Abstract
Background
Angiogenesis and inflammation are implicated in breast cancer prognosis; however, the role of individual germline variation in related genes is unknown.
Methods
A two-stage candidate pathway association study was conducted among 6,983 Chinese women. Stage 1 included 2,884 women followed for a median of 5.7 years; Stage 2 included 4,099 women followed for a median of 4.0 years. Cox proportional hazards regression was used to estimate the effects of genetic variants on disease-free survival (DFS) and overall survival (OS).
Results
Stage 1 included genotyping of 506 variants in 22 genes; analysis was conducted for 370 common variants. Nominally significant associations with DFS and/or OS were found for 20 loci in ten genes in Stage 1; variants in 19 loci were successfully genotyped and evaluated in Stage 2. In analyses of both study stages combined, nominally significant associations were found for nine variants in seven genes; none of these associations surpassed a significance threshold level corrected for the total number of variants evaluated in this study.
Conclusions
No association with survival was found for 370 common variants in 22 angiogenesis and inflammation pathway genes among Chinese women with breast cancer.
Impact
Our data do not support a large role for common genetic variation in 22 genes in breast cancer prognosis; research on angiogenesis and inflammation genes should focus on common variation in other genes, rare host variants, or tumor alterations.
Keywords: breast cancer survival, genetic variants, angiogenesis genes, inflammation pathway genes, Chinese women
Introduction
Breast cancer prognosis is largely determined by disease stage and tumor characteristics, such as estrogen receptor (ER) and human epidermal growth factor receptor 2 (HER2) status; however, considerable heterogeneity in disease outcome persists beyond categorization on such factors (1). As angiogenesis is critical for tumor growth (2) and inflammation can also promote cancer initiation and development (3), individual genetic variation in genes in these pathways may contribute to the variability of disease outcomes. Prior studies have reported associations between angiogenesis and inflammation related genes and breast cancer survival (4–9) but are generally limited by small sample size and/or lack of replication. Therefore, this study was undertaken in order to comprehensively evaluate genetic variants across 22 angiogenesis and inflammatory pathway genes for associations with breast cancer survival. To reduce the possibility of false positive findings, a two-stage study was undertaken in order to first identify, and then test for replication, associations with breast cancer survival. Genes evaluated included CCL2, CCL5, CCR2, COL18A1, FGFR4, FLT1, HIF1A, HPGD, IL1B, IL6, KDR, MMP1, MMP3, MMP7, MMP9, PLAU, PTGES, PTGIS, PTGS2, SERPINE1, THBS1, and VEGFA.
Subjects and Methods
Study Population
Breast cancer cases from the Shanghai Breast Cancer Study (SBCS), the Shanghai Breast Cancer Survival Study (SBCSS), and the Shanghai Women’s Health Study (SWHS) were evaluated. Study design and data collection procedures have been previously described for the SBCS (10), the SBCSS (11), and the SWHS (12, 13). Cancer diagnoses were histologically confirmed; clinical characteristics and treatment information were obtained by medical records abstraction. Breast cancer outcomes were determined by active follow up surveys and linkage with the Vital Statistics Registry database from the Shanghai Center for Disease Control and Prevention. Survival time was defined as beginning at the time of cancer diagnosis and ending at either relapse or breast cancer death for disease-free survival (DFS), any death for overall survival (OS), or else censored at the date of last contact. Approval was granted by all relevant institutional review boards; all participants provided informed consent.
Genotyping and SNP Selection
Twenty-two genes related to angiogenesis and inflammatory pathways were selected for study based on a literature review conducted at the initiation of this study. Genes included CCL2, CCL5, CCR2, COL18A1, FGFR4, FLT1, HIF1A, HPGD, IL1B, IL6, KDR, MMP1, MMP3, MMP7, MMP9, PLAU, PTGES, PTGIS, PTGS2, SERPINE1, THBS1, and VEGFA. Details on methods and quality control procedures have been previously described (13, 14). Briefly, DNA was extracted from either blood or buccal cell samples and analyzed by either of four genotyping platforms. Stage 1 genotyping was conducted by Affymetrix Targeted Genotyping for 1,062 breast cancer cases or the Affymetrix Genome-Wide Human SNP Array 6.0 for 2,918 breast cancer cases. Stage 2 genotyping was conducted with a custom-designed Illumina iSelect Beadchip for 1,613 breast cancer cases or the Sequenom iPLEX MassArray platform for 2,601 breast cancer cases. To maximize our coverage of genetic variation across genes, all genetic variants in these genes (±5 kb) that were genotyped by either of our Stage 1 genotyping platforms with minor allele frequencies (MAF) > 5% were evaluated. Variants with nominally significant Stage 1 associations with DFS or OS were evaluated for inclusion in Stage 2; only those with consistent directions of associations between DFS and OS in independent genetic loci (r2 <0.6) were selected for Stage 2.
Statistical Analysis
Analysis was limited to breast cancer cases with follow-up data available. Cox proportional hazards regression was used to evaluate associations between genetic variants and breast cancer outcomes using additive, dominant, and recessive models, with adjustment for age at diagnosis. Adjustment for study stage was included when appropriate using an indicator variable to adjust for unknown or unmeasured differences between the two study populations. Additional adjustment for disease stage and treatment (surgery, chemotherapy, radiotherapy, and tamoxifen) was also employed. Indicator variables were created for women with unknown information on these treatments. Sensitivity analyses were conducted by excluding either in situ breast cancer cases (N=192) or late stage (stages III and IV) breast cancer cases (N=698). Evaluation of the proportional hazards assumption was conducted using a test for interactions with survival times. Significance of statistical tests was based on two-tailed probability levels of 0.05; the Bonferroni correction was used to amend significance thresholds to address the issue of multiple comparisons. All analyses were performed using SAS 9.2 (SAS Institute, Cary, NC).
