Abstract
Human polynucleotide kinase (hPNK) is a 57.1-kDa monomeric protein with conserved motifs associated with phosphatase and kinase activities. hPNK catalyzes phosphorylation of 5′-DNA termini and dephosphorylation of 3′-DNA termini. Previous studies, employing cell-free systems, have suggested that hPNK participates in the repair of DNA strand breaks. To better define the cellular function of hPNK, a double-stranded small-interfering RNA molecule designed to stably target hPNK transcription was introduced into A549 human lung adenocarcinoma cells. The small-interfering RNA suppressed hPNK gene expression by at least 80–90%. These cells exhibited a 7-fold higher spontaneous mutation frequency based on the development of resistance to ouabain; elevated sensitivity to a broad range of genotoxic agents including γ-radiation, UVC radiation, methyl methanesulfonate, hydrogen peroxide, and camptothecin; and slower repair of radiation-induced DNA strand breaks. These findings underscore the importance of hPNK in the maintenance of DNA integrity after damage induced by endogenous and exogenous agents.
DNA is considered to be a primary cellular target for many cytotoxic and carcinogenic agents (1). DNA strand breaks, which constitute a major class of DNA damage, can be induced by endogenous and exogenous agents, either directly or as intermediates in the DNA excision-repair pathways (2). Prompt and efficient repair of strand breaks is fundamental for maintenance of genomic integrity. Frequently, strand-break termini, especially those generated by means of reactive oxygen species or as intermediates in repair of oxidative base damage, carry chemical modifications, such as 3′-phosphate, that preclude immediate gap filling and religation by DNA polymerases and ligases because these enzymes have an absolute requirement for 3′-OH and 5′-phosphate termini. Additional processing of such termini is, therefore, necessary before strand rejoining can be completed.
Human polynucleotide kinase (hPNK) is a bifunctional enzyme with DNA 5′-kinase and 3′-phosphatase activities (3, 4). The cDNA for hPNK encodes a 57.1-kDa monomeric protein (3–5). A role for mammalian PNK in DNA repair has been espoused ever since its discovery (6–8), and more recently, the need for such an enzyme has been reiterated by those examining the actions of topoisomerase I inhibitors (9, 10). Jilani et al. (3) showed that hPNK expression could partially rescue the sensitivity to oxidative damaging agents of the Escherichia coli DNA repair-deficient exonuclease III/endonuclease IV double mutant. A Schizosaccharomyces pombe knockout mutant of Pnk1, the fission yeast ortholog of hPNK, exhibited hypersensitivity to ionizing radiation and camptothecin (a topoisomerase I inhibitor), both of which induce DNA strand breaks (11). However, there are some significant differences between human and fission yeast repair systems; e.g., S. pombe has no ortholog of XRCC1, and Pnk1 lacks the 17-kDa N-terminal domain present in hPNK, which contains a putative forkhead-associated domain (12).
Data obtained from cell-free systems have suggested that mammalian PNK participates in single-strand break (SSB) repair (13, 14), in which its activity is enhanced by interaction with XRCC1, DNA polymerase β, and DNA ligase III (14). Repair synthesis by DNA polymerase β is enhanced dramatically by the presence of a 5′-phosphate group (15), as well as the absolute necessity for a 3′-hydroxyl group. Experiments with cell-free extracts also have suggested that hPNK participates in nonhomologous end joining of double-strand breaks (DSBs) by promoting phosphate replacement at damage termini (16). However, to date, no naturally occurring mutants of mammalian PNK have been isolated with which the phenotype of cells lacking this enzyme could be determined. Here, we describe the generation of human cells expressing markedly reduced levels of PNK, brought about by stable small-interfering RNA silencing, and we show that they display hypersensitivity to a wide range of exogenous genotoxic agents, slower repair of DNA strand breaks, and elevated spontaneous mutation frequency, presumably arising from endogenous DNA damage.
Materials and Methods
Cell Lines and Culture Conditions. A549 (human lung carcinoma cells) were obtained from the American Type Culture Collection. The cells were cultured in a 1:1 DMEM/nutrient mixture F12 supplemented with 10% FBS/50 units/ml penicillin/50 μg/ml streptomycin/2 mM l-glutamine/0.1 mM nonessential amino acids/1 mM sodium pyruvate and maintained at 37°C under 5% CO2 in a humidified incubator. All culture supplies were purchased from Gibco/BRL.
