used for discovery (Supplementary Table 8). However, the 1KG lead SNPs had similar minor allele frequency (R2=0.85), association p-value (R2=0.99) and explained variance (R2=0.96) to the HM2 SNPs (Supplementary Figure 3 a-c), with only one locus (CYP3A4/5) showing a significant improvement in association in the 1KGP dataset (Supplementary Figure 3 d,e).
Epistatic interactions in known heritability estimates
Frequent statistical interaction (epistasis) between genetic variants have been proposed to inflate heritability estimates by creating so-called phantom-heritability 44. While exhaustively testing interactions between SNPs genome-wide at all traits would be statistically intractable, it is nevertheless of interest to explore possible epistatic effects between genome-wide significant SNPs, particularly given their co-occurrence within tight metabolic networks. For the test for epistasis, we focused on all pairs of SNPs associated with the same metabolite at genome-wide significance (106 pairs of SNPs associated with 51 metabolites; Supplementary Table 9). The SNP-SNP interaction was defined as a departure from additive marginal effects and verified by ANOVA F-test comparing two models below with and without an interaction term:
and
The same covariates used in the primary discovery effort (age and sex in KORA; age, sex and batch in TwinsUK) were included in each model. For simplicity, in TwinsUK only unrelated individuals were included in the analysis. A Bonferroni corrected p-value of p=0.00047 (=0.05/106) was applied to declare significant epistasis. These tests revealed no evidence for interaction among the associated loci (P≤4.7×10−4 (=0.05/106)), suggesting a lack of strong interaction effects among the loci tested. Exceptions were variants at two loci (rs10469966 at NAT8 and rs4488133 at PYROXD2), for which a significant interaction was observed on metabolite X-12093 in TwinsUK (ANOVA F-test P=1.63×10−5), replicated in KORA (P=5.71×10−11, Figure 4, Supplementary Figure 4). Fitting the interaction model yielded a modest increase in explained trait variance compared to the additive model (R2INT=15.6% vs R2ADD=14.4% in TwinsUK; R2 =27.7% vs R2 INT ADD=24.2% in KORA).
Bioinformatic annotation of genes at metabolite loci
The 1,968 protein coding genes within 500 kb of any of the lead SNPs were systematically annotated to identify cases where the gene function could be linked biochemically to the associated metabolite (‘predicted causal genes’). First, each gene was annotated for its distance and linkage to the lead SNPs. A graph-based method was used to find potential paths from each gene to the linked metabolites with edges derived from the data in KEGG 17 and EHMN 45. Comprehensive text mining was then used to identify all PubMed abstracts mentioning a synonym of the gene and a synonym of the linked metabolite(s). The assembled evidence was then reviewed locus by locus. Possible causal genes at each locus were carefully reviewed in BRENDA 46 and in the primary literature, and the gene with highest functional plausibility was selected based on the preponderance of the evidence. Because of the rich literature in biochemistry and enzymology going back at least four decades, predicted causal genes could be identified for the vast majority of the loci with known metabolites. Where a predicted causal gene was not identified, the locus was annotated with the gene nearest to the metabolite-associated SNP.
Annotation of genes for inborn errors of metabolism
To annotate overlap with genes causative for inborn errors of metabolism, we retrieved from Orphanet (see URLs) a list of all 501 genes contributing to an “inborn error of metabolism” and compared it to the list of all genes contained within the 500kb flanking each of the 145 lead SNP. Of the 501 genes, 26 are in our list of 94 predicted causal genes. Against a background of 19,297 protein-coding genes, this corresponds to a highly significant enrichment (hypergeometric test p-value = 5.9×10−16). This is in fact the strongest p-value for any “branch” or subgroup within the Orphanet ontology.
Annotation of variants mapping to complex trait and disease loci
In order to identify cases where the metabolomic-associated SNPs had evidence for previous association with complex traits and diseases, we first retrieved all variant from the 1000 Genomes Project Pilot 1 having high linkage disequilibrium with the 145 sentinel SNPs (defined by r2≥0.8 in the CEU sample). These variants were then compared against the NHGRI GWAS Catalog (July 2013 release) with the search set to all available traits and diseases. These cases were annotated in Supplementary Table 4 and Supplementary Table 6 in the field “Complex trait association [PMID]”. Cases where the variant matched a metabolic trait association using the same rule were annotated in the “Metabolite association [PMID]” field, also in Supplementary Table 6.
