Abstract
Proteus mirabilis is a Gram-negative enteric bacterium that causes complicated urinary tract infections, particularly in patients with indwelling catheters. Sequencing of clinical isolate P. mirabilis HI4320 revealed the presence of 17 predicted chaperone-usher fimbrial operons. We classified these fimbriae into three groups by their genetic relationship to other chaperone-usher fimbriae. Sixteen of these fimbriae are encoded by all seven currently sequenced P. mirabilis genomes. The predicted protein sequence of the major structural subunit for 14 of these fimbriae was highly conserved (≥95 % identity), whereas three other structural subunits (Fim3A, UcaA and Fim6A) were variable. Further examination of 58 clinical isolates showed that 14 of the 17 predicted major structural subunit genes of the fimbriae were present in most strains (>85 %). Transcription of the predicted major structural subunit genes for all 17 fimbriae was measured under different culture conditions designed to mimic conditions in the urinary tract. The majority of the fimbrial genes were induced during stationary phase, static culture or colony growth when compared to exponential-phase aerated culture. Major structural subunit proteins for six of these fimbriae were detected using MS of proteins sheared from the surface of broth-cultured P. mirabilis, demonstrating that this organism may produce multiple fimbriae within a single culture. The high degree of conservation of P. mirabilis fimbriae stands in contrast to uropathogenic Escherichia coli and Salmonella enterica, which exhibit greater variability in their fimbrial repertoires. These findings suggest there may be evolutionary pressure for P. mirabilis to maintain a large fimbrial arsenal.
Introduction
Urinary tract infections (UTIs) are the most common bacterial infections in humans (Foxman, 2003). In 2006, UTIs were the cause of over eleven million medical consultations, as well as nearly half a million hospitalizations in the US alone (Litwin & Saigal, 2007; DeFrances et al., 2008). Worldwide, UTIs are also the most prevalent type of healthcare-associated infection (Tambyah, 2004). Most of these infections are associated with the use of urinary catheters (catheter-associated UTI) (Tambyah & Maki, 2000). In the US, an estimated 13 088 fatal healthcare-associated infections were due to UTIs in 2002 (Klevens et al., 2007).
Proteus mirabilis is a bacterial pathogen that is associated with complicated UTI, particularly during long-term urinary catheterization [reviewed by Nielubowicz & Mobley (2010) and Armbruster & Mobley (2012)]. P. mirabilis infection can result in cystitis and often progresses to pyelonephritis. This member of the Enterobacteriaceae produces many virulence factors (Nielubowicz & Mobley, 2010), including urease, which hydrolyses urea in the urine, thereby increasing local pH. The alkaline environment causes the formation of struvite and apatite crystals, which contribute to stone formation (Griffith et al., 1976; Mobley et al., 1995).
A critical component of P. mirabilis pathogenesis centres on fimbriae (also known as pili) and other adhesins, which mediate adherence to cells of the urinary tract, as well as catheters (Wray et al., 1986; Roberts et al., 1990; Jansen et al., 2004; Alamuri et al., 2010). Notably, in a survey of Gram-negative bacteria, this species was found to have the greatest ability to adhere to catheters (Roberts et al., 1990). Before sequencing and annotation of the P. mirabilis clinical isolate HI4320 genome (Pearson et al., 2008), five P. mirabilis-encoded fimbriae had been identified: mannose-resistant Proteus-like (MR/P), P. mirabilis fimbria (PMF), ambient temperature fimbria (ATF), urothelial cell adhesin (UCA) (or non-agglutinating fimbria, NAF) and P. mirabilis P-like pili (PMP) (Old & Adegbola, 1982; Wray et al., 1986; Bahrani et al., 1994; Massad et al., 1994a, b; Bijlsma et al., 1995). A sixth fimbria, MR/K, has been proposed based on the ability of P. mirabilis to agglutinate specific erythrocytes, but the genes responsible for this phenotype have not yet been identified (Old & Adegbola, 1982). Electron microscopy and immunoblot analyses have demonstrated that P. mirabilis can produce two fimbriae concurrently (Old & Adegbola, 1982; Adegbola et al., 1983; Bahrani & Mobley, 1993; Tolson et al., 1995; Zunino et al., 2007). However, those experiments were not designed to detect and identify a wider variety of fimbriae. Sequencing of the HI4320 genome led to the discovery of 17 potential chaperone-usher fimbrial operons, an additional 12 operons compared with what was known previously. Little is known about these predicted fimbriae, including whether or not they are functional adhesins.
Studies of fimbriae in other Gram-negative bacteria have shown that most fimbriae are generally not conserved within a given species. For example, in Escherichia coli, only 3 of 15 different fimbriae were detected in greater than 40 % of commensal isolates tested. Although more types of fimbriae are typically found in pathogenic E. coli isolates, only 5 of 15 fimbriae were present in greater than 60 % of urinary isolates (Spurbeck et al., 2011). Similarly, for Salmonella enterica, a species that infects a broad host range, there is considerable variation in the presence of fimbrial genes within different subspecies (Townsend et al., 2001). However, there is greater conservation of fimbrial genes within the human- and higher primate-specific S. enterica serovar Typhi. Other studies have suggested that four fimbriae of P. mirabilis (ATF, PMF, MR/P and UCA) are conserved in limited numbers of clinical or environmental isolates (Bahrani & Mobley, 1994; Gaastra et al., 1996; Massad et al., 1996; Zunino et al., 2000, 2003).
Since fimbriae often contribute to virulence, we sought to determine whether fimbriae encoded by P. mirabilis were broadly conserved or whether particular types were associated with specific isolation sites. Furthermore, we explored whether all 17 fimbrial operons are transcribed, and if so, how they are regulated. We have therefore analysed all 17 chaperone-usher fimbriae of P. mirabilis using DNA-, RNA- and protein-based approaches.
Methods
Bacterial strains and media.
A collection of clinical P. mirabilis strains was obtained during the early 1980s from two nursing homes in Baltimore, MD, USA, from patients with chronic indwelling catheters. Among these isolates was strain HI4320, which was isolated from the urine of a long-term (≥30 days) catheterized elderly woman with bacteriuria (Warren et al., 1982). NYU strains were collected from Tisch Hospital at New York University Langone Medical Center (NYULMC), NY, USA, from July 2012 to October 2012. The collection consists of tribe Proteeae (genera Proteus, Providencia and Morganella) strains identified by the Clinical Microbiology Laboratory during this time. All strains were grown in Luria Broth (LB; 10 g tryptone l−1, 5 g yeast extract l−1, 0.5 g NaCl l−1) or on LB solidified with 15 g agar l−1. Bacteria were routinely cultured at 37 °C.
To measure expression of fimbrial genes, P. mirabilis was cultured under the following conditions: exponential phase at 37 °C with aeration to an OD600 of 0.8. For experiments testing the effect of pH on fimbrial gene expression, MOPS was added to LB to a final concentration of 100 mM and the pH adjusted to 6.0 or 8.0. This buffering capacity was sufficient to maintain cultures near the target pH: at the time of collection, the pH 6.0 cultures were pH 6.10–6.13, while the pH 8.0 cultures were pH 7.86–7.89. Stationary-phase aerated cultures were harvested after 8 h (OD600 = 3.86 to 4.50) and stationary-phase static cultures were harvested after 24 h. Bacterial colonies were collected from the surface of LB agar plates using a sterile loop after incubation for 16 h. Once the appropriate time point or OD600 was reached, 500 µl (one volume) of cells was added to 1 ml (two volumes) of RNAprotect (Qiagen); colonies were added directly to RNAprotect. After 10 min, the bacteria were centrifuged for 10 min at 6000 g and the bacterial pellet was stored at −20 °C prior to RNA extraction.
Comparison of sequenced genomes.
blast (Altschul et al., 1990) was used to detect predicted fimbrial genes in six recently sequenced P. mirabilis genomes. These genomes were sequenced by several independent groups and are available in GenBank at the following accession numbers: ATCC 29906 (ACLE01000000), WGLW4 (AMGU01000000), WGLW6 (AMGT0100000), BB2000 (NC_010555.1), C05028 (ANBT01000000) and PR03 (AORN01000000). Of these, BB2000 and PR03 are published (Khalid et al., 2013; Sullivan et al., 2013). The Kyoto Encyclopedia of Genes and Genomes (KEGG) (Tanabe & Kanehisa, 2012) and xBASE (Chaudhuri et al., 2008) were used to analyse operon organization for HI4320, ATCC 29906 and BB2000, and genomic location of fimbrial operons for HI4320 and BB2000.
