Abstract
The extracellular regions of adhesion proteins of the Ig superfamily comprise multiple, tandemly arranged domains. We used directforce measurements to investigate how this modular architecture contributes to the adhesive interactions of the neural cell adhesion molecule (NCAM), a representative of this protein class. The extracellular region of NCAM comprises five immunoglobulin and two fibronectin domains. Previous investigations generated different models for the mechanism of homophilic adhesion that each use different domains. We use force measurements to demonstrate that NCAM binds in two spatially distinct configurations. Igdomain deletion mutants identified the domains responsible for each of the adhesive bonds. The measurements also confirmed the existence of a flexible hinge that alters the orientation of the adhesive complexes and the intermembrane distance. These results suggest that a combination of multiple bound states and internal molecular flexibility allows for sequentially synergistic bond formation and the ability to accommodate differences in intercellular space.
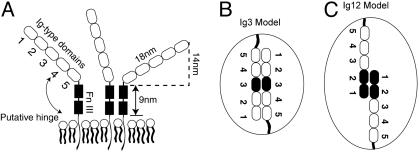
The neural cell adhesion molecule (NCAM) is one of the most studied members of the Ig domain superfamily of cell adhesion molecules (1) and contributes to a wide variety of cellular processes in developing and adult vertebrates (2). NCAM is a membrane anchored glycoprotein that mediates adhesion between cells through homophilic bonds formed between its extracellular domains, which comprise five tandem Ig domains (Ig1–5) and two juxtamembrane fibronectin type III (Fn III) domains (Fig. 1A). In addition, electron microscopy studies revealed a putative flexible, proline-rich hinge between the fifth Ig domain and first fibronectin domain (Fig. 1A) with an average hinge bend angle of 98 ± 45° (3, 4) (Fig. 1A).
Fig. 1.
(A) Schematic of NCAM showing five Ig domains, two fibronectin type III repeats, the membrane anchor, and the predicted hinge at the Ig5-Fn III junction. (B and C) Model showing the isologous Ig3 contacts (B) and the proposed adhesion between antiparallel Ig12 domains (C).
Earlier structural and biochemical studies aimed at identifying the extracellular fragments mediating NCAM binding generated two seemingly contradictory models for homophilic NCAM adhesion (1, 4–9). Bead aggregation studies using different NCAM fragments (5) and domain deletion studies with NCAM-transfected cells support a model in which adhesion requires the isologous interaction of the third Ig domain (Ig3) (6) (model 1; Fig. 1B). In support of this, Ig3 removal abrogates cell adhesion, and the active site of the Ig3 was mapped to a decapeptide sequence that, as a soluble peptide, inhibits cell–cell adhesion (6). This role of Ig3 was confirmed in subsequent studies, which also revealed that the homophilic bond may be augmented by weaker interactions by the flanking Ig domains (Fig. 1B) (5).
By contrast, NMR spectroscopy and surface plasmon resonance (SPR) studies did not detect self-association between recombinant Ig3 domains (7, 8), but SPR measurements rather identified an antiparallel association between the Ig1 and -2 domains (model 2; Fig. 1C). This finding was supported by crystallographic analysis of the Ig1 and -2 domains (7–9). However, mutations that abolish the association of these N-terminal domains in solution failed to abrogate NCAM-mediated adhesion between COS-7 cells (7).
A recently reported structure of Ig1–3 (10) appeared at first glance to resolve the apparent contradictions. This model (model 3; Fig. 2 C–E) proposes that Ig1 and -2 form a cis bond (Fig. 2C), instead of a trans bond. It also postulates the formation of two trans bonds that differ from models 1 and 2: namely, one involving antiparallel association between Ig1 and Ig3 (Fig. 2D) and a second involving the antiparallel association between Ig2 and Ig3 (Fig. 2E). Although this interpretation was supported by peptide inhibition of cell adhesion, two peptides used for this purpose shared substantial-to-complete overlap with those previously suggested to inhibit the trans bonds in Fig. 2 A and B (11).
Fig. 2.
