Abstract
It has generally been believed that the diffusion limit set by the nuclear pore for protein is 60kDa. We here studied the cellular localization of several artificial proteins and found that the diffusion limit set by the nuclear pore is not as small as previously thought. The results indicate that the maximal size of protein to diffuse through the nuclear pore complex could be quite larger than 60kDa, thus greatly extending the diffusion limit that the nuclear pore can accommodate.
Keywords: nuclear pore, protein nuclear translocation, diffusion limit
Introduction
The nucleus is separated from the cytoplasm by a double membrane called the nuclear envelope (NE) in eukaryotes. The NE is penetrated by nuclear pore complexes (NPCs), through which the cytoplasm communicates with the nucleus and permits exchange of contents between the two organelles. With improvements in detection methods and instrumentation, the structure of the NPC has been refined over the years. It has been shown that the overall structure is similar in different species and is thus believed to be conserved in all eukaryotes[1]. The NPC is a huge structure of around 120 million Daltons in size and is constructed from ~30 different proteins that are often called nucleoporins[2-4]. The canonical feature of the NPC includes the central plug/transporter(CP/T), three rings with cytoplasmic filaments attached to the cytoplasmic ring and a basket in the nuclear sides of the NPC. The diameter of the cytoplasmic, lumenal spoke and distal ring determined by the latest studies is around 125nm, 60nm and 40nm, respectively[5].
All nuclear proteins are made in the cytoplasm and must be translocated into the nucleus, while RNA products, including transfer RNA (tRNA), ribosomal RNA (rRNA) and messenger RNA (mRNA), are transcribed in the nucleus and subsequently exported to the cytoplasm for protein synthesis. Currently, the exchange of cytoplasmic and nuclear contents is believed to mainly involve two processes: 1) passive diffusion; and 2) an active process that is coupled to energy input. Both processes are mediated through NPCs. For more than two decades, it has generally been believed that for proteins to passively diffuse through the nuclear membrane, the limit set by the NPC is about 9~12nm in diameter[6-8]. This was later interpreted to allow for the diffusion of proteins with a maximal size of 60kDa[9-14]. Alternatively, proteins may shuttle between the cytoplasm and the nucleus in an active way that is mediated by nuclear localization signals (NLSs) or nuclear export signals (NESs). These signals are specifically recognized by corresponding adaptor proteins, dubbed as importins and exportins that chaperon the transported proteins into or out of the nucleus, respectively[12,15].
We have repeatedly observed the nuclear localization of a GFP3 oligomer protein, whose size is around 90kDa. This phenomenon contradicts the long-term view that the maximal size for protein diffusion through the nuclear pore is around 60kDa and it stimulates us to revisit the diffusion limit set by the NPC. For this study, the cellular localization of three artificial chimeric proteins was investigated. The results reveal that proteins with sizes from 90 to 110kDa are allowed to diffuse through the nuclear pore, which may vary with different proteins studied. Thus, the capability of the nuclear pore for allowing macromolecules to diffuse through is greatly increased. In addition, this study does not exclude that the nuclear pore allows the diffusion of proteins that are even larger than those used in this study.
Materials and Methods
Cell culture and transfection
Human cervical cancer cell line HeLa, prostate cancer cell line DU145 and lung cancer cell line A549 were cultured in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% FBS. Human melanoma cell line Colo38 was cultured in RPMI1640 supplemented with 10% FBS. Human colon cancer cell line HCT116 was cultured in McCoy’s 5a medium supplemented with 10% FBS. All cells were cultured at 37°C in a humidified incubator containing 6% CO2. Experiments involving transient transfection were conducted with exponentially growing cells. Transfections of cells were conducted using Lipofectamine 2000 as described by the manufacturer (Invitrogen).
