Abstract
By proteolytic modification of low abundant signaling proteins and membrane receptors, proteases exert potent posttranslational control over cell behavior at the postsecretion level. Hence, substrate discovery is indispensable for understanding the biological role of proteases in vivo. Indeed, matrix metalloproteinases (MMPs), long associated with extracellular matrix degradation, are increasingly recognized as important processing enzymes of bioactive molecules. MS is now the primary proteomic technique for detecting, identifying, and quantitating proteins in cells or tissues. Here we used isotopecoded affinity tag labeling and multidimensional liquid chromatography inline with tandem MS to identify MDA-MB-231 breast carcinoma cell proteins shed from the cell surface or the pericellular matrix and extracellular proteins that were degraded or processed after transfection with human membrane type 1-MMP (MT1-MMP). Potential substrates were identified as those having altered protein levels compared with the E240A inactive MT1-MMP mutant or vector transfectants. New substrates were biochemically confirmed by matrix-assisted laser desorption ionization–time-of-flight MS and Edman sequencing of cleavage fragments after incubation with recombinant soluble MT1-MMP in vitro. We report many previously uncharacterized substrates of MT1-MMP, including the neutrophil chemokine IL-8, secretory leukocyte protease inhibitor, pro-tumor necrosis factor α, death receptor-6, and connective tissue growth factor, indicating that MT1-MMP is an important signaling protease in addition to its traditionally ascribed roles in pericellular matrix remodeling. Moreover, the high-throughput and quantitative nature of isotope-coded affinity tag labeling combined with tandem MS sequencing is a previously undescribed degradomic screen for protease substrate discovery that should be generally adaptable to other classes of protease for exploring proteolytic function in complex and dynamic biological contexts.
In all living organisms, proteases exert high-order posttranslational control over a diverse range of cellular functions. Altered protease expression and substrate proteolysis are pivotal elements in the pathogenesis of many diseases (1) with 53 hereditary genetic diseases of proteolysis recognized (2). Indeed, proteases represent ≈10% of current drug targets (3). Elucidating the substrate repertoire of a protease is critical to understanding its biological role, but given the large number of proteases (>553) present in the human genome (2), this is a daunting task. Innovative approaches using combinatorial or positional scanning libraries of fluorogenic (4) and inhibitory peptides (5) and oriented (6) and phage display (7) peptide libraries can determine consensus protease cleavage sequences. However, bioinformatic identification of proteins containing these sequences followed by biochemical and in vivo validation of proteolytic susceptibility has led to the identification of relatively few biologically relevant new substrates. This is not surprising, because the majority of substrates in vivo are proteins and not peptides. Moreover, substrates must colocalize with proteases, spatially and temporally. Although useful, these techniques are inherently limited in their power, because they do not consider the influence of protein conformation, posttranslational modification, protease exosites, and substrate availability in vivo (8). Serial analysis of protein libraries and exosite scanning by yeast two-hybrid screens, although time consuming, have proven effective in identifying new protein substrates (9). Nonetheless, because proteases do not operate alone but more commonly in amplification cascades or regulatory circuits in the presence of a multitude of interacting proteins, substrates, and cleavage products, and often in distinct compartments, biologically relevant protease substrates in vivo may differ from theoretical activities inferred from in vitro experiments. In the face of such biochemical complexity, this emphasizes the need to identify protease-cleaved substrates and not just enzyme activities in cells, tissues, and whole organisms (10, 11). Thus, rapid techniques are needed to directly identify new substrates in biological settings and to quantitate differences in substrate processing as disease biomarkers and as surrogate markers of antiproteolytic drug treatment (3).
Proteomics offers the potential to identify protease substrates in complex biological samples in a system-wide approach that has been termed “degradomics” (12). However, apart from isolated reports (13), proteomic approaches for protease substrate identification have not been widely developed. In part, this may be due to difficulty in relating cleavage products with parent protein spots on 2D gels or to MS spectral peaks. Difficulties in quantification also hinder comparative analyses. Recently, techniques have been developed that incorporate stable isotopes into proteins by metabolic or posttranslational labeling (reviewed in ref. 14), thereby allowing MS quantification of the relative amounts of protein present in two samples (15). In isotope-coded affinity tag (ICAT) labeling (16), proteins in two samples that are to be compared are reduced and labeled by reductive alkylation of cysteines by using biotin-tagged reagents that are chemically identical but that differ in isotopic composition and mass. The two isotopically distinct labeled protein samples are then combined and thereafter treated identically, enabling quantitative comparisons to be made. Tryptic peptides are prepared, and the biotin-tagged cysteine-containing peptides only are avidin column purified for mass determination. Spectral peak analysis of the isotopically resolved identical peptides from the two sources enables relative quantitation. These proteins are then identified by tandem MS (MS/MS) sequencing in an iterative approach. ICAT has been recently refined by labeling with [13C]9 to avoid the slight alteration in hydrophobicity occurring in deuterium-labeled peptides and by the use of cleavable linkers to remove the biotin moiety after elution.
Matrix metalloproteinase (MMP)-14, or membrane type (MT)1-MMP, is one of 23 members of an important protease family historically associated with the cleavage of extracellular matrix proteins (17). MT1-MMP is constitutively activated by furin processing, is essential for normal growth and development (18), and is up-regulated in cancer and arthritis (19). MT1-MMP degrades fibrillar collagen and fibronectin (20, 21) and activates MMP-2 (gelatinase A) (22, 23), which is also heavily implicated in cancer metastasis. However, like most proteases, the breadth of potential in vivo substrates and biological role of MT1-MMP are largely unknown. This is important to address because, of all MMP gene knockouts, only the MT1-MMP-/- mouse exhibits strong developmental abnormalities (18).
