Abstract
Pseudomonas aeruginosa is an opportunistic pathogen that causes a multitude of infections. These infections can occur at almost any site in the body and are usually associated with a breach of the innate immune system. One of the prominent sites where P. aeruginosa causes chronic infections is within the lungs of cystic fibrosis patients. P. aeruginosa uses two-component systems that sense environmental changes to differentially express virulence factors that cause both acute and chronic infections. The P. aeruginosa AlgZR two component system is one of its global regulatory systems that affects the organism's fitness in a broad manner. This two-component system is absolutely required for two P. aeruginosa phenotypes: twitching motility and alginate production, indicating its importance in both chronic and acute infections. Additionally, global transcriptome analyses indicate that it regulates the expression of many different genes, including those associated with quorum sensing, type IV pili, type III secretion system, anaerobic metabolism, cyanide and rhamnolipid production. This review examines the complex AlgZR regulatory network, what is known about the structure and function of each protein, and how it relates to the organism's ability to cause infections.
Keywords: Pseudomonas aeruginosa, two-component regulation, AlgZR, alginate, twitching motility, LytTR family
Significance of P. aeruginosa
Pseudomonas aeruginosa is a ubiquitous, metabolically versatile, environmental organism with the ability to cause opportunistic infections in humans. Its >6 Mbp genome contains at least 5500 open reading frames (ORFs), encoding hundreds of virulence determinants (Wolfgang et al., 2003a,b; Winsor et al., 2011). P. aeruginosa is the predominant bacterial pathogen in cystic fibrosis (CF) patients, where colonization by P. aeruginosa in the CF lung is linked to a worsening disease prognosis (Henry et al., 1982, 1992; Nixon et al., 2001). It is also a significant cause of hospital-acquired infections (Almirante et al., 2012; Orsi et al., 2012; Horcajada et al., 2013; Khawaja et al., 2013; Simonetti et al., 2013), particularly in burn-wounds (Regules et al., 2008; Belba et al., 2013), and immunocompromised individuals (Gomes et al., 2011; Papagheorghe, 2012; Sousa et al., 2013). P. aeruginosa is able to form multiple types of biofilms, which allow them to persistently colonize a variety of surfaces, thereby making their eradication extremely challenging.
P. aeruginosa in the CF lung and the isolation of mucoid isolates
Respiratory P. aeruginosa infection in CF patients occurs intermittently between 6 months and 13 years of life, usually from an environmental reservoir (Johansen and Hoiby, 1992; Armstrong et al., 1995; Burns et al., 2001; Li et al., 2005; Ranganathan et al., 2013). Bacteria in the CF lung are constantly under stress due to: (i) the notably dehydrated airway mucus associated with this disease; (ii) bombardment from administered antibiotics; (iii) attack from host antibacterial compounds; and (iv) oxidative stress related to immune system assault (Hartl et al., 2012). Over time, bacteria in the CF lung, including P. aeruginosa, may adapt to these and other stresses and establish a chronic infection (Figure 1A). The hallmark phenotype of a chronic P. aeruginosa infection is overproduction of an exopolysaccharide matrix (Doggett et al., 1966). It is comprised of alginic acid (alginate), a polysaccharide consisting of a (1–4) linked β-D-mannuronate and α-L-guluronate copolymer. Strains over-expressing alginate exhibit a mucoid phenotype (Linker and Jones, 1964; Doggett, 1969; Evans and Linker, 1973).
Figure 1.
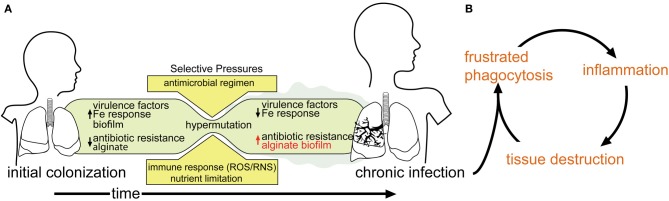
Model of P. aeruginosa infection within a CF patient. (A) As infection proceeds to a chronic state, phenotypes associated with chronic infection (e.g., mucoidy) predominate, while acute virulence factors are selected against. (B) The inability of the host-immune response to clear chronically established infection leads to a debilitating cycle of inflammation and tissue destruction.
Colonization by mucoid P. aeruginosa is associated with a significant decline in pulmonary function and disease outcome (Ballmann et al., 1998; Parad et al., 1999; Li et al., 2005). Alginate serves as a protective barrier for P. aeruginosa that resists opsonization and phagocytosis (Schwarzmann and Boring, 1971; Stiver et al., 1988; Pier et al., 1990; Leid et al., 2005), increases its resistance to some antibiotics (e.g., tobramycin) (Hentzer et al., 2001), as well as desiccation (Chang et al., 2007). Macrophages and neutrophils that are recruited through inflammatory chemotaxis are unable to clear the P. aeruginosa infection, and this “frustrated phagocytosis” leads to extensive auto-inflammatory lung damage (Pederson et al., 1992) (Figure 1B). Therefore, elucidating the molecular and biochemical mechanisms of alginate overproduction has been an area of active study for the last 40 years.
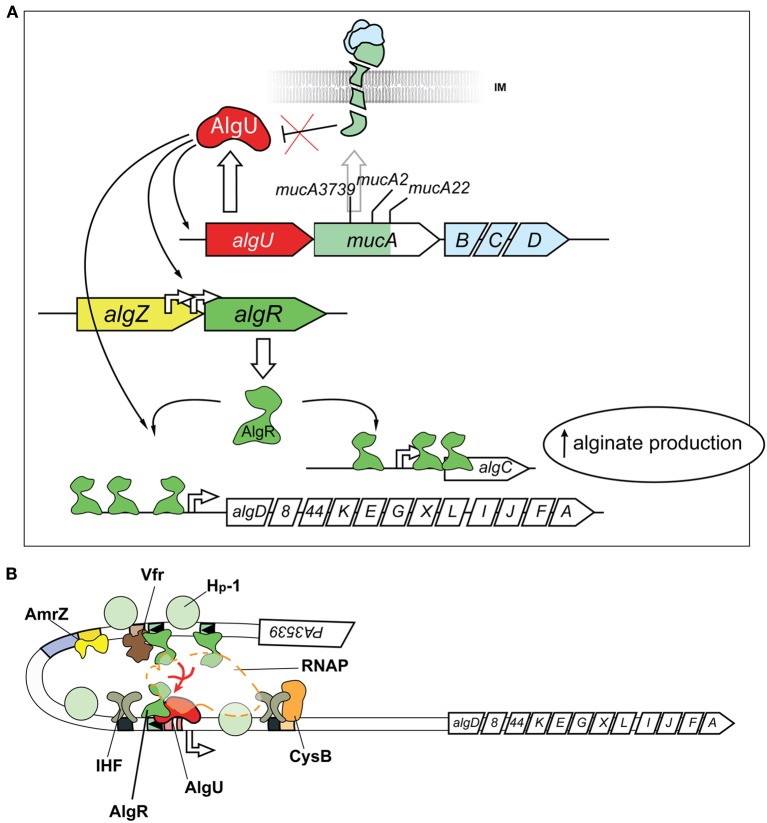
One of the mechanisms for mucoid conversion in P. aeruginosa is associated with an extracytoplasmic function (ECF) sigma/anti-sigma factor system, encoded by the algU/TmucABCD operon (PA0762–0766) (Figure 2A). The algU/TmucABCD system is analogous to the well-characterized ECF sigma factor class (σE) envelope stress response system in Escherchia coli (Rowley et al., 2006), which responds to environmental stressors, including reactive oxygen species, antimicrobial peptides, stationary phase, and carbon starvation that contribute to cell wall perturbations (Yu et al., 1995; Schurr et al., 1996; Yu et al., 1996; Barchinger and Ades, 2013).
Figure 2.
Post-Translational and Transcriptional controls for alginate production. (A) Post-translational activation of the alternative sigma factor AlgU (AlgT) is required for alginate production. Release of AlgU (AlgT) by MucA (depicted as a broken green transmembrane comma) through mutation (mucA3739, mucA2, mucA22) or degradation by proteases (see text) activates expression of itself, algR, and algD. AlgU (AlgT) and AlgR coordinate activation of the algD promoter along with other transcriptional activators (see B). Gene names are labeled within arrowed boxes, and pointed end of boxes indicate open coding direction. (B) Transcriptional model for algD promoter activation. At least seven different transcriptional regulators activate the algD promoter, causing a DNA looping of the promoter region. The following proteins bind and regulate algD transcription: (i) AlgR; (ii) AlgU (AlgT, α22), an alternative sigma factor and RpoE ortholog (iii) AmrZ, a positive regulator of the ribbon-helix-ribbon family of proteins (Baynham and Wozniak, 1996; Baynham et al., 1999, 2006; Pryor et al., 2012); (iv) Integration Host Factor (IHF), a histone-like protein (Mohr et al., 1992; Toussaint et al., 1993); (v) AlgP (AlgR3, Hp-1), C-terminus histone-like protein (Deretic and Konyecsni, 1990; Deretic et al., 1992a); (vi) the activator CysB, a LysR-like transcriptional regulator (Delic-Attree et al., 1997); and (vii) Vfr.
In non-mucoid P. aeruginosa, the MucA anti-sigma factor, encoded by the mucA gene, sequesters the stress-responsive AlgU/T sigma factor to the inner-membrane (Figure 2A) (Hershberger et al., 1995; Schurr et al., 1995, 1996; Xie et al., 1996). Sequestration of the sigma factor to the membrane prevents the transcriptionally competent AlgU/T- RNA polymerase (RNAP) complex from transcribing AlgU regulated genes, including the expression of alginate biosynthetic enzymes encoded on the PA3540–3551 operon, the algR (PA5261) gene (Rowen and Deretic, 2000) and many others (see below) (Firoved et al., 2002; Firoved and Deretic, 2003; Tart et al., 2006; Wood and Ohman, 2009).
Alginate overproduction is regulated at two general levels: at the post-translational level as a response to environmental stress (e.g., iron, phosphate, or carbon limitation), and at the genetic level from mutations in the chromosome. At the post-translational level, alginate biosynthesis is activated by the regulated proteolytic degradation of MucA. Several excellent reviews describing the details of post-translational regulation and biosynthesis of alginate overproduction are currently available (Govan and Deretic, 1996; Franklin et al., 2011; Damron and Goldberg, 2012; Okkotsu et al., 2013a; Wiens et al., 2014). At the genetic level, alginate overproduction occurs as a function of mutations in the chromosome. The P. aeruginosa mismatch repair (MMR) system and the 7,8-dihydro-8-oxo-deoxyguanine repair (GO) system are two levels of genomic repair that are commonly impaired during chronic infection. Mutations in these systems result in highly mutable strains (Oliver et al., 2000; Hogardt et al., 2007; Mena et al., 2008). There is an increase in hypermutable P. aeruginosa variants during chronic respiratory infections (Hogardt et al., 2007) thereby enabling a multitude of phenotypic variants to arise from an isogenic progenitor (Oliver et al., 2000). Another mechanism by which mutations potentially arise is through an epistatic effect of DinB (pol IV), an error-prone DNA polymerase, and MutS when the organism is exposed to hydrogen peroxide (Sanders et al., 2006, 2011). During chronic CF pulmonary infections, mutations most commonly arise in the mucA, mucB, and mucD genes resulting in premature stop-codons, frame-shifts, and missense mutations. The most common mucA mutation found in P. aeruginosa isolates from CF patients is a deletion in a homopolymeric guanine tract at △G430 of mucA (Martin et al., 1993b; Boucher et al., 1997; Anthony et al., 2002; Bragonzi et al., 2006; Ciofu et al., 2008). The consequence of these mutations is that MucA, MucB, or MucD either become non-functional, have reduced function or exhibit increased susceptibility to cellular proteases. These defects ultimately result in the release of AlgU/T to the cytoplasm and activation of the AlgU/T regulon, including its own expression, genes for alginate production, and production of lipoproteins (Firoved et al., 2002; Firoved and Deretic, 2003; Firoved et al., 2004; Wood et al., 2006).
Significance of AlgR: regulation of the algD promoter
As mentioned above, the release of AlgU to the cytoplasm increases its availability for activating the expression of the alginate biosynthetic enzymes encoded on the PA3540–3551 operon (Martin et al., 1993a; Devries and Ohman, 1994; Schurr et al., 1994). The alginate biosynthetic pathway was first discovered in the nitrogen-fixing diazotroph, Azotobacter vinelandii (Pindar and Bucke, 1975) and subsequently characterized in P. aeruginosa. In P. aeruginosa, the algD gene (PA3540) is the first ORF in an operon (PA3540–3551) comprised of 12 genes (algD, alg8, alg44, algK, algE, algG, algX, algL, algI, algJ, algF, and algA) encoding proteins involved in alginate biosynthesis, as well as its modification and export (Gacesa and Russell, 1990; Franklin et al., 2011; Okkotsu et al., 2013a). Transcription of the above genes is increased significantly in mucoid P. aeruginosa, leading to the over-production of alginate (Deretic et al., 1987a). Another gene, algC (PA5322), encoding a phosphomannomutase, is located distally to the algD locus, yet is still part of the alginate pathway (Zielinski et al., 1991).
The regulation of the algD promoter/alginate biosynthetic genes (PA3540–3551) is rather complex (Figure 2B). In addition to AlgU, the following proteins bind and regulate algD transcription: (a) AlgR (see below); (b) KinB/AlgB, a two component system where the AlgB (NtrC-family) response regulator positively regulates expression in a RpoN (σ54) dependent manner (Goldberg and Ohman, 1987; Wozniak and Ohman, 1991; Goldberg and Dahnke, 1992; Damron et al., 2012); (c) AmrZ, a positive regulator of the ribbon-helix-ribbon family of proteins (Baynham and Wozniak, 1996; Baynham et al., 1999, 2006; Pryor et al., 2012); (d) Integration Host Factor (IHF), a histone-like protein that binds to several regions on the promoter to induce DNA bending and activate transcription (Mohr et al., 1992; Toussaint et al., 1993); (e) AlgP (AlgR3), with a C-terminus histone-like element which also bends DNA and activates transcription (Deretic and Konyecsni, 1990; Deretic et al., 1992a); (f) the activator CysB, a LysR-like transcriptional regulator that is also a central regulator of cysteine metabolism (Delic-Attree et al., 1997); and (g) CRP (Vfr), a c-AMP dependent transcriptional regulator involved with the expression of several virulence genes (Devault et al., 1991). The reader is referred to extensive reviews for further details (Franklin et al., 2011; Okkotsu et al., 2013a).
AlgR was the first regulator discovered to be required for alginate over-production (Darzins and Chakrabarty, 1984). A P. aeruginosa chromosomal cosmid library was placed in chemically mutagenized non-mucoid strains, to screen for mucoid-rescue phenotypes (Darzins and Chakrabarty, 1984). A cosmid that restored alginate production in one particular mutant (strain Pa 8873) contained an ORF distal from the alginate biosynthetic genes (PA3540–3551). This 27.6 kDa protein was named AlgR (PA5261) for its ability to regulate alginate. AlgR contains an N-terminus with homology to CheY-like signaling transcriptional regulators (such as OmpR, NtrC) (Deretic et al., 1989), and a C-terminus with homology to LytR/YehT/AgrA family of DNA-binding transcriptional regulators (Galperin et al., 2001; Nikolskaya and Galperin, 2002; Sidote et al., 2008).
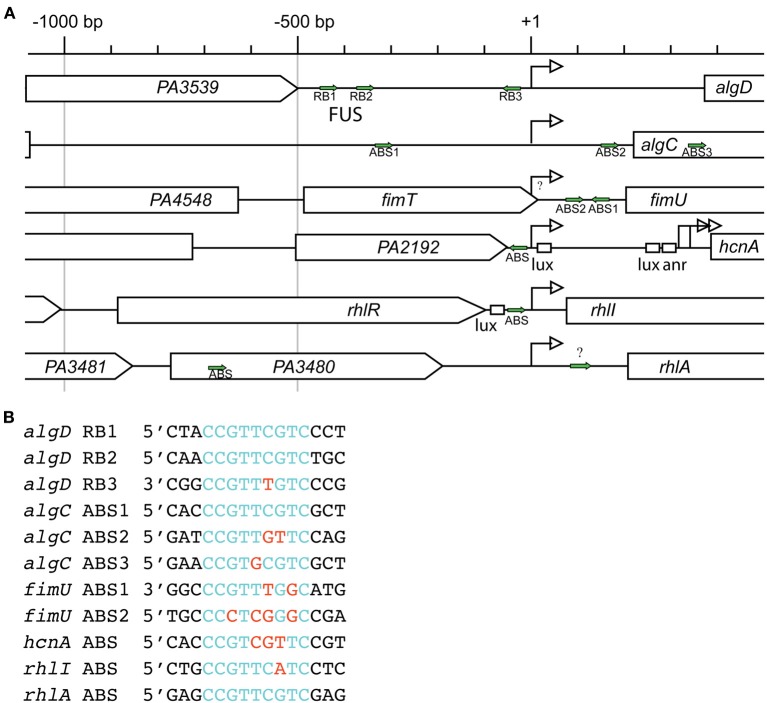
Transcription of algD is dependent on AlgR, as algR deletion in phenotypically mucoid backgrounds abrogated algD promoter activity as well as, alginate production (Deretic et al., 1987a, 1989; Mohr and Deretic, 1990). AlgR regulates the algD promoter through three distinct binding sites, termed RB1, RB2, and RB3 (Figures 3A,B). Electrophoretic mobility shift assays (EMSA) and DNase I footprinting have demonstrated two AlgR binding sequences in a region −332 bp upstream of the transcriptional start site, named the far-upstream site (FUS). The binding sites, named algD-RB1 located at −470 bp, and algD-RB2 located at −394 bp upstream of the transcriptional start site, share a core 9 bp consensus sequence (5′-CCGTTCGTC-3′) (Kato and Chakrabarty, 1991; Mohr et al., 1991). Additionally, a third AlgR binding sequence, algD-RB3 (5′-CCGTTTGTC-3′) is located −45 bp upstream of the transcriptional start site in the opposite orientation (Kato and Chakrabarty, 1991; Mohr et al., 1991, 1992). The occupation of all three AlgR binding sites is required for maximal algD transcription.