Plasmid constructs
The vector pEGFP-C1(from Clontech, Inc.) was used for GFP1 protein expression. The plasmid expressing GFP5 was as described previously[16]. The constructs expressing GFP2, GFP3 and GFP4 were generated by cutting the corresponding GFP portions from the parental vector for expression of GFP5 and ligating into pcDNA3.1+ vector (Invitrogen). In addition, the constructs expressing chimeric proteins based on Myc-ERK2 or Myc-PDK1 were made in the pcDNA3.1+ vector. Myc-ERK2 was created by polymerase chain reaction (PCR) using a 5′ oligonucleotide encoding the Myc-tag sequence (EQKLISEEDL) in addition to the ERK2 sequence. The resulting product was subcloned into EcoRV and Xba1 sites of the vector. Myc-PDK1 was provided by Dr. Alessi (University of Dundee, UK). All the constructs expressing chimeric proteins based on Myc-ERK2 or Myc-PDK1 were generated based on PCR method.
SDS-PAGE and Western blot
At 16 hours after transfection, cells were lysed in lysis buffer (150mM NaCl, 5mM EDTA, 50mM NaF, 1% Nonidet P-40, 1mM PMSF, 1mM Na3VO4) with brief vortexing. After incubation on ice for 30 min, the supernatants were collected by centrifugation at maximal speed for 10 min. Lysates were dissolved in 2x sample buffer (100mM Tris-HCl, pH6.8, 2% SDS, 20% glycerol, 0.03% bromophenol blue and 1.5% β-mercaptoethanol). Samples were subjected to SDS-PAGE followed by transferring to PVDF membranes. Membranes were blocked with TTBS (20mM Tris-HCl, pH7.0, 0.5M NaCl, 0.05% Tween-20) containing 5% nonfat milk for two hours. After blotting with specific primary antibodies and washing with TTBS, the membranes were then incubated with peroxidase-conjugated goat anti-mouse or anti-rabbit IgG (Jackson ImmunoResearch Laboratories, Inc) and the resulting signals were visualized by ECL detection (Amersham).
Immunofluorescence
Cells were grown on 6 well dishes containing coverslips, fixed in 2% formaldehyde/PBS at 4 °C for 1 hour, and permeabilized in 0.2% Triton/PBS for 10 min. After washing once with PBS, the cells were blocked with 10% horse serum/PBS for 40 min and washed once again with PBS. The coverslips were then incubated with 4 ug/ml primary antibody/PBS at room temperature for 1 hr followed by washing with PBS. The primary antibody against Myc tag was 9E10 from Santa Cruz Biotechnology, Inc. The samples were further blocked with rhodamine-conjugated goat anti-mouse immunoglobulin G antibody (Jackson ImmunoResearch Laboratories) for 1 hr (1:100 dilution). The coverslips were stained with 4, 6-diamidino-2-phenylindole (DAPI) and were washed twice with PBS. The coverslips were mounted on glass slides with Aquamount (Polyscience, Inc., Warrington,PA). Images were captured under fluorescent microscopy using the indicated filter with a Hamamatsu 16-bit digital camera mounted on a Zeiss Axioplan microscope using a 63x objective and processed with Adobe Photoshop software.
Results and Discussion
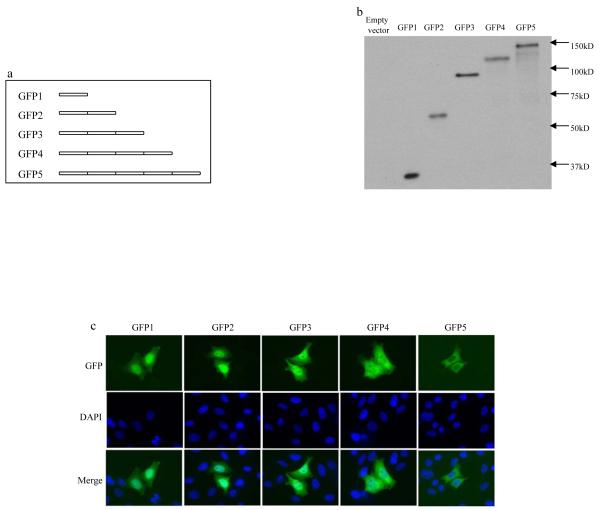
To determine the size limit for proteins that are able to diffuse through the nuclear pore, serial constructs for GFP oligomer fusion proteins were made, which include GFP1, GFP2, GFP3, GFP4 and GFP5. The GFPs are expressed in frame with each other and the number indicates the number of GFP proteins that are fused together (Fig. 1A). When these GFP fusion proteins were expressed, their molecular weights ranged from 28kDa to 140kDa (GFP1 to GFP5, respectively) (Fig. 1B). When the localization of these fusion proteins was determined in HeLa cells after transient transfection for 8 hours, it was found that GFP1, GFP2 and GFP3 were capable of translocating into the nucleus of essentially all the transfected cells, while GFP5 was predominantly localized in the cytoplasm (Fig. 1C). GFP4 protein was detected in the nucleus of most transfected cells (~90%), while it