Here we have developed a functional proteomic screen to identify new protease substrates in cell cultures. We hypothesized that the levels of individual proteins in conditioned culture medium would be differentially altered after cell transfection with MT1-MMP and thus would be amenable to ICAT and MS/MS identification. After proteolysis, secreted proteins may be reduced in amount if degraded or processed and subsequently cleared after reductions in stability, and other proteins may increase in quantity if proteolytically shed from the cell surface or released from the pericellular matrix. We report that ICAT is applicable to the proteomic analysis of proteolytic activity in complex cellular systems with the identification of multiple previously undescribed signaling molecules as substrates of MT1-MMP.
Experimental Procedures
Cell Culture. Human MDA-MB-231 breast carcinoma cells stably transfected with FLAG-tagged human MT1-MMP, a catalytically inactive MT1-MMP mutant (E240A) in which the active-site Glu is replaced, or pCR3.1 vector alone under G418 selection were as described (21). MT1-MMP expression was confirmed by Western blotting and flow cytometry. MT1-MMP activity was confirmed by zymographic analysis of MMP-2 activation when the transfected cells were incubated with human proMMP-2 expressed from Timp2-/- cells (23).
For protein analysis, confluent transfected MDA-MB-231 cells were grown in DMEM, 10% FBS, and G418 (1 mg/ml), washed with PBS, and incubated in DMEM alone to eliminate serum contamination. After 48 h, the medium was replaced with DMEM and ascorbic acid (50 μg/ml). The conditioned medium was harvested (48 h), protease inhibitors (6.7 μM EDTA, 1 mM PMSF, 10 μM leupeptin, and 1 μM pepstatin A) were immediately added, and the conditioned medium clarified by centrifugation (30 min, 1,000 × g) and filtration (0.22 μm). Amicon stirred cells and Centripreps (3-kDa cutoff, Millipore) were used to concentrate proteins 100–200×.
ICAT Labeling, Chromatography, and MS/MS. Conditioned medium proteins (100 μg) from MT1-MMP transfectants were labeled with isotopically heavy [13C]9-cleavable ICAT reagent (Applied Biosystems) (2 h, 37°C) for comparison with proteins from vector control or the E240A inactive MT1-MMP mutant-transfectant conditioned medium, which were labeled with the light [13C]0-ICAT reagent. Sample pairs were combined (MT1-MMP/vector or MT1-MMP/E240A), trypsin digested (18 h, 37°C), and fractionated on a cation-exchange column (4 × 15 mm) by elution with 350 mM KCl, pH 3.0, to remove the trypsin and free label. Labeled peptides were purified by using an avidin-affinity column (4 × 15 mm) and eluted with 20% acetonitrile/0.4% trifluoroacetic acid (TFA). Dried samples were incubated in 95% TFA (2 h, 37°C) to cleave the biotin tag from the ICAT-labeled peptides. The biotin tab was then cleaved with 95% TFA. Samples were fractionated by multidimensional liquid chromatography (MD-LC), first on a 500-μm i.d. × 15 mm BioX-SCX column (LC Packings, Sunnyvale, CA) and eluted as 50-μl fractions (25, 50, 75, 100, 150, 200, 250, 500, 1,000, or 2,000 mM ammonium acetate, pH 4.0). Eluted peptides were concentrated and desalted on a 300-μm i.d. × 1 mm PepMap nanotrapping column before loading onto a 75-μm i.d. × 15 cm PepMap nanoseparation column. Peptides were then fractionated by using an acetonitrile gradient (0–64%, 35 min) before entering the nanospray ionization source (New Objective, Woburn, MA). MS analysis of the separated peptides was performed by using a QStar PulsarI mass spectrometer (MDS-Sciex, Thornhill, ON, Canada). MS/MS fragmentation (2 s, 65–1,800 m/z) was performed on three of the most intense ions, as determined from a 1-s survey scan (300–1,500 m/z). ICAT ratios of isotopically heavy 13C9- to light 13C0-labeled tryptic peptides were determined by using proicat (Applied Biosystems) software and averaged for multiple peptides derived from a single parent protein. Using mascot (Matrix Science), proteins were identified from peptide sequences queried against the National Center for Biotechnology Information nonredundant protein database with human sequence filtering. Labeling and analysis were performed three times at the Genome BC Proteomic Centre (Victoria, BC, Canada).
Protease and Substrates. Soluble (s) human MT1-MMP lacking the transmembrane and cytoplasmic tail was expressed and purified (21). Human IL-8, growth-related oncogene (GRO)-α, and GRO-γ were chemically synthesized and purified (24). Human secretory leukocyte protease inhibitor (SLPI) (ICN) and human death receptor-6 fused via a Factor Xa site to the Fc region of IgG (R & D Systems) were purchased. Human connective tissue growth factor (CTGF) was a kind gift from D. Brigstock (Ohio State University, Columbus). Human pro-tumor necrosis factor (TNF)α fused to GST and a hydroxamate MMP inhibitor, BB2116, were provided by British Biotech (Oxford). Human tissue inhibitor of metalloproteinases (TIMP)-1 and -2 were expressed and purified (25). Monoclonal antibodies against human fibronectin (MAB1891, Chemicon) and death receptor-6 (MAB1441,R&D Systems) were purchased.
Assays. Recombinant and synthetic proteins were incubated with sMT1-MMP in 50 mM Tris·HCl/200 mM NaCl/5 mM CaCl2/3.8 mM NaN3/0.05% Brij 35, pH 7.4, for 18 h at 37°C. Reaction products were separated by Tris-glycine or -tricine SDS/PAGE and stained by Coomassie R-250 or Western blotted. The mass and sequence of the cleavage products were determined by matrix-assisted laser desorption ionization–time-of-flight (MALDI-TOF) MS and N-terminal Edman sequencing, respectively.
