Key Points
Adult T-cell leukemia (ATL) does not, as previously believed, result from the oligoclonal proliferation caused by HTLV-1 infection.
In both ATL patients and those with nonmalignant infection, the HTLV-1 provirus preferentially survives in vivo in acrocentric chromosomes.
Abstract
Adult T-cell leukemia/lymphoma (ATL) occurs in ∼5% of human T-lymphotropic virus type 1 (HTLV-1)–infected individuals and is conventionally thought to be a monoclonal disease in which a single HTLV-1+ T-cell clone progressively outcompetes others and undergoes malignant transformation. Here, using a sensitive high-throughput method, we quantified clonality in 197 ATL cases, identified genomic characteristics of the proviral integration sites in malignant and nonmalignant clones, and investigated the proviral features (genomic structure and 5′ long terminal repeat methylation) that determine its capacity to express the HTLV-1 oncoprotein Tax. Of the dominant, presumed malignant clones, 91% contained a single provirus. The genomic characteristics of the integration sites in the ATL clones resembled those of the frequent low-abundance clones (present in both ATL cases and carriers) and not those of the intermediate-abundance clones observed in 24% of ATL cases, suggesting that oligoclonal proliferation per se does not cause malignant transformation. Gene ontology analysis revealed an association in 6% of cases between ATL and integration near host genes in 3 functional categories, including genes previously implicated in hematologic malignancies. In all cases of HTLV-1 infection, regardless of ATL, there was evidence of preferential survival of the provirus in vivo in acrocentric chromosomes (13, 14, 15, 21, and 22).
Introduction
Human T-lymphotropic virus type 1 (HTLV-1) causes adult T-cell leukemia/lymphoma (ATL) in approximately 5% of HTLV-1–infected individuals. A further ∼5% of carriers develop an aggressive myelopathy known as HTLV-1–associated myelopathy (HAM) or other inflammatory diseases such as polymyositis. It remains uncertain why a minority develop aggressive clinical disease, typically decades following asymptomatic infection, whereas most infected individuals remain lifelong healthy carriers. The Shimoyama classification of ATL1 contains 4 subtypes: acute, lymphoma, chronic, and smoldering. These subtypes differ in the response to treatment and overall survival, but little is known about either viral or host molecular determinants of disease. Several host cytogenetic or molecular defects have been described, but no recurrent genetic lesions have been identified.
The major predictor of clinical disease is the proviral load (PVL), the percentage of HTLV-1–infected peripheral blood mononuclear cells (PBMCs). The PVL remains relatively constant over years within an individual, rising slowly over decades.2 However, the PVL varies widely between patients, ranging from <0.001% PBMCs to >100% (ie, >100 copies per 100 PBMCs); the risk of disease rises in carriers with a PVL >4% in Japan3 and in those with a PVL >10% in the United Kingdom.4 Nonetheless, there is overlap in the range of PVL seen between patients with disease and those that remain lifelong asymptomatic carriers, making individual patient prognosis difficult.
HTLV-1 appears to persist in chronic infection chiefly by mitotic proliferation of infected CD4+ T cells, although the ratio of this mitotic spread to de novo infection5 has not been rigorously estimated. Each clone of HTLV-1–infected cells can be identified by its particular integration site of the HTLV-1 provirus in the host genome6; the daughter cells of each clone share the same genomic integration site, and the frequency of these cells defines the abundance of a given clone. A majority of naturally infected cells in nonmalignant infection contain a single integrated provirus.7
ATL is characterized by monoclonal proliferation of CD4+CD25+ tumor cells. For many years, it has been believed that ATL arises following a steady progression from polyclonal infection of CD4+ T cells to an oligoclonal expansion and, many years later, following a series of undefined genetic or epigenetic events, malignant transformation of a previously abundant clone to a monoclonal tumor. However, there are indications that HTLV-1 clonality in ATL may be more complex. One or more abnormally abundant clones may underlie the largest, putatively malignant clone,6 and there are reports of “clonal succession” in which a malignant clone spontaneously regresses and an independent clone proliferates in its place.8
HTLV-1 expresses a transcriptional transactivator protein, Tax, which activates transcription of the HTLV-1 provirus and of many host genes9. Because Tax can immortalize rodent cells in vitro and Tax transgenic mice develop tumors, it has been widely accepted that Tax plays a role in leukemogenesis. This hypothesis is supported by the observations that Tax promotes DNA replication and cell-cycle progression, causes structural damage to host DNA, and inhibits DNA repair and cell-cycle checkpoints.9 Tax expression is lost in ∼40% of ATL cases, probably under selection from the strong anti-Tax cytotoxic T-lymphocyte (CTL) response,10 but the relation between Tax expression and ATL subtype and progression is unclear. There is also increasing evidence that another HTLV-1 gene, HBZ, plays a critical part in leukemogenesis.9
To summarize, the molecular mechanisms of oncogenesis of ATL, and in particular the mechanisms and role of selective oligoclonal proliferation, are incompletely understood. Here, in a large cohort of ATL patients and geographically matched asymptomatic HTLV-1 carriers, we used a quantitative high-throughput sequencing approach to test the hypothesis that the genomic environment flanking the proviral integration site is associated with malignant transformation of HTLV-1–infected clones and correlated the findings with both the clinical subtype of ATL and genetic and epigenetic modifications of the HTLV-1 provirus.