Figure 3.
AlgR regulated promoter complexity. (A) Comparison of AlgR binding site locations within its regulated gene promoters. Gene names are labeled within arrowed boxes, and pointed end of boxes indicate open coding direction. Promoters are aligned to the gene's transcriptional start sites (+1). Identified AlgR-binding sites (ABS) are indicated by green arrows. Lux, LasR or RhlR binding box; anr, Anr binding box; bp, base pairs. (B) Comparison of AlgR binding sites consensus sequences. Red letters, different nucleotides compared to consensus sequence 5′-CCGTTCGTC-3′ found on algD-RB1 and algD-RB2; Blue letters, nucleotides identical to consensus sequence 5′-CCGTTCGTC-3′ found on algD-RB1 and algD-RB2.
Promoter analyses show that the FUS site is required for algD expression, as deletion of the FUS results in a 10-fold reduction in promoter activity (Deretic et al., 1987b; Mohr et al., 1990a). A study using a algD transcriptional reporter showed that deletion of algD-RB1 resulted in a 70% reduction, and deletion of both algD-RB1/algD- RB2 resulted in a 99% reduction in algD promoter activity (Mohr et al., 1990a). A mutation in algD-RB3 also reduced algD transcription by approximately 30% of the wild type promoter (Mohr et al., 1992). As these binding sites are located at relatively far distances, DNA looping is a proposed mechanism by which AlgR and the other transcriptional regulators affect the activity of the algD promoter in the mucoid background (May et al., 1991; Schurr et al., 1993) (Figure 2B).
Additionally, binding affinity of AlgR to these AlgR binding sequences is also a contributing factor to algD regulation. Affinity of AlgR toward the two AlgR binding sequences in the FUS is higher (algD-RB1, Kd = 6.0 × 10−8; algD-RB2, Kd = 7.2 × 10−8) than toward the algD-RB3 (Kd = 3.7 × 10−7) (Mohr et al., 1992). The lower affinity is attributed to the single base difference within algD-RB3 (5′-CCGTTTGTC-3′, different base is underlined) compared to its consensus sequence (Mohr et al., 1992). Changing the sequence of algD-RB3 to a higher affinity sequence partially restored algD transcription, indicating that this third binding site is critical for modulating transcription (Mohr et al., 1992). Furthermore, overexpressing AlgR repressed algD transcription and alginate production, indicating that tight regulation of intracellular AlgR amounts is required for maximal algD promoter activity (Deretic and Konyecsni, 1989).
AlgR regulation of the algC promoter
AlgR regulates the expression of another enzyme in the alginate biosynthetic pathway, AlgC, encoded by the algC gene (PA5322). AlgC is a phosphohexomutase enzyme that has dual phosphomannomutase/phosphoglucomutase activity (Regni et al., 2002). AlgC is a multifunctional enzyme, responsible for converting mannose 6-phosphate to mannose 1-phosphate (a key step in both alginate and Psl polysaccharide biosynthetic pathways), and converting glucose 6-phosphate to glucose 1-phosphate, to provide a precursor for the core region of lipopolysaccharide (LPS) as well as the sugar moiety for rhamnolipids (Goldberg et al., 1993; Coyne et al., 1994; Olvera et al., 1999).
The utilization of a cosmid library in conjunction with a chemically mutagenized strain library led to the discovery of algC (Darzins and Chakrabarty, 1984; Zielinski et al., 1991). The algC gene is distal to the algD operon. Nevertheless, AlgR still acts as an activator of algC transcription, as the activity of a algC::lacZ transcriptional reporter decreased four-fold in an algR deletion background (Zielinski et al., 1991). Like the algD promoter, the algC promoter contains three AlgR binding sequences, termed ABS1-3, (Zielinski et al., 1991; Fujiwara et al., 1993) albeit their positions relative to the transcriptional start sites are different from those found in algD (Figure 3A). The algC-ABS1 is located at −94 bp upstream, and algC-ABS2 is located at +163 bp downstream of the algC transcriptional start site. The algC-ABS3 is located +391 bp of the transcriptional start site, within the algC ORF. While algC-ABS1 (5′-CCGTTCGTC-3′) and algC-ABS3 (5′-CCGTGCGTC-3′) are higher affinity ABSs, algC-ABS2 (5 ′-CCGTTGTTC-3′) has two base pair changes as compared to the consensus sequence, and algC-ABS2 displays lower affinity to AlgR.
There is evidence that algC-ABS1 acts like a eukaryotic enhancer element (Fujiwara et al., 1993). Placing the algC-ABS1 to either +532 bp downstream or −432 bp upstream of the transcriptional start site did not generally affect algC transcriptional reporter activity, as both conditions resulted in transcriptional reporter activity that was 126 or 87% activity of wild type, respectively. Replacing algC-ABS1 with its reverse-complement also retained reporter activity (98% activity). Yet, there was a reduction to 0.5% activity when algC-ABS1 was deleted. Additionally, all three binding sites were required for maximal algC::lacZ expression (Fujiwara et al., 1993). In summary, these data support the idea that the occupation of all three AlgR binding sequences is required for optimal expression from the algD and algC promoters.
Transcriptional regulation of AlgR
Concomitant with the activation of the algD promoter in mucoid P. aeruginosa, algR gene expression is also significantly increased (up to 50-fold) as compared to non-mucoid backgrounds (Deretic and Konyecsni, 1989; Kimbara and Chakrabarty, 1989; Deretic et al., 1990). In mucoid P. aeruginosa, algR expression is maximal at early stationary phase, coinciding with expression of several alginate biosynthesis genes including algA, algC, and algD (Leitao and Sa-Correia, 1995).
The region upstream of algR contains at least two transcriptional start sites (Martin et al., 1994; Wozniak and Ohman, 1994). AlgU acts on one of the promoters proximal (−73 bp) to the algR ATG. This proximal promoter contains an AlgU binding sequence (5′-GCACTT-N17-TCTGA) (Wozniak and Ohman, 1994; Firoved and Deretic, 2003). Therefore, as alluded to earlier, AlgR is part of the AlgU regulon and AlgR expression is responsive to membrane perturbations that result in the degradation of MucA, or through mutations in the mucA, mucB, or mucD genes (Figure 2A).
Another transcriptional start site, located at least 160 bp upstream of the algR ORF appears to be expressed constitutively (Mohr and Deretic, 1990; Mohr et al., 1990a); the expression from this algR promoter appears to not be affected by the mucoid status of the bacteria (Mohr et al., 1990a). This algR promoter is also independent of the nitrogen-responsive sigma factor RpoN (sigma-54) (Deretic et al., 1990; Mohr et al., 1990a), though, in one study, RpoN dependence was demonstrated in a rpoN gene deletion in a particular strain (Pa PAK-SN) (Kimbara and Chakrabarty, 1989). This algR promoter is responsive to increased osmolarity (Kimbara and Chakrabarty, 1989; Deretic et al., 1990; Mohr et al., 1990a) and nitrate (Deretic et al., 1990) in the mucoid background. However, the responsiveness is contingent on different muc alleles that result in the mucoid phenotype (Deretic et al., 1990). Incidentally, algR transcription is not responsive to changes in osmolarity or nitrate levels in the strain containing the mucA22 allele (which is most commonly isolated from CF patients) (Deretic et al., 1990). The reason for these different responses is unclear at this time.
AlgR orthologs in other bacteria
Inactivation of algR in mucoid P. aeruginosa isolates results in a loss of mucoidy (Deretic et al., 1990; Wozniak and Ohman, 1994). The algR gene is also required for alginate production in other Pseudomonas species. P. syringae is a plant pathogen responsible for dieback and canker disease in ornamental pears. Similar to P. aeruginosa, alginate production in P. syringae requires algR and the expression of the algD operon and algC gene. Alginate provides a medium for nutrient accumulation, water absorption and water retention, thus preventing desiccation of the organism (Denny, 1995). The algR gene is a virulence determinant for P. syringae pv. syringae FF5, as deletion of algR results in decreased necrosis in a tobacco leaf model of infection (Penaloza-Vazquez et al., 2004). However, consensus algR binding sites are absent on the algD promoter, and AlgR is not involved with the transcription of the algD gene (Fakhr et al., 1999). Instead, AlgR and the sigma factor RpoN, regulate algC transcription, as deletion of algR or rpoN results in decreased algC expression (Penaloza-Vazquez et al., 2004). Similarly, in P. putida WCS358, its AlgR homolog called PprA, activates algC but not algD transcription. Additionally, complementation with the pprA gene in a non-mucoid P. aeruginosa containing a algR deletion allele (Pa 8852) restores mucoidy (Venturi et al., 1995), supporting the idea that PprA is a functional ortholog of AlgR.
Through Southern hybridization, algR has been detected in A. vinelandii, Azomonas macrocytogenes, Xanthomonas campestris, and Serpens flexibilis (Fialho et al., 1990). However, algR has only been examined in A. vinelandii thus far. A. vinelandii is a free-living (unassociated with rhizobium), nitrogen-fixing, soil-borne organism, which has an alginate-encapsulated cyst as part of its lifecycle. Alginate serves to protect the cyst from desiccation, as well as to protect its nitrogenases from excess exogenous oxygen (Clementi, 1997). The A.v. AlgR ortholog, which is 79% identical to P.a. AlgR is required for encystment, as a algR mutant of A. vinelandii displays a 1000-fold reduction in its resistance against desiccation (Nunez et al., 1999). A.v. AlgR does not directly regulate the transcription of A.v.-algD, rather, alginate production is controlled through A.v.-AlgR activation of A.v.-algC (Nunez et al., 1999). These data suggest that the role of AlgR on exopolysaccharide production is conserved across genera, but the exact genes that it regulates are species-specific.
AlgR protein
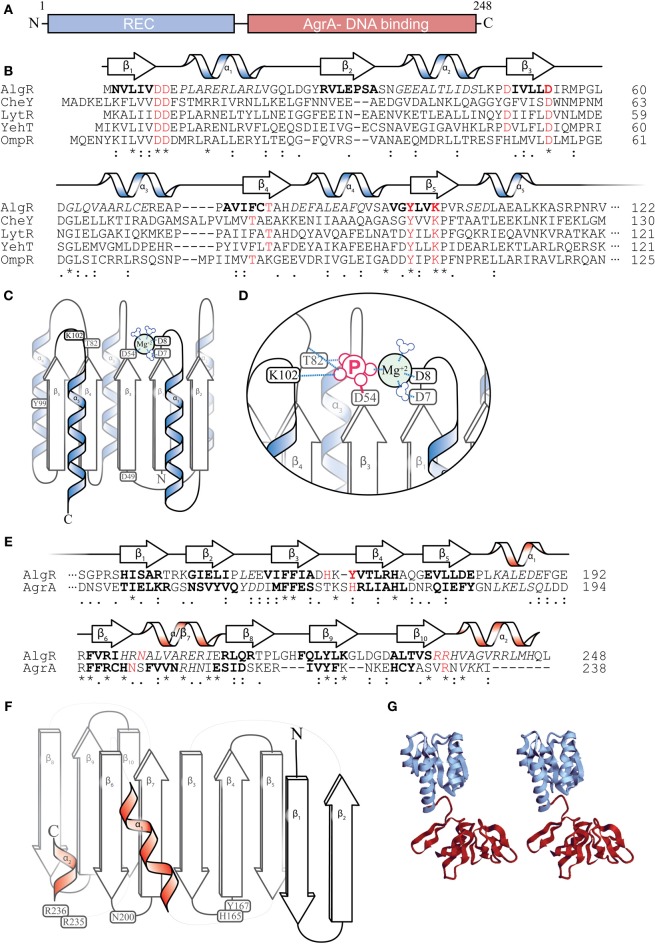
According to protein structure predictions, AlgR has two major structural domains: (i) an N-terminal CheY-like receiver (REC) domain (Deretic et al., 1989; Whitchurch et al., 2002; Kelley and Sternberg, 2009); and (ii) a carboxyl DNA-binding domain of the LytR/YehT/AgrA family of transcriptional regulators (Galperin et al., 2001; Nikolskaya and Galperin, 2002; Sidote et al., 2008) (Figures 4A,G). CheY is a well-studied signaling protein involved with the flagellar motor switch complex (Bren and Eisenbach, 2000). Phosphorylation of a conserved aspartate (Asp57) residue in CheY by its histidine kinase, CheA, leads to a clockwise rotation of the flagella. CheY and CheY-like domains contain a set of 5 alternating β-pleated sheets and α-helices (Figure 4B) (Bourret, 2010). The AlgR-REC domain contains the residues required for aspartyl- phosphorylation. While a crystal structure for AlgR has not been determined, various inferences could be made from its conserved residues. For instance, Asp7, Asp8, and Asp54 potentially form an acidic pocket that coordinates a divalent cation (Mg+2), which is required for catalyzing the phosphorylation of Asp54 (Figures 4C,D) (Whitchurch et al., 2002). Like many CheY-like domain containing response regulators (RR) (Lukat et al., 1992), AlgR is able to catalyze its own auto-phosphorylation in the presence of high-energy phospho-donors such as carbamyl phosphate and acetyl phosphate (Deretic et al., 1992b; Okkotsu et al., 2013b). A conservative substitution of the Asp54 residue of AlgR to an asparagine (D54N) eliminates its in vitro phosphorylation by an E. coli derived CheA HK, indicating that, like other RRs, this Asp54 residue is required for phospho-transfer (Whitchurch et al., 2002). Upon interaction with and phosphorylation by its cognate histidine kinase, the Asp54 could then covalently bind to phosphate and introduce a negative charge within the acid pocket of AlgR, displacing the Mg+2 ion (Figure 4D). Oxygen groups on the negative charged phosphate are proposed to interact with Thr82 (hydrogen bonding) and Lys102 (salt bridge) (Figure 4D), which would alter the conformation and stabilize the AlgR REC domain.
Figure 4.
AlgR Structural predictions. (A) Diagram depicting two major domains of the 249 amino acid AlgR polypeptide. (B) A ClustalW sequence alignment of the amino acid sequence of the N-terminus CheY-like receiver (REC) domain of AlgR and its orthologs. “*” = identical amino acids, “:” = conserved substitutions, “.” = semi-conserved substitutions. Red color denotes select highly conserved amino acids, most with functional importance for phosphorylation, italics indicate alpha helices, bold indicates beta sheets. The predicted secondary structure of AlgR according to Phyre2 is located above the sequence alignment. α = alpha helix, β = beta sheet. (C) An illustrated model depicting the location of conserved amino acids on the secondary structures of the REC domain of AlgR. Aspartate 7, 8, and 54 of CheY-like domains coordinate a magnesium ion, and are required for aspartyl-phosphate transfer from histidine kinase. (D) Once phosphorylated, residues threonine 82 and lysine 102 interact with phosphate ion, changing the conformation of the REC domain. (E) A ClustalW sequence alignment of AlgR and AgrA, with same annotation as (B). Red color denotes select conserved amino acids that are required for DNA interaction in AgrA, and may be required for AlgR interaction with DNA. (F) An illustrated model depicting the location of conserved amino acids on the secondary structures of the LytR/AgrA DNA-binding domain of AlgR. (G) a stereoscopic view of the full AlgR protein modeled, using CheY structure and S. aureus AgrA DNA binding domain.
The C-terminus of AlgR contains a DNA-binding domain in the LytR/YehT/AgrA family of transcriptional regulators (Galperin et al., 2001; Nikolskaya and Galperin, 2002; Sidote et al., 2008). This family is very uncommon, as it represents only 3% of all sequenced response regulators in bacteria. In contrast, winged-helix and helix-turn-helix domains are present in 30.1 and 16.9% of response regulators, respectively (Galperin, 2010). Helix-turn-helix (HTH) (e.g., NtrC) (Pelton et al., 1999) or winged-helix (e.g., OmpR) (Martinez-Hackert and Stock, 1997) motifs (Galperin, 2010) contain alpha-helices that embed in the major grooves of the DNA to make specific hydrogen bond contacts with the recognition sequence. On the other hand, the LytR/YehT/AgrA/AlgR family of transcriptional regulators utilizes unstructured loop regions to recognize DNA sequences (Sidote et al., 2008).
Though the crystal structure of AlgR has not been solved, secondary structure prediction based on the Phyre2 protein fold recognition server (http://www.sbg.bio.ic.ac.uk/phyre2) using the last 139 a.a. of AlgR (DNA-binding region) predicts that AlgR carboxyl terminus has at least 9 beta-pleated sheets, and demonstrates a 100% precision (E-value 2.4e–11) with the AgrA C-terminus domain (Figures 4E,F) (Sidote et al., 2008). AgrA is a Staphylococcus aureus RR that (together with AgrC HK) regulates the agr (accessory gene regulator) locus in S. aureus. The agr locus encodes a set of regulatory RNAs (RNAIII) that control S. aureus virulence factors in a temporal, cell-density (quorum) dependent manner (Novick, 2003). In AgrA, flexible loops (unstructured regions) between pairs of beta-pleated sheets make 2 or 3 specific contacts in successive major grooves (Sidote et al., 2008). The residues that make these interactions in AgrA are His169, Asn201, and Arg233. AlgR also has a conserved Asn200 and a pair of arginine residues (Arg235 and Arg236) that may correspond to those found in AgrA (Figure 4E). In addition to AgrA, another LytR-type response regulator from Bacillus cereus has been submitted to the RCSB Protein Data Bank (Osipiuk et al., to be published, PBD id: 3d6w) and will give additional insight into the structure of this family of regulators.
The LytR/YehT/AgrA/AlgR transcriptional regulators commonly affect the expression of virulence factors (Nikolskaya and Galperin, 2002). Some of the more well-studied regulators in this family include YehT of E. coli (Kraxenberger et al., 2012), AgrA of S. aureus (Koenig et al., 2004; Sidote et al., 2008), FsrA of Enterococcus faecalis (Del Papa and Perego, 2011), VirR of Clostridium perfringens (Cheung and Rood, 2000; Cheung et al., 2004), and PlnC and PlnD of Lactobacilus plantarum (Risoen et al., 1998, 2001; Straume et al., 2009). A common characteristic of these regulators is their mode of binding to DNA (Sidote et al., 2008). That is, they bind sequences that are between 8 and 13 bp long, arranged as two imperfect direct repeats approximately 12 bp apart (Risoen et al., 2001; Cheung et al., 2004; Koenig et al., 2004; Belete et al., 2008; Del Papa and Perego, 2011). The exact spacing between the direct repeats is crucial for gene transcription, though the molecular basis for this exact spacing is unknown (Sidote et al., 2008). According to the crystal structure of AgrA, large bends in the DNA are created to accommodate protein binding as direct repeats (Sidote et al., 2008). Interestingly, AlgR binding sites are not necessarily arranged as direct repeats separated by 12 bp, as seen in the algD, algC, and other AlgR-regulated promoters (Figures 3A,B). The reason for this is unclear at the moment, as further studies on AlgR are required.
