Abstract
Structurally diverse organophosphonate inhibitors targeting the active site of the enzyme were used to investigate the relationship of the active site and the dimer interface of wild-type protease in solution. Positional scanning synthetic combinatorial libraries revealed Kaposi's sarcoma-associated herpesvirus protease to be highly specific, even at sites distal to the peptide bond undergoing hydrolysis. Specificity results were used to synthesize a hexapeptide diphenylphosphonate inhibitor of Kaposi's sarcoma-associated herpesvirus protease. The transition state analog inhibitors covalently phosphonylate the active site serine, freezing the enzyme structure during catalysis. An NMR-based assay was developed to monitor the native monomer-dimer equilibrium in solution and was used to demonstrate the effect of protease inhibition on the quaternary structure of the enzyme. NMR, circular dichroism, and size exclusion chromatography analysis showed that active site inhibition strongly regulates the binding affinity of the monomer-dimer equilibrium at the spatially separate dimer interface of the protease, shifting the equilibrium to the dimeric form of the enzyme. Furthermore, inhibitor studies revealed that the catalytic cycles of the spatially separate active sites are independent. These results (i) provide direct evidence that peptide bond hydrolysis is integrally linked to the quaternary structure of the enzyme, (ii) establish a molecular mechanism of protease activation and stabilization during catalysis, and (iii) highlight potential implications of substoichiometric inhibition of the viral protease in developing herpesviral therapeutics.
Herpesviruses represent one of the most prevalent viral families worldwide, with roughly 100 viruses identified, affecting almost every animal species. The nine known human herpesviruses (HHV) are responsible for a variety of diseases, ranging from relatively harmless ailments, such as cold sores (Herpes simplex virus 1, HHV-1) and chicken pox (Varicella–Zoster virus, HHV-3), to life threatening illnesses caused by human cytomegalovirus, HHV-5, and Kaposi's sarcoma (KS)-associated herpesvirus (KSHV), HHV-8. In the United States, KS emerged as a serious problem in the early 1980s with the onset of AIDS and remains the most common neoplasm afflicting homosexual and bisexual men diagnosed with AIDS (1–3). Although the incidence of KS in this country has been reduced in part by AIDS-specific therapies such as highly active antiretroviral therapy (4, 5), the prevalence of KSHV infection in Africa has increased severely in recent years. In fact, since 1985, KS has emerged as the most common cancer in men and the second most common cancer in women in Uganda and Zimbabwe, reflecting nearly a 20-fold increase in incidence in these countries (6).
Despite their clinical diversity, all HHV use a homologous virally encoded maturational protease for the formation of infectious virions. The 25-kDa protease is expressed as an inactive monomer fused to the viral assembly protein, a capsid scaffolding protein. Upon formation of the immature viral capsid, the high local concentration of protease is thought to drive dimerization of monomers, which activates the protease and results in cleavage at the two natural proteolysis sites, the release site and maturation site, leading to formation of mature virions (7–9). Genetically modified herpes simplex virus mutants indicated that these protease cleavage events are essential for viral replication (10, 11). As a result, the protease has been implicated as a potential therapeutic target (12).
Although herpesvirus proteases are serine proteases, the crystal structures of herpes simplex virus, Varicella–Zoster virus, Epstein–Barr virus, human cytomegalovirus, and KSHV proteases reveal both a novel catalytic triad and a novel protein fold (13–20). The altered Ser-His-His catalytic triad results in a severely disabled protease with a catalytic efficiency (kcat/Km) approximately four orders of magnitude reduced when compared with digestive serine proteases, such as trypsin (21, 22). The proteases are active only as homodimers, yet despite burying nearly 2,500 Å2 of hydrophobic surface area upon dimerization, they interact with weak dissociation constants in the low micromolar range (23–25).
Several previous reports have provided circumstantial evidence supporting a link between activity and dimerization in herpesvirus proteases (18, 23, 26–29). The results in this report provide an indisputable link between the active site serine and the dimer interface of wild-type protease in solution during catalysis and a characterization of the individual active sites as catalytically independent. The findings also allow for conclusions addressing the contributions of substrate binding determinants to dimer stabilization, the molecular mechanism of activation and stabilization during the enzymatic reaction, and potential impacts of targeting the active site of the protease in drug development.
Materials and Methods
Recombinant Expression and Purification of KSHV Protease. A protease variant stable to autolysis (24), referred to in this paper as KSHV Pr, was recombinantly expressed in Escherichia coli and purified by following a previously described experimental protocol (30). Purified protease was stored in assay buffer (25 mM potassium phosphate, pH 8.0/150 mM potassium chloride/100 μM EDTA/1 mM 2-mercaptoethanol) at 4°C.
Positional Scanning Synthetic Combinatorial Library (PS-SCL) Analysis of KSHV Pr. KSHV Pr (200 μM) was added to 80 wells of a 96-well microtiter plate in assay buffer. A DMSO stock (1 μl) of a completely diverse PS-SCL (Y. Choe and C.S.C., unpublished results, and ref. 31) was added to each of the 80 wells, resulting in 8,000 compounds per well and 250 μM in total substrate per well. Substrate turnover was monitored for 1 h at 30°C as an increase in fluorescence by using an excitation wavelength of 380 nm, an emission wavelength of 460 nm, and a cutoff filter at 435 nm. The reaction rates remained linear over the entire 1-h interval.