Methods
Study subjects and control cell lines
Blood or lymph node samples were donated by 221 ATL patients and 75 asymptomatic HTLV-1 carriers (ACs) from the Kumamoto region of Japan, and DNA was extracted at the Institute for Viral Research, Kyoto University, Japan, with written consent in accordance with regulations defined by the Japanese Government and Kyoto University. This study was conducted in accordance with the Declaration of Helsinki. This study was approved by the UK National Research Ethics Service (reference 09/H0606/106). The chromosomal distribution of integration sites in the present cohort was compared with the distribution in samples from 2 previously described studies: individuals with natural (nonmalignant) HTLV-1 infection from Kagoshima, southern Japan,11,12 and cells infected with HTLV-1 in vitro.6,13 The rodent cell line Tarl2, containing a single copy of HTLV-1, was used for quantification of PVL. ATL control cell lines T-43 (methylated) and T-48 (unmethylated) were used as methylation controls.14
PVL quantification
PVL was measured by quantitative polymerase chain reaction (PCR) of tax and actin genes using ABI Fast SYBR green as per the manufacturer’s protocol (Applied Biosystems), using PCR primers as previously described15 and assuming a single copy of tax7 and 2 copies of actin per cell. Thermal cycling conditions were 95°C for 20 seconds and 40 cycles each of 95°C for 1 second followed by 60°C for 20 seconds. Standard curves were generated using serial dilutions of the cell line Tarl2, as previously described.6,13
Long-range PCR to identify defective proviruses
An internal control region at the 3′ end of the HTLV-1 genome was amplified for each ATL case, followed by a long-range PCR to identify defective proviruses based upon the length of the long-range PCR product as published by Tamiya et al.16 DNA was amplified using KOD Hot Start DNA polymerase (Toyoba, Novagen). Primers for the control PCR and cycling conditions were 5′-CTCTCACAGTGGGCTCGAGA-3′ and 5′-CAAAGACGTAGAGTTGAGCAAGC-3′, 95°C for 2 minutes, 30 cycles: 95°C for 20 seconds, 59°C for 10 seconds, and 70°C for 48 seconds, followed by 70°C for 5 minutes. The primers and cycling conditions for the long-range PCR were
5′-CTTAGAGCCTCCCAGTGAAAAACATTTCC-3′ and 5′-GATGCATGGTCCTGCAAGGATAACA-3′, 95°C for 2 minutes, 30 cycles: 95°C for 20 seconds, and 66°C for 175 seconds, followed by 72°C for 15 minutes. The PCR products were electrophoresed on a 1% agarose gel with expected product size of 2.85 kb for the control PCR and 6.5 kb for a complete long-range product. A long-range product shorter than 6.5 kb defines a type 1 defective provirus; failure to amplify any long-range product identifies a type 2 defective provirus.16
Exon 2 and exon 3 tax gene sequencing
Tax protein is 353 amino acids in length: exon 2 provides the methionine start codon, and the remaining amino acids are derived from exon 3. Exons 2 and 3 were sequenced in ATL samples with a complete provirus. Exon 2 was amplified using PCR products from long-range PCR using Phusion high-fidelity DNA polymerase (New England Biolabs [NEB]). Primers and cycling conditions were 5′-CCTCAGCAATAAACAAACCC-3′ and 5′-CAATTGTGAGAGTACAGCAG-3′, 98°C for 30 seconds, 20 cycles: 98°C for 5 s seconds, 51.5°C for 20 s seconds, and 72°C for 10 seconds, followed by 72°C for 5 minutes. PCR products were inspected on 2% agarose gel for product length (318 bp). Exon 3 was amplified from the control long-range PCR product using Phusion high-fidelity DNA polymerase (NEB). Primers and cycling conditions were 5′- ATACAAAGTTAACCATGCTT-3′ and 5′-AGACGTCAGAGCCTTAGTCT-3′, 98°C for 30 seconds, 20 cycles: 98°C for 5 seconds, 51.5°C for 10 seconds, and 72°C for 22 seconds, followed by 72°C for 5 minutes. PCR products were inspected on a 2% agarose gel for product length (1120 bp) and sequenced by Sanger sequencing using 6 different sequencing primers to capture the entire exon (5′-ATACAAAGTTAACCATGCTT-3′, 5′-CGTTATCGGCTCAGCTCTACA-3′, 5′-TTCCGTTCCACTCAACCCTC-3′, 5′- AGACGTCAGAGCCTTAGTCT-3′, 5′-GGGTTCCATGTATCCATTTC-3′, and 5′- GTCCAAATAAGGCCTGGAGT-3′).
Methylation-specific PCR (MS-PCR)
MS-PCR was undertaken on ATL samples with a complete provirus but without a nonsense mutation of the tax gene. Takeda et al17 showed that MS-PCR correlates with bisulfite sequencing PCR and with the methylation status of the promoter/enhancer Tax-response element-1 in the 5′ long terminal repeat (LTR). DNA was treated overnight with sodium bisulfite (Sigma) and purified using Zymo EZ Bisulfite DNA cleanup as per the manufacturer’s protocol (Zymo Research). DNA was amplified by heminested PCR using JumpStart RedTaq polymerase (Sigma). Primers for the first PCR reaction for methylated DNA were 5′-TTAAGTCGTTTTTAGGCGTTGAC-3′, 5′-AAAAAAATTTAACCCATTACC-3′ and for unmethylated DNA 5′- TTAAGTTGTTTTTAGGTGTTGAT-3′, 5′-AAAAAAATTTAACCCATTACC-3′. The thermal conditions for first PCR were 94°C for 2 minutes, 35 cycles: 94°C for 30 seconds, 53°C for 30 seconds, and 72°C for 2 minutes. Primers for the hemi-nested methylated PCR were 5′-GAGGTCGTTATTTACGTCGGTTGAGTC-3′, 5′-AAAAAAATTTAACCCATTACC-3′ and unmethylated PCR primers 5′-GAGGTTGTTATTTATGTTGGTTGAGTT-3′, 5′-AAAAAAATTTAACCCATTACC-3′. The cycling conditions for the second PCR were 94°C for 2 minutes, 30 cycles: 94°C for 30 seconds, 57°C for 30 seconds, and 72°C for 2 minutes, followed by 72°C for 5 minutes. The PCR product was inspected on a 2% agarose gel for length (428 bp). MS-PCR primers did not amplify unconverted HTLV-1 or host genomic DNA.