The putative histidine kinase for AlgR is encoded by AlgZ/FimS
AlgR belongs to the AlgZ/FimS-AlgR two-component regulatory system (TCS) (Deretic et al., 1989; Galperin et al., 2001). TCSs are bacterial sensing systems that couple environmental stimuli to adaptive responses (Stock et al., 2000; Gao and Stock, 2009). TCSs typically consist of a homodimeric histidine kinase (HK, sensor component) and a response regulator (RR, response component). AlgR's putative cognate histidine kinase is located upstream of the algR gene (Figure 2A), and is called algZ/fimS (PA5262) (Whitchurch et al., 1996; Yu et al., 1997). Another protein named AlgZ also exists in the literature for an AlgU/T-dependent DNA binding protein whose name has been changed to AmrZ to avoid future confusion (Baynham and Wozniak, 1996; Baynham et al., 1999, 2006). The ORF of algZ/fimS is 1077 nucleotides, encoding a 40.1 kDa polypeptide. However, the amino acid sequence of AlgZ does not show similarity to most histidine kinases. Accordingly, it belongs to a subset of non-canonical histidine kinases called the “unorthodox family of histidine kinases” (Kim and Forst, 2001). Other members of this atypical family, including LytS (Brunskill and Bayles, 1996), YehU (Kraxenberger et al., 2012), and the proposed AlgZ in the AlgZR system in Acinetobacter baumanii (ABAYE 3510) (Adams et al., 2008), are missing motifs normally required for ATP-mediated autophosphorylation.
A prototypical HK contains a membrane-bound sensor domain unique to the signals being perceived. Using secondary structure prediction analyses (Kyte and Doolittle, 1982; Engelman et al., 1986) and software (SOSUI, TMHMM 2.0) (Hirokawa et al., 1998; Sonnhammer et al., 1998; Krogh et al., 2001), the first 150 amino-terminal amino acids of AlgZ are predicted to fold as four transmembrane helices. This arrangement is similar to many membrane anchored histidine kinases, and strongly suggests AlgZ is localized to the membrane. However, the functional role of AlgZ's sensor domain is currently unknown, and is an area of active study by our lab.
Histidine kinases typically consist of a DHp domain (Dimerization and Histidine phosphotransfer) and a CA domain (Catalytic and ATP binding) (Kelley and Sternberg, 2009; Kallberg et al., 2012). In general, the DHp and CA domains house conserved motifs, termed “boxes” that are required for proper kinase function, known as the H, N, G1, F, and G2 boxes. The DHp domain accommodates the H box, a 9 amino acid sequence containing the phosphorylatable histidine central to phosphotransfer. The CA domain coordinates an ATP molecule through the N, G1, F, and G2 boxes to mediate the phosphotransfer reaction. The N box forms part of the ATP binding pocket, and the G1, F, and G2 boxes are found in a flexible lid-like structure which coordinates the ATP molecule within the pocket (Stock et al., 1989; Parkinson and Kofoid, 1992).
Secondary structure prediction software (Raptor X and Phyre2) now suggest that the cytoplasmic domain of AlgZ (amino acids 151–358) contains the kinase, but it was only after the characterization of its orthologs (e.g., S. aureus LytS and E. coli YehU) that AlgZ was considered to be part of this class of HKs. Part of the difficulty in categorizing AlgZ was due to the lack of sequence identity within the carboxyl-terminus. While the H box, having the sequence RPHFLFNSL (blue indicates conserved histidine residue, underline indicates canonical homology), shows reasonable identity among this group of proteins, the N box, LQPLLENALIYG (blue indicates conserved asparagine residue, underline indicates canonical homology) only retains 5 of the 12 amino acids found in the canonical sequence. The obvious similarities between AlgZ and canonical histidine kinases are limited to these two boxes, as AlgZ lacks identifiable G1, F, and G2 boxes that make up the ATP-coordinating flexible lid. Even among its most similar orthologs (e.g., LytS or YehU), AlgZ is unique as LytS contains both G boxes, and YehU contains one of the two. It is possible that AlgZ may bind and coordinate ATP in an atypical manner, or bind a related molecule like GTP, as recently identified in the Bacillus anthracis histidine kinase (BA2291) (Scaramozzino et al., 2009).
Because AlgR was first identified as a two-component regulator of alginate production, it was expected that its putative histidine kinase would play a regulatory role in alginate production (Deretic et al., 1989). However, as it turns out, AlgR phosphorylation may not be required for alginate production. Expression of the algRD54N allele (expressing the phospho-defective AlgR protein) in the mucoid strain FRD1 does not affect alginate production (Ma et al., 1998). In fact, deletion of algZ/fimS in PAO1, or its mucoid derivative PAO568, resulted in a two-fold increase in alginate production (Yu et al., 1997), suggesting that AlgR phosphorylation may not effect, or even inhibit alginate production in the mucoid background. There is currently limited characterization of AlgZ; neither direct interaction of AlgZ and AlgR, nor phospho-transfer between AlgZ and AlgR have been demonstrated. Therefore, its proposed role as the cognate HK for AlgR has been inferred from genetic evidence on its regulation of the fimU and hcnA promoters (Belete et al., 2008; Cody et al., 2009). As described below, twitching motility, requires AlgR and its HK, AlgZ/FimS (Whitchurch et al., 1996).
AlgZ/R controls type IVa pili at the fimUpilVWXY1E promoter
The discovery that AlgZ/FimS and AlgR are required for twitching motility (Whitchurch et al., 1996) prompted a search for AlgR regulated genes that control twitching motility (Lizewski et al., 2004). Twitching motility is mediated by type IVa pili. AlgR regulates type IVa pili function and twitching motility through transcriptional activation of the fimUpilVWXY1Y2E operon, as an algR deletion abrogated expression from this operon (Lizewski et al., 2004; Belete et al., 2008). Additionally, trans-complementation of the fimU operon into a PAO1 ΔalgR strain restored twitching motility, indicating AlgR controls twitching motility by regulating the expression of the prepillin genes (Lizewski et al., 2004). The fimUpilVWXY1Y2E genes encode prepillins (FimU, PilV, PilW, PilX, PilE) that incorporate into the type IVa pili, and a calcium-dependent retraction protein (PilY1). The pilY2 ORF is located in this operon but is proposed to be a pseudogene (Giltner et al., 2010).
All of the genes encoded in the operon fimUpilVWXY1Y2E contain a hydrophobic N-terminus leader sequence required for translocation to the periplasm for type IVa pili assembly (Russell and Darzins, 1994; Alm and Mattick, 1995; Alm et al., 1996; Alm and Mattick, 1996) FimU, PilV, PilW, PilX, and PilE incorporate and create a complex with the pilus fiber (Giltner et al., 2010). The PilY1 protein, encoded by pilY1 gene, is an ortholog of the Neisseria gonorrhoeae PilC adhesin protein (Rudel et al., 1995). PilY1 appears to be a multi- functional protein: pilY1 encodes a surface and outer-membrane associated integrin-binding protein (Heiniger et al., 2010; Johnson et al., 2012) that can inhibit PilT-mediated pilus retraction (Orans et al., 2010). Surface expression of type IVa pili requires a proper stoichiometric expression of the prepillins; the interaction/compatibility of the FimU, PilV, PilW, PilX, PilY1, and PilE with other pilus assembly components dictates the efficiency of pilus assembly and disassembly (Giltner et al., 2010, 2011). Therefore, the negative twitching motility phenotype observed by a strain containing a ΔalgR allele is proposed to be due to inefficient pilus assembly and extrusion.
AlgR phosphorylation is also required for fimUpilVWXY1Y2E operon activation. A strain lacking algZ/fimS (Whitchurch et al., 1996) or expressing a phospho-defective AlgR D54N loses twitching motility (Whitchurch et al., 2002) and expresses these prepilin genes at a lower level (Belete et al., 2008). More recently, a strain that expressed a gene encoding a phospho-mimetic isoform of AlgR (algRD54E) activated fimU transcription, while algZ deletion abrogated fimU transcription (Okkotsu et al., 2013b). As demonstrated by DNaseI Footprinting and EMSA, AlgR binds to an intergenic region between fimU and the upstream gene fimT at fimU-ABS1 (5′-CCGTTTGGC-3′), as well as fimU-ABS2 (5′-CCCTCGGGC-3′) (Figures 3A,B) (Belete et al., 2008). In vitro, phosphorylated AlgR displayed increased affinity for the fimU promoter region as compared to un-phosphorylated AlgR (Okkotsu et al., 2013b). Phosphorylation of the REC domain from other LytTR response regulators such as AgrA (Koenig et al., 2004) and both PlnC and PlnD (Risoen et al., 2001) also increased affinity for DNA. Together, these data indicate that AlgR phosphorylation by AlgZ increased AlgR affinity to the fimU promoter and activated its transcription. Though the signal for the AlgZ sensor kinase has not been determined, the signal, via AlgR phosphorylation, activates twitching motility by increasing the expression of these prepilins.
AlgR and the Rhl quorum sensing system
P. aeruginosa harboring the phosphorylation defective algRD54N allele was observed to have a defect in biofilm formation, as well as twitching motility (Whitchurch et al., 2002). Microbial biofilms are highly complex, multicellular communities. Organisms are embedded in a matrix that consists of proteins, polysaccharides, and extracellular DNA (eDNA). There are multiple steps in biofilm maturation: (1) initial attachment, (2) microcolony formation, (3) maturation, and (4) dispersion (Sauer et al., 2002; Stoodley et al., 2002). There is substantial evidence that quorum sensing is required for biofilm formation and virulence, both in vitro and in vivo (Davies et al., 1998; Rumbaugh et al., 2000; Bjarnsholt and Givskov, 2007; Murray et al., 2007). Quorum sensing is an auto-induction system that monitors cell density through the export of small molecules (Fuqua et al., 1994; Seed et al., 1995). P. aeruginosa has three main quorum sensing systems- the Las, Rhl, and PQS systems. The three systems, and the virulence factors they regulate are required for biofilm formation.
The LasI-LasR (Fuqua et al., 1994; Seed et al., 1995), and RhlI-RhlR (Brint and Ohman, 1995; Ochsner et al., 1995) quorum sensing pairs are LuxI-LuxR type regulators, where the LuxI component (LasI or RhlI) synthesize their respective N-acyl homoserine lactone (AHL) molecules (Passador et al., 1993; Winson et al., 1995), and the LuxR-type HTH-containing DNA-binding regulators (LasR and RhlR) sense the signals. LasI synthesizes N-(3-oxo-dodecanoyl)-L-homoserine lactone (3-oxo-C12-HSL), and RhlI synthesizes N-butanoyl-L-homoserine lactone (C4-HSL) from S-adenosylmethionine (SAM) and acyl-acyl carrier protein (ACP) from fatty acid biosynthesis (Parsek et al., 1999; Hoang et al., 2002). The LasR or RhlR regulators bind their respective signaling molecules, dimerize, and bind to lux-box sequences of their promoters to activate DNA transcription.
The PQS system produces 2-heptyl-3-hydroxy-4-quinolone (PQS) (Pesci et al., 1999) through a biosynthetic pathway encoded by the genes pqsABCDE and pqsH (D'Argenio et al., 2002; Gallagher et al., 2002; Deziel et al., 2004). These genes are also responsible for the production of up to 55 types of 4-hydroxyl-2-alkylquinolines (HAQs) (Lepine et al., 2004) that are similar to PQS and have antimicrobial properties (Lightbown and Jackson, 1956; Machan et al., 1992; Mahajan-Miklos et al., 1999). PQS induces the DNA-binding capacity of the LysR-type transcriptional regulator, PqsR/MvfR, to activate both the expression of the pqsABCDE (Wade et al., 2005) genes for auto-regulation, as well as genes responsible for the production of other HAQs (phnAB)(Deziel et al., 2004).
The interaction among the three QS circuits is complex. Studies have revealed a large amount of cross-talk among the three circuits (Dekimpe and Deziel, 2009) where LasR activates the pqsR/mvfR gene (Xiao et al., 2006), while RhlR represses pqsABCDE transcription (McGrath et al., 2004; Xiao et al., 2006). PQS and PqsR/MvfR can activate the Rhl quorum sensing system independently of the Las system (McKnight et al., 2000; Diggle et al., 2003) as well as virulence factors under RhlI/RhlR control (Deziel et al., 2005; Xiao et al., 2006). Adding to the complexity is an orphan regulator, QscR, which responds to 3-oxo-C12 HSL and represses quorum-sensing regulated factors including phenazine and hydrogen cyanide production, rhlI/rhlR and lasI expression (Chugani et al., 2001; Ledgham et al., 2003; Fuqua, 2006).
Deleting algR caused a defect in a 6-day biofilm, that was restored by algR complementation on a plasmid (Morici et al., 2007). Additionally, 765 genes were differentially regulated at least two-fold in a 6-day biofilm when PAO1 was compared to its isogenic algR deletion strain by global transcriptome analysis (Morici et al., 2007). According to this comparison, the majority of genes that showed differential expression included those in the Rhl quorum sensing system (Morici et al., 2007). Microarray analysis showed that AlgR repressed rhlABC, hcnAB, lecB, and genes in the biosynthetic pathway for pyocyanin production (phzC2-G2). As these genes are under the control of the Rhl quorum-sensing system, it was surmized that this regulation was through the rhlI gene, encoding the C4-homoserine lactone (HSL) autoinducer synthase. AlgR repressed rhlI transcription and bound the rhlI promoter in vitro directly at the rhlI-ABS (5′-CCGTTCATC-3′), located −28 bp upstream of the transcriptional start site (Figures 3A,B). Deletion of the AlgR binding site in the rhlI promoter showed a similar biofilm growth defect as in the algR deletion strain, providing additional evidence that AlgR suppressed the quorum sensing system at rhlI (Morici et al., 2007).
Another set of genes under the exclusive control of the Rhl QS system and highly regulated by AlgR, are the rhlAB and rhlC genes (Ochsner and Reiser, 1995; Croda-Garcia et al., 2011). These genes, encode key enzymes that are important for the production of the surface-active amphipathic glycolipids called rhamnolipids. Rhamnolipids are important wetting agents for group-coordinated swarming motility (Deziel et al., 2003), normal biofilm formation (Davey et al., 2003; Espinosa-Urgel, 2003), and are a virulence factor (McClure and Schiller, 1996; Jensen et al., 2007). In a biofilm, rhamnolipids are required for microcolony formation (Pamp and Tolker-Nielsen, 2007), fluid channel formation (Davey et al., 2003), and bacterial dispersion from a mature biofillm (Boles et al., 2005). The ΔalgR mutant was unable to form structured biofilms, and instead, developed into a flat mat of cells over the course of 6-days (Morici et al., 2007). Both rhlA promoter activity and rhamnolipid production increased in an algR deletion strain indicating that AlgR represses rhlA in biofilms. Additionally, AlgR bound to the rhlA promoter in vitro, at rhlA-ABS1 (5′-CCGTTCGTC-3), located −702 bp upstream of the rhlA transcriptional start site (Figures 3A,B). Therefore, the biofilm defect in the ΔalgR strain was attributed to an increase in rhamnolipid production that subsequently disrupted the biofilm structure.
More recently, it was demonstrated that over the course of an 11 day biofilm in non-mucoid (PAO1) and mucoid (PA17) strains, algR expression is negatively correlated with rhlA and rhlB expression. Specifically, a decrease in algR transcription in PAO1 at later time points (day 7 and beyond) was correlated with increased rhlA and rhlB transcription, where biofilm dispersion would occur (Wang et al., 2013a). It has also been demonstrated that AlgR phosphorylation modulates rhamnolipid production, as a phospho-defective isoform (algRD54N) was unable to express rhamnolipids, while a phospho-mimetic isoform (algRD54E) was able to express rhamnolipids. In support of this, rhamnolipid-dependent swarming motility was affected by the algRD54N allele, and complemented with a strain expressing the rhlAB genes (Okkotsu et al., 2013b). These results indicate that AlgR phosphorylation adds yet another level of regulation to rhamnolipid production. As mentioned above, type IVa pili expression and function (through the fimUpilVWXY1E genes) are dependent on AlgR (Lizewski et al., 2004). Type IVa pili are required for biofilm formation, as it is involved with initial micro-colony formation (O'Toole and Kolter, 1998; Heydorn et al., 2002). Type IVa pili also play a role in stalk and cap formation during biofilm establishment (Klausen et al., 2003; Wang et al., 2013b). Together, these results support an idea the AlgR levels may act to coordinate rhamnolipid production and twitching motility, possibly to aid in either early colonization or later biofilm dispersion.
AlgR represses the expression of genes involved with anaerobic metabolism
Defining the AlgR regulon is of great interest; in addition to regulating alginate production in mucoid P. aeruginosa, type IVa pili mediated twitching motility, and the Rhl quorum sensing and rhamnolipid production in non-mucoid cells, AlgR likely has an even larger role in the organism's physiology. By comparing an algR deletion strain to its parental wild type PAO1 strain, it was demonstrated that AlgR is involved with regulating hundreds of genes in logarithmic, stationary, and biofilm modes of growth (Lizewski et al., 2002; Morici et al., 2007), and regulating the expression of at least 47 proteins during logarithmic growth (Lizewski et al., 2002). In all, 95 genes during mid-log phase and 59 genes during stationary phase were differentially regulated two-fold or greater, and 885 genes were differentially expressed when algR was overexpressed (Lizewski et al., 2004). While the majority of genes regulated by AlgR in mid-log and stationary phase encode hypothetical proteins, many of the known genes indicate AlgR may be involved with repressing anaerobic metabolism genes.
