Abstract
We have developed an efficient, versatile, and user-friendly viral engineering and expression system that is based on in planta assembly of functional viral vectors from separate pro-vector modules. With this new system, instead of supplying a plant cell with a complete viral vector as a mature viral particle, an RNA or a linear DNA molecule, we use agrobacteria to deliver various modules that are assembled inside the cell with the help of a site-specific recombinase. The resulting DNA is transcribed, and undesired elements such as recombination sites are spliced out, generating a fully functional RNA replicon. The proposed protocol allows us, by simply treating a plant with a mixture of two or more agrobacteria carrying specific prefabricated modules, to rapidly and inexpensively assemble and test multiple vector/gene combinations, without the need to perform the various engineering steps normally required with alternative protocols. The process described here is very fast (expression requires 3–4 days); it provides very high protein yield (up to 80% of total soluble protein); more than before, it is carried out using in vivo manipulations; it is based on prefabricated genetic modules that can be developed/upgraded independently; and it is inherently scalable.
Heterologous gene expression in plants is a central component of the most experimental and industrial processes in plant biotechnology, including research and development studies as well as industrial scale production. All currently available expression methods suffer from various limitations, such as the long time frame necessary for stable transformation, the low yield obtained with transient expression systems (even in versions using suppressors of silencing), and the inability of transient expression systems to be scaled up. Theoretically, expression systems based on viral vectors hold a great promise (for reviews, see refs. 1–3). The tobacco mosaic virus (TMV)-based vectors developed by the company Large Scale Biology (4) have been successfully developed into a functional genomics engine with the highest possible throughput (5, 6), and similar vectors are being used for commercial-scale production of pharmaceutical proteins, some of which are currently undergoing clinical trials. Functional genomics using viral vectors is in fact so efficient that the steps other than those required for actual expression, such as vector engineering and in vitro RNA synthesis, become the time/effort bottlenecks.
Because one of the “unwanted” steps in generating RNA-based vectors is in vitro transcription, we selected Agrobacterium-mediated delivery (7) as the method of initiation of viral vector infection. Analysis of the different steps involved in vector delivery and infective amplicon formation (including T-DNA transfer, recombination inside the plant cell, transcription, optional splicing, export to the cytoplasm, replication, cell-to-cell and systemic movement, etc.) revealed that in the wild tobacco species Nicotiana benthamiana, the first four steps occur or can be induced with very high efficiency. We used these elements to design a robust and simple procedure that, in addition to replacing the test tube version of transcription by an in vivo process, allowed us to move earlier steps, such as gene and vector engineering, from in vitro to in planta. With this approach, instead of supplying the full vector as a linear DNA molecule, we use different agrobacteria to deliver various modules of the viral vector and of the gene of interest. Those components are assembled inside a plant cell with the help of a site-specific recombinase. The resulting DNA is transcribed and spliced, thus removing undesired elements such as recombination sites and leading to the creation of a perfect gene/vector sequence within a fully functional infective replicon. We show that this approach works with high fidelity with a number of different modules and that it leads to extremely high yield of expressed proteins.
Materials and Methods
Plasmid Constructs. Cloned cDNAs of the crucifer-infecting TMV (cr-TMV) (8) and of the turnip vein-clearing virus (TVCV) (9) were obtained from J. G. Atabekov (Moscow University, Moscow). A viral vector containing a GFP gene was made in several cloning steps. The resulting construct, pICH4351 (Fig. 1G), contains, in sequential order, a 787-bp fragment from the Arabidopsis actin 2 (ACT2) promoter (10) (GenBank accession no. AB026654, base pairs 57962–58748); the 5′ end of TVCV (GenBank accession no. BRU03387, base pairs 1–5455); a fragment of cr-TMV [GenBank accession no. Z29370, base pairs 5457–5677, with thymine 5606 changed to cytosine to remove the start codon of the coat protein (CP)]; sequences taa tcg ata act cga g; a loxP site (ata act tcg tat agc ata cat tat acg aag tta t); a synthetic GFP gene (11); cr-TMV 3′ nontranslated region (3′ NTR; GenBank accession no. Z29370, base pairs 6078–6312); and the nopaline synthase (Nos) terminator. The entire fragment was cloned between the T-DNA left and right borders of pICBV10, a pBIN19-derived binary vector.
Fig. 1.
