Abstract
Genome-wide association studies (GWAS) have identified at least 133 ulcerative colitis (UC) associated loci. The role of genetic factors in clinical practice is not clearly defined. The relevance of genetic variants to disease pathogenesis is still uncertain because of not characterized gene-gene and gene-environment interactions. We examined the predictive value of combining the 133 UC risk loci with genetic interactions in an ongoing inflammatory bowel disease (IBD) GWAS. The Wellcome Trust Case-Control Consortium (WTCCC) IBD GWAS was used as a replication cohort. We applied logic regression (LR), a novel adaptive regression methodology, to search for high order interactions. Exploratory genotype correlations with UC sub-phenotypes (extent of disease, need of surgery, age of onset, extra-intestinal manifestations and primary sclerosing cholangitis (PSC)) were conducted. The combination of 133 UC loci yielded good UC risk predictability (area under the curve [AUC] of 0.86). A higher cumulative allele score predicted higher UC risk. Through LR, several lines of evidence for genetic interactions were identified and successfully replicated in the WTCCC cohort. The genetic interactions combined with the gene-smoking interaction significantly improved predictability in the model (AUC, from 0.86 to 0.89, P=3.26E-05). Explained UC variance increased from 37% to 42% after adding the interaction terms. A within case analysis found suggested genetic association with PSC. Our study demonstrates that the LR methodology allows the identification and replication of high order genetic interactions in UC GWAS datasets. UC risk can be predicted by a 133 loci and improved by adding gene-gene and gene-environment interactions.
Keywords: ulcerative colitis, genetic polymorphism, IBD - genetics, IBD – clinical, primary sclerosing cholangitis
INTRODUCTION
While the precise etiology of ulcerative colitis (UC) remains elusive, both genetic and non-genetic factors are involved in its pathogenesis(Danese and Fiocchi, 2011). In terms of genetic factors, recent genome-wide association study (GWAS) meta-analyses have identified 23 UC-specific risk loci in addition to 110 loci that are associated with both UC and Crohn’s disease (CD) (Jostins et al., 2012). However, the value of these genetic variants in clinical practice, the role of gene-gene interactions, and the input of environmental factors are still unclear. Some of the non-genetic factors associated with UC include luminal microbiota, mucosal immune response, cigarette smoking and prior appendectomy. Smoking has been shown to have a protective effect against the development of UC and has been associated with a reduced risk of colectomy (Aldhous et al., 2007;Boyko et al., 1987;Mahid et al., 2006;Szamosi et al., 2010). Prior appendectomy for appendicitis has been linked to a lower risk of UC (Frisch et al., 2009;Hallas et al., 2004), but the effect of appendectomy on UC disease course remains inconclusive (Gardenbroek et al., 2012).
Analysis of single genetic markers has limited value in predicting risk for complex diseases such as inflammatory bowel disease (IBD), but the predictive value of combining multiple common genetic variants can be improved upon, especially when GWAS data are incorporated (Liu and Song, 2010;Wang et al., 2013;Wray et al., 2007;Yang et al., 2003). The development of cumulative UC genetic risk scores based on GWAS results to predict disease risk could provide better strategies for disease screening, guide extent of diagnostic testing, and potentially affect treatment choices.
In addition, only a limited proportion of total disease variance has been explained by the identified UC genetic loci (Jostins et al., 2012). Therefore, it is possible that defining gene-gene and gene-environment interactions could uncover missing variance left out from additive genetic models and could provide better UC risk predictive models. Applying the logic regression (LR) methodology, a new adaptive regression method to search for logical interactions of binary predictors (Ruczinski et al., 2003;Ruczinski et al., 2004;Schwender and Ruczinski, 2010), we recently successfully identified and replicated high order genetic interactions among five CD associated genetic loci, NOD2, ATG16L1, IL10/IL19, C13orf31 and chr21q (Wang et al., 2013).
The aims of this study are to assess the distribution and UC risk predictability of the 133 UC-associated meta-analysis loci, to explore high order genetic interactions using LR in two independent GWAS cohorts (a discovery cohort and a replication cohort), and to identify genotype-phenotype correlations. We also perform genetic and environmental association analyses taking into account UC sub-phenotypes and conduct exploratory gene-environment interactions.
MATERIALS AND METHODS
GWAS Datasets
Two GWAS datasets were used for this study, the Cleveland Clinic/University of Pittsburgh (CC/UP) IBD GWAS and the Wellcome Trust Case-Control Consortium (WTCCC) UC GWAS. The CC/UP GWAS dataset was used for the cumulative risk allele analysis, as the discovery dataset for evaluation of high order genetic interactions, and for the genotype-phenotype correlation analyses. The study design and data collection of this GWAS have been previously described (Achkar et al., 2012). Of note, the full GWAS has not yet been completed as the replication phase of the study is ongoing. However, we were able to pursue the current study as its main purposes were to predict UC risk using the 133 UC GWAS meta-analysis loci and to identify high order genetic interactions through a novel methodological approach. In brief, this GWAS consists of 566 UC cases and 1,436 unrelated healthy controls, all of non-Jewish, European ancestry, who were genotyped using the Illumina Human Omni1-Quad beadchip (Illumina, San Diego, CA, USA) at the Feinstein Institute for Medical Research of the North Shore-Long Island Jewish Health System. All participants gave written informed consent. Genotype imputation of this dataset was performed using 5-Mb regions across the whole genome with the BEAGLE imputation program (Browning and Browning, 2009). All but one of the 133 UC meta-analysis SNPs were imputed with good quality (R-squared >0.80) and with Hardy-Weinberg equilibrium (HWE) P-value > 1.0E-05 in controls. Single nucleotide polymorphism (SNP) rs6927022 (chromosome 6, base pair position 32,612,397) had poor imputation quality, so rs9272346 (chromosome 6, base pair position 32,604,372, located in HLA-DQA1) which is located 8 kb upstream and is in high linkage disequilibrium (R-squared 0.84) with rs6927022, was used instead.
The WTCCC2 UC GWAS (Affymetrix GeneChip 6.0 arrays) was used as the replication dataset for the genetic interaction analyses (Barrett et al., 2009). We downloaded the genotype data called by the CHIAMO algorithm (Marchini et al., 2007) for the UC samples and the shared controls (the 1958 Birth Cohort and UK Blood Service sample) from the WTCCC website. After excluding individuals with evidence of non-European ancestry or poor call rates, 2,361 UC cases and 5,417 controls remained. We applied the following quality control criteria to exclude SNPs: 1) HWE test P values <5.7E-07 in controls; 2) minor allele frequency <1% in UC cases and controls; 3) call rate <99%; and 4) plate relatedness (suggesting batch effect) provided by WTCCC. In total, 712,972 SNPs passed the quality control filters. Of the 133 UC meta-analysis SNPs, 71 had been successfully genotyped in the WTCCC2 dataset. The remaining 62 ungenotyped SNPs were imputed using the MaCH program (Li et al., 2009;Li et al., 2010) with 1000 Genomes Project-phase 1 haplotype data as reference. We used parameters of 60 iterations of the Markov sampler and 200 states. The 1000 Genomes reference panel was obtained from the University of Michigan Abecasis lab, version 20100804 (http://www.sph.umich.edu/csg/abecasis/MACH/download/1000G-2010-08.html). A total of 566 reference haplotypes for European ancestry served as the reference panel. Phenotypic data is not available for this dataset.
Phenotype information
In the CC/UP IBD GWAS cohort, all cases and 500 of the controls were administered questionnaires to assess demographic and environmental factors including ethnicity, history of appendectomy (prior to UC diagnosis for cases), and tobacco use (classified as current smoker, past smoker or never smoker at the time of diagnosis for UC subjects and at the time of study enrollment for controls). The remaining 936 controls did not have this information available as explained below. Medical records were reviewed to confirm UC diagnosis and to determine disease phenotypes using the validated NIDDK IBD Genetics Consortium modification of the Montreal Classification as previously described (Dassopoulos et al., 2007). Patients with indeterminate colitis were not included. Phenotypic data collected for UC subjects included age at diagnosis, maximal macroscopic extent of disease, history of surgical treatment for UC, development of dysplasia or cancer, and presence of extra-intestinal manifestations (EIM) including joint involvement (small joint, large joint, ankylosing spondylitis, sacroiliitis), eye involvement (iritis, uveitis, non-specific ocular inflammation), skin involvement (erythema nodosum, pyoderma gangrenonum), and primary sclerosing cholangitis (PSC).