could be sparsely detected to be localized in the cytoplasm (~10%). This implied that the size of GFP4 protein is very near to or exceeds the diffusion limit set by the nuclear pore, though most of the protein is still able to “squeeze” through the nuclear pore. In addition, it was found that in the cells transfected with GFP1, GFP2 and GFP3, most of them demonstrated a higher GFP nuclear staining when compared to that of the cytoplasm (Fig. 1C). These results are consistent with previous observations that the nucleus generally demonstrates a higher fluorescent staining than the cytoplasm of the small diffusion proteins in fixed cells[17,18]. In contrast, the distribution of GFP signal in the cytoplasm was very similar to that in the nucleus of the GFP4 transfectants and the distinction of the nucleus from the cytoplasm became unclear in many cells (Fig. 1C), suggesting that protein nuclear translocation is a relative slow process and becomes inefficient as the protein size increases to that of GFP4 protein. In a separate experiment (data not shown), cells were fixed every one half hour after transfection with GFP3 or GFP4. Due to the time it took DNA to enter the cells and its subsequent transcription and translation, the GFP signal was barely detectable after 2hs transfection. Only after 2.5hrs were the cells bearing the GFP signal clearly identified in both type of transfectants. GFP3 transfectants already demonstrated obvious nuclear localization by this time, indicating the nuclear translocation of GFP3 protein is a relatively efficient process. In contrast, the nuclear translocation of GFP4 protein could only be rarely identified after 3.5hrs and most GFP4 transfectants still demonstrated a cytoplasmic localization pattern, demonstrating that the nuclear translocation of GFP4 protein is a relative slow process.
Fig. 1.
Mapping of the largest chimeric GFP protein that is allowed to diffuse through the nuclear pore. (A) Schematic representation of chimeric GFP oligomer proteins. (B) Western blot showing expression and migration of chimeric GFP oligomer proteins. Lysates were collected at 16 hours after transfection with the indicated constructs in HEK293 cells and the molecular weights of the transfected proteins were determined by Western blot with mouse antibody against GFP. (C) The cellular localization of the chimeric GFP proteins was visualized at 8 hours after transient transfection with indicated constructs in HeLa cells.
It is well established that GFP1 protein is able to diffuse into the nucleus due to its small size and GFP1 itself does not contain an NLS, otherwise GFP5 will be translocated into the nucleus too. The above results indicated that in order for protein to passively diffuse through the NPC, its size does not need to be smaller than 60 kDa as established before. In contrast, large proteins, such as GFP3 and GFP4, whose size are around 90 and 110 kDa respectively, are able to diffuse into the nucleus, though the process becomes less efficient as the protein size increases to that of GFP4.
To extend this study, additional cell lines were investigated (HCT116, colo38, DU145 and A541 cells). Interestingly, although GFP3 was still effectively translocated into the nucleus and GFP5 was predominantly localized in the cytoplasm of all the cell lines studied, the efficiency of the GFP4 nuclear translocation varied among the different cell lines. For example, it appears that the GFP4 is much poorer at diffusing through the nuclear pore in HCT116 cells than in HeLa cells and is mainly restricted to the cytoplasm. The small discrepancy of the cellular distribution pattern of GFP4 in different cell lines reflects that there is a subtle difference on the diffusion limit set by the NPCs in different cells studied, which is consistent with previous observations[7].