The transcriptional regulator, Anr (anaerobic regulation of arginine catabolism and nitrate reduction), is a ortholog of E. coli FNR, and regulates the anaerobic denitrification system (Sawers, 1991; Ye et al., 1995; Winteler and Haas, 1996). Anr is a homodimeric, [4Fe-4S]+2 cluster-containing transcriptional regulator of the Fnr-Crp family. Anr is activated at low oxygen concentrations; increased oxygen can dissociate the homodimer and inactivate Anr (Yoon et al., 2007). During anaerobic growth, Anr, along with Dnr (a NO-sensing, heme-containing transcriptional regulator) and NarXL (NO−3 sensing TCS) regulate denitrification, and genes required for fermentation of arginine and pyruvate (Zimmermann et al., 1991; Castiglione et al., 2009).
According to global transcriptional analysis, there is an overlap between Anr-regulated genes and AlgR-repressed genes (Lizewski et al., 2004). Some of these include: (i) arcA (PA5171), part of the arcDABC operon encoding anaerobic arginine deiminase enzymes, which catabolizes arginine to generate ATP through substrate level phosphorylation under anaerobic conditions (Vander Wauven et al., 1984; Galimand et al., 1991); (ii) hemN (PA1546), which encodes an oxygen-independent copropophyrinogen III oxidase important for heme biosynthesis under anaerobiosis (Rompf et al., 1998) and essential for anaerobic growth (Filiatrault et al., 2006); (iii) ccoP2 (PA1555), ccoO2 (PA1556), and ccoN2 (PA1557), ccb3-2 type cytochrome oxidase components expressed at low oxygen concentrations (Comolli and Donohue, 2004); (iv) oprG (PA4067), which is an outer-membrane diffusion-driven specific transporter of hydrophobic molecules whose expression increases under anaerobic conditions in the presence of iron (McPhee et al., 2009; Touw et al., 2010), and; (v) hcnB (PA2194), a gene encoding hydrogen cyanide synthase responsible for the production of HCN, and a possible mediator of anaerobic growth (Zimmermann et al., 1991; Gallagher and Manoil, 2001; Cody et al., 2009).
There is substantial evidence to suggest that P. aeruginosa prefers to respire under microaerobic conditions (Schreiber et al., 2007). The CF lung is a low oxygen or anaerobic environment (Su and Hassett, 2012), and macrocolonies of P. aeruginosa found in the intraluminal space contain very low oxygen (2.5 mmHg, compared with 180 mmHg in the bronchial lumen) (Worlitzsch et al., 2002). Additionally, according to a survey of P. aeruginosa isolates from CF patients over time, organisms adapted for anaerobic metabolism are selected for during chronic infection (Hoboth et al., 2009). When “hypermutator” (late-CF infection) isolates were compared with their isogenic “non-mutator” (early-CF infection) isolates from the same patients, gene expression and/or protein production of anr, arcDABC, oprG, and hemN were increased in the chronic isolates as compared to the early isolates (Hoboth et al., 2009). Based on these results and the microarray data, AlgR appears to repress genes that are favorable for anaerobic growth in PAO1 when the organism is growing aerobically in logarithmic phase.
AlgR and hydrogen cyanide production
Among AlgR regulated genes within the Anr regulon, the hcnABC (PA2193–2195) genes are the best studied. Hydrogen cyanide (HCN) is a volatile, highly reactive molecule that inhibits respiratory cytochromes and metalloenzymes. HCN is produced primarily at microaerobic conditions at high-cell density (Castric, 1983). HCN production may serve to adapt P. aeruginosa to low oxygen environments by shutting down its own low-affinity cytochrome oxidases (Williams et al., 2007). P. aeruginosa resists self-poisoning by expressing cyanide-insensitive cytochromes (CIO) (Cunningham and Williams, 1995; Cunningham et al., 1997).
The hcnABC genes encode the synthase complex that creates HCN from glycine by oxidative decarboxylation (Castric, 1977; Laville et al., 1998). Expression of the hcnC gene contributes to killing in Drosophila melanogaster (Broderick et al., 2008) and Caenorhabditis elegans (Gallagher and Manoil, 2001) models of infection. HCN production may also be a clinically relevant virulence factor in CF patients. Cyanide in the CF sputum is associated with decreased pulmonary function as determined by forced expiratory volume (FEV) and forced vital capacity (FVC) (Ryall et al., 2008). Additionally, increased cyanide production was observed by 21 mucoid clinical CF isolates as compared to non-mucoid isolates from the same patient and sputum sample (Carterson et al., 2004). Detection of cyanide was proposed to be a potential in vivo biomarker for virulent, micro-aerobically growing P. aeruginosa (Sanderson et al., 2008). However, recent data suggest that HCN production may not be a biomarker for P. aeruginosa infection, as increased HCN levels in the bronchoalveolar lavage of CF patients was attributed to neutrophilic inflammation (Stutz et al., 2011), and HCN was also generated by salivary peroxidase in the oral cavity (Dummer et al., 2013).
Regulation of hcnABC expression is complex. The hcnA promoter is controlled by a number of factors, including Anr (Zimmermann et al., 1991), small RNA regulatory proteins GacA/RsmA (Pessi and Haas, 2001; Lapouge et al., 2007), quorum sensing regulators LasR and RhlR (Pessi and Haas, 2000), as well as AlgR (Carterson et al., 2004; Cody et al., 2009). The hcnA promoter contains three transcriptional start sites (T1–T3) (Pessi and Haas, 2000; Cody et al., 2009). Transcription from T1 is regulated by both LasR and RhlR, as this promoter region has corresponding luxα and luxβ binding sequences for these quorum-sensing regulators. Transcription from T2 is expressed microaerobically, and maximal transcription requires a combination of LasR, RhlR, and Anr proteins (Pessi and Haas, 2000).
In P. aeruginosa strain PAO1, T3 is located upstream of the luxβ box and is controlled by AlgR. The hcnA-ABS (5′-CCGTCGTTC-3′) is located −160 bp upstream of T3 (Figure 3A) (Carterson et al., 2004; Cody et al., 2009). This AlgR binding sequence is located immediately downstream of PA2192 (encoding a hypothetal protein), and exoY (a secreted adenylate cyclase). Interestingly, AlgR regulation of the hcnA gene is strain-dependent. Several P. aeruginosa strains contain chromosomal deletions of a region that would otherwise encode an AlgR binding site within the hcnA promoter region. Some strains (i.e., PA14, PAK) as well as several CF isolates have regions flanking exoY deleted (in the ORFs of PA2192 and PA2190) (Wolfgang et al., 2003a), which remove the AlgR-dependent hcnA-ABS from the chromosome. As a consequence, cyanide production in strains with such deletions are responsive to Anr and the quorum sensing regulators, but not responsive to AlgR (Cody et al., 2009).
Mucoid P. aeruginosa express and produce higher levels of hcnA and HCN as compared to isogenic non-mucoid strains (Firoved and Deretic, 2003; Carterson et al., 2004). AlgR positively regulates hcnA transcription in mucoid strains; inactivation of the algR gene decreases hcnA mRNA and HCN production (Carterson et al., 2004). In contrast, deletion of algR in non-mucoid strains resulted in increased HCN production, indicating that AlgR can represses hcnA in non-mucoid backgrounds (Cody et al., 2009). Additionally, AlgR phosphorylation is required for this repression, as non-mucoid strains expressing algRD54N displays increased HCN production. The reason for the apparent switch in phenotypes between mucoid and non-mucoid cells is currently unknown, but it may be due to effects of the other regulators that control the hcnA promoter (Cody et al., 2009). A possible system that could mediate this differential expression of hcnA is the RsmAYZ system, as RsmA has been demonstrated to post-translationally affect the expression of hcnA (Pessi and Haas, 2001; Pessi et al., 2001). As described below, AlgR regulation of the type III secretion system (T3SS) is likewise mediated through the RsmA/Y/Z system.
AlgR and the type III secretion system
The type III secretion system (T3SS) is a needle-like structure that injects effector proteins (e.g., toxins) into host cells. Bacterial surface-contact, and low calcium concentrations in laboratory settings induce the expression of the T3SS in non-mucoid P. aeruginosa (Iglewski et al., 1978; Vallis et al., 1999). Expression of some acute virulence factors, including the T3SS machinery, is inversely correlated with the mucoid status of the organism (Mohr et al., 1990b; Wu et al., 2004). The reason for the inverse correlation between acute virulence factors and mucoidy could be that down regulation of the expression of virulence factors helps the organism to evade the host immune response, and to concentrate its energy into alginate production.
AlgR plays an important role in suppressing the T3SS in the context of a mucoid (mucA) background (Wu et al., 2004; Yahr and Wolfgang, 2006; Jones et al., 2010; Intile et al., 2014). Using the main transcriptional regulator of the T3SS (encoded by exsA) and expression of T3SS effector protein ExoS as readouts, P. aeruginosa carrying a mucA22 allele (conferring mucoidy) exhibits suppressed T3SS expression as compared to non-mucoid organisms. Meanwhile, a mucA22ΔalgR double-deletion restored both exsA and exoS expression to wild type (in strain PAK) levels, and plasmid-expression of algR reduced exsA and exoS expression, indicating that AlgR represses these T3SS genes (Wu et al., 2004).
As discussed previously, the AlgU regulon is highly active in mucoid cells, leading to alginate biosynthesis and increased algR transcription (Mohr et al., 1990a, 1991, 1992; Martin et al., 1993b, 1994). However, AlgR-dependent suppression of the T3SS is unrelated to alginate production, as a mucA22ΔalgD allele has no affect on T3SS expression. These data indicate that AlgR is involved with T3SS modulation under conditions in which AlgU is activated (Wu et al., 2004).
Further studies showed that AlgR modulates T3SS expression indirectly through two mechanisms. The P. aeruginosa cAMP-responsive protein called virulence factor regulator (Vfr) is a cAMP-binding transcriptional regulator (West et al., 1994). Expression of cyaA or cyaB encoding adenylate cyclases increases intracellular cAMP levels, leading to higher Vfr activity, which results in activation of the T3SS (Wolfgang et al., 2003b). AlgR regulates the T3SS through control of Vfr. Deletion of both algZ and algR (a ΔalgZΔalgR strain) increased transcriptional activity from a Vfr-responsive promoter, while algZR expression from a plasmid decreased transcriptional activity from this promoter (Jones et al., 2010). These results indicate that AlgZR repression of the T3SS could be mediated through its repression of Vfr.
AlgR is also proposed to regulate the T3SS through the RsmAYZ small RNA system (composed of the regulatory protein RsmA, and the small RNAs RsmY and RsmZ) (Jones et al., 2010; Intile et al., 2014). RsmA is an RNA-binding translational regulator that normally represses translation of its target transcripts. The small RNAs encoded by rsmY and rsmZ typically inhibit RsmA activity by competitively binding to the protein, thereby repressing RsmA activity (and de-repressing translation). RsmA (CsrA) proteins can also activate gene expression by altering mRNA secondary structure, mRNA stability, and/or ribosomal recruitment (Wei et al., 2001; Patterson-Fortin et al., 2013).
It is suspected that AlgR indirectly represses the T3SS by regulating rsmA, rsmY, and rsmZ expression. The evidence for this pathway is that rsmA, rsmY, and rsmZ transcription increased when algR is over-expressed, and that expression of AlgR in the mucoid background is correlated with increased expression of rsmA, rsmY, and rsmZ (Jones et al., 2010; Intile et al., 2014). RsmA-regulated transcripts are usually repressed by free-RsmA (e.g., tssA1, encoding a type VI secretion protein). However, recent work showed that exsCEBA expression (encoding regulatory proteins for T3SS activation) was activated either directly or indirectly by free-RsmA. The authors propose that small changes in RsmA availability could explain the target gene's different responses to RsmA/RsmY/RsmZ (Jones et al., 2010; Intile et al., 2014). In other words, AlgR may activate RsmA, RsmY, and RsmZ (and because all three are expressed, RsmA is competitively bound and inhibited by RsmY and RsmZ leading to relatively decreased levels of free-RsmA and non-activation of the T3SS in the mucoid background (Jones et al., 2010; Intile et al., 2014). Altogether, AlgR represses the T3SS in the mucoid background through Vfr and RsmA/Y/Z. More work is required to determine whether AlgR directly regulates Vfr or RsmA/Y/Z, or if there is an indirect component to this regulation that ultimately results in repression of the T3SS in the mucoid background.
AlgR's repressive role on T3SS is in contrast to its activator role on alginate production. In addition, AlgR has dual functions as a repressor and activator of HCN production in non-mucoid and mucoid backgrounds, respectively. In light of recent evidence of AlgR's effect on T3SS through the RsmA/Y/Z system, and the fact that RsmA also regulates hcnA expression, we speculate that AlgR may play a key role in influencing the expression of other virulence factors such as HCN production through the RsmA/Y/Z system.
AlgR controls the expression of many virulence determinants
AlgR globally affects transcription of a broad range of virulence factors including alginate production, twitching motility, the Rhl quorum sensing system, rhamnolipid production, T3SS, biofilm formation and hydrogen cyanide production (Figure 5). Therefore, it was ascertained whether or not algR is required for virulence. A component of P. aeruginosa virulence is its ability to survive reactive oxygen intermediates. AlgR represses genes required for survival against oxidative stress (Lizewski et al., 2002). Strain PAO700 (algR::Gm) was more susceptible to hypochlorite treatment, but more resistant to both hydrogen peroxide and myeloperoxidase compared to its isogenic PAO1 parental strain. The algR mutant was more resistant to killing by murine macrophages and primary human neutrophils, indicating that AlgR may normally repress the genes that are necessary to survive exposure to primary host response cells (Lizewski et al., 2002). On a related note, it was also demonstrated that a transposon insertion in algR (in strain PA14) results in increased persistence (persistence in this study was defined by the ability to grow in 5 ug/mL of oflaxicin) (De Groote et al., 2009). Although the direct mechanism is unknown, it supports previous data indicating AlgR may also regulate the expression of antibiotic resistance genes (Lizewski et al., 2002).
Figure 5.
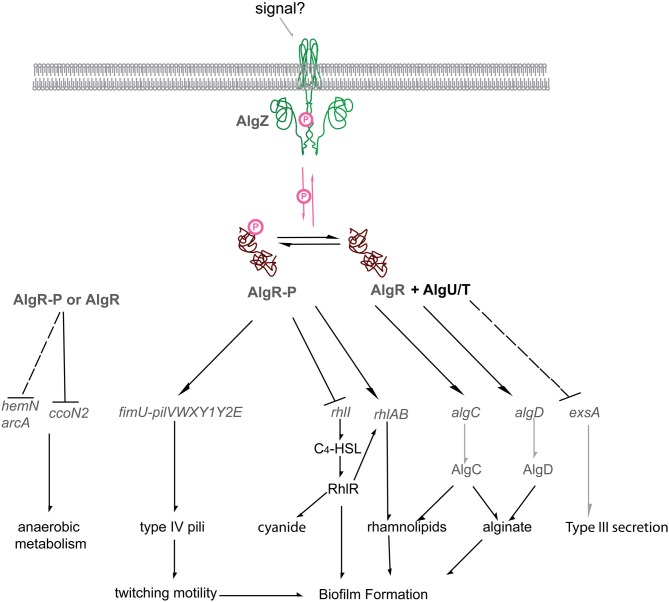
The P. aeruginosa AlgR regulon. Diagrammatic representation of the current AlgR regulon. Phosphorylated AlgR activates expression of the pseudopilin operon fimU pilVWXY1Y2, as well as modulates the expression of rhlA, but represses rhlI. Unphosphorylated AlgR activates expression of alginate biosynthetic genes algD and algC but represses the expression of the type III secretion system through RsmA/RsmY/RsmZ. Arrows, AlgR stimulated gene expression; Block arrows, AlgR repressed gene expression; AlgR-P, phosphorylated AlgR; AlgR, unphosphorylated AlgR; AlgR + AlgU/T, unphosphorylated AlgR and the alternative sigma factor AlgU (AlgT).
The algR gene was required for virulence in two different murine infection models and a competition assay. Strain PAO700 (algR::Gm) was tested in acute septicemia and acute pneumonia models of infection in C57BL/6j mice. Mice infected by intraperitoneal injection with PAO700 show increased survival as compared to mice infected with the wild type PAO1 strain. The same algR inactivated strain was cleared more quickly from murine lungs after acute infection. Co-infection with PAO700 and PAO1 resulted in decreased recovery of PAO700 as compared to wild type from the mice, suggesting an algR requirement for maximal virulence in these models. Interestingly, constitutive expression of algR decreased virulence as well suggesting that AlgR protein levels in P. aeruginosa need to be balanced for pathogenicity of the organism (Lizewski et al., 2002). The repression of virulence during increased AlgR expression is consistent with its regulation of the alginate system, as increased AlgR expression suppressed alginate production (Deretic and Konyecsni, 1989).
In all, AlgR appears to be a global regulator of P. aeruginosa virulence, and likely affects the organism's fitness in a broad manner. AlgR is also required for alginate production in a mucoid background, suggesting its importance in chronic pulmonary infections of CF victims. This response regulator is required for two phenotypes: twitching motility via fimU and alginate production through algD. However, global transcriptome analysis indicated that it controls many different genes, including those associated with quorum sensing, type IV pili, type III secretion system, anaerobic metabolism, cyanide and rhamnolipid production (Figure 5). Several questions remain: Does AlgR control these different P. aeruginosa genes directly or indirectly? What is/are the signal(s) to which AlgZ and ultimately AlgR respond? How is AlgR able to activate transcription regardless of its phosphorylation state? Current studies are underway to answer these questions.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Michael L. Vasil and Heath F. Damron for critical reading of the manuscript. This work was supported by grants from the National Institutes of Health NIAID AI1094487 to Michael J. Schurr, NIH Training grant 2T32 AI 052066 to Yuta Okkotsu.
