Abstract
Clostridium botulinum neurotoxins (BoNTs), the most potent toxins known, disrupt neurotransmission through proteolysis of proteins involved in neuroexocytosis. The light chains of BoNTs are unique zinc proteases that have stringent substrate specificity and require exceptionally long substrates. We have determined the crystal structure of the protease domain from BoNT serotype A (BoNT/A). The structure reveals a homodimer in a product-bound state, with loop F242–V257 from each monomer deeply buried in its partner's catalytic site. The loop, which acts as a substrate, is oriented in reverse of the canonical direction for other zinc proteases. The Y249–Y250 peptide bond of the substrate loop is hydrolyzed, leaving the Y249 product carboxylate coordinated to the catalytic zinc. From the crystal structure of the BoNT/A protease, detailed models of noncanonical binding and proteolysis can be derived which we propose are also consistent with BoNT/A binding and proteolysis of natural substrate synaptosome-associated protein of 25 kDa (SNAP-25). The proposed BoNT/A substrate-binding mode and catalytic mechanism are markedly different from those previously proposed for the BoNT serotype B.
Botulism is a commonly known lethal form of food poisoning caused by exotoxins from Clostridium botulinum. These exotoxins are neurotoxins (BoNTs) and the most deadly natural toxins known (1) (estimated lethal oral dose, 70 μg; ref. 2). BoNTs represent a significant threat as a potential bioweapon because of their extraordinary toxicity and because these proteins can be aerosolized (3). Botulism is caused by the toxin's interference with neurotransmitter exocytosis. After ingestion, the toxin enters the blood stream and is transported to the neuromuscular junction where it enters the presynaptic neuron and disrupts vesicle-membrane fusion by blocking the acetylcholine exocytosis pathway (4). The resulting cessation in neurotransmitter release leads to flaccid paralysis. Although very deadly, C. botulinum toxin is increasingly being used in medicine with applications ranging from the treatment of numerous muscular disorders to the popular cosmetic treatment (a.k.a. botox) for facial wrinkles. BoNT is also an important biomedical research tool used in basic research for examining mechanisms of vesicle transport, exocytosis, and endocytosis (1, 5).
BoNTs are synthesized in vivo as 150-kDa holotoxins that are subsequently cleaved to form disulfide-linked heterodimers containing an ≈50-kDa N-terminal light chain (LC) and the ≈100-kDa C-terminal heavy chain (4). The heavy chain consists of the C-terminal receptor-binding domain, which targets the toxin to presynaptic neurons, and the N-terminal translocation domain, which induces pore formation in acidic endosomes and translocates the LC into the cytosol. BoNT LCs are zinc proteases known to cleave proteins of the soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) complex which are critical for neurotransmitter exocytosis (1).
BoNT LCs belong to the thermolysin family of zinc proteases but are unique among zinc proteases due to their stringent specificities and their requirement for long substrates. BoNT LC minimum substrates range from 16 to >50 aa in length, depending on the isotype (6–9). In comparison, other zinc proteases can hydrolyze peptides as short as two amino acids. BoNT LC proteases are highly specific for their natural substrates but are tolerant of side-chain substitutions near the scissile bond in all but the amino acid adjacent to the scissile bond on the C-terminal side (the Pl′ position) (10, 11). The tolerance for substitutions at sites other than P1′ and the requirement for large substrates have led to two hypotheses: either the toxin stabilizes some transient substrate conformation before cleavage involving enzyme residues away from the catalytic site (7), or one or more cooperative subsites, or exosites, exist where the enzyme interacts with substrate remote from the catalytic site (7, 12). In support of the second hypothesis, SNARE secondary recognition (SSR) sequences have been found in all SNARE proteins and have been implicated as potential recognition sites for these exosite interactions (13).
BoNT serotype A (BoNT/A) may be unique among BoNTs. BoNT/A has been shown to cleave peptides as short as 16 aa that lack an SSR sequence. In addition, BoNT/A is very specific for R in the P1′ position, unlike other zinc proteases that require large hydrophobic residues in this position (14). To gain further insight into the structural basis underlying the unique properties Clostridium toxin zinc proteases (BotNT/A, in particular), such as stringent substrate specificity and long recognition motifs, we have determined the crystal structure of the recombinantly expressed LC from BoNT/A.
Methods
Expression and Purification. The stabilized, truncated (less 8 Nterminal and 32 C-terminal residues), and catalytically active BoNT/A LC has been cloned in pET30, overexpressed, and purified as described (15). The truncated form was used because of its enhanced stability and solubility while remaining catalytically active (16).