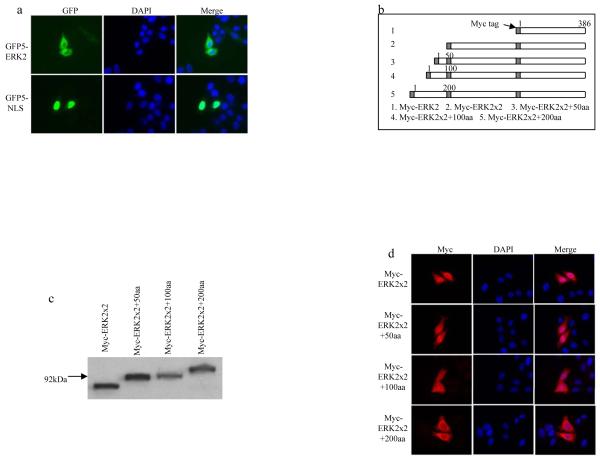
To further disprove that the nuclear pore only allows the diffusion of proteins with a maximal size of 60kD, the nuclear translocation of extracellular signal-regulated kinase 2 (ERK2) protein was studied. ERK2 is a member of mitogen-activated protein (MAP) kinase family that regulates many cellular events, such as cell proliferation and differentiation[19,20]. ERK2 has frequently been reported to be localized in the nucleus of many cell lines. Due to its small size (42kDa), it is generally believed that ERK2 can be transported into the nucleus by diffusion. Currently, it is not clear whether an additional mechanism contributes to ERK2 nuclear translocation. In recent studies of the nuclear translocation of ERK2, GFP was fused with ERK2 to generate a fusion protein larger than 60kDa (70kDa). Consequently, this fusion protein was interpreted as not being able to diffuse through the nuclear pore[21,22]. Interestingly, these studies found that the nuclear translocation of GFP-ERK2 is independent of any transport factors and ATP. Based on these results, we speculate that the underlying mechanism of GFP-ERK2 nuclear translocation is actually by diffusion. To test whether any other mechanism was responsible for ERK2 nuclear translocation, ERK2 was fused with GFP5, which is a cytoplasmic protein as shown above. If ERK2 could be transported into the nucleus via an active process, GFP5-ERK2 should be transported into the nucleus too. The results showed that the cellular localization of GFP5-ERK2 was predominantly localized in the cytoplasm. As a positive control, a classical NLS was able to concentrate GFP5 in the nucleus (Fig. 2A). Thus, the possibility that an additional mechanism was responsible for ERK2 nuclear translocation is excluded.
Fig. 2.
Mapping of the maximal size of protein based on ERK2 that is allowed to diffuse through the nuclear pore. (A) The cellular localization of the GFP5 fusion proteins was visualized at 8 hours after transient transfection with indicated constructs in HeLa cells. (B) Schematic representation of chimeric ERK2 fusion proteins. (C) Western blot showing expression and migration of chimeric ERK2 fusion proteins. Lysates were collected at 16 hours after transfection with the indicated constructs and the molecular weights of the transfected proteins were determined by Western blot with mouse antibody against Myc tag. (D) The cellular localization of the chimeric fusion proteins based on Myc-ERK2 was visualized at 8 hours after transient transfection with indicated constructs.
To further test the maximal size allowed to diffuse through the nuclear pore, serial chimeric Myc-tagged ERK2 fusion proteins were made, whereby various additional lengths of Myc-ERK2 were fused with one another at the N terminus (Fig. 2B). When expressed, the molecular weight of these chimeric proteins ranged from 86kDa to 110kDa (Fig. 2C). When the cellular localization of these proteins were studied (Fig. 2D), it was found that the Myc-ERK2×2 with a molecular weight of 86kDa was translocated into the nucleus in essentially all of the transfected cells, thus confirming that the nuclear pore allows the translocation of proteins larger than 60kDa. The extension of the fusion protein with an additional 50 amino acids(aa) (Myc-ERK2×2+50aa) did not change the nuclear localization pattern of this chimeric protein. In contrast, the addition of 100aa (Myc-ERK2×2+100aa) resulted in the restricted expression of protein in the cytoplasm in some cells (5~10%), though most protein could still be transported into the nucleus of the majority of the transfectants. The nuclear translocation efficiency was further greatly reduced in the protein containing additional 200aa (Myc-ERK2×2+200aa). These results indicated that the maximal size allowed to diffuse through the nuclear pore is between that of Myc-ERK2×2+100aa and Myc-ERK2×2+200aa (98 and 110kDa, respectively).