Transient expression by agroinfiltration of N. benthamiana leaves using nonviral expression cassettes (A–C) and viral constructs (D–F). Protoplasts from a leaf coinfiltrated with constructs containing GFP or DsRed under control of the 35S promoter viewed under blue (A) or red (B) light 4 dpi. (C) GUS staining of a leaf coinfiltrated with pICH11330 and pICH7900 (infiltration 1) or pICH11330 alone (infiltration 2) 3 dpi. (D) GFP expression in a leaf of a plant stably transformed with pICH10881 and coinfiltrated with pICH4851 and pICH6891 6 dpi. (E) GFP expression (7 dpi) in leaf sectors coinfiltrated with the following constructs: pICH4371 and pICH4461 together with pICH1754 (infiltration 1) or without (infiltration 2), pICH4851 and pICH6891 together with pICH10881 (infiltration 3) or without (infiltration 4), and pICH4351 alone (infiltration 5). (F) DsRed expression in a leaf infiltrated with pICH-NOP and pICH-RED, viewed under normal light. (G) Schematic representation of the T-DNA regions of some of the constructs used for infiltration.
pICH4371 and pICH4461 (Fig. 1G) are identical to pICH4351 except that sequences between the loxP site and the Nos terminator are missing in pICH4371 and pICH4461 lacks all sequences upstream of the loxP site. pICH4851 is similar to pICH4371 except that the loxP site was replaced by an attP site (gag tag tgc ccc aac tgg ggt aac ctt tga gtt ctc tca gtt ggg ggc gta g). pICH6891 is identical to pICH4461 except that the loxP site was replaced by an attB site (ctc gaa gcc gcg gtg cgg gtg cca ggg cgt gcc ctt ggg ctc ccc ggg cgc gta ctc cac ctc acc cat c) followed by 72 nt of the cr-TMV movement protein (MP) internal ribosome entry site sequences (nucleotides 4802–4874 of wild type cr-TMV) to serve as translational enhancer (12).
pIC7H900 and pICH1754 (Fig. 1G) contain the cre ORF cloned under control of the Hbt promoter (11) and the ACT2 promoter, respectively, in binary vector pICBV10. The cre ORF from pICH1754 was replaced by the PhiC31 integrase coding sequence (13) fused to the SV40 T-antigen nuclear localization signal at the C-terminal end (amino acids PKKKRKV) (14), resulting in construct pICH10881
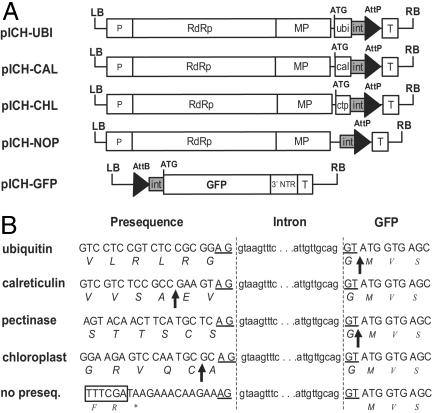
pICH-UBI (Fig. 2A) is identical to pICH4851 for the region including the ACT2 promoter and TVCV sequence. Sequences located 3′ consist of cr-TMV sequence (GenBank accession no. Z29370, base pairs 5457–5646, with thymine 5606 changed to cytosine), followed by sequence ata aga aac aag aaa cc, by Arabidopsis ubiquitin sequence (base pairs 525–750 of GenBank accession no. NM 116744, with nucleotides 528, 746, and 749 changed to G, C, and A, respectively), by intron sequence (gt aag ttt cat ttt cat aat tac aca aat tta gat ttg att ttt gtt, derived from the third intron of the petunia Psk7 gene, GenBank accession no. PHAJ4165), by attP sequence (gtg ccc caa ctg ggg taa cct ttg agt tct ctc agt tgg ggg cgt ag), and finally by the Nos terminator. pICH-CAL, pICH-CHL, and pICH-PEC are identical to pICH-UBI except that ubiquitin sequence was replaced, respectively, by Arabidopsis calreticulin sequence (15), by ribulose-1,5-bisphosphate carboxylase oxygenase (Rubisco) small subunit chloroplast targeting sequences (a consensus of dicot sequences), and finally by signal peptide sequences from an apple pectinase gene (GenBank accession no. L27743). pICH-NOP and pICH-CPF are similar to pICH-UBI except that sequences between the MP and downstream intron sequences were replaced by sequence a taa gaa aca aga aag and cta acg cag for pICH-NOP and pICH-CPF, respectively, and that cr-TMV thymine 5606 (coordinates relative to GenBank accession no. Z29370) was not changed to a cytosine in pICH-CPF (in contrast to other clones). pICH-GFP is identical to pICH6891 except that sequences between the attB site and the GFP start codon were replaced by intron sequence (ttt tat tac atg ttt gaa ctt caa caa ttt atg act ttt tgt tct tat tgt tgc agg t).
Fig. 2.