Results
A total of 6,983 Chinese women with breast cancer were included in the current analysis (Table 1). Stage 1 included a total of 2,884 women that were genotyped by either of our Stage 1 platforms; Stage 2 included a total of 4,099 women that were genotyped by either of our Stage 2 platforms. Stage 1 women were slightly younger than Stage 2 (means of 51.8 and 53.7 years, respectively), and were followed longer (means of 5.7 and 4.0 years, respectively). For all women, treatments included surgery (99.5%), chemotherapy (91.3%), tamoxifen (56.1%), and radiotherapy (32.0%). Of tumors with data available, 64.6% were ER positive, 59.4% were PR positive, and 29.2% were HER2 positive.
Table 1.
Clinical Characteristics of Study Population (N=6,983 Chinese Women)
| Characteristic* | Stage 1** | Stage 2** |
|---|---|---|
| Patients, N | 2,884 | 4,099 |
| Mean Follow-up Time, years | 5.7 (1.9) | 4.0 (1.4) |
| Age at Diagnosis, years | 51.8 (9.6) | 53.7 (10.1) |
| TNM Stage of Disease | ||
| 0–I | 869 (32.9) | 1,425 (37.7) |
| II | 1,477 (56.0) | 1,948 (51.6) |
| III–IV | 292 (11.1) | 406 (10.7) |
| Estrogen Receptor Status | ||
| Positive | 1,570 (65.1) | 2,527 (64.3) |
| Negative | 842 (34.9) | 1,406 (35.8) |
| Progesterone Receptor Status | ||
| Positive | 1,482 (61.6) | 2,271 (58.0) |
| Negative | 923 (38.4) | 1,647 (42.0) |
| Surgery | ||
| Yes | 2,853 (99.4) | 4,053 (99.5) |
| No | 16 (0.6) | 22 (0.5) |
| Chemotherapy | ||
| Yes | 2,646 (92.4) | 3,687 (90.5) |
| No | 218 (7.6) | 388 (9.5) |
| Radiotherapy | ||
| Yes | 883 (32.5) | 1,288 (31.6) |
| No | 1,837 (67.5) | 2,785 (68.4) |
| Tamoxifen | ||
| Yes | 1,432 (65.4) | 1,932 (50.8) |
| No | 758 (34.6) | 1,870 (49.2) |
Mean (standard error) or N (%) for each variable
Column percents may not sum to 100 due to rounding error
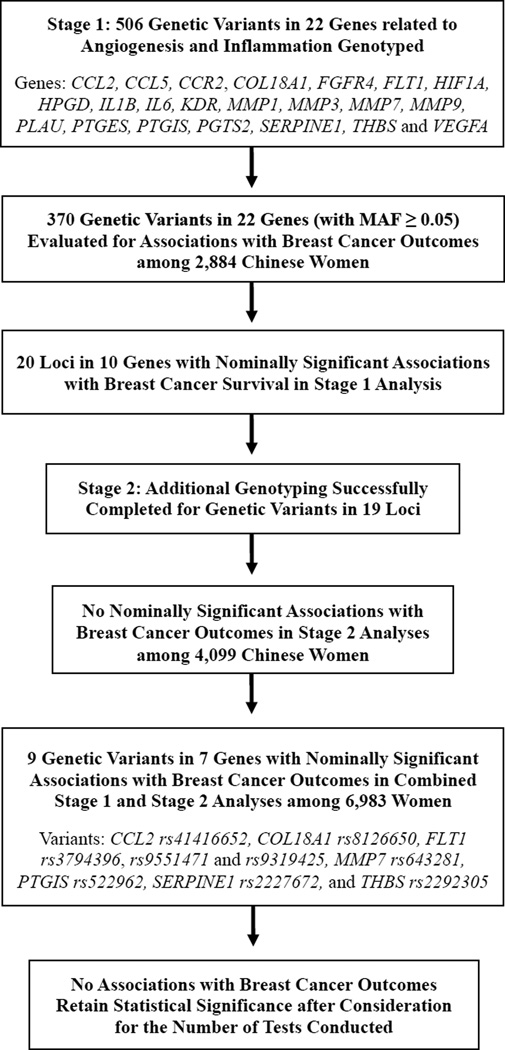
As shown in Figure 1, a total of 506 SNPs in 22 genes related to angiogenesis and inflammation were genotyped, and 370 variants with MAF ≥5% were evaluated for associations with breast cancer outcomes. Nominally significant associations with either DFS and/or OS were found for 20 loci in 10 genes in Stage 1 analyses (Table 2). Stage 2 genotyping was successful for variants in 19 loci; no significant associations with breast cancer survival outcomes were found in Stage 2 analyses. Nine variants in seven genes (CCL2 rs41416652, COL18A1 rs8126650, FLT1 rs3794396, rs9551471 and rs9319425, MMP7 rs643281, PTGIS rs522962, SERPINE1 rs2227672, and THBS rs2292305) had nominally significant associations with DFS and/or OS in analyses of the two stages combined. Bonferroni corrected P value thresholds for the total number of variants evaluated in the entire study, or just in Stage 2 are 0.00014 and 0.0026, respectively. The strongest association found was for rs8126650 and disease-free survival (P=0.008). Thus, no common genetic variants were significantly associated with breast cancer outcomes after considering the number of variants evaluated in this study.