Results
MT1-MMP Expression. FLAG-tagged MT1-MMP and the 44-kDa form were detected on transfected cells (Fig. 1 A and B), which activated exogenous proMMP-2 (Fig. 1C), confirming the presence of active MT1-MMP on the cell surface. Vector transfectants expressed only very low levels of endogenous MT1-MMP (Fig. 1B). No secreted active MMPs were detected by using a general MMP-quenched fluorescent substrate (25) (data not shown).
Fig. 1.
Characterization of cells expressing MT1-MMP and inactive mutant E240A MT1-MMP. (A) Cell lysates from MT1-MMP, E240A, and vector-transfected MDA-MB-231 cells were analyzed by SDS/PAGE (10%) and Western blotting. Full-length MT1-MMP (55 kDa) and the 44-kDa form are indicated. (B) Cells labeled with either α-FLAG (Sigma) or α-human MT1-MMP antibody (AB815, Chemicon) (dark line) or no primary antibody (light line) were analyzed by fluorescent flow cytometry on a FACScan. (C) Media from MT1-MMP and vector-transfected cells, with or without exogenous proMMP-2 (72 kDa) added, were analyzed by gelatin zymography. Fully activated (62-kDa) and activation intermediate (68-kDa) MMP-2 are indicated.
ICAT and MD-LC MS/MS Analysis. In comparing the relative abundance ratios of ICAT-labeled peptides from MT1-MMP transfectants with those from the vector or E240A MT1-MMP controls, we hypothesized that those proteins with an average labeled-peptide ICAT ratio <1.0 (MT1-MMP/control) had undergone MT1-MMP-mediated degradation or processing that triggered clearance; proteins with ratios >1.0 were hypothesized to have been shed from the cell membrane or pericellular matrix by MT1-MMP activity. Although ICAT is sensitive and highly reproducible (14), to reduce the probability of false positives, we analyzed only proteins with ICAT ratios <0.5 or >1.5, which typically numbered from 100 to 150 identified proteins. Without inline MD-LC fractionation, the number of proteins so identified was reduced to <100. Intracellular proteins were not further considered. A selective list of the biologically interesting proteins that may be potential MT1-MMP substrates was generated from the two data sets (Table 1) based on deduced roles in pathology and embryonic development. These included protease inhibitors (SLPI and skin-derived antileukoproteinase), chemokines (IL-8, GRO-α, GRO-γ, and macrophage migration inhibitory factor), cytokines (TNFα, CTGF), cell receptors (death receptor-6, neuropilin-1), latent transforming growth factor-binding protein-4S, and complement component-3. Extracellular matrix proteins (fibronectin, epidermal growth factor-containing fibulin-like extracellular matrix protein-1) and shed MT1-MMP and proMMP-1 (collagenase-1) were also elevated in the MT1-MMP cell-conditioned medium.
Table 1. Identification by ICAT and MS/MS analysis of cellular proteins in the conditioned medium of MDA-MB-231 cultures displaying altered abundance due to MT1-MMP activity.
| Protein* | MT1-MMP/control† mean ratio‡ | Peptide sequences§ |
|---|---|---|
| SLPI | 4.95 | CCMGMCGK |
| YKKPECQSDWQCPGK | ||
| EFEMP-1 | 3.90 | ADQVCINLR |
| CVNHYGGYLCLPK | ||
| DR-6 | 3.79¶ | VCSSCPVGTFTR |
| Fibronectin | 3.04 | EYLGAICSCTCFGGQR |
| GEWTCIAYSQLR | ||
| ISCTIANR | ||
| LLCQCLGFGSGHFR | ||
| TFYSCTTEGR | ||
| TYLGNALVCTCYGGSR | ||
| WCGTTQNYDADQK | ||
| YQCYCYGR | ||
| YSFCTDHTVLVQTR | ||
| MT1-MMP | 2.61 | CGVPDKFGAEIK∥ |
| WQHNEITFCIQNYTPK | ||
| ProMMP-1 | 2.05 | ACDSKLTFDAITTIR |
| CGVPDVAQFVLTEGNPR∥ | ||
| Neuropilin-1 | 1.90 | SPGFPEKYPNSLECTYIVFAPK |
| SKALP | 1.85 | CAMLNPPNR |
| CLKDTDCPGIK | ||
| CTGF | 1.74 | DGAPCIFGGTVYR |
| LTRP-4S | 1.73 | AGPDLASCLDVDECRER |
| DGGCSLPILR | ||
| IQQCPGTETAEYQSLCPHGR | ||
| TNFα | 1.29 | SWLPAGCETAILFPMR |
| TIMP-1 | 1.25 | EPGLCTWQSLR |
| FVYTPAMESVCGYFHR | ||
| LQSGTHCLWTDQLLQGSEK | ||
| MIF | 0.49 | LLCGLLAER |
| GRO-α | 0.37 | CQCLQTLQGIHPK |
| KACLNPASPIVK | ||
| SPGPHCAQTEVIATLK | ||
| IL-8 | 0.34 | ELCLDPK |
| LSDGRELCLDPK | ||
| VIESGPHCANTEIIVK | ||
| GRO-γ | 0.27 | CQCLQTLQGIHLK |
| SPGPHCAQTEVIATLK | ||
| C3 | 0.25 | NTMILEICTR |
| VFLDCCNYITELRR | ||
| VYAYYNLEESCTR |
SLPI, secretory leukocyte protease inhibitor; EFEMP-1, EGF-containing fibulin-like extracellular matrix protein-1; DR-6, death receptor-6; SKALP, skin derived antileukoproteinase; CTGF, connective tissue growth factor; LTBP-4S, latent transforming growth factor-binding protein-4S; TIMP-1, tissue inhibitor of metalloproteinase-1; MIF, macrophage migration inhibitory factor; GRO-α, growth related oncogene-α; GRO-γ, growth related oncogene-γ; C3, complement component 3