Crystallography. Rectangular crystals (0.5 × 0.25 × 0.25 mm) were grown by using hanging drop vapor diffusion (17) at 22°C in 6-μl drops, composed of 4 μl of protein solution [5 mg/ml LC/0.1% Triton X-100/25 mM Tris, pH 8.0/10% (vol/vol) glycerol/1 mM 2-mercaptoethanol] and 2 μl of well solution {Hampton Research Crystal Screen I, reagent 20 [0.2 M ammonium sulfate/0.1 M sodium acetate pH 4.6/25% (wt/vol) PEG 4000] plus 3% xylitol}, suspended over a 1-ml reservoir. X-ray data were collected on beamline 5.0.2 at the Advanced Light Source with an ADSC Quantum 4 CCD x-ray detector. Intensities were integrated and scaled with mosflm (18) and scala (19, 20), respectively. Crystals belong to spacegroup P21, with 53% (VM = 2.91) solvent (two molecules per asymmetric unit) and a native Patterson peak revealed a two fold noncrystallographic symmetry axis parallel to b. The structure was determined by molecular replacement with epmr (21) by using BoNT/A LC coordinates from the intact BoNT/A (3BTA) as the search model. Models were iteratively rebuilt into subsequent Shake&wARP maps (22) with xtalview (23) and refined with refmac5 (24) by using strong NCS restraints. The final model is missing residues 1–6, 199–206, and 393 from each monomer and residue 250 from molecule A of the dimer. Data collection and refinement statistics are summarized in Table 1. Further details are provided in the header of Protein Data Bank ID code 1E1H. Both coordinates and structure factors have been released.
Table 1. Data collection and refinement.
| Space group | P21 |
| Wavelength, Å | 1.100 |
| a, Å | 58.06 |
| b, Å | 94.26 |
| c, Å | 100.16 |
| β, ° | 103.52 |
| Resolution, Å | 1.8 |
| Unique* reflections | 93168(6918) |
| Redundancy | 3.3(1.9) |
| Rsym, %*† | 4.2(38.3) |
| <I/σ1>* | 7.9(1.7) |
| Molecules/asymmetric unit | 2 |
| Vm (Matthews coefficient) | 2.9 |
| R factor, % | 19.6 |
| Free R factor, % | 23.7 |
| Protein atoms | 6460 |
| Water atoms | 786 |
| rms deviation bond length, Å | 0.02 |
| rms deviation bond angle, ° | 1.85 |
| Coordinate error‡ | 0.127 |
| RSCC§ | 0.92 |
Values in parentheses for highest resolution bin (1.9-1.8 Å)
Rsym = Σj(Σj Ii - <Ii>)/Σi <Ii>
Estimated standard uncertainty, based on free R
Real space correlation coefficient against Shake&wARP map
Results and Discussion
Overall Structure of the BoNT/A Homodimer. The crystal structure of the recombinant BoNT/A LC reveals a homodimer with an extensive intermolecular interface (Fig. 1) burying ≈4,500-Å2 surface area of each molecule. With the exception of a few disordered solvent-exposed residues, the two copies of the molecule in the asymmetric unit (although packed differently) are identical (rms deviation = 0.16 Å on 396 Cα) for purposes of the following discussion. All secondary structural elements present in the BoNT/A LC crystal structure of the holotoxin (25, 26) are preserved in the structure of the recombinantly expressed BoNT/A LC (Fig. 2).
Fig. 1.
The BoNT/A LC dimer. The ribbon diagram of BoNT/A LC is shown overlaid with the translucent molecular surface. One monomer of the non-crystallographic dimer (Left) is shown with blue helices and orange β-strands; the second monomer (Right) is shown with cyan helices and red β-strands; the surface is colored by electrostatic potential (blue positive and red negative). This figure was generated with molscript (18), spock (34), and raster3d (35).
Fig. 2.
Comparison of BoNT/A LC structures from holotoxin and homodimer. (a) The superposition of the recombinant BoNT/A LC (red and light red) with the BoNT/A LC from holotoxin (25) (blue and light blue) is shown. Regions of greatest difference are highlighted with more saturated colors. This figure was generated with midas (36). (b) The molecular surface of BoNT/A LC from holotoxin is shown looking into the substrate-binding cleft. The variable loops are mapped to the surface by color (50s and 60s loops are combined and yellow, 160s loop magenta, 250s loop red, and catalytic loop cyan). (c) The molecular surface of BoNT/A LC from the homodimer is shown with the molecule in nearly the same orientation as in b and with the same color scheme. This figure was generated with spock (34) and raster3d (35).
A four-stranded antiparallel β-sheet at the center of the homodimer is formed between the 250s loops (F242–V257) exchanged between the dimer partners, and this arrangement contributes a major portion of the dimer interface by burying an extensive surface area of ≈1,000 Å2. Each 250s loop extends deeply into a cleft leading to the catalytic site of the homodimer partner, where the Y249–Y250 peptide bond of each loop is hydrolyzed and the resulting Y249 carboxylate remains coordinated to the catalytic zinc. The observed product-bound state shows that the BoNT/A 250s loop serves as substrate for BoNT/A LC catalytic autolysis.