Transfection Procedure. At 1 day before transfection, A549 cells were plated in 60-mm culture dishes so that they reached 50–70% confluency at the time of transfection. To obtain stable expression of the small-interfering RNA sequence and reduced expression of hPNK, an oligonucleotide (5′-AGAGATGACGGACTCCTCT-3′) directed against nucleotides 1391–1410 of the hPNK cDNA (GenBank accession no. AF125807) was designed. The duplex produced by annealing the forward and reverse oligonucleotides 5′-GATCCCCAGAGATGACGGACTCCTCTTTCAAGAGAAGAGGAGTCCGTCATCTCTTTTTTGGAA-3′ and 5′-AGCTTTTCCAAAAAAGAGATGACGGACTCCTCTTCTCTTGAAAGAGGAGTCCGTCATCTCTGGG-3′ was incorporated between BglII and HindIII sites in the pSUPER vector (17), which was then cotransfected into the A549 cells together with the pGFPneo plasmid carrying the Tn5 aminoglycoside phosphotransferase gene (neo) (pSUPER/pGFPneo, 5:1) by using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions with minor changes to obtain higher transfection efficiency. Briefly, the transfection complex containing 2.4 μg of pSUPER (with or without the inserted small-interfering RNA sequence) and 0.5 μg of pGFPneo plasmid DNA, and 10 μl of Lipofectamine 2000 was diluted in 600 μl of serum- and antibiotic-free DMEM and incubated at room temperature for 20 min before being added to each plate. After a 24-h incubation at 37°C, cells were trypsinized and passaged at a 1:10 dilution in complete growth medium. The next day, media from all plates was replaced with selective medium containing 300 μg/ml Geneticin (Gibco/BRL). Cells were grown in selective media for 2 weeks, and G418-resistant colonies were established in six-well plates, expanded, and cloned independently.
Preparation of Cell-Free Extract. After mild trypsinization, cells were collected by low-speed centrifugation, and the pellet was washed twice with PBS at room temperature. The cells were gently resuspended in ice-cold RIPA buffer (1× PBS/1% Nonidet P-40/0.1% SDS/0.5% sodium deoxycholate) containing a mixture of protease inhibitors (Sigma) (0.1 ml; 2 × 106 cells). The cells were held on ice for 30 min and then further disrupted by passage through a 21-gauge needle and held at 4°C for 30 min. Cell debris was removed by centrifugation at 12,000 × g for 10 min at 4°C. The supernatant typically contained 10 μg/μl protein. We used 50 μg of protein for each sample for Western blot analysis. Abs to human actin and p53 were purchased from Santa Cruz Biotechnology, XRCC1 Abs were obtained from NeoMarkers (Fremont, CA), and PNK mAbs were obtained from CytoStore (Calgary, AB, Canada). For partial purification, supernatant was dialyzed against buffer A (50 mM Tris·Cl, pH 7.5/150 mM NaCl/1 mM DTT/protease inhibitor mixture), and 500 μg of protein was loaded onto a Superdex-75 PC 3.2/30 gel-filtration column attached to a SMART micropurification system (Amersham Biosciences). The protein was eluted with buffer A in 80-μl fractions.
Kinase and Phosphatase Assays. DNA kinase activity was determined as described (4) by using micrococcal nuclease digested DNA as substrate and 10 μl of the major fraction (fraction 6) eluted from the gel filtration column (see above). For phosphatase activity (13), the assay measured the conversion of an oligonucleotide (5′-ATTACGAATGCCCACACCGCC-3′; p21p), bearing a 32P-label at the 5′ terminus as well as a 3′-phosphate, to the 3′-dephosphorylated 21-mer (p21). Reaction was carried out in 70 mM Tris·HCl, pH 7.6/10 mM MgCl2/5 mM DTT at 37°C for 60 min by using 10 μg of crude cell extract.
Clonogenic Cell Survival Assays. Exponentially growing stock cultures of A549, C-ter3, and pSuper control cells were subcultured in 60-mm culture dishes at densities of 200–600 cells per dish. The dishes were incubated overnight at 37°C to allow for cell attachment. The cells were then subjected to γ-radiation (60Co Gammacell; Atomic Energy of Canada Limited, Ottawa) or UVC radiation (G15T8 15W germicidal bulb; General Electric), or they were exposed to graded doses of methyl methanesulfonate (MMS; Sigma), hydrogen peroxide (Fisher Scientific), camptothecin (Sigma), and etoposide (Sigma) for 2 h at 37°C. (For UV exposure, the medium was removed, and the cells were washed with PBS and then drained before irradiation.) Drug-containing medium was then replaced with fresh DMEM/F12 medium, and the plates were incubated at 37°C for 2–3 weeks. Colonies were fixed in 70% ethanol, stained with 10% methylene blue, and counted. The surviving fraction was plotted as a function of dose and fitted by using prism software (version 3.03, GraphPad, San Diego).
