Abstract
Ribonucleotide reductases (RNRs) catalyze the conversion of nucleotides to deoxynucleotides in all organisms, providing the monomeric precursors required for DNA replication and repair. The class I RNRs are composed of two subunits; the R1 subunit contains the active site for nucleotide reduction and allosteric effector binding sites, whereas the R2 subunit houses the essential diirontyrosyl (Y·) radical cofactor. A major unresolved issue is the mechanism by which the tyrosyl radical on R2 (Y122, Escherichia coli numbering) reversibly generates the transient thiyl radical (S·) on R1 that initiates nucleotide reduction. This intersubunit radical initiation is postulated to occur through a defined pathway involving conserved aromatic amino acids (R2: Y122, W48, Y356; R1: Y731, Y730) over a long distance of 35 Å. A 20-mer peptide identical to the C-terminal tail of R2 (356–375) and containing Y356 is a competitive inhibitor with respect to R2, and it effectively blocks nucleotide reduction. We now report that a 21-mer peptide, in which a tryptophan has been incorporated at the N terminus of the 20th mer, can replace the R2 subunit and initiate nucleotide reduction by photoinitiated radical generation. The deoxynucleotide generated depends on the presence of allosteric effector and is pathway-dependent. Replacement of Y731 of R2 with phenylalanine prevents deoxynucleotide formation. These results provide direct evidence for the chemical competence of aromatic amino acid radicals and the importance of Y356 in R2 in the radical initiation process of the class I RNRs.
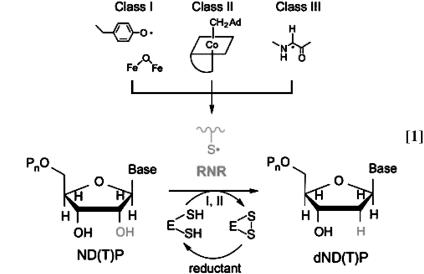
Ribonucleotide reductases (RNRs) catalyze the conversion of nucleotides to deoxynucleotides in all organisms, supplying a balanced pool of deoxynucleoside triphosphates required for DNA replication and repair. The central role of RNRs in nucleic acid metabolism, their exquisite control of free radical chemistry, and the proposed part they play in the conversion of an RNA to a DNA world, have fascinated scientists since their discovery (1). RNRs have been divided into three classes based on their different methods of radical initiation. The class I RNRs use a diiron-tyrosyl radical (·Y122, Escherichia coli numbering) cofactor, the class II RNRs use the homolysis of the carbon–cobalt bond of a B12 cofactor, and the class III RNRs use a glycyl radical generated by an S-adenosylmethionine/[4Fe4S] activase (1). In each case, the radical initiator generates a thiyl radical (S·) in a structurally homologous active site where the nucleotide is reduced to the corresponding deoxynucleotide concomitant with the oxidation of either two cysteines to a disulfide (class I and II) or formate to CO2 (class III) (see Eq. 1). Although much has been learned about the control over protein and nucleotide radical intermediates exerted by RNRs, a major unresolved problem is the mechanism of radical initiation by the class I enzyme (2).
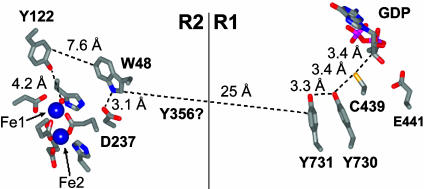
The class I RNRs are thought to be composed of a 1:1 complex of two homodimeric subunits, R1 and R2. In the class I RNR from E. coli, the stable ·Y122 is located on R2, whereas the cysteine (C439) that becomes the transient S· is contained in R1. In addition, R1 also contains the allosteric effector binding sites that determine both specificity (TTP, dGTP, dATP, and ATP) and rate (dATP and ATP) of nucleotide reduction. The radical initiation at C439 is thought to occur reversibly between the two subunits on every enzymatic turnover (3). The Y122 and C439 residues are proposed to be separated by 35 Å, based on a docking model generated from the individual crystal structures of R1 and R2 by using shape complementarity and electrostatics (4). The observed rate constants for RNR turnover are 2–10 s-1, depending on allosteric effector. These numbers are considerably faster than the rate of 10-4 to 10-9 s-1 calculated for a single electron transfer (ET) step over this distance based on Marcus theory, by using the expression for an activationless ET: kET = 1013exp(-βr) with r = 35 Å and β = 1.1–1.4 Å-1, respectively (5). If this model is correct, the long distance for radical initiation therefore requires the existence of amino acid radical intermediates to accommodate the observed turnover numbers of these enzymes. The putative pathway shown in Fig. 1 is based on the docking model and absolute conservation of all these residues across ≈140 sequences (4). Experimentally, the pathway has been examined by in vitro and in vivo mutagenesis studies of each of these residues (ref. 6 and references therein), which has shown these residues to be important for enzyme activity. These studies have not, however, provided any mechanistic insight into the radical initiation process. The three tyrosine residues from this pathway, Y356 in R2 and Y731 and Y730 in R1, stand out as they are not involved directly in either nucleotide reduction or biosynthesis of the diiron-Y· cofactor (7).