Schematic representation of intron-containing viral pro-vector modules. (A) T-DNA regions of 5′ modules. ubi, Arabidopsis ubiquitin sequence; cal, tobacco calreticulin apoplast targeting presequence; ctp, synthetic chloroplast targeting presequence; int, intron 5′ half; P, ACT2 promoter; T, nos terminator. (B) Product of recombination between 5′ modules and pICH-GFP (the sequence labeled “pectinase” is expected by recombination of the 5′ module pICH-PEC). Exon and intron sequences are displayed as uppercase and lowercase letters, respectively. Amino acids are shown in italics, and a stop codon is indicated by an asterisk. Protein cleavage sites are indicated with arrows. The end of the MP coding sequence is displayed in an open box.
pICH-GFPSYS (Fig. 3) was made by replacing cr-TMV 3′ NTR tRNA-like sequence (base pairs 6274–6312 from GenBank accession no. Z29370) by the 1005 3′ terminal sequence from tobacco mild green mosaic virus (TMGMV) U5 variant (base pairs 5498–6502) (4).
Fig. 3.
N. benthamiana plant expressing GFP in systemic leaves 14 days after infiltration of one of the lower leaves (hidden from view) with pICH-NOP, pICH-GFPSYS, and pICH10881. Int, intron 3′ half; 3′PK, cr-TMV pseudoknot-like motifs; sgp, TMGMV U5 subgenomic promoter; 3′ NTR, TMGMV U5 3′ untranslated region; T, Nos terminator.
Agroinfiltration Procedure. Two hundred microliters of overnight-grown Agrobacterium cultures were sedimented with a microcentrifuge at 8,000 rpm (6,000 × g) for 3 min. The pellet was resuspended in 1 ml of a solution containing 10 mM Mes (pH 5.5) and 10 mM MgSO4. Leaves of greenhouse-grown N. benthamiana plants were infiltrated by using a syringe without a needle.
RT-PCR. First strand cDNAs were synthesized from 5 μg total RNA treated with DNaseI by using the SuperScript First-Strand Synthesis System for RT-PCR (Invitrogen). PCR amplification was performed by using the following protocol: initial denaturation at 94°C for 3 min followed by 35 cycles of denaturation at 94°C for 30 sec, annealing at 65°C for 30 sec, elongation at 72°C for 1 min, and finally a 3-min extension at 72°C.
Protein Analysis. One hundred-milligram samples of N. benthamiana leaves infiltrated with Agrobacterium were ground in 300 μl of protein extraction buffer (50 mM sodium phosphate buffer, pH 7.0/5 mM 2-mercaptoethanol/10 mM EDTA/0.1% Triton X-100). Crude extracts were mixed with 2× loading buffer and separated by SDS/PAGE. Total soluble protein content was measured by Bradford assay. For quantification of GFP, dilutions of total soluble protein extracts were subjected to spectrofluorimetry with a Luminescence Spectrometer LS 50B (Perkin–Elmer) using excitation and emission wavelengths of 470 and 509 nm, respectively.
Isolation of N. benthamiana Mesophyll Protoplasts. Protoplasts were released from leaf tissue by incubation for 10–16 h at 25°C in the dark in a solution containing 0.4% cellulase Onozuka R-10 (Duchefa, Haarlem, The Netherlands), 0.4% Macerozyme R-10 (Duchefa), 0.5 M sucrose, 7 mM CaCl2, and 90 mM glycine. Released protoplasts were filtrated through a nylon mesh with a 50-μm pore diameter and resuspended in W5 medium (16).
Visualization of GFP and Red Fluorescent Protein from Discosoma (DsRED). Leaves expressing GFP were viewed under UV illumination generated by a B-100AP lamp (UVP, Upland, CA). Protoplasts were viewed with a microscope Leica DM IL with filter sets GFP plant fluorescence (excitation filter, 470/40 nm; barrier filter, 525/50 nm) or DsRED (excitation, 546/12 nm; barrier, 560 LP nm). Protoplasts were also viewed with a Zeiss Axioplan 2 microscope using GFP filter block.
Results
Multiple T-DNAs Can Be Efficiently Delivered to the Same Cell from Different Agrobacteria. Agrobacterium-mediated transfer of T-DNA to a plant cell is an efficient process, at least in some dicots (17–20). In N. benthamiana, infiltration of leaf tissue with an Agrobacterium suspension results in transient expression in nearly 100% of the mesophyll cells. To quantify the efficiency of codelivery of several T-DNAs into the same cell, we coinfiltrated N. benthamiana leaves with a mixture of two Agrobacterium cultures containing GFP and DsRed under control of the cauliflower mosaic virus 35S promoter and found that >95% of protoplasts isolated from infected areas expressed both proteins (Fig. 1 A and B). In subsequent studies (data not shown), we were able to show that constructs from up to five independent Agrobacterium cultures could be delivered to the same cell in a measurable way.
Transiently Expressed T-DNAs Can Recombine Efficiently in Plant Cells. It is known that T-DNAs can recombine after delivery to a plant cell (21). However, to be useful for gene assembly in planta, site-specific recombination of T-DNAs needs to be very efficient. To measure the ability of T-DNAs to recombine, we designed a test construct (pICH11330) that expresses glucuronidase synthase (GUS) after cre-mediated recombination (22) between two loxP sites (Fig. 1G). Coinfiltration of pICH11330 and a cre expression construct, pICH7900, resulted in GUS staining of the entire infiltrated area, whereas only weak background GUS activity could be detected in the absence of cre in the control infiltration (Fig. 1C). This experiment tells us that site-specific recombination of T-DNAs takes place efficiently in a majority of infiltrated cells.
