Abstract
Lysophosphatidic acid (LPA) is a growth factor for many cells including prostate and ovarian cancer-derived cell lines. LPA stimulates H2O2 production which is required for growth. However, there are significant gaps in our understanding of the spatial and temporal regulation of H2O2-dependent signaling and the way in which signals are transmitted following receptor activation. Herein, we describe the use of two reagents, DCP-Bio1 and DCP-Rho1, to evaluate the localization of active protein oxidation after LPA stimulation by detection of nascent protein sulfenic acids. We found that LPA stimulation causes internalization of LPA receptors into early endosomes that contain NADPH oxidase components and are sites of H2O2 generation. DCP-Rho1 allowed visualization of sulfenic acid formation, indicative of active protein oxidation, which was stimulated by LPA and decreased by an LPA receptor antagonist. Protein oxidation sites colocalized with LPAR1 and the endosomal marker EEA1. Concurrent with the generation of these redox signaling-active endosomes (redoxosomes) is the H2O2- and NADPH oxidase-dependent oxidation of Akt2 and PTP1B detected using DCP-Bio1. These new approaches therefore enable detection of active, H2O2-dependent protein oxidation linked to cell signaling processes. DCP-Rho1 may be a particularly useful protein oxidation imaging agent enabling spatial resolution due to the transient nature of the sulfenic acid intermediate it detects.
Keywords: protein oxidation, cysteine modifications, LPA signaling, proliferative signaling, lipid mediators
Lysophosphatidic acid (LPA) is a lipid growth factor that is found in high concentrations in the ascites fluid and plasma of ovarian cancer patients [1, 2] and acts as an autocrine mediator of prostate cancer cell growth [3] and migration [4]. LPA promotes proliferation, survival, and migration in prostate cancer cells through LPA receptor 1 (LPAR1)-mediated activation of NF-κB [5, 6], and multiple cell types respond to LPA through activation of protein kinase B/Akt and extracellular signal-related kinase (Erk) [1, 4, 7]. LPA receptors are members of the G-protein coupled receptor (GPCR) family and are strong positive regulators of metastatic phenotypes [4, 8-10]. A number of cancer cell types, including ovarian and prostate cancer-derived cell lines, exhibit elevated levels of both LPA synthetic enzymes and receptor proteins linked with LPA signaling [5, 6, 11]. Thus, alterations in LPA-dependent signaling appear to be key factors driving many metastatic cancers.
Reactive oxygen species (ROS), and particularly superoxide (O2*−) and H2O2, specifically generated by NADPH oxidase (Nox) complexes in response to growth factor stimulation are intimately involved in the regulation of signaling pathways downstream of receptor tyrosine kinases like epidermal growth factor receptor (EGFR) and platelet-derived growth factor receptor (PDGFR) [12-14]. PDGF was the original growth factor recognized in 1995 to require H2O2 as a signaling mediator for effective mitogenic responses [14]; later the same year, Chen et al. demonstrated that H2O2 is required for full activation of the MAP kinase pathway by LPA in HeLa cells [15]. A later study of HaCaT keratinocytes by Sekharam et al. demonstrated that H2O2 required for LPA-stimulated DNA synthesis was elicited by lipoxygenases and their products in this cell type. Another study with vascular smooth muscle cells in 2007 implicated Rac and Nox in ROS generation associated with LPA-induced proliferation, a process which is potentially linked to atherogenesis [16]. Our 2010 studies with SKOV3 ovarian cancer cells demonstrated that LPA stimulation of cell growth, ERK and Akt phosphorylation, and NF-κB activation all require the generation of ROS [7]. We also found that growth of the SKOV3 cells was blocked by the LPA receptor antagonist VPC32183 which shows the role of endogenously-produced LPA in cell growth and survival. The LPA-dependent signaling was also blocked by PEG-catalase, further indicating the importance of H2O2 in proliferative signaling.
While the evidence for ROS and H2O2 in various signaling pathways is becoming more established, questions regarding their spatial and temporal control remain. For example, how is an ROS “burst” achieved in the relevant location without nonspecific or toxic effects on other molecules? PMN have solved this dilemma in bacterial killing by concentrating superoxide and other ROS in phagosomes. However, it is less clear how the localization and specificity of ROS are controlled in non-phagocytic cells, particularly with highly diffusible H2O2. Some stimuli, like the cytokine interleukin 1β (IL-1β), require the formation of endosomes for the appropriate signaling output to be achieved, whereas others, such as EGF and PDGF, rely on endocytosis for specific subsets of downstream responses [17]. In those pathways that are dependent on both ROS generation and receptor endocytosis, luminal superoxide is generated by activated Nox in redox-active endosomes, subsequently forming H2O2 through dismutation [18]. The neutral H2O2 can diffuse through membranes and potentially oxidize proteins localized around the receptor-containing endosomes. These specialized ROS signaling endosomes were previously referred to by Oakley et al. as redoxosomes and we will adopt this nomenclature here [17].
While much has been learned in recent years about the roles protein oxidation by H2O2 may play in cell signaling, we are clearly at a very early stage in achieving a molecular understanding of how H2O2-mediated oxidation influences cell signaling. The overall effects of Nox activation and H2O2 production are often characterized as promoting the downstream signaling outputs, but the molecular effects may be better described as “shaping” the overall context through which signal transduction interactions take place. It is now well documented that H2O2 can enhance phosphorylation cascades through the oxidative inactivation of protein tyrosine phosphatases (PTPs) [19, 20]. However, not every oxidation will inhibit a protein and/or have overall signal-promoting effects. As with phosphorylation, oxidation events will almost certainly have quite distinct effects depending on the protein and site of oxidation as well as the timing, the subcellular location, and the specific oxidation product(s) formed.
In H2O2-mediated oxidation, the most likely targets in proteins are cysteine residues. This two-electron chemistry results in the formation of a cysteine sulfenic acid (R-SOH) on the protein and the reduction of H2O2 to H2O. Sulfenic acids then readily react with proximal thiols to generate disulfide bonds with cysteine thiols in proteins or with glutathione. In the absence of thiols, R-SOH may be stabilized or form a variety of other products; further reaction with additional peroxide molecules can form the irreversibly oxidized sulfinic and sulfonic acids, or a proximal nucleophilic amine or amide nitrogen can attack the SOH and reversibly form a sulfenamide and H2O. Thus, crosslinks, conformational changes and/or reorganized local structures can result from the oxidation of cysteine residues [21]. It is now well established that cysteine thiols vary greatly in their reactivity to oxidants and electrophiles [22, 23]. The reactivity of cysteines toward H2O2 is minimal for protonated thiols in the absence of catalysts like metals, greater for (deprotonated) thiolates (to a maximum of ~20 M−1 s−1 in small molecules), and in some cases, like the specialized active sites of peroxiredoxins (Prxs), as fast as 107 – 108 M−1 s−1, accelerated through strategic positioning of active site threonine and arginine residues [24, 25]. There are kinetic arguments that abundant antioxidant proteins like Prxs and perhaps glutathione peroxidases would outcompete other proteins as “targets” for intracellular H2O2 [25, 26], but the limitations in our ability to assess signaling-relevant protein oxidation, particularly in situ in cells, have made this difficult to assess. The goal in this area continues to be to establish signaling proteins and pathways influenced by oxidation as it occurs in cells, and to determine the molecular and kinetic details associated with these changes in different cellular settings.
In an effort to determine molecular changes in cells associated with localized production of signaling-relevant H2O2, we describe herein the use of recently-developed chemical trapping reagents designed to label sulfenic acids in situ in cells as they are formed. These reagents, among a set of new chemical probes recently introduced by us and others [22, 27-30], have the potential to report on the status of protein redox states not only from proteins labeled during lysis and analyzed by Western blot or mass spectrometry, but also after labeling intact cells, followed by biochemical or imaging analysis to evaluate sites of protein oxidation relative to locations of other cellular components. In addition to our affinity probe, DCP-Bio1 (for identification of oxidized proteins), we describe here the use of a fluorescent oxidation-sensing probe, DCP-Rho1, for visualization of sites of protein oxidation within both ovarian and prostate cancer-derived cells. Thus, for the first time, we have been able to demonstrate the colocalization of protein oxidation sites with internalized LPA receptors as well as an early endosome marker, consistent with the LPA-induced formation of receptor-bearing endosomes associated with activated Nox components (i.e., redoxosomes). By targeting the first intermediary product, sulfenic acid, for labeling by our fluorescent probe, our approach may allow a degree of temporal resolution as nascent sulfenic acids are detected whereas subsequently formed disulfide-bonded proteins are not. Furthermore, our data demonstrate the dependence of the observed protein oxidation on LPA-mediated receptor activation, Nox activation, and H2O2 generation based on treatment with targeted pharmacological agents. Notably, this new linkage of protein oxidation to GPCR activation and subsequent redoxosome formation supports the view that targets of oxidation should be colocalized with sites of ROS formation.
MATERIALS AND METHODS
Reagents and Antibodies
Primary antibodies for Western blots to p44/42 MAP Kinase (ERK), phospho-p44/42 MAP Kinase (Thr202/Tyr204) (p-ERK), and phospho-SAPK/JNK (Thr183/Tyr185) (p-JNK) were from Cell Signaling Technology. Antibodies to EEA1, SHP-2, Akt2, β-actin and p47-phox were from Santa Cruz Biotechnology, and the antibody toward LPAR1 and the corresponding blocking peptide were purchased from Cayman Chemical Company. AlexaFluor fluorescent secondary antibodies, DCFH diacetate, RPMI 1640 medium, Lipofectamine, and Opti-MEM I + Glutamax media were from Invitrogen. The pNiFty-SEAP reporter plasmid was from InvivoGen; the plasmid for expression of HyPer was from Evrogen. PEG-catalase was from Sigma, and apocynin and anti-PTP1B antibodies were from Calbiochem. Antibody to Salmonella typhimurium AhpC was purified from rabbit serum. Diphenyleneiodonium (DPI) chloride was from Calbiochem. Fetal bovine serum was from Lonza. Nitrocellulose membranes were from Schleicher and Schuell and Western Lightning chemiluminescence reagent was from Perkin Elmer. VPC32183 and alkyl- and acyl-linked 18:1 lysophosphatidic acid [1-(9Z-octadecenyl)-2-hydroxy-sn-glycero-3-phosphate (ammonium salt), and 1-oleoyl-2-hydroxy-sn-glycero-3-phosphate (sodium salt), respectively] were from Avanti Polar Lipids, Inc.