T-cell receptor (TCR) gene rearrangement studies
TCR-γ gene rearrangement studies were undertaken in the Imperial Molecular Pathology Laboratory, Hammersmith Hospital (London, United Kingdom) using the established BIOMED-2 protocol followed by heteroduplex analysis and/or GeneScanning. GeneScan analysis was performed on an ABI 3130 genetic analyzer using GeneMapper 4.0 (Life Technologies).
Integration site mapping and quantification
The high-throughput protocol for identification and quantification of proviral integration sites was carried out as previously described.6 Mapped integration sites were compared with a set of randomly generated in silico genomic sites (n = 175 505) as previously reported.13
Bioinformatic annotation of genomic environment
Transcription units and cytosine guanine dinucleotide island data were retrieved from the National Center for Biotechnology Information (ftp.ncbi.nih.gov/gene/) and University of California, Santa Cruz tables, respectively. Epigenetic marks were annotated according to primary CD4+ T-cell chromatin immunoprecipitation sequencing data published by Barski et al.18 Transcription factor binding sites were obtained from published data sets (supplemental Figure 1 available at the Blood Web site) from chromatin immunoprecipitation sequencing experiments on primary human CD4+ T cells or other primary human cells or cell lines, as previously described.13 Cancer-associated gene data sets are defined by Sadelain et al.19 Annotated genomic positions were compared with the integration site data using the hiAnnotator R package kindly provided by N. Malani and F. Bushman (University of Pennsylvania; http://malnirav.github.com/hiAnnotator).
Diversity estimator
The diversity estimator (DivE)20 was used to estimate the total number of clones in addition to those observed. DivE involves fitting many mathematical models to nested subsamples of individual-based rarefaction curves. Estimates from the best-performing models are aggregated to produce the final estimate.20 DivE requires an estimate of the number of cells in the blood; because the absolute PBMC count for each case was unknown, DivE estimates were calculated for each patient over 2 orders of magnitude of variation in the PBMC count (3 × 109/L, 50 × 109/L, and 500 × 109/L).
Statistical analysis
Statistical analysis was carried out using R version 2.15.2 (http://www.R-project.org/). The oligoclonality index (OCI; Gini coefficient)6,21 was calculated using the R reldist package22 (http://CRAN.R-project.org/package=reldist). Two-tailed nonparametric tests (Mann Whitney U, Fisher’s exact, χ2) were used for all comparisons. Bonferroni’s correction for multiple testing was applied where appropriate. To test the hypothesis that 2 observed HTLV-1 integration sites were present in 1 T-cell clone, we used the Gaussian approximation to the binomial distribution (supplemental Figure 3). To identify clusters of integration sites or genomic hotspots of integration, we used R software developed by Presson et al23 (http://www.biomedcentral.com/1471-2105/12/367). Functional categories of genes were analyzed through the use of Ingenuity Pathway Analysis (Ingenuity Systems; http://www.ingenuity.com).
Results
The ATL samples are derived from a representative cohort
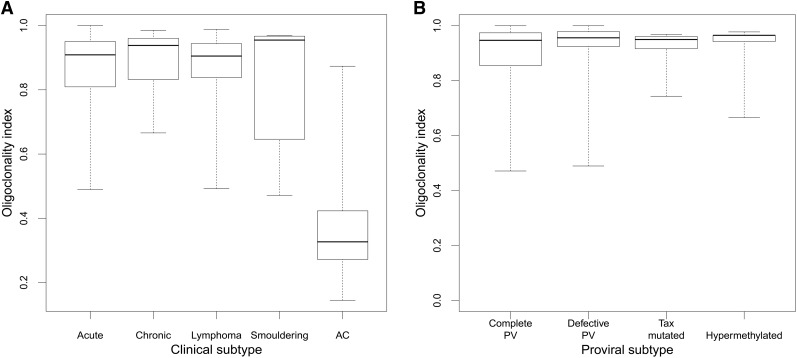
We analyzed 197 cases of ATL; patients’ characteristics are detailed in supplemental Figure 4. Systematic analysis of the proviral structure showed a complete provirus in 46% of cases, putatively capable of Tax expression; 39% of cases contained a defective provirus, 7% contained a nonsense mutation of the tax gene, and 8% contained a hypermethylated promoter in the 5′ LTR. There was no significant difference in OCI between ATL clinical subtypes (median OCI = 0.91) or by proviral subtype (median OCI = 0.91); the median OCI in asymptomatic carriers was 0.33, in the range previously reported6 (Figure 1).
Figure 1.
OCI by clinical and proviral subtype. (A) Median OCI of the ACs was 0.33 (range, 0.14-0.87), and median OCI for the ATL (all subtypes combined) was 0.91 (range, 0.47-1.0). There was no difference in OCI between ATL clinical subtypes. (B) There was no difference in OCI between the different mechanisms of proviral silencing. PV, provirus.
The median absolute number of HTLV-1+ T-cell clones (estimated by the DivE technique) in the circulation in ACs was 9054. The median number of clones in the ATL cases was 1741 (assuming PBMC = 3 × 109/L) or 2154 (assuming PBMC = 50 × 109/L). These results show that although the white cell count may vary over an order of magnitude between individuals with ATL, the estimated number of distinct clones underlying the malignant clone remains relatively stable (∼2000).