Control protein was from vector transfected cells unless stated otherwise
The relative abundance of the 13C9-labeled peptides from the MT1-MMP transfected cell proteins compared with the 13C0-labeled peptides from control cell protein were averaged to give a mean ratio
All peptides containing multiple Cys residues were quantitatively labeled by the ICAT reagent
Control protein was from inactive MT1-MMP mutant transfected cells
These peptides represent the Cys-switch motif of the MMP prodomain indicating increased expression of the zymogen form of these MMPs and that furin activation of MT1-MMP may occur on the cell surface
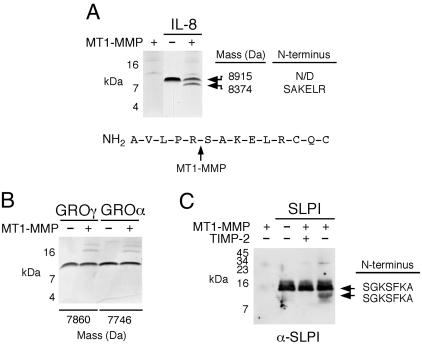
Chemokine Processing. To test the hypothesis that ICAT can identify potential protease substrates, we biochemically analyzed a number of the proteins from Table 1 for cleavage by sMT1-MMP using recombinant or synthetic proteins as substrates. Because chemokines are a recently recognized class of MMP substrates (9, 26–28), it was of interest that the chemokines IL-8, GRO-α, and GRO-γ showed decreased levels on MT1-MMP expression. N-terminal sequencing confirmed the proteolytic removal of the first five residues of IL-8 by sMT1-MMP in vitro to generate a truncated form beginning with the sequence SAKELR (Fig. 2A). However, SDS/PAGE and MALDI-TOF MS showed no change in mass for GRO-α and -γ after incubation with sMT1-MMP (Fig. 2B). Complement component-3, which displayed a similar ICAT ratio to GRO-γ (Table 1), was also not susceptible to MT1-MMP proteolysis in vitro (data not shown), revealing that MT1-MMP expression can decrease the levels of these proteins by indirect means.
Fig. 2.
MT1-MMP processing of IL-8 and SLPI. (A–C) CXC chemokines or SLPI were incubated with sMT1-MMP for 18 h at 37°C. Samples were then separated on 15% Tris-tricine gels under nonreducing conditions. (A) MT1-MMP cleavage of IL-8 and sequence of the cleavage site. (B) MT1-MMP did not cleave GRO-α or -γ as analyzed by SDS/PAGE. MALDI-TOF MS revealed identical masses for both chemokines in the presence or absence of MT1-MMP. (C) SLPI was detected by Western blotting by using α-SLPI antibody. SLPI cleavage was blocked by TIMP-2 added with sMT1-MMP or after preincubation (not shown). N-terminal sequences are shown.
Sheddase Activity. Levels of the secretory leukocyte protease inhibitor, SLPI, were greatly increased on MT1-MMP expression (Table 1). Biochemical assays revealed that sMT1-MMP cleaved SLPI in a TIMP-2-inhibitable manner to generate a single 12.3-kDa product with an N terminus of SGKSFKA (Fig. 2C). This sequence is identical to the intact inhibitor, and so cleavage occurred near the C terminus, which may release the cleaved SLPI from association with the cell layer proteins.
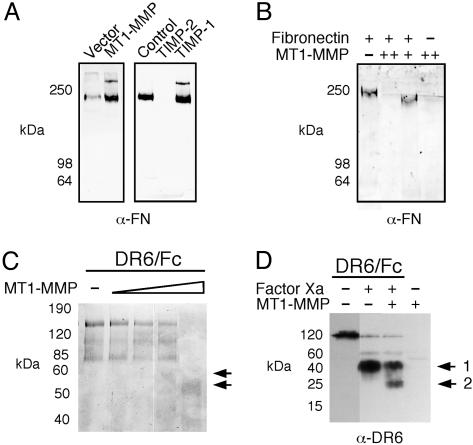
Medium levels of fibronectin, a known substrate of sMT1-MMP (20), were also significantly elevated upon MT1-MMP expression (Table 1), implying that MT1-MMP may shed fibronectin from the cell surface. This was confirmed by gelatin-Sepharose affinity purification and quantitation of fibronectin from the conditioned medium of the transfectants (Fig. 3A). Fibronectin shedding was inhibited by adding TIMP-2 but not -1 to the cultures. Because TIMP-1 spares MT1-MMP in its inhibitory profile, this confirms the specific requirement for MT1-MMP activity in the proteolytic release of fibronectin from the cell surface. Shed immunoreactive protein migrated with the expected molecular mass (220 kDa + DTT) of the full-length protein (Fig. 3A); cleaved fibronectin fragments were never observed in the conditioned medium; and the ICAT-labeled peptides 67TYLGNALVCTCYGGSR84 and 2199LLCQCLGFGSGHFR2214 (Table 1), which are proximal to the N and C termini of the molecule, displayed similar abundance ratios to the other fibronectin peptides. This indicates that the solubilized form of fibronectin is intact and that overexpressed MT1-MMP did not appear to degrade fibronectin in a cellular context, despite its activity on this protein in biochemical assays (Fig. 3B). Although processing of fibronectin cannot be discounted as a shedding mechanism, this indicates that cleavage of a fibronectin-binding cell surface molecule by MT1-MMP is the mechanism for release.
