The preponderance of mammalian resistance to infection is inherited rather than acquired. Even without lymphoid cells, mammals still protect themselves. They respond violently to bacteria, fungi, and viruses; or, more precisely, to specific molecular components of these organisms. Most of the molecular targets for recognition have been known for decades (1). However, only recently have the receptors and pathways for innate immune sensing been elucidated, and at that, only in part. The molecular basis of lipopolysaccharide recognition was established by positional cloning in 1998, with the identification of Toll-like receptor 4 (TLR4) as a highly specific, nonredundant receptor for lipopolysaccharide (2). This identification set the stage for the use of reverse genetic methods to establish the sensing functions of TLRs 1–3 and 5–9. Mouse TLR7 (3) and human TLRs 7 and 8 (4) sense imidazoquinolines, which are guanosine-based drugs that induce an antiviral response in vivo. TLRs 7 and 8 are close phylogenetic relatives that arose from a recent X-linked duplication event, and in the mouse, it is believed that TLR8 is biologically inactive, because animals lacking TLR7 are entirely unresponsive to imidazoquinolines. However, the natural (microbial) ligand(s) for TLRs 7 and 8 have remained a subject of considerable puzzlement. In what is surely a landmark piece of work, in a recent issue of PNAS Lund et al. (5) have provided the answer. Their data are concordant with those from two other laboratories, adduced independently (6, 7). We now have a more complete picture of what the TLRs do and are also left with several important questions, as discussed below.
TLRs 7, 8, and 9 form an evolutionary cluster (8), and TLR9 is a sensor for unmethylated DNA (9). TLR3, although evolutionarily distant from TLRs 7, 8, and 9, is a sensor for double-stranded (ds)RNA (10). TLRs 3, 7, 8, and 9 all seem to be located within the endosomes (11–13), and are targeted to the endosomes by structural features of the cytoplasmic domain (12). Stimulation of TLRs 7 or 9 causes a type I IFN response (Fig. 1).
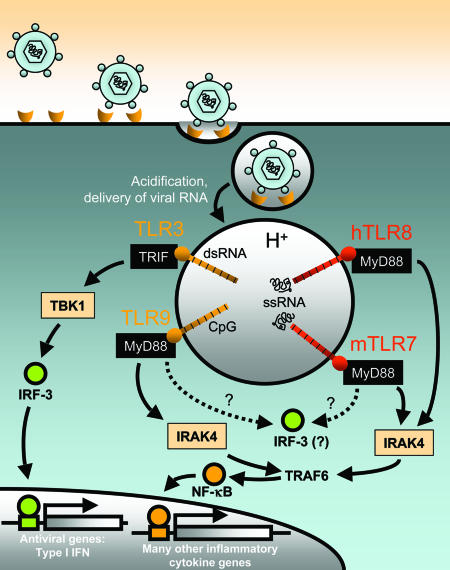
Fig. 1.
What is the role of mTLR7 in the antiviral response? Many ssRNA viruses (including VSV and influenza viruses) engage host cell receptors that trigger endocytosis. Once within endosomes, these enveloped virions fuse with the membrane to release their capsids into the cytosol. However, maturation and acidification of the endosomal vesicle may damage some viral particles, leading to ssRNA release. Human TLR8 (hTLR8) and mouse TLR7 (mTLR7), which are only expressed within endosomal membranes, recognize ssRNA [especially poly(U) and poly(U/G) motifs in the case of hTLR8], which triggers activation. Their associated signaling pathways involve myeloid differentiation factor 88 (MyD88), IL-1 receptor-associated kinase 4 (IRAK4), and tumor necrosis factor receptor-associated factor 6 (TRAF6), which lead to NF-κB activation and inflammatory cytokine production. mTLR7 and hTLR8 might also activate a MyD88-independent pathway involving IFN regulatory factor 3 (IRF-3), or perhaps another IRF family member. This activation leads to the expression of type I IFNs. mTLR7 may also recognize ssRNA derived from viruses that release directly their capsids into the cytosol through plasma membrane fusion (data not shown). The mechanism by which ssRNA would find its way to the endosome in this case is not clear. TLRs 3 and 9 are also activated by viral nucleic acids: dsRNA- and DNA-bearing unmethylated CpG motifs, respectively.