A Functional cr-TMV-Based Viral Vector Can Be Delivered by Agroin-filtration. Next we constructed a viral vector that could be delivered by Agrobacterium infiltration. This vector is based on cr-TMV (8), which can also infect tobacco and Arabidopsis. For cloning reasons, the 5′ end of the viral vector, including the RNA-dependent RNA polymerase (RdRp) and part of the MP, was derived from a closely related tobamovirus, TVCV (9), which shares 98% homology with cr-TMV at the amino acid level (for the RdRp). The remaining part of the vector, consisting of part of the MP, the CP subgenomic promoter, and the 3′ NTR, is derived from cr-TMV. The CP ORF of cr-TMV was replaced by GFP. Therefore, the resulting vector is not expected to move systemically, but it should still be able to move cell-to-cell because it contains a MP. In one specific modification, a loxP site was inserted between the MP and GFP to mimic the recombination product that we would expect after cre-mediated recombination of the separate modules that we intended to make next. Finally, the viral construct was cloned in a binary vector between the ACT2 promoter and the Nos terminator. The resulting construct, pICH4351 (Fig. 1G), was transformed into Agrobacterium strain GV3101 and infiltrated into N. benthamiana leaves. At 3 days postinfiltration (dpi), numerous foci of green fluorescence could be seen under UV light. These spots grew as viral RNA replicated and became confluent (Fig. 1E, infiltration 5), and intense fluorescence could be seen for >2 weeks without any apparent sign of silencing.
Functional Viral Vectors Can Be Assembled in Planta from Pro-Vectors Delivered by Agrobacterium. Next, pICH4351 was split into two constructs, pICH4371 and pICH4461 (Fig. 1G). pICH4371 contains the 5′ part of the viral vector including the RNA-dependent RNA polymerase (RdRP) and MP genes, the CP subgenomic promoter (which overlaps with the MP), and a loxP site. The 3′ module contains a loxP site, the gene of interest (in this case GFP), and the 3′ end of the viral vector. The gene of interest cannot be expressed in the 3′ module alone, even when RdRp is provided in trans, because the construct lacks a subgenomic promoter. Both modules were introduced into Agrobacterium GV3101. Coinfiltration of N. benthamiana leaves with both modules together with a cre-expression construct (pICH1754; Fig. 1G) resulted in appearance of green fluorescence foci 3 dpi (Fig. 1E, infiltration 1). No fluorescence could be detected when either one of the three modules was omitted. We also infiltrated N. benthamiana plants stably transformed with a cre-expression construct, with Agrobacterium cultures containing pICH4371 and pICH4461. Efficient recombination and viral amplification was obtained with transformants expressing cre, whereas no recombination event or viral amplification could be detected by infiltration of nontransformed plants (data not shown).
In Planta Pro-Vector Assembly Can Be Achieved Using Alternative Recombination Systems. To show that assembly of a functional viral vector in planta is not restricted to one recombination system, the loxP sites in pICH4351 and pICH4371 were replaced by the Streptomyces phage C31 integrase recombination sites AttP and AttB (13, 23) resulting in plasmids pICH4851 and pICH6891, respectively (Fig. 1G). Coinfiltration of both constructs and of pICH10881 (a binary vector containing the integrase under control of the ACT2 promoter) resulted in GFP-expressing foci 4 days later (Fig. 1E, infiltrations 3). Control infiltrations lacking one of the three constructs did not yield any GFP expression. The integrase in pICH10881 is fused at the C-terminal end to the SV40 T-antigen nuclear localization signal (NLS) (described for use in mammalian cells by Andreas et al. in ref. 14). A version of integrase without a NLS was also functional but less efficient. We also generated transgenic N. benthamiana plants expressing integrase under control of the ACT2 promoter and found that the recombination efficiency was essentially the same as when integrase was provided transiently [Fig. 1D; GFP-expressing foci are not yet confluent as the picture was taken at 6 dpi, but they became confluent later].