Synthesis and Kinetic Analysis of Individual 7-amino-4-carbamoylmethylcoumarin (ACC) Substrates. Single ACC substrates were synthesized and purified as described (31). Concentrated KSHV Pr was diluted into assay buffer and incubated at room temperature for 1 h to ensure monomer–dimer equilibrium was established. Aliquots of KSHV Pr (1 μM) were placed in individual wells of a 96-well microtiter plate. Peptide substrate stocks were prepared in DMSO and added to each protease-containing well. After initiation of substrate hydrolysis, reaction rates were monitored by an increase in fluorescence over 60 min at 30°C, as described above.
Synthesis of Biotinyl-Pro-Val-Tyr-tBug-Gln-AlaP-(OPh)2. Diphenyl [α-aminoethyl] phosphonate [H-AlaP-(OPh)2] was synthesized as reported by Oleksyszyn (32). Biotinyl-Pro-Val-Tyr-tBug-Gln-OH (tBug, tert-butylglycine) was synthesized by standard fluorenylmethoxycarbonyl chemistry. Biotin was incorporated to monitor protease labeling by a streptavidin–horseradish peroxidase blot (33). The peptide was purified by reversed-phase HPLC and characterized by mass spectrometry.
Free peptide (230 mg, 0.27 mmol), PyBOP (140 mg, 0.27 mmol), and diisopropylethylamine (104 mg, 0.81 mmol) were dissolved in dimethylformamide (2 ml). H-AlaP-(OPh)2 (75 mg, 0.27 mmol) was added, and the reaction was stirred for 18 h at room temperature. Solvent was removed, and the resulting oil was purified by reversed-phase HPLC, yielding pure product, as confirmed by mass spectrometry. Active and inactive diastereomers were separated by an additional reversed-phase HPLC purification. Stocks of biotinyl-Pro-Val-Tyr-tBug-Gln-AlaP-(OPh)2 were prepared in DMSO and stored at -20°C.
Characterization of Protease Inhibition. Inhibition was monitored based on the method of Kitz and Wilson (34). Before addition of inhibitor, KSHV Pr was diluted to 10 μM in activity buffer and incubated at room temperature for 1 h to establish the monomer–dimer equilibrium. Upon addition of inhibitor (100× in DMSO), aliquots of the reaction solution were removed at various time points, and remaining protease activity was monitored (at 2.5 μM protease) as described above.
Inhibitor titration experiments were performed by incubation of a range of biotinyl-Pro-Val-Tyr-tBug-Gln-AlaP-(OPh)2 (2.3 - 20 μM) with KSHV Pr (20 μM) for 60 h. KSHV Pr was incubated for 3 h at room temperature before addition of inhibitor. Following complete inhibition, reactions were diluted to a final protease concentration of 200 nM, incubated for 2 h at room temperature, and characterized by monitoring remaining activity as described above. The experimental data are an average of four repetitions. Methods for determination of protease concentration and derivation of theoretical activity curve are described in Supporting Text, which is published on the PNAS web site.
Diisopropyl fluorophosphate (DFP) inhibition was carried out by using KSHV Pr (20 μM) in assay buffer at room temperature. As a result of the high rate of hydrolysis in buffer, DFP was added in 100 μM aliquots every 16 h for 64 h. After each addition, a portion of the inhibition reaction was removed, diluted to 6 μM, incubated at room temperature for 1 h, and subsequently analyzed by size exclusion chromatography. Biotinyl-Pro-Val-Tyr-tBug-Gln-AlaP-(OPh)2 (100 μM) was then added to the original inhibition reaction, and the solution was incubated at room temperature for an additional 72 h before size exclusion analysis.
FPLC Analysis of Quaternary Structure of the Protease. KSHV Pr (20 μM) was incubated with inhibitor (95 μM) or DMSO at 25°C until no residual protease activity was detectable. Aliquots of enzyme were removed from each reaction and diluted to final concentrations of 5 μM and 300 nM. Before size exclusion analysis, samples were incubated for 1 h at room temperature. Monomer–dimer equilibrium of the protease was examined at both concentrations with a Superdex 75 analytical column (Amersham Pharmacia) equilibrated in assay buffer. KSHV Pr dimers and monomers eluted at ≈10.5 ml and 12 ml, respectively, as originally reported by Pray et al. (24).
NMR and Circular Dichroism Spectroscopy. NMR experiments were measured on a Bruker 500 MHz Avance NMR instrument equipped with a 5-mm triple-resonance cryoprobe with a z-axis gradient coil. NMR spectra were recorded by using 500-μl solutions in H2O/2H2O (93%/7%)/25 mM potassium phosphate, pH 8.0/0.1 mM EDTA/1 mM 2-mercaptoethanol at 27°C. 13C-1H heteronuclear single quantum coherence spectra of KSHV Pr labeled with methyl 13C methionine (25 μM) with and without inhibitor were recorded in 2.5 h with 64 scans, 128 and 512 complex t1 and t2 increments with 3,000-Hz F1 and 7,002-Hz F2 spectral widths, respectively. The 13C-1H heteronuclear single quantum coherence spectrum of KSHV Pr Met197L labeled with methyl 13C methionine was recorded with four scans by using the same increments and spectral widths as KSHV Pr spectra.