Fig. 3.
MT1-MMP shedding of fibronectin and death receptor-6. (A) MT1-MMP and vector-transfected MDA-MB-231 cells were incubated in serum-free DMEM with or without inhibitors (TIMP-1 and -2, 100 nM). Shed fibronectin was then purified from the 48-h media by using a gelatin-Sepharose column and eluted with 8 M urea in PBS before analysis by SDS/PAGE (5%) and Western blotting with α-fibronectin monoclonal antibody. (B) Fibronectin was incubated with sMT1-MMP (+, ++) in vitro for18hat37°C. (C) DR6/Fc fusion protein was treated with increasing amounts of sMT1-MMP and separated by SDS/PAGE (10%). Cleavage products are indicated by arrows. (D) DR6/Fc was cleaved with Factor Xa (5 h, 37°C) before digestion with sMT1-MMP (18 h, 37°C). Samples were then electrophoresed and Western blotted by using α-DR6 antibody. Factor Xa (1) and MT1-MMP (2) cleaved-DR6/Fc products are shown.
Death receptor-6 is a member of the TNF receptor family and consists of a 350-residue ectodomain and a cytoplasmic death domain (29). Using a death receptor-6 fusion protein, DR6/Fc, as a substrate for in vitro assays with sMT1-MMP, we found loss of intact protein and generation of two immunoreactive fragments at 55 and 50 kDa (Fig. 3C). To verify that cleavage occurred within the ectodomain and not the Fc region, DR6/Fc was first treated with Factor Xa to cleave a susceptible site between the ectodomain and the Fc region. Upon incubation with sMT1-MMP in vitro, a 25-kDa product was generated from the 40-kDa ectodomain confirming susceptibility of DR6 to MT1-MMP proteolysis (Fig. 3D).
Cytokine Substrates. The inflammatory cytokine, TNFα, had a MT1-MMP/vector ICAT ratio of 1.29 that indicated shedding to the medium. Although this was below our arbitrary 1.5 ICAT ratio cutoff, proTNFα can be shed by MMP-7 (30). Therefore, the susceptibility of proTNFα to sMT1-MMP processing was investigated by using a proTNFα/GST fusion protein as substrate. This was cleaved by sMT1-MMP to generate a product corresponding to the correct size of mature TNFα (≈16 kDa) (Fig. 4A). N-terminal sequencing revealed that, in addition to cleavage at the Ala-76—Val-77 bond, which generates the fully active TNFα, three other bonds in close proximity in the proregion of TNFα were also processed by sMT1-MMP in vitro (Fig. 4A).
Fig. 4.
MT1-MMP processing of proTNFα and CTGF. (A) ProTNFα/GST fusion protein was incubated with sMT1-MMP (18 h, 37°C) with or without BB2116. Samples were separated by SDS/PAGE (5–15%). MT1-MMP cleavage sites within the TNFα proregion were determined by N-terminal sequencing as schematically shown. Mature TNFα commences at VRSSSRT. (B) CTGF was treated with sMT1-MMP (+, ++)for 18 h at 37°C and analyzed by SDS/PAGE (15%) and MALDI-TOF MS. MT1-MMP cleavage sites were located by N-terminal sequencing to be in the CTGF linker. The IGF-binding protein module (IGFBP), von Willebrand factor type 1C module (vWFC-1), thrombospondin 1 module (TSP-1), and the cysteine-knot-containing C-terminal module (CYS) of CTGF are indicated. Insufficient protein was present to sequence the 18.8-kDa fragment. No sequence was attainable for intact CTGF, indicative of a modified N terminus.
Last, we analyzed CTGF, a member of the CCN (CTGF/cysteine-rich 61/nephroblastoma overexpressed) family of extracellular matrix-associated signaling molecules (31) for cleavage by sMT1-MMP. Three distinct lower molecular mass fragments were generated (Fig. 4B). Sequencing of the 17.5- and 16.2-kDa sMT1-MMP cleavage products revealed an identical N terminus, YRLEDT, corresponding to cleavage at the Ala-181—Tyr-182 bond (Fig. 4B).
Discussion
High-throughput protein quantitation and identification by ICAT and MD-LC MS/MS are powerful techniques that we have adapted for the proteomic investigation of protease function and substrate discovery in a dynamic cellular milieu. In MDA-MB-231 cells, we specifically identified a number of proteins that were biochemically confirmed as novel substrates of MT1-MMP, thereby validating the utility of ICAT MD-LC MS/MS for substrate discovery in cell-based systems. The majority of substrates discovered to date have been from serial approaches using available proteins or hypothesis-driven intuition using time-consuming biochemical and genetic approaches or peptide libraries and bioinformatics (12). The alternative of using proteomic techniques to screen for novel protease substrates is advantageous because of its coverage on a system-wide scale, speed, and resolution of even complex biological samples. However, to date there have been few proteomic studies except for using 2D-PAGE and stable isotope dilution (13). In comparison to gel-based analyses, LC MS approaches provide more extensive and rapid proteome coverage, the abundance of isotopically labeled peptides can be quantitated, and cell membrane proteins are amenable for MS analysis (reviewed in ref. 14). Nonetheless, protein abundance sets detection limits, and until improvements are made in sample preparation and chromatography before MS, it is not yet possible to localize cleaved substrates or to analyze whole tissues by ICAT because of the added sample complexity and variability. Moreover, differences in protein abundance as measured by ICAT must be interpreted with caution; some cleaved substrates may not change levels, and as we have found the levels of some uncleaved proteins may alter just by expression of the transfected gene, an important caveat for interpreting any study using protease-transfected cells. Thus, ICAT and MD-LC MS/MS should be considered as a screen for potential protease substrates that must be confirmed biochemically and in vivo when possible but that offers general applicability to other protease classes for functional proteomic investigation of protease function in complex samples.