DCP-Bio1 and DCP-Rho1 were synthesized as described previously [29].
Cell Culture and Treatments
PC3 and SKOV3 cells (from ATCC stocks) were grown, maintained, and, unless otherwise indicated, treated at 37°C with 5% CO2 in RPMI 1640 medium supplemented with 10% fetal bovine serum, L-glutamine, penicillin and streptomycin. LPA, supplied in chloroform, was dried under a stream of nitrogen, resuspended to a concentration of 1 mM in phosphate buffered saline (PBS) containing 1% fatty acid free bovine serum albumin (BSA), then diluted into culture medium to indicated concentrations. VPC32183 was resuspended and stored at a concentration of 1 mM in PBS containing 3% fatty acid free BSA before dilution into culture medium.
DCF Fluorescence Analyses
For ROS measurement by DCF fluorescence, cells were pre-incubated with serum-containing media (and, where indicated, 400 U/mL PEG-catalase) overnight, then treated or not with 1 μM VPC32183, 1 μM DPI, or 500 nM apocynin for 30 min prior to incubation with or without 100 nM alkyl-LPA (18:1) for 30 min. Cells were incubated with 50 μM DCFH diacetate for the final 10 min of LPA treatment, then washed and visualized using an Olympus inverted epi-fluorescence microscope with FITC filters.
Western Blotting
For Western blotting, cells were plated at 2.5 × 105 cells per dish in 35 mm dishes, treated or not with pharmacological agents, then washed with cold, calcium-free PBS, scraped into lysis buffer (50 mM Tris-HCl, 100 mM NaCl, 2 mM EDTA, 0.1% SDS, 0.5% sodium deoxycholate, 1 mM PMSF, 10 μg/mL aprotinin, 10 μg/mL leupeptin, 50 mM NaF, and 1 mM sodium vanadate), sonicated, and centrifuged to remove cell debris. Protein concentration was measured (Pierce BCA protein assay) and samples (typically 40 μg protein/lane) were resolved on 10 or 12% SDS polyacrylamide gels, then transferred to nitrocellulose membranes, probed with protein-specific antibodies and visualized using Western Lightning chemiluminescence reagent.
DCP-Bio1 Labeling and Affinity Capture
PC3 or SKOV3 cells (~5 × 105) were grown in 100 mm plates for 48 h as described above, and preincubated with 400 U/ml PEG-catalase (added the night before) in some cases, or in other cases with reagents (1 μM VPC32183, 500 nM apocynin, or 20 mM N-acetylcysteine) added 30 min prior to LPA treatment. After incubation of 100 nM alkyl-LPA with the cells for 30 min, DCP-Bio1 was added with the lysis buffer to chemically trap sulfenic acid proteins using previously described methods [31] with slight modifications. Lysis buffer was freshly prepared without phosphatase inhibitors or EDTA (50 mM Tris-HCl, 100 mM NaCl, 0.1% SDS, 0.5% sodium deoxycholate, 1 mM PMSF, 10 μg/ mL aprotinin, and 10 μg/ mL leupeptin) and supplemented with 1 mM DCP-Bio1, 10 mM NEM, 10 mM iodoacetamide, 100 μM DTPA and 200 U/mL catalase. Lysis buffer (150 μL/plate) was added to each plate, then cell samples were scraped from the plates, transferred to Eppendorf tubes, incubated on ice for 30 min and stored at −80°C. For affinity capture and elution of the labeled proteins [32], unreacted DCP-Bio1 was removed immediately upon thawing using a BioGel P6 spin column, then samples were assayed for protein concentration, diluted into 2M urea to 1.6 mg/mL protein, and supplemented with pre-biotinylated AhpC (1 μg/500 μg of total lysate) to control for the efficiency of the affinity capture and elution procedures. Samples were precleared with Sepharose CL-4B beads (Sigma), then applied to plugged columns containing high capacity streptavidin-agarose beads from Pierce and incubated overnight at 4 °C. Multiple stringent washes of the beads were performed (at least 4 column volumes and two washes each) using, in series, 1% SDS, 4M urea, 1M NaCl, 10 mM DTT, 50 mM ammonium bicarbonate and water, before elution with 50 mM Tris-HCl, pH 8, containing 2% SDS and 1 mM EDTA (1 μL/4 μL starting lysate) [32]. Samples were stored at −80°C as needed and analyzed by Western blot as described above.
Immunofluorescence
For immunofluorescence studies, 5 × 104 cells were plated in 2 mL of media in 2-well chambered coverslips and incubated for 48 h, then treated (or not) with LPA for 30 min in the presence or absence of signaling or ROS modulators as described for the DCF and DCP-Bio1 labeling procedures, rinsed with PBS, and fixed with 10% formalin for 20 min. Cells were washed 3 times with 0.1 mM glycine in PBS containing 2% fetal bovine serum, then permeablized for 15 min with 0.1% Triton-X100 in PBS (PBS-T). After washing 3 times with PBS-T, samples were blocked with 2% BSA in PBS-T for one hour, then incubated with primary antibodies (1:100 dilution) overnight at 4 °C in 2 % BSA in PBS-T. After washing with PBS-T, AlexaFluor-labeled secondary antibodies diluted 1:1000 were added and incubated for 2 h at 24 °C, then washed again with PBS-T and imaged. Labeled cells were visualized by confocal microscopy using a Zeiss LSM510 laser scanning confocal microscope with a 63X water-corrected lens, or a Zeiss LSM710 laser scanning confocal microscope for cells prepared in conjunction with DCP-Rho1 labeling (see below). Imaging of LPAR1 employed a goat anti-rabbit AlexaFluor488 secondary antibody with excitation at 488 nm and emission at 505 – 550 nm; p47-phox or EEA1 imaging (when combined with LPAR1 labeling) employed donkey anti-goat AlexaFluor546 secondary antibodies detected with excitation at 543 nm and a long pass 560 nm emission filter.
DCP-Rho1 labeling and visualization
Cells (5 × 104) were plated in chambered coverslips and grown for 48 hours. Where indicated, cells were pre-incubated with 400 U/ml PEG-catalase overnight, or with 1 μM VPC32183, 1 μM DPI, or 500 nM apocynin for 30 min, followed by treatment with or without 100 nM alkyl-LPA for 30 min. DCP-Rho1 at 10 μM was added for the final 10 min of LPA stimulation, then cells were washed extensively with RPMI containing 10% fetal bovine serum, then PBS, before fixation in 10% formalin and subsequent immunofluorescence analyses for other cellular components as described above. Cells were imaged using a Zeiss LSM 710 confocal microscope, with excitation at 488 nm and emission at 493 – 547 nm (for the LPAR1, β-actin or EEA1 signals) or excitation at 561 nm and emission at 566-703 nm (for the DCP-Rho1 signal). For acquisition of the fluorescence images, the gain, laser power and other settings were held constant between conditions, intensity values were kept in the linear range, and the absence of crossover and bleedthrough between channels was confirmed. As controls for specific fluorescence labeling of the cells, one set of samples was simultaneously incubated with both the DCP-Rho1 and 50 μM dimedone, a non-fluorescent compound which also reacts with sulfenic acids (Supplementary Fig. 1b). Another set was incubated with the synthetic precursor of DCP-Rho1, DCP-Rho1SP (Supplementary Fig. 1a, showing the protecting group at the reactive center) under the same conditions as the DCP-Rho1 labeling experiments (Supplementary Fig. 1b).
Image Analysis and Quantitation
Analysis of the overall fluorescence intensity of the DCFH diacetate-treated samples was conducted using Image J software. For DCP-Rho1-labeled samples, Volocity software (Perkin-Elmer) was used to extract fluorescence intensity values for manually-defined cell and background areas (with cell boundaries confirmed by DIC). To correct for background fluorescence, an area-adjusted background intensity level was subtracted from the total intensity measured for each cell and expressed on a per cell basis [33]. One way Analysis of Variance (ANOVA) followed by Tukey’s Honestly Significant Difference (HSD) for Post-Hoc Analysis was conducted for the multiple treatments used in the DCF fluorescence analysis and DCP-Rho1 intensity analysis.
Image J (version 1.44p) software [Just Another Colocalisation Plugin (JACoP)] [34] was used to evaluate colocalization of LPAR1 with EEA1 and p47, and of DCP-Rho1 fluorescence with LPAR1, EEA1 and β-actin. After setting thresholds (using the same percentages of selected pixels across images and experiments), the Manders’ coefficients were determined from the summed intensities of those pixels in the red channel (e.g., for the DCP-Rho1 signal) for which the other (green) signal is also above the threshold, divided by the summed intensities of all red pixels (defined as M2 according to [34]) (Fig. 4d). The converse of M2, M1, gives the fraction of colocalized green pixel summed intensities. Statistical analyses were conducted by Students’ two-tailed t-test for the effects of LPA on Manders’ colocalization coefficients.