Detection of fimbrial genes.
Genomic DNA was extracted from the strains from the 1980s isolates using the Generation capture column kit (Qiagen) and PCR was performed using primers targeting the predicted major structural subunits for all 17 fimbriae. Colony PCR was performed to detect genes in NYU clinical isolates. In both cases, negative or inconclusive results were tested at least twice, and if results remained inconclusive, genomic DNA was extracted and PCR was performed with genomic DNA as the template. All strains with negative results were rescreened using a second set of primers targeting the same genes. Primers used are listed in Table S1 (available in the online Supplementary Material).
Relatedness of isolates.
Primers that uniquely target the mrpA and flaA (PMI1620) genes were used to PCR amplify these genes from five NYU clinical isolates. A total of 82 and 83 % of each gene was amplified, respectively. Each PCR product was sequenced (Genewiz). The resulting sequences were aligned by the clustal w method and plotted on a phylogenetic tree using megalign software (dnastar v. 10.1.1).
RNA extraction, reverse transcriptase PCR and quantitative reverse transcriptase PCR (qRT-PCR).
The RNeasy minikit (Qiagen) was used to extract RNA with the following modification: bacteria were incubated with 3 mg lysozyme ml−1 instead of 1 mg lysozyme ml−1, and placed on a vortex at a low setting at room temperature for 15 min. RNA was then treated with DNase (Ambion) according to the manufacturer’s instructions.
The Superscript First-Strand synthesis system (Invitrogen) was used to generate cDNA from RNA according to the manufacturer’s instructions. PCR using primers targeting rpoA (RNA polymerase A) with cDNA templates with or without reverse transcriptase treatment was used to confirm lack of genomic contamination. cDNA was then purified and concentrated using a plasmid miniprep kit (Zymo) with the following modifications: PB buffer (Qiagen) was used to bind cDNA to the columns, and steps 9–11 from the Zymo plasmid miniprep kit were performed according to the manufacturer’s instructions.
qRT-PCR was performed to test the expression of the 17 fimbriae in duplicate 25 µl reactions using 30 ng cDNA template, 150 nM each primer (Table S1) and 12.5 µl Maxima SYBR Green/fluorescein qPCR master mix (Thermo Scientific). Amplification of specific genes was performed over 40 cycles with a CFX Connect thermal cycler (Bio-Rad). No-template controls and melting curve analysis were used to confirm the absence of genomic DNA and determine the presence of primer dimers. Data were analysed using the threshold cycle (2−ΔΔCT) method (Livak & Schmittgen, 2001) and normalized to rpoA. At least three independent experiments were analysed for each condition; for stationary-aerated culture and stationary-static culture, four or five experiments were conducted, respectively. Statistical analysis was performed using a two-tailed t-test with GraphPad Prism software, v. 5, on log2 transformed data.
Mass spectrometry.
P. mirabilis HI4320 cultured overnight at 37 °C with aeration was diluted 1 : 100 into LB and grown at 37 °C with aeration for 8 h. Extracellular proteins were sheared from the cell surface by vigorously shaking the culture for 5 min. Cells were pelleted by centrifugation; supernatants were then sterilized with a 0.22 µM pore filter (Millipore) and concentrated ~100-fold using a centrifugal filtration unit with a 10 kDa filter (Millipore). To dissociate assembled fimbriae, samples were mixed 1 : 1 with HCl-acidified water (pH 1.8), boiled for 15 min and neutralized using NaOH.
Sheared proteins were sent to the NYULMC Proteomic Core for MS. Proteins were reduced with 10 mM DTT and alkylated with 25 mM iodoacetamide and loaded into a 4–12 % Bis-Tris gel (Life Technologies). The gel was run for 1.5 min at 200 V, fixed with 16 % methanol in 1 % acetic acid and stained with GelCode blue stain reagent (Thermo Scientific). The portion of the gel that contained all proteins was excised, destained and trypsin digested (200 ng) overnight. Then, peptides were extracted using R2 20 µm Poros beads (Life Technologies) as described elsewhere (Cotto-Rios et al. 2012). Approximately 7 µg of the peptide mixture was loaded onto a Acclaim PepMap 100 precolumn (75 µm×2 cm, C18, 3 µm, 100 Å; Thermo Scientific) that was connected to an EASY-Spray, PepMap RSLC column (75 µm×25 cm, C18, 2 µm, 100 Å; Thermo Scientific) with a 5 µm emitter using the autosampler of an EASY-nLC 1000 (Thermo Scientific). Peptides were gradient eluted from the column directly into a Q Exactive mass spectrometer (Thermo Scientific) using a 120 min gradient from 2 % solvent B to 40 % solvent B. Solvent A was 2 % acetonitrile in 0.5 % acetic acid and solvent B was 90 % acetonitrile in 0.5 % acetic acid. High-resolution MS1 spectra were acquired with a resolution of 70 000, an AGC target of 1e6, with a maximum ion time of 120 ms and scan range of 400 to 1500 m/z. Following each MS1, 20 data-dependent high-resolution HCD MS2 spectra were acquired. All MS2 spectra were collected using the following instrument parameters: resolution of 17 500, AGC target of 5e4, maximum ion time of 250 ms, one microscan, 2 m/z isolation window, fixed first mass of 150 m/z, 30 s exclusion list and NCE of 27. The MS/MS spectra were searched against a Uniprot database constructed from P. mirabilis HI4320 and P. mirabilis ATCC 29906 using Sequest within Proteome Discoverer. Only proteins with two or more high-quality peptide matches were included in the analysis.
Results
Greek classification
Chaperone-usher fimbriae are protein structures located on the surface of Gram-negative bacteria, and are named after the periplasmic chaperone and outer membrane usher proteins that are required for folding and assembly of the fimbria. These fimbriae have been classified into nine groups (α, β, γ1, γ2, γ3, γ4, κ, π and σ) by evolutionary descent using chaperone sequence, gene order within fimbrial operons and conserved protein domains in the major structural subunits (Nuccio & Bäumler, 2007). Using this method, we provide here the Greek classification for the 17 fimbriae encoded by P. mirabilis (Tables 1 and S2). All except Fim14 can be classified into three clades (γ1,, γ2, π). These three clades all have chaperones of the FGS subfamily (short F1-G1 loop; Hung et al., 1996), and major fimbrial subunits with the conserved domains PFAM00419 and COG3539. The majority of the fimbriae encoded by P. mirabilis belong to either the γ1 or the π clade (Table 1). The γ1 clade operon structure consists at minimum of a major subunit, followed by a chaperone, an usher and a tip adhesin. Eight of the fimbriae encoded by P. mirabilis belong to the γ1 clade (Table 1). The prototypical member of this clade is the type I fimbria of E. coli, which is associated with UTI [reviewed by Hannan et al. (2012)]. Seven P. mirabilis fimbriae belong to the π clade (Table 1). The core genetic organization for operons of this clade consists of a major fimbrial subunit and an usher, followed by a chaperone. The prototypical member of this clade is the uropathogenic E. coli P fimbria, which contributes to pyelonephritis [reviewed by Lane & Mobley (2007)]. Fim12 is a member of the γ2 clade, which is identified by the presence of three chaperones encoded within the operon. The fim14 operon does not encode a chaperone and therefore cannot be categorized using the Greek classification system.
Table 1. Greek classification of P. mirabilis fimbriae.
| Class | Fimbriae* | Operon organization† |
| γ1 | UCA, Fim6, Fim7, Fim8, Fim10, ATF, Fim16, Fim17 | M, C, U |
| γ2 | Fim12 | M, C, C, C, U |
| π | MR/P', MR/P, Fim3, Fim5, PMF, PMP, Fim15 | M, U, P |
Fimbria 14 not determined – lacks chaperone.
M, Major subunit; C, chaperone; U, usher; minor subunits and tip adhesins are not shown.