The pool of potential substrates available to MT1-MMP expressed in MDA-MB231 cells will contain overlapping candidates with normal mammary epithelium as well as unique targets. The majority of proteins identified as potential MT1-MMP substrates were not extracellular matrix molecules. Rather, the levels of a variety of signaling proteins were specifically modulated on expression of MT1-MMP. Of these, IL-8, GRO-α, and GRO-γ are important for the regulation of leukocyte migration during wound healing (32, 33). MT1-MMP is also known to participate in wound healing and is expressed by monocytes. IL-8, GRO-α, and GRO-γ belong to the subgroup of CXC chemokine family members that contain the ELR motif near the N terminus, which is essential for neutrophil chemotactic activity (34). MT1-MMP processing of IL-8 removes the first five residues, which is more active than the untruncated form. In contrast to MMP-9, which processes a different site in IL-8 and also GROα (28), MT1-MMP does not cleave GRO-α or -γ, demonstrating selective susceptibility of CXC chemokines to MMP cleavage. Therefore, the reduction in the levels of these chemokines appears to be an indirect upstream response to the expression of MT1-MMP but biologically may be as important a consequence of MT1-MMP activity.
A number of protease inhibitors, including the plasma serine protease inhibitor SLPI, are also essential for normal wound repair. SLPI suppresses monocyte MMP production and is resistant to neutrophil collagenase (35). Here, ICAT identified SLPI as a potential MT1-MMP substrate, which we biochemically confirmed was processed near its C terminus. Because the inhibitory site of SLPI lies within the C domain (36), this reveals a previously undescribed intersection between the serine and metalloproteases in which the activity of a serine protease inhibitor is predicted to be abrogated on MT1-MMP proteolysis.
Expression of recombinant proteases may occur at levels greater than that seen in some normal tissues, thereby favoring proteolysis of less preferred or low abundant substrates. Although sMT1-MMP can degrade fibronectin in vitro, we found that in cell culture, MT1-MMP activity led only to the shedding of intact fibronectin from the MDA-MB-231 cell surface, suggesting that the levels of the recombinant MT1-MMP expressed were not artificially extreme or generated cleavage artifacts. This result not only highlights the importance of biochemically confirming proteomic data but equally important is the biological validation of biochemically determined proteolytic activity with cell-based and in vivo studies at the genetic or proteomic levels. Although the mode of fibronectin release is not clear, it has been reported that MT1-MMP degrades transglutaminase, a fibronectin coreceptor, thereby reducing both the adhesion and migration of cells on fibronectin (37).
We also found that MT1-MMP cleaved the ectodomain of death receptor-6, a transmembrane receptor that regulates apoptosis through its cytoplasmic death domain and activation of the TRADD/FADD/caspase-8 pathway (29). Although the purpose of our study was to develop a novel proteomic approach for protease substrate discovery and not to fully explore here the biological ramifications of the cleavage of newly identified substrates, it is reasonable to hypothesize that MT1-MMP proteolysis of the ectodomain may regulate death receptor-6 signaling and apoptosis. Ligands that bind death receptor-6 belong to the TNF family of cytokines, one of which, TRANCE, is cleaved by MT1-MMP (38). Although ADAM-17 is the enzyme thought to be primarily responsible for the generation of mature TNFα from a membrane-bound precursor, particularly under lipopolysaccharide (LPS) stimulation (39, 40), MMP-7 can also process TNFα (30). Here, we extend the repertoire of MMPs that can cleave proTNFα by showing that MT1-MMP cleaves at the Ala-76—Val-77 bond within the proregion of TNFα to generate mature fully active TNFα, a potentially important mechanism in non-LPS-induced inflammation.
CTGF was also identified as a potential MT1-MMP substrate by ICAT that we then showed was cleaved by sMT1-MMP in vitro at the Ala-181—Tyr-182 bond. This site is within the linker that connects the insulin-like growth factor-binding protein domain and von Willebrand factor type-1C domain of CTGF with the thrombospondin-1 and C-terminal domains (Fig. 3B). Cleavage here potentially disengages matrix-binding domains from the signaling functions of CTGF, leading to release from the cell layer. Hashimoto et al. (41) also recently described CTGF cleavage by soluble MMPs 1, 3, 7, and 13 at Met-194-Ile-195 within the CTGF linker, with a minor cleavage site at Ala-181-Tyr-182. CTGF is unstable in vivo, being converted into low molecular mass forms (10–20 kDa) by an unidentified protease (42). Interestingly, conditioned medium from serum-stimulated mouse fibroblasts did not degrade CTGF (43), suggesting that a cell surface protease was responsible for CTGF processing. These results are consistent with our data demonstrating that the cell surface MT1-MMP can process CTGF in cell culture to generate ≈17-kDa fragments.