References
- Adams M. D., Goglin K., Molyneaux N., Hujer K. M., Lavender H., Jamison J. J., et al. (2008). Comparative genome sequence analysis of multidrug-resistant Acinetobacter baumannii. J. Bacteriol. 190, 8053–8064 10.1128/JB.00834-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alm R. A., Hallinan J. P., Watson A. A., Mattick J. S. (1996). Fimbrial biogenesis genes of Pseudomonas aeruginosa: pilW and pilX increase the similarity of type 4 fimbriae to the GSP protein-secretion systems and pilY1 encodes a gonococcal PilC homologue. Mol. Microbiol. 22, 161–173 10.1111/j.1365-2958.1996.tb02665.x [DOI] [PubMed] [Google Scholar]
- Alm R. A., Mattick J. S. (1995). Identification of a gene, pilV, required for type 4 fimbrial biogenesis in Pseudomonas aeruginosa, whose product possesses a pre-pilin-like leader sequence. Mol. Microbiol. 16, 485–496 10.1111/j.1365-2958.1995.tb02413.x [DOI] [PubMed] [Google Scholar]
- Alm R. A., Mattick J. S. (1996). Identification of two genes with prepilin-like leader sequences involved in type 4 fimbrial biogenesis in Pseudomonas aeruginosa. J. Bacteriol. 178, 3809–3817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almirante B., Limon E., Freixas N., Gudiol F., Program V. I. (2012). Laboratory-based surveillance of hospital-acquired catheter-related bloodstream infections in Catalonia. Results of the VINCat Program (2007–2010). Enferm. Infecc. Microbiol. Clin. 30Suppl. 3, 13–19 10.1016/S0213-005X(12)70091-5 [DOI] [PubMed] [Google Scholar]
- Anthony M., Rose B., Pegler M. B., Elkins M., Service H., Thamotharampillai K., et al. (2002). Genetic analysis of Pseudomonas aeruginosa isolates from the sputa of Australian adult cystic fibrosis patients. J. Clin. Microbiol. 40, 2772–2778 10.1128/JCM.40.8.2772-2778.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong D. S., Grimwood K., Carzino R., Carlin J. B., Olinsky A., Phelan P. D. (1995). Lower respiratory infection and inflammation in infants with newly diagnosed cystic fibrosis. BMJ 310, 1571–1572 10.1136/bmj.310.6994.1571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballmann M., Rabsch P., Von Der Hardt H. (1998). Long-term follow up of changes in FEV1 and treatment intensity during Pseudomonas aeruginosa colonisation in patients with cystic fibrosis. Thorax 53, 732–737 10.1136/thx.53.9.732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barchinger S. E., Ades S. E. (2013). Regulated proteolysis: control of the Escherichia coli sigma(E)-dependent cell envelope stress response. Subcell. Biochem. 66, 129–160 10.1007/978-94-007-5940-4_6 [DOI] [PubMed] [Google Scholar]
- Baynham P. J., Brown A. L., Hall L. L., Wozniak D. J. (1999). Pseudomonas aeruginosa AlgZ, a ribbon-helix-helix DNA-binding protein, is essential for alginate synthesis and algD transcriptional activation. Mol. Microbiol. 33, 1069–1080 10.1046/j.1365-2958.1999.01550.x [DOI] [PubMed] [Google Scholar]
- Baynham P. J., Ramsey D. M., Gvozdyev B. V., Cordonnier E. M., Wozniak D. J. (2006). The Pseudomonas aeruginosa ribbon-helix-helix DNA-binding protein AlgZ (AmrZ) controls twitching motility and biogenesis of type IV pili. J. Bacteriol. 188, 132–140 10.1128/JB.188.1.132-140.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baynham P. J., Wozniak D. J. (1996). Identification and characterization of AlgZ, an AlgT-dependent DNA- binding protein required for Pseudomonas aeruginosa algD transcription. Mol. Microbiol. 22, 97–108 10.1111/j.1365-2958.1996.tb02659.x [DOI] [PubMed] [Google Scholar]
- Belba M. K., Petrela E. Y., Belba A. G. (2013). Epidemiology of infections in a burn unit, Albania. Burns 39, 1456–1467 10.1016/j.burns.2013.03.013 [DOI] [PubMed] [Google Scholar]
- Belete B., Lu H., Wozniak D. J. (2008). Pseudomonas aeruginosa AlgR regulates type IV pilus biosynthesis by activating transcription of the fimU-pilVWXY1Y2E operon. J. Bacteriol. 190, 2023–2030 10.1128/JB.01623-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjarnsholt T., Givskov M. (2007). The role of quorum sensing in the pathogenicity of the cunning aggressor Pseudomonas aeruginosa. Anal. Bioanal. Chem. 387, 409–414 10.1007/s00216-006-0774-x [DOI] [PubMed] [Google Scholar]
- Boles B. R., Thoendel M., Singh P. K. (2005). Rhamnolipids mediate detachment of Pseudomonas aeruginosa from biofilms. Mol. Microbiol. 57, 1210–1223 10.1111/j.1365-2958.2005.04743.x [DOI] [PubMed] [Google Scholar]
- Boucher J. C., Yu H., Mudd M. H., Deretic V. (1997). Mucoid Pseudomonas aeruginosa in cystic fibrosis: characterization of muc mutations in clinical isolates and analysis of clearance in a mouse model of respiratory infection. Infect. Immun. 65, 3838–3846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourret R. B. (2010). Receiver domain structure and function in response regulator proteins. Curr. Opin. Microbiol. 13, 142–149 10.1016/j.mib.2010.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bragonzi A., Wiehlmann L., Klockgether J., Cramer N., Worlitzsch D., Doring G., et al. (2006). Sequence diversity of the mucABD locus in Pseudomonas aeruginosa isolates from patients with cystic fibrosis. Microbiology 152, 3261–3269 10.1099/mic.0.29175-0 [DOI] [PubMed] [Google Scholar]
- Bren A., Eisenbach M. (2000). How signals are heard during bacterial chemotaxis: protein-protein interactions in sensory signal propagation. J. Bacteriol. 182, 6865–6873 10.1128/JB.182.24.6865-6873.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brint J. M., Ohman D. E. (1995). Synthesis of multiple exoproducts in Pseudomonas aeruginosa is under the control of RhlR-RhlI, another set of regulators in strain PAO1 with homology to the autoinducer-responsive LuxR-LuxI family. J. Bacteriol. 177, 7155–7163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broderick K. E., Chan A., Balasubramanian M., Feala J., Reed S. L., Panda M., et al. (2008). Cyanide produced by human isolates of Pseudomonas aeruginosa contributes to lethality in Drosophila melanogaster. J. Infect. Dis. 197, 457–464 10.1086/525282 [DOI] [PubMed] [Google Scholar]
- Brunskill E. W., Bayles K. W. (1996). Identification and molecular characterization of a putative regulatory locus that affects autolysis in Staphylococcus aureus. J. Bacteriol. 178, 611–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns J. L., Gibson R. L., McNamara S., Yim D., Emerson J., Rosenfeld M., et al. (2001). Longitudinal assessment of Pseudomonas aeruginosa in young children with cystic fibrosis. J. Infect. Dis. 183, 444–452 10.1086/318075 [DOI] [PubMed] [Google Scholar]
- Carterson A. J., Morici L. A., Jackson D. W., Frisk A., Lizewski S. E., Jupiter R., et al. (2004). The transcriptional regulator AlgR controls cyanide production in Pseudomonas aeruginosa. J. Bacteriol. 186, 6837–6844 10.1128/JB.186.20.6837-6844.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castiglione N., Rinaldo S., Giardina G., Cutruzzola F. (2009). The transcription factor DNR from Pseudomonas aeruginosa specifically requires nitric oxide and haem for the activation of a target promoter in Escherichia coli. Microbiology 155, 2838–2844 10.1099/mic.0.028027-0 [DOI] [PubMed] [Google Scholar]
- Castric P. A. (1977). Glycine metabolism by Pseudomonas aeruginosa: hydrogen cyanide biosynthesis. J. Bacteriol. 130, 826–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castric P. A. (1983). Hydrogen cyanide production by Pseudomonas aeruginosa at reduced oxygen levels. Can. J. Microbiol. 29, 1344–1349 10.1139/m83-209 [DOI] [PubMed] [Google Scholar]
- Chang W. S., Van De Mortel M., Nielsen L., Nino De Guzman G., Li X., Halverson L. J. (2007). Alginate production by Pseudomonas putida creates a hydrated microenvironment and contributes to biofilm architecture and stress tolerance under water-limiting conditions. J. Bacteriol. 189, 8290–8299 10.1128/JB.00727-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung J. K., Dupuy B., Deveson D. S., Rood J. I. (2004). The spatial organization of the VirR boxes is critical for VirR-mediated expression of the perfringolysin O gene, pfoA, from Clostridium perfringens. J. Bacteriol. 186, 3321–3330 10.1128/JB.186.11.3321-3330.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung J. K., Rood J. I. (2000). The VirR response regulator from Clostridium perfringens binds independently to two imperfect direct repeats located upstream of the pfoA promoter. J. Bacteriol. 182, 57–66 10.1128/JB.182.1.57-66.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chugani S. A., Whiteley M., Lee K. M., D'Argenio D., Manoil C., Greenberg E. P. (2001). QscR, a modulator of quorum-sensing signal synthesis and virulence in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 98, 2752–2757 10.1073/pnas.051624298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciofu O., Lee B., Johannesson M., Hermansen N. O., Meyer P., Hoiby N. (2008). Investigation of the algT operon sequence in mucoid and non-mucoid Pseudomonas aeruginosa isolates from 115 Scandinavian patients with cystic fibrosis and in 88 in vitro non-mucoid revertants. Microbiology 154, 103–113 10.1099/mic.0.2007/010421-0 [DOI] [PubMed] [Google Scholar]
- Clementi F. (1997). Alginate production by Azotobacter vinelandii. Crit. Rev. Biotechnol. 17, 327–361 10.3109/07388559709146618 [DOI] [PubMed] [Google Scholar]
- Cody W. L., Pritchett C. L., Jones A. K., Carterson A. J., Jackson D., Frisk A., et al. (2009). Pseudomonas aeruginosa AlgR controls cyanide production in an AlgZ dependent manner. J. Bacteriol. 191, 2993–3002 10.1128/JB.01156-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comolli J. C., Donohue T. J. (2004). Differences in two Pseudomonas aeruginosa cbb3 cytochrome oxidases. Mol. Microbiol. 51, 1193–1203 10.1046/j.1365-2958.2003.03904.x [DOI] [PubMed] [Google Scholar]
- Coyne M. J., Russell K. S., Coyle C. L., Goldberg J. B. (1994). The Pseudomonas aeruginosa algC gene encodes phosphoglucomutase, required for the synthesis of a complete lipopolysaccharide core. J. Bacteriol. 176, 3500–3507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croda-Garcia G., Grosso-Becerra V., Gonzalez-Valdez A., Servin-Gonzalez L., Soberon-Chavez G. (2011). Transcriptional regulation of Pseudomonas aeruginosa rhlR: role of the CRP orthologue Vfr (virulence factor regulator) and quorum-sensing regulators LasR and RhlR. Microbiology 157, 2545–2555 10.1099/mic.0.050161-0 [DOI] [PubMed] [Google Scholar]
- Cunningham L., Pitt M., Williams H. D. (1997). The cioAB genes from Pseudomonas aeruginosa code for a novel cyanide-insensitive terminal oxidase related to the cytochrome bd quinol oxidases. Mol. Microbiol. 24, 579–591 10.1046/j.1365-2958.1997.3561728.x [DOI] [PubMed] [Google Scholar]
- Cunningham L., Williams H. D. (1995). Isolation and characterization of mutants defective in the cyanide-insensitive respiratory pathway of Pseudomonas aeruginosa. J. Bacteriol. 177, 432–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damron F. H., Goldberg J. B. (2012). Proteolytic regulation of alginate overproduction in Pseudomonas aeruginosa. Mol. Microbiol. 84, 595–607 10.1111/j.1365-2958.2012.08049.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damron F. H., Owings J. P., Okkotsu Y., Varga J. J., Schurr J. R., Goldberg J. B., et al. (2012). Analysis of the Pseudomonas aeruginosa regulon controlled by the sensor kinase KinB and sigma factor RpoN. J. Bacteriol. 194, 1317–1330 10.1128/JB.06105-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Argenio D. A., Calfee M. W., Rainey P. B., Pesci E. C. (2002). Autolysis and autoaggregation in Pseudomonas aeruginosa colony morphology mutants. J. Bacteriol. 184, 6481–6489 10.1128/JB.184.23.6481-6489.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzins A., Chakrabarty A. M. (1984). Cloning of genes controlling alginate biosynthesis from mucoid cystic fibrosis isolate of Pseudomonas aeruginosa. J. Bacteriol. 159, 9–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey M. E., Caiazza N. C., O'Toole G. A. (2003). Rhamnolipid surfactant production affects biofilm architecture in Pseudomonas aeruginosa PAO1. J. Bacteriol. 185, 1027–1036 10.1128/JB.185.3.1027-1036.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies D. G., Parsek M. R., Pearson J. P., Iglewski B. H., Costerton J. W., Greenberg E. P. (1998). The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280, 295–298 10.1126/science.280.5361.295 [DOI] [PubMed] [Google Scholar]
- De Groote V. N., Verstraeten N., Fauvart M., Kint C. I., Verbeeck A. M., Beullens S., et al. (2009). Novel persistence genes in Pseudomonas aeruginosa identified by high-throughput screening. FEMS Microbiol. Lett. 297, 73–79 10.1111/j.1574-6968.2009.01657.x [DOI] [PubMed] [Google Scholar]
- Dekimpe V., Deziel E. (2009). Revisiting the quorum-sensing hierarchy in Pseudomonas aeruginosa: the transcriptional regulator RhlR regulates LasR-specific factors. Microbiology 155, 712–723 10.1099/mic.0.022764-0 [DOI] [PubMed] [Google Scholar]
- Delic-Attree I., Toussaint B., Garin J., Vignais P. M. (1997). Cloning, sequence and mutagenesis of the structural gene of Pseudomonas aeruginosa CysB, which can activate algD transcription. Mol. Microbiol. 24, 1275–1284 10.1046/j.1365-2958.1997.4121799.x [DOI] [PubMed] [Google Scholar]
- Del Papa M. F., Perego M. (2011). Enterococcus faecalis virulence regulator FsrA binding to target promoters. J. Bacteriol. 193, 1527–1532 10.1128/JB.01522-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denny T. P. (1995). Involvement of bacterial polysaccharides in plant pathogenesis. Annu. Rev. Phytopathol. 33, 173–197 10.1146/annurev.py.33.090195.001133 [DOI] [PubMed] [Google Scholar]
- Deretic V., Dikshit R., Konyecsni W. M., Chakrabarty A. M., Misra T. K. (1989). The algR gene, which regulates mucoidy in Pseudomonas aeruginosa, belongs to a class of environmentally responsive genes. J. Bacteriol. 171, 1278–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V., Gill J. F., Chakrabarty A. M. (1987a). Gene algD coding for GDPmannose dehydrogenase is transcriptionally activated in mucoid Pseudomonas aeruginosa. J. Bacteriol. 169, 351–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V., Gill J. F., Chakrabarty A. M. (1987b). Pseudomonas aeruginosa infection in cystic fibrosis:nucleotide sequence and transcriptional regulation of the algD gene. Nuc. Acids. Res. 15, 4567–4581 10.1093/nar/15.11.4567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V., Govan J. R., Konyecsni W. M., Martin D. W. (1990). Mucoid Pseudomonas aeruginosa in cystic fibrosis: mutations in the muc loci affect transcription of the algR and algD genes in response to environmental stimuli. Mol. Microbiol. 4, 189–196 10.1111/j.1365-2958.1990.tb00586.x [DOI] [PubMed] [Google Scholar]
- Deretic V., Hibler N. S., Holt S. C. (1992a). Immunocytochemical analysis of AlgP (Hp1), a histonelike element participating in control of mucoidy in Pseudomonas aeruginosa. J. Bacteriol. 174, 824–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V., Konyecsni W. M. (1989). Control of mucoidy in Pseudomonas aeruginosa: transcriptional regulation of algR and identification of the second regulatory gene, algQ. J. Bacteriol. 171, 3680–3688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V., Konyecsni W. M. (1990). A procaryotic regulatory factor with a histone H1-like carboxy-terminal domain: clonal variation of repeats within algP, a gene involved in regulation of mucoidy in Pseudomonas aeruginosa. J. Bacteriol. 172, 5544–5554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V., Leveau J. H., Mohr C. D., Hibler N. S. (1992b). In vitro phosphorylation of AlgR, a regulator of mucoidy in Pseudomonas aeruginosa, by a histidine protein kinase and effects of small phospho-donor molecules. Mol. Microbiol. 6, 2761–2767 10.1111/j.1365-2958.1992.tb01455.x [DOI] [PubMed] [Google Scholar]
- Devault J. D., Hendrickson W., Kato J., Chakrabarty A. M. (1991). Environmentally regulated algD promoter is responsive to the cAMP receptor protein in Escherichia coli. Mol. Microbiol. 5, 2503–2509 10.1111/j.1365-2958.1991.tb02096.x [DOI] [PubMed] [Google Scholar]
- Devries C. A., Ohman D. E. (1994). Mucoid-to-nonmucoid conversion in alginate-producing Pseudomonas aeruginosa often results from spontaneous mutations in algT, encoding a putative alternate sigma factor, and shows evidence for autoregulation. J. Bacteriol. 176, 6677–6687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deziel E., Gopalan S., Tampakaki A. P., Lepine F., Padfield K. E., Saucier M., et al. (2005). The contribution of MvfR to Pseudomonas aeruginosa pathogenesis and quorum sensing circuitry regulation: multiple quorum sensing-regulated genes are modulated without affecting lasRI, rhlRI or the production of N-acyl-L-homoserine lactones. Mol. Microbiol. 55, 998–1014 10.1111/j.1365-2958.2004.04448.x [DOI] [PubMed] [Google Scholar]