Enzymatic autolysis of BoNT/A LC has been reported (27) and, to demonstrate that autolysis observed in the crystal structure does indeed occur in vitro, the following experiments were carried out. Recombinant BoNT/A LC solutions were buffered to pH 7.5, 6.5, 5.5, and 4.5, incubated at 4°C for 72 h, and assayed by SDS/PAGE. Incubation at pH 7.5 yielded a single 50-kDa species, whereas at pH 6.5 to 4.5 two additional bands (≈26 and ≈20 kDa), becoming the dominant species at the lowest pH, were observed. Dissolving a crystal of the recombinant BoNT/A LC, crystallized at pH 4.6, with SDS and 2-mercaptoethanol yielded bands of the same size observed in the in vitro incubation experiments. These results demonstrate that the stabilized, truncated BoNT/A LC is acid-labile and that the breakdown is due to enzymatic autolysis.
Substrate Loop-Binding Cleft. Extension of the 250s substrate loop from each monomer into the catalytic cleft of the other molecule is concomitant with large rearrangements of several loops surrounding the catalytic site. The 50s, 170s, 200s, 250s, and 370s loops (Fig. 2) that form the boundaries of a large cleft in the enzyme surface have all undergone significant shifts compared with BoNT/A LC in the holotoxin. The 250s loop forming the substrate-binding cleft has moved ≈12 Å at its end and ≈8 Å near its middle and above the mouth of the cleft. The 370s loop (also referred to as the catalytic loop) undergoes a small shift, and the 170s loop relaxes to a position partially filling the end of the substrate-binding cleft. The net effect of all these rearrangements is to widen the cleft and to shorten it slightly. The loop rearrangements are likely due in part to the loss of interactions between the LC and the hydrophobic belt of the translocation domain (25). The 200s loop, which is packed against the back of the translocation domain in the holotoxin, is mostly disordered in the homodimer LC structure. The residues of the zinc-binding motif (HEXXH 222–226 and E261), the residues comprising the base of the cleft, and the entire periphery of the molecule remain essentially unaltered.
The substrate-binding cleft bounded by the mobile loops is very large with a narrow inlet just above the catalytic loop (Fig. 2 B and C). The cleft is ≈20 Å long and increases in width from ≈9 Å at the inlet above the catalytic loop to >12 Å at the back near the catalytic zinc. The cleft varies in depth from ≈9 Å at its shallowest point at the inlet to nearly 20 Å below the catalytic zinc site. Nearly all residues lining the basin of the cleft are hydrophobic, with the notable exceptions of the zinc ion, the zinc-binding motif, and conserved R362 near the bottom of the cleft.
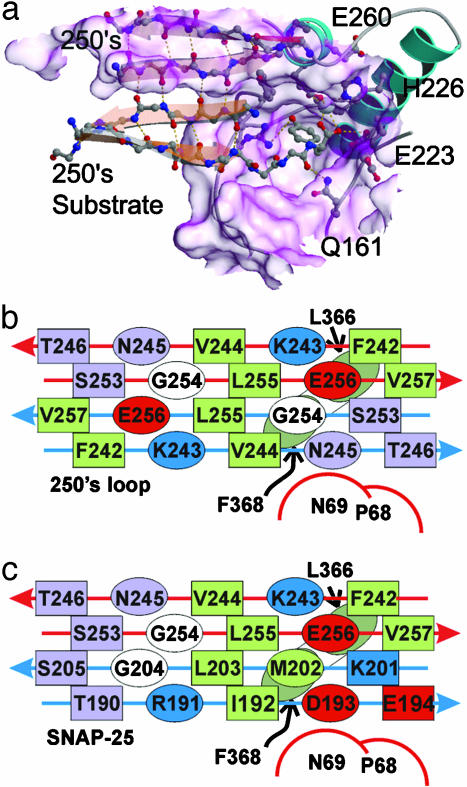
Interactions Between the 250s Loop and the Substrate-Binding Cleft. The 250s loop bound to the substrate-binding cleft is held in place by extensive interactions. The Y249–Y250 bond has been hydrolyzed and the product Y249 carboxylate remains coordinated with the catalytic zinc. The side chain of Y249 in the P1 position points toward the base of the cleft near the deepest point and forms hydrogen bonds with E350 and R362. The four residues opposite the hydrolyzed scissile bond (P1′–P4′) have moved away from the catalytic zinc and have begun leaving the substrate-binding cleft. The remainder of the substrate loop bound to the cleft (K243–Y249, G254–V257) adopts a β-conformation and forms half of a four-stranded intermolecular β-sheet within the homodimer (Fig. 3A).
Fig. 3.
Substrate binding. (a) The substrate-binding cleft and active site region of the homodimer are shown for one monomer occupied by the 250s loop from the second monomer. Nonspecific hydrogen bonds and zinc coordination are highlighted with red dashes. β-Strands are shown superimposed with 250s loop and pseudosubstrate backbone atoms to emphasize secondary structure interactions. This figure was generated with molscript (18), spock (34), and raster3d (35). (b) A schematic diagram of the interaction between the enzyme and pseudosubstrate shows the distribution of hydrophobic, hydrophilic, and charged residues in the intermolecular β-sheet, the catalytic loop, and the 60s loop. Ovals represent residues pointing out of the page, squares represent residues pointing into the page; green residues are hydrophobic, blue are positive, red are negative, and light purple are hydrophilic uncharged. (c) A schematic diagram similar to the figure in b shows the hypothetical arrangement of residues for the natural substrate (SNAP-25) mapped on to the intermolecular β-sheet observed in the homodimer. The sequence was mapped by aligning the scissile bonds.