Fig. 1.
Radical transfer pathway and distances generated from the docking model of the R1 and R2 subunits of the class I RNR from E. coli. The last 35 amino acids of the C-terminal tail of R2, in which Y356 resides, are thermally labile and undetectable in available crystal structures. Thus, the distance between W48 on R2 and Y731 in R1 (25 Å) is based solely on the docking model of a 1:1 complex of R1 and R2 (4). [Reprinted with permission from ref. 27 (Copyright 2003, American Chemical Society).]
In only the case of the class II AdoCbl RNR has thiyl radical formation been shown to occur in a chemically and kinetically competent fashion (8). Although the mechanism of S· generation in the class I enzyme remains to be clarified, the overlay of the active sites of the three classes of RNRs reveals that the axial ligand of adenosylcobalamin (5′-deoxyadenosine) of the class II enzyme and glycyl radical of the class III RNR (Eq. 1), both involved in direct hydrogen atom abstraction to generate S·, are superimposable on Y730 and Y731 in R1 (9, 10). This superposition provides the most compelling evidence for the involvement of these conserved tyrosines in the radical initiation pathway. The role of Y356, which is proposed to serve as the radical conduit between R2 and R1, is the focus of this study.
Many ET processes in biological systems involve metal-based cofactors spaced 10–15 Å apart and are physiologically unidirectional (11). In the case of the class I RNRs, there are no metal-based cofactors involved in the radical transfer process and consequently, aromatic amino acids must function in this capacity. The redox potentials of the amino acids along the pathway (tyrosine, tryptophan, and cysteine) require changes in protonation state to become transiently oxidized (12) and control the directionality of radical propagation, implicating proton-coupled ET (2, 13). Whereas the role of aromatic amino acid radical intermediates in long-range charge transport has recently been recognized in model proteins (14) and in photoreactivation of the flavin-dependent DNA photolyase (15, 16), the radical initiation in class I RNRs is unique because of its exceptionally long distance and its physiological relevance.
No direct evidence for this pathway (Fig. 1) or the proposed radical intermediates in the pathway exists, because the radical initiation step in the E. coli RNR is kinetically masked by rate-limiting conformational changes (3). To avoid the problem of conformational gating associated with substrate and effector binding to R1, to address the direct involvement of Y356, and to eventually monitor directly the kinetics of the radical initiation process, we have undertaken studies in which the Y356→Y731→Y730→C439 pathway of the R2–R1 complex (Fig. 1) has been isolated. This approach exploits the weak interaction between R1 and R2 (0.2 μM) and the unusual ability of a 20-mer peptide derived from the C-terminal tail of R2 to prevent subunit association. This 20-mer peptide (residues 356–375) contains the essential Y356 (17) and has been shown to be a competitive inhibitor (Ki ≈ 20 μM, where Ki = KD for competitive inhibition) of nucleotide reduction by binding to R1 in place of R2 (18). The peptide also facilitates the crystallization of the R1 subunit; the crystal structure of the R1-peptide complex is shown in Fig. 2. Although Y356 is not located in the structure because of thermal disorder at the N terminus of the peptide, the orientation of the peptide places it in the vicinity of Y731 (Fig. 2). In this study, we describe modification of the 20-mer peptide with a phototrigger, X, adjacent to Y356 that has provided a method for activating radical initiation on a synthetic peptide complexed to R1 in the presence of substrate and effector (Fig. 3). In this way, specific entryway into the radical initiation pathway of R1 through transient ·Y356 generation can be achieved, bypassing the complexities attendant to the R2 pathway and the associated conformational changes (2). We now report the chemical competence of aromatic amino acid radical intermediates and consequently the importance of Y356 in the conserved pathway (Fig. 1) of the radical initiation process catalyzed by class I RNRs.
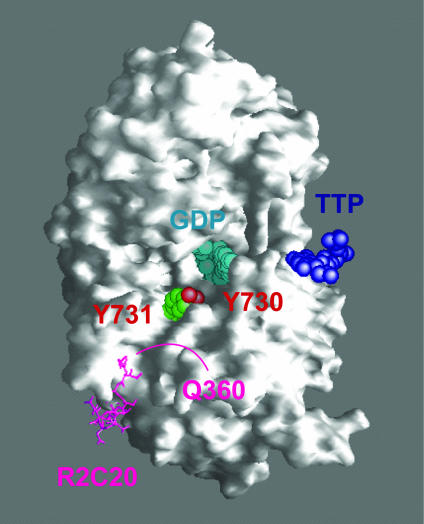
Fig. 2.