The substrates identified here reveal that MT1-MMP may be an important cell signaling protease and add to recent studies describing new MMP substrates that are redefining our outlook of the biological role of this protease family (12). Traditionally, MMPs have been viewed as enzymes of catabolism being primarily involved in the degradation and turnover of the extracellular matrix (17). MMPs are now known to process a diverse range of extracellular proteins other than those of the matrix (44). Like many proteases, MMPs regulate and refine many aspects of protein function, including activity, localization, shedding, exposure of cryptic binding sites, and release of neoproteins thereby achieving precise control over many cell processes. Thus, elucidating the substrate repertoires or degradomes of proteases is essential for understanding their in vivo roles. By establishing the physiological function of a protease, its role in pathology can be more readily ascertained. This in turn is a necessary prerequisite for drug target validation. No one technique will provide complete coverage of the protease substrate landscape, but overall MD-LC ICAT MS/MS provides a powerful functional degradomic approach that renders the quantitative analysis of proteolysis in defined cell cultures feasible, and one that should be generally applicable to elucidate the biological role of other proteases.
Acknowledgments
We thank Derek Smith for help with the ICAT data and Suzanne Perry for N-terminal sequencing and MALDI-TOF. E.M.T. is supported by a Canadian Institutes of Health Research Strategic Training Fellow Award in Cell Signals and a Canadian Arthritis Network Trainee Award. C.M.O. is supported by a Canada Research Chair in Metalloproteinase Biology. This work was supported by grants from Protein Engineering National Centres of Excellence National Cancer Institute of Canada and the Canadian Institutes of Health Research.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ICAT, isotope-coded affinity tag; MS/MS, tandem MS; MD-LC, multidimensional liquid chromatography; MMP, matrix metalloproteinase; MT, membrane type; sMT, soluble MT; TIMP, tissue inhibitor of metalloproteinases; GRO, growth-related oncogene; SLPI, secretory leukocyte protease inhibitor; CTGF, connective tissue growth factor; TNF, tumor necrosis factor; MALDI-TOF, matrix-assisted laser desorption ionization–time-of-flight.
References
- 1.Barrett, A. J., Rawlings, N. D. & Woessner, J. F. (1998) Handbook of Proteolytic Enzymes (Academic, San Diego).
- 2.Puente, X. S., Sanchez, L. M., Overall, C. M. & LÓpez-Otín, C. (2003) Nat. Rev. Genet. 4, 544-558. [DOI] [PubMed] [Google Scholar]
- 3.Fingleton, B. (2003) Expert Opin. Ther. Targets 7, 385-397. [DOI] [PubMed] [Google Scholar]
- 4.Harris, J. L., Backes, B. J., Leonetti, F., Mahrus, S., Ellman, J. A. & Craik, C. S. (2000) Proc. Natl. Acad. Sci. USA 97, 7754-7759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nazif, T. & Bogyo, M. (2001) Proc. Natl. Acad. Sci. USA 98, 2967-2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Turk, B. E., Huang, L. L., Piro, E. T. & Cantley, L. C. (2001) Nat. Biotechnol. 19, 661-667. [DOI] [PubMed] [Google Scholar]
- 7.Deng, S. J., Bickett, D. M., Mitchell, J. L., Lambert, M. H., Blackburn, R. K., Carter, H. L., Neugebauer, J., Pahel, G., Weiner, M. P. & Moss, M. L. (2000) J. Biol. Chem. 275, 31422-31427. [DOI] [PubMed] [Google Scholar]
- 8.Overall, C. M., McQuibban, G. A. & Clark-Lewis, I. (2002) Biol. Chem. 383, 1059-1066. [DOI] [PubMed] [Google Scholar]
- 9.McQuibban, G. A., Gong, J. H., Tam, E. M., McCulloch, C. A., Clark-Lewis, I. & Overall, C. M. (2000) Science 289, 1202-1206. [DOI] [PubMed] [Google Scholar]
- 10.Overall, C. M., Wiebkin, O. W. and Thonard, J. C. (1987) J. Periodont. Res. 22, 81-88. [DOI] [PubMed] [Google Scholar]
- 11.Hughes, C. E., Caterson, B., Fosang, A. J., Roughley, P. J. & Mort, J. S. (1995) Biochem. J. 305, 799-804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lopéz-Otín, C. & Overall, C. M. (2002) Nat. Rev. Mol. Cell Biol. 3, 509-519. [DOI] [PubMed] [Google Scholar]
- 13.Guo, L., Eisenman, J. R., Mahimkar, R. M., Peschon, J. J., Paxton, R. J., Black, R. A. & Johnson, R. S. (2002) Mol. Cell. Proteomics 1, 30-36. [DOI] [PubMed] [Google Scholar]
- 14.Abersold, R. & Mann, M. (2003) Nature 422, 198-207. [DOI] [PubMed] [Google Scholar]
- 15.De Leenheer, A. P. & Thienpont, L. M. (1992) Mass Spectrom. Rev. 11, 249-307. [Google Scholar]