Annotation of variants mapping to expression QTLs
A thorough analysis of the eQTL signals at the lead SNPs was conducted as follows. For each lead metabolomic SNP, we first retrieved all SNPs with high linkage disequilibrium (r2≥0.8) in the 1000 Genomes pilot phase (CEU population). Each lead SNP and its proxies were then used as baits to search the MuTHER Project expression database 29 and a published liver eQTL dataset 28. All significant cis-eQTLs within a 1Mb window centered on the lead SNP were retrieved from these dataset, and the best eQTL p-value in each tissue was noted. A total of 57 lead SNPs identified cis-eQTLs in at least one of three tissues searched under the nominal or permutation p-value<0.001 (Supplementary Table 10). For additional information see the Supplementary Information.
Mendelian randomization analysis on causal metabolite pathways
The relative normalized gene expression values for all eQTLs identified in the previous paragraph were retrieved for 484 unrelated TwinsUK participants with available genotype data, and with gene expression and metabolite data were measured at the same time point by the MuTHER project 29 in three tissues (fat, skin and lymphoblastoid cell lines or LCL).
Mendelian randomization 47 analysis was used to test the hypothesis that changes in expression levels (GE) would cause metabolite level changes (MET), using genotype of the lead SNP as an instrumental variable. The causal effect from GE to MET was estimated by the Wald ratio method 16 as the ratio of SNP effects (i.e. instrumental variable effects) on GE and MET as follows:
where is the coefficient for the linear regression model of MET on SNP, and is the coefficient for the linear regression model of GE on SNP. The covariate adjustments (age, sex and batch effects for MET; age, sex and probe batches for GE) were made prior to analysis. To test the causal effect of 32 lead SNPs of the loci where annotated causal genes overlap with MuTHER eQTLs, a Bonferroni corrected 99.85% confidence interval (~=1-0.05/32) for the causal effect from GE to MET was obtained from 10,000 permutations, and the null hypothesis on no causation was rejected if the confidence interval did not cross zero (Supplementary Table 11).
Drug associations
A list of approved drugs by the Food and Drug Administration (FDA) and/or the European Medicines Agency (EMA) was retrieved from the ChEMBL database 48. A total of 132 unique genes reported in Supplementary Table 4, and including either predicted causal genes (94 unique genes) or genes nearest to the associated SNP, were considered. Genes were classified as drug efficacy targets (DT), metabolising enzymes (ME) and/or transporters (TP) according with their role in clinical pharmacology. For the genes classified as drug targets, the therapeutic annotation was extracted from the current version of the approved prescribing information, summary of product characteristics (SPC) and the primary literature. For the genes classified as metabolizing enzymes and transporters, the annotation was compiled from the information present in 49,50. The mapping of Uniprot IDs to gene locus was retrieved from ENSEMBL Biomart, release 70 51, and the mapping of Uniprot IDs to ChEMBL IDs from ChEMBL14 48.
Further mapping of the 132 genes to drugs was also conducted using Citeline Pharmaprojects Pipeline (see URLs, accessed on July 1, 2013) to annotate targets for drugs in other stages of development. For each metabolic locus, Pipeline was searched using the metabolic locus’ gene symbol as search criteria for the Biological Target field. From the search results, information gathered was the drug’s name, and the drug’s global status of drug development. Entries for drug targets corresponding to launched drugs were checked for consistency against the previous set of FDA- and/or EMA-approved drugs, and information was retained from the initial set. Only information for drug targets corresponding to the following categories was retained from this latter dataset (excluding ‘launched’): (i) In development (preclinical, Phase I, Phase II, Phase III, pre-registration, registration); (ii) no development (suspended, withdrawn, no development reported).
A full description of Methods used in this study is available in the Supplementary Note.
Supplementary Material
Acknowledgements
For TwinsUK, the Genotyping Facilities at the Wellcome Trust Sanger Institute and CIDR/NIH for SNP genotyping. The KORA-Study Group consists of A. Peters (speaker), J. Heinrich, R. Holle, R. Leidl, C. Meisinger, K. Strauch, and their co-workers, who are responsible for the design and conduct the KORA studies. For KORA, P. Lichtner, G. Eckstein, G. Fischer, T. Strom, the Helmholtz Zentrum München genotyping staff, the field staff the MONICA/KORA Augsburg studies. G. Fischer (KORA) and G. Surdulescu (TwinsUK) for sample handling; H. Chavez (KORA) and D. Hodgkiss (TwinsUK) for sample shipment. We are grateful to the MuTHER investigators for the transcriptomic data. Finally, we wish to express our appreciation to all study participants of the TwinsUK and KORA studies for donating their blood and time.