Surface rendition of the R1–R2C20 peptide complex generated from the crystal structure (25) with substrate (GDP, cyan) and specificity effector (TTP, blue) bound. The first four amino acids of R2C20 (fuchsia), including Y356, are thermally labile in the structure. Crystallization of the E. coli R1 required the presence of the C-terminal peptide of R2 (residues 356–375); however, only residues 360–375 are detectable. The distance between residue 360 of R2C20 and Y731 of R1 is 16 Å (Cα–Cα), which can be spanned by the four disordered amino acids (residues 356–359) of the peptide.
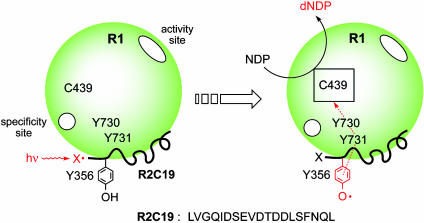
Fig. 3.
Strategy for the photochemical triggering of the Y731→ Y730→ C439 radical initiation pathway in the E. coli R1. NDP, nucleoside diphosphate substrate. dNDP, deoxynucleoside diphosphate product. X, W. This strategy exploits the weak interaction between R1 and R2 (0.2 μM) and the ability of peptides to the C terminus of R2 to completely inhibit nucleotide reduction in a competitive fashion.
Experimental Procedures
Peptide Synthesis and Characterization. Peptides were synthesized by methods previously described (17). N-acetylation was carried out by shaking in 0.5 M acetic anhydride/0.5 M diisopropylethylamine in N, N-dimethylformamide for 1 h before removal of the peptide from the solid-phase support. Each peptide was purified by semipreparative RP-HPLC (Waters XTerra MS C-18, 19 × 100 mm) by using a linear gradient of 10–65% acetonitrile versus 0.1 M ammonium bicarbonate, pH 8.0 (flow rate, 10 ml/min) over 45 min. Purified peptides were then dialyzed against 5 mM potassium phosphate, pH 7.0, by using cellulose ester 1,000 molecular weight cut-off tubing (Spectra/Por CE) at 4°C. Purified peptides were analyzed by analytical HPLC (Waters XTerra MS C-18, 4.6 × 100 mm) by using a linear gradient of 10–65% acetonitrile versus 0.1 M ammonium bicarbonate, pH 8.0 (flow rate, 1.0 ml/min) over 45 min and by matrix-assisted laser desorption ionization time-of-f light (MALDI-TOF) MS. Ac-WY-LVGQIDSEVDTDDLSFNQL (= Ac-WY-R2C19): HPLC: tR (retention time) = 18.9 min. MALDI-TOF MS: MWcalc (calculated molecular mass) = 2,499 Da, MWobs (observed molecular mass) = 2,498 Da. Ac-WFLVGQIDSEVDTDDLSFNQL (= Ac-WF-R2C19): HPLC: tR = 20.3 min. MALDI-TOF MS: MWcalc = 2,484 Da, MWobs = 2,484 Da. Ac-Y-LVGQIDSEVDTDDLSFNQL (= Ac-Y-R2C19): HPLC: tR = 14.2 min. MALDI-TOF MS: MWcalc = 2,313 Da, MWobs = 2,315 Da. Ac-F-LVGQIDSEVDTDDLSFNQL (= Ac-F-R2C19): HPLC: tR = 16.1 min. MALDI-TOF MS: MWcalc = 2,297 Da, MWobs = 2,297 Da. See supporting information, which is published on the PNAS web site, for HPLC traces.
Isolation and Preparation of Prereduced R1. R1 was isolated by standard procedures (19) and further purified by anion exchange with a POROS HQ/20 column (12–14 mg of R1 loaded per run, 16 mm × 100 mm) with a linear gradient of 0–700 mM NaCl over 30 min (4 ml/min). The fractions containing R1 were pooled and concentrated by Amicon using a PM30 membrane. The purified R1 (40 μM) was treated with hydroxyurea (20 mM) for 60 min to reduce the ·Y122 present in the small amounts of R2 that copurify with R1. R1 was then prereduced with DTT (20 mM) for 15 min. The R1 was then passed through a G-25 column preequilibrated with assay buffer. The protein-containing fractions were pooled and concentrated by using a YM30 Centriprep to a final protein concentration of 80 μM. After this treatment, the specific activity of the R1 was reduced to 1,800 nmol·min-1·mg-1.