Figure 1.
Study Overview
Table 2.
Two Stage Candidate Pathway Analysis of Angiogenesis and Inflammation Variants and Breast Cancer Survival
| Disease Free Survival (DFS) | Overall Survival (OS) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene, SNP, and Information |
Genotyping Method** |
HR (95% CI)*** | P values**** | HR (95% CI)*** | P values**** | ||||||||
| Cases / Events | Heterozygotes | Homozygotes | Allelic | Dominant | Recessive | Cases / Events | Heterozygotes | Homozygotes | Allelic | Dominant | Recessive | ||
| CCL2 rs41416652 (T/C, 44.1) | |||||||||||||
| Study Stage 1 | Affy 6.0 | 2,000 / 497 | 0.98 (0.80–1.20) | 0.86 (0.66–1.11) | 0.272 | 0.541 | 0.217 | 2,448 / 394 | 1.02 (0.82–1.27) | 0.71 (0.52–0.97) | 0.060 | 0.483 | 0.012 |
| Study Stage 2 | iSelect | 1,292 / 225 | 0.91 (0.68–1.22) | 0.78 (0.53–1.14) | 0.202 | 0.324 | 0.260 | 1,324 / 197 | 1.08 (0.79–1.48) | 0.98 (0.65–1.47) | 0.989 | 0.748 | 0.718 |
| Combined | NA | 3,292 / 722 | 0.96 (0.82–1.13) | 0.83 (0.67–1.03) | 0.106 | 0.308 | 0.092 | 3,772 / 591 | 1.05 (0.87–1.25) | 0.79 (0.62–1.01) | 0.130 | 0.736 | 0.022 |
| COL18A1 rs6518240 (A/G, 8.7) | |||||||||||||
| Study Stage 1 | Affy 6.0 | 1,999 / 496 | 0.99 (0.78–1.27) | 1.82 (0.75–4.39) | 0.667 | 0.858 | 0.183 | 2,448 / 393 | 1.04 (0.79–1.36) | 2.97 (1.32–6.66) | 0.225 | 0.464 | 0.009 |
| Study Stage 2 | iSelect | 1,301 / 226 | 1.04 (0.74–1.47) | 2.11 (0.52–8.47) | 0.587 | 0.701 | 0.299 | 1,333 / 198 | 0.95 (0.65–1.39) | 0.00 (no events) | 0.606 | 0.707 | 0.969 |
| Combined | NA | 3,300 / 722 | 1.01 (0.83–1.24) | 1.94 (0.92–4.09) | 0.472 | 0.679 | 0.081 | 3,781 / 591 | 1.02 (0.82–1.27) | 2.17 (0.97–4.85) | 0.442 | 0.655 | 0.060 |
| COL18A1 rs8126650 (G/T, 26.0) | |||||||||||||
| Study Stage 1 | Affy 6.0 | 1,895 / 453 | 0.84 (0.69–1.03) | 0.63 (0.41–0.97) | 0.013 | 0.029 | 0.072 | 2,326 / 364 | 0.80 (0.64–1.00) | 0.60 (0.37–0.99) | 0.008 | 0.015 | 0.091 |
| Study Stage 2 | Sequenom | 2,152 / 265 | 0.86 (0.67–1.11) | 0.90 (0.54–1.51) | 0.319 | 0.251 | 0.868 | 2,344 / 207 | 0.85 (0.64–1.14) | 1.19 (0.70–2.04) | 0.779 | 0.448 | 0.376 |
| Combined | NA | 4,047 / 718 | 0.85 (0.73–0.99) | 0.72 (0.52–1.00) | 0.009 | 0.013 | 0.110 | 4,670 / 571 | 0.81 (0.68–0.97) | 0.77 (0.54–1.11) | 0.015 | 0.011 | 0.325 |
| FLT1 rs3794396 (G/C, 6.1) | |||||||||||||
| Study Stage 1 | Affy 6.0 | 1,993 / 492 | 1.26 (0.97–1.63) | 3.58 (1.34–9.58) | 0.016 | 0.038 | 0.013 | 2,440 / 391 | 1.30 (0.98–1.73) | 4.04 (1.51–10.83) | 0.010 | 0.028 | 0.007 |
| Study Stage 2 | iSelect | 1,292 / 225 | 1.02 (0.69–1.52) | 0.86 (0.12–6.11) | 0.961 | 0.931 | 0.874 | 1,324 / 197 | 1.00 (0.65–1.54) | 0.00 (no events) | 0.688 | 0.844 | 0.976 |
| Combined | NA | 3,285 / 717 | 1.19 (0.96–1.48) | 2.22 (0.92–5.36) | 0.037 | 0.062 | 0.083 | 3,764 / 588 | 1.21 (0.95–1.53) | 2.03 (0.76–5.44) | 0.052 | 0.076 | 0.172 |
| FLT1 rs7326277 (T/C, 31.3) | |||||||||||||
| Study Stage 1 | Targeted | 733 / 282 | 1.12 (0.88–1.44) | 0.69 (0.45–1.08) | 0.429 | 0.883 | 0.057 | 821 / 223 | 1.12 (0.86–1.48) | 0.52 (0.30–0.90) | 0.166 | 0.834 | 0.010 |
| Study Stage 2 | iSelect | 1,298 / 225 | 0.94 (0.71–1.25) | 1.06 (0.69–1.64) | 0.998 | 0.797 | 0.676 | 1,330 / 197 | 0.94 (0.70–1.27) | 1.10 (0.70–1.74) | 0.906 | 0.854 | 0.579 |