Most of the H-bond interactions at the homodimer interface result from the intermolecular β-sheet backbone H-bond network. There are only four side chain–main chain hydrogen-bonding interactions (T246 γO-N367 N, N247 δN-D369 O, N247 δO-V169 N, and N245 δN-Q66 O), and only three specific side chain–side chain interactions (S253 γO-Y365 ηO, Y249 ηO-R362 ηN, and E256 εO-E251 εO). The homodimer is also stabilized by an extensive network of intermolecular van der Waals interactions. The bottom of the 250s loop presents a hydrophobic face that interacts with a cluster of hydrophobic residues at the base of the substrate-binding cleft thus forming a large hydrophobic nucleus at the inlet to the cleft (Fig. 3B). Binding does not depend on SSR sequence exosite cooperative binding, because the 250s loop does not contain an SSR sequence. This observation, along with existing literature describing turnover of truncated substrate lacking an SSR sequence (14), suggests, for at least this BoNT serotype, that SSR sequence exosite cooperativity is not necessary for substrate binding and hydrolysis. However, further stabilization of the BoNT/A substrate complex through SSR sequence exosite binding cannot be ruled out for the full-length substrate.
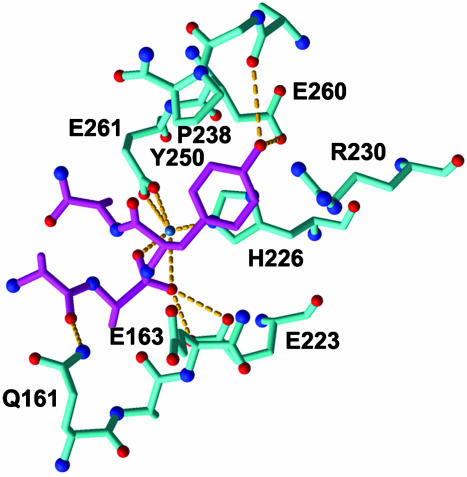
The S1′ Specificity Pocket. A notable pocket is in the wall of the substrate-binding cleft just above the catalytic zinc. This pocket is formed between the 170s loop and the base of the 250s loop. Aside from the cavernous pocket at the base of the cleft occupied by the Y249 side chain, this is the only large, pocket-like feature on the surface of the substrate-binding cleft. The location of this pocket suggests that it might bind the P1′ side chain and may be the S1′ specificity-determining pocket. Before hydrolysis of the Y249–Y250 peptide bond, Y250 would have to be at least proximal to this pocket and the pocket provides good shape and chemical complimentarity for a tyrosine side chain. Additional stabilizing H-bonding and π-stacking interactions are also possible (Fig. 4).
Fig. 4.
The S1′ specificity pocket. A ball-and-stick representation of a modeled interaction between P1′ and S1′ for the homodimer with Y250 in the proposed S1′ pocket. Hydrogen bonds and ionic interactions are highlighted with yellow dashes. This figure was generated with midas (36).
Hydrolysis of the Substrate Loop. The observation of BoNT/A LC in its product-bound state provides detailed information about the mechanism of BoNT/A LC hydrolysis of the 250s loop. Although the substrate-binding clefts are very different, a remarkable similarity exists between the catalytic residues in the BoNT/A product-bound state and thermolysin complexes (28) with product analogues (Fig. 5A). The zinc-coordinated residues and catalytic glutamate superimpose nearly identically and BoNT/A Q161 is in a similar location to thermolysin N112, which is important for stabilizing reaction intermediates.
Fig. 5.
Catalytic mechanism. (a) A stereo view of thermolysin residues [yellow, with side chain O atoms, red, and side chain N atoms, blue; PDB 1TMN (28)] involved in catalysis is shown superimposed on the homologous residues from BoNT/A (shown in cyan). P1 and P2 of the cleavage product from pseudosubstrate are also shown with the product carboxylate coordinated to the catalytic zinc. BoNT/A LC Y365 (magenta) is shown in the conformation from holotoxin. This figure was generated with midas (36). (b) A schematic illustration of the proposed catalytic mechanism of 250s loop hydrolysis in the BoNT/A homodimer is shown. This figure was generated with isis draw.