In 91% of ATL cases, a single copy of HTLV-1 is integrated into the host genome
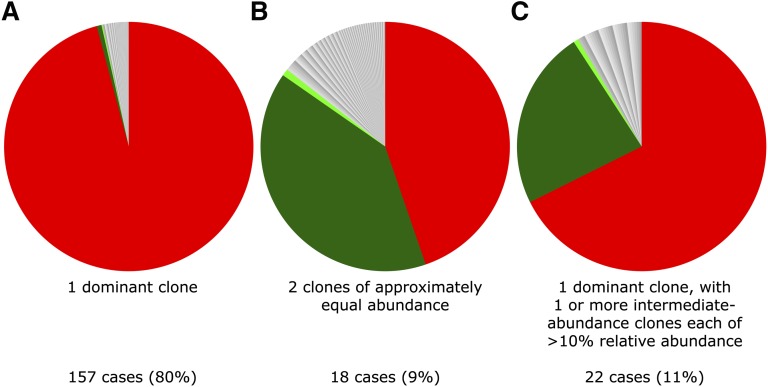
In our protocol, the quasi-random DNA shearing by sonication allows unbiased, quantitative detection of proviruses6,24 and therefore enabled us to quantify the presence of 2 abundant integration sites in an ATL tumor (Figure 2). In 157 out of 197 samples (80%), as expected, a single dominant proviral integration site was observed, with a median relative abundance of 99.4% of the PVL (range, 35% to 100%). However, in 40 out of 197 samples (20%), the presence of only a single provirus was less certain, because >1 abundant integration site was observed. In each of these 40 cases, there was 1 “large” ATL integration site with relative abundance >35% and an additional site with a relative abundance >10%. The question arises whether these represented 2 proviruses in 1 malignant clone or if there were 2 distinct abnormally expanded clones. If a single malignant clone contains 2 proviruses, then each will be present at the same frequency, assuming a steady kinetic state and no recent reinfection with a second provirus, and the clone will carry a single TCR gene rearrangement. Alternatively, if there are 2 large independent clones, then the 2 integration sites will differ in abundance and 2 distinct TCR gene rearrangements will be detected. We found no significant difference in the abundance of 2 integration sites in 18 cases (9.1% of cohort) (supplemental Figure 3), suggesting the presence of 2 proviruses in a single tumor clone. In 22 cases (11% of the cohort), we observed a large ATL clone and a second clone of abnormal but significantly lower abundance. TCR-γ gene rearrangement analysis of these 40 samples confirmed a monoclonal population in 7 out of 40 cases (3.1%); this technique may underestimate monoclonality, owing to the possibility of a second rearranged TCR-γ allele. To conclude, a single dominant provirus was detected in 91% of cases, whereas in 9% of tumors there was evidence of 2 proviruses. These results are consistent with the finding of multiple proviruses in 11% of cases reported by Tamiya et al16 using low-throughput techniques.
Figure 2.
Examples of 3 typical clonal structures of ATL cases. Each sector in the pie charts depicts the relative abundance of the respective integration site. (A) Typical “monoclonal” ATL tumor sample; PVL = 63% (relative abundance of dominant clone = 97% of PVL). (B) Two equally abundant integration sites (relative abundance respectively 44% and 39% of PVL); PVL = 9%. (C) ATL with dominant clone and additional intermediate-abundance clone (relative abundance respectively 67% and 23% of PVL); PVL = 241%.
Binning of clones into small, intermediate, or large
In subsequent analysis, each clone was binned according to its relative abundance, ie, the proportion of the subject’s PVL occupied by that clone. Each ATL case contained at least 1 abundant clone with a relative abundance >35% (n = 217 clones) that was defined as large; “small” clones were defined as those of relative abundance <1% (n = 5925), and such clones constitute the great bulk of PVL in nonmalignant HTLV-1 infection.6,13 Clones (n = 90) that constituted between 1% and 35% of PVL were classified as intermediate abundance. Clones (n = 16 909) identified in the AC cohort were analyzed together, because only 4 of these clones fulfilled the large-clone classification (supplemental Figure 2).