Circular dichroism spectra were acquired on a Jasco 710 spectropolarimeter with a 0.1-cm pathlength sample cuvette. Inhibitor and DMSO-treated protease samples (30 μM) were dialyzed against assay buffer to remove DMSO and excess inhibitor. Melting curves were generated by monitoring the elipticity at 222 nm as a function of temperature. Temperature was varied from 10 to 90°C at a rate of 0.5°C·min-1.
Results
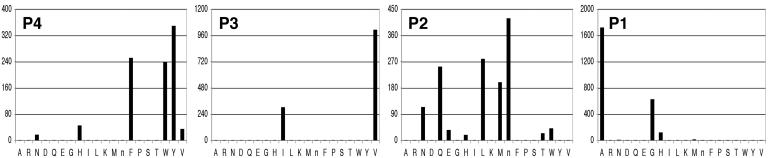
Substrate Profiling of KSHV Pr by Using PS-PCLs and Assay Development. The PS-SCL results reveal KSHV Pr to be a highly specific protease (Fig. 1). As expected, based on the strongly conserved P1-alanine seen in all herpesvirus protease cleavage site sequences, alanine is the preferred amino acid at the P1 position. Despite the lack of an obvious substrate-binding pocket at the S4 position upon examination of the crystal structure of KSHV Pr, a dramatic preference for aromatic residues is demonstrated at P4.
Fig. 1.
Substrate specificity profile of KSHV Pr. Complete diversity PS-SCL results for each subsite, P4–P1, are shown. The y axis reflects picomolar concentrations of free ACC generated per second upon enzymatic hydrolysis. The x axis reveals the spatially addressed amino acid at each position, and “n” represents norleucine, a methionine isostere. In vivo protease cleavage sites are the release site (YLKA), maturation site (RLEA), and dimer disruption site (AIDA).
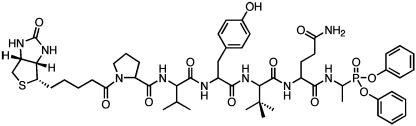
Given the ability of the protease to cleave the tetrameric ACC substrates in the PS-SCL library, individual ACC substrates were synthesized to develop an improved enzyme activity assay. Although the optimal sequence from the library was Tyr (P4), Val (P3), Nle (P2), Ala (P1), poor solubility limited the utility of this extremely hydrophobic substrate. Accordingly, norleucine was replaced by glutamine to improve solubility. Additionally, previous results suggest a preference for tert-butylglycine at the P3 position, so this information was also incorporated into the modified substrate (35). Kinetic analysis of the tetrameric ACC substrate revealed Michaelis–Menten kinetics with a Km of 81 ± 11 μM and a kcat of 0.022 ± 0.001 s-1, a dramatic improvement over the intramolecularly quenched substrate previously used in enzymatic assays (24). Furthermore, the vastly increased solubility of the tetrapeptide substrate (high micromolar) as compared to the intramolecularly quenched substrate (low micromolar) renders it far superior for use in enzymatic activity assays. In addition to assay development, substrate specificity information was used to design a potent peptide-based organophosphonate inhibitor. Analysis of a natural protease cleavage site (release site) resulted in incorporation of proline and valine at the P6 and P5 positions, respectively, to increase binding affinity of the inhibitor (Fig. 2).
Fig. 2.
Chemical structure of the optimized biotinyl-Pro-Val-Tyr-tBug-Gln-AlaP-(OPh)2 inhibitor.
Covalent Modification of the Active Site Serine by an (α-Aminoalkyl)-phosphonate Diphenyl Ester. Incubation of KSHV Pr with the diphenylphosphonate revealed time-dependent inhibition of enzymatic activity. A streptavidin blot identified the biotinylated inhibitor bound to KSHV Pr, whereas no inhibitor was detected bound to a point mutant removing the catalytic serine of the protease (S114A/S204G) (data not shown).
Because of the low activity of the protease, enzyme concentration could not be significantly decreased without approaching the limit of detection of the assay. Therefore, inhibitor concentrations were limited as well. Kinetic analysis, even at the lowest concentrations of inhibitor, indicated that the inhibitor concentration is still well above the Ki (data not shown). As a result, a kobs/[I] of 6 M-1·s-1 was determined as a lower limit for kinact/Ki.
To determine whether the enzyme reactivated because of hydrolysis of the phosphonylated protease, KSHV Pr (30 μM) was incubated with inhibitor (100 μM) in activity assay buffer until all enzyme activity was removed. Excess inhibitor was removed by size exclusion chromatography, and activity assays with aliquots of the protease solution revealed no significant recovery of activity over 100 h.