Measurement of Steady-State Turnover Numbers. R1 (0.1 μM, specific activity = 2,700 nmol·min-1·mg-1), R2 (0.5 μM, specific activity = 7,000 nmol·min-1·mg-1), thioredoxin (50 μM), thioredoxin reductase (1.0 μM), CDP (1.0 mM), and NADPH (0.2 mM) were incubated with either ATP (1.6 mM), TTP (100 μM), or no effector. Enzymatic activity was then measured by the decrease in A340 nm.
Isolation and Preparation of Prereduced Y731F-R1. The Y731F-R1 mutant was isolated by the same manner as the WT R1, omitting the DEAE anion-exchange chromatographic purification step in the standard procedure. The mutant was then subjected to anion-exchange FPLC and treated with hydroxyurea and DTT as described for WT R1. The Y731F-R1 was measured by the radioactive assay for 2′-deoxycytosine (dC) to contain ≈2% contaminating WT R1 activity (3).
Single Turnover Assays for Photoinitiated Nucleotide Reduction in R1–Peptide Complexes. Prereduced R1 (30 μM, specific activity = 1,800 nmol·min-1·mg-1), peptide (20 μM or 200 μM), ATP (1.6 mM) or TTP (100 μM), and [14C]-CDP (0.75 mM, 1.8 × 107 cpm/μmol) were incubated in either a 1.0-cm or 1.0-mm pathlength sealed quartz cell in 50 mM Hepes/15 mM MgSO4/1 mM EDTA, pH 7.6. Samples were irradiated at 30°C for 30 min by using a 1,000-W high-pressure Hg-Xe lamp. The irradiation beam was passed through 285-nm long pass and 90% neutral density filters and a collimating lens before entering the sample chamber. Samples were quenched with 2.0% HClO4 (0.5 V) and neutralized to pH 7.0 with 0.5 M KOH. The precipitated protein was pelleted by centrifugation and washed twice with buffer. dC (50 nmol) was added to the washes, which were combined with the supernatant. The phosphates of the nucleotide were removed with alkaline phosphatase, and the resulting dC was separated from cytidine (20). The dC was then isolated by RP-HPLC on an Alltech Associates nucleotide–nucleoside C-18 column (4.6 mm × 250 mm) by using a linear gradient of 0–50% methanol versus 4 mM potassium phosphate, pH 7.0, over 30 min (flow rate, 1 ml/min) and quantitated against carrier recovery. The identity of the product was further corroborated by enzymatic conversion of dC to 2′-deoxyuridine (dU). dC (≈20 nmol) was incubated with cytidine deaminase (0.025 units) in 50 mM Tris·HCl, pH 8.0, for 30 min at 25°C followed by heat inactivation of the enzyme by boiling for 1 min. dC carrier (20 nmol) was then added to the sample. dC and dU were separated by RP-HPLC on an Alltech Associates nucleotide–nucleoside C-18 column (4.6 × 250 mm) by using 4 mM potassium phosphate, pH 7.0, buffer versus methanol. The pump flow program consisted of three steps: an isocratic 0% methanol phase from 0 to 5 min, a linear gradient of 0–10% methanol from 5 to 15 min, and a separating linear gradient of 10–20% methanol from 15 to 35 min. The flow rate was held constant at 1.0 ml/min. The 14C was quantitatively recovered in the dU peak for each sample.
Results
Development of the Assay for Photoinitiated Nucleotide Reduction. The 20-mer C-terminal tail of R2 (356–375) was synthesized by solid-phase methods (17) with an additional N-terminal tryptophan residue (Ac-WY-R2C19 = Ac-WY-LVGQIDSEVDTD-DLSFNQL) as the phototrigger for radical formation at Y356 (Fig. 3). Irradiation of tryptophan is well known to lead to its photoionization, generating both a cation (·WH+) and a neutral (W·) radical (21). These species are both capable of oxidizing the adjacent Y356 to a tyrosyl radical, which occurs on a microsecond time scale in WY dipeptides (22). The chemical competence of the R1–peptide complex to carry out radical initiation at C439 was assayed by measuring deoxynucleotide formation upon photolysis, which requires formation of ·C439. Because generation of ·WH+ and W· on the peptide is carried out under UV irradiation (λ > 285 nm), the activity of R1 under the photolytic conditions was tested and found to be unchanged. An additional concern was the removal of small amounts of R2 that always copurifies with R1. This contamination would lead to high background levels of dCDP formation, given the high concentrations of R1 (30 μM) in our experiments. Thus subsequent to the dATP affinity column purification, R1 was further purified by anion-exchange FPLC to remove the contaminating R2. After the additional chromatographic step of R1, it was also treated with high concentrations of hydroxyurea (20 mM for extended incubation times) to inactivate any residual R2 activity, by reducing the essential ·Y122. A third strategy to reduce background dCDP production was to carry out the reaction under single turnover conditions (by omitting a disulfide-reducing system, Eq. 1). Multiple turnover conditions would enhance any dCDP production from traces of residual active R2 during the extended period used in the photolysis reaction. Theoretically, as many as four dNDPs could be produced due to the C-terminal pair of cysteines on R1 that function by disulfide interchange to re-reduce the active site disulfide (C225–C462) produced concomitant with nucleotide reduction (23). Studies under single turnover conditions, in which R1 is prereduced with DTT and no external reductant is present in the assay mixture, have revealed that 3.0 to 3.5 dNDPs are routinely produced per R1 homodimer (3, 23). Under our conditions to purify R1 by using an additional FPLC step and extended treatment with hydroxyurea, 2.8 ± 0.3 dCDPs were observed per R1 homodimer.