STATISTICAL ANALYSIS
Cumulative allele scores of the 133 UC GWAS loci
Risk alleles were designated as the alleles reported to be associated with UC in the IBD GWAS meta-analysis and the Immunochip replication paper (Jostins et al., 2012). Odds ratios (ORs) and 95% confidence intervals (CI) were calculated to estimate single locus effects for risk alleles and genotypes. These analyses were implemented in the Golden Helix SVS software suite 7 (Golden Helix, Bozeman, MT).
The cumulative allele score was determined by summing up the number of risk alleles (in dominant mode) across the 133 UC SNPs for each study subject with complete genotype information. Estimated UC risk based on cumulative allele score was conducted through conventional logistic regression model using SAS 9.2/Genetics package PROC LOGISTIC procedures (SAS Institute, Cary, NC).
Exploring high order genetic interactions using LR among 133 UC genes
LR (R package Logic.Reg ver. 1.4.10) was used to search for models of high order SNP interactions. Each of the 133 UC SNPs was recoded into two binary predictors in dominant and recessive genetic modes. Simulated annealing, a stochastic search algorithm with increase of model size (i.e. the complexity of SNP logical combinations), implemented in LR was used to search for models composed of logical SNP interactions called ‘Trees’ which could explain the disease risk better than single SNP models. Through the randomization processes, a test for different model sizes can be used to determine an optimal model size and, in combination with the greedy algorithm, a best model can then be identified (Ruczinski et al., 2003;Ruczinski et al., 2004;Schwender and Ruczinski, 2010). To assess the significance of genetic interactions contributed from the Trees, we used the following two approaches: 1) we evaluated the UC association of each Tree after excluding the marginal effects of the SNPs from which the Tree was composed. If the adjusted Tree association remained significant (P<0.05), this would support the presence of genetic interactions between the SNPs in each Tree (Wang et al., 2013); 2) within each Tree, we examined the significance of effect modification (i.e., interaction) through the conventional logistic regression model which includes pairwise SNPs and their interaction terms. The interaction is considered significant if the SNP interaction term P value is less than 0.05.
Model predictability and assessment of explained phenotype variance
We then applied standard logistic regression modeling to investigate the predictive accuracy of models derived from the 133 UC SNPs with and without including the SNP interactions (i.e. Trees). Discriminative accuracy was evaluated using the area under the receiver operating characteristic (ROC) curves (AUC). AUCs were calculated for the predicted risks of the logistic regression models, the cumulative allele score, and the linear predictor values of the logistic regression models. AUCs of different models were statistically compared through the SAS 9.2 GPLOT procedures (SAS Institute, Cary, NC). Goodness-of-fit tests between the models of 133 UC SNPs with and without genetic interactions (i.e. Trees) were conducted through the PROC LOGISTIC and ROC functions in SAS 9.2.
The estimate of the proportion of disease variation explained by a model can be regarded as a measure of goodness of fit to the data and can be done using the McFaddens’ likelihood-based pseudo R-squared provided by the SAS PROC LOGISTIC procedure (Makowsky et al., 2011;Stokes ME et al., 2000).
UC-phenotypes, environmental factors and genotype correlations
In the CC/UP cohort, a total of 566 UC and 1,436 controls subjects were included. Among the 1,436 controls, 936 were older controls from an Alzheimer GWAS using the same Illumina Human Omni1-Quad beadchip (Kamboh et al., 2012) for whom age and gender at study entry were the only phenotypic data available. Otherwise, detailed demographic information, history of tobacco use, history of appendectomy, and UC phenotypic data were available for 504 UC cases and 500 controls and further analyses were performed in this subgroup. Two sample t-tests were used for continuous variables if the appropriate normality assumptions were valid. Nominal data were analyzed using the χ2 test or Fisher’s exact test. A two-tailed P <0.05 was considered significant.
To assess the associations between tobacco use, history of appendectomy (before UC diagnosis for cases and before study entry for controls) and risk of UC, adjustment of the exposure ascertainment time difference between cases and controls (i.e. age at diagnosis in cases and age at study entry in controls) is necessary and was therefore conducted during the association analyses.
We then applied LR in the subgroup of 504 UC subjects and 500 controls with information for the 133 UC SNPs and for smoking and appendectomy history and explored possible high order gene-environment interactions.