Imaging of H2AX Phosphorylation. The introduction of DSBs by ionizing radiation was monitored by the protocol of Furuta et al. (18) with some modifications. We seeded ≈1 × 105 cells onto each glass coverslip, retained them in 35-mm tissue culture dishes, and incubated them overnight at 37°Cin a CO2 incubator. On the next day, the cells were exposed to 5 Gy γ-radiation and incubated for various times up to 24 h. The cells were then fixed in 2% paraformaldehyde in PBS for 5 min, washed with PBS and 50:50 methanol/PBS solution, and permeabilized with 100% methanol for 20 min at -20°C. After removing the methanol, the cells were washed with PBS and blocked with 5% milk powder at room temperature. A mAb to anti-phospho-histone H2AX (Ser-139) (Upstate, Charlottesville, VA) diluted 500-fold in 5% milk powder was placed on the coverslips and incubated for 1 h in the dark. The coverslips were washed twice with PBS alone and PBS containing 0.1% Tween 20. Alexa Fluor 488 goat anti-mouse secondary Ab (Molecular Probes) diluted 1:500 in 5% milk powder was added to the coverslips and incubated at room temperature for 1 h in the dark. The coverslips were again washed twice with PBS and PBS/Tween 20 and finally rinsed with water. The coverslips were mounted on a microscope slide by using 95% glycerol in PBS containing 3 μg/ml 4′,6-diamidino-2-phenylindole. Slides were stored at 4°C in the dark. Phosphorylated H2AX foci were viewed with a LSM510 laser-scanning confocal microscope (Zeiss) mounted on a Axiovert 100M microscope (Zeiss). Images were taken with a ×40 (NA 1.3) objective, following the Nyquist sampling requirement, with the same instrument settings for different slides. The average integrated intensity per nucleus was determined by using metamorph offline 6.1 software (Universal Imaging, Downingtown, PA). Briefly, images were background-corrected first, and a mask was then generated over the nucleus of each cell by using a 4′,6-diamidino-2-phenylindole counter stain. Integrated intensity of the H2AX stain in each nucleus was calculated within the masked volume.
Determination of SSBs Plus Alkali-Labile Sites by Single-Cell Gel Electrophoresis. For each slide, 1 × 105 cells were trypsinized and mixed with 0.1% molten low-melting-point agarose at 42°C at a 1:10 ratio (10 μl cells per 100 μl of agarose). The mixture (50 μl) was spread immediately on a comet slide (Trevigen, Gaithersburg, MD) and kept at 4°C for 40 min to allow the agarose to solidify. Slides were immersed in prechilled lysis solution (2.5 M NaCl/100 mM Na2·EDTA, pH 10/10 mM Tris base/1% SDS/1% Triton X-100), kept on ice for 40 min, and then immersed in an alkaline solution (300 mM NaOH/1 mM Na2·EDTA) for 30 min at 23°C. Slides were then placed in an electrophoresis apparatus filled with fresh alkaline solution and run at 1 V/cm and ≈300 mA at 4°C for 40 min. The slides were then washed in 70% ethanol for 5 min. The DNA was stained with SYBR Green I (Molecular Probes) and viewed with an AxioScope 2 fluorescence microscope (Zeiss). For each data point a minimum of 100 random cell images were visually analyzed and categorized according to the National Institutes of Health listserv (Comet Assay Interest Group web site; http://cometassay.com/introduction.htm) and Kobayashi et al. (19).
Determination of Spontaneous Mutation Frequency. On the day before ouabain selection, stock cultures of A549, C-ter3, and pSup control cells were trypsinized and pooled, and 2 × 107 cells from each cell line were reseeded at a density of 1 × 106 cells per 100-mm plate in DMEM/F12 medium supplemented with 10% FCS and antibiotics. (Additional aliquots of the cell suspension were diluted and replated in triplicate at a density of 200–400 cells per 60-mm dish for the determination of plating efficiency.) The next day, mutation selection was carried out by replacing the medium on all of the plates with selective medium (DMEM/F12) containing 0.1 μM ouabain, 15% FCS, and antibiotics. The cells were maintained in selective medium for 21–28 days until mutant colonies developed. The mutant colonies were fixed and stained with 10% methylene blue. The numbers of ouabain-resistant colonies were counted, and the spontaneous mutation frequency was calculated as the number of mutant colonies divided by the plating efficiency per 107 cells.