Large ATL clones have the same genomic characteristics as small (nonmalignant) clones, whereas intermediate-sized clones have unique genomic characteristics
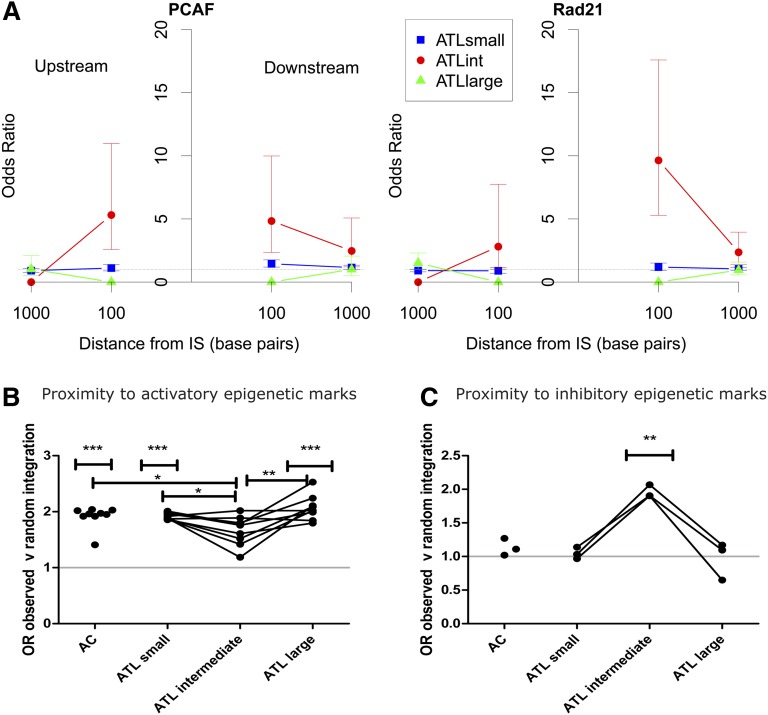
The intermediate-abundance clones observed in 24% of cases (48/197) in addition to the large (presumed malignant) clone were larger (ie, had a greater absolute abundance) than any clones previously observed in AC or HAM/tropical spastic paraparesis cohorts.6,13 Because progressive oligoclonal proliferation has been postulated to precede malignant transformation, we tested the hypothesis that there is a stepwise progression in the frequency of integration site characteristics from low-abundance clones through intermediate-abundance to large ATL clones. The results showed that the large, presumed malignant ATL clones had integration site characteristics indistinguishable from those of the low-abundance clones present in ACs and in patients with ATL. In contrast, the integration sites present in the intermediate-abundance clones in ATL patients, which are not observed in nonmalignant infection, differed from both the low- and high-abundance clones in each genomic attribute examined (Figure 3). Specifically, the intermediate-abundance clones lacked the associations observed in the ACs and in the low-abundance and high-abundance clones seen in ATL, with either transcriptional orientation or proximity to transcription start sites, cytosine guanine dinucleotide islands, or activatory epigenetic marks. Instead, the intermediate-abundance clones showed an association with proximity to inhibitory epigenetic marks (Figure 3C) and specific transcription-factor binding sites (TFBSs) within 100 bp upstream or downstream of the integration site, notably binding sites for P300/CBP-associated factor (odds ratio [OR] = 4.78), Rad 21 (part of the cohesin complex) (OR = 4.08), and ZNF263 (OR = 5.57). These effects disappeared at 1 kb from the integration site (Figure 3A). Integration in proximity to these specific TFBSs was identified in 8 out of 197 tumor samples (4.1% of cohort).
Figure 3.
Intermediate-abundance clones in ATL cases contained proviruses with distinct genomic marks. (A) The OR of integration in proximity to specific TFBSs compared with AC is illustrated for 2 TFBSs, P300/CBP-associated factor binding sites (PCAFbsites) and Rad21. (See supplemental Figure 1 for full list of TFBSs tested). The y-axis shows the OR compared with ACs. The x-axis shows the distance in base pairs (logarithmic scale) from the integration site upstream (left-hand side) or downstream (right-hand side). “Upstream” and “downstream” are defined with respect to the sense strand of the HTLV-1 provirus. The junction of the x-axis and y-axis represents the integration site. There were no independent TFBS predictors for small clones (blue squares) or large clones (green triangles) in ATL cases compared with ACs (OR = 1) or when compared with each other or to random data sets (not illustrated). Independent TFBSs associated with intermediate-abundance clones in ATL cases (red circles) (PCAFbsites, Rad21) at 100 bp upstream or downstream compared with ACs are illustrated. (B) OR of integration in proximity to activatory epigenetic marks compared with random sites. AC, small, and large clones in ATL cases showed a significant bias toward integration in proximity to activatory epigenetic marks. There was no such bias in the intermediate-abundance clones. (C) OR of integration in proximity to inhibitory epigenetic marks compared with random. AC, small, and large clones in ATL cases showed no bias toward integration in proximity to inhibitory marks compared with random sites, whereas intermediate-abundance clones showed a bias toward inhibitory epigenetic marks (see supplemental Figure 1 for details of epigenetic marks tested). IS, integration site.
There are no hotspots of integration associated with large ATL clones
All data sets were further annotated to investigate the proximity of the integrated provirus to the nearest cancer-associated gene. The frequency of integration was significantly higher than random expectation within 10 kb of oncogenes in clones from ACs and low-abundance clones from ATL patients, and within 150 kb in the large ATL clones; this association was not observed in the intermediate-abundance clones. We conclude that integration in proximity to these cancer-related genes confers a survival advantage in vivo but does not play a significant role in leukemogenesis per se. The use of the powerful bioinformatic method of Presson et al23 confirmed that there were no significant hotspots of integration associated with ATL.
The ontology of the nearest downstream gene was associated with the malignant clone in 6% of ATL cases
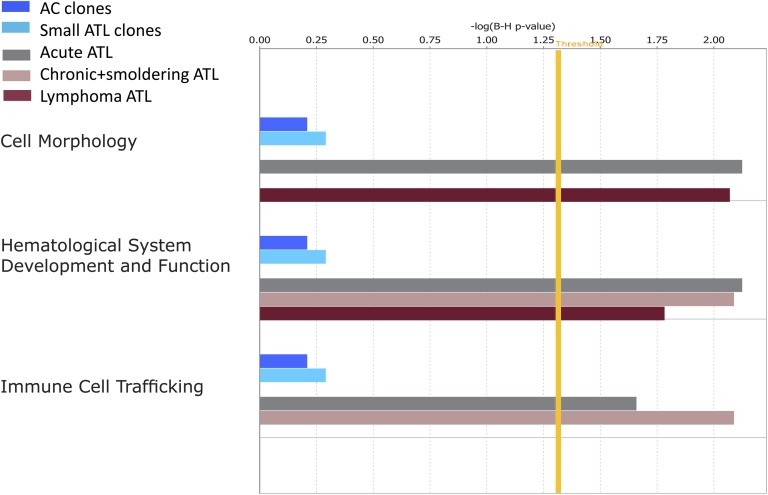
As a further test of the hypothesis that HTLV-1 proviral integration near host genes in a certain functional category confers a proliferative advantage on the infected T-cell clone, we used Ingenuity Pathway Analysis software to analyze the ontology of the nearest host genes upstream and downstream of each integration site. The results showed a significant overrepresentation of genes in 3 cellular pathways (“cell morphology,” “immune cell trafficking,” and “hematological system development and function”) in the large ATL (“malignant”) clones, but not in either the low- or intermediate-abundance clones (Figure 4). The 11 genes responsible for this significant association (CD46, ITGA4, DPYSL2, RAP2A, CASP8, CDKN2A, GTF2I, TACR1, BCL2, IL6ST, and HGF) accounted for 11 ATL cases (5.8% of the cohort) of different clinical subtypes. Furthermore, these effects were only seen in the nearest host gene downstream, regardless of its transcriptional orientation relative to the provirus. The median distance from the integration site to these nearest genes was 13.7 kb (range, 0.6-294 kb) compared with a median distance of 122.3 kb from all integration sites to the nearest cancer-associated gene (P = .009, Mann Whitney U test).