- Deziel E., Lepine F., Milot S., He J., Mindrinos M. N., Tompkins R. G., et al. (2004). Analysis of Pseudomonas aeruginosa 4-hydroxy-2-alkylquinolines (HAQs) reveals a role for 4-hydroxy-2-heptylquinoline in cell-to-cell communication. Proc. Natl. Acad. Sci. U.S.A. 101, 1339–1344 10.1073/pnas.0307694100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deziel E., Lepine F., Milot S., Villemur R. (2003). rhlA is required for the production of a novel biosurfactant promoting swarming motility in Pseudomonas aeruginosa: 3-(3-hydroxyalkanoyloxy)alkanoic acids (HAAs), the precursors of rhamnolipids. Microbiology 149, 2005–2013 10.1099/mic.0.26154-0 [DOI] [PubMed] [Google Scholar]
- Diggle S. P., Winzer K., Chhabra S. R., Worrall K. E., Camara M., Williams P. (2003). The Pseudomonas aeruginosa quinolone signal molecule overcomes the cell density-dependency of the quorum sensing hierarchy, regulates rhl-dependent genes at the onset of stationary phase and can be produced in the absence of LasR. Mol. Microbiol. 50, 29–43 10.1046/j.1365-2958.2003.03672.x [DOI] [PubMed] [Google Scholar]
- Doggett R. G. (1969). Incidence of mucoid Pseudomonas aeruginosa from clinical sources. Appl. Microbiol. 18, 936–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doggett R. G., Harrison G. M., Stillwell R. N., Wallis E. S. (1966). An atypical Pseudomonas aeruginosa associated with cstic fibrosis of the pancreas. J. Pediatry 68, 215–221 10.1016/S0022-3476(66)80152-X [DOI] [Google Scholar]
- Dummer J., Storer M., Sturney S., Scott-Thomas A., Chambers S., Swanney M., et al. (2013). Quantification of hydrogen cyanide (HCN) in breath using selected ion flow tube mass spectrometry—HCN is not a biomarker of Pseudomonas in chronic suppurative lung disease. J. Breath. Res. 7:017105 10.1088/1752-7155/7/1/017105 [DOI] [PubMed] [Google Scholar]
- Engelman D. M., Steitz T. A., Goldman A. (1986). Identifying nonpolar transbilayer helices in amino acid sequences of membrane proteins. Annu. Rev. Biophys. Biophys. Chem. 15, 321–353 10.1146/annurev.bb.15.060186.001541 [DOI] [PubMed] [Google Scholar]
- Espinosa-Urgel M. (2003). Resident parking only: rhamnolipids maintain fluid channels in biofilms. J. Bacteriol. 185, 699–700 10.1128/JB.185.3.699-700.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans L. R., Linker A. (1973). Production and characterization of the slime polysaccharide of Pseudomonas aeruginosa. J. Bacteriol. 116, 915–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakhr M. K., Penaloza-Vazquez A., Chakrabarty A. M., Bender C. L. (1999). Regulation of alginate biosynthesis in Pseudomonas syringae pv. syringae. J. Bacteriol. 181, 3478–3485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fialho A. M., Zielinski N. A., Fett W. F., Chakrabarty A. M., Berry A. (1990). Distribution of alginate gene sequences in the Pseudomonas rRNA homology group I-Azomonas-Azotobacter lineage of superfamily B procaryotes. Appl. Environ. Microbiol. 56, 436–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filiatrault M. J., Picardo K. F., Ngai H., Passador L., Iglewski B. H. (2006). Identification of Pseudomonas aeruginosa genes involved in virulence and anaerobic growth. Infect. Immun. 74, 4237–4245 10.1128/IAI.02014-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firoved A. M., Boucher J. C., Deretic V. (2002). Global genomic analysis of AlgU (sigma(E))-dependent promoters (Sigmulon) in Pseudomonas aeruginosa and implications for inflammatory processes in cystic fibrosis. J. Bacteriol. 184, 1057–1064 10.1128/jb.184.4.1057-1064.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firoved A. M., Deretic V. (2003). Microarray analysis of global gene expression in mucoid Pseudomonas aeruginosa. J. Bacteriol. 185, 1071–1081 10.1128/JB.185.3.1071-1081.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firoved A. M., Ornatowski W., Deretic V. (2004). Microarray analysis reveals induction of lipoprotein genes in mucoid Pseudomonas aeruginosa: implications for inflammation in cystic fibrosis. Infect. Immun. 72, 5012–5018 10.1128/IAI.72.9.5012-5018.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin M. J., Nivens D. E., Weadge J. T., Howell L. P. (2011). Biosynthesis of the Pseudomonas aeruginosa extracellular polysaccharides Alginate, Pel and Psl. Front. Cell. Infect.Microbiol. 2:167 10.3389/fmicb.2011.00167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara S., Zielinski N. A., Chakrabarty A. M. (1993). Enhancer-like activity of AlgR1-binding site in alginate gene activation: positional, orientational, and sequence specificity. J. Bacteriol. 175, 5452–5459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuqua C. (2006). The QscR quorum-sensing regulon of Pseudomonas aeruginosa: an orphan claims its identity. J. Bacteriol. 188, 3169–3171 10.1128/JB.188.9.3169-3171.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuqua W. Q., Winans S. C., Greenberg E. P. (1994). Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J. Bacteriol. 176, 269–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gacesa P., Russell N. J. (1990). The structure and properties of alginate, in PseudomonasInfections and Alginates: Biochemistry, Genetics and Pathology, eds Gacesa P., Russell N. J. (London: Chapman and Hall; ), 29–49 [Google Scholar]
- Galimand M., Gamper M., Zimmermann A., Haas D. (1991). Positive FNR-like control of anaerobic arginine degradation and nitrate respiration in Pseudomonas aeruginosa. J. Bacteriol. 173, 1598–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher L. A., Manoil C. (2001). Pseudomonas aeruginosa PAO1 Kills Caenorhabditis elegans by cyanide poisoning. J. Bacteriol. 183, 6207–6214 10.1128/JB.183.21.6207-6214.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher L. A., McKnight S. L., Kuznetsova M. S., Pesci E. C., Manoil C. (2002). Functions required for extracellular quinolone signaling by Pseudomonas aeruginosa. J. Bacteriol. 184, 6472–6480 10.1128/JB.184.23.6472-6480.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin M. Y. (2010). Diversity of structure and function of response regulator output domains. Curr. Opin. Microbiol. 13, 150–159 10.1016/j.mib.2010.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin M. Y., Nikolskaya A. N., Koonin E. V. (2001). Novel domains of the prokaryotic two-component signal transduction systems. FEMS Microbiol. Lett. 203, 11–21 10.1111/j.1574-6968.2001.tb10814.x [DOI] [PubMed] [Google Scholar]
- Gao R., Stock A. M. (2009). Biological insights from structures of two-component proteins. Annu. Rev. Microbiol. 63, 133–154 10.1146/annurev.micro.091208.073214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giltner C. L., Habash M., Burrows L. L. (2010). Pseudomonas aeruginosa minor pilins are incorporated into type IV pili. J. Mol. Biol. 398, 444–461 10.1016/j.jmb.2010.03.028 [DOI] [PubMed] [Google Scholar]
- Giltner C. L., Rana N., Lunardo M. N., Hussain A. Q., Burrows L. L. (2011). Evolutionary and functional diversity of the Pseudomonas type IVa pilin island. Environ. Microbiol. 13, 250–264 10.1111/j.1462-2920.2010.02327.x [DOI] [PubMed] [Google Scholar]
- Goldberg J. B., Dahnke T. (1992). Pseudomonas aeruginosa AlgB, which modulates the expression of alginate, is a member of the NtrC subclass of prokaryotic regulators. Mol. Microbiol. 6, 59–66 10.1111/j.1365-2958.1992.tb00837.x [DOI] [PubMed] [Google Scholar]
- Goldberg J. B., Hatano K., Pier G. B. (1993). Synthesis of lipopolysaccharide O side chains by Pseudomonas aeruginosa PAO1 requires the enzyme phosphomannomutase. J. Bacteriol. 175, 1605–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg J. B., Ohman D. E. (1987). Construction and characterization of Pseudomonas aeruginosa algB mutants: role of algB in high-level production of alginate. J. Bacteriol. 169, 1593–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes M. Z., Machado C. R., Da Conceicao Mde S., Ortega J. A., Neves S. M., Lourenco M. C., et al. (2011). Outbreaks, persistence, and high mortality rates of multiresistant Pseudomonas aeruginosa infections in a hospital with AIDS-predominant admissions. Braz. J. Infect. Dis. 15, 312–322 10.1016/S1413-8670(11)70198-2 [DOI] [PubMed] [Google Scholar]
- Govan J. R., Deretic V. (1996). Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol. Rev. 60, 539–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl D., Gaggar A., Bruscia E., Hector A., Marcos V., Jung A., et al. (2012). Innate immunity in cystic fibrosis lung disease. J. Cyst. Fibros. 11, 363–382 10.1016/j.jcf.2012.07.003 [DOI] [PubMed] [Google Scholar]
- Heiniger R. W., Winther-Larsen H. C., Pickles R. J., Koomey M., Wolfgang M. C. (2010). Infection of human mucosal tissue by Pseudomonas aeruginosa requires sequential and mutually dependent virulence factors and a novel pilus-associated adhesin. Cell Microbiol. 12, 1158–1173 10.1111/j.1462-5822.2010.01461.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry R. L., Dorman D. C., Brown J. M., Mellis C. (1982). Mucoid Pseudomonas aeruginosa in cystic fibrosis. Austr. Paediatr. J. 18, 43–45 [DOI] [PubMed] [Google Scholar]
- Henry R. L., Mellis C. M., Petrovic L. (1992). Mucoid Pseudomonas aeruginosa is a marker of poor survival in cystic fibrosis. Pediatr. Pulmonol. 12, 158–161 10.1002/ppul.1950120306 [DOI] [PubMed] [Google Scholar]
- Hentzer M., Teitzel G. M., Balzer G. J., Heydorn A., Molin S., Givskov M., et al. (2001). Alginate overproduction affects Pseudomonas aeruginosa biofilm structure and function. J. Bacteriol. 183, 5395–5401 10.1128/JB.183.18.5395-5401.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershberger C. D., Ye R. W., Parsek M. R., Xie Z.-D., Chakrabarty A. M. (1995). The algT (algU) gene of Pseudomonas aeruginosa, a key regulator involved in alginate biosynthesis, encodes an alternative s factor (sE). Proc. Natl. Acad. Sci. U.S.A. 92, 7941–7945 10.1073/pnas.92.17.7941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heydorn A., Ersboll B., Kato J., Hentzer M., Parsek M. R., Tolker-Nielsen T., et al. (2002). Statistical analysis of Pseudomonas aeruginosa biofilm development: impact of mutations in genes involved in twitching motility, cell-to-cell signaling, and stationary-phase sigma factor expression. Appl. Environ. Microbiol. 68, 2008–2017 10.1128/AEM.68.4.2008-2017.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa T., Boon-Chieng S., Mitaku S. (1998). SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics 14, 378–379 10.1093/bioinformatics/14.4.378 [DOI] [PubMed] [Google Scholar]
- Hoang T. T., Sullivan S. A., Cusick J. K., Schweizer H. P. (2002). Beta-ketoacyl acyl carrier protein reductase (FabG) activity of the fatty acid biosynthetic pathway is a determining factor of 3-oxo-homoserine lactone acyl chain lengths. Microbiology 148, 3849–3856 [DOI] [PubMed] [Google Scholar]
- Hoboth C., Hoffmann R., Eichner A., Henke C., Schmoldt S., Imhof A., et al. (2009). Dynamics of adaptive microevolution of hypermutable Pseudomonas aeruginosa during chronic pulmonary infection in patients with cystic fibrosis. J. Infect. Dis. 200, 118–130 10.1086/599360 [DOI] [PubMed] [Google Scholar]
- Hogardt M., Hoboth C., Schmoldt S., Henke C., Bader L., Heesemann J. (2007). Stage-specific adaptation of hypermutable Pseudomonas aeruginosa isolates during chronic pulmonary infection in patients with cystic fibrosis. J. Infect. Dis. 195, 70–80 10.1086/509821 [DOI] [PubMed] [Google Scholar]
- Horcajada J. P., Shaw E., Padilla B., Pintado V., Calbo E., Benito N., et al. (2013). Healthcare-associated, community-acquired and hospital-acquired bacteraemic urinary tract infections in hospitalized patients: a prospective multicentre cohort study in the era of antimicrobial resistance. Clin. Microbiol. Infect. 19, 962–968 10.1111/1469-0691.12089 [DOI] [PubMed] [Google Scholar]
- Iglewski B. H., Sadoff J., Bjorn M. J., Maxwell E. S. (1978). Pseudomonas aeruginosa exoenzyme S: an adenosine diphosphate ribosyltransferase distinct from toxin A. Proc. Natl. Acad. Sci. U.S.A. 75, 3211–3215 10.1073/pnas.75.7.3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Intile P. J., Diaz M. R., Urbanowski M. L., Wolfgang M. C., Yahr T. L. (2014). The AlgZR two-component system recalibrates the RsmAYZ posttranscriptional regulatory system to inhibit expression of the Pseudomonas aeruginosa type III secretion system. J. Bacteriol. 196, 357–366 10.1128/JB.01199-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen P. O., Bjarnsholt T., Phipps R., Rasmussen T. B., Calum H., Christoffersen L., et al. (2007). Rapid necrotic killing of polymorphonuclear leukocytes is caused by quorum-sensing-controlled production of rhamnolipid by Pseudomonas aeruginosa. Microbiology 153, 1329–1338 10.1099/mic.0.2006/003863-0 [DOI] [PubMed] [Google Scholar]
- Johansen H. K., Hoiby N. (1992). Seasonal onset of initial colonisation and chronic infection with Pseudomonas aeruginosa in patients with cystic fibrosis in Denmark. Thorax 47, 109–111 10.1136/thx.47.2.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M. D., Garrett C. K., Bond J. E., Coggan K. A., Wolfgang M. C., Redinbo M. R. (2012). Pseudomonas aeruginosa PilY1 binds integrin in an RGD- and calcium-dependent manner. PLoS ONE 6:e29629 10.1371/journal.pone.0029629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones A. K., Fulcher N. B., Balzer G. J., Urbanowski M. L., Pritchett C. L., Schurr M. J., et al. (2010). Activation of the Pseudomonas aeruginosa AlgU regulon through mucA mutation inhibits cAMP/Vfr signaling. J. Bacteriol. 21, 5709–5717 10.1128/JB.00526-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallberg M., Wang H., Wang S., Peng J., Wang Z., Lu H., et al. (2012). Template-based protein structure modeling using the RaptorX web server. Nat. Protoc. 7, 1511–1522 10.1038/nprot.2012.085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato J., Chakrabarty A. M. (1991). Purification of the regulatory protein AlgR1 and its binding in the far upstream region of the algD promoter in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 88, 1760–1764 10.1073/pnas.88.5.1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley L. A., Sternberg M. J. (2009). Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4, 363–371 10.1038/nprot.2009.2 [DOI] [PubMed] [Google Scholar]
- Khawaja A., Zubairi A. B., Durrani F. K., Zafar A. (2013). Etiology and outcome of severe community acquired pneumonia in immunocompetent adults. BMC Infect. Dis. 13:94 10.1186/1471-2334-13-94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Forst S. (2001). Genomic analysis of the histidine kinase family in bacteria and archaea. Microbiology 147, 1197–1212 [DOI] [PubMed] [Google Scholar]
- Kimbara K., Chakrabarty A. M. (1989). Control of alginate synthesis in Pseudomonas aeruginosa: regulation of the algR1 gene. Biochem. Biophys. Res. Com. 164, 601–608 10.1016/0006-291X(89)91502-7 [DOI] [PubMed] [Google Scholar]
- Klausen M., Heydorn A., Ragas P., Lambertsen L., Aaes-Jorgensen A., Molin S., et al. (2003). Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants. Mol. Microbiol. 48, 1511–1524 10.1046/j.1365-2958.2003.03525.x [DOI] [PubMed] [Google Scholar]
- Koenig R. L., Ray J. L., Maleki S. J., Smeltzer M. S., Hurlburt B. K. (2004). Staphylococcus aureus AgrA binding to the RNAIII-agr regulatory region. J. Bacteriol. 186, 7549–7555 10.1128/JB.186.22.7549-7555.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraxenberger T., Fried L., Behr S., Jung K. (2012). First insights into the unexplored two-component system YehU/YehT in Escherichia coli. J. Bacteriol. 194, 4272–4284 10.1128/JB.00409-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A., Larsson B., Von Heijne G., Sonnhammer E. L. (2001). Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580 10.1006/jmbi.2000.4315 [DOI] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. (1982). A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157, 105–132 10.1016/0022-2836(82)90515-0 [DOI] [PubMed] [Google Scholar]
- Lapouge K., Sineva E., Lindell M., Starke K., Baker C. S., Babitzke P., et al. (2007). Mechanism of hcnA mRNA recognition in the Gac/Rsm signal transduction pathway of Pseudomonas fluorescens. Mol. Microbiol. 66, 341–356 10.1111/j.1365-2958.2007.05909.x [DOI] [PubMed] [Google Scholar]
- Laville J., Blumer C., Von Schroetter C., Gaia V., Defago G., Keel C., et al. (1998). Characterization of the hcnABC gene cluster encoding hydrogen cyanide synthase and anaerobic regulation by ANR in the strictly aerobic biocontrol agent Pseudomonas fluorescens CHA0. J. Bacteriol. 180, 3187–3196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledgham F., Ventre I., Soscia C., Foglino M., Sturgis J. N., Lazdunski A. (2003). Interactions of the quorum sensing regulator QscR: interaction with itself and the other regulators of Pseudomonas aeruginosa LasR and RhlR. Mol. Microbiol. 48, 199–210 10.1046/j.1365-2958.2003.03423.x [DOI] [PubMed] [Google Scholar]
- Leid J. G., Willson C. J., Shirtliff M. E., Hassett D. J., Parsek M. R., Jeffers A. K. (2005). The exopolysaccharide alginate protects Pseudomonas aeruginosa biofilm bacteria from IFN-gamma-mediated macrophage killing. J. Immunol. 175, 7512–7518 10.4049/jimmunol.175.11.7512 [DOI] [PubMed] [Google Scholar]
- Leitao J. H., Sa-Correia I. (1995). Growth-phase-dependent alginate synthesis, activity of biosynthetic enzymes and transcription of alginate genes in Pseudomonas aeruginosa. Arch. Microbiol. 163, 217–222 [DOI] [PubMed] [Google Scholar]