Results and Discussion
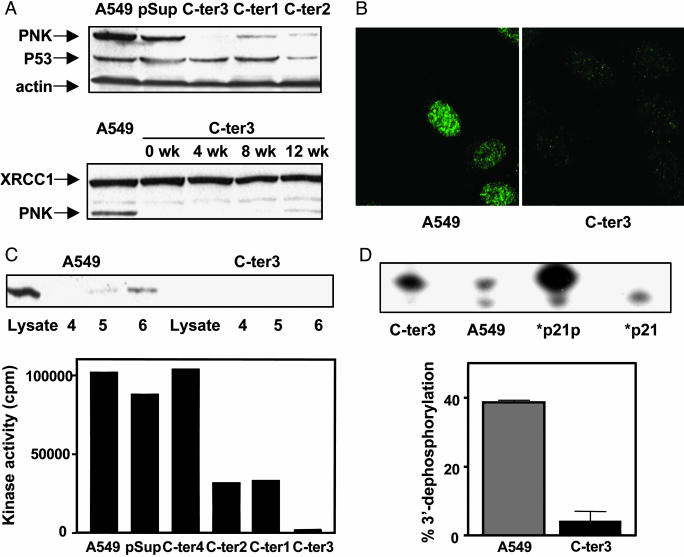
To reduce hPNK expression in a human cell line, we incorporated an oligonucleotide sequence directed against nucleotides 1391–1410 (5′-AGAGATGACGGACTCCTCT-3′) of the hPNK cDNA (GenBank accession no. AF125807) in the pSUPER vector (17) and transfected the construct into A549 human lung adenocarcinoma cells. Cotransfection with a plasmid carrying a neo expression cassette and selection with G418 enabled us to isolate individual clones of hPNK-depleted A549 cells. Reduction of hPNK in these stable transfectants was evident by Western blot analysis (Fig. 1A) and was not accompanied by suppression of its associated protein, XRCC1, or p53. Similarly, the expression of several other repair-related proteins, including DNA ligase III, DNA polymerase β, DNA PKCs, and p21, was unaffected by loss of hPNK expression (data not shown). One clone in particular, C-ter3, showed a marked reduction (≈90%) of hPNK, which was confirmed by immunofluorescence (Fig. 1B). Fig. 1 A indicates also that suppression of hPNK was stable, being lost only gradually over a period of weeks of continuous cell culture in the absence of G418 after initial selection. DNA kinase activity in several of the G418 resistant clones was measured in cell-free extracts fractionated by gel filtration (Fig. 1C). The results reflected the level of hPNK revealed by the Western blot analysis. When residual 3′-phosphatase activity in cell-free extracts was quantified by using a duplex oligonucleotide substrate containing a DNA nick with a 3′-phosphate, we observed that the C-ter3 cells had ≈10% of the wild-type activity (Fig. 1D). A similar result was obtained with a substrate containing a one-nucleotide gap (data not shown). Because the phosphatase assay used crude cell extract under conditions suitable for other potential 3′-phosphatases, such as APE1 (20), this result implies that hPNK is the dominant 3′-phosphatase in A549 cells and is consistent with the previous observation that PNK is the major 3′-DNA phosphatase associated with chromatin in mammalian cells (21). This result also agrees with experiments that demonstrate that other mammalian DNA-repair enzymes, including APE1, have poor 3′-phosphatase activity (L. Wiederhold, T. K. Hazra, T. Izumi, and S. Mitra, unpublished data).
Fig. 1.
Suppression of hPNK gene expression in the human A549 cell line. (A) Upper Western blot shows expression of hPNK, p53, and actin in A549 control, vector only (pSup), and three G418 resistant clones: C-ter1–3 cells. Lower Western blot shows the stability of down-regulation of hPNK in the C-ter3 cells over a period of 12 weeks. XRCC1 expression was used as a control. (B) Immunofluorescence of hPNK in A549 control and C-ter3 cells. (C) hPNK DNA kinase activity in fractionated cell extracts isolated from A549 cells, vector-only controls (pSup), and several G418-resistant clones. Confirmation that fraction 6 obtained from C-ter3 cells lacks hPNK is shown in the Western blot. (D) hPNK 3′-phosphatase activity in crude cell extracts of wild-type A549 and C-ter3 cells. The duplex substrate consisting of three oligonucleotides, a 45-mer complementary to a 24-mer, and a 5′-labeled 21-mer bearing a 3′-phosphate (p21p) to mimic a single-strand nick has been fully described (13). The lanes marked p21p and p21 indicate the starting material and a marker for the dephosphorylated product, respectively.
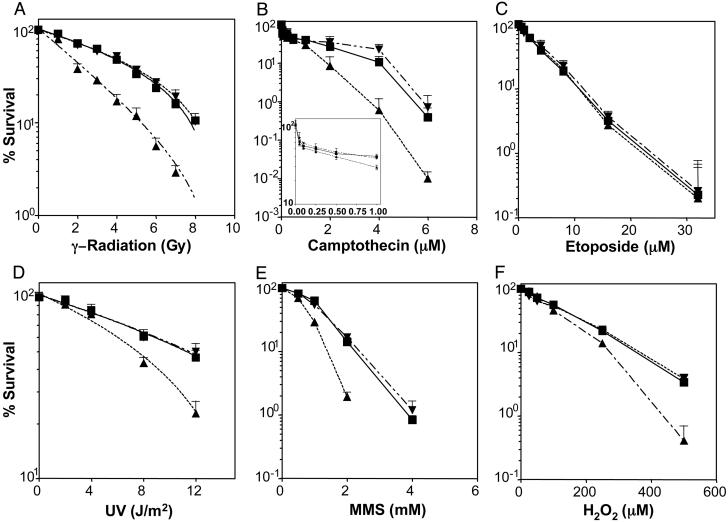
Cellular sensitivity of the C-ter3 cells to a series of genotoxic agents was determined by the clonogenic survival assay (Fig. 2). Loss of expression of hPNK resulted in elevated sensitivity to γ-radiation, camptothecin, the alkylating agent MMS, hydrogen peroxide, and UVC radiation, but not to etoposide (a topoisomerase II inhibitor). The significant hypersensitivity (≈2-fold) of C-ter3 cells to γ-radiation (Fig. 2 A) was not unexpected because the majority of radiation-induced 3′-strand-break termini possess a phosphate or phosphoglycolate residue (22), and recent observations (23) suggest that hPNK plays a role in the removal of 3′-phosphoglycolate residues as well as 3′-phosphate groups.