Exact Protein Fusions Are Obtained by Removal of Recombination Sites by Splicing. One of the goals of designing this system is to have the capability of making protein fusions by in-frame recombination of 5′ and 3′ modules. One application would be the ability to target genes to different subcellular compartments by coinfiltration of 3′ modules with 5′ modules chosen from a library of clones with different targeting specificities. To make exact protein fusions, it is necessary to remove recombination site sequence from the product of recombination. cr-TMV replicates in the cytoplasm and never enters the nucleus. However, passage through the nucleus, resulting from Agrobacterium-mediated delivery of the vector, gives us the opportunity to remove unwanted sequences by splicing. Therefore, recombination site sequences were flanked by intron sequences. Three criteria were used for design of an optimal intron (24): (i) a size close to the average size of plant introns (80–140 nt; a very large intron that would prevent vector replication if unspliced might also have been an alternative), (ii) a low GC content, and (iii) sequences of the splice sites close to the consensus. The first criterion is easy to fulfill, because numerous introns of all sizes are present in genome databases. Therefore, we chose a 104-nt intron (from petunia), which, with the addition of the recombination site (and a few modifications; see below), amounts to 160 nt. To respect the second criterion, we chose very T-rich sequences to flank the recombination sequences, and we made sure that no cryptic splice site was present in the intron near the GC-rich sequences of the recombination site. Finally, sequences of the splice sites were modified to fit exactly the consensus for dicot plants (24).
As a first example, a 5′ pro-vector module containing ubiquitin sequences was made. Ubiquitin fusions are used in biotechnology applications for the production of proteins starting with any amino acid of choice (except proline), because they are precisely cleaved by host proteases immediately after the last amino acid of ubiquitin (25). The last amino acid of ubiquitin is a glycine, which is fortunate, because one of the codons of glycine is GGT, which corresponds to part of the consensus found in exon sequences at splice site junctions: AG-intron-GT. The amino acid preceding the ubiquitin terminal amino acid is another glycine, which allows, by choosing the sequence GGA G-intron-GT, to respect the consensus (Fig. 2B). Two constructs were therefore made: a 5′ pro-vector module, pICH-UBI, and a 3′ pro-vector module, pICH-GFP (Fig. 2 A and B). These constructs were coinfiltrated with an integrase construct, and numerous GFP foci were obtained 4 days later (Fig. 4A). To analyze the efficiency and accuracy of splicing, RT-PCR was performed from RNA isolated from the infiltrated area. A single fragment of the expected size was amplified (Fig. 4B), and sequencing revealed that splicing occurred accurately as hoped. Splicing was also shown to take place as expected for several other modules (described in A Library of 5′ Modules with Different Specificities).
Fig. 4.
Targeted expression of GFP using different 5′ pro-vector modules. (A) GFP expression in N. benthamiana leaves 12 dpi. (a–e) Leaves viewed under UV illumination. (f–j) Mesophyll protoplasts viewed under blue light. (a and f) Uninfiltrated leaf. (b and g) Cytosolic localization of GFP provided by ubiquitin fusion (pICH-UBI). (c and h) Targeting to the secretory pathway with calreticulin module (pICH-CAL). (d and i) Targeting to the chloroplast (pICH-CHL). (e and j) No presequence for expression in the cytosol (pICH-NOP). (Scale bar, 10 μM.) (B) RT-PCR analysis of recombined pro-vector modules for ubiquitin-GFP, calreticulin-GFP, and CTP-GFP fusions. (C) Coomassie-stained gel (SDS/PAGE) containing total soluble protein extracts from leaves inoculated with the GFP 3′ module in combination with pICH-UBI (lane 2), pICH-CAL (lane 3), pICH-CHL (lane 4), pICH-CPF (lane 5), pICH-NOP (lane 6), and modules for His tag fusion without a cleavage site (lane 7) or with an enterokinase cleavage site (lane 8). Lane 1 shows noninfiltrated leaf tissue.
A Library of 5′ Modules with Different Specificities. Additional 5′ pro-vector modules were made for expression in the chloroplast (pICH-CHL) and the apoplast (pICH-CAL). A clone lacking any presequence (pICH-NOP) was made for expression in the cytosol (Fig. 2 A and B). Coinfiltration of all clones with the same 3′ GFP-containing module resulted in GFP expression in the appropriate subcellular compartment, as shown by observation of protoplasts under blue light (Fig. 4A). For apoplast expression, observation of the infiltrated areas under UV light revealed only very weak fluorescence (not even visible on a picture, Fig. 4A). This finding has been reported before (26) and shows that little or no expression is taking place in the cytosol, as expected. However, fluorescence could clearly be detected in the apoplast under blue light (data not shown). Analysis of expressed proteins by SDS/PAGE and Coomassie staining showed that all had the predicted molecular weight (Fig. 4C). In the case of the cleavable fusions (ubiquitin, apoplast, and chloroplast targeting presequences), no unprocessed form could be detected.
In contrast to the ubiquitin module described above, which leads to a processed protein lacking any extra amino acid at the N-terminal end, the chloroplast and calreticulin fusion modules lead to processed proteins with two and three extra amino acids, respectively (Fig. 2B). However, it is possible to create 5′ modules that result in a processed protein lacking any extra amino acids. One such module was made by using an apoplast targeting sequence from an apple pectinase gene (Fig. 2B).