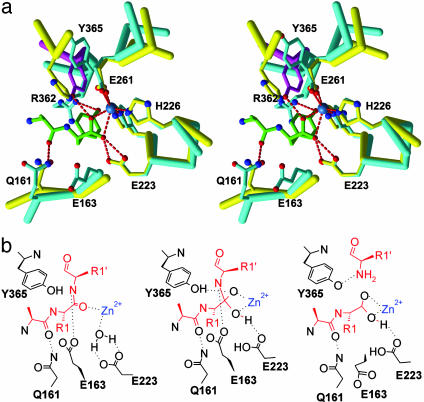
In the mechanism we propose, E223 of BoNT/A acts as the general base (like E143 of thermolysin) initiating BoNT/A-catalyzed proteolysis by creating a nucleophilic hydroxyl which attacks the polarized carbonyl of the scissile bond. In thermolysin (28) the activated complex is stabilized by an extensive network of interactions (zinc, H231 δN, Y157 ζO, and E143 interact with oxygens of the scissile bond tetrahedral carbon, and N112 δO and A113 O accept hydrogen bonds from the scissile nitrogen). BoNT/A appears to have nearly as extensive an arrangement of hydrogen-bonding donors and acceptors that stabilize its activated complex, even though the scissile bond is in reverse of the canonical orientation and the details of the interactions are different.
In contrast to thermolysin N112, which directly stabilizes a reaction intermediate by H-bonding with the scissile amide, BoNT/A Q161, in a position similar to thermolysin N112, cannot hydrogen bond with either the carbonyl or amide of the scissile peptide bond because the substrate is bound in the reverse orientation. Q161 can, however, stabilize reaction intermediate complexes by hydrogen bonding to the carbonyl oxygen of P2. Another BoNT/A LC residue, E163, appears to be ideally positioned to stabilize the reaction intermediates by hydrogen bonding with the backbone nitrogen of the scissile bond (Fig. 5A).
With substrate bound in the noncanonical orientation, a different proton shuttle for promoting the leaving group is also needed. For thermolysin, the catalytic base that initiates hydrolysis, E143, accepts a hydrogen bond from the protonated oxygen resulting from nucleophilic attack and shuttles a proton to the scissile nitrogen to promote the leaving group. With substrate in the reverse orientation, E223 in the BoNT/A LC is not close enough to the leaving amide to serve this dual role. The strictly conserved residue Y365, although >8 Å away from the scissile bond in the homodimer structure, would be positioned within hydrogen-bonding distance of the product carboxylate with a simple χ1 torsion, which, in fact, brings the tyrosine side chain to the conformation observed in the BoNT/A LC structure from holotoxin (Fig. 5A). In this position, Y365 may act as the proton shuttle to protonate the amide nitrogen and promote the leaving group. Conserved BoNT serotype B (BoNT/B) R369 was previously proposed to stabilize the tetrahedral intermediate through interaction with the scissile bond carbonyl (26). Homologous BoNT/A R362 adopts a nearly extended conformation pointing toward the catalytic site and is still ≈8 Å away from the product carboxylate. R362 does hydrogen bond to the Y365 backbone carbonyl, possibly stabilizing or anchoring the pivot point of Y365 for catalysis.
BoNT/A LC Binding and Hydrolysis of Synaptosome-Associated Protein of 25 kDa (SNAP-25). The BoNT/A LC homodimer structure reveals considerable insight into the structure–function relationship of BoNT/A LC autolysis of the 250s loop, and a number of details indicate that the structure also provides a useful model for binding and hydrolysis of its natural substrate SNAP-25 by the BoNT/A LC. Although it becomes apparent on aligning the scissile bond of the 250s loop and SNAP-25 (Table 2) that the sequences are very different, the pattern of hydrophobic and hydrophilic residues is similar between the two sequences (Fig. 3 b and c). This similarity may be sufficient to allow SNAP-25 binding to BoNT/A, because most of the intermolecular interactions are nonspecific, consistent with BoNT/As tolerance of substrate side-chain substitutions (11). Examining the SNAP-25 sequence mapped on to the intermolecular β-sheet in the homodimer (Fig. 3c) suggests that the SNAP-25 sequence is compatible with the formation of an intermolecular β-sheet with BoNT/A. In fact, SNAP-25 could provide several additional stabilizing side chain–side chain interactions not present in the homodimer complex. SNAP-25 R191, for example, could form a salt bridge with E256 and SNAP-25 D193 could interact with N69 or form a salt bridge with nearby K243. SNAP-25 E194 and K201 would both point into the cleft and would be juxtaposed such that they could form an intramolecular salt bridge, further stabilizing the β-sheet.
Table 2. SNAP-25 alignment with 250s loop.
| SNAP-25 | |||||||||||||||||
| 1 | 9 | 0 | 2 | 0 | 0 | 2 | 0 | 5 | |||||||||
| T | R | I | D | E | A | N | Q | R | A | T | K | M | L | G | S | ||
| F | K | V | N | T | N | A | Y | Y | E | M | S | G | L | E | V | ||
| 2 | 4 | 2 | 2 | 5 | 0 | 2 | 5 | 7 | |||||||||
| 250s loop | |||||||||||||||||
Aside from the intermolecular β-sheet formation, the insertion of the P1′ residue side chain into the S1′ pocket appears to be the primary substrate-binding interaction, and it may be the key factor that determines specificity. Previous studies have reported that substitutions at the P1′ position greatly reduce BoNT/A LC catalysis, unless P1′ is either tyrosine or arginine (10). The proposed S1′ specificity pocket described above is compatible with binding of Y250 before hydrolysis in the homodimer complex (Fig. 4). Arginine in the P1′ prime position of SNAP-25 would fill this pocket with even greater complementarity, presumably forming a strong buried salt bridge with E260 and a hydrogen bond with the backbone of I236.