| Combined | NA | 2,031 / 507 | 1.03 (0.86–1.24) | 0.85 (0.62–1.15) | 0.537 | 0.937 | 0.230 | 2,151 / 420 | 1.04 (0.85–1.27) | 0.77 (0.54–1.09) | 0.351 | 0.805 | 0.108 |
| FLT1 rs9551471 (A/G, 20.9) | |||||||||||||
| Study Stage 1 | Affy 6.0 | 1,991 / 497 | 0.86 (0.71–1.04) | 0.69 (0.42–1.14) | 0.045 | 0.065 | 0.204 | 2,440 / 394 | 1.00 (0.81–1.23) | 0.92 (0.56–1.53) | 0.829 | 0.902 | 0.752 |
| Study Stage 2 | Sequenom | 3,355 / 472 | 0.86 (0.71–1.05) | 0.87 (0.54–1.39) | 0.152 | 0.126 | 0.695 | 3,448 / 383 | 0.95 (0.76–1.18) | 1.02 (0.62–1.70) | 0.758 | 0.669 | 0.871 |
| Combined | NA | 5,346 / 969 | 0.86 (0.75–0.99) | 0.77 (0.55–1.09) | 0.016 | 0.019 | 0.227 | 5,888 / 777 | 0.98 (0.84–1.14) | 0.96 (0.67–1.37) | 0.728 | 0.736 | 0.858 |
| FLT1 rs9319425 (T/C, 49.8) | |||||||||||||
| Study Stage 1 | Targeted | 733 / 282 | 1.07 (0.81–1.41) | 0.89 (0.64–1.24) | 0.527 | 0.961 | 0.265 | 821 / 223 | 1.02 (0.75–1.38) | 0.66 (0.45–0.97) | 0.047 | 0.435 | 0.012 |
| Study Stage 2 | iSelect | 1,304 / 226 | 1.07 (0.78–1.48) | 1.04 (0.71–1.53) | 0.834 | 0.695 | 0.953 | 1,336 / 198 | 1.06 (0.75–1.50) | 1.01 (0.67–1.52) | 0.951 | 0.789 | 0.861 |
| Combined | NA | 2,037 / 508 | 1.07 (0.86–1.31) | 0.94 (0.73–1.21) | 0.660 | 0.814 | 0.331 | 2,157 / 421 | 1.03 (0.82–1.29) | 0.79 (0.60–1.05) | 0.116 | 0.637 | 0.035 |
| FLT1 rs9513116 (G/A, 37.4) | |||||||||||||
| Study Stage 1 | Targeted | 734 / 283 | 0.86 (0.67–1.11) | 1.00 (0.71–1.43) | 0.690 | 0.350 | 0.606 | 821 / 225 | 0.72 (0.54–0.95) | 0.81 (0.54–1.21) | 0.088 | 0.025 | 0.830 |
| Study Stage 2 | iSelect | 1,280 / 222 | 0.91 (0.68–1.21) | 1.10 (0.75–1.63) | 0.875 | 0.722 | 0.415 | 1,313 / 193 | 1.01 (0.74–1.37) | 1.15 (0.75–1.75) | 0.604 | 0.798 | 0.506 |
| Combined | NA | 2,014 / 505 | 0.89 (0.74–1.08) | 1.07 (0.82–1.38) | 0.956 | 0.424 | 0.309 | 2,134 / 418 | 0.85 (0.69–1.04) | 0.96 (0.72–1.29) | 0.436 | 0.177 | 0.717 |
| MMP1 rs1939008 (A/G, 43.3) | |||||||||||||
| Study Stage 1 | Affy 6.0 | 1,992 / 493 | 0.94 (0.77–1.15) | 1.20 (0.94–1.52) | 0.230 | 0.895 | 0.045 | 2,436 / 392 | 1.02 (0.81–1.28) | 1.16 (0.88–1.53) | 0.324 | 0.591 | 0.257 |
| Study Stage 2 | iSelect | 1,300 / 226 | 1.10 (0.83–1.47) | 0.77 (0.51–1.16) | 0.369 | 0.949 | 0.092 | 1,332 / 198 | 1.06 (0.77–1.44) | 0.77 (0.50–1.19) | 0.349 | 0.858 | 0.142 |
| Combined | NA | 3,292 / 719 | 0.99 (0.84–1.17) | 1.05 (0.86–1.30) | 0.678 | 0.932 | 0.521 | 3,768 / 590 | 1.03 (0.85–1.23) | 1.02 (0.81–1.29) | 0.843 | 0.782 | 0.978 |
| MMP1 rs470215 (A/G, 8.1) | |||||||||||||
| Study Stage 1 | Affy 6.0 | 1,996 / 497 | 1.17 (0.93–1.49) | 2.14 (1.01–4.52) | 0.046 | 0.093 | 0.053 | 2,442 / 394 | 1.14 (0.88–1.49) | 1.94 (0.80–4.69) | 0.142 | 0.215 | 0.154 |
| Study Stage 2 | iSelect | 1,291 / 225 | 1.17 (0.83–1.65) | 0.57 (0.08–4.10) | 0.556 | 0.439 | 0.562 | 1,323 / 197 | 1.18 (0.82–1.70) | 0.68 (0.10–4.89) | 0.512 | 0.424 | 0.683 |
| Combined | NA | 3,287 / 722 | 1.16 (0.96–1.42) | 1.55 (0.77–3.11) | 0.062 | 0.084 | 0.245 | 3,765 / 591 | 1.15 (0.93–1.43) | 1.43 (0.64–3.20) | 0.137 | 0.161 | 0.413 |
| MMP7 rs11568818 (T/C, 8.9) | |||||||||||||
| Study Stage 1 | Targeted | 739 / 284 | 1.02 (0.75–1.40) | 4.31 (1.37–13.55) | 0.437 | 0.666 | 0.013 | 827 / 225 | 1.04 (0.73–1.47) | 6.04 (1.92–19.05) | 0.320 | 0.567 | 0.002 |