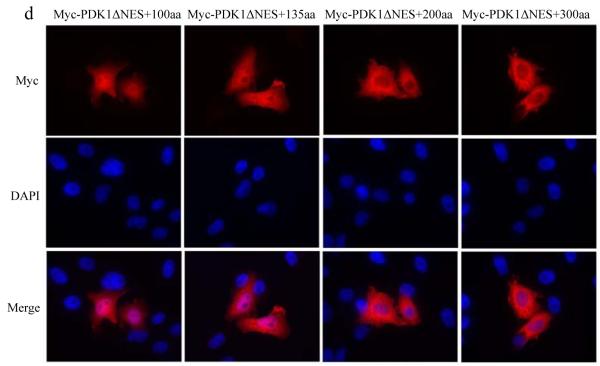
Lastly, as another confirmation that nuclear pore is able to allow the diffusion of proteins with a size of ~ 90kDa, serial artificial proteins based on 3′-phosphoinositide-dependent kinase 1 (PDK1) were made. PDK1 is a ~70kDa protein comprised of a Ser/Thr kinase domain near the N terminus and a C-terminal pleckstrin homology (PH) domain[23]. PDK1 has been proposed as a master AGC kinase that is able to phosphorylate AKT, p70 ribosomal S6 kinase, protein kinase C(S6K), serum- and glucocorticoid-stimulated protein kinase (SGK) and p90 ribosomal S6 kinase(RSK) [24]. It has been found that PDK1 is mainly localized in the cytoplasm due to the presence of a nuclear export signal. Consistent with previous results[25], PDK1 was redistributed to both the nucleus and the cytoplasm after a small deletion of this nuclear export signal in PDK1(Myc-PDK1ΔNES) (Fig. 3A). To determine whether the nuclear translocation of Myc-PDK1ΔNES is mediated by a nuclear localization signal, PDK1ΔNES was fused with the cytoplasmic protein, GFP5. It was found that GFP5-PDK1ΔNES is primarily localized in the cytoplasm, thus the possibility that PDK1 contains a nuclear localization signal was excluded (data not shown). In addition, the nuclear localization of Myc-PDK1ΔNES implies that the nuclear pore at least allows the diffusion of Myc-PDK1ΔNES protein, which is a ~70kDa protein. To further map the maximal size of protein that is based on the PDK1ΔNES and is allowed to diffuse across the nuclear pore, various lengths of the N-terminal Myc-PDK1ΔNES were fused to the N-terminus of another Myc-PDK1ΔNES (Fig.3B). These constructs expressed fusion proteins ranging from 83kDa to 120kDa (Fig. 3C). When the cellular localization of these proteins was determined (Fig. 3D), it was found that the fusion proteins containing an additional 100 or 135 amino acids to the N-terminus of Myc-PDK1ΔNES (Myc-PDK1ΔNES+100aa or Myc-PDK1ΔNES+135aa) (83kDa and 88kDa respectively), were transported into the nucleus of all the transfected cells. In contrast, the addition of 200 amino acids, which corresponds to a fusion protein of ~92kDa, restricted the fusion protein mainly to the cytoplasm and similar results were observed when additional 300 or 400 (not shown) amino acids were fused to Myc-PDK1ΔNES. These results indicated that in order to diffuse through the nuclear pore, the maximal size of the fusion proteins derived from PDK1 is between that of Myc-PDK1ΔNES+135aa and Myc-PDK1ΔNES+200aa, which are 88kDa and 92kDa respectively. Again, these results confirmed the previous observations that proteins of ~90kD could diffuse through the nuclear pore.
Fig. 3.
Mapping of the maximal size of protein based on PDK1 that is allowed to diffuse through the nuclear pore. (A) The cellular localization of the wt or mutant PDK1 protein was visualized at 8 hours after transient transfection with indicated constructs. (B) Schematic representation of chimeric PDK1 fusion proteins. (C) Western blots showing expression and migration of chimeric PDK1 fusion proteins. Lysates were collected at 16 hours after transfection with the indicated constructs and the molecular weights of the transfected proteins were determined by Western blot with mouse antibody against Myc tag. (D) The cellular localization of the chimeric fusion proteins based on Myc-PDK1ΔNES was visualized at 8 hours after transient transfection with indicated constructs. a: Myc-PDK1ΔNES+100aa; b: Myc-PDK1ΔNES+135aa; c: Myc-PDK1ΔNES+200aa; d: Myc-PDK1ΔNES+300aa.