Finally, for the 504 UC subjects with phenotypic disease data, we conducted within case genetic association analyses between the 133 SNPs and different clinical behaviors: macroscopic left sided vs. extensive disease; need for surgery vs. no need for surgery; age of diagnosis <20 years vs. ≥ 20 years; EIM positive vs. EIM negative; PSC positive vs. PSC negative. To obtain P-values that take into account multiple testing, we performed a random permutation test with 10,000 replications.
RESULTS
Individual effects of 133 SNPs on UC risk
The UC-associated risk for each of the 23 UC-specific and 110 IBD-common (UC and CD) SNPs in the CC/UP dataset is shown in Supplementary Table 1. The association patterns of effect sizes (ORs) and association directions are similar to those in the recent UC meta-analysis study (Jostins et al., 2012), suggesting that our dataset is representative of those included in the meta-analysis which consisted of a total of 10,920 UC patients and 15,977 controls (Jostins et al., 2012).
Cumulative allele score analysis
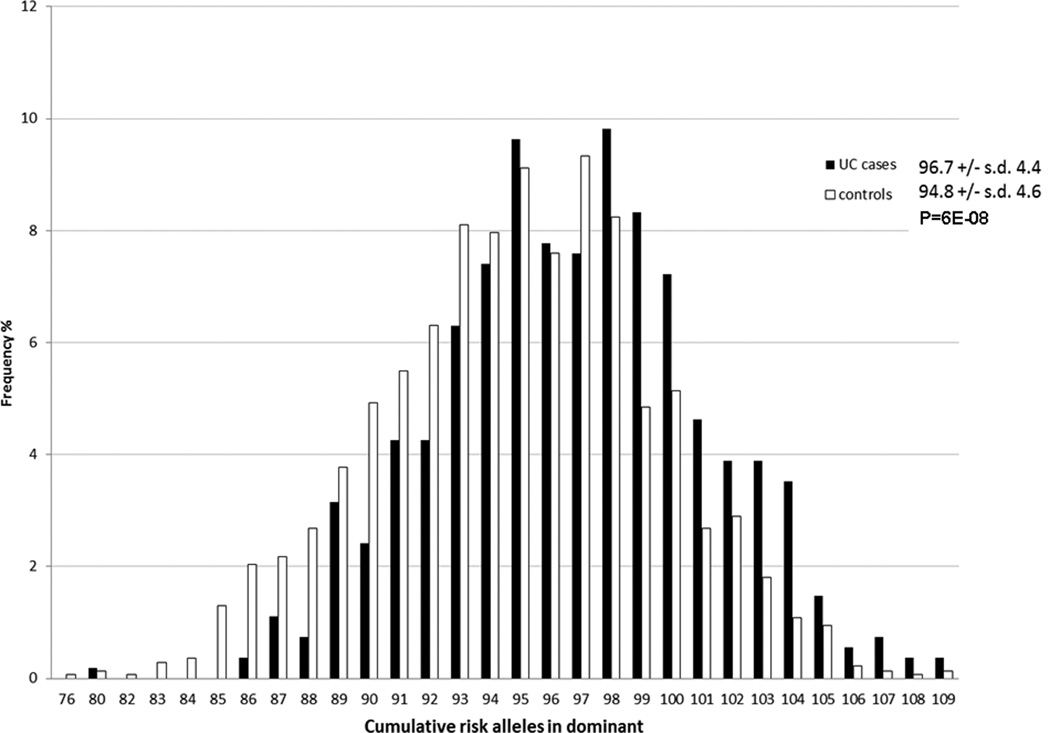
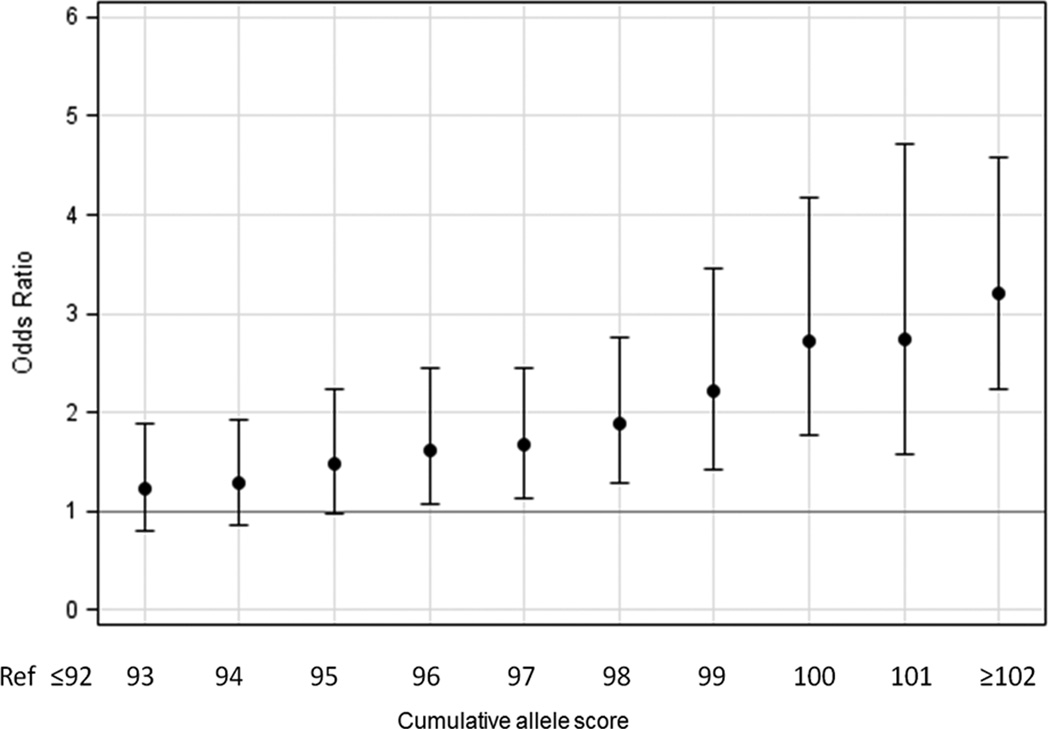
The difference in average cumulative allele scores between UC subjects (96.8 ± S.D. 4.4; range: 80 to 109) and controls (94.8 ± S.D. 4.6; range: 76 to 109) was relatively small (Figure 1) but statistically significant (P=6.0E-08). Figure 2 shows the ORs associated with increasing cumulative allele scores for UC subjects compared to a reference group of UC subjects with ≤ 92 risk alleles, which represented the lowest 20% of the total sample in a logistic regression model.
Figure 1.
Histograms of cumulative allele scores in UC vs. controls in CC/UP cohort Odds Ratios (ORs) and 95% C.I. for CD according to the number of risk alleles carried in CC/UP cohort
Figure 2.
Odds Ratios (ORs) and 95% C.I. for UC according to the number of risk alleles carried in CC/UP cohort
Logic regression-identified genetic interactions in CC/UP and replicated in WTCCC
Applying LR to the 133 UC SNPs in the CC/UP GWAS dataset, four Trees (Supplementary Figures 2A–2D) were identified through the LR randomization model comparison process (Wang et al., 2013):
Tree1: [rs6927022.recc|HLA.DQA1.B1/DRA.B1 and (rs670523.domc|UBQLN4/RIT1 or rs7134599.recc|IFNG/IL26/IL22)]
Tree2: (rs7134599.recc|IFNG/IL26/IL22 or rs561722.domc|REXO2)
Tree3: [(rs11209026.domc|IL23R/IL12RB2 or rs561722.domc|REXO2) or rs3749171.dom|GPR35]
Tree4: [rs3197999.dom|MST1 or (rs7911264.rec|near KIF11 and rs2823286.dom|near USP25)]
(*footnote: ‘.dom’ refers to dominant genetic mode; ‘.rec’ refers to recessive genetic mode; ‘c’ refers to the complement of the SNP risk allele, i.e. not carrying the risk allele)
To test whether the LR-identified SNP interactions (i.e. Trees) added additional information beyond that of the individual SNPs from which they were composed, we examined each Tree’s UC association by excluding the marginal effects of the SNPs from which the Tree was composed. If the adjusted Tree association remained significant (P<0.05), this would support the presence of genetic interactions between the SNPs in each Tree (Wang et al., 2013). As shown in Table 1, all four tested Trees (Trees1–4) in the discovery CC/UP cohort remained significant (nominal P ≤ 0.005) after adjusting for the SNPs from which the Trees were composed. We then examined Trees1–4 in the replicate WTCCC cohort to determine if they provided additional information beyond the SNPs from which they were composed. Tree1 achieved modest statistical significance (P=0.07) while Trees2–4 remained significant (P<0.05) after adjusting for the SNPs from which the Trees were composed (Table 1). These results further support and highlight the significance of the genetic interactions implicated in the Trees.
Table 1.
Assessment of the significance of SNP interactions (Trees) before and after adjusting for the marginal effects of SNPs from which the Trees were composed in the discovery CC/UP cohort and the WTCCC cohort
| Discovery cohort CC/UP | Replicate cohort WTCCC | |||
|---|---|---|---|---|
| Before adjusting for SNPs OR (95% C.I.) P value |
After adjusting for SNPs OR (95% C.I.) P value |
Before adjusting for SNPs OR (95% C.I.) P value |
After adjusting for SNPs OR (95% C.I.) P value |
|
|
Tree1 HLA-DQA1, RIT1/UBQLN4, and IFNG/IL22IL26/IL26 |
0.52 (0.42–0.64) 2.92E-09 |
0.45 (0.27–0.76) 0.002 |
0.67 (0.60–0.75) 1.01E-12 |
0.79 (0.62–1.02) 0.07 |
|
Tree2 IFNG/IL26/IL22, REXO2 |
1.63 (1.34–1.98) 1.10E-06 |
2.15 (1.27–3.62) 0.004 |
1.17 (1.06–1.29) 0.001 |
1.34 (1.04–1.74) 0.02 |
|
Tree3 IL23R/IL12RB2, REXO2, and GPR35 |
11.06 (3.48–35.1) 2.00E-09 |
8.35 (2.47–28.2) 0.0006 |
2.33 (1.72–3.14) 2.05E-09 |
1.45 (1.01–2.08) 0.04 |
|
Tree4 MST1, KIF11, USP25 |
0.56 (0.46–0.69) 3.35E-08 |
0.51 (0.35–0.75) 0.0005 |
0.79 (0.71–0.87) 7.05E-06 |
0.81 (0.67–0.97) 0.02 |
|
Tree5 (gene-environment) HLA.DQA1, CALM3, TRIB1, IL2/IL21, and smoking |
0.38 (0.29–0.49) 7.49E-13 |
0.24 (0.12–0.47) 2.55E-05 |
NA | NA |
To further elucidate the effect modification (i.e. interaction) of a SNP by another SNP within the same Tree, we examined the significance of pairwise SNP interactions within each Tree through a conventional logistic regression model which includes single SNPs and their interaction terms. Using this method, we again found significant SNP interactions for Tree1–4 and successfully replicated these findings in the replicate cohort (Supplementary Figures 2A–2D).