- 16.Gygi, S. P., Rist, B., Gerber, S. A., Turecek, F., Gelb, M. H. & Aebersold, R. (1999) Nat. Biotechnol. 17, 994-999. [DOI] [PubMed] [Google Scholar]
- 17.Nagase, H. & Woessner, J. F. (1999) J. Biol. Chem. 274, 21491-21494. [DOI] [PubMed] [Google Scholar]
- 18.Holmbeck, K., Bianco, P., Caterina, J., Yamada, S., Kromer, M., Kuznetsov, S. A., Mankani, M., Robey, P. G., Poole, A. R., Pidoux, I., et al. (1999) Cell 99, 81-92. [DOI] [PubMed] [Google Scholar]
- 19.Seiki, M. (2003) Cancer Lett. 194, 1-11. [DOI] [PubMed] [Google Scholar]
- 20.Ohuchi, E., Imai, K., Fujii, Y., Sato, H., Seiki, M. & Okada, Y. (1997) J. Biol. Chem. 272, 2446-2451. [DOI] [PubMed] [Google Scholar]
- 21.Tam, E. M., Wu, Y. I., Butler, G. S., Stack, M. S. & Overall, C. M. (2002) J. Biol. Chem. 277, 39005-39014. [DOI] [PubMed] [Google Scholar]
- 22.Sato, H., Takino, T., Okada, Y., Cao, J., Shinagawa, A., Yamamoto, E. & Seiki, M. (1994) Nature 370, 61-65. [DOI] [PubMed] [Google Scholar]
- 23.Morrison, C. J., Butler, G. S., Bigg, H. F., Roberts, C. R., Soloway, P. D. & Overall, C. M. (2001) J. Biol. Chem. 276, 47402-47410. [DOI] [PubMed] [Google Scholar]
- 24.Clark-Lewis, I., Vo, L., Owen, P. & Anderson, J. (1997) Methods Enzymol. 287, 233-250. [DOI] [PubMed] [Google Scholar]
- 25.Bigg, H. F., Morrison, C. J., Butler, G. S., Bogoyevitch, M. A., Wang, Z., Soloway, P. D. & Overall, C. M. (2001) Cancer Res. 61, 3610-3618. [PubMed] [Google Scholar]
- 26.Zhang, K., McQuibban, G. A., Silva, C., Butler, G. S., Johnston, J. B., Holden, J., Clark-Lewis, I., Overall, C. M. & Power, C. (2003) Nat. Neurosci. 6, 1064-1071. [DOI] [PubMed] [Google Scholar]
- 27.Balbín, M., Fueyo, A., Tester, A. M., Pendás, A. M., Pitiot, A. S., Astudillo, A., Overall, C. M., Shapiro, S. & LÓpez-Otín, C. (2003) Nat. Genet. 35, 252-257. [DOI] [PubMed] [Google Scholar]
- 28.Van den Steen, P. E., Proost, P., Wuyts, A., Van Damme, J. & Opdenakker, G. (2000) Blood 96, 2673-2681. [PubMed] [Google Scholar]
- 29.Pan, G., Bauer, J. H., Haridas, V., Wang, S., Liu, D., Yu, G., Vincenz, C., Aggarwal, B. B., Ni, J. & Dixit, V. M. (1998) FEBS Lett. 431, 351-356. [DOI] [PubMed] [Google Scholar]
- 30.Haro, H., Crawford, H. C., Fingleton, B., Shinomiya, K., Spengler, D. M. & Matrisian, L. M. (2000) J. Clin. Invest. 105, 143-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brigstock, D. R. (1999) Endocr. Rev. 20, 189-206. [DOI] [PubMed] [Google Scholar]
- 32.Olson, T. S. & Ley, K. (2002) Am. J. Physiol. 283, R7-R28. [DOI] [PubMed] [Google Scholar]
- 33.Ashcroft, G. S., Lei, K., Jin, W., Longenecker, G., Kulkarni, A. B., Greenwell-Wild, T., Hale-Donze, H., McGrady, G., Song, X. Y. & Wahl, S. M. (2000) Nat. Med. 6, 1147-1153. [DOI] [PubMed] [Google Scholar]
- 34.Van Damme, J., Decock, B., Conings, R., Lenaerts, J. P., Opdenakker, G. & Billiau, A. (1989) Eur. J. Immunol. 19, 1189-1194. [DOI] [PubMed] [Google Scholar]
- 35.Henry, M. T., McMahon, K., Costello, C., Fitzgerald, M. X. & O'Connor, C. M. (2002) Exp. Lung Res. 28, 85-97. [DOI] [PubMed] [Google Scholar]
- 36.Grutter, M. G., Fendrich, G., Huber, R. & Bode, W. (1988) EMBO J. 7, 345-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Belkin, A. M., Akimov, S. S., Zaritskaya, L. S., Ratnikov, B. I., Deryugina, E. I. & Strongin, A. Y. (2001) J. Biol. Chem. 276, 18415-18422. [DOI] [PubMed] [Google Scholar]
- 38.Schlondorff, J., Lum, L. & Blobel, C. P. (2001) J. Biol. Chem. 276, 14665-14674. [DOI] [PubMed] [Google Scholar]
- 39.Black, R. A., Rauch, C. T., Kozlosky, C. J., Peschon, J. J., Slack, J. L., Wolfson, M. F., Castner, B. J., Stocking, K. L., Reddy, P., Srinivasan, S., et al. (1997) Nature 385, 729-733. [DOI] [PubMed] [Google Scholar]
- 40.Moss, M. L., Jin, S. L., Milla, M. E., Bickett, D. M., Burkhart, W., Carter, H. L., Chen, W. J., Clay, W. C., Didsbury, J. R., Hassler, D., et al. (1997) Nature 385, 733-736. [DOI] [PubMed] [Google Scholar]
- 41.Hashimoto, G., Inoki, I., Fujii, Y., Aoki, T., Ikeda, E. & Okada, Y. (2002) J. Biol. Chem. 277, 36288-36295. [DOI] [PubMed] [Google Scholar]
- 42.Ball, D. K., Surveyor, G. A., Diehl, J. R., Steffen, C. L., Uzumcu, M., Mirando, M. A. & Brigstock, D. R. (1998) Biol. Reprod. 59, 828-835. [DOI] [PubMed] [Google Scholar]
- 43.Kireeva, M. L., Latinkic, B. V., Kolesnikova, T. V., Chen, C., Yang, G., Abler, A. & Lau, L. F. (1997) Exp. Cell Res. 233, 63-77. [DOI] [PubMed] [Google Scholar]
- 44.Egeblad, M. & Werb, Z. (2002) Nat. Rev. Cancer 2, 161-174. [DOI] [PubMed] [Google Scholar]