| Study Stage 2 | iSelect | 1,289 / 223 | 0.96 (0.67–1.39) | 0.60 (0.08–4.28) | 0.699 | 0.768 | 0.614 | 1,321 / 195 | 0.91 (0.61–1.36) | 0.75 (0.11–5.39) | 0.590 | 0.607 | 0.791 |
| Combined | NA | 2,028 / 507 | 0.99 (0.78–1.26) | 1.69 (0.63–4.52) | 0.774 | 0.915 | 0.298 | 2,148 / 420 | 0.97 (0.75–1.27) | 2.25 (0.84–6.04) | 0.744 | 0.960 | 0.106 |
| MMP7 rs643281 (G/A, 9.9) | |||||||||||||
| Study Stage 1 | Affy 6.0 | 2,002 / 496 | 1.09 (0.87–1.37) | 2.35 (1.17–4.74) | 0.109 | 0.240 | 0.019 | 2,450 / 394 | 1.08 (0.84–1.38) | 1.97 (0.88–4.42) | 0.249 | 0.392 | 0.107 |
| Study Stage 2 | iSelect | 1,300 / 226 | 1.33 (0.97–1.83) | 0.92 (0.23–3.70) | 0.132 | 0.092 | 0.840 | 1,332 / 198 | 1.09 (0.77–1.57) | 1.11 (0.28–4.49) | 0.616 | 0.610 | 0.900 |
| Combined | NA | 3,302 / 722 | 1.16 (0.97–1.40) | 1.74 (0.93–3.25) | 0.032 | 0.057 | 0.101 | 3,782 / 592 | 1.08 (0.88–1.32) | 1.56 (0.78–3.15) | 0.260 | 0.360 | 0.223 |
| PTGES rs10448290 (C/T, 20.7) | |||||||||||||
| Study Stage 1 | Affy 6.0 | 1,958 / 487 | 0.93 (0.77–1.13) | 0.62 (0.37–1.02) | 0.094 | 0.224 | 0.071 | 2,406 / 385 | 0.92 (0.74–1.14) | 0.51 (0.27–0.97) | 0.065 | 0.183 | 0.047 |
| Study Stage 2 | iSelect | 1,299 / 225 | 0.99 (0.75–1.31) | 0.81 (0.38–1.73) | 0.704 | 0.819 | 0.589 | 1,331 / 197 | 1.14 (0.85–1.53) | 0.96 (0.45–2.05) | 0.557 | 0.434 | 0.813 |
| Combined | NA | 3,257 / 712 | 0.95 (0.81–1.11) | 0.67 (0.44–1.01) | 0.099 | 0.239 | 0.066 | 3,737 / 582 | 0.99 (0.83–1.17) | 0.63 (0.39–1.03) | 0.215 | 0.492 | 0.066 |
| PTGIS rs522962 (T/C, 27.4) | |||||||||||||
| Study Stage 1 | Affy 6.0 | 1,903 / 455 | 0.81 (0.67–0.99) | 0.81 (0.56–1.18) | 0.042 | 0.027 | 0.520 | 2,335 / 365 | 0.83 (0.67–1.04) | 0.87 (0.57–1.32) | 0.146 | 0.096 | 0.760 |
| Study Stage 2 | iSelect | 1,298 / 225 | 0.82 (0.62–1.09) | 1.15 (0.72–1.82) | 0.683 | 0.305 | 0.339 | 1,330 / 197 | 0.95 (0.70–1.28) | 1.32 (0.82–2.13) | 0.557 | 0.950 | 0.204 |
| Combined | NA | 3,201 / 680 | 0.82 (0.69–0.96) | 0.92 (0.69–1.24) | 0.062 | 0.018 | 0.964 | 3,665 / 562 | 0.88 (0.74–1.05) | 1.03 (0.75–1.41) | 0.455 | 0.224 | 0.599 |
| SERPINE1 rs2227672 (G/T, 8.9) | |||||||||||||
| Study Stage 1 | Targeted | 738 / 284 | 0.92 (0.67–1.25) | 3.27 (1.04–10.26) | 0.967 | 0.785 | 0.040 | 826 / 225 | 1.01 (0.72–1.42) | 4.75 (1.51–14.95) | 0.444 | 0.704 | 0.008 |
| Study Stage 2 | iSelect | 1,304 / 226 | 1.20 (0.86–1.67) | 0.66 (0.09–4.72) | 0.417 | 0.336 | 0.656 | 1,336 / 198 | 1.24 (0.87–1.77) | 1.47 (0.36–5.95) | 0.200 | 0.210 | 0.625 |
| Combined | NA | 2,042 / 510 | 1.03 (0.82–1.29) | 1.62 (0.61–4.35) | 0.601 | 0.710 | 0.339 | 2,162 / 423 | 1.10 (0.86–1.40) | 2.53 (1.05–6.14) | 0.168 | 0.289 | 0.043 |
| THBS1 rs2292305 (A/G, 32.1) | |||||||||||||
| Study Stage 1 | Targeted | 734 / 281 | 0.95 (0.75–1.21) | 0.58 (0.35–0.95) | 0.071 | 0.284 | 0.033 | 821 / 223 | 0.93 (0.71–1.21) | 0.32 (0.16–0.67) | 0.009 | 0.130 | 0.003 |
| Study Stage 2 | iSelect | 1,297 / 224 | 0.97 (0.74–1.29) | 0.97 (0.62–1.50) | 0.838 | 0.834 | 0.917 | 1,328 / 197 | 0.99 (0.73–1.33) | 1.05 (0.66–1.65) | 0.917 | 0.998 | 0.820 |
| Combined | NA | 2,031 / 505 | 0.97 (0.81–1.16) | 0.75 (0.54–1.05) | 0.159 | 0.397 | 0.096 | 2,149 / 420 | 0.96 (0.79–1.17) | 0.66 (0.45–0.96) | 0.075 | 0.292 | 0.034 |
| VEGFA rs3024994 (C/T, 5.4) | |||||||||||||