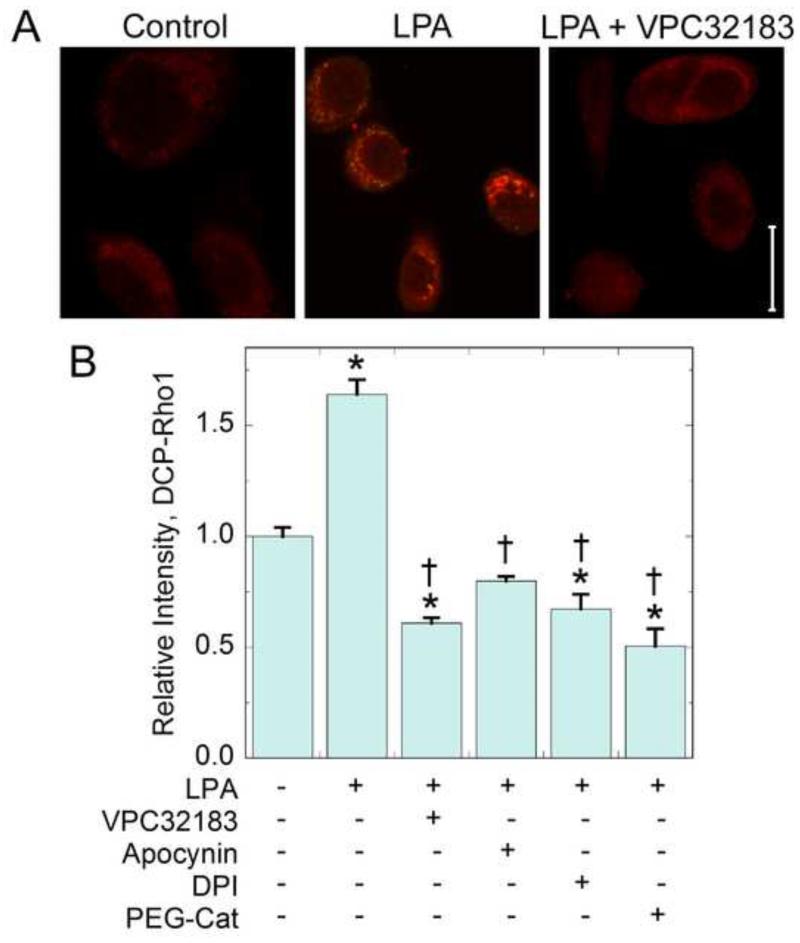
Figure 4.
Protein oxidation (detected by DCP-Rho1) is increased in response to LPA stimulation. SKOV3 cells grown in 10% fetal bovine serum were pretreated for 30 minutes with the LPA receptor antagonist VPC32183 (1 μM, 30 min), apocynin (500 nM, 30 min), DPI (1 μM, 30 min), or with PEG-catalase (400 U/ml, overnight). Cells were then stimulated with with 100 nM alkyl-LPA for 30 min. During the last 10 minutes, 10 μM DCP-Rho1 was added. Fixed and permeabilized cells were washed to remove unreacted probe. (a) Images of control, LPA stimulated, and LPA plus VPC32183 treated cells represent oxidized proteins labeled with DCP-Rho1. The scale bar represents 20 μm. (b) DCP-Rho1 total intensity after various treatments (means ± standard error) was analyzed using Volocity software as described in Methods (N = 48 cells from three independent experiments).The data were then analyzed using one way ANOVA and Tukey’s test for significance. Asterisks indicate means are significantly different from the untreated control; daggers indicate means are significantly different from the LPA treated sample.
RESULTS
LPA Stimulation of SKOV3 and PC3 Cells Results in Nox-Mediated Hydrogen Peroxide Production
In previous work with SKOV3 cells we established that both Nox activity and H2O2 generation were required for LPA-dependent NF-κB activation as well as ERK and Akt phosphorylation [7]. To extend this observation to other cancers we tested PC3 prostate cancer cells and found that they also exhibited increased ERK and JNK phosphorylation in response to LPA (Supplementary Fig. 2a) as well as increased NF-κB-mediated transcription (Supplementary Fig. 2b). As with the SKOV3 cells, exogenous alkyl-linked LPA [1-(9Z-octadecenyl)-2-hydroxy-sn-glycero-3-phosphate] was more effective in activating these pathways than was the corresponding acyl-linked LPA (1-oleoyl-2-hydroxy-sn-glycero-3-phosphate) which had comparable effects at higher concentrations.
LPA is released by activated platelets and therefore present in serum [35], and is synthesized by cancer cells like SKOV3 and PC3 even in the absence of serum; because of this, these cells are continuously undergoing LPA stimulation. In support of this, addition of VPC32183, a receptor antagonist of LPA receptors 1 and 3 (LPAR1 and LPAR3), to serum-deprived or serum-supplemented SKOV3 cells inhibits their growth based on decreased cell number. On the other hand, addition of exogenous LPA further stimulates cell growth in both conditions [7]. As ROS generation tracked with growth effects in SKOV3 cells treated with LPA or VPC32183 in these experiments [7] and has also been observed in other LPA-treated cancer cells, as well [15, 36], we began our assessments of ROS effects in LPA-stimulated PC3 cells with experiments that utilized fluorescence detection of 2′,7′-dichlorodihydrofluorescein (DCFH) diacetate oxidation to dichlorofluorescein (DCF). As with SKOV3 cells, LPA stimulation of PC3 cells led to a strong increase in the DCF fluorescence (Fig. 1) which was blocked by the receptor antagonist, VPC32183. This LPA-linked increase in DCF fluorescence was also blocked by two Nox inhibitors, diphenyleneiodonium (DPI), which is a general inhibitor of flavoproteins including Noxs and nitric oxide synthases, and apocynin, which inhibits p47 recruitment to Nox2 complexes [37] (Fig. 1b). These data strongly suggest that both receptor activation and Nox activation (likely Nox2 based on the inhibition by apocynin) are responsible for ROS generation in LPA-stimulated SKOV3 and PC3 cells.
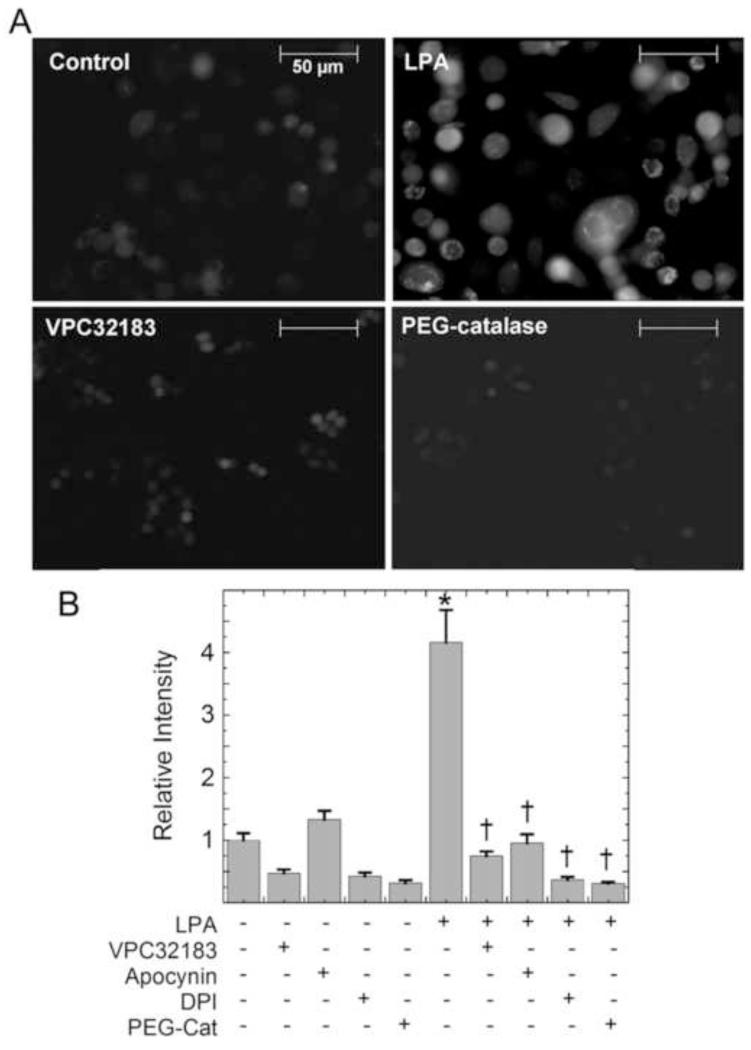
Figure 1.
LPA stimulates NADPH oxidase-dependent production of H2O2 in PC3 cells. (a) PC3 cells were pretreated for 30 minutes with the LPA receptor antagonist VPC32183 (1 μM), apocynin (500 nM), or DPI (1 μM), or overnight with PEG-catalase (400 U/mL), prior to 30 min stimulation with 100 nM alkyl-LPA. Cells were incubated with 50 μM DCFH diacetate for the final 10 min of LPA stimulation, washed, and visualized for DCF fluorescence. Images shown are representative of at least six separate experiments. (b) The relative DCF fluorescence was determined using ImageJ software and averages reflect the total intensity from 3 experiments (n = 48 cells total). The data were then analyzed using one way ANOVA and Tukey’s test for significance. The asterisk indicates the mean is significantly different from the untreated control; daggers indicate means are significantly different from the LPA-treated sample.