Funding. Part of this work was funded by Pfizer Worldwide Research and Development. For TwinsUK, the study was funded by the Wellcome Trust; European Community’s Seventh Framework Programme (FP7/2007-2013). The study also receives support from the National Institute for Health Research (NIHR) BioResource Clinical Research Facility and Biomedical Research Centre based at Guy’s and St Thomas’ NHS Foundation Trust and King’s College London. Tim Spector is holder of an ERC Advanced Principal Investigator award. SNP Genotyping was performed by The Wellcome Trust Sanger Institute and National Eye Institute via NIH/CIDR. The KORA research platform (KORA: Kooperative Gesundheitsforschung in der Region Augsburg) and the MONICA Augsburg studies were initiated and financed by the Helmholtz Zentrum München -National Research Center for Environmental Health, which is funded by the German Federal Ministry of Education, Science, Research and Technology and by the State of Bavaria. This study was supported by a grant from the German Federal Ministry of Education and Research (BMBF) to the German Center for Diabetes Research (DZD e.V.). The German National Genome Research Network financed part of this work (NGFNPlus: 01GS0823). Computing resources have been made available by the Leibniz Supercomputing Centre of the Bavarian Academy of Sciences and Humanities (HLRB project h1231) and the DEISA Extreme Computing Initiative (project PHAGEDA). Part of this research was supported within the Munich Center of Health Sciences (MC Health) as part of LMUinnovativ. SYS is supported by a Post-Doctoral Research Fellowship from the Oak Foundation. FJT is supported by an ERC starting grant (“LatentCauses”). JK is supported by the German Research Foundation (SPP 1395 “InKoMBio”) and by a grant from the German Helmholtz Postdoc Programme. KS is supported by ‘Biomedical Research Program’ funds at Weill Cornell Medical College in Qatar, a program funded by Qatar Foundation. CG is supported by the European Union’s Seventh Framework project MIMOmics (FP7-Health-F5-2012-305280) and RFBR-Helmholtz research group program. NS is supported by the Wellcome Trust (Grant Codes WT098051 and WT091310) and the European Commission (EUFP7 EPIGENESYS Grant Code 257082 and BLUEPRINT Grant Code HEALTH-F5-2011-282510). JBR & VF are supported by the Canadian Institutes of Health Research, Fonds du Recherche du Science Québec, and the Québec Consortium for Drug Discovery.
The Pfizer colleagues dedicate this manuscript to the memory of our friend Phoebe Roberts whose passion for text-mining, molecular biology and drug discovery contributed to the identification of causal genes in this research and to our work in general.
Footnotes
Disclosures and competing financial interests. MVM and RPM are employees of Metabolon Inc. EF, CH, VW, DZ, PR, LX, SJ, JT and MJB are full time employees and share holders of Pfizer.
- A supporting online website summarizing genetic associations and biologic and disease annotation from this study, and a searchable version of the metabolic network, is accessible from http://gwas.eu/si;
- A fully annotated version of the GGM network in Cytoscape format is available for download from http://metabolomics.helmholtz-muenchen.de/gwa/si/network/SI_network.cys;
- Genome-wide association statistics are freely accessible through the Metabolomics GWAS server at http://metabolomics.helmholtz-muenchen.de/gwas;
- Gene expression data is available from http://www.muther.ac.uk/.
- The raw metabolomic measurements can be accessed, together with the genome-wide genotype data, through application from the respective data access committees (http://epi.gsf.de/kora-gen/seiten/angebot_regel_e.php?page=Services and www.twinsuk.ac.uk/data-access)
- The Orphanet a list of all 501 genes contributing to an inborn error of metabolism can be found at http://www.orpha.net/consor/cgi-bin/Disease_Classif.php?lng=EN&data_id=150&PatId=10507&search=Disease_Classif_Simple&new=1.
- The Citeline Pharmaprojects Pipeline can be found at http://www.citeline.com/products/pharmaprojects/.