| Study Stage 1 | Affy 6.0 | 1,994 / 496 | 1.31 (1.02–1.69) | 0.67 (0.09–4.76) | 0.066 | 0.046 | 0.665 | 2,443 / 393 | 1.14 (0.84–1.54) | 0.79 (0.11–5.60) | 0.493 | 0.446 | 0.799 |
| Study Stage 2 | iSelect | 1,300 / 225 | 1.00 (0.65–1.56) | 0.00 (no events) | 0.822 | 0.920 | 0.973 | 1,331 / 198 | 0.81 (0.49–1.35) | 0.00 (no events) | 0.338 | 0.375 | 0.976 |
| Combined | NA | 3,294 / 721 | 1.23 (0.98–1.53) | 0.49 (0.07–3.48) | 0.138 | 0.094 | 0.460 | 3,774 / 591 | 1.04 (0.80–1.35) | 0.59 (0.08–4.21) | 0.901 | 0.824 | 0.597 |
| VEGFA rs3025035 (C/T, 14.6) | |||||||||||||
| Study Stage 1 | Targeted | 728 / 278 | 1.17 (0.90–1.53) | 1.12 (0.53–2.39) | 0.263 | 0.227 | 0.855 | 816 / 219 | 1.36 (1.02–1.81) | 1.64 (0.81–3.35) | 0.019 | 0.023 | 0.258 |
| Study Stage 2 | iSelect | 1,305 / 225 | 0.78(0.56–1.07) | 1.34 (0.60–3.04) | 0.358 | 0.199 | 0.391 | 1,337 / 197 | 0.81 (0.58–1.14) | 0.95 (0.35–2.58) | 0.302 | 0.249 | 0.997 |
| Combined | NA | 2,033 / 503 | 0.98 (0.80–1.20) | 1.19 (0.68–2.07) | 0.877 | 0.973 | 0.525 | 2,153 / 416 | 1.07 (0.86–1.34) | 1.29 (0.72–2.30) | 0.336 | 0.412 | 0.423 |
| VEGFA rs6905288 (A/G, 26.7) | |||||||||||||
| Study Stage 1 | Affy 6.0 | 2,000 / 497 | 0.95 (0.79–1.14) | 0.74 (0.50–1.10) | 0.183 | 0.348 | 0.158 | 2,449 / 394 | 0.92 (0.75–1.13) | 0.57 (0.35–0.94) | 0.048 | 0.168 | 0.034 |
| Study Stage 2 | iSelect | 1,300 / 226 | 0.89 (0.67–1.18) | 1.24 (0.80–1.94) | 0.842 | 0.692 | 0.232 | 1,332 / 198 | 0.93 (0.69–1.25) | 1.25 (0.78–2.02) | 0.705 | 0.915 | 0.276 |
| Combined | NA | 3,300 / 723 | 0.93 (0.80–1.09) | 0.92 (0.68–1.23) | 0.344 | 0.328 | 0.682 | 3,781 / 592 | 0.92 (0.78–1.09) | 0.81 (0.58–1.14) | 0.163 | 0.222 | 0.304 |
SNP information includes (Major / Minor allele, and minor allele frequecy) as determined by all available genotyped breast cancer cases
Genotyping Methods: Stage 1 genotyping by Affymetrix Targeted genotyping among 1,062 cases from the SBCS (Targeted) or the Affymetrix Genome Wide Array 6.0 among 2,918 cases from the SBCS (Affy 6.0); Stage 2 genotyping by Illumina iSelect Beadchip among 1,613 cases from the SBCS and SBCSS (iSelect) or by a Sequenom iPLEX platform among 2,601 cases from the SBCSS and SWHS (Sequenom)
Hazard Ratios (HR) and 95% Confidence Intervals (CI) from Cox Proportional Hazards Regression, including adjustment for age at diagnosis, and study stage when appropriate; Major allele homozygotes are referent, estimates are for heterozygotes and minor allele homozygotes
P values from tests for allelic associations (trend), dominant associations, and recessive associations (bold values denote significance at ≤ 0.05)
These analyses included a small number of in situ breast cancer cases (N=192); when excluded from analyses, nominal significance was gained for two variants (rs3794396 and rs470215). Analyses after excluding late stage (III and IV) breast cancer cases were also conducted (N=698). Nominal significance was attenuated for six variants (rs41416652, rs3794396, rs9551471, rs9319425, rs522962, and rs2227672) and gained for two variants (rs470215 and rs643281) when late stage patients were excluded. One of these associations (MMP7 rs643281 and disease-free survival) resulted in a P value of 0.0017; this surpassed our significance threshold for the number of variants evaluated in Stage 2, but not for the total number of variants evaluated in the entire study.