Presence of the 17 fimbriae in sequenced P. mirabilis strains
Following the original sequencing of the P. mirabilis HI4320 genome (Pearson et al., 2008), the genomes of six new P. mirabilis strains [ATCC 29906, BB2000 (Sullivan et al., 2013), WGLW4, WGLW6, C05028 and PR03 (Khalid et al., 2013)] have been sequenced and deposited in GenBank. These strains were obtained from diverse locations: ATCC 29906 is the P. mirabilis type strain; BB2000 is a laboratory strain (Belas et al., 1991) that has been the basis of several swarming-motility and strain-identity studies (e.g. Belas et al., 1991, 1998; Gibbs et al., 2008); WGLW4 was isolated from the urine of a human subject; WGLW6 was isolated from the stool of a mouse; C05028 was isolated from the stool of a patient with diarrhoea during an outbreak in Shenzhen, China; and PR03 was found in the blood of a patient with septicaemia in Putrajaya, Malaysia. blast was performed to determine the percentage identity of the predicted amino acid sequence of the major structural subunit for all 17 fimbriae in comparison to HI4320 (Tables 2 and S2). Interestingly, 13 of the 17 fimbriae had ≥99 % identity with HI4320; an additional fimbria was ≥95 % identical. The remaining three fimbriae (Fim3A, UcaA and Fim6A) had greater diversity in amino acid identity. The only fimbrial major subunit that was not found in all sequenced strains was Fim3A, which was absent from BB2000 and C05028.
Table 2. Amino acid identity of predicted fimbrial major structural subunits compared to HI4320.
| Strain | MrpA' | MrpA | Fim3A | UcaA | Fim5A | Fim6A | Fim7A | Fim8A | PmfA | Fim10A | PmpA | Fim12A | AtfA | Fim14A | Fim15A | Fim16A | Fim17A |
| ATCC 29906 | 100 % | 100 % | 75 % | 82 % | 100 % | 99 % | 100 % | 100 % | 100 % | 100 % | 100 % | 100 % | 100 % | 100 % | 99 % | 100 % | 100 % |
| BB2000 | 100 % | 100 % | – | 96 % | 100 % | 51 % | 95 % | 100 % | 100 % | 100 % | 100 % | 100 % | 100 % | 100 % | 100 % | 99 % | 100 % |
| WGLW4 | 100 % | 100 % | 100 % | 41 % | 99 % | 98 % | 97 % | 100 % | 99 % | 100 % | 100 % | 100 % | 100 % | 99 % | 99 % | 100 % | 100 % |
| WGLW6 | 100 % | 100 % | 100 % | 95 % | 100 % | 95 % | 99 % | 100 % | 99 % | 100 % | 100 % | 99 % | 99 % | 100 % | 99 % | 100 % | 100 % |
| C05028 | 100 % | 100 % | – | 82 % | 99 % | 100 % | 100 % | 100 % | 100 % | 100 % | 100 % | 100 % | 100 % | 100 % | 100 % | 100 % | 100 % |
| PR03 | 100 % | 100 % | 76 % | 82 % | 100 % | 90 % | 100 % | 100 % | 100 % | 100 % | 100 % | 100 % | 100 % | 100 % | 99 % | 99 % | 100 % |
Three of these sequenced genomes (HI4320, ATCC 29906 and BB2000) have been annotated, allowing us to examine the organization of the fimbrial operons in these strains. Further genomic comparison showed that all 17 fimbrial operons initially identified in HI4320, including 11 fimbria-associated mrpJ-type transcriptional regulators (Pearson & Mobley, 2008), were also present in ATCC 29906. In BB2000, 16 of the operons were present; the fim3 operon was missing except for an orphaned putative adhesin gene. The location of fimbrial operons in the bacterial chromosome was conserved between HI4320 and BB2000 with the exception of the uca operon, which in both strains was found adjacent to phage genes.
Detection of the 17 fimbriae in 1980s P. mirabilis isolates
Primers designed to recognize the predicted major structural subunit of each of the 17 fimbriae (the first gene in the operon which encodes a fimbrial structural gene; Table S2) were designed using the genomic sequence from HI4320. We examined 10 P. mirabilis isolates obtained from two nursing homes in Baltimore over 30 years ago (Warren et al., 1982). From Fig. 1 and Table 3, it can be seen that the presence of the 17 fimbriae is highly conserved in these strains. Three out of the ten clinical isolates tested positive for all of the fimbriae. The only genes that were not detected in all 10 strains were fim3A (detected in 3 of 10) and fim5A (6 of 10).
Fig. 1.
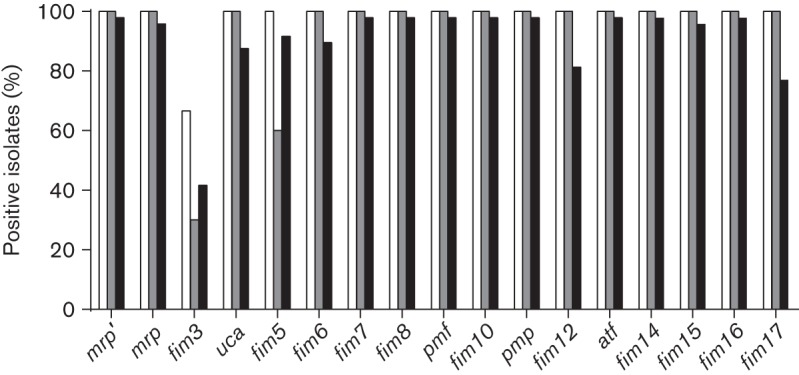
Conservation of fimbriae encoded by P. mirabilis. The percentage of isolates with each fimbria is shown. For the sequenced strains (white bars), fimbriae were identified using blast. For the 1980s isolates (grey bars) and hospital isolates (NYU) (black bars), fimbrial genes were detected by PCR.
Table 3. PCR detection of predicted fimbrial major subunit genes in 1980s isolates.
mrpA’, mrpA, ucaA, fim6A, fim7A, fim8A, pmfA, fim10A, pmpA, fim12A, atfA, fim14A, fim15A, fim16A and fim17A were detected in all 10 isolates.
| Strain | HI4320 | NI114 #800 | TA507 #1249 | MC118 #1296 | HA135 #2537 | RU107 #34 | MA112 #348 | HU119 #798 | HA139 #3871 | RO106 #23 | FE513 #4244 |
| fim3A | + | − | + | − | − | − | + | − | − | + | − |
| fim5A | + | − | + | − | + | − | + | − | + | + | + |
Conservation of the 17 fimbriae in the NYU collection
Clinical isolates of Proteus sp., Providencia sp. and Morganella sp. [collectively known as tribe Proteeae (O'Hara et al., 2000)] were collected from Tisch Hospital at NYULMC from 17 July 2012 to 23 October 2012. A total of 205 strains were collected during this time (166 P. mirabilis, 8 Proteus vulgaris, 3 Proteus penneri, 7 Providencia stuartii, 3 Providencia rettgeri and 18 Morganella morganii). Of the 166 P. mirabilis strains, 124 (75 %) were obtained from urine samples, 22 (13 %) from wounds or abscesses, 7 (4 %) from sputum, 5 (3 %) from incision or drainage sites, 2 from the genital tract (1 %) and 6 (4 %) from other sites.