Activation of Nox generates superoxide which is rapidly dismutated to H2O2 either nonenzymatically or by superoxide dismutase. DCF fluorescence increases can be caused by various ROS, including products of H2O2 and/or self-propagating redox cycling reactions induced by the DCF radical once formed, and must be interpreted with caution [38]. To help evaluate whether or not H2O2 was a key oxidant elicited by LPA signaling, we assessed the ability for a specific cell permeable scavenger of H2O2, PEG-catalase, to modulate the DCF fluorescence observed upon LPA addition. As the DCF fluorescence signal observed after LPA addition was decreased by addition of PEG-catalase, H2O2 is likely involved in LPA-induced processes leading to DCFH oxidation. Further proof that H2O2 levels are strongly affected by LPA signaling was obtained using the H2O2-specific redox sensor HyPer [13], which showed increases in fluorescence intensity from 5-30 minutes after LPA stimulation (Supplementary Fig. 3).
LPA Receptor (LPAR1) Undergoes LPA-Induced Internalization and Colocalizes With Early Endosomes and p47phox
Endocytic trafficking of LPA-stimulated LPAR1 occurs through both clathrin- and β-arrestin-mediated pathways and plays multiple roles in signal transduction and desensitization [39-41], and although not previously evaluated, receptor internalization could be linked with the ROS generation observed above. To confirm that exogenous LPA led to increased receptor internalization in PC3 cells, we exposed intact cells to a low concentration of trypsin and performed a western blot of the resulting lysate to evaluate the amount of un-cleaved LPAR1, which represents the protected, internalized fraction of the receptor (Supplementary Fig. 4). We observed that PC3 cells treated with exogenous LPA in either the presence or absence of serum respond by significantly increasing the fraction of internalized LPA receptor (Supplementary Fig. 4).
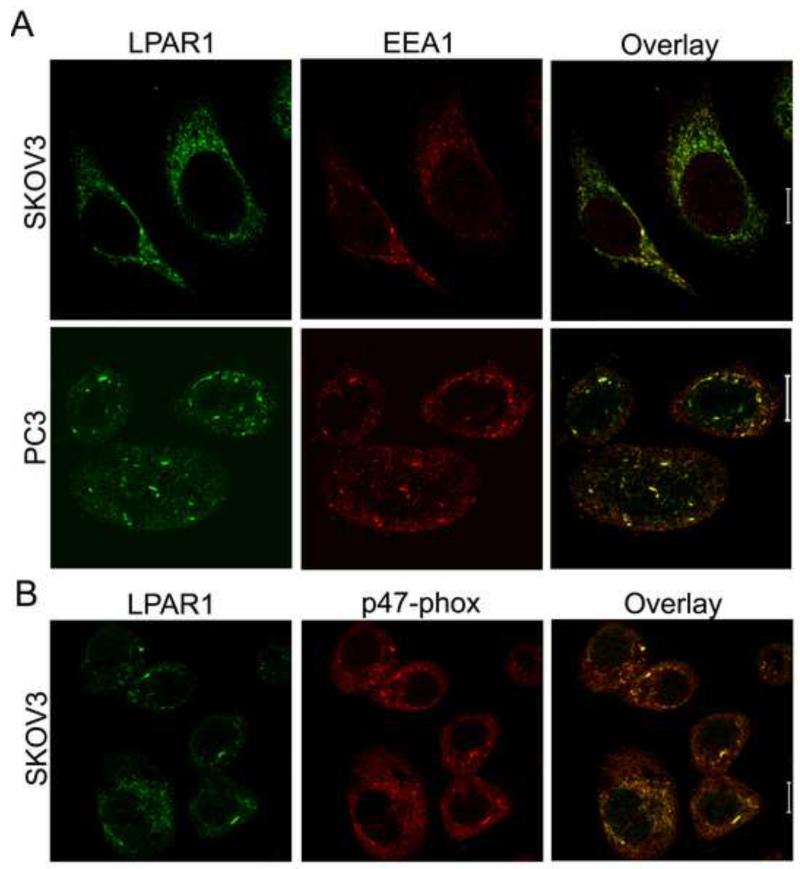
In experiments designed to evaluate other components associated with LPA-stimulated LPAR1 internalization in PC3 cells, immunofluorescence images collected with anti-LPAR1 antibody (see Supplementary Fig. 5 for control images with blocking peptide) show extensive association with an early endosomal marker, EEA1, both before and after exogenous LPA addition (Fig. 2a, with a Manders’ colocalization coefficient of ~0.8 for the fraction of summed EEA1 pixel intensities colocalized with LPAR1). Furthermore, the LPAR1 signal significantly colocalizes (colocalization coefficient of ~0.85) with p47phox (Fig. 2b), one of the cytosolic components recruited during Nox2 activation which serves an organizing role in bringing p67phox and p40phox to join Nox2 (gp91phox) and p22 at the membrane [42]. Thus, endosomes forming during LPA-stimulated LPA receptor internalization contain activated Nox complexes, forming the ROS-generating endosomes referred to as redoxosomes [17].
Figure 2.
LPAR1 receptor activated by LPA localizes with an early endosomal marker (EEA1) and p47phox, a component of NADPH oxidase complexes. SKOV3 or PC3 cells were treated with 100 nM LPA for 30 min, then fixed, permeabilized, and analyzed by immunofluorescence staining for (a) LPAR1 and EEA1, or (b) LPAR1 and p47phox, by confocal microscopy (Zeiss LSM 510). The scale bars represent 10 μm.
Evidence Using the Sulfenic Acid Probe DCP-Rho1 that LPA Induces Protein Oxidation Around LPAR1-Containing Redoxosomes
Elevated H2O2 associated with receptor-containing redoxosomes resulting from signaling by growth factors is likely to cause local protein oxidation, but a direct link between redoxosome formation and protein oxidation has not been established. To detect actively-occurring protein oxidation, we turned to our recently-developed chemical biology approaches for labeling oxidized proteins at sulfenic acid sites [29, 31, 32, 43-46]. Two of these probes, DCP-Bio1 and DCP-Rho1, covalently tether biotin or a rhodamine fluorophore, respectively, to proteins bearing sulfenic acid modifications for subsequent affinity capture or fluorescence detection (Fig. 3). Both DCP-Bio1 and DCP-Rho1 are cell permeable and can be used to label sulfenic acid-containing proteins either in intact cells or at the time of lysis [31, 32]. In experiments described below, we have found that after labeling intact cells with DCP-Rho1, cells can be fixed, permeabilized, washed extensively to remove unreacted probe, then visualized to observe the amount and location of sulfenic acid-containing proteins.
Figure 3.
Reagents for covalently trapping sulfenic acids (R-SOH) in cellular proteins. Both DCP-Bio1 and DCP-Rho1 covalently attach to proteins at sites of sulfenic acid modification and can be used to detect and/or affinity capture oxidized proteins in cells or during lysis.
Using DCP-Rho1 imaging after labeling of intact SKOV3 cells, we observed a significant increase in DCP-Rho1 intensities of SKOV3 cells following treatment with LPA (Fig. 4) indicating that protein oxidation is increased in SKOV3 cells in response to LPA stimulation. In contrast, when cells were treated with the receptor antagonist, VPC32183, the signal was decreased to a level significantly below that of untreated cells (Fig. 4b), demonstrating the involvement of LPA receptors (likely LPAR1 or LPAR3) in mediating the LPA-dependent enhancement of protein oxidation. Lower DCP-Rho1 fluorescence was also detected when cells were pretreated with apocynin, DPI or PEG-catalase, further supporting the role of Nox proteins and hydrogen peroxide in mediating protein oxidation in this system. Importantly, control experiments using (i) pretreatment of cells with excess dimedone prior to DCP-Rho1 addition, or (ii) replacement of DCP-Rho1 with its non-reactive synthetic precursor (DCP-Rho1SP) demonstrated little or no fluorescent labeling of the cells, highlighting the chemical specificity of the DCP-Rho1 labeling (Supplementary Fig. 1).
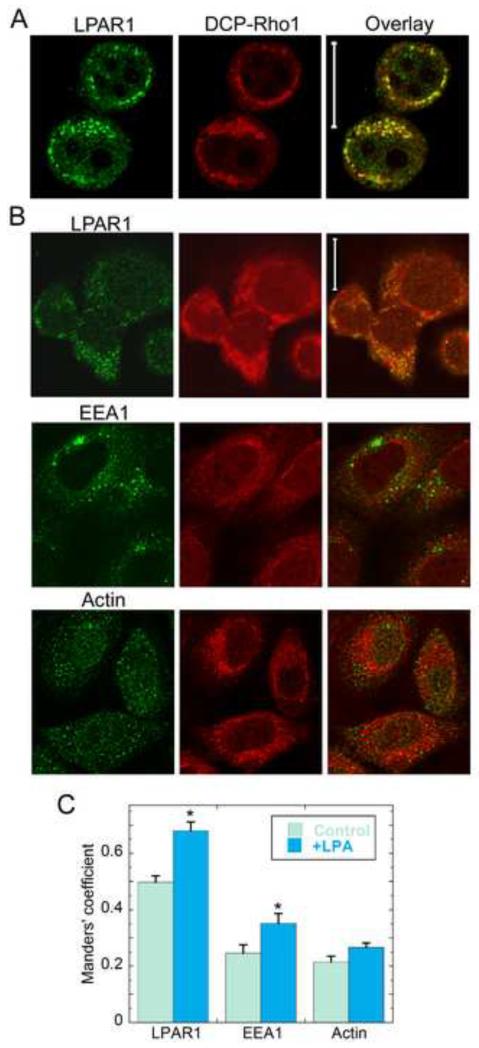
To test the hypothesis that enhanced protein oxidation due to LPA signaling was localized to the region surrounding the LPAR1-containing endosomes, we evaluated the degree to which the DCP-Rho1 fluorescence colocalized with intracellular markers for β-actin, early endosomes (EEA1), and LPAR1 in cells that were either untreated or treated with exogenous LPA (Fig. 5). DCP-Rho1 images and immunostaining for LPAR1 showed a strong correlation between sites of protein oxidation and LPAR1-positive pixels in both PC3 (Fig. 5a) and SKOV3 cells (Fig. 5b), although the oxidation signal is typically more widespread across the cell; this is expected since redox-linked processes are always active in living cells. To quantitatively assess colocalization of the red and green signals, the Manders’ coefficient was used to compute the fraction of summed DCP-Rho1 pixel intensities that colocalized with the green signal (LPAR1, EEA1 or β-actin); thresholds were set in order to select the same percentages of positive pixels across all images and experiments. By this measure, the protein oxidation signal was shown to be significantly correlated with pixels that were positive for LPAR1 in untreated cells (at 50%), and the correlation increased to almost 70% upon addition of exogenous LPA (Fig. 5c). Analysis of these data using the conversely-defined Manders’ coefficient (assessing the fraction of green signal that colocalized with DCP-Rho1) gave very similar results (Supplementary Fig. 6a). These findings were also corroborated in a separate analysis which showed that the pattern with which DCP-Rho1 intensity increased or decreased across the cell was highly correlated with the pattern of LPAR intensity (Supplementary Fig. 6b). In contrast, only ~20% of the DCP-Rho1 signal colocalized with the green signal after counterstaining for β-actin; importantly, the addition of LPA did not significantly change this relationship (Fig. 5c). By this same approach, a modestly larger fraction of the DCP-Rho1 fluorescence colocalized with EEA1 (~25%) which is representative of a heterogeneous population of early endosomes; importantly, this fraction was significantly increased by the addition of LPA (to 35%), consistent with a greater fraction of the endosomes being ROS-producing redoxosomes. Together, these data indicate that stimulation and internalization of LPAR1 by LPA leads to the generation of H2O2 within and around these redoxosomes (presumably through the activation and recruitment of Nox complexes) and leads to a high relative concentration of oxidized proteins in and around these endosomes.