All regression models included adjustment for age at diagnosis, and study stage when appropriate; results were materially unaltered when additional adjustment for disease stage and treatment (surgery, chemotherapy, radiotherapy, and tamoxifen) were included. The proportional hazards assumption was evaluated for all genetic variants that were analyzed in Stage 2; all but one (MMP7 rs643281) were found to be compatible with the proportional hazards assumption.
Discussion
This large two-stage candidate pathway study comprehensively evaluated genetic variants in genes related to angiogenesis and inflammation pathways on breast cancer outcomes. Based on Stage 1 results, variants in 10 genes were selected for additional evaluation; however, no associations were replicated in Stage 2. In analyses of all women combined, nominally significant associations were found for nine genetic variants in seven genes; however, no associations retained statistical significance after considering the total number of variants evaluated.
Prior studies on germline variants in angiogenesis or inflammation related genes and breast cancer survival are limited. In one small study, an IL6 variant was associated with markers of poor prognosis and a VEGFA variant was associated with markers of favorable prognosis (4). Another small study reported an association between a VEGFA variant and reduced disease-free survival (5). A mid-sized study found no association between a variant in PTGS2 and breast cancer survival, but a significant association for an IL10 variant (6). Another mid-size study found no association between variants in MMP1, MMP2, MMP3, MMP9, and MMP13 and breast cancer survival (7), but a significant association between a SERPINE1 (PAI1) variant and worse survival (8). One larger study found no association between variants in the KDR and POSTN genes and breast cancer prognosis (9). Notably, a very large two-stage study failed to show replicated associations with breast cancer survival for the majority of nine variants previously reported to be associated with breast cancer survival, including variants in the SERPINE1, TGFB1, and VEGFA genes (15). Thus, without replication, it is likely that many of the previously reported associations with breast cancer survival may actually be false positive findings.
In addition to a two-stage study design, strengths of this study include a large sample size, genetically homogenous population (Han Chinese), and prospective investigation of disease outcomes. A limitation of this investigation is that variants in only 22 genes were evaluated; other genes related to angiogenesis and inflammation were not included in this study. However, inclusion of more genes or variants would also increase the significance threshold to account for multiple comparisons. Without consideration for adapting the significance threshold, this study, despite being very large, was somewhat underpowered to detect small effect sizes due to the low number of deaths that occurred (N=808). Given our total sample size, this study had greater than 80% power to detect an HRs of 1.20, 1.18, and 1.16 for variants with MAFs of 0.20, 0.25, and 0.30, respectively. Another limitation of this study is that only Chinese women were included; results may not be generalizable to other ethnic groups or populations.
In conclusion, this study is the first and largest two-stage candidate pathway study to examine associations between genetic variants in genes related to angiogenesis and inflammation in relation to breast cancer survival. Results indicate that common genetic variants within 22 angiogenesis and inflammation related genes (CCL2, CCL5, CCR2, COL18A1, FGFR4, FLT1, HIF1A, HPGD, IL1B, IL6, KDR, MMP1, MMP3, MMP7, MMP9, PLAU, PTGES, PTGIS, PTGS2, SERPINE1, THBS1, and VEGFA) are unlikely to play a major role in breast cancer survival among Chinese women.
Acknowledgments
The authors wish to thank the participants and research staff of the SBCS, SBCSS, and the SWHS for their contributions and commitment to this project, Regina Courtney for DNA preparation, and Bethanie Hull and Samantha Stansel for assistance with the submission of this manuscript.
Grant Support: This research was supported by grants from the NIH/NCI, including R01CA124558 and R01CA64277 (PI: W Zheng), R01CA90899 (PI: XO Shu) as well as a grant from the DAMD 170210607 (PI: XO Shu). Dr. Beeghly-Fadiel is supported in part by a grant from the NIH/NICHHD (5K12 HD043483-09; PI: K Hartmann). The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. DNA extraction and genotyping was conducted at the Survey and Biospecimen and Functional Genomic Resource Laboratory, at the Vanderbilt Microarray Shared Resource, which is supported in part by the Vanderbilt Ingram Center (P30CA68485).