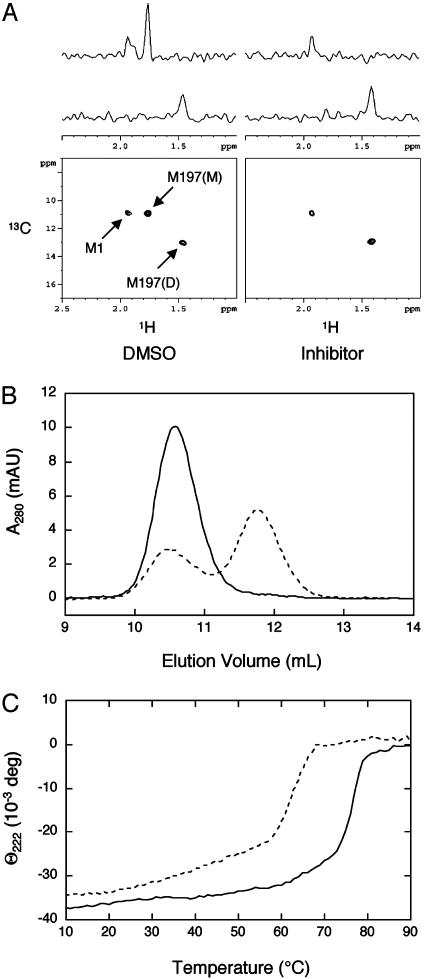
Determination of Monomer–Dimer Equilibrium Upon Protease Inhibition. Other than the N terminus of the protein, only one methionine is present in KSHV Pr. Its location in the middle of the dimer interface made it a potentially attractive probe for monitoring the oligomerization state of the protease. As predicted, heteronuclear single quantum coherence analysis of a sample of KSHV Pr labeled with methyl 13C methionine revealed three peaks in the protease spectrum corresponding to the N terminus, Met197 (monomer), and Met197 (dimer). These assignments were confirmed by a Met197L variant (36), eliminating two of the peaks, and by increasing protease concentration and observing a concomitant increase in the dimeric peak. As seen in Fig. 3A, a comparison of inhibitor- or DMSO-treated protease reveals the disappearance of the M197 (monomer) peak. The NMR experiments demonstrate that upon protease inhibition, the equilibrium is shifted dramatically toward dimeric protease. Gel filtration of the inhibited sample confirmed the dramatic dimer stabilization (Fig. 3B).
Fig. 3.
Inhibition of KSHV Pr stabilizes the dimeric conformation. (A) Heteronuclear single quantum coherence spectra of KSHV Pr labeled with methyl 13C methionine after incubation with or without inhibitor. Signals reveal the solution oligomeric state of protease: M1, N terminus; M197(M), monomer; M197(D), dimer. (B) Size exclusion chromatography of KSHV Pr. After treatment with inhibitor (solid trace) or DMSO (dashed trace), KSHV Pr (5 μM) was incubated for 1 h before analysis on a Superdex 75 analytical column. (C) Circular dichroism temperature melts. Elipticity of KSHV Pr (30 μM) after incubation with (solid trace) or without (dashed trace) inhibitor was monitored at 222 nm as a function of temperature.
To compare relative protease stability, KSHV Pr was subjected to thermal denaturation in the presence and absence of the inhibitor (Fig. 3C). Strikingly, a 13°C increase in Tm was observed in the inhibited dimer. In native (DMSO-treated) protease, a steady loss in elipticity occurred from 20°C to 57°C, which was followed by a rapid loss in elipticity. The gradual loss in elipticity was attributed to a dimer–monomer transition, whereas the sharp transition represented global unfolding. However, the inhibited dimer shifts the dimer–monomer transition nearly 30°C, to 65°C. It appears that the dimer is the predominant species, even at 50°C, and if an inhibited monomeric species exists, it is very short lived.
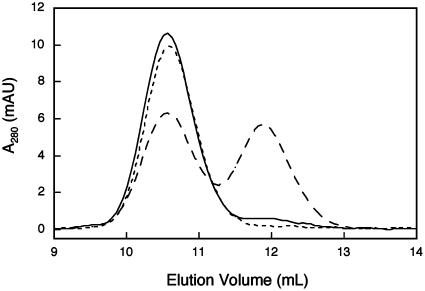
KSHV Pr inhibition was also observed after incubation with the smaller organophosphate, DFP. A small population of monomeric protease remained after inhibition of the enzyme with DFP, even following multiple additions of DFP. This population was not inhibited monomer, reflective of a higher Kd of the DFP-inhibited dimer, but rather was uninhibited protease at a concentration well below the dimerization constant. Accordingly, given the high rate of hydrolysis of DFP in water and the exceedingly low population of uninhibited dimers remaining in solution, most of the DFP added to the reaction simply hydrolyzed before further protease inhibition. This was confirmed by addition of the significantly more stable diphenylphosphonate to the sample, which resulted in complete inhibition of the remaining protease (Fig. 4).
Fig. 4.
DFP induces stabilization of the KSHV Pr dimer. Protease (6 μM) was incubated with DMSO (long-dashed trace), DFP (solid trace), or DFP followed by diphenylphosphonate (short-dashed trace). Samples were equilibrated at room temperature for 1 h before analysis.