Figure 4.
Functional classification of gene ontologies overrepresented among the large ATL clones. Functional categories significantly overrepresented among the random, AC, ATL small, intermediate, and large ATL clones as analyzed by Ingenuity software using the Ingenuity Pathways Knowledge Base (IPKB) gene population as baseline. Horizontal bars are only visible where there was a statistical overrepresentation of the pathway compared with the IPKB. Because there were no overrepresented pathways involving the random integration sites or intermediate-abundance clones in ATL cases, the bars are not visible. The vertical yellow threshold represents the line of statistical significance (P < .05) after correction (Benjamini-Hochberg) for multiple testing. The numbers of searchable genes for comparison with the IPKB were random (n = 96 706), AC (n = 5679), ATL small (n = 1628), ATL intermediate (n = 87), or ATL large (acute n = 141, lymphoma n = 31, chronic and smoldering n = 38).
The HTLV-1 provirus preferentially survives in acrocentric chromosomes in vivo
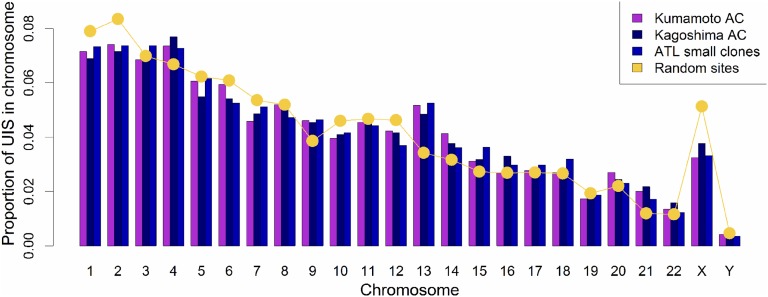
Meekings et al25 reported that the frequency of HTLV-1 proviruses in chromosome 13 was significantly higher in vivo than expected by chance, but the biological significance of this observation was uncertain. Here, using our quantitative, high-throughput technique, we observed a significant excess of integrations in chromosomes 13, 14, 15, and 21 compared with random and in vitro data sets. This excess was seen in all infected individuals and was not confined to those with ATL. There was a trend toward excess integrations in chromosome 22, but this was not statistically significant (Figure 5). These findings were validated with a second cohort of independent AC samples from the Kagoshima region of Japan. The chromosomal distribution of proviruses in the intermediate-abundance and large ATL clones was not significantly different from random, perhaps because of the small number of clones (n = 307).
Figure 5.
Preferential survival of HTLV-1 in vivo in chromosomes 13, 14, 15, and 21. The proportion of unique integration sites (UIS) per chromosome is shown for 2 independent AC data sets (Kumamoto and Kagoshima) and the small clones in ATL cases. The yellow line shows the frequency of sites in the random data set. There were an increased number of integrations in chromosomes 13, 14, 15, and 21 in the clones of asymptomatic carriers and small clones in ATL cases compared with random. The bias remained in chromosomes 13 and 15 when compared with a previously reported data set6 of integration sites from Jurkat cells infected in vitro with HTLV-1.
Discussion
Oligoclonal proliferation of HTLV-1–infected T-cells is a cardinal feature of HTLV-1 infection. It has long been believed that this oligoclonal proliferation is primarily responsible for maintaining the high PVL of HTLV-1, which is the strongest correlate of risk of both the inflammatory (HAM) and malignant (ATL) diseases. However, we recently showed that the PVL correlates with the total number of infected clones, but not with the degree of oligoclonal proliferation as measured by the OCI.6,13 Here, we show that ATL is frequently accompanied by a population of abnormally abundant HTLV-1–infected T-cell clones underlying the largest, putatively malignant clone. This observation suggested that such intermediate-abundance clones might represent an intermediate stage of malignant transformation between the low-abundance clones and the fully transformed, largest clone. However, we found that the host genomic attributes of the integration site in the large ATL clones closely resembled those of the low-abundance clones present both in ATL patients and in those with nonmalignant infection, whereas the integration site characteristics of the intermediate-abundance clones differed from both the low- and high-abundance clones and from the clones observed in nonmalignant cases of HTLV-1 infection. We conclude that the malignant clone does not arise from the intermediate-abundance clones but instead from the low-abundance clones. This conclusion is consistent with the observations that the low-abundance clones constitute the bulk of the PVL in HTLV-1 infection6 and that the risk of ATL is correlated with the PVL.2,4 We have also observed cases in which the malignant clone emerges from the large population of low-abundance clones, not from the preexisting oligoclonally expanded population.26 Finally, this conclusion is also consistent with our recent observation27 of highly oligoclonal proliferation and a small total number of clones in human T-lymphotropic virus type 2 infection, which does not cause malignant disease.