Figure 5.
Intense signals for protein oxidation are centered around endosomes containing LPA Receptor 1. PC3 and SKOV3 cells were treated with 100 nM alkyl-LPA for 30 min; for the last 10 minutes, cells were treated with 10 μM DCP-Rho1 and washed extensively to remove unreacted DCP-Rho1 before fixation, permeabilization and immunofluorescent staining for LPAR1, EEA1 or β-actin as described in Methods. (a) PC3 cell images (collected with a Zeiss LSM510 confocal microscope) show overlap (yellow) between signals from DCP-Rho1 (red) and LPAR1 (green). (b) SKOV3 cell images (collected with a Zeiss LSM710 confocal microscope) show overlap between DCP-Rho1 and a second signal (green) representing LPAR1, EEA1, or β-actin. The scale bars represent 20 μm for all images. (c) Colocalization analysis of images in (b) using the JACoP plugin in Image J [34] to obtain the Manders’ coefficient as described in Methods. Additional replicates of images of cells treated or untreated with exogenous LPA were included in the analysis. Threshold values for DCP-Rho1 were set to highlight a similar number of pixels as selected for the green channel and to highlight only the regions with the most intense signal. Asterisks indicate means that are statistically different from the non-LPA treated control (Students’ t-test with p<0.0005 for LPAR1, p=0.032 for EEA1, and p=0.057 for β-actin).
LPA-Stimulated Hydrogen Peroxide Oxidizes Key Signaling Proteins
Having established that protein oxidation in proximity of LPAR1-containing endosomes is upregulated by LPA and Nox activation, we set out to determine whether several oxidatively-regulated signaling proteins known to be involved in LPA-mediated growth signaling were part of the redox-sensitive signaling networks regulated by redoxosome formation. For this we chose two protein tyrosine phosphatases, PTP1B and SHP-2, that are known to be inhibited by oxidation to sulfenic acid, its reversible condensation product sulfenamide, and/or subsequent disulfide-bonded products. These two phosphatases are also involved in LPA-mediated growth signaling [47-50]. We also chose an isoform of Akt, Akt2, that is inhibited by oxidation in a way that functionally supports PDGF-regulated glucose uptake in NIH 3T3 cells [45]. Importantly, all three proteins have previously been shown to be labeled by DCP-Bio1 or closely-related dimedone-based probes [32, 45, 51].
For labeling of cellular proteins with a biotinylated sulfenic acid trapping agent and affinity capture of proteins undergoing active oxidation, we lysed the cells in the presence of DCP-Bio1 (on ice for 30-60 min) with precautions taken to minimize artifactual protein oxidation [31, 32]. To suppress thiol-disulfide rearrangements, sulfenic acid-thiol reactions, and post-lysis oxidations, cells were harvested into lysis buffer containing catalase to scavenge H2O2 and both iodoacetamide and N-ethlymaleimide to covalently trap reactive thiol groups. This protocol was shown to prevent the post-lysis oxidation of OxyR, a highly peroxide-sensitive bacterial transcription factor used in our earlier work to assess and minimize artifactual oxidation during lysis of cultured cells [31, 32]. PC3 cells grown in the presence of serum were labeled with DCP-Bio1 without treatment and after addition of LPA for 30 min. The labeled proteins were then affinity captured using streptavidin beads, washed extensively to remove any associated unlabeled proteins, and subjected to identification of the affinity-captured proteins by Western blot.
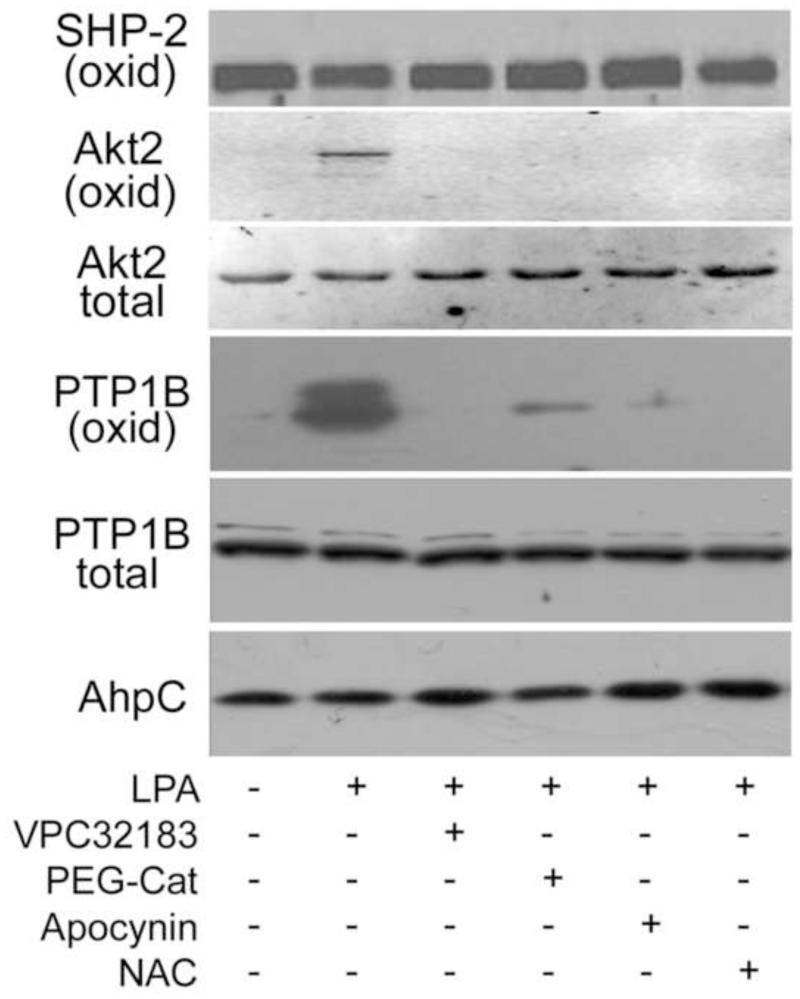
Using DCP-Bio1, PC3 cells show strong labeling of oxidized PTP1B protein after LPA stimulation for 30 minutes (Fig. 6). We also observe that DCP-Bio1 incorporation is significantly increased in Akt2 at 30 min after LPA stimulation. This signal likely corresponds to oxidation of Cys124 in the linker region between the catalytic and regulatory domains, as previously demonstrated in PDGF signaling [45]. In both Akt2 and PTP1B, the labeling is not observed if receptor antagonist or redox modulators are added to the cells (Fig. 6), suggesting that we are catching a biologically meaningful oxidation event. Interestingly, SHP-2 is significantly labeled by this reagent, indicating that dimedone is able to capture the sulfenic acid in spite of its propensity to form subsequent disulfides [47]. This result indicates that SHP-2 is undergoing active redox cycling during normal, growth in these cells. However, unlike PTP1B and Akt2, the degree of SHP-2 labeling does not change with any of the treatments, suggesting that its oxidation is not directly mediated by LPA-stimulated ROS. Lack of response of SHP-2 sulfenic acid levels to LPA signaling and ROS-suppressing treatments strongly suggests distinct localization of these three proteins, with only PTP1B and Akt2 undergoing localized oxidation proximal to the LPAR1- and Nox-containing redoxosomes. SHP-2 is well known for its association with other signaling and scaffolding proteins involved in receptor tyrosine kinase signaling [52] and in this system appears not to be colocalized with the redoxosomes.
Figure 6.
DCP-Bio1 trapping of sulfenic acid indicates that LPA stimulates PTP1B and Akt2 oxidation in situ. PC3 cells were pretreated before stimulation with LPA for 30 min as described in Figure 4, or with N-acetylcysteine (NAC, 20 mM) for 30 min. To covalently trap sulfenic acid-modified cysteines, DCP-Bio1 was added to lysis buffer as described in Methods and incubated for 30 min at 4 °C and biotinylated proteins were affinity captured as described in Methods. Western blots of these affinity-captured samples were performed to detect PTP1B, Akt2, and SHP-2. Western blots of the starting lysate indicated that total protein amounts did not change during the experiment (data not shown for SHP-2). Prelabeled biotinylated AhpC was added based on protein concentrations prior to affinity capture and used as a procedural control for the biotin-based affinity capture, elution and gel loading steps.