Phenotype-genotype correlations in the CC/UP cohort
In the CC/UP cohort, there were a total of 566 UC subjects and 1,436 controls. Among the controls, 936 were controls recruited from an Alzheimer GWAS study (Kamboh et al., 2012) and these subjects were older than the remaining 500 healthy controls who were recruited as part of IBD studies (age at study entry: 75.5 ± 6.3 vs. 46.1 ± 14.2, P<1.0E-08). The cumulative allele scores between older controls (mean 94.8, S.D. 4.5) and younger controls (mean 94.8, S.D. 4.7) were nearly identical (P=0.85) suggesting that these two groups of controls are comparable for the purposes of our study. History of tobacco use and appendectomy were not available for the Alzheimer GWAS controls. Therefore, detailed demographic information, history of tobacco use and appendectomy, and UC phenotype information were available in a subset of 504 UC cases and 500 controls.
The mean age at study entry for this subset of UC cases and controls was similar (44.3 ± 14.8 vs. 46.1 ± 13.9, P=0.05) (Table 2). There were more males among UC cases compared to controls (56% vs. 44%, P=0.0002). The mean age at diagnosis for UC cases was 33.3 years (S.D. 14.5). Among UC subjects, phenotype classifications were as follow: extent of disease (left sided 28.4%, extensive 71.6%); need for colectomy 28.1%; and EIMs involving joints (6.3%), eyes (2.9%), skin (2.4%), and PSC (6.5%).
Table 2.
Demographic and phenotypic information of UC cases and controls in CC/UP cohort
| UC (n=504) | Controls (n=500) | P value | |
|---|---|---|---|
| Mean age at diagnosis (years) | 33.3 +/− S.D.14.5 | -- | -- |
| Mean age at study entry (years) | 44.3 +/− S.D. 14.8 | 46.1 +/− S.D. 13.9 | 0.05 |
| Gender: Male (%) | 56.0% | 44.2% | 0.0002 |
| Appendectomy (before IBD diagnosed) | 13 (2.6%) | 73 (14.0%) | 9.74E-15 |
| UC sub-phenotypes: | -- | -- | |
| Extent: Extensive (%) | 71.6% | ||
| Surgery (%) | 28.1% | ||
| Extra-GI manifestations: | |||
| Any (%) | 16.2% | ||
| Joints (%) | 6.3% | ||
| Skin (%) | 2.4% | ||
| Eyes (%) | 2.9% | ||
| PSC (%) | 6.5% | ||
| Tobacco use (before UC diagnosed): | |||
| Current/Past/Never | 26/128/350 | 80/126/291 | 3.66E-08 |
| Family history of IBD in first degree relative | 113 (24.8%) | 0 (0%) | 1.39E-28 |
After adjustment for gender and age, and using the never smoking subjects as the reference group, current smoking was found to be associated with a reduced risk of UC (OR: 0.26, 95% CI: 0.15–0.45, P=8.09E-07) and conversely, past smoking was associated with an increased risk of UC (OR: 1.53, 95% CI: 1.05–2.25, P=0.02). A history of appendectomy before the diagnosis of UC was associated with a reduced risk of UC (OR: 0.45, 95% CI: 0.22–0.92, P=0.02) after adjustment for gender and age.
i) Exploring genes and environment interactions
We explored gene-gene and gene-environment (smoking and prior appendectomy) interactions using LR within the subset of 504 UC cases and 500 controls. One Tree (Tree5, see Supplementary Figure 2E), composed of smoking (including current and past smokers at the time of diagnosis) and four genetic loci, was identified through LR and was found to be significantly associated with UC risk (OR 0.38, 95% CI: 0.29–0.49, P=7.49E-13).
Tree5: {rs6927022.recc|HLA.DQA1.B1/DRA.B1 and [(rs1126510.recc|CALM3 or smoking) and (rs921720.recc|TRIB1 or rs7657746.dom|IL2/IL21)]}
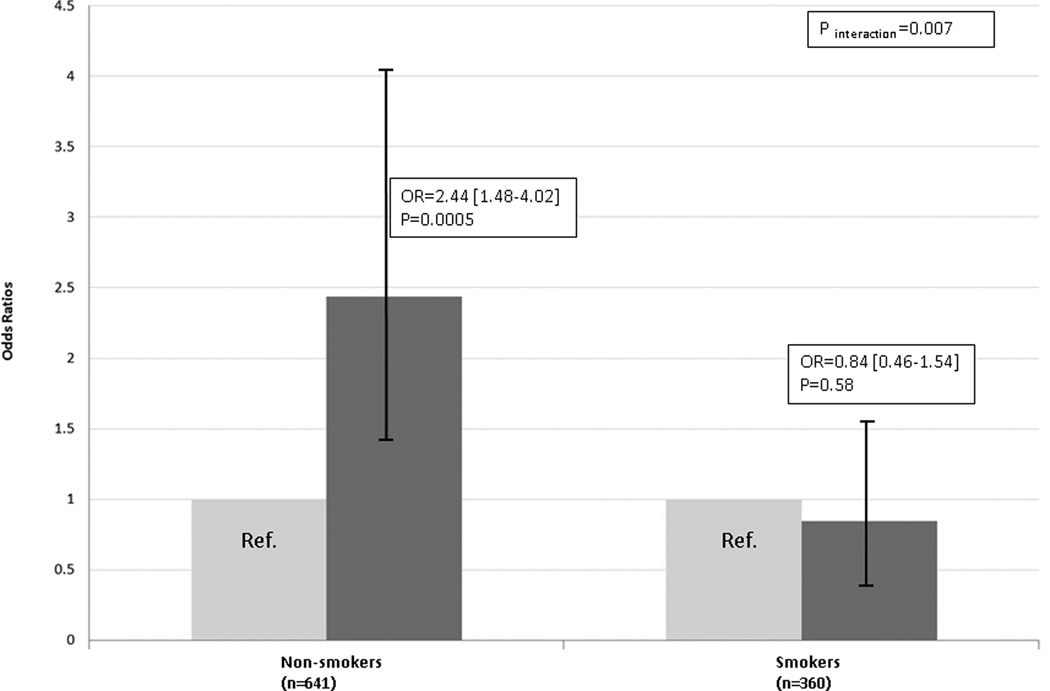
After adjusting for the four SNPs from which the Tree was composed, tobacco use and the exposure ascertainment time difference between cases and controls, the identified interactions between the four genetic loci and tobacco use remained highly significant (adjusted OR 0.24, 95% CI: 0.12–0.47, P=2.55E-05)(Table 1). We also examined the significance of interaction through conventional logistic regression modeling and found significant genetic interactions and gene-smoking interactions in Tree5 (Supplementary Figure 2E). We could not test this Tree in the WTCCC dataset for replication because that dataset does not include information about smoking. However, a very intriguing smoking-gene interaction was identified in the discovery CC/UP cohort. Among smokers, the risk variant SNP rs1126510 (in recessive mode) in CALM3 was not associated with risk of UC (OR: 0.84, 95% CI: 0.46–1.54, P=0.58). However, this genetic association was significantly increased among those who never smoked (OR: 2.44, 95% CI: 1.48–4.02, P=0.0005). In other words, the genetic effect of CALM3 was significantly modified by the exposure of smoking (Pinteraction =0.007) (Figure 3).
Figure 3.
Stratified analysis of genetic effect of CALM3 (SNP rs1126510, in recessive mode) on UC risk by the exposure of smoking
We further assessed the model predictability of the133 UC loci in this subset of 504 UC cases and 500 controls with and without including the genetic interactions (Trees1–4) and gene-smoking interaction (Tree5). The AUC increased from 86% to 89%, corresponding to an increase in explained UC variance from 37% to 42% (P=3.26E-05), after adding the interactions terms (Tree1–5).
ii) Correlations between genotype and UC sub-phenotypes
We next performed a within case analysis of the 504 UC subjects evaluating sub-phenotypes. Analyses for colectomy vs. no colectomy, extensive vs. left-sided disease, age at diagnosis <20 years vs. ≥ 20 years, and EIM vs. no EIM did not achieve statistical significance after multiple testing correction (data not shown). However, for UC with associated PSC versus UC without PSC, two SNPs remained significant after correcting for multiple testing: 1) rs38904 (chromosome 7, in the genetic locus of WNT2,CFTR; allele G vs. A, OR 2.78, 95% CI:1.62–4.76, nominal P=0.0001; permutated P=0.01); 2) rs11209026 (chromosome 1, in IL23R; allele A vs. G, OR 4.08, 95% CI: 1.87–8.92, nominal P=0.0001; permutated P=0.02).