Fig. 2.
Survival curves of A549, vector-only, and knock-down cells treated with different genotoxic agents. ▪, A549; ▾, vector-only control; ▴, C-ter3. Cells were exposed to various doses and concentrations of DNA-damaging agents, as indicated. Each data point represents the mean values from a minimum of three independent determinations ± SEM.
The difference in response to the two topoisomerase inhibitors can be explained on the basis of the mechanisms of strand cleavage induced by topoisomerase I and II and the repair pathways that overcome topoisomerase inhibition. Topoisomerase I cleavage of a phosphodiester bond generates a 5′-OH terminus and covalent linkage of the protein to the phosphate group at the 3′ terminus (24). Addition of camptothecin blocks topoisomerase I action at this stage. Repair of these topoisomerase I “dead-end” complexes is initiated by hydrolysis of the topoisomerase-phosphate bond, in a reaction catalyzed by tyrosyl-DNA phosphodiesterase, to yield 3′-phosphate and 5′-OH termini (25), i.e., termini requiring processing by PNK. Strand cleavage by topoisomerase II, on the other hand, produces 3′-OH termini and covalent attachment of the enzyme to the 5′-phosphoryl group (24). Etoposide extends the lifetime of these intermediates by inhibiting the religation step (24). The fate of the blocked intermediates has not been fully elucidated but similar hydrolysis of the enzyme–phosphate bond would yield termini that do not require further processing before ligation. Furthermore, the most likely route of repair of topoisomerase II-blocked termini is by means of homologous recombination, which may not necessitate hydrolysis of the enzyme-DNA complex (26). Close examination of Fig. 2B indicates a biphasic response to camptothecin in both the control and PNK knock-down cells, which most likely reflects the heightened sensitivity of cells in S phase (27). The aberrant response of C-ter3 cells to camptothecin was confined to drug doses >1 μM, which implies that hPNK does not play a role in the survival of camptothecin-treated cells in S phase.
The sensitivity of C-ter3 cells to MMS was unanticipated because this agent does not generate strand breaks directly. However, strand breaks with 3′-phosphates may arise as repair intermediates in a PNK-dependent base excision-repair (BER) pathway (L. Wiederhold, T. K. Hazra, T. Izumi, and S. Mitra, unpublished data). MMS methylates DNA bases, which are removed subsequently by DNA glycosylases, such as 3-methy-ladenine-DNA glycosylase, to generate abasic sites. The recently identified DNA glycosylases, NEIL1 and NEIL2 (28), can catalyze βδ-elimination at the resultant abasic sites, producing strand breaks with 3′-phosphate termini requiring PNK for complete repair. Our results suggest that such a pathway competes with the traditional APE1-dependent BER pathway and that a deficiency of PNK can result in the accumulation of cytotoxic strand-break intermediates.
Also unanticipated was the ≈1.6-fold greater sensitivity to UVC radiation displayed by the C-ter3 cells (Fig. 2D). The major lesions generated by UVC radiation are cyclobutane pyrimidine dimers and 6,4-photoproducts, which are repaired by means of the nucleotide excision-repair pathway (29). Because there is no evidence that either 3′-phosphate or 5′-OH termini are produced in this repair pathway (29), the UV sensitivity of the C-ter3 cells implies that UVC can induce DNA strand breaks with either or both of these terminal groups in human cells. Again, such strand breaks might be induced directly or through an alternative repair pathway for UV-induced base damage. However, direct induction of SSBs by UVC is relatively low. Peak and Peak (30) determined that exposure of human P3 teratocarcinoma cells to a dose of 10 J/m2 254-nm light directly generated 140 SSB per cell, whereas irradiation with 1-Gy x-rays yielded 840 SSB. Thus, directly induced strand breaks are unlikely to account for the observed hypersensitivity of C-ter3 cells to UVC radiation, leaving open the possibility for repair-mediated strand scission that is not part of the NER pathway. This repair may be in response to base damage other than UV dimers (e.g., oxidative DNA damage) and would suggest that such damage contributes to a low but measurable extent to UVC-induced cytotoxicity. Interestingly, EM9 hamster cells, which express a truncated form of XRCC1, display ≈1.8-fold elevated sensitivity to 254-nm light (31) and show a deficiency in strand rejoining after UVC irradiation that is corrected in XRCC1-complemented cells (32).