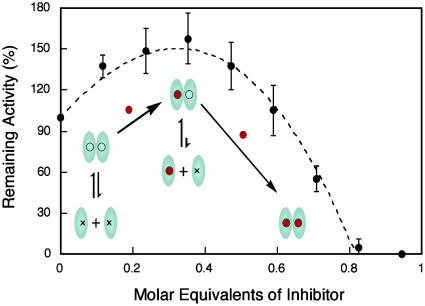
Protease Active Sites Are Capable of Simultaneous Substrate Processing. Upon incubation of various molar equivalents of inhibitor with high concentrations of protease, three protease species were obtained: uninhibited, singly inhibited, and doubly inhibited dimers. Given the increased stability of the inhibited dimer, remaining activity of the singly inhibited dimer was examined by diluting the inhibited protease sample, thereby minimizing contributions from the uninhibited dimer. The remaining activity profile clearly demonstrates that singly inhibited protease was still active, even though one active site was locked in a transitionstate conformation (Fig. 5 and Supporting Text). In fact, singly inhibited protease showed increased activity relative to the uninhibited protease at low concentrations as a result of the augmented stability of the singly inhibited species. Titration of the remaining active site with increased molar equivalents of inhibitor resulted in a complete loss of protease activity. The experimental data fits well to a theoretical model based on two fully active, independent active sites.
Fig. 5.
Inhibitor titration reveals independent active sites. Protease (20 μM) was incubated with various molar equivalents of inhibitor followed by dilution of the protease to 200 nM. Remaining protease activity was monitored as described. Experimental data fit well to theoretical data (dashed trace) generated by using a Kd of 1.8 μM as described in Supporting Text. Kinetic schemes show the effect of inhibitor (red circles) on KSHV Pr equilibrium (x, inactive protease; ○, active protease). Although negligible, the equilibrium between doubly inhibited protease and inhibited monomers was omitted for clarity.
Discussion
As a result of their critical role in the viral life cycle, there has been significant interest in herpesvirus proteases as therapeutic targets. Indirect evidence has associated dimerization with activity in herpesvirus proteases; however, a direct link between the active site and the dimer interface has not been illustrated. Furthermore, the effect of substrate binding determinants and oxyanion formation on dimerization is also unknown. In the present work, we clearly demonstrate the link between the active site and dimer interface and elucidate the importance of substrate structural features on enzymatic catalysis. We also show the independence of catalytic activity of the two active sites within an intact dimer. Taken together, the data suggest several important impacts of active site inhibition of the protease and supports the hypothesis that an alternative mechanism of protease inhibition may also be effective as a therapeutic strategy.
To address the association of the active site and dimer interface, an active site inhibitor was synthesized. Organophosphonates provide insight into the catalytic mechanism as a result of their structural similarity to the tetrahedral transition state upon covalent modification of the active site serine (37, 38). Accordingly, upon covalent inhibition of KSHV Pr, the enzyme is locked in a conformation arguably most relevant to catalysis. Development of a transition-state analog inhibitor has allowed for a glimpse into the solution properties of the enzyme during the process of peptide bond hydrolysis. In the protein–protein interaction required for activation, one monomer acts essentially as a cofactor for the other monomer, repositioning and stabilizing the opposite active site, nearly 20 Å away, for catalysis. However, this repositioning is transient, given that dimers and monomers are in constant exchange. Upon covalent modification of the active site serine by a phosphonate inhibitor, the monomer–dimer equilibrium is dramatically shifted toward dimer, demonstrating a clear structural link between the oxyanion hole and the dimer interface.
To analyze the contribution of binding determinants to enzyme stabilization, an inhibitor was synthesized containing an optimized peptide sequence for KSHV Pr. A PS-SCL was used to assess protease specificity, providing an exhaustive search of potential substrates. The PS-SCL substrates provide a completely diverse profiling of amino acid preference at each position, P4–P1, in the context of a tetrapeptide substrate. KSHV Pr proved to be a highly specific protease with distinct specificity, even at the P4 position, where a strict aromatic preference was observed, despite the lack of an obvious binding pocket upon examination of the crystal structure.
In an effort to separate the importance of oxyanion formation from substrate binding, the effects of inhibition by DFP on quaternary structure were examined. Upon inhibition with DFP, the protease exhibited the same structural stability as seen with diphenylphosphonates. Because DFP is devoid of any structural binding determinants, it appears that generation of the oxyanion alone is sufficient for stabilization. Although not directly transferable to substrate hydrolysis, this result strongly suggests that generation of the oxyanion hole, as opposed to the substrate-binding pockets, are the predominant factors in protease activation and stabilization.
Similarly, it is unlikely that the inhibitors are capable of inducing dimerization by binding to and reacting with the monomer. This is reflected by the difficulty in attaining complete inhibition of the protease with DFP. This situation indicates that DFP completely hydrolyzes before reacting with the protease, presumably as a result of the low levels of dimer present in the low concentration of uninhibited protease remaining. If the inhibitor were capable of inducing dimerization by reacting with the monomer, the difficulty in obtaining complete inhibition would not exist. This model is confirmed by the inhibition of the remaining protease upon addition of a diphenylphosphonate, which is significantly less susceptible to hydrolysis. Additionally, no active-site labeling or dimer stabilization is observed upon incubation of the diphenylphosphonate with a catalytically inactive protease variant with the active site serine removed or upon incubation with a monomeric variant (M197D) containing a complete catalytic triad (data not shown). These findings suggest that substrate binding of monomers in vivo does not induce the active form of the enzyme and that catalysis only occurs upon binding to the active dimeric form of the protease.