DISCUSSION
Studies of ROS have traditionally focused on the harmful or damaging effects of these oxidizing molecules, and indeed, cells must properly respond to high levels of endogenous or exogenous ROS as a stress or toxin in order to survive. But while ROS are notorious for the roles they play in development and/or progression of numerous diseases including cancer, diabetes and neurodegenerative diseases, they are also generated by regulated proteins and processes and are important and often essential in shaping the course of signal transduction networks in cells. Yet significant questions are raised by this relatively newly recognized role for ROS. For example, how is the specificity of these oxidants maintained so that non-specific and toxic effects can be minimized while appropriate signals are sent? How are the ROS involved in specific signaling pathways generated at high enough concentration inside the cell to oxidize relatively slowly-reacting targets without being rapidly scavenged by the oxidant defense systems present throughout the cell? One attractive model for the localization of ROS generation and the efficient transmission of redox-dependent signals is through the generation of redox-active signaling endosomes. These redoxosomes provide a site where proteins in and around these ROS generators are concentrated and serve as targets for the ROS produced locally (Fig. 7). We report here a novel approach which demonstrates the generation of redoxosomes during the course of LPA-directed signal transduction as sites of ROS generation and now, imaged for the first time, as sites of localized protein oxidation. Our observations thus demonstrate redoxosome formation as a mechanism for localizing signaling proteins to be subjected to oxidation and for the efficient transmission of redox-dependent signals allowing LPA, and likely many other types of redoxosome-dependent signaling molecules, to shape the responses that ensue.
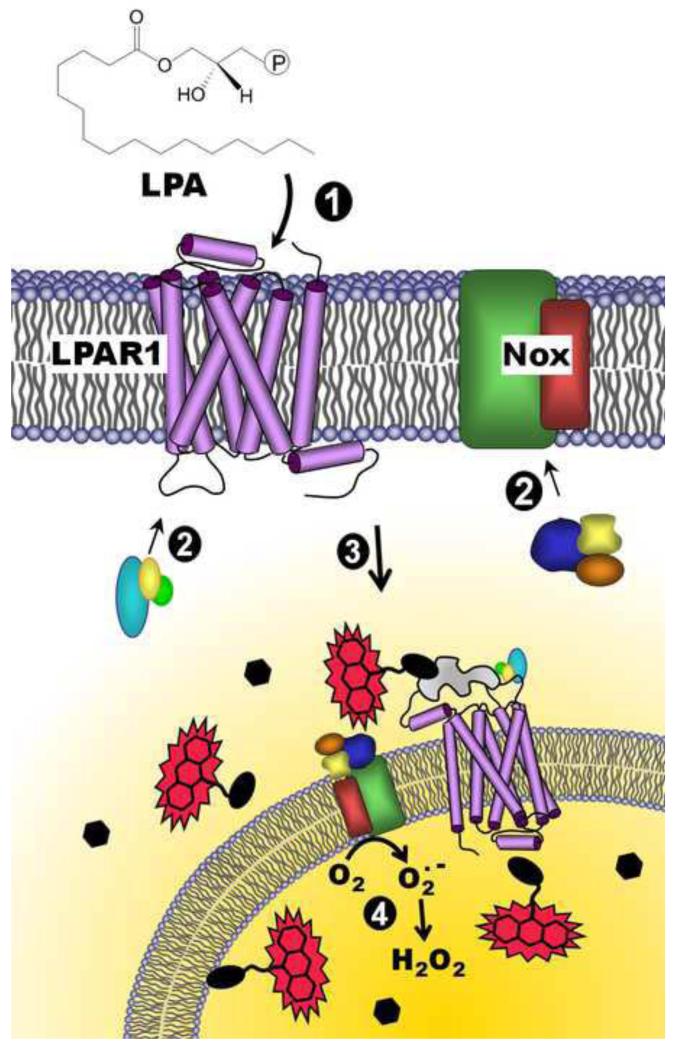
Figure 7.
Model for redox-dependent signaling through LPAR1 and depiction of the DCP-Rho1 labeling results demonstrating localized oxidation of proteins. Lysophosphatidic acid (LPA) binds to the extracellular portion of the LPA receptor (e.g. LPAR1) (1), resulting in the recruitment of G-proteins to the receptor (2, left) and co-stimulation of NADPH oxidase(s) (Nox) through association of cytosolic organizer and activator proteins (2, right). Together, activated LPAR1 and Nox complexes are internalized (3), and superoxide is produced by Nox inside the endosome (4). H2O2 generated by the spontaneous or enzyme-catalyzed dismutation of superoxide, which can diffuse out of the endosome, can then oxidize thiolate (R-S−)-containing proteins within or near these “redoxosomes”, generating the sulfenic acid modification (R-SOH). Upon addition of a labeling reagent, DCP-Rho1, which targets sulfenic acids, proteins containing this modification are covalently modified and associated with the rhodamine fluorophore, allowing detection following fixation, permeabilization and extensive washing to remove unbound DCP-Rho1. Black ellipses represent redox-sensitive proteins, and hexagons represent proteins which are insensitive to oxidation by H2O2. The irregular gray shape represents a scaffolding protein such as β-arrestin.
Demonstration of Akt2 and PTP1B oxidation resulting from LPA-induced endosome signaling and H2O2 generation provides an indication that peroxide defense enzymes like peroxiredoxins and glutathione peroxidases are not the exclusive recipients of oxidizing equivalents generated through Nox activation. Because sulfenic acid detection picks up only the initial and transient oxidation state formed upon direct oxidation of cysteine with H2O2, our evidence indicates that these signaling proteins are serving as direct targets for this oxidant rather than becoming oxidized secondarily by an intervening peroxidase, which would instead generate disulfides through thiol-disulfide exchange. Peroxiredoxins and glutathione peroxidase activities, while very high under ideal circumstances, may not be efficiently recycled during oxidative bursts, and/or they may be rendered less active or inactive by modifications like phosphorylation, as demonstrated by the Rhee group [53], or even oxidant hypersensitivity, as put forward in the “floodgate” hypothesis by our group [54]. It seems likely that the various direct and indirect models for redox regulation of signaling proteins may be true in certain circumstances and contribute to different extents in various signaling networks and cellular conditions. Further clarification of the relative importance of various redox signaling models should become possible with the rapid development and improvement in chemical biology tools to test these models.
Previous studies using DCP-Bio1 and similar cysteine sulfenic acid-targeted reagents have helped link protein oxidation to such processes as T-cell and B-cell proliferation and differentiation [43, 55], angiogenesis [44, 46], insulin and PDGF responses [45, 56], and tyrosine kinase signaling through EGF receptor [51]. In our view, the targeting of this often transient intermediate in protein oxidation makes it ideal not only for capturing information about the specific sites within proteins that are directly modified by oxidants through affinity-based approaches, but also for reporting on active protein oxidation processes within cells. We propose that the temporal resolution provided by using our novel compound, DCP-Rho1, to detect newly-formed sulfenic acids present near sites of ROS generation leads to the spatial resolution of the signal we observe. Thus, the signal tapers off in more distant locations as the maturation of the oxidation product yields (primarily) disulfide bonds. Localization of proteins in signaling complexes is a common theme for regulating such posttranslational modifications as phosphorylation and acetylation. Localized ROS generation has also been a developing theme in the literature [57], now supported by our new data. While we identify two signaling proteins, Akt2 and PTP1B, as endosomal H2O2 targets functionally altered by oxidation, there are undoubtedly additional important players yet to be identified. Because other GPCR signaling pathways have also been shown to involve ROS and share many of the same kinases and phosphatases, our findings of localized protein oxidation occurring proximal to LPA receptor-containing endosomes may have much broader implications for GPCR-linked signaling mechanisms of therapeutic interest. Indeed, the strong connections of GPCRs and receptor tyrosine kinases to proliferative signaling associated with cancers [52, 58], as well as the high levels of ROS observed in cancer cells [59], make it imperative to better understand how ROS regulate and modulate proliferative and metastatic pathways in normal and pathological settings.
Supplementary Material
Highlights.
-
■
LPA stimulates the production of H2O2 around LPAR1-containing endosomes
-
■
Endosomally-localized H2O2 is linked to reversible oxidation of Akt2 and PTP1B
-
■
Protein oxidation, visualized in situ using DCP-Rho1, is localized at endosomes
-
■
DCP-Rho1 enables spatial resolution of oxidation by capturing transient R-SOH
Acknowledgements
This work was supported by grants from the National Institutes of Health to L.W.D. (RO1 CA142838) and L.B.P. (R01 GM050389, R33 CA126659, and R33 CA177461 to L.B.P. and Cristina M. Furdui). Pilot funding was also provided to L.W.D. through the National Institutes of Health grant P50 AT002782. The authors thank Herman Odens and Sarah Knaggs for synthesizing the DCP-Rho1, Erika Bechtold and Rajeswari Mukherjee for synthesizing the DCP-Bio1, Glenn Marrs and Ken Grant for assistance with confocal microscopy, Scott Wood and Jennifer Gentry for helpful suggestions, and Richard Loeser and Cristina Furdui for critical reading of the manuscript. Purchase of the Zeiss LSM 710 confocal microscope was made possible by National Science Foundation MRI award numbers 0722926 and 1039755.