Substrate Requirements for Binding to BoNT/A LC. The substrate-binding clefts for the BoNT proteases are markedly different from the binding site of thermolysin, implying, along with the BoNT LCs extraordinary substrate specificities, a markedly different substrate–enzyme interaction for BoNT zinc proteases compared with thermolysin. The binding cleft of thermolysin is narrow and shallow and traverses the length of the enzyme on one face, accommodating substrates in an extended conformation (28). In contrast, the substrate-binding cleft of the BoNT/A LC is wide and deep and traverses approximately half the length of the enzyme. A steep wall also exists at the end of the substrate-binding groove, where the catalytic zinc is embedded. The shape of the BoNT/A cleft prevents peptide substrates from laying across the active site in an extended conformation. Similarly, the binding cleft of the serotype B LC also has a deep binding cavity (discussed below) that would require a peptide to assume a loop- or turn-like conformation to reach the catalytic site (26).
From our analysis of the BoNT/A LC substrate loop binding, details have emerged that allow us to postulate a set of guidelines for BoNT/A LC protease–substrate interaction. Based on our analysis of the BoNT/A LC homodimer, we propose the following: A substrate should be able to adopt a β-sheet conformation to form an intermolecular β-sheet with the 250s loop; the scissile bond must be presented to the catalytic zinc at the end of a β-turn; the amino acid at the P1′ position must complement the S1′ pocket; and a hydrophobic cluster of residues must exist at the appropriate distance from the scissile bond to complement the hydrophobic residues at the base of the cleft near the inlet. Such a model for substrate binding is compatible with the hypothesis (12) that BoNT LCs stabilize a transient substrate conformation through interactions with LC residues distant from the catalytic site that contribute to stabilization. This mode of substrate binding does not rely on cooperative exosite stabilization.
A Proposed Model for General C. botulinum Toxin Substrate Binding. The interesting question arises whether the postulated rules for substrate binding to BoNT/A LC can be extended to other serotypes and thus serve as a general model for clostridial neurotoxin zinc protease binding.
Comparison of BoNT/A LC with the structure of the BoNT/B LC (26) shows that the structural elements we propose to be important for substrate binding are still present. The distribution of chemical properties, the approximate shape, and the approximate size of the binding cleft are similar. Both substrate-binding clefts are pliable and large (wide and deep) compared with other known zinc proteases and, as with BoNT/A, the BoNT/B 250s loop adopts a β-sheet conformation, at least for a short section nearest the 170s loop. As with BoNT/A, the BoNT/B 250s loop undergoes a large movement on dissociation from the holotoxin to expose an inlet to the binding cleft. In addition, both proposed S1 pockets for BoNT/B and BoNT/A are very large, deep, hydrophilic, and likely promiscuous. As in BoNT/A, a large pocket exists in the wall of the BoNT/B substrate-binding cleft just between the 170s loop and the base of the 250s loop. This pocket is similar in size to the proposed S1′ pocket for BoNT/A, but it has different chemical characteristics, consistent with differing BoNT/B specificity for P1′ side chains. The lysine residue at the back of the BoNT/B LC P1′ pocket, for example, would disfavor binding of the arginine of the scissile bond of the BoNT/A substrate SNAP-25. If the synaptobrevin-II (Sb2) peptide substrate were bound to the BoNT/B LCs in the conformation we propose, an SSR sequence would be appropriately positioned to interact directly with the BoNT/B substrate-binding cleft.
Although the structural features of the BoNT/A and BoNT/B LC binding clefts are remarkably similar and indicate that our “substrate rules” are compatible with at least the B serotype LC structure, our postulated substrate-binding model differs substantially from a previously proposed model for BoNT/B LC substrate binding (26). The previous model is based on a structure derived by flash-soaking of BoNT/B crystals with the 38-residue Sb2 peptide and suggests that Sb2 is bound in a random coil conformation containing a hairpin at the scissile bond. This model of BoNT/B-Sb2 interaction includes exosite stabilization and purports to explain SSR sequence cooperative binding by induced fit recognition by means of the 50s and 200s loops of the SSR-containing portion of Sb2.