We therefore propose that the major determinant of the risk of ATL is the absolute number of clones: the larger the number, the greater the chance of malignant transformation. It is likely that the number of HTLV-1–infected clones present in an individual during chronic infection is determined chiefly by the efficiency of the host’s CTL response to the virus, which in turn is determined by the HLA and killer immunoglobulin-like receptor genotype.10,28
The observation that the abnormally expanded intermediate-abundance clones seen in patients with ATL do not share genomic characteristics with either the polyclonal background or the malignant clones suggests that the intermediate-abundance clones arise as a consequence of ATL development and are not causative. One possibility is that these clones survive and proliferate as a consequence of the severely impaired immune response in ATL. The malignant clones in ATL use well-described mechanisms to silence Tax, either before or after malignant transformation, which allows them to escape the immunodominant CTL response and so confers a survival advantage. Once the malignant clone has emerged, the resulting immune impairment may allow the intermediate-abundance clones to survive despite continued expression of viral genes.
As expected, we did not identify any hotspots of integration, although analysis of the ontology of flanking genes demonstrated a functional overrepresentation of certain genes that are known to be dysregulated in many leukemias. This effect was significant only in the large (presumed malignant) ATL clones and only when considering the ontology of the nearest host gene downstream; there was no effect of the upstream host gene. Further, these specific genes lay very close (median 13.7 kb) to the provirus, suggesting a mechanistic interaction between the provirus and the downstream gene. Although the associations reported here between ATL and individual genes and genomic features account for a small proportion of the observed cases of ATL, these results indicate that transcriptional interactions between the provirus and the flanking host genome influence the risk of malignant transformation. Vogelstein recently estimated that each tumor-driver mutation contributes a survival advantage of ∼0.4% to a clone29; the HTLV-1 genomic integration site may contribute a similar advantage.26
A further unexpected observation was the preferential survival in vivo of the HTLV-1 provirus in the acrocentric chromosomes 13, 14, 15, 21, and (although not reaching formal significance) 22. Throughout most of the cell cycle, these chromosomes are physically associated with the nucleolus, and they encode the machinery of the ribosome on the short (p) arm. Because the HTLV-1 proviral integration sites are found only in the long (q) arm of these chromosomes, we postulate that the selective advantage enjoyed by these clones derives not from the proviral integration near the ribosome-coding genes but rather from the physical location of the provirus-containing chromatin in the nucleus, perhaps by coupling proviral transcription to transcription of the acrocentric chromosomes. Experiments are underway to test this hypothesis.
Acknowledgments
The authors thank Nirav Malani and Frederic D. Bushman at the department of Microbiology, University of Pennsylvania, Philadelphia, PA for the list of random integration sites and for developing software packages and the Core Genomics Laboratory at the MRC Clinical Sciences Centre, Hammersmith Hospital, London, United Kingdom. The authors thank Aileen Rowan and Yorifumi Satou for many helpful discussion and comments and the patient donors in Japan.
This work was funded by Leukaemia and Lymphoma Research and the Wellcome Trust.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: L.B.C., G.P.T., M.M., and C.R.M.B. conceived and designed the experiments; M.M. performed the clinical diagnosis; L.B.C. performed the experiments; M.V. and L.F. performed and interpreted TCR studies; L.B.C. analyzed the data; A.M., H.N., and D.J.L. contributed to the bioinformatic and statistical analysis, tools, and data sets; and L.B.C. and C.R.M.B. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Charles R. M. Bangham, Wright-Fleming Institute, Imperial College, Norfolk Place, London, W2 1PG, United Kingdom; e-mail: c.bangham@imperial.ac.uk.
References
- 1.Shimoyama M. Diagnostic criteria and classification of clinical subtypes of adult T-cell leukaemia-lymphoma. A report from the Lymphoma Study Group (1984-87). Br J Haematol. 1991;79(3):428–437. doi: 10.1111/j.1365-2141.1991.tb08051.x. [DOI] [PubMed] [Google Scholar]
- 2.Sasaki D, Doi Y, Hasegawa H, et al. High human T cell leukemia virus type-1(HTLV-1) provirus load in patients with HTLV-1 carriers complicated with HTLV-1-unrelated disorders. Virol J. 2010;7:81. doi: 10.1186/1743-422X-7-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iwanaga M, Watanabe T, Utsunomiya A, et al. Joint Study on Predisposing Factors of ATL Development investigators. Human T-cell leukemia virus type I (HTLV-1) proviral load and disease progression in asymptomatic HTLV-1 carriers: a nationwide prospective study in Japan. Blood. 2010;116(8):1211–1219. doi: 10.1182/blood-2009-12-257410. [DOI] [PubMed] [Google Scholar]
- 4.Demontis MA, Hilburn S, Taylor GP. Human T cell lymphotropic virus type 1 viral load variability and long-term trends in asymptomatic carriers and in patients with human T cell lymphotropic virus type 1-related diseases. AIDS Res Hum Retroviruses. 2013;29(2):359–364. doi: 10.1089/AID.2012.0132. [DOI] [PubMed] [Google Scholar]