Abbreviations
- LPA
lysophosphatidic acid
- LPAR
LPA receptor
- ERK
extracellular signal-related kinase, p44/42 MAP kinase
- GPCR
G-protein coupled receptor
- ROS
reactive oxygen species
- Nox
NADPH oxidase
- EGF
epidermal growth factor
- EGFR
EGF receptor
- PDGF
platelet-derived growth factor
- PDGFR
PDGF receptor
- PTP
protein tyrosine phosphatases
- DCP-Bio1
3-(2,4-dioxocyclohexyl)propyl 5-((3aR,6S,6aS)-hexahydro-2-oxo-1H-thieno[3,4-d]imidazol-6-yl)pentanoate
- DCP-Rho1
Rhodamine B [4-[3-(2,4-dioxocyclohexyl)propyl]carbamate]piperazine amide
- DCFH-DA
dichlorodihydrofluorescein diacetate
- DCF
dichlorofluorescein
- PEG
polyethylene glycol
- DPI
diphenyleneiodonium
- BSA
bovine serum albumin
- PBS
phosphate-buffered saline
- EDTA
ethylenediaminetetraacetic acid
- PMSF
Phenylmethylsulphonyl fluoride
- NEM
N-ethylmaleimide
- DTPA
diethylene triamine pentaacetic acid
- DTT
1,4-dithio-DL-threitol
- SDS
Sodium dodecyl sulfate
- DIC
differential interference contrast
- NAC
N-acetylcysteine
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Additional Information. Supplementary information is available, including additional methods used to collect data shown in Supplementary figures, as well as additional results referred to above.
REFERENCES
- [1].Bian D, Su S, Mahanivong C, Cheng RK, Han Q, Pan ZK, Sun P, Huang S. Lysophosphatidic acid stimulates ovarian cancer cell migration via a Ras-MEK kinase 1 pathway. Cancer Res. 2004;64:4209–4217. doi: 10.1158/0008-5472.CAN-04-0060. [DOI] [PubMed] [Google Scholar]
- [2].Umezu-Goto M, Tanyi J, Lahad J, Liu S, Yu S, Lapushin R, Hasegawa Y, Lu Y, Trost R, Bevers T, Jonasch E, Aldape K, Liu J, James RD, Ferguson CG, Xu Y, Prestwich GD, Mills GB. Lysophosphatidic acid production and action: validated targets in cancer? J Cell Biochem. 2004;92:1115–1140. doi: 10.1002/jcb.20113. [DOI] [PubMed] [Google Scholar]
- [3].Xie Y, Gibbs TC, Mukhin YV, Meier KE. Role for 18:1 lysophosphatidic acid as an autocrine mediator in prostate cancer cells. J Biol Chem. 2002;277:32516–32526. doi: 10.1074/jbc.M203864200. [DOI] [PubMed] [Google Scholar]
- [4].Hao F, Tan M, Xu X, Han J, Miller DD, Tigyi G, Cui MZ. Lysophosphatidic acid induces prostate cancer PC3 cell migration via activation of LPA(1), p42 and p38alpha. Biochim Biophys Acta. 2007;1771:883–892. doi: 10.1016/j.bbalip.2007.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Guo R, Kasbohm EA, Arora P, Sample CJ, Baban B, Sud N, Sivashanmugam P, Moniri NH, Daaka Y. Expression and function of lysophosphatidic acid LPA1 receptor in prostate cancer cells. Endocrinology. 2006;147:4883–4892. doi: 10.1210/en.2005-1635. [DOI] [PubMed] [Google Scholar]
- [6].Raj GV, Sekula JA, Guo R, Madden JF, Daaka Y. Lysophosphatidic acid promotes survival of androgen-insensitive prostate cancer PC3 cells via activation of NF-kappaB. Prostate. 2004;61:105–113. doi: 10.1002/pros.20083. [DOI] [PubMed] [Google Scholar]
- [7].Saunders JA, Rogers LC, Klomsiri C, Poole LB, Daniel LW. Reactive oxygen species mediate lysophosphatidic acid induced signaling in ovarian cancer cells. Free Radic Biol Med. 2010;49:2058–2067. doi: 10.1016/j.freeradbiomed.2010.10.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Liu S, Murph M, Panupinthu N, Mills GB. ATX-LPA receptor axis in inflammation and cancer. Cell Cycle. 2009;8:3695–3701. doi: 10.4161/cc.8.22.9937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Yu S, Murph MM, Lu Y, Liu S, Hall HS, Liu J, Stephens C, Fang X, Mills GB. Lysophosphatidic acid receptors determine tumorigenicity and aggressiveness of ovarian cancer cells. J Natl Cancer Inst. 2008;100:1630–1642. doi: 10.1093/jnci/djn378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Goldsmith ZG, Ha JH, Jayaraman M, Dhanasekaran DN. Lysophosphatidic acid stimulates the proliferation of ovarian cancer cells via the gep proto-oncogene Galpha(12) Genes Cancer. 2011;2:563–575. doi: 10.1177/1947601911419362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Gibbs TC, Rubio MV, Zhang Z, Xie Y, Kipp KR, Meier KE. Signal transduction responses to lysophosphatidic acid and sphingosine 1-phosphate in human prostate cancer cells. Prostate. 2009;69:1493–1506. doi: 10.1002/pros.20994. [DOI] [PubMed] [Google Scholar]
- [12].Malinouski M, Zhou Y, Belousov VV, Hatfield DL, Gladyshev VN. Hydrogen peroxide probes directed to different cellular compartments. PloS One. 2011;6:e14564. doi: 10.1371/journal.pone.0014564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Markvicheva KN, Bogdanova EA, Staroverov DB, Lukyanov S, Belousov VV. Imaging of intracellular hydrogen peroxide production with HyPer upon stimulation of HeLa cells with epidermal growth factor. Methods Mol Biol. 2009;476:76–83. doi: 10.1007/978-1-59745-129-1_6. [DOI] [PubMed] [Google Scholar]
- [14].Sundaresan M, Yu ZX, Ferrans VJ, Irani K, Finkel T. Requirement for generation of H2O2 for platelet-derived growth factor signal transduction. Science. 1995;270:296–299. doi: 10.1126/science.270.5234.296. [DOI] [PubMed] [Google Scholar]
- [15].Chen Q, Olashaw N, Wu J. Participation of reactive oxygen species in the lysophosphatidic acid-stimulated mitogen-activated protein kinase kinase activation pathway. J Biol Chem. 1995;270:28499–28502. doi: 10.1074/jbc.270.48.28499. [DOI] [PubMed] [Google Scholar]
- [16].Kaneyuki U, Ueda S, Yamagishi S, Kato S, Fujimura T, Shibata R, Hayashida A, Yoshimura J, Kojiro M, Oshima K, Okuda S. Pitavastatin inhibits lysophosphatidic acid-induced proliferation and monocyte chemoattractant protein-1 expression in aortic smooth muscle cells by suppressing Rac-1-mediated reactive oxygen species generation. Vascular Pharmacol. 2007;46:286–292. doi: 10.1016/j.vph.2006.11.002. [DOI] [PubMed] [Google Scholar]
- [17].Oakley FD, Abbott D, Li Q, Engelhardt J. Signaling Components of Redox Active Endosomes: The Redoxosomes. Antioxid Redox Signal. 2009;11:1313–1333. doi: 10.1089/ars.2008.2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Mishina NM, Tyurin-Kuzmin PA, Markvicheva KN, Vorotnikov AV, Tkachuk VA, Laketa V, Schultz C, Lukyanov S, Belousov VV. Does cellular hydrogen peroxide diffuse or act locally? Antioxid Redox Signal. 2011;14:1–7. doi: 10.1089/ars.2010.3539. [DOI] [PubMed] [Google Scholar]
- [19].Lou YW, Chen YY, Hsu SF, Chen RK, Lee CL, Khoo KH, Tonks NK, Meng TC. Redox regulation of the protein tyrosine phosphatase PTP1B in cancer cells. Febs J. 2008;275:69–88. doi: 10.1111/j.1742-4658.2007.06173.x. [DOI] [PubMed] [Google Scholar]
- [20].Rhee SG. Cell signaling. H2O2, a necessary evil for cell signaling. Science. 2006;312:1882–1883. doi: 10.1126/science.1130481. [DOI] [PubMed] [Google Scholar]
- [21].Poole LB, Nelson KJ. Discovering mechanisms of signaling-mediated cysteine oxidation. Curr Opin Chem Biol. 2008;12:18–24. doi: 10.1016/j.cbpa.2008.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Furdui CM, Poole LB. Chemical approaches to detect and analyze protein sulfenic acids. Mass Mass Spectrom Rev. 2014;33:126–146. doi: 10.1002/mas.21384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Paulsen CE, Carroll KS. Cysteine-mediated redox signaling: Chemistry, biology, and tools for discovery. Chem Rev. 2013;113:4633–4679. doi: 10.1021/cr300163e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Hall A, Nelson K, Poole LB, Karplus PA. Structure-based insights into the catalytic power and conformational dexterity of peroxiredoxins. Antioxid Redox Signal. 2011;15:795–815. doi: 10.1089/ars.2010.3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Winterbourn CC. Reconciling the chemistry and biology of reactive oxygen species. Nat Chem Biol. 2008;4:278–286. doi: 10.1038/nchembio.85. [DOI] [PubMed] [Google Scholar]
- [26].Adimora NJ, Jones DP, Kemp ML. A model of redox kinetics implicates the thiol proteome in cellular hydrogen peroxide responses. Antioxid Redox Signal. 2010;13:731–743. doi: 10.1089/ars.2009.2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Charles RL, Schröder E, May G, Free P, Gaffney PR, Wait R, Begum S, Heads RJ, Eaton P. Protein sulfenation as a redox sensor: proteomics studies using a novel biotinylated dimedone analogue. Mol Cell Proteomics. 2007;6:1473–1484. doi: 10.1074/mcp.M700065-MCP200. [DOI] [PubMed] [Google Scholar]