We also explored gene-gene and gene-environment interactions using LR across the different sub-phenotypes defined above within the 504 UC cases, but no significant interactions were identified.
DISCUSSION
Using a case-control GWAS dataset, we found good predictability of UC risk by combining the 133 GWAS meta-analysis SNPs (AUC of 0.86). Higher cumulative allele scores predicted greater UC risk but, interestingly, our study also highlights that the absolute difference in cumulative allele scores between UC cases and controls, although statistically significant, is relatively small (96.8 ± 4.4 versus 94.8 ± 4.6; P=6.0E-08). This small difference implicates influences beyond those at the level of single genetic loci including unidentified rare genetic variants and the involvement of gene-gene and gene-environment interactions. Supporting this conclusion, by taking a novel approach of applying LR to our GWAS dataset, we identified high order genetic interactions (Trees) among the 133 UC loci and found that risk prediction improved by adding these interactions to the 133 UC SNP model. Importantly, high order SNP interactions were successfully replicated in a second large independent WTCCC cohort, thus validating the LR methodology. Furthermore, exploratory gene-environment interaction analysis identified interactions between genes (HLA-DQA1, CALM3, TRIB1, and IL2/IL21) and smoking in the discovery cohort. Our results confirm improved UC risk predictability (AUC increase from 86% to 89%, P=3.26E-05) and better explanation of disease variance (pseudo R-squared increase from 37% to 42%) by adding the gene-gene and gene-environment interactions we identified through LR.
Searching for possible gene-gene and gene-environment interactions can be approached using statistical approaches or functional studies (Berzuini et al., 2012). The identification of genetic and environmental factors interacting mechanistically may be more useful than simply defining statistical interactions when trying to understand which factors are parts of the biological mechanisms that influence UC susceptibility. To assess whether the LR-identified interactions could add additional information beyond those from the individual predictors in the Trees models, we examined the excess risk from the Trees in a logistic regression model by eliminating the marginal effects of the individual predictors that made up the Trees and found that all four tested Trees remained significant. This approach is analogous to testing mechanistic interactions to identify underlying causal mechanisms (Berzuini et al., 2012). In addition, we examined the significance of pairwise SNP interactions within each Tree through conventional logistic regression modeling which includes single predictors and their interaction terms and again found significant SNPs interactions with replication for Trees1–4. Once novel genetic interactions are identified through LR, the next challenge relates to interpreting and proving that the statistically identified genetic interactions are of importance at a biological level. We highlight Tree1 and Tree5 as excellent examples of biological plausibility for the identified genetic interactions.
Tree1, composed of three genetic loci in HLA-DQA1, RIT1/UBQLN4, and IFNG/IL26/IL22, highlights known pathways of host response to microbial organisms. The major histocompatibility complex (MHC) region containing the human leukocyte antigen (HLA) genes was the first identified UC associated genetic locus(Satsangi et al., 1996;Toyoda et al., 1993) and includes known encoding genes regulating the immune response to antigens. Our group previously reported that variation at HLA-DRβ1, amino acid 11 in the P6 pocket of the HLA-DR complex antigen binding cleft is a major determinant associated with UC (Achkar et al., 2012). HLA-DRβ1 and HLA-DQA1 encode for the β- and α-chains respectively of class II HLA molecules. UBQLN4, at chromosome 1q21, plays a role in the regulation of proteasomal protein degradation (Riley et al., 2004). Interestingly, two genes PSMB9 and PSMB8, which encode for proteasome subunits are located in the same haplotype block of the HLA class II region (HLA-DRβ1 and -DQβ1) (Deng et al., 1995). Furthermore, the IFNG/IL26/IL22 gene combination of Tree 1 is particularly interesting as the three genes are closely located on chromosome 12 and all their products are essential to mucosal immunity. IFN-γ, the product of IFNG (interferon-gamma) displays potent immunoregulatory function and induces expression of HLA class II molecules on epithelial cells, as observed in IBD colonic epithelium (Selby et al., 1983). IL-26, an IL-10 family cytokine, is produced by Th17 cells and is present in increased amounts in inflamed colonic tissue of IBD patients (Dambacher et al., 2009). A genetic variant in IL26 (rs2870946) was found to be strongly associated with UC (Silverberg et al., 2009). IL-22 is often co-expressed with IL-26 by activated Th17 cells and, together with IL-17, is a major product of innate lymphoid cells (Donnelly et al., 2010). More importantly, both IL-22 and IL-17 are essential to the innate immune defense against enteric bacterial antigens (Rubino et al., 2012), and IL-22 maintains the epithelial barrier and restores barrier integrity in a mouse model of UC (Sugimoto et al., 2008). Thus, when the genetic combinations making up Tree 1 are taken together, they not only appear to be functionally related, but also intimately involved in an integrated epithelial inflammatory response. This interpretation fits well in our current knowledge of UC pathogenesis and lends considerable support to the LR analytic approach. Clearly, follow up functional work will be necessary, but the methodology we have used can identify relevant pathway based approaches to unravel the underlying pathogenesis of UC.
Similar to our findings, prior studies and a meta-analysis demonstrate that current smoking protects against development of UC while former smoking is associated with increased risk of UC (Timmer, 2003). Prior epigenetic studies highlight the potential role of gene-environment interactions in UC (Hasler et al., 2012;Quigley, 2012). In our gene-environment analysis, we found evidence for interactions between genes (HLA-DQA1, CALM3, TRIB1, and IL2/IL21) and tobacco use and again, there is evidence to support biological plausibility of these findings. Studies of cardiovascular risk in rheumatoid arthritis patients have demonstrated interaction between HLA-DRB1 and smoking through high-affinity binding between protein citrullination induced by smoking and the epitope P4 pocket of HLA-DRB1 with subsequent exaggerated T-cell activation (Hill et al., 2003;Kallberg et al., 2007). Calmodulin, encoded by CALM3, mediates the control of different protein kinases. Experimental studies show that nicotine exposure up-regulates the calcium/calmodulin-dependent protein kinase II in mice(Damaj, 2000) and variants in the TRIB1 (G protein coupled receptor) gene can regulate protein kinase signaling cascades (Kiss-Toth et al., 2004). IL-2 is required for T-cell proliferation and is considered to be a key component of the adaptive immune response. Prior studies show an influence of both cigarette smoking(Ouyang et al., 2000) and genetic variation (Hoffmann et al., 2001) on IL-2 expression. Furthermore, animal model supports the concept that smoking exposure can induce Th17 cells to promotes CD8+ T cell cytotoxic effect through the increase of IL-21 production (Duan et al., 2012). Based on these lines of evidence we conclude that our finding of interaction between genetic factors and smoking is biologically plausible and warrants further investigation in UC pathogenesis studies.
In our CC/UP GWAS, 37% of UC disease variance was explained by the 133 UC loci and increased further to 42% after adding the high order gene-gene (Trees1–4) and gene-environment interactions (Tree 5) into the 133 UC loci model. Even further explanation of phenotypic variance would be expected through the search of more complicated interaction models within or between IBD related biological pathways such as IL23/ Th17, TNF-α, and NF-κB signaling pathways (Vaishnava et al., 2008). Investigating interactions between genes, biological pathways, environmental factors, and the host microbiome(Morgan et al., 2012) would provide the ideal integrated approach to understand IBD pathogenesis.