- 5.Overbaugh J, Bangham CR. Selection forces and constraints on retroviral sequence variation. Science. 2001;292(5519):1106–1109. doi: 10.1126/science.1059128. [DOI] [PubMed] [Google Scholar]
- 6.Gillet NA, Malani N, Melamed A, et al. The host genomic environment of the provirus determines the abundance of HTLV-1-infected T-cell clones. Blood. 2011;117(11):3113–3122. doi: 10.1182/blood-2010-10-312926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cook LB, Rowan AG, Melamed A, Taylor GP, Bangham CR. HTLV-1-infected T cells contain a single integrated provirus in natural infection. Blood. 2012;120(17):3488–3490. doi: 10.1182/blood-2012-07-445593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsukasaki K, Tsushima H, Yamamura M, et al. Integration patterns of HTLV-I provirus in relation to the clinical course of ATL: frequent clonal change at crisis from indolent disease. Blood. 1997;89(3):948–956. [PubMed] [Google Scholar]
- 9.Matsuoka M, Jeang KT. Human T-cell leukemia virus type 1 (HTLV-1) and leukemic transformation: viral infectivity, Tax, HBZ and therapy. Oncogene. 2011;30(12):1379–1389. doi: 10.1038/onc.2010.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bangham CR. CTL quality and the control of human retroviral infections. Eur J Immunol. 2009;39(7):1700–1712. doi: 10.1002/eji.200939451. [DOI] [PubMed] [Google Scholar]
- 11.Nagai M, Usuku K, Matsumoto W, et al. Analysis of HTLV-I proviral load in 202 HAM/TSP patients and 243 asymptomatic HTLV-I carriers: high proviral load strongly predisposes to HAM/TSP. J Neurovirol. 1998;4(6):586–593. doi: 10.3109/13550289809114225. [DOI] [PubMed] [Google Scholar]
- 12.Jeffery KJ, Usuku K, Hall SE, et al. HLA alleles determine human T-lymphotropic virus-I (HTLV-I) proviral load and the risk of HTLV-I-associated myelopathy. Proc Natl Acad Sci USA. 1999;96(7):3848–3853. doi: 10.1073/pnas.96.7.3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Melamed A, Laydon DJ, Gillet NA, Tanaka Y, Taylor GP, Bangham CR. Genome-wide determinants of proviral targeting, clonal abundance and expression in natural HTLV-1 infection. PLoS Pathog. 2013;9(3):e1003271. doi: 10.1371/journal.ppat.1003271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nosaka K, Maeda M, Tamiya S, Sakai T, Mitsuya H, Matsuoka M. Increasing methylation of the CDKN2A gene is associated with the progression of adult T-cell leukemia. Cancer Res. 2000;60(4):1043–1048. [PubMed] [Google Scholar]
- 15.Kwok S, Ehrlich G, Poiesz B, Kalish R, Sninsky JJ. Enzymatic amplification of HTLV-I viral sequences from peripheral blood mononuclear cells and infected tissues. Blood. 1988;72(4):1117–1123. [PubMed] [Google Scholar]
- 16.Tamiya S, Matsuoka M, Etoh K, et al. Two types of defective human T-lymphotropic virus type I provirus in adult T-cell leukemia. Blood. 1996;88(8):3065–3073. [PubMed] [Google Scholar]
- 17.Takeda S, Maeda M, Morikawa S, et al. Genetic and epigenetic inactivation of tax gene in adult T-cell leukemia cells. Int J Cancer. 2004;109(4):559–567. doi: 10.1002/ijc.20007. [DOI] [PubMed] [Google Scholar]
- 18.Barski A, Cuddapah S, Cui K, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129(4):823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 19.Sadelain M, Papapetrou EP, Bushman FD. Safe harbours for the integration of new DNA in the human genome. Nat Rev Cancer. 2012;12(1):51–58. doi: 10.1038/nrc3179. [DOI] [PubMed] [Google Scholar]
- 20.Laydon DJ, Melamed A, Sim A, et al. Quantification of HTLV-1 clonality and TCR diversity. PLOS Comput Biol. doi: 10.1371/journal.pcbi.1003646. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gini C. Variabilita e Mutuabilita. Contributo allo Studio delle Distribuzioni e delle Relazioni Statistiche. Bologna, Italy: C. Cuppini; 1912. [Google Scholar]
- 22.Handcock M, Morris M. Relative Distribution Methods in the Social Sciences. New York, NY: Springer-Verlag; 1999. [Google Scholar]
- 23.Presson AP, Kim N, Xiaofei Y, Chen IS, Kim S. Methodology and software to detect viral integration site hot-spots. BMC Bioinformatics. 2011;12:367. doi: 10.1186/1471-2105-12-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berry CC, Gillet NA, Melamed A, Gormley N, Bangham CR, Bushman FD. Estimating abundances of retroviral insertion sites from DNA fragment length data. Bioinformatics. 2012;28(6):755–762. doi: 10.1093/bioinformatics/bts004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meekings KN, Leipzig J, Bushman FD, Taylor GP, Bangham CR. HTLV-1 integration into transcriptionally active genomic regions is associated with proviral expression and with HAM/TSP. PLoS Pathog. 2008;4(3):e1000027. doi: 10.1371/journal.ppat.1000027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bangham CRM, Cook LB, Melamed A. HTLV-1 clonality in adult T-cell leukaemia and non-malignant HTLV-1 infection [published online ahead of print December 6, 2013]. Semin Cancer Biol. doi: 10.1016/j.semcancer.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Melamed A, Witkover AD, Laydon DJ, et al. Clonality of HTLV-2 in natural infection. PLoS Pathog. 2014;10(3):e1004006. doi: 10.1371/journal.ppat.1004006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Macnamara A, Rowan A, Hilburn S, et al. HLA class I binding of HBZ determines outcome in HTLV-1 infection. PLoS Pathog. 2010;6(9):e1001117. doi: 10.1371/journal.ppat.1001117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339(6127):1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]