We selected 48 P. mirabilis isolates for screening by PCR to determine the presence of the predicted major structural subunit genes for each of the 17 fimbriae. The isolates tested were the first 40 urine isolates plus an additional 8 isolates obtained from diverse anatomical sites (Table S3). Results from the NYU strains were consistent with the findings from the nursing home isolates collected in the 1980s (Fig. 1). The majority of the fimbriae were conserved; that is, 47 out of 48 clinical isolates tested positive for at least 13 of the 17 fimbriae (Fig. 2; Table S3). Sixteen isolates were positive for all 17 fimbriae. The fim3A gene was only detected in 20 of 48 (42 %) isolates. Fimbriae that were less conserved in the NYU P. mirabilis strains compared with the 1980s isolates were ucaA (42/48), fim6A (43/48), fim12A (39/48) and fim17A (37/48). All others were detected in at least 44 of 48 strains (92 %). In contrast to the 1980s collection, the fim5A gene was highly conserved (44/48) in the NYU collection. The presence of specific fimbrial genes did not correlate with the anatomical site of P. mirabilis isolation (Table S3). The mrpA gene, which is generally conserved and required to establish infection in a mouse model of UTI (Bahrani et al., 1994; Jansen et al., 2004), was not detected in two P. mirabilis clinical isolates (4 %). Also of note is isolate NYU032 in which 14 fimbrial genes were not detected. Although this isolate was identified as P. mirabilis by the clinical laboratory, we further tested this strain to confirm its identity. Primers for rpoA (specific for the genus Proteus) and flhD (specific to P. mirabilis based on currently available genome sequences) yielded PCR products with genomic DNA, and this strain swarmed on a hard agar surface (indicative of Proteus sp.; data not shown). We also screened one P. vulgaris blood isolate and one M. morganii urine isolate for the presence of the 17 fimbriae. The P. vulgaris isolate was positive for mrpA, ucaA and atfA genes, while none of the fimbrial genes were detected in the M. morganii strain.
Fig. 2.
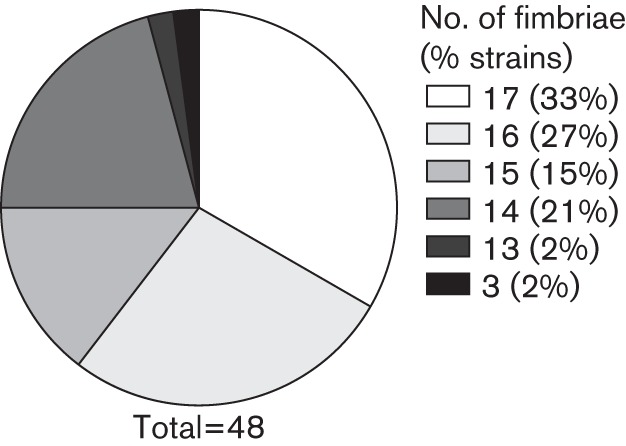
The majority of fimbriae are highly conserved in recent clinical isolates. The percentage of NYU P. mirabilis isolates encoding given numbers of the 17 fimbriae is depicted. A total of 48 isolates were screened.
In order to gauge the genetic diversity of the clinical isolates, we chose five representative strains for further analysis. Four of these strains (NYU009, NYU037, NYU052 and NYU066) were urine isolates collected in different weeks. The fifth isolate, NYU011, was collected the same week as NYU009, but was isolated from an abscess. We sequenced the MrpA- and FlaA (flagellin)-encoding genes for each of these strains and compared the sequences to the available P. mirabilis genomes. As expected, the mrpA sequence was 100 % identical in all strains examined, with the exception of a single synonymous mutation in NYU011. In contrast, while flaA was conserved at the 5′ and 3′ ends, there was a highly variable region corresponding to nucleotides 531–785 of HI4320 flaA. Based on alignment of the sequences, the relatedness of these flaA genes was plotted as a phylogenetic tree (Fig. 3). With the exception of NYU037 and NYU052 (which clustered with WGLW4 and HI4320), the flaA sequences from the NYU collection did not cluster together; rather, they were distributed among flaA from the sequenced P. mirabilis genomes.
Fig. 3.
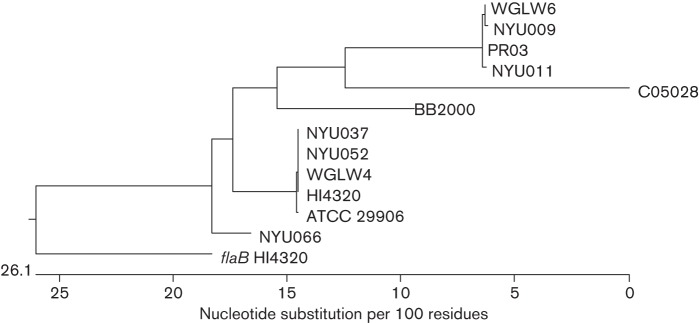
Phylogenetic tree of flaA sequences. The clustal w algorithm was used to align flaA from five NYU clinical isolates and the sequenced P. mirabilis genomes. The related flaB gene (PMI1619) from HI4320 was included as a reference, and clustered separately from the flaA sequences.
Expression of 17 fimbriae in HI4320
To determine whether all 17 fimbrial operons are transcribed, qRT-PCR was performed with cDNA derived from HI4320 cultured under different conditions (exponential-phase aerated, stationary aerated, stationary static, pH 6.0, pH 8.0 and colonies on an agar surface). We detected transcript for the major structural genes of all 17 fimbriae (Fig. 4). During exponential-phase aerated culture, ucaA was the most abundant transcript, followed by pmfA and fim8A. The least abundant transcripts were fim6A and fim10A (mean cycle threshold 30.2 and 31.8, respectively).
Fig. 4.
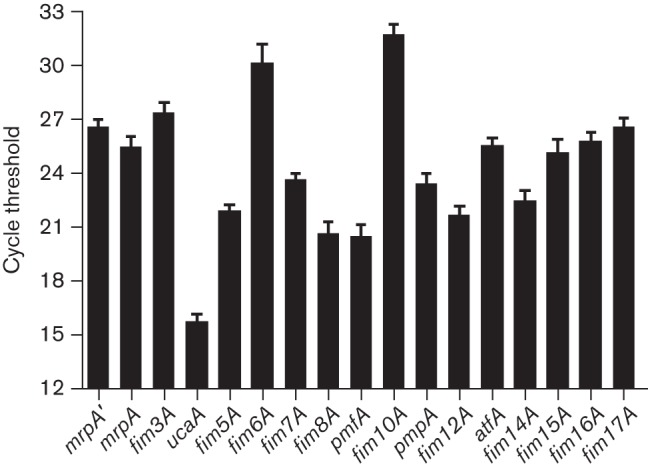
Expression of fimbrial genes during exponential-phase aerated culture. The cycle threshold of detection for each of the predicted major structural subunit genes is depicted; a lower value indicates more abundant transcript. The data are the result of eight experiments. The error bars represent sem.
To evaluate differential expression of fimbrial genes under specific culture conditions, gene expression was compared to that from exponential-phase aerated culture (Table 4). Experimental conditions were chosen to address an environment that P. mirabilis might encounter during infection. The bladder is a reduced-oxygen environment, which was mimicked here by culturing bacteria to stationary phase and/or statically (without shaking). In addition, stationary-phase static culture is a long-established method for increasing fimbriation in different bacterial species including P. mirabilis (Old & Duguid, 1970; Old & Adegbola, 1982; Bahrani & Mobley, 1994; Lane et al., 2009). Agar-grown colonies were used to assess bacterial growth on a surface, such as on the bladder epithelium or a urinary catheter. Urine in a healthy individual is normally slightly acidic (pH 5.5–6.5) (Simerville et al., 2005). However, P. mirabilis in urine rapidly alkalinizes its environment via urease activity, and so we tested the effect of pH on fimbrial gene expression.
Table 4. Fold change in expression of fimbrial genes during different culture conditions compared to exponential-phase aerated culture.