References
- 1.Garrod AE. Inborn Factors in Disease. Oxford University Press; 1931. [Google Scholar]
- 2.Kettunen J, et al. Genome-wide association study identifies multiple loci influencing human serum metabolite levels. Nature Genetics. 2012;44:269–276. doi: 10.1038/ng.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sabatine MS, et al. Metabolomic identification of novel biomarkers of myocardial ischemia. Circulation. 2005;112:3868–75. doi: 10.1161/CIRCULATIONAHA.105.569137. [DOI] [PubMed] [Google Scholar]
- 4.Holmes E, et al. Human metabolic phenotype diversity and its association with diet and blood pressure. Nature. 2008;453:396–400. doi: 10.1038/nature06882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sreekumar A, et al. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature. 2009;457:910–914. doi: 10.1038/nature07762. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 6.Bictash M, et al. Opening up the “Black Box”: Metabolic phenotyping and metabolome-wide association studies in epidemiology. Journal of clinical epidemiology. 2010 doi: 10.1016/j.jclinepi.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Backshall A, Sharma R, Clarke SJ, Keun HC. Pharmacometabonomic profiling as a predictor of toxicity in patients with inoperable colorectal cancer treated with capecitabine. Clin Cancer Res. 2011;17:3019–28. doi: 10.1158/1078-0432.CCR-10-2474. [DOI] [PubMed] [Google Scholar]
- 8.Wang TJ, et al. Metabolite profiles and the risk of developing diabetes. Nature Medicine. 2011;17:448–453. doi: 10.1038/nm.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suhre K, Gieger C. Genetic variation in metabolic phenotypes: study designs and applications. Nat Rev Genet. 2012;13:759–69. doi: 10.1038/nrg3314. [DOI] [PubMed] [Google Scholar]
- 10.Gieger C, et al. Genetics meets metabolomics: a genome-wide association study of metabolite profiles in human serum. PLoS Genetics. 2008;4:e1000282. doi: 10.1371/journal.pgen.1000282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suhre K, et al. Human metabolic individuality in biomedical and pharmaceutical research. Nature. 2011;477:54–60. doi: 10.1038/nature10354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kettunen J, et al. Genome-wide association study identifies multiple loci influencing human serum metabolite levels. Nat Genet. 2012;44:269–76. doi: 10.1038/ng.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nicholson G, et al. Human metabolic profiles are stably controlled by genetic and environmental variation. Molecular systems biology. 2011;7:525. doi: 10.1038/msb.2011.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanseau P, et al. Use of genome-wide association studies for drug repositioning. Nat Biotechnol. 2012;30:317–20. doi: 10.1038/nbt.2151. [DOI] [PubMed] [Google Scholar]
- 15.Krumsiek J, et al. Mining the unknown: a systems approach to metabolite identification combining genetic and metabolic information. PLoS Genet. 2012;8:e1003005. doi: 10.1371/journal.pgen.1003005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lawlor DA, Harbord RM, Sterne JA, Timpson N, Davey Smith G. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat Med. 2008;27:1133–63. doi: 10.1002/sim.3034. [DOI] [PubMed] [Google Scholar]
- 17.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Research. 2012;40:D109–14. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frazer KA, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–61. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pe’er I, Yelensky R, Altshuler D, Daly MJ. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet Epidemiol. 2008;32:381–5. doi: 10.1002/gepi.20303. [DOI] [PubMed] [Google Scholar]
- 20.Rhee EP, et al. A genome-wide association study of the human metabolome in a community-based cohort. Cell Metab. 2013;18:130–43. doi: 10.1016/j.cmet.2013.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Illig T, et al. A genome-wide perspective of genetic variation in human metabolism. Nature Genetics. 2010;42:137–141. doi: 10.1038/ng.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suhre K, et al. A genome-wide association study of metabolic traits in human urine. Nature Genetics. 2011;43:565–569. doi: 10.1038/ng.837. [DOI] [PubMed] [Google Scholar]
- 23.Nicholson G, et al. A genome-wide metabolic QTL analysis in Europeans implicates two loci shaped by recent positive selection. PLoS Genetics. 2011;7:e1002270. doi: 10.1371/journal.pgen.1002270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shrawder E, Martinez-Carrion M. Evidence of phenylalanine transaminase activity in the isoenzymes of aspartate transaminase. J Biol Chem. 1972;247:2486–92. [PubMed] [Google Scholar]
- 25.Lee HC, et al. Caenorhabditis elegans mboa-7, a member of the MBOAT family, is required for selective incorporation of polyunsaturated fatty acids into phosphatidylinositol. Mol Biol Cell. 2008;19:1174–84. doi: 10.1091/mbc.E07-09-0893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu CA, et al. Overexpression of proline oxidase induces proline-dependent and mitochondria-mediated apoptosis. Mol Cell Biochem. 2007;295:85–92. doi: 10.1007/s11010-006-9276-6. [DOI] [PubMed] [Google Scholar]