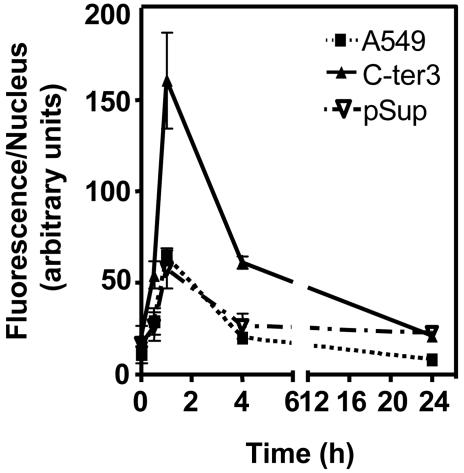
DNA DSBs are considered to contribute significantly toward genomic instability and cell lethality induced by ionizing radiation (33). Most, if not all, radiation-induced DSBs would require processing before strand rejoining. In light of our observation that C-ter3 cells display elevated sensitivity to ionizing radiation (Fig. 2 A) and the fact that in vitro hPNK can act efficiently at DSB to restore 5′-phosphate groups and remove 3′-phosphate groups (16, 23), we examined the effect of reduction of hPNK expression on DSB repair in A549 cells. For this analysis, we made use of the recent observations that the introduction of DSBs by ionizing radiation induces phosphorylation of serine 139 of histone H2AX, and the measurement of the phosphorylated histone (γ-H2AX) is a useful means by which to monitor the repair of DSB (18, 34, 35). Accordingly, we monitored the level of γ-H2AX before and after irradiation (5 Gy) in wild-type, control, and hPNK knock-down cells (Fig. 3 and see Fig. 6, which is published as supporting information on the PNAS web site). All of the cell lines exhibited a sharp increase in the level of γ-H2AX up to 1 h after exposure to ionizing radiation. Although the cell lines displayed the same pattern of H2AX phosphorylation over time, phosphorylation in the C-ter3 cells was greater (≈3-fold at 1 h after irradiation) and more prolonged than in the parental A549 or the vector-only control cells. Nonetheless, delay in DSB repair in C-ter3 cells appeared to be transient because the γ-H2AX level was similar to that in the control cells after 24 h, indicating that either the residual hPNK in the cells was sufficient to eventually process all of the modified termini or that an alternative repair pathway was used by the cells that bypassed the need for hPNK. Further study will be required to examine the fidelity of strand rejoining in the hPNK knock-down cells.
Fig. 3.
Quantification of the average integrated fluorescence intensity per nucleus due to phosphorylation of histone H2AX as a function of time after 5-Gy irradiation. [Fig. 6 shows cells stained with a nuclear stain (4′,6-diamidino-2-phenylindole) and Abs to phosphorylated histone H2AX (γ-H2AX) at various times after irradiation.]
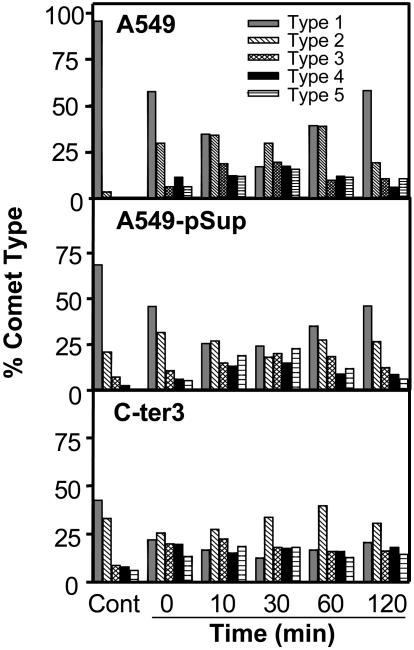
We used single-cell gel electrophoresis (comet assay) to also compare the relative level of SSBs plus alkali-labile sites in unirradiated and irradiated (5 Gy) cells. Fig. 4 indicates a higher basal level of these lesions in unirradiated C-ter3 cells than in either A549 or vector-only transfected cells. After irradiation, the level of measurable damage (based on the increased level of type 3–5 comets and concomitant drop of type 1 and 2 comets) appeared to maximize at 30 min after irradiation. In the wild-type and vector-only transfected cells this maximal level had been reduced by 50–60% by 120 min after irradiation, but the reduction of type 3–5 comets in C-ter3 cells was only ≈20%.
Fig. 4.
SSBs plus alkali-labile sites before (Cont) and at the indicated times after 5-Gy irradiation. Comets were categorized according to the National Institutes of Health listserv (Comet Assay Interest Group web site) and Kobayashi et al. (19), type 1 comets having the least damage and type 5 comets having the most damage.