P is the probability result of a two-tailed t-test (values below 0.050 are shown in bold).
| Condition* | Value | mrpA' | mrpA | fim3A | ucaA | fim5A | fim6A | fim7A | fim8A | pmfA | fim10A | pmpA | fim12A | atfA | fim14A | fim15A | fim16A | fim17A |
| Stationary static | Mean | 205 | 8943 | 145 | 23.47 | 16.99 | 2992 | 7.53 | 4.28 | 37.41 | 21.06 | 3.646 | 73.44 | 201.3 | 5.66 | 203 | 120.7 | 428.9 |
| sd | 213.3 | 14946 | 170.7 | 17.62 | 13.25 | 5591 | 7.71 | 6.19 | 31.51 | 26.11 | 2.39 | 61.86 | 126.9 | 6.87 | 282.7 | 102.3 | 588.5 | |
| P | 0.099 | 0.252 | 0.132 | 0.046 | 0.054 | 0.298 | 0.131 | 0.302 | 0.061 | 0.161 | 0.069 | 0.059 | 0.024 | 0.204 | 0.185 | 0.059 | 0.179 | |
| Stationary aerated | Mean | 32.36 | 80.42 | 50.66 | 5.14 | 5.48 | 10.53 | 1.01 | 50.84 | 4.11 | 27.87 | 5.69 | 26.74 | 21.44 | 18.55 | 91.33 | 8.57 | 130.6 |
| sd | 14.29 | 39.01 | 31.50 | 2.65 | 3.86 | 6.09 | 0.603 | 34.85 | 2.56 | 17.86 | 3.29 | 16.27 | 11.98 | 14.99 | 65.02 | 3.52 | 48.34 | |
| P | 0.022 | 0.027 | 0.051 | 0.052 | 0.103 | 0.052 | 0.988 | 0.065 | 0.093 | 0.057 | 0.065 | 0.051 | 0.042 | 0.101 | 0.069 | 0.023 | 0.013 | |
| Plate | Mean | 17.35 | 46.92 | 15.21 | 7.21 | 4.58 | 12.28 | 2.29 | 111.1 | 0.876 | 4.56 | 131.9 | 19.28 | 9.40 | 19.15 | 177.6 | 5.10 | 69.31 |
| sd | 20.05 | 60.94 | 11.63 | 6.76 | 2.67 | 7.62 | 1.44 | 93.59 | 0.285 | 1.60 | 129.3 | 11.96 | 5.91 | 18.93 | 264.7 | 5.57 | 74.41 | |
| P | 0.293 | 0.322 | 0.169 | 0.253 | 0.146 | 0.124 | 0.261 | 0.179 | 0.528 | 0.061 | 0.222 | 0.118 | 0.133 | 0.239 | 0.367 | 0.331 | 0.253 | |
| pH 6 | Mean | 1.67 | 0.847 | 0.574 | 0.485 | 0.737 | 0.450 | 0.554 | 0.504 | 1.02 | 0.427 | 3.05 | 1.26 | 3.96 | 4.88 | 4.33 | 1.00 | 3.07 |
| sd | 1.22 | 0.596 | 0.417 | 0.256 | 0.499 | 0.300 | 0.306 | 0.354 | 1.17 | 0.215 | 1.93 | 0.854 | 2.67 | 2.62 | 3.57 | 0.654 | 2.03 | |
| P | 0.444 | 0.700 | 0.219 | 0.074 | 0.457 | 0.087 | 0.127 | 0.136 | 0.975 | 0.044 | 0.209 | 0.646 | 0.195 | 0.124 | 0.248 | 0.997 | 0.220 | |
| pH 8 | Mean | 2.55 | 1.51 | 2.74 | 1.07 | 1.79 | 1.55 | 1.40 | 1.02 | 2.69 | 2.04 | 6.01 | 3.53 | 9.28 | 26.85 | 1.93 | 1.05 | 6.85 |
| sd | 1.50 | 0.266 | 1.16 | 0.278 | 0.942 | 0.537 | 0.464 | 0.321 | 0.872 | 0.521 | 7.43 | 3.00 | 7.09 | 16.99 | 1.26 | 0.925 | 4.39 | |
| P | 0.215 | 0.081 | 0.121 | 0.694 | 0.286 | 0.216 | 0.278 | 0.915 | 0.079 | 0.074 | 0.363 | 0.282 | 0.180 | 0.119 | 0.329 | 0.934 | 0.147 |
For stationary static, n = 5 independent experiments; for stationary aerated, n = 4; for all other experiments, n = 3.
Fimbrial genes were generally induced during stationary-phase culture, and even more so during static culture (Table 4). The most highly induced gene during static culture was mrpA (8943-fold mean), with the fold change in five experiments ranging from 70- to 35 108-fold. Fimbrial genes were also induced in colonies, with the exception of fim7A and pmfA. At pH 8.0, fim10A and fim14A were induced (2.04-fold and 27-fold, respectively); there was little change in expression for the other genes at pH 6.0 or 8.0.
Multiple fimbrial proteins are produced in a single culture
Given that quantitative PCR detected transcription of all 17 major fimbrial subunits, we were curious to discover which fimbrial proteins are translated in wild-type P. mirabilis. To enrich for fimbrial production while limiting the amount of cell lysis, P. mirabilis was cultured at 37 °C with aeration for 8 h. Proteins were identified by MS after enrichment for extracellular proteins by first shearing extracellular proteins from the surface and then concentrating the soluble proteins. This enrichment technique has historically been used to identify specific fimbriae produced by P. mirabilis (Wray et al., 1986; Bahrani & Mobley, 1993; Massad et al., 1994a; Bijlsma et al., 1995). We detected 232 P. mirabilis proteins, the most abundant of which were FlaA (the primary flagellin) and PmfA (the major subunit of the PMF fimbria). The proteins detected included those from the cytoplasm, periplasm and inner membrane, suggesting that some cell lysis occurred during growth and the shearing process; the 50 most abundant proteins detected are listed in Table S4. In total, we detected components from six different fimbriae (Table 5). The PMF fimbria was most abundant, with the major fimbrial subunit (PmfA), tip adhesin (PmfE) and chaperone (PmfD) identified. Fim8 was the second most abundant fimbria found; the major fimbrial subunit (Fim8A), a minor fimbrial subunit (Fim8F) and the tip adhesin (Fim8E) were detected. The remaining fimbriae were of lower abundance, and with the exception of UCA, only the putative major subunit was detected. The peptides detected for each fimbrial protein are listed in Table S5.
Table 5. Detection of fimbrial proteins using MS.
Proteins from concentrated P. mirabilis shear preparations were identified by searching detected peptides against a P. mirabilis database. Measurements of percent age coverage, the number of unique peptides detected and the number of total peptides allows an approximation of the relative quantity of protein present.
| Protein ID | ORF | Gene | Annotation | Coverage (%) | Unique peptides | Total peptides |
| PMF | ||||||
| Q04681 | PMI1877 | pmfA | Major fimbrial subunit | 81.0 | 12 | 573 |
| P53522 | PMI1880 | pmfE | Putative tip adhesin | 52.7 | 12 | 53 |
| P53520 | PMI1879 | pmfD | Fimbrial chaperone protein | 9.1 | 2 | 2 |
| Fimbria 8 | ||||||
| B4EXA4 | PMI1469 | fim8A | Fimbrial subunit | 61.3 | 8 | 75 |
| B4EXA0 | PMI1465 | fim8E | Putative fimbrial adhesin | 25.6 | 6 | 6 |
| B4EX99 | PMI1464 | fim8F | Fimbrial subunit | 12.4 | 2 | 2 |
| MR/P | ||||||
| Q03011 | PMI0263 | mrpA | Major MR/P fimbrial protein | 54.9 | 6 | 30 |
| UCA | ||||||
| B4EV68 | PMI0536 | ucaA | Major UCA fimbrial subunit | 42.0 | 5 | 21 |
| B4EV67 | PMI0535 | ucaB | Fimbrial chaperone | 15.8 | 3 | 3 |
| B4EV66 | PMI0534 | ucaC | Fimbrial usher protein | 7.4 | 5 | 5 |
| Fimbria 14 | ||||||
| B4F012 | PMI3002 | fim14A | Putative fimbrial protein | 48.3 | 8 | 15 |
| ATF | ||||||
| B4EYH3 | PMI2728 | atfA | Major fimbrial subunit | 22.0 | 3 | 14 |
Discussion
The initial discovery that P. mirabilis encodes 17 chaperone-usher fimbrial operons (Pearson et al., 2008) was surprising because, to our knowledge, no other bacterial species encodes more fimbriae within a single genome. Here we have shown that the chaperone-usher fimbriae encoded by P. mirabilis are highly conserved among strains collected from diverse sources. This finding is unusual compared with other bacterial species with large repertoires of fimbrial adhesins: studies with fimbrial genes in S. enterica and E. coli have shown that the majority of fimbrial genes are highly variable and only a few fimbrial operons are present in all the strains tested (Townsend et al., 2001; Gibbs et al., 2008; Wurpel et al., 2013). Among species that possess multiple fimbriae, the distribution of these fimbriae often correlates with isolation from a particular niche. For example, the P fimbria, which mediates adherence to kidney epithelia, is more prevalent in uropathogenic E. coli compared with faecal commensal or pathogenic E. coli associated with other disease sites (Källenius et al., 1981). In contrast, we found that fimbrial genes in P. mirabilis are highly conserved regardless of location isolated. Not only were these fimbriae conserved when isolated from different parts of the body, but also the fimbrial genes were highly conserved within strains isolated in different parts of the world and within strains collected 30 years apart. The high degree of conservation within these genes indicates that fimbrial genes are important for the life cycle of P. mirabilis and have been maintained by evolutionary pressure. An alternative explanation is that P. mirabilis is more clonal than E. coli or S. enterica. However, we do not believe this to be the case: P. mirabilis is found in a variety of environments and animal hosts (O'Hara et al., 2000), and our comparison of the major flagellin gene flaA shows that some genetic sequences are variable in this species.