Consequently, this study reveals that the nuclear pore allows the diffusion of protein with size of 90~110kDa, which is different from the long established view that proteins need to be smaller than 60kDa to diffuse through the nuclear pore. Due to the different structures that various proteins may adopt in vivo, there is no doubt that the maximal size of the protein that is allowed to diffuse through the nuclear pore could be different. For the same reason, currently we are not excluding the possibility that the nuclear pore allows the diffusion of proteins that are even larger than 110kDa as in this study, which have a more compact structure than those of the chimeric proteins used in this study. Another possibility is that the protein is asymmetric shaped, such as rod-shaped and therefore diffuses through the nuclear pore end on[26]. Moreover, as mentioned previously, this issue could be further complicated by that subtle differences of the nuclear pore complex may exist between cells originated from different types or species
Protein nuclear translocation has been intensively studied since 1960s. In the early period of these studies, most investigations were done by using colloidal gold particles, dextrans or the limited choices of proteins, such as insulin, ovalbumin and bovine serum albumin (BSA)[6-8,26-31]. Based on the nuclear translocation studies on colloidal gold particles and dextrans[6-8], the maximal diameter of the nuclear pore was estimated to be around 9~12nm. During this similar period, BSA (67kDa) has been frequently used in the studies of protein nuclear translocation and was reported to be very inefficiently transported into the nucleus[6,26,30,31]. Although other proteins with a similar size as BSA (60~100kDa) have seldom been tested in this early period and contradicting cases have only sporadically been reported since then[32,33], it gradually becomes a general view that the maximal size of protein to diffuse through the nuclear pore is around 60kDa. The demonstration in this study that the NPC allows diffusion of proteins of various sizes larger than 60kDa significantly extends the diffusion limit set by the nuclear pore. It suggests that many proteins do not require a nuclear localization/export signal to diffuse through the nuclear pore, which may be regarded unlikely before. Accordingly, this study may explain the observations of the carrier and energy independent nuclear translocation of some molecules, such as GFP-ERK2 (70kDa)[21,22]. On the other hand, the demonstrations of interaction between ERK2 and nucleoporins in those studies suggest that the protein diffusion through the nuclear pore could be further aided by the protein interaction with nuclear pore components.
Interestingly, although it has been frequently demonstrated that BSA is transported into the nucleus in a very poor manner, the size of BSA is estimated be around 7nm[6,30], which is still less than the maximal size (9~12nm) that is generally believed as the diffusion limit of the NPC (9~12nm). This suggests that other factors besides the protein size may contribute to BSA cytoplasmic retention. On the other hand, at the time when those studies were performed, the nuclear translocation/export mechanism of the cells was very poorly understood. For example, it has not been addressed whether these proteins contain an NES. In fact, the issue of whether the proteins contain an NLS/NES was not generally realized and was neglected in most protein nuclear translocation studies before the mid 1980s. In addition, several factors may participate in influencing protein cellular localization and contribute to the discrepancies of the diffusion size among different proteins: 1) by forming dimer or oligomer complexes between individual subunits; 2) protein compartmentalized retention caused by forming a complex with other proteins or other high order structures, such as cytoskeleton; 3) physical compatibility between nuclear pore components and the protein to be transported, such as charges and hydrophobicity. Furthermore, the cellular localization of protein may be complicated by: 1) the association with other proteins that undergo nuclear import/export; 2) the nucleus/cytoplasm-associated protein degradation.
Acknowledgements
We are grateful to Dr.Lazebnik (Cold Spring Harbor Laboratory, NY) for the GFP5 construct, Dr. Cobb (University of Texas Southwestern Medical Center at Dallas, TX) for ERK2 cDNA, and Dr. Alessi (University of Dundee, UK) for Myc-PDK1 construct. This work was supported by National Institute of Health Grant CA034432 and CA054807 (to M.G.B).