Both the TLR3 → Trif pathway (14, 15) and the TLR9 → MyD88 pathway are required for effective responses to mouse cytomegalovirus infection (16). Because TLRs 3 and 9 sense nucleoside-based ligands and are required for effective antiviral defense, because TLRs 3, 7, 8, and 9 are located within the same cellular compartment, and because TLRs 7 and 8 also sense nucleoside-based molecules, it was logical to posit that TLRs 7 and 8 might also detect viral infections; but which class of viruses and what molecules?
By using TLR7-deficient mice, Lund et al. (5) demonstrated that single stranded (ss)RNA viruses [either vesicular stomatitis virus (VSV; a rhabdovirus) or influenza virus (an orthomyxovirus)] stimulate type I IFN responses through TLR7. The authors also showed MyD88 dependence through the use of MyD88-deficient mice. By contrast, responses to the dsDNA viruses HSV1 and HSV2 do not require TLR7. Regardless of the route of internalization of an ssRNA virion: whether by plasma membrane fusion (as for VSV-RSV-F or Sendai virus), or through endosome membrane fusion (as for VSV or influenza virus), sensing occurs within the endosome because acidification of the endosomal vacuole is required for a response.
A Spatial View of Self and Nonself
Any immune system must discriminate between molecules of self and nonself, and it has long been believed that the innate immune system has solved this problem by targeting molecules that simply do not exist in the host. ssRNA obviously does exist in the host, and indeed, is very abundant. The notion of “pattern recognition,” never particularly robust, is once more challenged by the fact that ssRNA presents no obvious “molecular pattern” to be recognized. Heil et al. (6) observe that U-rich or U/G-rich oligonucleotides (but not A/G-rich oligonucleotides), presented as a complex with cationic lipids (but not in free form), induce recognition by means of mouse TLR7 and human TLR8. However, this tendency may not be sufficient to permit discrimination between viral and host ssRNA. Diebold et al. (7) note that all forms of ssRNA tested, including mouse splenocyte RNA and in vitro-transcribed mRNA encoding GFP, induce TLR7-dependent signaling. Hence, the consensus view holds that intracellular location of ssRNA, rather than ssRNA structure per se, is the key determinant of mouse TLR7- and human TLR8-mediated cell activation. Similar puzzles have been confronted before. For example, host RNAs are inevitably double-stranded to some extent. The ability of TLR3 to detect dsRNA thus poses an analogous issue of specificity. The work of Karikó et al. (17) suggests that in vitro-transcribed mRNAs can indeed stimulate TLR3, provided that they are presented to cells in a manner likely to cause uptake via the endosomal pathway.
Because acidification of the vacuole is required to permit detection even when viruses have deposited RNA into the cytoplasm by fusion with the plasma membrane, it is possible that cells have a mechanism for targeting capsids to the endosome. On the other hand, not much virus needs to enter the endosomes by means of phagocytosis to elicit a response, even when such agents as Sendai virus or VSV-RSV-F are used as inducers. Assuming that the endosomal compartment is 1 μm in diameter and is spherical, its volume is ≈5.2e-19 liters, and a single viral ssRNA molecule would have a 3-μM concentration within the organelle. It is believed that TLRs directly engage microbial inducers such as lipopolysaccharide (18, 19) and unmethylated DNA (20), and whereas the affinity of the interaction is not known, it is entirely possible that strong activation of one or perhaps many TLR complexes might result from such a polyvalent stimulus, particularly if the inducer were hydrolyzed to yield dozens of fragments, each capable of triggering a response.
Because any ssRNA molecule that finds its way to the phagosome is likely to elicit an innate immune response, whether it has arisen from the host transcriptional machinery or from a virus, it is immediately clear that bad things might befall the host as a result of aberrations in the ssRNA sensing system. The recent demonstration that host DNA plays a part in the pathogenesis of autoimmunity because it activates TLR9 (21, 22) throws the question into relief: can ssRNA also enhance an autoimmune response? Can dsRNA do so? Are TLRs 3, 7, 8, and 9 key purveyors of the “innate immune component” of autoimmunity? It is also likely that the adaptive immune response to mRNA, elicited by transfection of dendritic cells (23, 24), owes its effectiveness at least partly to the adjuvant effect of the mRNA itself.