Although it appears difficult to reconcile the differences between the two proposed models, the source of the disagreement may be one of any of a number of factors. Significant yet unrecognized serotype differences might exist, despite the high overall structural similarities between the two isoforms. One can also concede that the BoNT/A LC–250s loop homodimer complex may differ markedly from the BoNT/A SNAP-25 complex, which would however lead to the otherwise unsupported hypothesis that BoNT/A uses at least two substantially different mechanisms to bind and hydrolyze proteins. We have previously pointed out (29) and further supported (22) that, based on experimental evidence, the Sb2 peptide in the BoNT/B LC structure must have extraordinarily low occupation; and low occupation and a possible mixture of states have been conceded by the authors (30). We thus conclude that, likely due to the nature of the rapid-flash-soaking experiments, we do not have a clear picture of Sb2-BoNT/B LC interactions. The observation that the BoNT/B LC structure is essentially identical in the native and complexed forms is also consistent with the native structure being the predominant form in the reported BoNT/B-Sb2 cocomplex structure. Although small inhibitor complex structures of BoNT/B have been published (31–33), further structural studies of the BoNT LCs in complex with their natural substrates will be required to progress toward a full understanding of BoNT toxicity by proteolysis of their specific SNARE target peptides.
Conclusions
Most of the recombinant BoNT/A LC structure superimposes well with the LC as determined in the holotoxin. The most notable differences are several loops, proximal to the catalytic site, which undergo significant rearrangements. Comparison of holotoxin-associated and free LC of BoNT/B reveals that many of the same loops also rearrange, even in the absence of substrate. Thus, it is likely that the most substantial rearrangements occur on dissociation of the LC from the rest of the toxin. Both substrate specificity and the unique requirement of large polypeptide substrate for BoNT zinc proteases appear to result from recognition of a large strand-turn-strand secondary structure motif. Similarity between BoNT/A and BoNT/B substrate-binding clefts suggests such a secondary motif recognition may be a general feature of substrate binding for the BoNT zinc proteases, and that the differential serotype substrate selectivity is imparted by relatively few side-chain specificity-binding pockets, in particular, the S1′ pocket. The catalytic mechanism is similar in detail to the mechanism proposed for thermolysin. Although the scissile bond is in reverse orientation, the main catalytic components are conserved or homologous between BoNTs and thermolysin. With the scissile bond reversed, however, the catalytic glutamate of BoNT/A LC cannot act as a proton shuttle to protonate the leaving group as is the case for thermolysin. Instead, the conserved Y365 in BoNT/A is proposed to protonate the leaving group during catalysis.
Acknowledgments
This work was supported by U.S. Department of Energy at Lawrence Livermore National Laboratory Contract No. W-7405-ENG-48.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: BoNT, Clostridium botulinum neurotoxin; BoNT/A, BoNT serotype A; BoNT/B, BoNT serotype B; SNARE, soluble N-ethylmaleimide-sensitive factor attachment protein receptor; SSR, SNARE secondary recognition; LC, light chain; SNAP-25, synaptosome-associated protein of 25 kDa.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 1E1H).
References
- 1.Lalli, G., Bohnert, S., Deinhardt, K., Verastegui, C. & Schiavo, G. (2003) Trends Microbiol. 11, 431-437. [DOI] [PubMed] [Google Scholar]
- 2.Whitby, M., Street, A. C., Ruff, T. A. & Fenner, F. (2002) Med. J. Aust. 176, 431-433. [DOI] [PubMed] [Google Scholar]
- 3.Arnon, S. S., Schechter, R., Inglesby, T. V., Henderson, D. A., Bartlett, J. G., Ascher, M. S., Eitzen, E., Fine, A. D., Hauer, J., Layton, M., et al. (2001) J. Am. Med. Assoc. 285, 1059-1070. [DOI] [PubMed] [Google Scholar]
- 4.Montecucco, C. & Schiavo, G. (1995) Q. Rev. Biophys. 28, 423-472. [DOI] [PubMed] [Google Scholar]