One additional complexity of our experimental design requires comment. Ideally, the photolysis experiments would be performed with each R1 homodimer saturated with two peptides. However, the low solubility of R1 (R1 aggregation becomes an issue at concentrations >50 μM) in combination with the low affinity of the peptide (KD ≈ 20 μM) constrained our options. The major concern initially was to limit the amount of free peptide, which could interact nonspecifically with R1. Therefore, the photolytic assays were typically carried out with 30 μM R1 and 20 μM peptide present. Under these conditions, 70% of the peptide and 23% of the R1 is bound, assuming a model of independent monomers (18). Later experiments were also performed with saturating peptide concentrations (200 μM), in which 27% of the peptide and 88% of the R1 is bound. Under these conditions, as described below, the amount of dCDP produced scaled with the amount of R1–peptide complex formed. Thus, we believe that the excess free peptide in solution is not problematic and the experiments carried out with the two different concentrations of peptide may be directly compared.
The R1–Peptide Complex Is Chemically Competent for Nucleotide Reduction in the Absence of R2. With the photolysis assay in place, we turned our attention to addressing the chemical competence of the R1–peptide complex to carry out nucleotide reduction by using light initiation. Prereduced R1 was incubated with Ac-WY-R2C19 in the presence of substrate, CDP, and ATP (the most efficient effector) and photolyzed (λ > 285 nm) for 30 min at 30°C. The reaction was then stopped by acid quenching and neutralized. The phosphates of the nucleotides were removed by using alkaline phosphatase, and the nucleoside substrate was removed from the deoxynucleoside product by the method of Steeper and Steuart (20). dCDP production was then quantitated as dC by RP-HPLC. After this step, dC was converted enzymatically to dU by using cytidine deaminase and quantitated by RP-HPLC to confirm the identity of the product. In each case, dU was recovered quantitatively from the initial dC. The photoreaction of R1 with Ac-WY-R2C19 and ATP as effector was found to produce a significant amount of dCDP product (Table 1, effector dependence 1); 6.3% dCDP is observed per equivalent of R1 added (see the supporting information for further discussion of dCDP stoichiometry). When the peptide concentration was increased 10-fold, the amount of dCDP produced increased 3.1-fold to 19.8% (Table 1, effector dependence 2), scaling with the amount of R1–peptide complex (3.8-fold increase). The observed product must be derived from ·C439 generation in the R1 active site, because there is no chemical precedence for direct nucleotide reduction in any system studied to date (24). The additional control experiments performed are summarized below. No dCDP formation above the background of small amounts of incompletely Y·-reduced R2 (0.32–0.48%; Table 1, controls 1, 5, and 6) was observed in dark reactions at either peptide concentration used, indicating that this reaction is light-dependent. Furthermore, no product was detected when substrate was irradiated solely in the presence of the peptide (Table 1, control 4). Other controls in which R1 was irradiated in the absence of peptide are consistent with low levels of contaminating R2 (Table 1, controls 2 and 3).
Table 1. Light-initiated single turnover experiments in R1-peptide complexes.