A number of 5′ pro-vector modules were also made to allow attachment of a purification tag (with or without a cleavage sequence) to the polypeptide of interest. These modules were tested and found to provide the expected fusions (Fig. 4C). In the case of His tags, we also demonstrated that the expressed proteins could be successfully purified by using the tag (data not shown). Finally, a clone was made (pICH-CPF) that results in fusion of the expressed protein to the 17 N-terminal amino acids of cr-TMV CP.
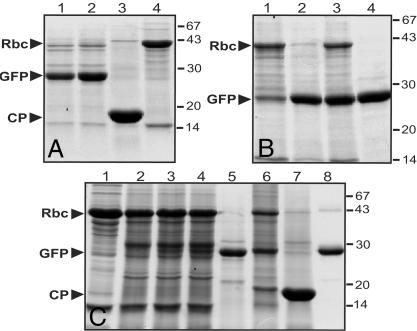
A High-Yield Potential for Expressed Proteins. Protein content in the infiltrated areas was analyzed by SDS/PAGE and Coomassie staining and by Western blot (data not shown). Although we expected a high amount of GFP protein due the high level of green fluorescence in infiltrated leaves, we were still surprised to find concentrations of GFP of up to 80% of total soluble protein or 5 g/kg of fresh weight biomass (Fig. 5). At 14 dpi, even the normally most abundant plant protein, the large subunit of Rubisco, was reduced and replaced by GFP. The same observation was also made with DsRed (data not shown).
Fig. 5.
Coomassie-stained polyacrylamide gels showing protein expressed with the viral pro-vector system. (A) Total soluble proteins extracted from leaves infiltrated with pICH-NOP, pICH-GFP, and integrase (lane 1), from leaves infiltrated with pICH4351 (lane 2), from a systemic leaf containing WT cr-TMV virus (lane 3), and from uninfiltrated leaf tissue (lane 4). (B) Extracts from leaves infiltrated with pICH-NOP, pICH-GFP, and integrase 7 dpi (lane 1), 10 dpi (two different infiltrations 2 and 3), and 17 dpi (lane 4). (C) Extracts from uninfiltrated tissue (lane 1), from leaves infiltrated with a 35S-GFP cassette without suppressor of silencing (lane 2) or together with 35S-HcPro (lane 3) or 35S-p19 (lane 4), pICH-NOP infiltrated with integrase and pICH-GFP (lane 5) or pICH-GFPSYS (lane 6), WT cr-TMV virus (lane 7), and recombinant GFP (lane 8). Rbc, Rubisco large subunit.
We also compared the potential of our system with a recently described expression system, which uses suppressors of silencing to increase the yield of protein obtained by transient expression (17). Whereas the most efficient suppressor of silencing described to date, the p19 protein of the tomato bushy stunt virus, provided a dramatic increase of GFP fluorescence in infiltrations with a normal expression cassette, analysis of Coomassie-stained gels revealed that still far less protein was obtained than with viral vectors (Fig. 5). We also investigated the effect of the suppressors of silencing, p19 or HcPro (27), on the amount of protein produced using our viral constructs, but we did not find any increase.
Different Genes Cloned in 3′ Modules Can Be Expressed by Coinfiltration with the Same 5′ Pro-Vector Module. To illustrate that these vectors also work with other genes, we have replaced GFP in pICH-GFP by coding sequences of the reporter gene DsRed (28). Coinfiltration of this clone (pICH-RED) with pICH-NOP and integrase led to DsRed expression in such amount that it became visible in normal light (Fig. 1F).
3′ Pro-Vector Modules Containing CP Allow Systemic Movement of Assembled Vectors. An additional 3′ module was made, pICH-GFPSYS, by replacing part of the cr-TMV 3′ NTR in pICH10570 by a fragment from the TMGMV U5 virus, which comprises the CP subgenomic promoter, the CP ORF and 3′ NTR sequences. The same approach, developed by Large Scale Biology with a vector built from TMV variant U1, was reported to provide efficient systemic movement while preventing unwanted recombination events between duplicated subgenomic promoter and 3′ NTR sequences by using heterologous sequences from TMGMV U5 (4). Such an approach was not guaranteed to work for our vector, because TMGMV is more closely related to TMV U1 than to cr-TMV (29). Despite this difference, pICH-GFPSYS was found to be functional and to allow systemic movement of viral RNA (Fig. 3), even when launched from a modular system.
Discussion
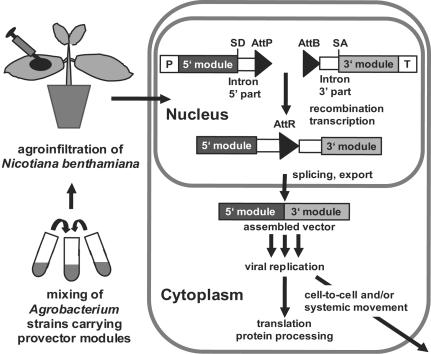
The protocol and vectors described here represent a simple, robust, inexpensive, and versatile system for amplifying and expressing nucleotide sequences in green plants (summarized in Fig. 6). The system is very fast (expression requires just 3–14 days), reaching the speed of prokaryotic systems while providing eukaryotic (plant-specific) posttranslational capabilities. The adopted engineering strategy of deconstructing and reconstructing the viral RNA vector provides enhanced versatility and efficiency, because numerous gene/vector combinations can be tested without the need to actually make each individual variant construct. Such capability can be useful for functional genomic studies, e.g., by allowing studied proteins to be targeted to different subcellular compartments or fused to a variety of tags or coding sequences fragments.