In addition to evaluating genetic and interaction factors, we performed a genotype-phenotype correlation analysis in the UC only subgroup. To address multiple testing corrections, we performed a random permutation test with 10,000 replicates for each of the five genotype-phenotype correlations. With this correction, the only significant suggestive associations were for two genetic loci, WNT2/CFTR and IL23R, with PSC. PSC, characterized by progressive fibro-obliterative inflammation of the biliary tract is strongly associated with IBD, but only 3%–7% of IBD (mostly UC) patients will develop PSC (Fausa et al., 1991). It is relevant for clinicians to know which IBD patients have a higher chance of developing this extra-intestinal complication. In our analysis, there were significant associations between PSC and a genetic locus in CFTR region (rs38904, chr7q31, OR 2.78, 95% CI: 1.62–4.76, permutated P=0.01). Recent studies suggest that CFTR variants may play a role in the development of PSC (McGill et al., 1996) and there are CFTR abnormalities demonstrated by molecular and functional analyses which may contribute to the development of PSC in a subset of patients with IBD (Sheth et al., 2003). We also found suggestive association of PSC risk among UC patients and an IL23R SNP (rs11209026, chr1p31, OR 4.08, 95% CI: 1.87–8.92, permutated P=0.02). Interestingly, our PSC associated findings were not reported as PSC risk loci in recent GWAS studies, but this is likely due to the fact that those studies compared UC patients with PSC to population healthy controls while our study compared UC patients with PSC to UC patients without PSC (Ellinghaus et al., 2013;Folseraas et al., 2012;Liu JZ et al., 2013). Our findings in PSC warrant further study in larger samples.
Potential limitations of this study include the relatively small sample size in our CC/UP GWAS (566 UC and 1,436 controls), which may explain why many of the 133 UC SNPs did not achieve genome-wide level of significance. Detailed clinical phenotypic information and environmental factors were limited to a smaller subset (504 UC and 500 controls), which may limit the generalizability of study findings. Also, the assumptions of dominant and recessive genetic modes while searching for the genetic interactions using LR may not exactly represent the complex hereditary patterns of IBD loci. Finally, despite using a high imputation accuracy program like MaCH (Li et al., 2009;Li et al., 2010), several factors can still affect imputation results, including the density of genotype platform in the experimental samples, level of linkage disequilibrium in different genetic regions, minor allele frequency of the marker being imputed, and degree of genetic relationship or population heterogeneity between the experimental and reference populations.
In conclusion, using the LR approach, we found higher disease risk with increasing UC risk allele burden, evidence for high order genetic interactions (Trees1–4), biologically plausible gene-smoking interactions, and improved disease predictability. Potential future applications of this approach include verifying these findings in a larger population-based cohort, defining the biological significance of suggested interactions in cell line or animal models, and searching for further interactions using meta-genomic approaches that integrate genes, biological pathways, environmental factors, and the host microbiome.
Supplementary Material
Acknowledgements
We thank the patients and the controls for participating in this study. We acknowledge the Feinstein Institute for Medical Research of the North Shore-Long Island Jewish Health System for Illumina Genotyping BeadChip processing. This study makes use of data generated by the Wellcome Trust Case Control Consortium. A full list of the investigators who contributed to the generation of the data is available from www.wtccc.org.uk. Funding for the project was provided by the Wellcome Trust under award 076113.
Funding:
T32 DK083251, NIH - NATIONAL INSTITUTE OF DIABETES AND DIGESTIVE AND KIDNEY DISEASES (NIDDK)(M-H W, CF); DK068112 (J-PA), DK062420 (RHD) and DK076025 (RHD); AG030653 (MIK); a Crohn’s & Colitis Foundation of America Senior Research Award (RHD); and funds generously provided by Kenneth and Jennifer Rainin, the Wesley Roj and Douglas Durham Roj Endowed Fund, and Gerald and Nancy Goldberg.
Footnotes
Contributorship:
M-H W conceived, designed and executed the study, performed the statistical analyses, and drafted the manuscript; C F helped design the study and drafted the manuscript; X Z helped with statistical analyses; S R helped generate the imputation of genotype data; M.I. K helped design the study and contributed genotype data from a different GWAS; N R helped collect and generate data; R.H. D conceived, designed and executed the study, and drafted the manuscript; J.P. A conceived, designed and executed the study, and drafted the manuscript.
Competing interest: None
Reference List
- Achkar JP, Klei L, Bakker PIW, Bellone G, Rebert N, Scott R, Lu Y, Regueiro M, Brzezinski A, Kamboh MI, Fiocchi C, Devlin B, Trucco M, Ringquist S, Roeder K, Duerr RH. Amino acid position 11 of HLA-DR[beta]1 is a major determinant of chromosome 6p association with ulcerative colitis. Genes Immun. 2012;13:245–252. doi: 10.1038/gene.2011.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldhous MC, Drummond HE, Anderson N, Baneshi MR, Smith LA, Arnott ID, Satsangi J. Smoking habit and load influence age at diagnosis and disease extent in ulcerative colitis. Am J Gastroenterol. 2007;102:589–597. doi: 10.1111/j.1572-0241.2007.01065.x. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Lee JC, Lees CW, Prescott NJ, Anderson CA, Phillips A, Wesley E, Parnell K, Zhang H, Drummond H, Nimmo ER, Massey D, Blaszczyk K, Elliott T, Cotterill L, Dallal H, Lobo AJ, Mowat C, Sanderson JD, Jewell DP, Newman WG, Edwards C, Ahmad T, Mansfield JC, Satsangi J, Parkes M, Mathew CG, Donnelly P, Peltonen L, Blackwell JM, Bramon E, Brown MA, Casas JP, Corvin A, Craddock N, Deloukas P, Duncanson A, Jankowski J, Markus HS, Mathew CG, McCarthy MI, Palmer CN, Plomin R, Rautanen A, Sawcer SJ, Samani N, Trembath RC, Viswanathan AC, Wood N, Spencer CC, Barrett JC, Bellenguez C, Davison D, Freeman C, Strange A, Donnelly P, Langford C, Hunt SE, Edkins S, Gwilliam R, Blackburn H, Bumpstead SJ, Dronov S, Gillman M, Gray E, Hammond N, Jayakumar A, McCann OT, Liddle J, Perez ML, Potter SC, Ravindrarajah R, Ricketts M, Waller M, Weston P, Widaa S, Whittaker P, Deloukas P, Peltonen L, Mathew CG, Blackwell JM, Brown MA, Corvin A, McCarthy MI, Spencer CC, Attwood AP, Stephens J, Sambrook J, Ouwehand WH, McArdle WL, Ring SM, Strachan DP. Genome-wide association study of ulcerative colitis identifies three new susceptibility loci, including the HNF4A region. Nat Genet. 2009;41:1330–1334. doi: 10.1038/ng.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berzuini, Dawid P, Zhang H, Parkes M. Analysis of interaction for identifying causal mechanisms. In: Berzuini C, Dawid P, Bernardinelli L, editors. Causality: Statistical Perspectives and Applications. Wiley; 2012. pp. 192–207. [Google Scholar]
- Boyko EJ, Koepsell TD, Perera DR, Inui TS. Risk of ulcerative colitis among former and current cigarette smokers. N Engl J Med. 1987;316:707–710. doi: 10.1056/NEJM198703193161202. [DOI] [PubMed] [Google Scholar]