| No. | Sample | Condition | Effector | dC/% R1 | σ/% R1* |
|---|---|---|---|---|---|
| Effector dependence | |||||
| 1. | R1 + Ac-WY-R2C19 | Light | ATP | 6.3 | 1.53 |
| 2. | R1 + Ac-WY-R2C19 (× 10)† | Light | ATP | 19.8 | 4.38 |
| 3. | R1 + Ac-WY-R2C19 | Light | TTP | 2.4 | 0.007 |
| 4. | R1 + Ac-WY-R2C19 | Light | None | 0.68 | -‡ |
| Y356 pathway dependence | |||||
| 1. | R1 + Ac-WF-R2C19 | Light | ATP | 2.7 | 0.14 |
| 2. | R1 + Ac-Y-R2C19 | Light | ATP | 2.3 | 0.06 |
| 3. | R1 + Ac-F-R2C19 | Light | ATP | 0.75 | 0.52 |
| R1-Y731 pathway dependence | |||||
| 1. | R1Y731F + Ac-WY-R2C19 | Light | ATP | 0.55 | 0.07 |
| 2. | R1Y731F + Ac-WY-R2C19 (× 10)† | Light | ATP | 0.75 | 0.18 |
| 3. | R1Y731F | Dark | ATP | 0.17 | 0.07 |
| 4. | R1Y731F | Light | ATP | 0.21 | 0.11 |
| 5. | R1Y731F† | Light | ATP | 0.16 | 0.03 |
| 6. | R1Y731F + Ac-WY-R2C19 | Dark | ATP | 0.07 | 0.05 |
| Controls | |||||
| 1. | R1 | Dark | ATP | 0.44 | 0.32 |
| 2. | R1 | Light | ATP | 0.62 | 0.07 |
| 3. | R1† | Light | ATP | 0.31 | 0.23 |
| 4. | Ac-WY-R2C19 | Light | ATP | n.d. | - |
| 5. | R1 + Ac-WY-R2C19 | Dark | ATP | 0.48 | 0.28 |
| 6. | R1 + Ac-WY-R2C19 (× 10)† | Dark | ATP | 0.32 | 0.23 |
n.d., not detected.
σ is the standard deviation measured for two to four experiments
1.0-mm pathlength cell (all other experiments were carried out with a 1.0-cm pathlength cell)
Experiment performed only once, because of protein precipitation
In an effort to understand the basis for the stoichiometry of dCDP produced, a number of additional controls were performed. First, a time-course study was run to determine whether continued irradiation would produce additional deoxynucleotide. Studies revealed that the product formation is maximized within 15 min of irradiation, with no further dCDP production for up to 90 min of photolysis (data not shown). The assay components were tested for photodegradation under the assay conditions. R1, CDP, and peptide were analyzed after the photoreaction. The R1 activity was not significantly altered, nor were the thiols oxidized during photolysis; in addition electrospray ionization MS of the photolyzed R1 showed no evidence of modification. The CDP was analyzed by ion-pairing RP-HPLC and found to be unaffected by photolysis. The peptide, however, was found to be partially decomposed (45%) by using RP-HPLC and UV-visible spectroscopy. Thus, our working hypothesis is that the peptide may be the limiting reagent in the photoreaction with R1 (see supporting information).
Allosteric Effector Dependence. The efficiency of the dCDP production in the R1–peptide complex is quite remarkable considering that ability of the peptide to place R1 in the appropriate conformation to initiate formation of ·C439 is not a given. Thus, it was very interesting to observe a clear difference between an activity/specificity effector (ATP, 6.3%), a specificity effector (TTP, 2.4%), and no effector (0.68%) (Table 1, effector dependence 1, 3, and 4, respectively). These results in this set of experiments (end product analysis) follow the trends observed by steady-state kinetic studies of R1 in combination with R2, in which the turnover numbers for ATP, TTP, and no effector are 11.3 s-1, 7.5 s-1, and 2.1 s-1, respectively. As noted above, ATP can bind to activity and specificity sites, whereas TTP binds only to the specificity site within R1 (25); thus the variance in activity most likely arises from distinct conformations of R1 in these three cases.
Sequence Dependence for Radical Initiation. We next turned our attention to examine the role of individual amino acids in the radical initiation pathway of R1 (Fig. 1). Three additional peptides, Ac-WF-R2C19, Ac-Y-R2C19, and Ac-F-R2C19, were synthesized to probe the specificity of ·Y356 as an entryway into the R1 pathway. Competitive inhibition studies with R2 show that these three peptides bind to R1 with a similar affinity as Ac-WY-R2C19 (see supporting information). Both Ac-WF-R2C19 (2.7%) and Ac-Y-R2C19 (2.3%) were competent to turnover CDP to dCDP (Table 1, Y356 pathway dependence 1 and 2, respectively). In the crystal structure of R1 with the 20-mer peptide bound (Fig. 2), only the last 16 amino acids (360 to 375) are visible, suggesting that the N terminus on which Y356 is located is fluxional and unstructured (26). Thus, the observed dCDP can be attributed to the ability of Ac-WF-R2C19 to oxidize the solvent-exposed Y731 directly when the adjacent Y356 is deleted. As for Ac-Y-R2C19, it is known that direct UV irradiation of Y can also generate Y· at lower quantum efficiencies§ and accordingly can also directly promote radical initiation in R1. The observation that Y in the peptide leads to dCDP formation, whereas Y731 in R1 does so much less efficiently, possibly suggests a conformational contribution by the R2 C-terminal peptide toward chemically competent radical transfer. The results with these peptides also aid in setting a lower limit on the time required to enter the proton-coupled ET pathway of R1, as it must occur before quenching of either W· or Y· (microsecond time scale) (22). Being a redox inactive peptide, photolysis of Ac-F-R2C19 should not yield any dCDP above background; the value observed (0.75%) is within the experimental error of the control experiment (Table 1, Y356 pathway dependence 3 as compared to controls 1, 2, and 4–6). These results indicate that light-mediated generation of either W· or Y· within the R2 peptide plays an essential role in nucleotide reduction and, hence, radical initiation at C439.