Fig. 6.
Schematic overview of in planta assembly of viral pro-vector modules. SD, splice donor site; SA, splice acceptor site.
Because the amplifiable vector is an RNA molecule, we are able to apply different recombination techniques at the DNA level, while still being able to “clean” the final sequence at the RNA level. In separate experiments, we also tested the possibility of assembly of viral modules at the RNA level using transsplicing ribozymes and found that this, too, was working (P. Ivanov, S.M., V.K., and Y.G., unpublished observations).
We have used the system described here to successfully express proteins of different classes, in addition to GFP, DsRed, and GUS, such as human cytokines, human hormones, single-chain antibodies, bacterial and plant proteins, and enzymes. The yields that we obtained varied, but, in the case of nontoxic proteins, levels of 0.5–1 g/kg of fresh weight biomass were achievable without special optimization, and the system yielded enough protein for research purposes with highly toxic proteins such as restriction enzymes or DNases.
Among the advantages offered by the viral vector system described here is the extremely high yield potential, which approaches the highest theoretically possible level. Leaf per se is not the best system for protein production, with a protein content in uninfected mature N. benthamiana leaf of 8–10 g/kg of fresh weight biomass, 50% of that being Rubisco. However, this limitation is compensated for by the exceptional ability of TMV to redirect cellular synthesis to viral proteins in infected cells. By using the easily scorable proteins GFP and DsRed in our vectors, we were stunned by the absolute amount of protein that was obtained routinely, with up to 5 g of expressed protein per kg of fresh weight biomass. Thus, the high yield that can be obtained with this system (up to 80% of total protein) will also simplify and reduce the costs of the upstream and downstream components of the technology process.
When trying to use the same vectors in tobacco (Nicotiana tabacum) as a host, we initially encountered several problems, the most important one being the low efficiency of release of the initial transcript from the nucleus to the cytoplasm. By rational engineering of the vector backbone to minimize the occurrence of undesired posttranscriptional events such as abnormal RNA processing, we made improved versions that work in tobacco as well as in N. benthamiana (S.M., C. Thoeringer, V.K., and Y.G., unpublished observations) and assume that similar modifications, using these or other viruses, can be developed for expression in other plant species as well.
In an ideal protein production process, especially in case of pharmaceutical proteins, the scale-up (commercial) version of the protein expression system has to be essentially the same as the version used to express the protein during early research and preclinical and clinical trials. Our system fulfills that requirement, because it can be scaled up in a number of ways, one of which, namely the protocol using the systemic version of the viral vector, is already available. A potentially more powerful and universal scale-up process would be one based on controlled activation of an encrypted version of a replicon present on a plant chromosome of a production host. Such an approach, currently in development in our laboratory, allows for simultaneous activation of vector replication, expression, and cell-to-cell movement in multiple independent cells of different organs, thus effectively replacing the need for systemic movement.
Acknowledgments
We thank Dr. Gerd Hause for help with microscopy, Dr. Stefan Werner for making construct pICH11330, Dr. Gregor Benning for his contribution for coexpression of nonviral constructs in protoplasts, Dr. Maxim Skulachev for making the ACT2 promoter-viral vector sequence, and Maxim Vasilenko for stable transformation of N. benthamiana with pICH10881.
Abbreviations: ACT2, Arabidopsis actin 2; TMV, tobacco mosaic virus; cr-TMV, crucifer-infecting TMV; DsRED, red fluorescent protein from Discosoma; GUS, glucuronidase synthase; MP, movement protein; TMGMV, tobacco mild green mosaic virus; CP, coat protein; NTR, nontranslated region; Nos, nopaline synthase.