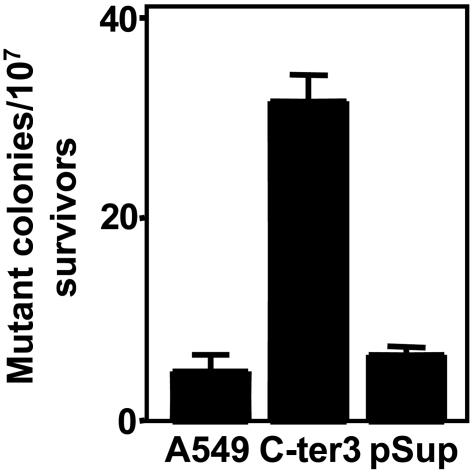
Finally, to evaluate the effects of reduced expression of hPNK on spontaneous mutation frequency, we compared C-ter3 cells with the controls for the development of resistance to ouabain (36). This drug binds competitively to the K+ ion binding domain of the plasma membrane Na+/K+ pump, thereby inhibiting ATP production (37). Fig. 5 reveals that the mutation frequency giving rise to ouabain resistance of the hPNK-deficient cells was ≈7-fold higher than that of the parental cells or vector-only controls. Thus, it can be inferred that hPNK is required for the protection of cells from endogenous DNA damage.
Fig. 5.
Influence of hPNK on spontaneous mutation frequency. Mutation of the plasma membrane Na+/K+ pump was determined by ouabain resistance. Cells were cultured in complete media containing 0.1 μM ouabain for 3–4 weeks until ouabain-resistant colonies could be visualized. The plot represents the mean values from three independent determinations ± SEM.
Results presented here, together with data obtained from cell-free systems (13, 14, 16), attest to a significant role for mammalian PNK in the cellular response to genotoxic stress from a diverse array of sources and implies its participation in several repair pathways. In addition to the potential importance hPNK expression level may have with regard to exposure to environmental carcinogens, it could also influence clinical outcome. Ionizing radiation, alkylating agents, and topoisomerase I inhibitors, such as camptothecin, are widely used in the treatment of cancer. Cancer cells expressing low levels of hPNK should be more sensitive to these forms of therapy. If so, the assessment of hPNK levels may have predictive value and the enzyme may be a useful target for enhancing treatment with these agents.
Supplementary Material
Acknowledgments
We thank Dr. Reuven Agami (The Netherlands Cancer Institute, Amsterdam) for providing the pSUPER vector; Dr. Xuejun Sun and Gerry Barron (Cell Imaging Facility, Cross Cancer Institute) for assistance with immunofluorescence; and Drs. Keith Caldecott (University of Sussex, Brighton, U.K.) and Sankar Mitra (University of Texas Medical Branch, Galveston) for reviewing the manuscript. This work was supported by a grant from the Canadian Institutes of Health Research (to M.W.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: hPNK, human polynucleotide kinase; MMS, methyl methanesulfonate; DSB, double-strand break; SSB, single-strand break.
References
- 1.Friedberg, E. C., Walker, G. C. & Siede, W. (1995) DNA Repair and Mutagenesis (ASM Press, Washington, D.C.), pp. 1-58.
- 2.Hoeijmakers, J. H. (2001) Nature 411, 366-374. [DOI] [PubMed] [Google Scholar]
- 3.Jilani, A., Ramotar, D., Slack, C., Ong, C., Yang, X. M., Scherer, S. W. & Lasko, D. D. (1999) J. Biol. Chem. 274, 24176-24186. [DOI] [PubMed] [Google Scholar]
- 4.Karimi-Busheri, F., Daly, G., Robins, P., Canas, B., Pappin, D. J., Sgouros, J., Miller, G. G., Fakhrai, H., Davis, E. M., Le Beau, M. M., et al. (1999) J. Biol. Chem. 274, 24187-24194. [DOI] [PubMed] [Google Scholar]
- 5.Mani, R. S., Karimi-Busheri, F., Fanta, M., Cass, C. E. & Weinfeld, M. (2003) Biochemistry 42, 12077-12084. [DOI] [PubMed] [Google Scholar]
- 6.Teraoka, H., Mizuta, K., Sato, F., Shimoyachi, M. & Tsukada, K. (1975) Eur. J. Biochem. 58, 297-302. [DOI] [PubMed] [Google Scholar]
- 7.Kleppe, K. & Lillehaug, J. R. (1979) Adv. Enzymol. 48, 245-275. [DOI] [PubMed] [Google Scholar]
- 8.Zimmerman, S. B. & Pheiffer, B. H. (1981) in The Enzymes, ed. Boyer, P. D. (Academic, New York), Vol. 14, pp. 315-329. [Google Scholar]