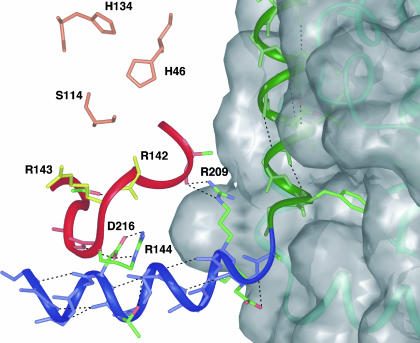
We suggest the mechanism for stabilization involves four structural elements: the active site serine, the loop supporting the oxyanion hole, helix 6, and helix 5 (Fig. 6). Phosphonylation of the active site serine by the inhibitor yields a tetrahedral phosphonate adduct, placing a strong negative charge in the oxyanion hole. Based on previous mutagenesis data and crystal structures with inhibitor bound, it is anticipated that this conformation is stabilized by the formation of hydrogen bonds to Arg-143, through a water molecule, and the backbone amide of Arg-142, located on the oxyanion hole loop (19, 26, 39, 40). This interaction provides additional stabilization of the oxyanion hole loop not present in the native dimer. The oxyanion hole loop, in turn, establishes hydrogen bonds and electrostatic interactions with residues in helix 6, including an interaction from Arg-144 to Asp-216 and two H-bonds from Arg-209 to the carbonyl of Ala-139 (18). Helix 6, stabilized by H-bonds to the oxyanion hole loop and by intrahelix H-bonds, is directly connected to helix 5, the main dimer interface contact, which is stabilized by intrahelix H-bonds, as well as significant intermolecular contacts to the adjacent monomer. These results provide a potential pathway for protease activation and stabilization and are consistent with crystallographic data on cytomegalovirus protease mutants (26).
Fig. 6.
Structural model for protease stabilization upon inhibition. Catalytic Ser-114, His-46, and His-134 are colored orange; oxyanion residues Arg-143 and Arg-142 are colored yellow (density for Arg-142 unsolved in crystal structure); and critical sidechains are colored by atom (oxygen, red; nitrogen, blue; carbon, green). All others are represented only as a backbone chain for clarity.
Assuming that all herpesvirus proteases undergo a similar mechanism of activation, conservation of the proposed structural connection between the active site and dimer interface was examined. Among α, β, and γ HHV proteases, the oxyanion hole loop is nearly identical, with 8 of 9 residues functionally conserved. The α herpesviruses appear to have diverged from the β and γ herpesviruses in helix 6, resulting in only 3 of 14 functionally conserved residues among the entire family. Despite the structural diversity in most of helix 6, Arg-209 is strictly conserved among all herpesviruses. Perhaps the most striking difference is the absence of Asp-216 in the α herpesviruses, one of the main intersubunit contacts seen in KSHV Pr. Interestingly, the α herpesvirus proteases contain an Arg at this position, and the corresponding Arg (144) in the oxyanion hole loop seen in KSHV Pr, is a Val in the α-herpesviruses, eliminating a potentially unfavorable charge–charge interaction. Examination of the crystal structure of herpes simplex virus 2 confirms that the Arg establishes stabilizing intersubunit H-bonds with the oxyanion hole loop. The high conservation of the oxyanion hole loop and the rational differences seen in helix 6 support the hypothesis that the extended network of intersubunit H-bonds and electrostatic interactions established upon formation of the transition state may contribute significantly to the exceptional stabilization of the dimer upon inhibition.
Given the intimate association of the active site to the dimer interface, we sought to address the possibility of communication between active sites during catalysis. A number of dimeric proteins with spatially separate active sites have been reported in which binding of substrate induces asymmetry in the dimer, preventing substrate binding at the second active site (41, 42). Inhibitor titration results demonstrate that the active sites of KSHV Pr support catalysis independently and do not exhibit half-of-sites reactivity. This finding has significant implications for therapeutically targeting the protease with transition-state analog inhibitors.
Although many of the details regarding the fate of the protease during capsid maturation remain unclear, a singly inhibited stabilized protease may alter many steps in the process. Substoichiometric amounts of inhibitor stabilize the dimer and result in singly inhibited active protease that could affect capsid processing, DNA encapsidation, and, ultimately, viral replication. The development of a selective peptide based transition state inhibitor provides the opportunity to determine the efficacy at inhibiting viral replication.
Considering the reduced catalytic activity and the shallow active site of herpesvirus proteases and the difficulty in achieving high inhibitor concentrations inside the capsid, alternative mechanisms of protease inhibition may be more successful. The direct link between transition-state stabilization and dimerization has been established, and this link highlights the possibility of inhibiting activity by preventing association of monomers. By using the assays developed in this report, it is possible that the dimer interface, with increased surface area and deep grooves, will provide a target more amenable to inhibitor development.