- [28].Leonard SE, Reddie KG, Carroll KS. Mining the thiol proteome for sulfenic acid modifications reveals new targets for oxidation in cells. ACS Chem Biol. 2009;4:783–799. doi: 10.1021/cb900105q. [DOI] [PubMed] [Google Scholar]
- [29].Poole LB, Klomsiri C, Knaggs SA, Furdui CM, Nelson KJ, Thomas MJ, Fetrow JS, Daniel LW, King SB. Fluorescent and affinity-based tools to detect cysteine sulfenic acid formation in proteins. Bioconjug Chem. 2007;18:2004–2017. doi: 10.1021/bc700257a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Rehder DS, Borges CR. Possibilities and pitfalls in quantifying the extent of cysteine sulfenic acid modification of specific proteins within complex biofluids. BMC Biochem. 2010;11:25. doi: 10.1186/1471-2091-11-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Klomsiri C, Nelson KJ, Bechtold E, Soito L, Johnson LC, Lowther WT, Ryu SE, King SB, Furdui CM, Poole LB. Use of dimedone-based chemical probes for sulfenic acid detection: evaluation of conditions affecting probe incorporation into redox-sensitive proteins. Methods Enzymol. 2010;473:77–94. doi: 10.1016/S0076-6879(10)73003-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Nelson KJ, Klomsiri C, Codreanu SG, Soito L, Liebler DC, Rogers LC, Daniel LW, Poole LB. Use of dimedone-based chemical probes for sulfenic acid detection; methods to visualize and identify labeled proteins. Methods Enzymol. 2010;473:95–115. doi: 10.1016/S0076-6879(10)73004-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Swedlow JR. Quantitative fluorescence microscopy and image deconvolution. Methods in Cell Biol. 2007;81:447–465. doi: 10.1016/S0091-679X(06)81021-6. [DOI] [PubMed] [Google Scholar]
- [34].Bolte S, Cordelieres FP. A guided tour into subcellular colocalization analysis in light microscopy. J Microsc. 2006;224:213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- [35].Durieux ME, Lynch KR. Signalling properties of lysophosphatidic acid. Trends Pharmacol Sci. 1993;14:249–254. doi: 10.1016/0165-6147(93)90021-b. [DOI] [PubMed] [Google Scholar]
- [36].Sekharam M, Cunnick JM, Wu J. Involvement of lipoxygenase in lysophosphatidic acid-stimulated hydrogen peroxide release in human HaCaT keratinocytes. Biochem J. 2000;3:751–758. 346 Pt. [PMC free article] [PubMed] [Google Scholar]
- [37].Stefanska J, Pawliczak R. Apocynin: molecular aptitudes. Mediators Inflamm. 2008;2008:106507. doi: 10.1155/2008/106507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Kalyanaraman B, Darley-Usmar V, Davies KJ, Dennery PA, Forman HJ, Grisham MB, Mann GE, Moore K, Roberts LJ, 2nd, Ischiropoulos H. Measuring reactive oxygen and nitrogen species with fluorescent probes: challenges and limitations. Free Radic Biol Med. 2012;52:1–6. doi: 10.1016/j.freeradbiomed.2011.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Urs NM, Jones KT, Salo PD, Severin JE, Trejo J, Radhakrishna H. A requirement for membrane cholesterol in the beta-arrestin- and clathrin-dependent endocytosis of LPA1 lysophosphatidic acid receptors. J Cell Sci. 2005;118:5291–5304. doi: 10.1242/jcs.02634. [DOI] [PubMed] [Google Scholar]
- [40].Wang DA, Lorincz Z, Bautista DL, Liliom K, Tigyi G, Parrill AL. A single amino acid determines lysophospholipid specificity of the S1P1 (EDG1) and LPA1 (EDG2) phospholipid growth factor receptors. J Biol Chem. 2001;276:49213–49220. doi: 10.1074/jbc.M107301200. [DOI] [PubMed] [Google Scholar]
- [41].Murph MM, Nguyen GH, Radhakrishna H, Mills GB. Sharpening the edges of understanding the structure/function of the LPA1 receptor: expression in cancer and mechanisms of regulation. Biochim Biophys Acta. 2008;1781:547–557. doi: 10.1016/j.bbalip.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Nauseef WM. Biological roles for the NOX family NADPH oxidases. J Biol Chem. 2008;283:16961–16965. doi: 10.1074/jbc.R700045200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Michalek RD, Nelson KJ, Holbrook BC, Yi JS, Stridiron D, Daniel LW, Fetrow JS, King SB, Poole LB, Grayson JM. The requirement of reversible cysteine sulfenic acid formation for T cell activation and function. J Immunol. 2007;179:6456–6467. doi: 10.4049/jimmunol.179.10.6456. [DOI] [PubMed] [Google Scholar]
- [44].Oshikawa J, Urao N, Kim HW, Kaplan N, Razvi M, McKinney R, Poole LB, Fukai T, Ushio-Fukai M. Extracellular SOD-derived H2O2 promotes VEGF signaling in caveolae/lipid rafts and post-ischemic angiogenesis in mice. PloS one. 2010;5:e10189. doi: 10.1371/journal.pone.0010189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Wani R, Qian J, Yin L, Bechtold E, King SB, Poole LB, Paek E, Tsang AW, Furdui CM. Isoform-specific regulation of Akt by PDGF-induced reactive oxygen species. Proc Natl Acad Sci U S A. 2011;108:10550–10555. doi: 10.1073/pnas.1011665108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Kaplan N, Urao N, Furuta E, Kim SJ, Razvi M, Nakamura Y, McKinney RD, Poole LB, Fukai T, Ushio-Fukai M. Localized cysteine sulfenic acid formation by vascular endothelial growth factor: role in endothelial cell migration and angiogenesis. Free Radic Res. 2011;45:1124–1135. doi: 10.3109/10715762.2011.602073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Chen CY, Willard D, Rudolph J. Redox regulation of SH2-domain-containing protein tyrosine phosphatases by two backdoor cysteines. Biochemistry. 2009;48:1399–1409. doi: 10.1021/bi801973z. [DOI] [PubMed] [Google Scholar]
- [48].Salmeen A, Andersen JN, Myers MP, Meng TC, Hinks JA, Tonks NK, Barford D. Redox regulation of protein tyrosine phosphatase 1B involves a sulphenyl-amide intermediate. Nature. 2003;423:769–773. doi: 10.1038/nature01680. [DOI] [PubMed] [Google Scholar]
- [49].Cunnick JM, Dorsey JF, Munoz-Antonia T, Mei L, Wu J. Requirement of SHP2 binding to Grb2-associated binder-1 for mitogen-activated protein kinase activation in response to lysophosphatidic acid and epidermal growth factor. J Biol Chem. 2000;275:13842–13848. doi: 10.1074/jbc.275.18.13842. [DOI] [PubMed] [Google Scholar]
- [50].Huang RY, Wen CC, Liao CK, Wang SH, Chou LY, Wu JC. Lysophosphatidic acid modulates the association of PTP1B with N-cadherin/catenin complex in SKOV3 ovarian cancer cells. Cell Biol Int. 2012;36:833–841. doi: 10.1042/CBI20110687. [DOI] [PubMed] [Google Scholar]
- [51].Paulsen CE, Truong TH, Garcia FJ, Homann A, Gupta V, Leonard SE, Carroll KS. Peroxide-dependent sulfenylation of the EGFR catalytic site enhances kinase activity. Nat Chem Biol. 2011;8:57–64. doi: 10.1038/nchembio.736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Grossmann KS, Rosario M, Birchmeier C, Birchmeier W. The tyrosine phosphatase Shp2 in development and cancer. Adv Cancer Res. 2010;106:53–89. doi: 10.1016/S0065-230X(10)06002-1. [DOI] [PubMed] [Google Scholar]
- [53].Woo HA, Yim SH, Shin DH, Kang D, Yu DY, Rhee SG. Inactivation of peroxiredoxin I by phosphorylation allows localized H2O2 accumulation for cell signaling. Cell. 2010;140:517–528. doi: 10.1016/j.cell.2010.01.009. [DOI] [PubMed] [Google Scholar]
- [54].Wood ZA, Poole LB, Karplus PA. Peroxiredoxin evolution and the regulation of hydrogen peroxide signaling. Science. 2003;300:650–653. doi: 10.1126/science.1080405. [DOI] [PubMed] [Google Scholar]
- [55].Crump KE, Juneau DG, Poole LB, Haas KM, Grayson JM. The reversible formation of cysteine sulfenic acid promotes B-cell activation and proliferation. Eur J Immunol. 2012;42:2152–2164. doi: 10.1002/eji.201142289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Wani R, Bharathi NS, Field J, Tsang AW, Furdui CM. Oxidation of Akt2 kinase promotes cell migration and regulates G1-S transition in the cell cycle. Cell Cycle. 2011;10:3263–3268. doi: 10.4161/cc.10.19.17738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Ushio-Fukai M. Compartmentalization of redox signaling through NADPH oxidase-derived ROS. Antioxid Redox Signal. 2009;11:1289–1299. doi: 10.1089/ars.2008.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Wu J, Xie N, Zhao X, Nice EC, Huang C. Dissection of aberrant GPCR signaling in tumorigenesis--a systems biology approach. Cancer Genomics Proteomics. 2012;9:37–50. [PubMed] [Google Scholar]
- [59].Sosa V, Moline T, Somoza R, Paciucci R, Kondoh H, ME LL. Oxidative stress and cancer: an overview. Ageing Res Rev. 2013;12:376–390. doi: 10.1016/j.arr.2012.10.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.