Abbreviations
- CCL2
chemokine (C-C motif) ligand 2
- CCL5
chemokine (C-C motif) ligand 5
- CCR2
chemokine (C-C motif) receptor 2
- COL18A1
collagen, type XVIII, alpha 1
- DFS
disease-free survival
- DNA
deoxyribonucleic acid
- ER
estrogen receptor
- FGFR4
fibroblast growth factor receptor 4
- FLT1
fms-related tyrosine kinase 1
- HER2
human epidermal growth factor receptor 2
- HIF1A
hypoxia inducible factor 1, alpha subunit
- HPGD
hydroxyprostaglandin dehydrogenase
- IL1B
interleukin 1, beta
- IL6
interleukin 6
- IL10
interleukin 10
- kb
kilobase
- KDR
kinase insert domain receptor
- MAF
minor allele frequency
- MMP1
matrix metallopeptidase 1
- MMP2
matrix metallopeptidase 2
- MMP3
matrix metallopeptidase 3
- MMP7
matrix metallopeptidase 7
- MMP9
matrix metallopeptidase 9
- MMP13
matrix metallopeptidase 13
- OS
overall survival
- PLAU
plasminogen activator, urokinase
- POSTN
periostin, osteoblast specific factor
- PR
progesterone receptor
- PTGES
prostaglandin E synthase
- PTGIS
prostaglandin I2 synthase
- PTGS2
prostaglandin-endoperoxide synthase 2
- SBCS
Shanghai Breast Cancer Study
- SBCSS
Shanghai Breast Cancer Survival Study
- SERPINE1
serpin peptidase inhibitor, clade E, member 1 (previously known as PAI1)
- SWHS
Shanghai Women’s Health Study
- THBS1
thrombospondin 1
- TGFB1
transforming growth factor, beta 1
- VEGFA
vascular endothelial growth factor A
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest Disclosure: The authors declare that they have no potential conflicts of interest to disclose.
References
- 1.Bertos NR, Park M. Breast cancer - one term, many entities? J Clin Invest. 2011;121(10):3789–3796. doi: 10.1172/JCI57100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miller KD, Dul CL. Breast cancer: the role of angiogenesis and antiangiogenic therapy. Hematol Oncol Clin North Am. 2004;18(5):1071–1086. ix. doi: 10.1016/j.hoc.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 3.DeNardo DG, Coussens LM. Inflammation and breast cancer. Balancing immune response: crosstalk between adaptive and innate immune cells during breast cancer progression. Breast Cancer Res. 2007;9(4):212. doi: 10.1186/bcr1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith KC, Bateman AC, Fussell HM, Howell WM. Cytokine gene polymorphisms and breast cancer susceptibility and prognosis. Eur J Immunogenet. 2004;31(4):167–173. doi: 10.1111/j.1365-2370.2004.00462.x. [DOI] [PubMed] [Google Scholar]
- 5.Maae E, Andersen RF, Steffensen KD, et al. Prognostic impact of VEGFA germline polymorphisms in patients with HER2-positive primary breast cancer. Anticancer Res. 2012;32(9):3619–3627. [PubMed] [Google Scholar]
- 6.Gerger A, Renner W, Langsenlehner T, et al. Association of interleukin-10 gene variation with breast cancer prognosis. Breast Cancer Res Treat. 2010;119(3):701–705. doi: 10.1007/s10549-009-0417-y. [DOI] [PubMed] [Google Scholar]
- 7.Lei H, Hemminki K, Altieri A, et al. Promoter polymorphisms in matrix metalloproteinases and their inhibitors: few associations with breast cancer susceptibility and progression. Breast Cancer Res Treat. 2007;103(1):61–69. doi: 10.1007/s10549-006-9345-2. [DOI] [PubMed] [Google Scholar]
- 8.Lei H, Hemminki K, Johansson R, et al. PAI-1 -675 4G/5G polymorphism as a prognostic biomarker in breast cancer. Breast Cancer Res Treat. 2008;109(1):165–175. doi: 10.1007/s10549-007-9635-3. [DOI] [PubMed] [Google Scholar]
- 9.Forsti A, Jin Q, Altieri A, et al. Polymorphisms in the KDR and POSTN genes: association with breast cancer susceptibility and prognosis. Breast Cancer Res Treat. 2007;101(1):83–93. doi: 10.1007/s10549-006-9265-1. [DOI] [PubMed] [Google Scholar]
- 10.Zheng W, Long J, Gao YT, et al. Genome-wide association study identifies a new breast cancer susceptibility locus at 6q25.1. Nat Genet. 2009;41(3):324–328. doi: 10.1038/ng.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shu XO, Zheng Y, Cai H, et al. Soy food intake and breast cancer survival. JAMA. 2009;302(22):2437–2443. doi: 10.1001/jama.2009.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng W, Chow WH, Yang G, et al. The Shanghai Women's Health Study: rationale, study design, and baseline characteristics. Am J Epidemiol. 2005;162(11):1123–1131. doi: 10.1093/aje/kwi322. [DOI] [PubMed] [Google Scholar]
- 13.Long J, Cai Q, Sung H, et al. Genome-wide association study in east Asians identifies novel susceptibility loci for breast cancer. PLoS Genet. 2012;8(2):e1002532. doi: 10.1371/journal.pgen.1002532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng W, Long J, Gao YT, et al. Genome-wide association study identifies a new breast cancer susceptibility locus at 6q25.1. Nat Genet. 2009;41(3):324–328. doi: 10.1038/ng.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beeghly-Fadiel A, Zheng W, Lu W, et al. Replication study for reported SNP associations with breast cancer survival. J Cancer Res Clin Oncol. 2012 doi: 10.1007/s00432-012-1174-6. [DOI] [PMC free article] [PubMed] [Google Scholar]