Challenges That Remain
Both TLR7 and TLR9 seem to be capable of initiating type I IFN synthesis in a MyD88-dependent manner. However, other TLRs that activate MyD88 clearly cannot do so. TLR2, for example, depends on MyD88 and MAL to signal (25, 26), but does not activate the IFN-β gene (27). TLR4 also activates MyD88 and MAL (25, 26), but absent the adapter Trif (14, 15), cannot activate IFN-β gene transcription either.
As already mentioned, TLRs 7 and 8 are encoded by X-linked genes. Hypomorphic mutations affecting these genes would be phenotypically exposed at a very high frequency in the population because males would often be hemizygous for them. Are some males vulnerable to viral infection as a result of such mutations? Very possibly, although it may be that such mutations are relatively rare. Moreover, in humans (unlike mice), TLRs 7 and 8 may have at least partially redundant functionality, each covering for the absence of the other should it occur. Both human TLR7 and human TLR8 sense imidazoquinolines, for example. But what, if anything, does TLR8 sense in mice? Also, what is the natural ligand for human TLR7? It is very uncommon to find truly orthologous proteins with entirely different functions in humans and in mice, and TLR7 and TLR8 became paralogs long before mice and humans diverged from a common ancestor. Have these particular TLRs acquired divergent function in the years since speciation occurred? Or are mouse TLR8 and human TLR7 each on the way to becoming degenerate pseudogenes? Questions like these will engage workers in the TLR field for many years to come.
See companion article on page 5598 in issue 15 of volume 101.
References
- 1.Beutler, B. & Rietschel, E. T. (2003) Nat. Rev. Immunol. 3, 169-176. [DOI] [PubMed] [Google Scholar]
- 2.Poltorak, A., He, X., Smirnova, I., Liu, M.-Y., Van Huffel, C., Du, X., Birdwell, D., Alejos, E., Silva, M., Galanos, C., et al. (1998) Science 282, 2085-2088. [DOI] [PubMed] [Google Scholar]
- 3.Hemmi, H., Kaisho, T., Takeuchi, O., Sato, S., Sanjo, H., Hoshino, K., Horiuchi, T., Tomizawa, H., Takeda, K. & Akira, S. (2002) Nat. Immunol. 3, 196-200. [DOI] [PubMed] [Google Scholar]
- 4.Jurk, M., Heil, F., Vollmer, J., Schetter, C., Krieg, A. M., Wagner, H., Lipford, G. & Bauer, S. (2002) Nat. Immunol. 3, 499. [DOI] [PubMed] [Google Scholar]
- 5.Lund, J. M., Alexopoulou, L., Sato, A., Karow, M., Adams, N. C., Gale, N. W., Iwasaki, A. & Flavell, R. A. (2004) Proc. Natl. Acad. Sci. USA 101, 5598-5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heil, F., Hemmi, H., Hochrein, H., Ampenberger, F., Kirschning, C., Akira, S., Lipford, G., Wagner, H. & Bauer, S. (2004) Science 303, 1526-1529. [DOI] [PubMed] [Google Scholar]
- 7.Diebold, S. S., Kaisho, T., Hemmi, H., Akira, S. & Reis e Sousa, C. (2004) Science 303, 1529-1531. [DOI] [PubMed] [Google Scholar]
- 8.Du, X., Poltorak, A., Wei, Y. & Beutler, B. (2000) Eur. Cytokine Network 11, 362-371. [PubMed] [Google Scholar]
- 9.Hemmi, H., Takeuchi, O., Kawai, T., Kaisho, T., Sato, S., Sanjo, H., Matsumoto, M., Hoshino, K., Wagner, H., Takeda, K., et al. (2000) Nature 408, 740-745. [DOI] [PubMed] [Google Scholar]