The abbreviations used are
- NPC
nuclear pore complex
- ERK2
extracellular signal-regulated kinase 2
- PDK1
3′-phosphoinositide-dependent kinase
- aa
amino acid(s)
- NLS
nuclear localization signal
- NES
nuclear export signal
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Stoffler D, Fahrenkrog B, Aebi U. The nuclear pore complex: from molecular architecture to functional dynamics. Curr Opin Cell Biol. 1999;11:391–401. doi: 10.1016/S0955-0674(99)80055-6. [DOI] [PubMed] [Google Scholar]
- [2].Reichelt R, Holzenburg A, Buhle EL, Jr., Jarnik M, Engel A, Aebi U. Correlation between structure and mass distribution of the nuclear pore complex and of distinct pore complex components. J Cell Biol. 1990;110:883–94. doi: 10.1083/jcb.110.4.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Cronshaw JM, Krutchinsky AN, Zhang W, Chait BT, Matunis MJ. Proteomic analysis of the mammalian nuclear pore complex. J Cell Biol. 2002;158:915–27. doi: 10.1083/jcb.200206106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Rout MP, Aitchison JD, Suprapto A, Hjertaas K, Zhao Y, Chait BT. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J Cell Biol. 2000;148:635–51. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Beck M, Forster F, Ecke M, Plitzko JM, Melchior F, Gerisch G, Baumeister W, Medalia O. Nuclear pore complex structure and dynamics revealed by cryoelectron tomography. Science. 2004;306:1387–90. doi: 10.1126/science.1104808. [DOI] [PubMed] [Google Scholar]
- [6].Paine PL. Nucleocytoplasmic movement of fluorescent tracers microinjected into living salivary gland cells. J Cell Biol. 1975;66:652–7. doi: 10.1083/jcb.66.3.652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Peters R. Nucleo-cytoplasmic flux and intracellular mobility in single hepatocytes measured by fluorescence microphotolysis. Embo J. 1984;3:1831–6. doi: 10.1002/j.1460-2075.1984.tb02055.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Feldherr CM. The Effect of the Electron-Opaque Pore Material on Exchanges through the Nuclear Annuli. J Cell Biol. 1965;25:43–53. doi: 10.1083/jcb.25.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Nigg EA. Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature. 1997;386:779–87. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- [10].Peters R. Fluorescence microphotolysis to measure nucleocytoplasmic transport and intracellular mobility. Biochim Biophys Acta. 1986;864:305–59. doi: 10.1016/0304-4157(86)90003-1. [DOI] [PubMed] [Google Scholar]
- [11].Silver PA. How proteins enter the nucleus. Cell. 1991;64:489–97. doi: 10.1016/0092-8674(91)90233-o. [DOI] [PubMed] [Google Scholar]
- [12].Mattaj IW, Englmeier L. Nucleocytoplasmic transport: the soluble phase. Annu Rev Biochem. 1998;67:265–306. doi: 10.1146/annurev.biochem.67.1.265. [DOI] [PubMed] [Google Scholar]
- [13].Gorlich D. Transport into and out of the cell nucleus. Embo J. 1998;17:2721–7. doi: 10.1093/emboj/17.10.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Weis K. Regulating access to the genome: nucleocytoplasmic transport throughout the cell cycle. Cell. 2003;112:441–51. doi: 10.1016/s0092-8674(03)00082-5. [DOI] [PubMed] [Google Scholar]
- [15].Gorlich D, Kutay U. Transport between the cell nucleus and the cytoplasm. Annu Rev Cell Dev Biol. 1999;15:607–60. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- [16].Faleiro L, Lazebnik Y. Caspases disrupt the nuclear-cytoplasmic barrier. J Cell Biol. 2000;151:951–9. doi: 10.1083/jcb.151.5.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Harootunian AT, Adams SR, Wen W, Meinkoth JL, Taylor SS, Tsien RY. Movement of the free catalytic subunit of cAMP-dependent protein kinase into and out of the nucleus can be explained by diffusion. Mol Biol Cell. 1993;4:993–1002. doi: 10.1091/mbc.4.10.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Wen W, Meinkoth JL, Tsien RY, Taylor SS. Identification of a signal for rapid export of proteins from the nucleus. Cell. 1995;82:463–73. doi: 10.1016/0092-8674(95)90435-2. [DOI] [PubMed] [Google Scholar]