We further note that our results for fimbrial conservation in clinical isolates are likely an underestimate for the presence of these genes. We used two primer sets for each gene to increase the likelihood of detecting a gene if it were present, but the possibility remains that the fimbriae are more ubiquitous than we are reporting here. A lack of detection by PCR for specific fimbrial genes in the two clinical isolate collections may be due to variable sequences within the primer binding sites of these fimbrial genes and not due to the lack of their presence. In particular, the Fim3A, UcaA and Fim6A proteins had considerable amino acid variability in the seven sequenced P. mirabilis strains. Of the 17 fimbrial operons, only fim3 was not detected in a majority of isolates. Although we designed primer sets to target conserved regions of the fimbrial genes (by aligning the sequences from the six sequenced genomes), there is not a sufficiently conserved region of fim3A that would allow universal detection by PCR. Therefore, this fimbria is likely present in a greater percentage of strains than we reported here. The diversity in UCA is especially interesting since this fimbria has been implicated in adherence to shed human uroepithelial cells (Wray et al., 1986) and as a contributor to virulence in a mouse model of UTI (Pellegrino et al., 2013).
We had anticipated using fimbrial distribution to predict which proteins might be associated with colonization of particular niches. Instead, it may be possible that while most of these fimbriae are ubiquitous in P. mirabilis, amino acid variations within a given fimbria confer an advantage in specific environments. This phenomenon has already been observed for the type I fimbria, which is present in the vast majority of E. coli isolates (Mobley et al., 2009). Specific amino acid sequences of the FimH adhesin are more likely to be found in uropathogenic E. coli compared with faecal E. coli, and these variations alter binding characteristics (Sokurenko et al., 1995, 1998). These binding properties may be tied to the need for uropathogens to adhere in the face of urine flow, or to mediate adherence to specific displays of mannose (e.g. monomannose vs trimannose) (Sokurenko et al., 1997; Schembri & Klemm, 2001). In our current study, we used a PCR assay to look for the presence of the predicted major structural subunit genes of these fimbriae, because we expected a large proportion of fimbrial operons would be completely absent from many isolates. However, if fimbrial presence is conserved while binding affinity varies, we would expect to find variation in the tip adhesins for these fimbriae.
We detected transcripts corresponding to the predicted major structural subunit of all 17 fimbriae, as well as proteins from six predicted fimbrial major subunits. However, we do not yet know whether the remaining fimbrial transcripts are translated, nor do we know whether all of the detected subunits are assembled into mature fimbriae at the cell surface. In addition to the 17 fimbrial operons described here, the P. mirabilis HI4320 genome contains 13 additional putative fimbrial structural genes that are not organized with genes encoding chaperones or ushers (Pearson et al., 2008); however, we did not examine these orphans in this study. Our finding of fimbrial gene upregulation during static or stationary-phase culture is consistent with previous reports on fimbrial production by P. mirabilis and other species (Old & Duguid, 1970; Blomfield et al., 1991; Lane et al., 2009). In particular, MR/P fimbriae are assembled in greater numbers during controlled oxygen restriction, as well as during infection of mice (Lane et al., 2009). Expression of fim10A and fim14A was induced under alkaline conditions, which would occur in the urinary tract due to urease activity during UTI. Consistent with this idea, another P. mirabilis virulence factor, the Proteus toxic agglutinin, has previously been shown to have increased activity at basic pH (Alamuri & Mobley, 2008).
Our results show for the first time that a single population of P. mirabilis can produce six major fimbrial subunits that likely results in the assembly of six distinct fimbriae on the cell surface. We also detected proteins from Fim8 and Fim14 for the first time. Others have previously reported detection of MR/P, PMF, UCA, ATF and PMP protein (Adegbola et al., 1983; Wray et al., 1986; Bahrani et al., 1993, 1994; Massad et al., 1994a; Bijlsma et al., 1995; Cook et al., 1995), although not necessarily all at the same time. Electron microscopy has shown that a single P. mirabilis bacterium can express at least two distinct types of fimbriae, which correspond to two unique haemagglutination patterns (Old & Adegbola, 1982; Adegbola et al., 1983). Analysis of MR/P and ATF revealed that both proteins are produced in a single culture grown statically for 48 h (Zunino et al., 2007). Our data do not distinguish between a single bacterium expressing up to six distinct fimbriae and population-wide variation in gene expression resulting in different bacteria presenting different fimbriae on their surface. Because tools (e.g. antibodies) are developed to study these proteins, this question may be addressed.
The presence of Fim14A is particularly interesting, as the fim14 operon lacks a chaperone and its usher in HI4320 is a pseudogene with an internal frameshift. Notably, the usher gene appears intact in the annotated ATCC 29906 and BB2000 genomes, but the chaperone is entirely absent. A chaperone and usher are essential for exporting fimbrial subunits to the extracellular space, so it is possible that Fim14A is not exported, but instead is cytoplasmic protein that we detected due to cell lysis. A more intriguing possibility is that Fim14A is exported to the bacterial surface via interaction with another fimbrial chaperone/usher or with an orphan chaperone/usher. It has been shown that in the absence of the native chaperone and usher, E. coli type 1 and F1C fimbriae can be functionally assembled by a heterologous chaperone and usher pair (Klemm et al., 1994, 1995). Although we are unaware of any instances in which a fimbria is natively assembled by a heterologous chaperone and usher pair, the identification of Fim14A as a putatively exported protein and fim14B as a gene important for pathogenesis (Himpsl et al., 2008) suggests that Fim14 is assembled into a functional fimbria.
In summary, we have documented the surprisingly high degree of conservation of the 17 chaperone-usher fimbrial operons encoded by P. mirabilis. It would be interesting to further investigate conservation of other, non-fimbrial genes encoded by diverse P. mirabilis isolates to determine the heterogeneity of virulence factors. For example, genes involved in individual strain self-recognition are not conserved when the two sequenced strains HI4320 and BB2000 are compared (Sullivan et al., 2013). Future studies will dissect the roles of specific fimbriae in colonization and disease toward the goal of designing better treatments or vaccines against P. mirabilis-mediated UTI.
Acknowledgements
We are grateful to Dr Herbert Lepor, Dr Philip Tierno and Benjamin See for their assistance in collection of tribe Proteeae clinical isolates. We thank Dr Beatrix Ueberheide for mass spectrometry discussions and Dr Xue-Ru Wu, Dr Tung-Tien Sun and members of the Pearson lab for critical reading of the manuscript. We also thank the residents of the Old Public Health Building at NYULMC for their gracious accommodation of our laboratory in the wake of Hurricane Sandy. This work was supported by US Public Health Service grant no. AI083743.
Abbreviations:
- NYULMC
New York University Langone Medical Center
- qRT-PCR
quantitative reverse transcriptase PCR
- UTI
urinary tract infection
Footnotes
Five supplementary tables are available with the online version of this paper.