- 10.Alexopoulou, L., Holt, A. C., Medzhitov, R. & Flavell, R. A. (2001) Nature 413, 732-738. [DOI] [PubMed] [Google Scholar]
- 11.Heil, F., Ahmad-Nejad, P., Hemmi, H., Hochrein, H., Ampenberger, F., Gellert, T., Dietrich, H., Lipford, G., Takeda, K., Akira, S., et al. (2003) Eur. J. Immunol. 33, 2987-2997. [DOI] [PubMed] [Google Scholar]
- 12.Nishiya, T. & DeFranco, A. L. (2004) J. Biol. Chem., in press. [DOI] [PubMed]
- 13.Lee, J., Chuang, T. H., Redecke, V., She, L., Pitha, P. M., Carson, D. A., Raz, E. & Cottam, H. B. (2003) Proc. Natl. Acad. Sci. USA 100, 6646-6651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoebe, K., Du, X., Georgel, P., Janssen, E., Tabeta, K., Kim, S. O., Goode, J., Lin, P., Mann, N., Mudd, S., et al. (2003) Nature 424, 743-748. [DOI] [PubMed] [Google Scholar]
- 15.Yamamoto, M., Sato, S., Hemmi, H., Hoshino, K., Kaisho, T., Sanjo, H., Takeuchi, O., Sugiyama, M., Okabe, M., Takeda, K., et al. (2003) Science 301, 640-643. [DOI] [PubMed] [Google Scholar]
- 16.Tabeta, K., Georgel, P., Janssen, E., Du, X., Hoebe, K., Crozat, K., Mudd, S., Shamel, L., Sovath, S., Goode, J., et al. (2004) Proc. Natl. Acad. Sci. USA 101, 3516-3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kariko, K., Ni, H., Capodici, J., Lamphier, M. & Weissman, D. (2004) J. Biol. Chem. 279, 12542-12550. [DOI] [PubMed] [Google Scholar]
- 18.Poltorak, A., Ricciardi-Castagnoli, P., Citterio, A. & Beutler, B. (2000) Proc. Natl. Acad. Sci. USA 97, 2163-2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lien, E., Means, T. K., Heine, H., Yoshimura, A., Kusumoto, S., Fukase, K., Fenton, M. J., Oikawa, M., Qureshi, N., Monks, B., et al. (2000) J. Clin. Invest. 105, 497-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bauer, S., Kirschning, C. J., Hacker, H., Redecke, V., Hausmann, S., Akira, S., Wagner, H. & Lipford, G. B. (2001) Proc. Natl. Acad. Sci. USA 98, 9237-9242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leadbetter, E. A., Rifkin, I. R., Hohlbaum, A. M., Beaudette, B. C., Shlomchik, M. J. & Marshak-Rothstein, A. (2002) Nature 416, 603-607. [DOI] [PubMed] [Google Scholar]
- 22.Viglianti, G. A., Lau, C. M., Hanley, T. M., Miko, B. A., Shlomchik, M. J. & Marshak-Rothstein, A. (2003) Immunity 19, 837-847. [DOI] [PubMed] [Google Scholar]
- 23.Heiser, A., Maurice, M. A., Yancey, D. R., Wu, N. Z., Dahm, P., Pruitt, S. K., Boczkowski, D., Nair, S. K., Ballo, M. S., Gilboa, E., et al. (2001) J. Immunol. 166, 2953-2960. [DOI] [PubMed] [Google Scholar]
- 24.Boczkowski, D., Nair, S. K., Snyder, D. & Gilboa, E. (1996) J. Exp. Med. 184, 465-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horng, T., Barton, G. M., Flavell, R. A. & Medzhitov, R. (2002) Nature 420, 329-333. [DOI] [PubMed] [Google Scholar]
- 26.Yamamoto, M., Sato, S., Hemmi, H., Sanjo, H., Uematsu, S., Kaisho, T., Hoshino, K., Takeuchi, O., Kobayashi, M., Fujita, T., et al. (2002) Nature 420, 324-329. [DOI] [PubMed] [Google Scholar]
- 27.Kawai, T., Takeuchi, O., Fujita, T., Inoue, J., Muhlradt, P. F., Sato, S., Hoshino, K. & Akira, S. (2001) J. Immunol. 167, 5887-5894. [DOI] [PubMed] [Google Scholar]