- 5.Turton, K., Chaddock, J. A. & Acharya, K. R. (2002) Trends Biochem. Sci. 27, 552-558. [DOI] [PubMed] [Google Scholar]
- 6.Shone, C. C., Quinn, C. P., Wait, R., Hallis, B., Fooks, S. G. & Hambleton, P. (1993) Eur. J. Biochem. 217, 965-971. [DOI] [PubMed] [Google Scholar]
- 7.Cornille, F., Martin, L., Lenoir, C., Cussac, D., Roques, B. P. & Fournie-Zaluski, M. C. (1997) J. Biol. Chem. 272, 3459-3564. [DOI] [PubMed] [Google Scholar]
- 8.Shone, C. C. & Roberts, A. K. (1994) Eur. J. Biochem. 225, 263-270. [DOI] [PubMed] [Google Scholar]
- 9.Yamasaki, S., Binz, T., Hayashi, T., Szabo, E., Yamasaki, N., Eklund, M., Jahn, R. & Niemann, H. (1994) Biochem. Biophys. Res. Commun. 200, 829-835. [DOI] [PubMed] [Google Scholar]
- 10.Vaidyanathan, V. V., Yoshino, K., Jahnz, M., Dorries, C., Bade, S., Nauenburg, S., Niemann, H. & Binz, T. (1999) J. Neurochem. 72, 327-337. [DOI] [PubMed] [Google Scholar]
- 11.Schmidt, J. J., Stafford, R. G. & Bostian, K. A. (1998) FEBS Lett. 435, 61-64. [DOI] [PubMed] [Google Scholar]
- 12.Wictome, M. & Shone, C. C. (1998) Symp. Ser. Soc. Appl. Microbiol. 27, 87S-97S. [DOI] [PubMed] [Google Scholar]
- 13.Rossetto, O., Schiavo, G., Montecucco, C., Poulain, B., Deloye, F., Lozzi, L. & Shone, C. C. (1994) Nature 372, 415-416. [DOI] [PubMed] [Google Scholar]
- 14.Schmidt, J. J. & Bostian, K. A. (1997) J. Protein Chem. 16, 19-26. [DOI] [PubMed] [Google Scholar]
- 15.Kadkhodayan, S., Knapp, M. S., Schmidt, J. J., Fabes, S. E., Rupp, B. & Balhorn, R. (2000) Protein Expression Purif. 19, 125-130. [DOI] [PubMed] [Google Scholar]
- 16.Kurazono, H., Mochida, S., Binz, T., Eisel, U., Quanz, M., Grebenstein, O., Wernars, K., Poulain, B., Tauc, L. & Niemann, H. (1992) J. Biol. Chem. 267, 14721-14729. [PubMed] [Google Scholar]
- 17.McPherson, A. (1982) Preparation andAnalysis of Protein Crystals (Wiley, New York).
- 18.Kraulis, P. J. (1991) J. Appl. Crystallogr. 24, 946-950. [Google Scholar]
- 19.Kabsch, W. (1988) J. Appl. Crystallogr. 21, 916-924. [Google Scholar]
- 20.Collaborative Computational Project No. 4 (CCP4) (1994) Acta Crystallogr. D 50, 760-763. [DOI] [PubMed] [Google Scholar]
- 21.Kissinger, C. R., Gehlhaar, D. K. & Fogel, D. B. (1999) Acta Crystallogr. D 55, 484-491. [DOI] [PubMed] [Google Scholar]
- 22.Reddy, V., Swanson, S. M., Segelke, B., Kantardjieff, K. A., Sacchettini, J. C. & Rupp, B. (2003) Acta Crystallogr. D 59, 2200-2210. [DOI] [PubMed] [Google Scholar]
- 23.McRee, D. E. (1999) J. Struct. Biol. 125, 156-165. [DOI] [PubMed] [Google Scholar]
- 24.Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997) Acta Crystallogr. D 53, 240-255. [DOI] [PubMed] [Google Scholar]
- 25.Lacy, D. B., Tepp, W., Cohen, A. C., DasGupta, B. R. & Stevens, R. C. (1998) Nat. Struct. Biol. 5, 898-902. [DOI] [PubMed] [Google Scholar]
- 26.Hanson, M. A. & Stevens, R. C. (2000) Nat. Struct. Biol. 7, 687-692. [DOI] [PubMed] [Google Scholar]
- 27.Ahmed, S. A., Byrne, M. P., Jensen, M., Hines, H. B., Brueggemann, E. & Smith, L. A. (2001) J. Protein Chem. 20, 221-231. [DOI] [PubMed] [Google Scholar]
- 28.Holden, H. M., Tronrud, D. E., Monzingo, A. F., Weaver, L. H. & Matthews, B. W. (1987) Biochemistry 26, 8542-8553. [DOI] [PubMed] [Google Scholar]
- 29.Rupp, B. & Segelke, B. (2001) Nat. Struct. Biol. 8, 663-664. [DOI] [PubMed] [Google Scholar]
- 30.Hanson, M. A. & Stevens, R. C. (2001) Nat. Struct. Biol. 8, 664. [DOI] [PubMed] [Google Scholar]
- 31.Eswaramoorthy, S., Kumaran, D. & Swaminathan, S. (2002) Biochemistry 41, 9795-9802. [DOI] [PubMed] [Google Scholar]
- 32.Hanson, M. A., Oost, T. K., Sukonpan, C., Rich, D. H. & Stevens, R. C. (2000) J. Am. Chem. Soc. 122, 11268-11269. [Google Scholar]
- 33.Hanson, M. A., Oost, T. K., Rich, D. H., Stevens, R. C. & Sukonpan, C. (2002) J. Am. Chem. Soc. 124, 10248-10248. [Google Scholar]
- 34.Christopher, J. A. (1997) spock, The Structural Properties Observation and Calculation Kit (The Center for Macromolecular Design, Texas A&M University, College Station, TX).
- 35.Merritt, E. A. & Bacon, D. J. (1997) Methods Enzymol. 277, 505-524. [DOI] [PubMed] [Google Scholar]
- 36.Ferrin, T. E., Huang, C. C., Jarvis, L. E. & Langridge, R. (1988) J. Mol. Graphics. 6, 13-27. [Google Scholar]