- 27.Krumsiek J, Suhre K, Illig T, Adamski J, Theis FJ. Gaussian graphical modeling reconstructs pathway reactions from high-throughput metabolomics data. BMC Syst Biol. 2011;5:21. doi: 10.1186/1752-0509-5-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schadt EE, et al. Mapping the genetic architecture of gene expression in human liver. PLoS Biol. 2008;6:e107. doi: 10.1371/journal.pbio.0060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grundberg E, et al. Mapping cis- and trans-regulatory effects across multiple tissues in twins. Nature Genetics. 2012;44:1084–1089. doi: 10.1038/ng.2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim DK, et al. The human T-type amino acid transporter-1: characterization, gene organization, and chromosomal location. Genomics. 2002;79:95–103. doi: 10.1006/geno.2001.6678. [DOI] [PubMed] [Google Scholar]
- 31.Kaper T, et al. Nanosensor detection of an immunoregulatory tryptophan influx/kynurenine efflux cycle. PLoS Biol. 2007;5:e257. doi: 10.1371/journal.pbio.0050257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mootha VK, Hirschhorn JN. Inborn variation in metabolism. Nature Genetics. 2010;42:97–98. doi: 10.1038/ng0210-97. [DOI] [PubMed] [Google Scholar]
- 33.Kottgen A, et al. New loci associated with kidney function and chronic kidney disease. Nature Genetics. 2010;42:376–384. doi: 10.1038/ng.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Illig T, et al. A genome-wide perspective of genetic variation in human metabolism. Nat Genet. 2010;42:137–41. doi: 10.1038/ng.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xie W, et al. Genetic variants associated with glycine metabolism and their role in insulin sensitivity and type 2 diabetes. Diabetes. 2013;62:2141–50. doi: 10.2337/db12-0876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kikuchi G, Motokawa Y, Yoshida T, Hiraga K. Glycine cleavage system: reaction mechanism, physiological significance, and hyperglycinemia. Proc Jpn Acad Ser B Phys Biol Sci. 2008;84:246–63. doi: 10.2183/pjab/84.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
Online References
- 37.Moayyeri A, Hammond CJ, Hart DJ, Spector TD. The UK Adult Twin Registry (TwinsUK Resource) Twin Res Hum Genet. 2013;16:144–149. doi: 10.1017/thg.2012.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wichmann HE, Gieger C, Illig T. KORA-gen--resource for population genetics, controls and a broad spectrum of disease phenotypes. Gesundheitswesen. 2005;67(Suppl 1):S26–30. doi: 10.1055/s-2005-858226. [DOI] [PubMed] [Google Scholar]
- 39.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin--rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 40.Kutalik Z, Whittaker J, Waterworth D, Beckmann JS, Bergmann S. Novel method to estimate the phenotypic variation explained by genome-wide association studies reveals large fraction of the missing heritability. Genet Epidemiol. 2011;35:341–9. doi: 10.1002/gepi.20582. [DOI] [PubMed] [Google Scholar]
- 41.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–1. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boker SM, et al. OpenMx: An Open Source Extended Structural Equation Modeling Framework. Psychometrika. 2011 doi: 10.1007/s11336-010-9200-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Veihelmann A, Krombach F, Refior HJ, Messmer K. [Influence of selective versus nonselective inhibitors of nitric oxide synthases on synovial microcirculation of the knee joint of the mouse in vivo] Langenbecks Arch Chir Suppl Kongressbd. 1998;115:197–201. [PubMed] [Google Scholar]
- 44.Zuk O, Hechter E, Sunyaev SR, Lander ES. The mystery of missing heritability: Genetic interactions create phantom heritability. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:1193–1198. doi: 10.1073/pnas.1119675109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hao T, Ma HW, Zhao XM, Goryanin I. Compartmentalization of the Edinburgh Human Metabolic Network. BMC Bioinformatics. 2010;11:393. doi: 10.1186/1471-2105-11-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schomburg I, et al. BRENDA in 2013: integrated reactions, kinetic data, enzyme function data, improved disease classification: new options and contents in BRENDA. Nucleic Acids Res. 2013;41:D764–72. doi: 10.1093/nar/gks1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith GD, Ebrahim S. ‘Mendelian randomization’: can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol. 2003;32:1–22. doi: 10.1093/ije/dyg070. [DOI] [PubMed] [Google Scholar]
- 48.Gaulton A, et al. ChEMBL: a large-scale bioactivity database for drug discovery. Nucleic Acids Res. 2012;40:D1100–7. doi: 10.1093/nar/gkr777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Parkinson A, Ogilvie BW. Biotransformation of Xenobiotics. In: Doull C.a., editor. Toxicology: the basic science of poisons. 7th ed McGraw-Hill; 2008. [Google Scholar]
- 50.Giacomini KM, et al. Membrane transporters in drug development. Nat Rev Drug Discov. 2010;9:215–36. doi: 10.1038/nrd3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kinsella RJ, et al. Ensembl BioMarts: a hub for data retrieval across taxonomic space. Database. 2011;2011:bar030. doi: 10.1093/database/bar030. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.