References
- Adegbola R. A., Old D. C., Senior B. W. (1983). The adhesins and fimbriae of Proteus mirabilis strains associated with high and low affinity for the urinary tract. J Med Microbiol 16, 427–431 10.1099/00222615-16-4-427 [DOI] [PubMed] [Google Scholar]
- Alamuri P., Mobley H. L. (2008). A novel autotransporter of uropathogenic Proteus mirabilis is both a cytotoxin and an agglutinin. Mol Microbiol 68, 997–1017 10.1111/j.1365-2958.2008.06199.x [DOI] [PubMed] [Google Scholar]
- Alamuri P., Löwer M., Hiss J. A., Himpsl S. D., Schneider G., Mobley H. L. (2010). Adhesion, invasion, and agglutination mediated by two trimeric autotransporters in the human uropathogen Proteus mirabilis. Infect Immun 78, 4882–4894 10.1128/IAI.00718-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990). Basic local alignment search tool. J Mol Biol 215, 403–410 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Armbruster C. E., Mobley H. L. T. (2012). Merging mythology and morphology: the multifaceted lifestyle of Proteus mirabilis. Nat Rev Microbiol 10, 743–754 10.1038/nrmicro2890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahrani F. K., Mobley H. L. (1993). Proteus mirabilis MR/P fimbriae: molecular cloning, expression, and nucleotide sequence of the major fimbrial subunit gene. J Bacteriol 175, 457–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahrani F. K., Mobley H. L. T. (1994). Proteus mirabilis MR/P fimbrial operon: genetic organization, nucleotide sequence, and conditions for expression. J Bacteriol 176, 3412–3419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahrani F. K., Cook S., Hull R. A., Massad G., Mobley H. L. (1993). Proteus mirabilis fimbriae: N-terminal amino acid sequence of a major fimbrial subunit and nucleotide sequences of the genes from two strains. Infect Immun 61, 884–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahrani F. K., Massad G., Lockatell C. V., Johnson D. E., Russell R. G., Warren J. W., Mobley H. L. (1994). Construction of an MR/P fimbrial mutant of Proteus mirabilis: role in virulence in a mouse model of ascending urinary tract infection. Infect Immun 62, 3363–3371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belas R., Erskine D., Flaherty D. (1991). Transposon mutagenesis in Proteus mirabilis. J Bacteriol 173, 6289–6293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belas R., Schneider R., Melch M. (1998). Characterization of Proteus mirabilis precocious swarming mutants: identification of rsbA, encoding a regulator of swarming behavior. J Bacteriol 180, 6126–6139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bijlsma I. G. W., van Dijk L., Kusters J. G., Gaastra W. (1995). Nucleotide sequences of two fimbrial major subunit genes, pmpA and ucaA, from canine-uropathogenic Proteus mirabilis strains. Microbiology 141, 1349–1357 10.1099/13500872-141-6-1349 [DOI] [PubMed] [Google Scholar]
- Blomfield I. C., McClain M. S., Princ J. A., Calie P. J., Eisenstein B. I. (1991). Type 1 fimbriation and fimE mutants of Escherichia coli K-12. J Bacteriol 173, 5298–5307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri R. R., Loman N. J., Snyder L. A. S., Bailey C. M., Stekel D. J., Pallen M. J. (2008). xBASE2: a comprehensive resource for comparative bacterial genomics. Nucleic Acids Res 36 (Database), D543–D546 10.1093/nar/gkm928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook S. W., Mody N., Valle J., Hull R. (1995). Molecular cloning of Proteus mirabilis uroepithelial cell adherence (uca) genes. Infect Immun 63, 2082–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotto-Rios X. M., Békés M., Chapman J., Ueberheide B., Huang T. T. (2012). Deubiquitinases as a signaling target of oxidative stress. Cell Reports 2, 1475–1484 10.1016/j.celrep.2012.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFrances C. J., Lucas C. A., Buie V. C., Golosinskiy A. (2008). 2006 National Hospital Discharge Survey; National Health Statistics Reports, no. 5. Hyattsville, MD: National Center for Health Statistics. [PubMed] [Google Scholar]
- Foxman B. (2003). Epidemiology of urinary tract infections: incidence, morbidity, and economic costs. Dis Mon 49, 53–70 10.1067/mda.2003.7 [DOI] [PubMed] [Google Scholar]
- Gaastra W., van Oosterom R. A., Pieters E. W., Bergmans H. E., van Dijk L., Agnes A., ter Huurne H. M. (1996). Isolation and characterisation of dog uropathogenic Proteus mirabilis strains. Vet Microbiol 48, 57–71 10.1016/0378-1135(95)00133-6 [DOI] [PubMed] [Google Scholar]
- Gibbs K. A., Urbanowski M. L., Greenberg E. P. (2008). Genetic determinants of self identity and social recognition in bacteria. Science 321, 256–259 10.1126/science.1160033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith D. P., Musher D. M., Itin C. (1976). Urease. The primary cause of infection-induced urinary stones. Invest Urol 13, 346–350 [PubMed] [Google Scholar]
- Hannan T. J., Totsika M., Mansfield K. J., Moore K. H., Schembri M. A., Hultgren S. J. (2012). Host-pathogen checkpoints and population bottlenecks in persistent and intracellular uropathogenic Escherichia coli bladder infection. FEMS Microbiol Rev 36, 616–648 10.1111/j.1574-6976.2012.00339.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himpsl S. D., Lockatell C. V., Hebel J. R., Johnson D. E., Mobley H. L. (2008). Identification of virulence determinants in uropathogenic Proteus mirabilis using signature-tagged mutagenesis. J Med Microbiol 57, 1068–1078 10.1099/jmm.0.2008/002071-0 [DOI] [PubMed] [Google Scholar]
- Hung D. L., Knight S. D., Woods R. M., Pinkner J. S., Hultgren S. J. (1996). Molecular basis of two subfamilies of immunoglobulin-like chaperones. EMBO J 15, 3792–3805 [PMC free article] [PubMed] [Google Scholar]
- Jansen A. M., Lockatell V., Johnson D. E., Mobley H. L. (2004). Mannose-resistant Proteus-like fimbriae are produced by most Proteus mirabilis strains infecting the urinary tract, dictate the in vivo localization of bacteria, and contribute to biofilm formation. Infect Immun 72, 7294–7305 10.1128/IAI.72.12.7294-7305.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Källenius G., Svenson S. B., Hultberg H., Möllby R., Helin I., Cedergren B., Winberg J. (1981). Occurrence of P-fimbriated Escherichia coli in urinary tract infections. Lancet 318, 1369–1372 10.1016/S0140-6736(81)92797-5 [DOI] [PubMed] [Google Scholar]
- Khalid M. I. H., Teh L. K., Lee L. S., Zakaria Z. A., Salleh M. Z. (2013). Genome sequence of Proteus mirabilis strain PR03, isolated from a local hospital in Malaysia. Genome Announc 1, e00327-13 10.1128/genomeA.00327-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm P., Christiansen G., Kreft B., Marre R., Bergmans H. (1994). Reciprocal exchange of minor components of type 1 and F1C fimbriae results in hybrid organelles with changed receptor specificities. J Bacteriol 176, 2227–2234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm P., Jørgensen B. J., Kreft B., Christiansen G. (1995). The export systems of type 1 and F1C fimbriae are interchangeable but work in parental pairs. J Bacteriol 177, 621–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klevens R. M., Edwards J. R., Richards C. L., Jr, Horan T. C., Gaynes R. P., Pollock D. A., Cardo D. M. (2007). Estimating health care-associated infections and deaths in U.S. hospitals, 2002. Public Health Rep 122, 160–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane M. C., Mobley H. L. (2007). Role of P-fimbrial-mediated adherence in pyelonephritis and persistence of uropathogenic Escherichia coli (UPEC) in the mammalian kidney. Kidney Int 72, 19–25 10.1038/sj.ki.5002230 [DOI] [PubMed] [Google Scholar]