- Lepine F., Milot S., Deziel E., He J., Rahme L. G. (2004). Electrospray/mass spectrometric identification and analysis of 4-hydroxy-2-alkylquinolines (HAQs) produced by Pseudomonas aeruginosa. J. Am. Soc. Mass. Spectrom. 15, 862–869 10.1016/j.jasms.2004.02.012 [DOI] [PubMed] [Google Scholar]
- Li Z., Kosorok M. R., Farrell P. M., Laxova A., West S. E., Green C. G., et al. (2005). Longitudinal development of mucoid Pseudomonas aeruginosa infection and lung disease progression in children with cystic fibrosis. JAMA 293, 581–588 10.1001/jama.293.5.581 [DOI] [PubMed] [Google Scholar]
- Lightbown J. W., Jackson F. L. (1956). Inhibition of cytochrome systems of heart muscle and certain bacteria by the antagonists of dihydrostreptomycin: 2-alkyl-4-hydroxyquinoline N-oxides. Biochem. J. 63, 130–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linker A., Jones R. S. (1964). A polysaccharide resembling alginic acid from a Pseudomonas micro-organism. Nature 204, 187–188 10.1038/204187a0 [DOI] [PubMed] [Google Scholar]
- Lizewski S. E., Lundberg D. S., Schurr M. J. (2002). The transcriptional regulator AlgR is essential for Pseudomonas aeruginosa pathogenesis. Infect. Immun. 70, 6083–6093 10.1128/IAI.70.11.6083-6093.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizewski S. E., Schurr J. R., Jackson D. W., Frisk A., Carterson A. J., Schurr M. J. (2004). Identification of AlgR-regulated genes in Pseudomonas aeruginosa by use of microarray analysis. J. Bacteriol. 186, 5672–5684 10.1128/JB.186.17.5672-5684.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukat G. S., McCleary W. R., Stock A. M., Stock J. B. (1992). Phosphorylation of bacterial response regulator proteins by low molecular weight phospho-donors. Proc. Natl. Acad. Sci. U.S.A. 89, 718–722 10.1073/pnas.89.2.718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S., Selvaraj U., Ohman D. E., Quarless R., Hassett D. J., Wozniak D. J. (1998). Phosphorylation-independent activity of the response regulators AlgB and AlgR in promoting alginate biosynthesis in mucoid Pseudomonas aeruginosa. J. Bacteriol. 180, 956–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machan Z. A., Taylor G. W., Pitt T. L., Cole P. J., Wilson R. (1992). 2-Heptyl-4-hydroxyquinoline N-oxide, an antistaphylococcal agent produced by Pseudomonas aeruginosa. J. Antimicrob. Chemother. 30, 615–623 10.1093/jac/30.5.615 [DOI] [PubMed] [Google Scholar]
- Mahajan-Miklos S., Tan M. W., Rahme L. G., Ausubel F. M. (1999). Molecular mechanisms of bacterial virulence elucidated using a Pseudomonas aeruginosa-Caenorhabditis elegans pathogenesis model. Cell 96, 47–56 10.1016/S0092-8674(00)80958-7 [DOI] [PubMed] [Google Scholar]
- Martin D. W., Holloway B. W., Deretic V. (1993a). Characterization of a locus determining the mucoid status of Pseudomonas aeruginosa: AlgU shows sequence similarities with a Bacillus sigma factor. J. Bacteriol. 175, 1153–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D. W., Schurr M. J., Mudd M. H., Govan J. R. W., Holloway B. W., Deretic V. (1993b). Mechanism of conversion to mucoidy in Pseudomonas aeruginosa infecting cystic fibrosis patients. Proc. Natl. Acad. Sci. U.S.A. 90, 8377–8381 10.1073/pnas.90.18.8377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D. W., Schurr M. J., Yu H., Deretic V. (1994). Analysis of promoters controlled by the putative sigma factor AlgU regulating conversion to mucoidy in Pseudomonas aeruginosa: relationship to sigma E and stress response. J. Bacteriol. 176, 6688–6696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Hackert E., Stock A. M. (1997). The DNA-binding domain of OmpR: crystal structures of a winged helix transcription factor. Structure 5, 109–124 10.1016/S0969-2126(97)00170-6 [DOI] [PubMed] [Google Scholar]
- May T. B., Shinabarger D., Maharaj R., Kato J., Chu L., Devault J. D., et al. (1991). Alginate synthesis by Pseudomonas aeruginosa: a key pathogenic factor in chronic pulmonary infections of cystic fibrosis patients. Clin. Microbiol. Rev. 4, 191–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure C. D., Schiller N. L. (1996). Inhibition of macrophage phagocytosis by Pseudomonas aeruginosa rhamnolipids in vitro and in vivo. Curr. Microbiol. 33, 109–117 10.1007/s002849900084 [DOI] [PubMed] [Google Scholar]
- McGrath S., Wade D. S., Pesci E. C. (2004). Dueling quorum sensing systems in Pseudomonas aeruginosa control the production of the Pseudomonas quinolone signal (PQS). FEMS Microbiol. Lett. 230, 27–34 10.1016/S0378-1097(03)00849-8 [DOI] [PubMed] [Google Scholar]
- McKnight S. L., Iglewski B. H., Pesci E. C. (2000). The Pseudomonas quinolone signal regulates rhl quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 182, 2702–2708 10.1128/JB.182.10.2702-2708.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPhee J. B., Tamber S., Bains M., Maier E., Gellatly S., Lo A., et al. (2009). The major outer membrane protein OprG of Pseudomonas aeruginosa contributes to cytotoxicity and forms an anaerobically regulated, cation-selective channel. FEMS Microbiol. Lett. 296, 241–247 10.1111/j.1574-6968.2009.01651.x [DOI] [PubMed] [Google Scholar]
- Mena A., Smith E. E., Burns J. L., Speert D. P., Moskowitz S. M., Perez J. L., et al. (2008). Genetic adaptation of Pseudomonas aeruginosa to the airways of cystic fibrosis patients is catalyzed by hypermutation. J. Bacteriol. 190, 7910–7917 10.1128/JB.01147-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr C. D., Deretic V. (1990). Gene-scrambling mutagenesis: generation and analysis of insertional mutations in the alginate regulatory region of Pseudomonas aeruginosa. J. Bacteriol. 172, 6252–6260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr C. D., Hibler N. S., Deretic V. (1991). AlgR, a response regulator controlling mucoidy in Pseudomonas aeruginosa, binds to the FUS sites of the algD promoter located unusually far upstream from the mRNA start site. J. Bacteriol. 173, 5136–5143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr C. D., Leveau J. H., Krieg D. P., Hibler N. S., Deretic V. (1992). AlgR-binding sites within the algD promoter make up a set of inverted repeats separated by a large intervening segment of DNA. J. Bacteriol. 174, 6624–6633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr C. D., Martin D. W., Konyecsni W. M., Govan J. R., Lory S., Deretic V. (1990a). Role of the far-upstream sites of the algD promoter and the algR and rpoN genes in environmental modulation of mucoidy in Pseudomonas aeruginosa. J. Bacteriol. 172, 6576–6580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr C. D., Rust L., Albus A. M., Iglewski B. H., Deretic V. (1990b). Expression patterns of genes encoding elastase and controlling mucoidy: co-ordinate regulation of two virulence factors in Pseudomonas aeruginosa isolates from cystic fibrosis. Mol. Microbiol. 4, 2103–2110 10.1111/j.1365-2958.1990.tb00571.x [DOI] [PubMed] [Google Scholar]
- Morici L. A., Carterson A. J., Wagner V. E., Frisk A., Schurr J. R., Zu Bentrup K. H., et al. (2007). Pseudomonas aeruginosa AlgR represses the Rhl quorum-sensing system in a biofilm-specific manner. J. Bacteriol. 189, 7752–7764 10.1128/JB.01797-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray T. S., Egan M., Kazmierczak B. I. (2007). Pseudomonas aeruginosa chronic colonization in cystic fibrosis patients. Curr. Opin. Pediatry 19, 83–88 10.1097/MOP.0b013e3280123a5d [DOI] [PubMed] [Google Scholar]
- Nikolskaya A. N., Galperin M. Y. (2002). A novel type of conserved DNA-binding domain in the transcriptional regulators of the AlgR/AgrA/LytR family. Nucleic Acids Res. 30, 2453–2459 10.1093/nar/30.11.2453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon G. M., Armstrong D. S., Carzino R., Carlin J. B., Olinsky A., Robertson C. F., et al. (2001). Clinical outcome after early Pseudomonas aeruginosa infection in cystic fibrosis. J. Pediatry 138, 699–704 10.1067/mpd.2001.112897 [DOI] [PubMed] [Google Scholar]
- Novick R. P. (2003). Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 48, 1429–1449 10.1046/j.1365-2958.2003.03526.x [DOI] [PubMed] [Google Scholar]
- Nunez C., Moreno S., Soberon-Chavez G., Espin G. (1999). The Azotobacter vinelandii response regulator AlgR is essential for cyst formation. J. Bacteriol. 181, 141–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochsner U. A., Reiser J. (1995). Autoinducer-mediated regulation of rhamnolipid biosurfactant synthesis in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 92, 6424–6428 10.1073/pnas.92.14.6424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochsner U. A., Vasil A. I., Vasil M. L. (1995). Role of the ferric uptake regulator of Pseudomonas aeruginosa in the regulation of siderophores and exotoxin A expression: purification and activity on iron-regulated promoters. J. Bacteriol. 177, 7194–7201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okkotsu Y., Pritchett C., Schurr M. J. (2013a). Regulation of exopolysaccharide biosynthesis in Pseudomonas aeruginosa, in Regulation of Bacterial Virulence, eds Vasil M. L., Darwin A. J. (Washington, DC: ASM Press; ), 171–189 [Google Scholar]
- Okkotsu Y., Tieku P., Fitzsimmons L. F., Churchill M. E., Schurr M. J. (2013b). Pseudomonas aeruginosa AlgR phosphorylation modulates rhamnolipid production and motility. J. Bacteriol. 195, 5499–5515 10.1128/JB.00726-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver A., Canton R., Campo P., Baquero F., Blazquez J. (2000). High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science 288, 1251–1254 10.1126/science.288.5469.1251 [DOI] [PubMed] [Google Scholar]
- Olvera C., Goldberg J. B., Sanchez R., Soberon-Chavez G. (1999). The Pseudomonas aeruginosa algC gene product participates in rhamnolipid biosynthesis. FEMS Microbiol. Lett. 179, 85–90 10.1111/j.1574-6968.1999.tb08712.x [DOI] [PubMed] [Google Scholar]
- Orans J., Johnson M. D., Coggan K. A., Sperlazza J. R., Heiniger R. W., Wolfgang M. C., et al. (2010). Crystal structure analysis reveals Pseudomonas PilY1 as an essential calcium-dependent regulator of bacterial surface motility. Proc. Natl. Acad. Sci. U.S.A. 107, 1065–1070 10.1073/pnas.0911616107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsi G. B., Franchi C., Marrone R., Giordano A., Rocco M., Venditti M. (2012). Laboratory confirmed bloodstream infection aetiology in an intensive care unit: eight years study. Ann. Ig. 24, 269–278 [PubMed] [Google Scholar]
- O'Toole G. A., Kolter R. (1998). Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30, 295–304 10.1046/j.1365-2958.1998.01062.x [DOI] [PubMed] [Google Scholar]
- Pamp S. J., Tolker-Nielsen T. (2007). Multiple roles of biosurfactants in structural biofilm development by Pseudomonas aeruginosa. J. Bacteriol. 189, 2531–2539 10.1128/JB.01515-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papagheorghe R. (2012). Bloodstream infections in immunocompromised hosts. Roum. Arch. Microbiol. Immunol. 71, 87–94 [PubMed] [Google Scholar]
- Parad R. B., Gerard C. J., Zurakowski D., Nichols D. P., Pier G. B. (1999). Pulmonary outcome in cystic fibrosis is influenced primarily by mucoid Pseudomonas aeruginosa infection and immune status and only modestly by genotype. Infect. Immun. 67, 4744–4750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson J. S., Kofoid E. C. (1992). Communication modules in bacterial signaling proteins. Annu. Rev. Genet. 26, 71–112 10.1146/annurev.ge.26.120192.000443 [DOI] [PubMed] [Google Scholar]
- Parsek M. R., Val D. L., Hanzelka B. L., Cronan J. E., Jr., Greenberg E. P. (1999). Acyl homoserine-lactone quorum-sensing signal generation. Proc. Natl. Acad. Sci. U.S.A. 96, 4360–4365 10.1073/pnas.96.8.4360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passador L., Cook J. M., Gambello M. J., Rust L., Iglewski B. H. (1993). Expression of Pseudomonas aeruginosa virulence genes requires cell-to-cell communication. Science 260, 1127–1130 10.1126/science.8493556 [DOI] [PubMed] [Google Scholar]
- Patterson-Fortin L. M., Vakulskas C. A., Yakhnin H., Babitzke P., Romeo T. (2013). Dual posttranscriptional regulation via a cofactor-responsive mRNA leader. J. Mol. Biol. 425, 3662–3677 10.1016/j.jmb.2012.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson S. S., Høiby N., Espersen F. (1992). Role of alginate in infection with mucoid Pseudomonas aeruginosa in cystic fibrosis. Thorax 47, 6–13 10.1136/thx.47.1.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelton J. G., Kustu S., Wemmer D. E. (1999). Solution structure of the DNA-binding domain of NtrC with three alanine substitutions. J. Mol. Biol. 292, 1095–1110 10.1006/jmbi.1999.3140 [DOI] [PubMed] [Google Scholar]
- Penaloza-Vazquez A., Fakhr M. K., Bailey A. M., Bender C. L. (2004). AlgR functions in algC expression and virulence in Pseudomonas syringae pv. syringae. Microbiology 150, 2727–2737 10.1099/mic.0.27199-0 [DOI] [PubMed] [Google Scholar]
- Pesci E. C., Milbank J. B., Pearson J. P., McKnight S., Kende A. S., Greenberg E. P., et al. (1999). Quinolone signaling in the cell-to-cell communication system of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 96, 11229–11234 10.1073/pnas.96.20.11229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pessi G., Haas D. (2000). Transcriptional control of the hydrogen cyanide biosynthetic genes hcnABC by the anaerobic regulator ANR and the quorum-sensing regulators LasR and RhlR in Pseudomonas aeruginosa. J. Bacteriol. 182, 6940–6949 10.1128/JB.182.24.6940-6949.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pessi G., Haas D. (2001). Dual control of hydrogen cyanide biosynthesis by the global activator GacA in Pseudomonas aeruginosa PAO1. FEMS Microbiol. Lett. 200, 73–78 10.1111/j.1574-6968.2001.tb10695.x [DOI] [PubMed] [Google Scholar]
- Pessi G., Williams F., Hindle Z., Heurlier K., Holden M. T., Camara M., et al. (2001). The global posttranscriptional regulator RsmA modulates production of virulence determinants and N-acylhomoserine lactones in Pseudomonas aeruginosa. J. Bacteriol. 183, 6676–6683 10.1128/JB.183.22.6676-6683.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pier G. B., Small G. J., Warren H. B. (1990). Protection against mucoid Pseudomonas aeruginosa in rodent models of endobronchial infections. Science 249, 537–540 10.1126/science.2116663 [DOI] [PubMed] [Google Scholar]
- Pindar D. F., Bucke C. (1975). The biosynthesis of alginic acid by Azotobacter vinelandii. Biochem. J. 152, 617–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryor E. E., Jr., Waligora E. A., Xu B., Dellos-Nolan S., Wozniak D. J., Hollis T. (2012). The transcription factor AmrZ utilizes multiple DNA binding modes to recognize activator and repressor sequences of Pseudomonas aeruginosa virulence genes. PLoS Pathog. 8:e1002648 10.1371/journal.ppat.1002648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranganathan S. C., Skoric B., Ramsay K. A., Carzino R., Gibson A. M., Hart E., et al. (2013). Geographical differences in first acquisition of Pseudomonas aeruginosa in cystic fibrosis. Ann. Am. Thorac. Soc. 10, 108–114 10.1513/AnnalsATS.201209-077OC [DOI] [PubMed] [Google Scholar]
- Regni C., Tipton P. A., Beamer L. J. (2002). Crystal structure of PMM/PGM: an enzyme in the biosynthetic pathway of P. aeruginosa virulence factors. Structure 10, 269–279 10.1016/S0969-2126(02)00705-0 [DOI] [PubMed] [Google Scholar]
- Regules J. A., Glasser J. S., Wolf S. E., Hospenthal D. R., Murray C. K. (2008). Endocarditis in burn patients: clinical and diagnostic considerations. Burns 34, 610–616 10.1016/j.burns.2007.08.002 [DOI] [PubMed] [Google Scholar]
- Risoen P. A., Havarstein L. S., Diep D. B., Nes I. F. (1998). Identification of the DNA-binding sites for two response regulators involved in control of bacteriocin synthesis in Lactobacillus plantarum C11. Mol. Gen. Genet. 259, 224–232 [DOI] [PubMed] [Google Scholar]
- Risoen P. A., Johnsborg O., Diep D. B., Hamoen L., Venema G., Nes I. F. (2001). Regulation of bacteriocin production in Lactobacillus plantarum depends on a conserved promoter arrangement with consensus binding sequence. Mol. Genet. Genomics 265, 198–206 10.1007/s004380000397 [DOI] [PubMed] [Google Scholar]
- Rompf A., Hungerer C., Hoffmann T., Lindenmeyer M., Romling U., Gross U., et al. (1998). Regulation of Pseudomonas aeruginosa hemF and hemN by the dual action of the redox response regulators Anr and Dnr. Mol. Microbiol. 29, 985–997 10.1046/j.1365-2958.1998.00980.x [DOI] [PubMed] [Google Scholar]
- Rowen D. W., Deretic V. (2000). Membrane-to-cytosol redistribution of ECF sigma factor AlgU and conversion to mucoidy in pseudomonas aeruginosa isolates from cystic fibrosis patients. Mol. Microbiol. 36, 314–327 10.1046/j.1365-2958.2000.01830.x [DOI] [PubMed] [Google Scholar]
- Rowley G., Spector M., Kormanec J., Roberts M. (2006). Pushing the envelope: extracytoplasmic stress responses in bacterial pathogens. Nat. Rev. Microbiol. 4, 383–394 10.1038/nrmicro1394 [DOI] [PubMed] [Google Scholar]