Supplementary Material
Acknowledgments
We thank Sami Mahrus, Dr. Jeohoong Sun, and Dr. Christopher Eggers for helpful discussions and critical review of the manuscript and Prof. Jon Ellman for generously supplying the ACC resin used to synthesize the protease substrate. This work was supported by Research Training in Chemistry and Chemical Biology National Institutes of Health Training Grant GM64337-01 (to A.B.M) and National Institutes of Health Grant GM56531 (to P.O.M. and C.S.C.).
Abbreviations: HHV, human herpesvirus; KSHV Pr, Kaposi's sarcoma-associated herpesvirus protease; PS-SCL, positional scanning synthetic combinatorial libraries; DFP, diisopropyl fluorophosphate; ACC, 7-amino-4-carbamoylmethylcoumarin; H-Alap-(OPh)2, diphenyl [α-aminoethyl] phosphonate; tBug, tert-butylglycine.
References
- 1.Goedert, J. J. (2000) Semin. Oncol. 27, 390-401. [PubMed] [Google Scholar]
- 2.Gottlieb, G. J., Ragaz, A., Vogel, J. V., Friedman-Kien, A., Rywlin, A. M., Weiner, E. A. & Ackerman, A. B. (1981) Am. J. Dermatopathol. 3, 111-114. [DOI] [PubMed] [Google Scholar]
- 3.Hymes, K. B., Cheung, T., Greene, J. B., Prose, N. S., Marcus, A., Ballard, H., William, D. C. & Laubenstein, L. J. (1981) Lancet 2, 598-600. [DOI] [PubMed] [Google Scholar]
- 4.Parisi, S. G., Mazzi, R., Sarmati, L., Carolo, G., Uccella, I., Rianda, A., Nicastri, E., Concia, E. & Andreoni, M. (2002) AIDS 16, 1089-1091. [DOI] [PubMed] [Google Scholar]
- 5.Tam, H. K., Zhang, Z. F., Jacobson, L. P., Margolick, J. B., Chmiel, J. S., Rinaldo, C. & Detels, R. (2002) Int. J. Cancer 98, 916-922. [DOI] [PubMed] [Google Scholar]
- 6.Dedicoat, M. & Newton, R. (2003) Br. J. Cancer 88, 1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Welch, A. R., Woods, A. S., McNally, L. M., Cotter, R. J. & Gibson, W. (1991) Proc. Natl. Acad. Sci. USA 88, 10792-10796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weinheimer, S. P., McCann, P. J., III, O'Boyle, D. R., II, Stevens, J. T., Boyd, B. A., Drier, D. A., Yamanaka, G. A., DiIanni, C. L., Deckman, I. C. & Cordingley, M. G. (1993) J. Virol. 67, 5813-5822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sheaffer, A. K., Newcomb, W. W., Brown, J. C., Gao, M., Weller, S. K. & Tenney, D. J. (2000) J. Virol. 74, 6838-6848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Preston, V. G., Coates, J. A. & Rixon, F. J. (1983) J. Virol. 45, 1056-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao, M., Matusick-Kumar, L., Hurlburt, W., DiTusa, S. F., Newcomb, W. W., Brown, J. C., McCann, P. J., III, Deckman, I. & Colonno, R. J. (1994) J. Virol. 68, 3702-3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Waxman, L. & Darke, P. L. (2000) Antiviral Chem. Chemother. 11, 1-22. [DOI] [PubMed] [Google Scholar]
- 13.Chen, P., Tsuge, H., Almassy, R. J., Gribskov, C. L., Katoh, S., Vanderpool, D. L., Margosiak, S. A., Pinko, C., Matthews, D. A. & Kan, C. C. (1996) Cell 86, 835-843. [DOI] [PubMed] [Google Scholar]
- 14.Tong, L., Qian, C., Massariol, M. J., Bonneau, P. R., Cordingley, M. G. & Lagacé, L. (1996) Nature 383, 272-275. [DOI] [PubMed] [Google Scholar]
- 15.Shieh, H. S., Kurumbail, R. G., Stevens, A. M., Stegeman, R. A., Sturman, E. J., Pak, J. Y., Wittwer, A. J., Palmier, M. O., Wiegand, R. C., Holwerda, B. C. & Stallings, W. C. (1996) Nature 383, 279-282. [DOI] [PubMed] [Google Scholar]
- 16.Qiu, X., Culp, J. S., DiLella, A. G., Hellmig, B., Hoog, S. S., Janson, C. A., Smith, W. W. & Abdel-Meguid, S. S. (1996) Nature 383, 275-279. [DOI] [PubMed] [Google Scholar]