- Lane M. C., Li X., Pearson M. M., Simms A. N., Mobley H. L. (2009). Oxygen-limiting conditions enrich for fimbriate cells of uropathogenic Proteus mirabilis and Escherichia coli. J Bacteriol 191, 1382–1392 10.1128/JB.01550-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litwin M. S., Saigal C. S. (2007). Urologic Diseases in America. Washington, D.C.: Government Printing Office [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Massad G., Bahrani F. K., Mobley H. L. (1994a). Proteus mirabilis fimbriae: identification, isolation, and characterization of a new ambient-temperature fimbria. Infect Immun 62, 1989–1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massad G., Lockatell C. V., Johnson D. E., Mobley H. L. (1994b). Proteus mirabilis fimbriae: construction of an isogenic pmfA mutant and analysis of virulence in a CBA mouse model of ascending urinary tract infection. Infect Immun 62, 536–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massad G., Fulkerson J. F., Jr, Watson D. C., Mobley H. L. (1996). Proteus mirabilis ambient-temperature fimbriae: cloning and nucleotide sequence of the aft gene cluster. Infect Immun 64, 4390–4395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mobley H. L., Island M. D., Hausinger R. P. (1995). Molecular biology of microbial ureases. Microbiol Rev 59, 451–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mobley H. L. T., Donnenberg M. S., Hagan E. C. (2009). Uropathogenic Escherichia coli. In EcoSal – Escherichia coli and Salmonella: Cellular and Molecular Biology. Edited by Böck A., Curtiss R., III, Kaper J. B., Karp P. D., Neidhardt F. C., Nyström T., Slauch J. M., Squires C. L., Ussery D. Washington, D.C.: American Society for Microbiology [Google Scholar]
- Nielubowicz G. R., Mobley H. L. (2010). Host-pathogen interactions in urinary tract infection. Nat Rev Urol 7, 430–441 10.1038/nrurol.2010.101 [DOI] [PubMed] [Google Scholar]
- Nuccio S. P., Bäumler A. J. (2007). Evolution of the chaperone/usher assembly pathway: fimbrial classification goes Greek. Microbiol Mol Biol Rev 71, 551–575 10.1128/MMBR.00014-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hara C. M., Brenner F. W., Miller J. M. (2000). Classification, identification, and clinical significance of Proteus, Providencia, and Morganella. Clin Microbiol Rev 13, 534–546 10.1128/CMR.13.4.534-546.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Old D. C., Adegbola R. A. (1982). Haemagglutinins and fimbriae of Morganella, Proteus and Providencia. J Med Microbiol 15, 551–564 10.1099/00222615-15-4-551 [DOI] [PubMed] [Google Scholar]
- Old D. C., Duguid J. P. (1970). Selective outgrowth of fimbriate bacteria in static liquid medium. J Bacteriol 103, 447–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson M. M., Mobley H. L. (2008). Repression of motility during fimbrial expression: identification of 14 mrpJ gene paralogues in Proteus mirabilis. Mol Microbiol 69, 548–558 10.1111/j.1365-2958.2008.06307.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson M. M., Sebaihia M., Churcher C., Quail M. A., Seshasayee A. S., Luscombe N. M., Abdellah Z., Arrosmith C., Atkin B. & other authors (2008). Complete genome sequence of uropathogenic Proteus mirabilis, a master of both adherence and motility. J Bacteriol 190, 4027–4037 10.1128/JB.01981-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrino R., Scavone P., Umpiérrez A., Maskell D. J., Zunino P. (2013). Proteus mirabilis uroepithelial cell adhesin (UCA) fimbria plays a role in the colonization of the urinary tract. Pathog Dis 67, 104–107 10.1111/2049-632X.12027 [DOI] [PubMed] [Google Scholar]
- Roberts J. A., Fussell E. N., Kaack M. B. (1990). Bacterial adherence to urethral catheters. J Urol 144, 264–269 [DOI] [PubMed] [Google Scholar]
- Schembri M. A., Klemm P. (2001). Biofilm formation in a hydrodynamic environment by novel fimh variants and ramifications for virulence. Infect Immun 69, 1322–1328 10.1128/IAI.69.3.1322-1328.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simerville J. A., Maxted W. C., Pahira J. J. (2005). Urinalysis: a comprehensive review. Am Fam Physician 71, 1153–1162 [PubMed] [Google Scholar]
- Sokurenko E. V., Courtney H. S., Maslow J., Siitonen A., Hasty D. L. (1995). Quantitative differences in adhesiveness of type 1 fimbriated Escherichia coli due to structural differences in fimH genes. J Bacteriol 177, 3680–3686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokurenko E. V., Chesnokova V., Doyle R. J., Hasty D. L. (1997). Diversity of the Escherichia coli type 1 fimbrial lectin. Differential binding to mannosides and uroepithelial cells. J Biol Chem 272, 17880–17886 10.1074/jbc.272.28.17880 [DOI] [PubMed] [Google Scholar]
- Sokurenko E. V., Chesnokova V., Dykhuizen D. E., Ofek I., Wu X. R., Krogfelt K. A., Struve C., Schembri M. A., Hasty D. L. (1998). Pathogenic adaptation of Escherichia coli by natural variation of the FimH adhesin. Proc Natl Acad Sci U S A 95, 8922–8926 10.1073/pnas.95.15.8922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spurbeck R. R., Stapleton A. E., Johnson J. R., Walk S. T., Hooton T. M., Mobley H. L. T. (2011). Fimbrial profiles predict virulence of uropathogenic Escherichia coli strains: contribution of Ygi and Yad fimbriae. Infect Immun 79, 4753–4763 10.1128/IAI.05621-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan N. L., Septer A. N., Fields A. T., Wenren L. M., Gibbs K. A. (2013). The complete genome sequence of Proteus mirabilis strain BB2000 reveals differences from the P. mirabilis reference strain. Genome Announc 1, e00024-13 10.1128/genomeA.00024-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tambyah P. A. (2004). Catheter-associated urinary tract infections: diagnosis and prophylaxis. Int J Antimicrob Agents 24 (Suppl. 1), S44–S48 10.1016/j.ijantimicag.2004.02.008 [DOI] [PubMed] [Google Scholar]
- Tambyah P. A., Maki D. G. (2000). Catheter-associated urinary tract infection is rarely symptomatic: a prospective study of 1,497 catheterized patients. Arch Intern Med 160, 678–682 [DOI] [PubMed] [Google Scholar]
- Tanabe M., Kanehisa M. (2012). Using the KEGG database resource. Curr Protoc Bioinformatics 38, 1.12.1–1.12.43 [DOI] [PubMed] [Google Scholar]
- Tolson D. L., Barrigar D. L., McLean R. J., Altman E. (1995). Expression of a nonagglutinating fimbria by Proteus mirabilis. Infect Immun 63, 1127–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend S. M., Kramer N. E., Edwards R., Baker S., Hamlin N., Simmonds M., Stevens K., Maloy S., Parkhill J. & other authors (2001). Salmonella enterica serovar Typhi possesses a unique repertoire of fimbrial gene sequences. Infect Immun 69, 2894–2901 10.1128/IAI.69.5.2894-2901.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren J. W., Tenney J. H., Hoopes J. M., Muncie H. L., Anthony W. C. (1982). A prospective microbiologic study of bacteriuria in patients with chronic indwelling urethral catheters. J Infect Dis 146, 719–723 10.1093/infdis/146.6.719 [DOI] [PubMed] [Google Scholar]
- Wray S. K., Hull S. I., Cook R. G., Barrish J., Hull R. A. (1986). Identification and characterization of a uroepithelial cell adhesin from a uropathogenic isolate of Proteus mirabilis. Infect Immun 54, 43–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurpel D. J., Beatson S. A., Totsika M., Petty N. K., Schembri M. A. (2013). Chaperone-usher fimbriae of Escherichia coli. PLoS ONE 8, e52835 10.1371/journal.pone.0052835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zunino P., Geymonat L., Allen A. G., Legnani-Fajardo C., Maskell D. J. (2000). Virulence of a Proteus mirabilis ATF isogenic mutant is not impaired in a mouse model of ascending urinary tract infection. FEMS Immunol Med Microbiol 29, 137–143 10.1111/j.1574-695X.2000.tb01516.x [DOI] [PubMed] [Google Scholar]
- Zunino P., Sosa V., Allen A. G., Preston A., Schlapp G., Maskell D. J. (2003). Proteus mirabilis fimbriae (PMF) are important for both bladder and kidney colonization in mice. Microbiology 149, 3231–3237 10.1099/mic.0.26534-0 [DOI] [PubMed] [Google Scholar]
- Zunino P., Sosa V., Schlapp G., Allen A. G., Preston A., Maskell D. J. (2007). Mannose-resistant Proteus-like and P. mirabilis fimbriae have specific and additive roles in P. mirabilis urinary tract infections. FEMS Immunol Med Microbiol 51, 125–133 10.1111/j.1574-695X.2007.00285.x [DOI] [PubMed] [Google Scholar]