If the crystal structure of the peptide bound to R1 (Fig. 2) corresponds to that in solution, it is not surprising that the peptide has a looser geometric requirement for radical initiation because of its intrinsic lack of structure in the vicinity of Y356. We would expect, however, the stringency of radical initiation to be much higher within R1 itself. If radical initiation is indeed pathway-specific, mutation of Y731 to a phenylalanine should inhibit nucleotide reduction if no other pathway exists that could circumvent Y731. To test this hypothesis, the Y731F-R1 mutant was prepared and examined for competence for dCDP production. Assays on the mutant itself revealed that the enzyme turned over at ≈2% of WT R1, suggesting the presence of a small amount of contaminating WT R1. The Y731F-R1 mutant, when incubated with CDP, ATP, and 20 μM peptide, reduced dCDP formation to almost background levels (Table 1, R1-Y731 pathway dependence 1 and 2 compared to 3–5). Importantly, a 10-fold increase in peptide concentration to 200 μM, in contrast to WT R1 (Table 1, effector dependence 1 and 2), did not result in increased dCDP production. If the observed dCDP were related to activity intrinsic to the R1Y731F–peptide complex, whether direct Y730 oxidation or tunneling through Y731F, we would expect the dCDP formation to scale to 2.1% ± 0.27%. Thus, the very small amount of dCDP observed is likely due to the contaminating WT R1 (2%) in the sample. Although we cannot establish conclusively if the peptide can bypass Y731 during the radical initiation process, it is our belief that nucleotide reduction is blocked by mutation of the essential Y731 to phenylalanine.
Discussion
Our results demonstrate that a small peptide can replace R2 of RNR and initiate nucleotide reduction by light-mediated amino acid radical generation in an effector-dependent manner. Radical initiation in the R1–peptide complex depends on both Y731 within R1 and an aromatic amino acid within the peptide. These results have important implications for the radical initiation process of RNR. They (i) provide the first direct experimental support for the long-range, intersubunit pathway shown in Fig. 1, (ii) demonstrate the chemical competence of aromatic amino acid radical intermediates in RNR catalysis, (iii) establish the intermediacy of Y356, although structurally yet undefined, as a radical conduit between the R1 and R2 subunits, and (iv) may provide the methodology to kinetically resolve radical intermediates in R1. In addition, the ability of a small peptide to replace the entire R2 subunit reproduces the radical initiation process of monomeric class II RNRs, in which the small molecule adenosylcobalamin initiates thiyl radical formation directly on the R1 equivalent. Thus, by controlling the radical initiation pathway, we have succeeded in converting class I RNR to its simpler class II counterpart.
More generally, these results underscore the importance of conserved pathways for biological radical transport and radical hopping in enzymatic systems (2). Here in class I RNRs we build on previous studies of long-range radical hopping in DNA and protein-based systems and link radical initiation over a long pathway to enzyme catalysis.
Supplementary Material
Figure 1.
Acknowledgments
We thank Dr. Daniel DeOliveira (Dyax, Boston) and Dr. John Mc-Namara (Applied Biosystems) for advice on solid-phase peptide synthesis, Prof. Richard Wolfenden (University of North Carolina, Chapel Hill) for generously providing cytidine deaminase, Dr. Jay Winkler (California Institute of Technology, Pasadena) for his helpful discussions, and Dr. Jie Ge for providing valuable suggestions, molecular modeling expertise, and [14C]-CDP. This work was supported by a National Science Foundation predoctoral fellowship (to M.C.Y.C.), Merck/MIT Collaboration predoctoral fellowships (to M.C.Y.C. and C.S.Y.), and National Institutes of Health Grants GM29595 (to J.S.) and GM47274 (to D.G.N.).
Abbreviations: RNR, ribonucleotide reductase; ET, electron transfer; MALDI-TOF, matrix-assisted laser desorption/ionization–time of flight; dC, 2′-deoxycytosine; dU, 2′-deoxyuridine.