- 9.Yang, S.-W., Burgin, A. B. Jr., Huizenga, B. N., Robertson, C. A., Yao, K. C. & Nash, H. A. (1996) Proc. Natl. Acad. Sci. USA 93, 11534-11539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nitiss, J. L. & Wang, J. C. (1996) Mol. Pharmacol. 50, 1095-1102. [PubMed] [Google Scholar]
- 11.Meijer, M., Karimi-Busheri, F., Huang, T. Y., Weinfeld, M. & Young, D. (2002) J. Biol. Chem. 277, 4050-4055. [DOI] [PubMed] [Google Scholar]
- 12.Caldecott, K. W. (2003) Cell 112, 7-10. [DOI] [PubMed] [Google Scholar]
- 13.Karimi-Busheri, F., Lee, J., Tomkinson, A. E. & Weinfeld, M. (1998) Nucleic Acids Res. 26, 4395-4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Whitehouse, C. J., Taylor, R. M., Thistlethwaite, A., Zhang, H., Karimi-Busheri, F., Lasko, D. D., Weinfeld, M. & Caldecott, K. W. (2001) Cell 104, 107-117. [DOI] [PubMed] [Google Scholar]
- 15.Chagovetz, A. M., Sweasy, J. B. & Preston, B. D. (1997) J. Biol. Chem. 272, 27501-27504. [DOI] [PubMed] [Google Scholar]
- 16.Chappell, C., Hanakahi, L. A., Karimi-Busheri, F., Weinfeld, M. & West, S. C. (2002) EMBO J. 21, 2827-2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brummelkamp, T. R., Bernards, R. & Agami, R. (2002) Science 296, 550-553. [DOI] [PubMed] [Google Scholar]
- 18.Furuta, T., Takemura, H., Liao, Z. Y., Aune, G. J., Redon, C., Sedelnikova, O. A., Pilch, D. R., Rogakou, E. P., Celeste, A., Chen, H. T., et al. (2003) J. Biol. Chem. 278, 20303-20312. [DOI] [PubMed] [Google Scholar]
- 19.Kobayashi, H., Sugiyama, C., Morikawa, Y., Hayashi, M. & Sofuni, T. (1995) MMS Commun. 3, 103-115. [Google Scholar]
- 20.Wilson III, D. M. (2003) J. Mol. Biol. 330, 1027-1037. [DOI] [PubMed] [Google Scholar]
- 21.Habraken, Y. & Verly, W. G. (1988) Eur. J. Biochem. 171, 59-66. [DOI] [PubMed] [Google Scholar]
- 22.Henner, W. D., Rodriguez, L. O., Hecht, S. M. & Haseltine, W. A. (1983) J. Biol. Chem. 258, 711-713. [PubMed] [Google Scholar]
- 23.Inamdar, K. V., Pouliot J. J., Zhou, T., Lees-Miller, S. P., Rasouli-Nia, A. & Povirk, L. F. (2002) J. Biol. Chem. 277, 27162-27168. [DOI] [PubMed] [Google Scholar]
- 24.Wang, J. C. (1996) Annu. Rev. Biochem. 65, 635-692. [DOI] [PubMed] [Google Scholar]
- 25.Pouliot, J. J., Pouliot J. J., Yao, K. C., Robertson, C. A. & Nash, H. A. (1999) Science 286, 552-555. [DOI] [PubMed] [Google Scholar]
- 26.Lundin, C., Schultz, N., Arnaudeau, C., Mohindra, A., Hansen, L. T. & Helleday, T. (2003) J. Mol. Biol. 328, 521-535. [DOI] [PubMed] [Google Scholar]
- 27.Borovitskaya, A. E. & D'Arpa, P. (1998) Oncol. Res. 10, 271-276. [PubMed] [Google Scholar]
- 28.T. K. Hazra, T. K., Izumi, T., Boldogh, I., Imhoff, B., Kow, Y. W., Jaruga, P., Dizdaroglu, M. & Mitra, S. (2002) Proc. Natl. Acad. Sci. USA 99, 3523-3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Batty, D. P. & Wood, R. D. (2000) Gene 241, 193-204. [DOI] [PubMed] [Google Scholar]
- 30.Peak, J. G. & Peak, M. J. (1991) Mutat. Res. 246, 187-191. [DOI] [PubMed] [Google Scholar]
- 31.Thompson, L. H., Brookman, K. W., Dillehay, L. E., Carrano, A. V., Mazrimas J. A., Mooney, C. L. & Minkler, J. L. (1982) Mutat. Res. 95, 427-440. [DOI] [PubMed] [Google Scholar]
- 32.Nocentini, S. (1999) Radiat. Res. 151, 423-432. [PubMed] [Google Scholar]
- 33.Thompson, L. H. & Schild, D. (2002) Mutat. Res. 509, 49-78. [DOI] [PubMed] [Google Scholar]
- 34.Sedelnikova, O. A., Rogakou, E. P., Panyutin, I. G. & Bonner, W. M. (2003) Radiat. Res. 158, 486-492. [DOI] [PubMed] [Google Scholar]
- 35.Rothkamm, K. & Löbrich, M. (2003) Proc. Natl. Acad. Sci. USA 100, 5057-5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cole, J., Arlett, C. F. & Green, M. H. (1976) Mutat. Res. 41, 377-386. [DOI] [PubMed] [Google Scholar]
- 37.Lamb, J. F. & McCall, D. (1971) J. Physiol. (London) 213, 57P-58P. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.