- Browning, Browning SR. A Unified Approach to Genotype Imputation and Haplotype-Phase Inference for Large Data Sets of Trios and Unrelated Individuals. 2009;84:210–223. doi: 10.1016/j.ajhg.2009.01.005. edn. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damaj MI. The involvement of spinal Ca(2+)/calmodulin-protein kinase II in nicotine-induced antinociception in mice. Eur J Pharmacol. 2000;404:103–110. doi: 10.1016/s0014-2999(00)00579-3. [DOI] [PubMed] [Google Scholar]
- Dambacher J, Beigel F, Zitzmann K, De Toni EN, Goke B, Diepolder HM, Auernhammer CJ, Brand S. The role of the novel Th17 cytokine IL-26 in intestinal inflammation. Gut. 2009;58:1207–1217. doi: 10.1136/gut.2007.130112. [DOI] [PubMed] [Google Scholar]
- Danese S, Fiocchi C. Ulcerative colitis. N Engl J Med. 2011;365:1713–1725. doi: 10.1056/NEJMra1102942. [DOI] [PubMed] [Google Scholar]
- Dassopoulos T, Nguyen GC, Bitton A, Bromfield GP, Schumm LP, Wu Y, Elkadri A, Regueiro M, Siemanowski B, Torres EA, Gregory FJ, Kane SV, Harrell LE, Franchimont D, Achkar JP, Griffiths A, Brant SR, Rioux JD, Taylor KD, Duerr RH, Silverberg MS, Cho JH, Steinhart AH. Assessment of reliability and validity of IBD phenotyping within the National Institutes of Diabetes and Digestive and Kidney Diseases (NIDDK) IBD Genetics Consortium (IBDGC) Inflamm Bowel Dis. 2007;13:975–983. doi: 10.1002/ibd.20144. [DOI] [PubMed] [Google Scholar]
- Deng GY, Muir A, Maclaren NK, She JX. Association of LMP2 and LMP7 genes within the major histocompatibility complex with insulin-dependent diabetes mellitus: population and family studies. Am J Hum Genet. 1995;56:528–534. [PMC free article] [PubMed] [Google Scholar]
- Donnelly RP, Sheikh F, Dickensheets H, Savan R, Young HA, Walter MR. Interleukin-26: an IL-10-related cytokine produced by Th17 cells. Cytokine Growth Factor Rev. 2010;21:393–401. doi: 10.1016/j.cytogfr.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan MC, Huang Y, Zhong XN, Tang HJ. Th17 Cell Enhances CD8 T-Cell Cytotoxicity via IL-21 Production in Emphysema Mice. Mediators Inflamm. 2012;2012:898053. doi: 10.1155/2012/898053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellinghaus D, Folseraas T, Holm K, Ellinghaus E, Melum E, Balschun T, Laerdahl JK, Shiryaev A, Gotthardt DN, Weismuller TJ, Schramm C, Wittig M, Bergquist A, Bjornsson E, Marschall HU, Vatn M, Teufel A, Rust C, Gieger C, Wichmann HE, Runz H, Sterneck M, Rupp C, Braun F, Weersma RK, Wijmenga C, Ponsioen CY, Mathew CG, Rutgeerts P, Vermeire S, Schrumpf E, Hov JR, Manns MP, Boberg KM, Schreiber S, Franke A, Karlsen TH. Genome-wide association analysis in Primary sclerosing cholangitis and ulcerative colitis identifies risk loci at GPR35 and TCF4. Hepatology. 2013 doi: 10.1002/hep.25977. [DOI] [PubMed] [Google Scholar]
- Fausa O, Schrumpf E, Elgjo K. Relationship of inflammatory bowel disease and primary sclerosing cholangitis. Semin Liver Dis. 1991;11:31–39. doi: 10.1055/s-2008-1040420. [DOI] [PubMed] [Google Scholar]
- Folseraas T, Melum E, Rausch P, Juran BD, Ellinghaus E, Shiryaev A, Laerdahl JK, Ellinghaus D, Schramm C, Weismuller TJ, Gotthardt DN, Hov JR, Clausen OP, Weersma RK, Janse M, Boberg KM, Bjornsson E, Marschall HU, Cleynen I, Rosenstiel P, Holm K, Teufel A, Rust C, Gieger C, Wichmann HE, Bergquist A, Ryu E, Ponsioen CY, Runz H, Sterneck M, Vermeire S, Beuers U, Wijmenga C, Schrumpf E, Manns MP, Lazaridis KN, Schreiber S, Baines JF, Franke A, Karlsen TH. Extended analysis of a genome-wide association study in primary sclerosing cholangitis detects multiple novel risk loci. J Hepatol. 2012;57:366–375. doi: 10.1016/j.jhep.2012.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisch M, Pedersen BV, Andersson RE. Appendicitis, mesenteric lymphadenitis, and subsequent risk of ulcerative colitis: cohort studies in Sweden and Denmark. BMJ. 2009;338:b716. doi: 10.1136/bmj.b716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardenbroek TJ, Eshuis EJ, Ponsioen CI, Ubbink DT, D'Haens GR, Bemelman WA. The effect of appendectomy on the course of ulcerative colitis: a systematic review. Colorectal Dis. 2012;14:545–553. doi: 10.1111/j.1463-1318.2011.02600.x. [DOI] [PubMed] [Google Scholar]
- Hallas J, Gaist D, Sorensen HT. Does appendectomy reduce the risk of ulcerative colitis? Epidemiology. 2004;15:173–178. doi: 10.1097/01.ede.0000112208.22339.4a. [DOI] [PubMed] [Google Scholar]
- Hasler R, Feng Z, Backdahl L, Spehlmann ME, Franke A, Teschendorff A, Rakyan VK, Down TA, Wilson GA, Feber A, Beck S, Schreiber S, Rosenstiel P. A functional methylome map of ulcerative colitis. Genome Res. 2012;22:2130–2137. doi: 10.1101/gr.138347.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill JA, Southwood S, Sette A, Jevnikar AM, Bell DA, Cairns E. Cutting edge: the conversion of arginine to citrulline allows for a high-affinity peptide interaction with the rheumatoid arthritis-associated HLA-DRB1*0401 MHC class II molecule. J Immunol. 2003;171:538–541. doi: 10.4049/jimmunol.171.2.538. [DOI] [PubMed] [Google Scholar]
- Hoffmann SC, Stanley EM, Darrin CE, Craighead N, DiMercurio BS, Koziol DE, Harlan DM, Kirk AD, Blair PJ. Association of cytokine polymorphic inheritance and in vitro cytokine production in anti-CD3/CD28-stimulated peripheral blood lymphocytes. Transplantation. 2001;72:1444–1450. doi: 10.1097/00007890-200110270-00019. [DOI] [PubMed] [Google Scholar]
- Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, Lee JC, Schumm LP, Sharma Y, Anderson CA, Essers J, Mitrovic M, Ning K, Cleynen I, Theatre E, Spain SL, Raychaudhuri S, Goyette P, Wei Z, Abraham C, Achkar JP, Ahmad T, Amininejad L, Ananthakrishnan AN, Andersen V, Andrews JM, Baidoo L, Balschun T, Bampton PA, Bitton A, Boucher G, Brand S, Buning C, Cohain A, Cichon S, D'Amato M, De JD, Devaney KL, Dubinsky M, Edwards C, Ellinghaus D, Ferguson LR, Franchimont D, Fransen K, Gearry R, Georges M, Gieger C, Glas J, Haritunians T, Hart A, Hawkey C, Hedl M, Hu X, Karlsen TH, Kupcinskas L, Kugathasan S, Latiano A, Laukens D, Lawrance IC, Lees CW, Louis E, Mahy G, Mansfield J, Morgan AR, Mowat C, Newman W, Palmieri O, Ponsioen CY, Potocnik U, Prescott NJ, Regueiro M, Rotter JI, Russell RK, Sanderson JD, Sans M, Satsangi J, Schreiber S, Simms LA, Sventoraityte J, Targan SR, Taylor KD, Tremelling M, Verspaget HW, De VM, Wijmenga C, Wilson DC, Winkelmann J, Xavier RJ, Zeissig S, Zhang B, Zhang CK, Zhao H, Silverberg MS, Annese V, Hakonarson H, Brant SR, Radford-Smith G, Mathew CG, Rioux JD, Schadt EE, Daly MJ, Franke A, Parkes M, Vermeire S, Barrett JC, Cho JH. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallberg H, Padyukov L, Plenge RM, Ronnelid J, Gregersen PK, van der Helm-van Mil AH, Toes RE, Huizinga TW, Klareskog L, Alfredsson L. Gene-gene and gene-environment interactions involving HLA-DRB1, PTPN22, and smoking in two subsets of rheumatoid arthritis. Am J Hum Genet. 2007;80:867–875. doi: 10.1086/516736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamboh MI, Demirci FY, Wang X, Minster RL, Carrasquillo MM, Pankratz VS, Younkin SG, Saykin AJ, Jun G, Baldwin C, Logue MW, Buros J, Farrer L, Pericak-Vance MA, Haines JL, Sweet RA, Ganguli M, Feingold E, Dekosky ST, Lopez OL, Barmada MM. Genome-wide association study of Alzheimer's disease. Transl Psychiatry. 2012;2:e117. doi: 10.1038/tp.2012.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss-Toth E, Bagstaff SM, Sung HY, Jozsa V, Dempsey C, Caunt JC, Oxley KM, Wyllie DH, Polgar T, Harte M, O'neill LA, Qwarnstrom EE, Dower SK. Human tribbles, a protein family controlling mitogen-activated protein kinase cascades. J Biol Chem. 2004;279:42703–42708. doi: 10.1074/jbc.M407732200. [DOI] [PubMed] [Google Scholar]