Footnotes
References
- 1.Jordan, A. & Reichard, P. (1998) Annu. Rev. Biochem. 67, 71-98. [DOI] [PubMed] [Google Scholar]
- 2.Stubbe, J., Nocera, D. G., Yee, C. S. & Chang, M. C. Y. (2003) Chem. Rev. 103, 2167-2201. [DOI] [PubMed] [Google Scholar]
- 3.Ge, J., Yu, G., Ator, M. A. & Stubbe, J. (2003) Biochemistry 42, 10071-10083. [DOI] [PubMed] [Google Scholar]
- 4.Uhlin, U. & Eklund, H. (1994) Nature 370, 533-539. [DOI] [PubMed] [Google Scholar]
- 5.Page, C. C., Moser, C. C., Chen, X. & Dutton, L. (1999) Nature 402, 47-52. [DOI] [PubMed] [Google Scholar]
- 6.Ekberg, M., Birgander, P. & Sjöberg, B.-M. (2003) J. Bacteriol. 185, 1167-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stubbe, J. & van der Donk, W. A. (1998) Chem. Rev. 98, 705-762. [DOI] [PubMed] [Google Scholar]
- 8.Licht, S., Gerfen, G. J. & Stubbe, J. (1996) Science 271, 477-481. [DOI] [PubMed] [Google Scholar]
- 9.Stubbe, J., Ge, J. & Yee, C. S. (2001) Trends Biochem. Sci. 26, 93-99. [DOI] [PubMed] [Google Scholar]
- 10.Sintchak, M., Arjara, G., Kellogg, B. A., Stubbe, J. & Drennan, C. L. (2002) Nat. Struct. Biol. 9, 293-300. [DOI] [PubMed] [Google Scholar]
- 11.Gray, H. B. & Winkler, J. R. (2003) Q. Rev. Biophys. 36, 341-372. [DOI] [PubMed] [Google Scholar]
- 12.Tommos, C., Skalicky, J. J., Pilloud, D. L., Wand, S. J. & Dutton, P. L. (1999) Biochemistry 38, 9495-9507. [DOI] [PubMed] [Google Scholar]
- 13.Cukier, R. I. & Nocera, D. G. (1998) Annu. Rev. Phys. Chem. 49, 337-369. [DOI] [PubMed] [Google Scholar]
- 14.Di Bilio, A. J., Crane, B. R., Wehbi, W. A., Kiser, C. N., Abu-Omar, M. M., Carlos, R. M., Richards, J. H., Winkler, J. R. & Gray, H. B. (2001) J. Am. Chem. Soc. 123, 3181-3182. [DOI] [PubMed] [Google Scholar]
- 15.Aubert, C., Vos, M. H., Mathis, P., Eker, A. P. M. & Brettel, K. (2000) Nature 405, 586-590. [DOI] [PubMed] [Google Scholar]
- 16.Sancar, A. (2003) Chem. Rev. 103, 2203-2237. [DOI] [PubMed] [Google Scholar]
- 17.Yee, C. S., Seyedsayamdost, M. R., Chang, M. C. Y., Nocera, D. G. & Stubbe, J. (2003) Biochemistry, 42, 14541-14552. [DOI] [PubMed] [Google Scholar]
- 18.Climent, I., Sjöberg, B.-M. & Huang, C. Y. (1991) Biochemistry 30, 5164-5171. [DOI] [PubMed] [Google Scholar]
- 19.Salowe, S. P. & Stubbe, J. (1986) J. Bacteriol. 165, 363-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Steeper, J. R. & Steuart, C. D. (1970) Anal. Biochem. 43, 123-130. [DOI] [PubMed] [Google Scholar]
- 21.Baugher, J. F. & Grossweiner, L. I. (1977) J. Phys. Chem. 81, 1349-1354. [Google Scholar]
- 22.Sloper, R. W. & Land, E. J. (1980) Photochem. Photobiol. 32, 687-689. [Google Scholar]
- 23.Mao, S. S., Holler, T. P., Yu, G. X., Bollinger, J. M., Booker, S., Johnston, M. I. & Stubbe, J. (1992) Biochemistry 31, 9733-9743. [DOI] [PubMed] [Google Scholar]
- 24.Licht, S. & Stubbe, J. (1999) in Comprehensive Natural Products Chemistry, eds. Barton, S. D., Nakanishi, K., Meth-Cohn, O. & Poulter, C. D. (Elsevier Science, New York), Vol. 5, pp. 163-203. [Google Scholar]
- 25.Eriksson, M., Uhlin, U., Ramaswamy, S., Ekberg, M., Regnström, K., Sjöberg, B.-M. & Eklund, H. (1997) Structure 5, 1077-1092. [DOI] [PubMed] [Google Scholar]
- 26.Lycksell, P.-O. & Sahlin, M. (1995) FEBS Lett. 368, 441-444. [DOI] [PubMed] [Google Scholar]
- 27.Yee, C. S., Chang, M. C. Y., Ge, J., Nocera, D. G. & Stubbe, J. (2003) J. Am. Chem. Soc. 125, 10506-10507. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.