References
- 1.Turpen, T. H. (1999) Philos. Trans. R. Soc. London B Biol. Sci. 354, 665-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yusibov, V., Shivprasad, S., Turpen, T. H., Dawson, W. & Koprowski, H. (1999) Curr. Top. Microbiol. Immunol. 240, 81-94. [DOI] [PubMed] [Google Scholar]
- 3.Pogue, G. P., Lindbo, J. A., Garger, S. J. & Fitzmaurice, W. P. (2002) Annu. Rev. Phytopathol. 40, 45-74. [DOI] [PubMed] [Google Scholar]
- 4.Shivprasad, S., Pogue, G. P., Lewandowski, D. J., Hidalgo, J., Donson, J., Grill, L. K. & Dawson, W. O. (1999) Virology 255, 312-323. [DOI] [PubMed] [Google Scholar]
- 5.Fitzmaurice, W. P., Holzberg, S., Lindbo, J. A., Padgett, H. S., Palmer, K. E., Wolfe, G. M. & Pogue, G. P. (2002) OMICS 6, 137-151. [DOI] [PubMed] [Google Scholar]
- 6.Escobar, N. M., Haupt, S., Thow, G., Boevink, P., Chapman, S. & Oparka, K. (2003) Plant Cell 15, 1507-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turpen, T. H., Turpen, A. M., Weinzettl, N., Kumagai, M. H. & Dawson, W. O. (1993) J. Virol. Methods 42, 227-239. [DOI] [PubMed] [Google Scholar]
- 8.Dorokhov, Y. L., Ivanov, P. A., Novikov, V. K., Agranovsky, A. A., Morozov, S., Efimov, V. A., Casper, R. & Atabekov, J. G. (1994) FEBS Lett. 350, 5-8. [DOI] [PubMed] [Google Scholar]
- 9.Lartey, R. T., Lane, L. C. & Melcher, U. (1994) Arch. Virol. 138, 287-298. [DOI] [PubMed] [Google Scholar]
- 10.An, Y. Q., McDowell, J. M., Huang, S., McKinney, E. C., Chambliss, S. & Meagher, R. B. (1996) Plant J. 10, 107-121. [DOI] [PubMed] [Google Scholar]
- 11.Chiu, W., Niwa, Y., Zeng, W., Hirano, T., Kobayashi, H. & Sheen, J. (1996) Curr. Biol. 6, 325-330. [DOI] [PubMed] [Google Scholar]
- 12.Skulachev, M. V., Ivanov, P. A., Karpova, O. V., Korpela, T., Rodionova, N. P., Dorokhov, Y. L. & Atabekov, J. G. (1999) Virology 263, 139-154. [DOI] [PubMed] [Google Scholar]
- 13.Thorpe, H. M. & Smith, M. C. (1998) Proc. Natl. Acad. Sci. USA 95, 5505-5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andreas, S., Schwenk, F., Kuter-Luks, B., Faust, N. & Kuhn, R. (2002) Nucleic Acids Res. 30, 2299-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Borisjuk, N., Sitailo, L., Adler, K., Malysheva, L., Tewes, A., Borisjuk, L. & Manteuffel, R. (1998) Planta 206, 504-514. [DOI] [PubMed] [Google Scholar]
- 16.Medgyesy, P., Menczel, L. & Maliga, P. (1980) Mol. Gen. Genet. 179, 693-696. [Google Scholar]
- 17.Janssen, B. J. & Gardner, R. C. (1990) Plant Mol. Biol. 14, 61-72. [DOI] [PubMed] [Google Scholar]
- 18.Kapila, J., De Rycke, R., Van Montagu, M. & Angenon, G. (1997) Plant Sci. 122, 101-108. [Google Scholar]
- 19.Yang, Y., Li, R. & Qi, M. (2000) Plant J. 22, 543-551. [DOI] [PubMed] [Google Scholar]
- 20.Voinnet, O., Rivas, S., Mestre, P. & Baulcombe, D. (2003) Plant J. 33, 949-956. [DOI] [PubMed] [Google Scholar]
- 21.Vergunst, A. C., Jansen, L. E. & Hooykaas, P. J. (1998) Nucleic Acids Res. 26, 2729-2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dale, E. C. & Ow, D. W. (1990) Gene 91, 79-85. [DOI] [PubMed] [Google Scholar]
- 23.Groth, A. C., Olivares, E. C., Thyagarajan, B. & Calos, M. P. (2000) Proc. Natl. Acad. Sci. USA 97, 5995-6000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simpson, G. G. & Filipowicz, W. (1996) Plant Mol. Biol. 32, 1-41. [DOI] [PubMed] [Google Scholar]
- 25.Baker, R. T. (1996) Curr. Opin. Biotechnol. 7, 541-546. [DOI] [PubMed] [Google Scholar]
- 26.Boevink, P., Martin, B., Oparka, K., Santa Cruz, S. & Hawes, C. (1999) Planta 208, 392-400. [DOI] [PubMed] [Google Scholar]
- 27.Brigneti, G., Voinnet, O., Li, W. X., Ji, L. H., Ding, S. W. & Baulcombe, D. C. (1998) EMBO J. 17, 6739-6746. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Matz, M. V., Fradkov, A. F., Labas, Y. A., Savitsky, A. P., Zaraisky, A. G., Markelov, M. L. & Lukyanov, S. A. (1999) Nat. Biotechnol. 17, 969-973. [DOI] [PubMed] [Google Scholar]
- 29.Lartey, R. T., Voss, T. C. & Melcher, U. (1996) Mol. Biol. Evol. 13, 1327-1338. [DOI] [PubMed] [Google Scholar]