- Li Y, Willer C, Sanna S, Abecasis G. Genotype imputation. Annu Rev Genomics Hum Genet. 2009;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Willer CJ, Ding J, Scheet P, Abecasis GaR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;34:816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JZ, Hov JR, Folserras T. Dense genotyping of immune-related disease regions identifies nine new risk loci for primary sclerosing cholangitis. Nat Genet. 2013 doi: 10.1038/ng.2616. (in press): [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Song Y. Building Genetic Scores to Predict Risk of Complex Diseases in Humans: Is It Possible? Diabetes. 2010;59:2729–2731. doi: 10.2337/db10-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahid SS, Minor KS, Soto RE, Hornung CA, Galandiuk S. Smoking and inflammatory bowel disease: a meta-analysis. Mayo Clin Proc. 2006;81:1462–1471. doi: 10.4065/81.11.1462. [DOI] [PubMed] [Google Scholar]
- Makowsky R, Pajewski NM, Klimentidis YC, Vazquez AI, Duarte CW, Allison DB, de los CG. Beyond missing heritability: prediction of complex traits. PLoS Genet. 2011;7:e1002051. doi: 10.1371/journal.pgen.1002051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- McGill JM, Williams DM, Hunt CM. Survey of cystic fibrosis transmembrane conductance regulator genotypes in primary sclerosing cholangitis. Dig Dis Sci. 1996;41:540–542. doi: 10.1007/BF02282335. [DOI] [PubMed] [Google Scholar]
- Morgan XC, Tickle TL, Sokol H, Gevers D, Devaney KL, Ward DV, Reyes JA, Shah SA, LeLeiko N, Snapper SB, Bousvaros A, Korzenik J, Sands BE, Xavier RJ, Huttenhower C. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012;13:R79. doi: 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang Y, Virasch N, Hao P, Aubrey MT, Mukerjee N, Bierer BE, Freed BM. Suppression of human IL-1beta, IL-2, IFN-gamma, and TNF-alpha production by cigarette smoke extracts. J Allergy Clin Immunol. 2000;106:280–287. doi: 10.1067/mai.2000.107751. [DOI] [PubMed] [Google Scholar]
- Quigley EM. Epigenetics: filling in the 'heritability gap' and identifying gene-environment interactions in ulcerative colitis. Genome Med. 2012;4:72. doi: 10.1186/gm373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley BE, Xu Y, Zoghbi HY, Orr HT. The effects of the polyglutamine repeat protein ataxin-1 on the UbL-UBA protein A1Up. J Biol Chem. 2004;279:42290–42301. doi: 10.1074/jbc.M406284200. [DOI] [PubMed] [Google Scholar]
- Rubino SJ, Geddes K, Girardin SE. Innate IL-17 and IL-22 responses to enteric bacterial pathogens. Trends Immunol. 2012;33:112–118. doi: 10.1016/j.it.2012.01.003. [DOI] [PubMed] [Google Scholar]
- Ruczinski I, Kooperberg C, LeBlanc L. Logic Regression. Journal of Computational and Graphical Statistics. 2003;12:475–511. [Google Scholar]
- Ruczinski I, Kooperberg C, LeBlanc L. Exploring interactions in high-dimensional genomic data: an overview of Logic Regression, with applications. Journal of Multivariate Analysis. 2004;90:178–195. [Google Scholar]
- Satsangi J, Welsh KI, Bunce M, Julier C, Farrant JM, Bell JI, Jewell DP. Contribution of genes of the major histocompatibility complex to susceptibility and disease phenotype in inflammatory bowel disease. Lancet. 1996;347:1212–1217. doi: 10.1016/s0140-6736(96)90734-5. [DOI] [PubMed] [Google Scholar]
- Schwender H, Ruczinski I. Logic regression and its extensions. Adv Genet. 2010;72:25–45. doi: 10.1016/B978-0-12-380862-2.00002-3. [DOI] [PubMed] [Google Scholar]
- Selby WS, Janossy G, Mason DY, Jewell DP. Expression of HLA-DR antigens by colonic epithelium in inflammatory bowel disease. Clin Exp Immunol. 1983;53:614–618. [PMC free article] [PubMed] [Google Scholar]
- Sheth S, Shea JC, Bishop MD, Chopra S, Regan MM, Malmberg E, Walker C, Ricci R, Tsui LC, Durie PR, Zielenski J, Freedman SD. Increased prevalence of CFTR mutations and variants and decreased chloride secretion in primary sclerosing cholangitis. Hum Genet. 2003;113:286–292. doi: 10.1007/s00439-003-0963-z. [DOI] [PubMed] [Google Scholar]
- Silverberg MS, Cho JH, Rioux JD, McGovern DP, Wu J, Annese V, Achkar JP, Goyette P, Scott R, Xu W, Barmada MM, Klei L, Daly MJ, Abraham C, Bayless TM, Bossa F, Griffiths AM, Ippoliti AF, Lahaie RG, Latiano A, Pare P, Proctor DD, Regueiro MD, Steinhart AH, Targan SR, Schumm LP, Kistner EO, Lee AT, Gregersen PK, Rotter JI, Brant SR, Taylor KD, Roeder K, Duerr RH. Ulcerative colitis-risk loci on chromosomes 1p36 and 12q15 found by genome-wide association study. Nat Genet. 2009;41:216–220. doi: 10.1038/ng.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes ME, Davis C, Koch G. Categorical data analysis using the SAS system. second edn. SAS Institute Inc.; 2000. [Google Scholar]
- Sugimoto K, Ogawa A, Mizoguchi E, Shimomura Y, Andoh A, Bhan AK, Blumberg RS, Xavier RJ, Mizoguchi A. IL-22 ameliorates intestinal inflammation in a mouse model of ulcerative colitis. J Clin Invest. 2008;118:534–544. doi: 10.1172/JCI33194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szamosi T, Banai J, Lakatos L, Czegledi Z, David G, Zsigmond F, Pandur T, Erdelyi Z, Gemela O, Papp M, Papp J, Lakatos PL. Early azathioprine/biological therapy is associated with decreased risk for first surgery and delays time to surgery but not reoperation in both smokers and nonsmokers with Crohn's disease, while smoking decreases the risk of colectomy in ulcerative colitis. Eur J Gastroenterol Hepatol. 2010;22:872–879. doi: 10.1097/MEG.0b013e32833036d9. [DOI] [PubMed] [Google Scholar]
- Timmer A. Environmental influences on inflammatory bowel disease manifestations. Lessons from epidemiology. Dig Dis. 2003;21:91–104. doi: 10.1159/000073242. [DOI] [PubMed] [Google Scholar]
- Toyoda H, Wang SJ, Yang HY, Redford A, Magalong D, Tyan D, McElree CK, Pressman SR, Shanahan F, Targan SR. Distinct associations of HLA class II genes with inflammatory bowel disease. Gastroenterology. 1993;104:741–748. doi: 10.1016/0016-5085(93)91009-7. [DOI] [PubMed] [Google Scholar]
- Vaishnava S, Behrendt CL, Ismail AS, Eckmann L, Hooper LV. Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proc Natl Acad Sci U S A. 2008;105:20858–20863. doi: 10.1073/pnas.0808723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang MH, Fiocchi C, Zhu X, Duerr RH, Ripke S, Achkar JP. A Novel Approach to Detect Cumulative Genetic Effects and Genetic Interactions in Crohn's Disease. Inflamm Bowel Dis. 2013 doi: 10.1097/MIB.0b013e31828706a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray NR, Goddard ME, Visscher PM. Prediction of individual genetic risk to disease from genome-wide association studies. Genome Research. 2007;17:1520–1528. doi: 10.1101/gr.6665407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Khoury MJ, Botto L, Friedman JM, Flanders WD. Improving the prediction of complex diseases by testing for multiple disease-susceptibility genes. Am J Hum Genet. 2003;72:636–649. doi: 10.1086/367923. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.