- [19].Widmann C, Gibson S, Jarpe MB, Johnson GL. Mitogen-activated protein kinase: conservation of a three-kinase module from yeast to human. Physiol Rev. 1999;79:143–80. doi: 10.1152/physrev.1999.79.1.143. [DOI] [PubMed] [Google Scholar]
- [20].Bogoyevitch MA, Court NW. Counting on mitogen-activated protein kinases--ERKs 3, 4, 5, 6, 7 and 8. Cell Signal. 2004;16:1345–54. doi: 10.1016/j.cellsig.2004.05.004. [DOI] [PubMed] [Google Scholar]
- [21].Whitehurst AW, Wilsbacher JL, You Y, Luby-Phelps K, Moore MS, Cobb MH. ERK2 enters the nucleus by a carrier-independent mechanism. Proc Natl Acad Sci U S A. 2002;99:7496–501. doi: 10.1073/pnas.112495999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Matsubayashi Y, Fukuda M, Nishida E. Evidence for existence of a nuclear pore complex-mediated, cytosol-independent pathway of nuclear translocation of ERK MAP kinase in permeabilized cells. J Biol Chem. 2001;276:41755–60. doi: 10.1074/jbc.M106012200. [DOI] [PubMed] [Google Scholar]
- [23].Alessi DR, et al. 3-Phosphoinositide-dependent protein kinase-1 (PDK1): structural and functional homology with the Drosophila DSTPK61 kinase. Curr Biol. 1997;7:776–89. doi: 10.1016/s0960-9822(06)00336-8. [DOI] [PubMed] [Google Scholar]
- [24].Mora A, Komander D, van Aalten DM, Alessi DR. PDK1, the master regulator of AGC kinase signal transduction. Semin Cell Dev Biol. 2004;15:161–70. doi: 10.1016/j.semcdb.2003.12.022. [DOI] [PubMed] [Google Scholar]
- [25].Lim MA, Kikani CK, Wick MJ, Dong LQ. Nuclear translocation of 3′-phosphoinositide-dependent protein kinase 1 (PDK-1): a potential regulatory mechanism for PDK-1 function. Proc Natl Acad Sci U S A. 2003;100:14006–11. doi: 10.1073/pnas.2335486100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Bonner WM. Protein migration and accumulation in nuclei. In: Busch H, editor. The cell nucleus. Academic Press; New York: 1978. pp. 97–148. [Google Scholar]
- [27].Feldherr CM, Marshall JM., Jr The use of colloidal gold for studies of intracellular exchanges in the ameba Chaos chaos. J Cell Biol. 1962;12:640–5. doi: 10.1083/jcb.12.3.640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Feldherr CM. The nuclear annuli as pathways for nucleocytoplasmic exchanges. J Cell Biol. 1962;14:65–72. doi: 10.1083/jcb.14.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Horowitz SB, Moore LC. The nuclear permeability, intracellular distribution, and diffusion of inulin in the amphibian oocyte. J Cell Biol. 1974;60:405–15. doi: 10.1083/jcb.60.2.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Paine PL, Feldherr CM. Nucleocytoplasmic exchange of macromolecules. Exp Cell Res. 1972;74:81–98. doi: 10.1016/0014-4827(72)90483-1. [DOI] [PubMed] [Google Scholar]
- [31].Bonner WM. Protein migration into nuclei. I. Frog oocyte nuclei in vivo accumulate microinjected histones, allow entry to small proteins, and exclude large proteins. J Cell Biol. 1975;64:421–30. doi: 10.1083/jcb.64.2.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Stacey DW, Allfrey VG. Microinjection studies of protein transit across the nuclear envelope of human cells. Exp Cell Res. 1984;154:283–92. doi: 10.1016/0014-4827(84)90687-6. [DOI] [PubMed] [Google Scholar]
- [33].Stoffler D, Schwarz-Herion K, Aebi U, Fahrenkrog B. Getting across the nuclear pore complex: new insights into nucleocytoplasmic transport. Can J Physiol Pharmacol. 2006;84:499–507. doi: 10.1139/y06-001. [DOI] [PubMed] [Google Scholar]