- 17.Qiu, X., Janson, C. A., Culp, J. S., Richardson, S. B., Debouck, C., Smith, W. W. & Abdel-Meguid, S. S. (1997) Proc. Natl. Acad. Sci. USA 94, 2874-2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reiling, K. K., Pray, T. R., Craik, C. S. & Stroud, R. M. (2000) Biochemistry 39, 12796-12803. [DOI] [PubMed] [Google Scholar]
- 19.Hoog, S. S., Smith, W. W., Qiu, X., Janson, C. A., Hellmig, B., McQueney, M. S., O'Donnell, K., O'Shannessy, D., DiLella, A. G., Debouck, C. & Abdel-Meguid, S. S. (1997) Biochemistry 36, 14023-14029. [DOI] [PubMed] [Google Scholar]
- 20.Buisson, M., Hernandez, J. F., Lascoux, D., Schoehn, G., Forest, E., Arlaud, G., Seigneurin, J. M., Ruigrok, R. W. & Burmeister, W. P. (2002) J. Mol. Biol. 324, 89-103. [DOI] [PubMed] [Google Scholar]
- 21.Register, R. B. & Shafer, J. A. (1997) J. Virol. 71, 8572-8581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khayat, R., Batra, R., Massariol, M. J., Lagacé, L. & Tong, L. (2001) Biochemistry 40, 6344-6351. [DOI] [PubMed] [Google Scholar]
- 23.Buisson, M., Valette, E., Hernandez, J. F., Baudin, F., Ebel, C., Morand, P., Seigneurin, J. M., Arlaud, G. J. & Ruigrok, R. W. (2001) J. Mol. Biol. 311, 217-228. [DOI] [PubMed] [Google Scholar]
- 24.Pray, T. R., Nomura, A. M., Pennington, M. W. & Craik, C. S. (1999) J. Mol. Biol. 289, 197-203. [DOI] [PubMed] [Google Scholar]
- 25.Cole, J. L. (1996) Biochemistry 35, 15601-15610. [DOI] [PubMed] [Google Scholar]
- 26.Batra, R. K., Khayat, R. & Tong, L. (2001) Nat. Struct. Biol. 8, 810-817. [DOI] [PubMed] [Google Scholar]
- 27.Darke, P. L., Cole, J. L., Waxman, L., Hall, D. L., Sardana, M. K. & Kuo, L. C. (1996) J. Biol. Chem. 271, 7445-7449. [DOI] [PubMed] [Google Scholar]
- 28.Margosiak, S. A., Vanderpool, D. L., Sisson, W., Pinko, C. & Kan, C. C. (1996) Biochemistry 35, 5300-5307. [DOI] [PubMed] [Google Scholar]
- 29.Schmidt, U. & Darke, P. L. (1997) J. Biol. Chem. 272, 7732-7735. [DOI] [PubMed] [Google Scholar]
- 30.Unal, A., Pray, T. R., Lagunoff, M., Pennington, M. W., Ganem, D. & Craik, C. S. (1997) J. Virol. 71, 7030-7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harris, J. L., Backes, B. J., Leonetti, F., Mahrus, S., Ellman, J. A. & Craik, C. S. (2000) Proc. Natl. Acad. Sci. USA. 97, 7754-7759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oleksyszyn, J., Subotkowska, L. & Mastalerz, P. (1979) Synthesis 12, 985-986. [Google Scholar]
- 33.Thornberry, N. A., Peterson, E. P., Zhao, J. J., Howard, A. D., Griffin, P. R. & Chapman, K. T. (1994) Biochemistry 33, 3934-3940. [DOI] [PubMed] [Google Scholar]
- 34.Kitz, R. & Wilson, I. B. (1962) J. Biol. Chem. 237, 3245-3249. [PubMed] [Google Scholar]
- 35.Ogilvie, W., Bailey, M., Poupart, M. A., Abraham, A., Bhavsar, A., Bonneau, P., Bordeleau, J., Bousquet, Y., Chabot, C., Duceppe, J. S., et al. (1997) J. Med. Chem. 40, 4113-4135. [DOI] [PubMed] [Google Scholar]
- 36.Shimba, N., Nomura, A. M., Marnett, A. B. & Craik, C. S. (2004) J. Virol., in press. [DOI] [PMC free article] [PubMed]
- 37.Sampson, N. S. & Bartlett, P. A. (1991) Biochemistry 30, 2255-2263. [DOI] [PubMed] [Google Scholar]
- 38.Oleksyszyn, J. & Powers, J. C. (1991) Biochemistry 30, 485-493. [DOI] [PubMed] [Google Scholar]
- 39.Liang, P. H., Brun, K. A., Feild, J. A., O'Donnell, K., Doyle, M. L., Green, S. M., Baker, A. E., Blackburn, M. N. & Abdel-Meguid, S. S. (1998) Biochemistry 37, 5923-5929. [DOI] [PubMed] [Google Scholar]
- 40.Khayat, R., Batra, R., Qian, C., Halmos, T., Bailey, M. & Tong, L. (2003) Biochemistry 42, 885-891. [DOI] [PubMed] [Google Scholar]
- 41.Biemann, H. P. & Koshland, D. E., Jr. (1994) Biochemistry 33, 629-634. [DOI] [PubMed] [Google Scholar]
- 42.Xiao, B., Singh, S. P., Nanduri, B., Awasthi, Y. C., Zimniak, P. & Ji, X. (1999) Biochemistry 38, 11887-11894. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.