- Rudel T., Scheurerpflug I., Meyer T. F. (1995). Neisseria PilC protein identified as type-4 pilus tip-located adhesin. Nature 373, 357–359 10.1038/373357a0 [DOI] [PubMed] [Google Scholar]
- Rumbaugh K. P., Griswold J. A., Hamood A. N. (2000). The role of quorum sensing in the in vivo virulence of Pseudomonas aeruginosa. Microbes. Infect. 2, 1721–1731 10.1016/S1286-4579(00)01327-7 [DOI] [PubMed] [Google Scholar]
- Russell M. A., Darzins A. (1994). The pilE gene product of Pseudomonas aeruginosa, required for pilus biogenesis, shares amino acid sequence identity with the N-termini of type 4 prepilin proteins. Mol. Microbiol. 13, 973–985 10.1111/j.1365-2958.1994.tb00489.x [DOI] [PubMed] [Google Scholar]
- Ryall B., Davies J. C., Wilson R., Shoemark A., Williams H. D. (2008). Pseudomonas aeruginosa, cyanide accumulation and lung function in CF and non-CF bronchiectasis patients. Eur. Respir. J. 32, 740–747 10.1183/09031936.00159607 [DOI] [PubMed] [Google Scholar]
- Sanders L. H., Devadoss B., Raja G. V., O'Connor J., Su S., Wozniak D. J., et al. (2011). Epistatic roles for Pseudomonas aeruginosa MutS and DinB (DNA Pol IV) in coping with reactive oxygen species-induced DNA damage. PLoS ONE 6:e18824 10.1371/journal.pone.0018824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders L. H., Rockel A., Lu H., Wozniak D. J., Sutton M. D. (2006). Role of Pseudomonas aeruginosa dinB-encoded DNA polymerase IV in mutagenesis. J. Bacteriol. 188, 8573–8585 10.1128/JB.01481-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson K., Wescombe L., Kirov S. M., Champion A., Reid D. W. (2008). Bacterial cyanogenesis occurs in the cystic fibrosis lung. Eur. Respir. J. 2, 329–333 10.1183/09031936.00152407 [DOI] [PubMed] [Google Scholar]
- Sauer K., Camper A. K., Ehrlich G. D., Costerton J. W., Davies D. G. (2002). Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J. Bacteriol. 184, 1140–1154 10.1128/jb.184.4.1140-1154.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawers R. G. (1991). Identification and molecular characterization of a transcriptional regulator from Pseudomonas aeruginosa PAO1 exhibiting structural and functional similarity to the FNR protein of Escherichia coli. Mol. Microbiol. 5, 1469–1481 10.1111/j.1365-2958.1991.tb00793.x [DOI] [PubMed] [Google Scholar]
- Scaramozzino F., White A., Perego M., Hoch J. A. (2009). A unique GTP-dependent sporulation sensor histidine kinase in Bacillus anthracis. J. Bacteriol. 191, 687–692 10.1128/JB.01184-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber K., Krieger R., Benkert B., Eschbach M., Arai H., Schobert M., et al. (2007). The anaerobic regulatory network required for Pseudomonas aeruginosa nitrate respiration. J. Bacteriol. 189, 4310–4314 10.1128/JB.00240-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schurr M. J., Martin D. W., Mudd M. H., Deretic V. (1994). Gene cluster controlling conversion to alginate-overproducing phenotype in Pseudomonas aeruginosa: functional analysis in a heterologous host and role in the instability of mucoidy. J. Bacteriol. 176, 3375–3382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schurr M. J., Martin D. W., Mudd M. H., Hibler N. S., Boucher J. C., Deretic V. (1993). The algD promoter: regulation of alginate production by Pseudomonas aeruginosa in cystic fibrosis. Cel. Mol. Biol. Res. 39, 371–376 [PubMed] [Google Scholar]
- Schurr M. J., Yu H., Martinez-Salazar J. M., Boucher J. C., Deretic V. (1996). Control of AlgU, a member of the sigma E-like family of stress sigma factors, by the negative regulators MucA and MucB and Pseudomonas aeruginosa conversion to mucoidy in cystic fibrosis. J. Bacteriol. 178, 4997–5004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schurr M. J., Yu H., Martinez-Salazar J. M., Hibler N. S., Deretic V. (1995). Biochemical characterization and posttranslational modification of AlgU, a regulator of stress response in Pseudomonas aeruginosa. Biochem. Biophysic. Res. Comm. 216, 874–880 10.1006/bbrc.1995.2703 [DOI] [PubMed] [Google Scholar]
- Schwarzmann S., Boring J. R. (1971). Antiphagocytic effect of slime from a mucoid strain of P. aeruginosa. Infect. Immun. 3, 762–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seed P. C., Passador L., Iglewski B. H. (1995). Activation of the Pseudomonas aeruginosa lasI gene by LasR and the Pseudomonas autoinducer PAI: an autoinduction regulatory hierarchy. J. Bacteriol. 177, 654–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidote D. J., Barbieri C. M., Wu T., Stock A. M. (2008). Structure of the Staphylococcus aureus AgrA LytTR domain bound to DNA reveals a beta fold with an unusual mode of binding. Structure 16, 727–735 10.1016/j.str.2008.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonetti A., Ottaiano E., Diana M. V., Onza C., Triassi M. (2013). Epidemiology of hospital-acquired infections in an adult intensive care unit: results of a prospective cohort study. Ann. Ig 25, 281–289 [DOI] [PubMed] [Google Scholar]
- Sonnhammer E. L., Von Heijne G., Krogh A. (1998). A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 6, 175–182 [PubMed] [Google Scholar]
- Sousa D., Justo I., Dominguez A., Manzur A., Izquierdo C., Ruiz L., et al. (2013). Community-acquired pneumonia in immunocompromised older patients: incidence, causative organisms and outcome. Clin. Microbiol. Infect. 19, 187–192 10.1111/j.1469-0691.2012.03765.x [DOI] [PubMed] [Google Scholar]
- Stiver H. G., Zachidniak K., Speert D. P. (1988). Inhibition of polymorphonuclear leukocyte chemotaxis by the mucoid exopolysaccharide of Pseudomonas aeruginosa. Clin. Invest. Med. 11, 247–252 [PubMed] [Google Scholar]
- Stock A. M., Robinson V. L., Goudreau P. N. (2000). Two-component signal transduction. Annu. Rev. Biochem. 69, 183–215 10.1146/annurev.biochem.69.1.183 [DOI] [PubMed] [Google Scholar]
- Stock J. B., Ninfa A. J., Stock A. M. (1989). Protein phosphorylation and the regulation of adaptive responses in bacteria. Microbiol. Rev. 53, 450–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoodley P., Sauer K., Davies D. G., Costerton J. W. (2002). Biofilms as complex differentiated communities. Annu. Rev. Microbiol. 56, 187–209 10.1146/annurev.micro.56.012302.160705 [DOI] [PubMed] [Google Scholar]
- Straume D., Johansen R. F., Bjoras M., Nes I. F., Diep D. B. (2009). DNA binding kinetics of two response regulators, PlnC and PlnD, from the bacteriocin regulon of Lactobacillus plantarum C11. BMC Biochem. 10:17 10.1186/1471-2091-10-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stutz M. D., Gangell C. L., Berry L. J., Garratt L. W., Sheil B., Sly P. D., et al. (2011). Cyanide in bronchoalveolar lavage is not diagnostic for Pseudomonas aeruginosa in children with cystic fibrosis. Eur. Respir. J 37, 553–558 10.1183/09031936.00024210 [DOI] [PubMed] [Google Scholar]
- Su S., Hassett D. J. (2012). Anaerobic Pseudomonas aeruginosa and other obligately anaerobic bacterial biofilms growing in the thick airway mucus of chronically infected cystic fibrosis patients: an emerging paradigm or “Old Hat”? Expert. Opin. Ther. Targets 16, 859–873 10.1517/14728222.2012.708025 [DOI] [PubMed] [Google Scholar]
- Tart A. H., Blanks M. J., Wozniak D. J. (2006). The AlgT-dependent transcriptional regulator AmrZ (AlgZ) inhibits flagellum biosynthesis in mucoid, nonmotile Pseudomonas aeruginosa cystic fibrosis isolates. J. Bacteriol. 188, 6483–6489 10.1128/JB.00636-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toussaint B., Delic-Attree I., Vignais P. M. (1993). Pseudomonas aeruginosa contains an IHF-like protein that binds to the algD promoter. Biochem. Biophys. Res. Commun. 196, 416–421 10.1006/bbrc.1993.2265 [DOI] [PubMed] [Google Scholar]
- Touw D. S., Patel D. R., Van Den Berg B. (2010). The crystal structure of OprG from Pseudomonas aeruginosa, a potential channel for transport of hydrophobic molecules across the outer membrane. PLoS ONE 5:e15016 10.1371/journal.pone.0015016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallis A. J., Yahr T. L., Barbieri J. T., Frank D. W. (1999). Regulation of ExoS production and secretion by Pseudomonas aeruginosa in response to tissue culture conditions. Infect. Immun. 67, 914–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Wauven C., Pierard A., Kley-Raymann M., Haas D. (1984). Pseudomonas aeruginosa mutants affected in anaerobic growth on arginine: evidence for a four-gene cluster encoding the arginine deiminase pathway. J. Bacteriol. 160, 928–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venturi V., Otten M., Korse V., Brouwer B., Leong J., Weisbeek P. (1995). Alginate regulatory and biosynthetic gene homologs in Pseudomonas putida WCS358: correlation with the siderophore regulatory gene pfrA. Gene 155, 83–88 10.1016/0378-1119(94)00868-S [DOI] [PubMed] [Google Scholar]
- Wade D. S., Calfee M. W., Rocha E. R., Ling E. A., Engstrom E., Coleman J. P., et al. (2005). Regulation of Pseudomonas quinolone signal synthesis in Pseudomonas aeruginosa. J. Bacteriol. 187, 4372–4380 10.1128/JB.187.13.4372-4380.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Yu B., Tian D., Ni M. (2013a). Rhamnolipid but not motility is associated with the initiation of biofilm seeding dispersal of Pseudomonas aeruginosa strain PA17. J. Biosci. 38, 149–156 10.1007/s12038-012-9297-0 [DOI] [PubMed] [Google Scholar]
- Wang S., Parsek M. R., Wozniak D. J., Ma L. Z. (2013b). A spider web strategy of type IV pili-mediated migration to build a fibre-like Psl polysaccharide matrix in Pseudomonas aeruginosa biofilms. Environ. Microbiol. 15, 2238–2253 10.1111/1462-2920.12095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei B. L., Brun-Zinkernagel A. M., Simecka J. W., Pruss B. M., Babitzke P., Romeo T. (2001). Positive regulation of motility and flhDC expression by the RNA-binding protein CsrA of Escherichia coli. Mol. Microbiol. 40, 245–256 10.1046/j.1365-2958.2001.02380.x [DOI] [PubMed] [Google Scholar]
- West S. E., Sample A. K., Runyen-Janecky L. J. (1994). The vfr gene product, required for Pseudomonas aeruginosa exotoxin A and protease production, belongs to the cyclic AMP receptor protein family. J. Bacteriol. 176, 7532–7542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitchurch C. B., Alm R. A., Mattick J. S. (1996). The alginate regulator AlgR and an associated sensor FimS are required for twitching motility in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A 93, 9839–9843 10.1073/pnas.93.18.9839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitchurch C. B., Erova T. E., Emery J. A., Sargent J. L., Harris J. M., Semmler A. B., et al. (2002). Phosphorylation of the Pseudomonas aeruginosa response regulator AlgR is essential for type IV fimbria-mediated twitching motility. J. Bacteriol. 184, 4544–4554 10.1128/JB.184.16.4544-4554.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiens J. R., Vasil A. I., Schurr M. J., Vasil M. L. (2014). Iron-regulated expression of alginate production, mucoid phenotype, and biofilm formation by Pseudomonas aeruginosa. MBio 5, e01010–e01013 10.1128/mBio.01010-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams H. D., Zlosnik J. E., Ryall B. (2007). Oxygen, cyanide and energy generation in the cystic fibrosis pathogen Pseudomonas aeruginosa. Adv. Microb. Physiol. 52, 1–71 10.1016/S0065-2911(06)52001-6 [DOI] [PubMed] [Google Scholar]
- Winson M. K., Camara M., Latifi A., Foglino M., Chhabra S. R., Daykin M., et al. (1995). Multiple N-acyl-L-homoserine lactone signal molecules regulate production of virulence determinants and secondary metabolites in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 92, 9427–9431 10.1073/pnas.92.20.9427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winsor G. L., Lam D. K., Fleming L., Lo R., Whiteside M. D., Yu N. Y., et al. (2011). Pseudomonas Genome Database: improved comparative analysis and population genomics capability for Pseudomonas genomes. Nucleic Acids Res. 39, D596–D600 10.1093/nar/gkq869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winteler H. V., Haas D. (1996). The homologous regulators ANR of Pseudomonas aeruginosa and FNR of Escherichia coli have overlapping but distinct specificities for anaerobically inducible promoters. Microbiology 142(Pt 3), 685–693 10.1099/13500872-142-3-685 [DOI] [PubMed] [Google Scholar]
- Wolfgang M. C., Kulasekara B. R., Liang X., Boyd D., Wu K., Yang Q., et al. (2003a). Conservation of genome content and virulence determinants among clinical and environmental isolates of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 100, 8484–8489 10.1073/pnas.0832438100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfgang M. C., Lee V. T., Gilmore M. E., Lory S. (2003b). Coordinate regulation of bacterial virulence genes by a novel adenylate cyclase-dependent signaling pathway. Dev. Cell 4, 253–263 10.1016/S1534-5807(03)00019-4 [DOI] [PubMed] [Google Scholar]
- Wood L. F., Leech A. J., Ohman D. E. (2006). Cell wall-inhibitory antibiotics activate the alginate biosynthesis operon in Pseudomonas aeruginosa: roles of sigma (AlgT) and the AlgW and Prc proteases. Mol. Microbiol. 62, 412–426 10.1111/j.1365-2958.2006.05390.x [DOI] [PubMed] [Google Scholar]
- Wood L. F., Ohman D. E. (2009). Use of cell wall stress to characterize sigma 22 (AlgT/U) activation by regulated proteolysis and its regulon in Pseudomonas aeruginosa. Mol. Microbiol. 72, 183–201 10.1111/j.1365-2958.2009.06635.x [DOI] [PubMed] [Google Scholar]
- Worlitzsch D., Tarran R., Ulrich M., Schwab U., Cekici A., Meyer K. C., et al. (2002). Effects of reduced mucus oxygen concentration in airway Pseudomonas infections of cystic fibrosis patients. J. Clin. Invest. 109, 317–325 10.1172/JCI0213870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak D. J., Ohman D. E. (1991). Pseudomonas aeruginosa AlgB, a two-component response regulator of the NtrC family, is required for algD transcription. J. Bacteriol. 173, 1406–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak D. J., Ohman D. E. (1994). Transcriptional analysis of the Pseudomonas aeruginosa genes algR, algB, and algD reveals a hierarchy of alginate gene expression which is modulated by algT. J. Bacteriol. 176, 6007–6014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W., Badrane H., Arora S., Baker H. V., Jin S. (2004). MucA-mediated coordination of type III secretion and alginate synthesis in Pseudomonas aeruginosa. J. Bacteriol. 186, 7575–7585 10.1128/JB.186.22.7575-7585.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao G., He J., Rahme L. G. (2006). Mutation analysis of the Pseudomonas aeruginosa mvfR and pqsABCDE gene promoters demonstrates complex quorum-sensing circuitry. Microbiology 152, 1679–1686 10.1099/mic.0.28605-0 [DOI] [PubMed] [Google Scholar]
- Xie Z. D., Hershberger C. D., Shankar S., Ye R. W., Chakrabarty A. M. (1996). Sigma factor-anti-sigma factor interaction in alginate synthesis: inhibition of AlgT by MucA. J. Bacteriol. 178, 4990–4996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahr T. L., Wolfgang M. C. (2006). Transcriptional regulation of the Pseudomonas aeruginosa type III secretion system. Mol. Microbiol. 62, 631–640 10.1111/j.1365-2958.2006.05412.x [DOI] [PubMed] [Google Scholar]
- Ye R. W., Haas D., Ka J. O., Krishnapillai V., Zimmermann A., Baird C., et al. (1995). Anaerobic activation of the entire denitrification pathway in Pseudomonas aeruginosa requires Anr, an analog of Fnr. J. Bacteriol. 177, 3606–3609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon S. S., Karabulut A. C., Lipscomb J. D., Hennigan R. F., Lymar S. V., Groce S. L., et al. (2007). Two-pronged survival strategy for the major cystic fibrosis pathogen, Pseudomonas aeruginosa, lacking the capacity to degrade nitric oxide during anaerobic respiration. Embo. J. 26, 3662–3672 10.1038/sj.emboj.7601787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Boucher J. C., Hibler N. S., Deretic V. (1996). Virulence properties of Pseudomonas aeruginosa lacking the extreme- stress sigma factor AlgU (sigmaE). Infect. Immun. 64, 2774–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Mudd M., Boucher J. C., Schurr M. J., Deretic V. (1997). Identification of the algZ gene upstream of the response regulator algR and its participation in control of alginate production in Pseudomonas aeruginosa. J. Bacteriol. 179, 187–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Schurr M. J., Deretic V. (1995). Functional equivalence of Escherichia coli sE and Pseudomonas aeruginosa AlgU: E. coli rpoE restores mucoidy and reduces sensitivity to reactive oxygen intermediates in algU mutants of P. aeruginosa. J. Bacteriol. 177, 3259–3268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielinski N. A., Chakrabarty A. M., Berry A. (1991). Characterization and regulation of the Pseudomonas aeruginosa algC gene encoding phosphomannomutase. J. Biol. Chem. 266, 9754–9763 [PubMed] [Google Scholar]
- Zimmermann A., Reimmann C., Galimand M., Haas D. (1991). Anaerobic growth and cyanide synthesis of Pseudomonas aeruginosa depend on anr, a regulatory gene homologous with fnr of Escherichia coli. Mol. Microbiol. 5, 1483–1490 10.1111/j.1365-2958.1991.tb00794.x [DOI] [PubMed] [Google Scholar]