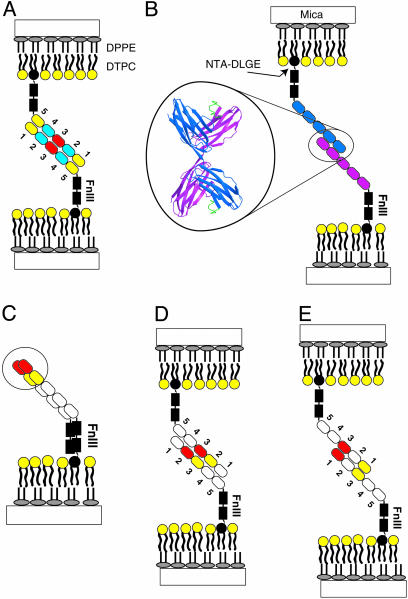
Illustration of hypothetical protein–protein interactions in surface force measurements. The proteins are supported on planar lipid bilayers containing NTA-TRIG-DLGE lipids via engineered C-terminal histidine tags. The supported bilayers consist of a primary monolayer of 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine (DPPE) and a superficial lipid monolayer containing 25 mol % 1,2-ditridecanoyl-sn-glycero-3-phosphocoline (DTPC) and 75 mol % -(9-(2,3-bis(dodecyloxy)propyl)-3,6,9-trioxanonyl-1-oxycarboxylamino)-2-(di(carboxylmethyl)amino)-hexanoic acid (NTA-TRIG-DLGE). Current models predict that the proteins could adhere via Ig3 domains (A)or by antiparallel Ig12 contacts (B). In an alternative model, the outer Ig12 domains form cis bonds (C), and both antiparallel Ig1–Ig3 (D) and antiparallel Ig2–Ig3 (E) form trans bonds
An important distinguishing difference between these models is the membrane separations spanned by the adhesive complexes (Fig. 2). Measurements of the distance-dependence of the attractive and repulsive forces between membrane-bound NCAM extracellular domains could thereby directly test these models. The surface force apparatus (SFA) is widely used to quantify the distance dependence of interaction potentials between materials such as lipid membranes (12, 13). The SFA technique is one of the most sensitive instruments for quantifying weak forces between two surfaces that act over large distances (14). Unlike atomic force microscopy, the absolute distance is determined within ± 0.1 nm in situ by optical interferometry (14, 15) and is more than adequate to distinguish between the proposed NCAM-binding models. Recent SFA studies investigated the structure–structure–function relationships of classic cadherin (16–18), the adhesion proteins CD2 and CD48 (19), and the dynamic properties of tethered receptor–ligand interactions (20).
In this report, we used surface force measurements to assess the molecular basis of the adhesive properties of NCAM ectodomains. The distance dependence of the interactions between the full-length NCAM ectodomains and between NCAM deletion mutants directly demonstrated that the protein forms two distinct adhesive bonds that each involves different Ig domains. These findings simultaneously reconcile several apparently contradictory experimental findings and validate two of the current models as contributing to spatially and molecularly distinct NCAM bonds. They also provide direct evidence that the hinge at the NCAM Ig5–Fn junction can impact the configuration of the protein–protein complex, thereby altering the membrane–membrane distance spanned by that bond.
Materials and Methods
Materials. 1,2-Ditridecanoyl-sn-glycero-3-phosphocholine (DTPC) and 1,2-Dipalmitoyl-sn-glycero-3-phosphoethanolamine (DPPE) were from Avanti Polar Lipids, and 6-[9-[2,3-bis(dodecyloxy)propyl]-3,6,9-trioxanonyl-1-oxycarboxylamino]-2-[di(carboxymethyl)-amino]-hexanoic acid (NTA-TRIG-DLGE) was custom synthesized by Northern Lipids (Vancouver). Hepes was from Sigma. Sodium nitrate, nickel sulfate, 11-mercaptoundecanoic, 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloric acid (EDC), and N-hydroxysulfosuccinimide (NHS) were from Aldrich. The soluble peptides were synthesized at the Protein Science Facility in the Biotechnology Center of the University of Illinois at Urbana–Champaign.
Plasmid Construction. NCAM constructs are derived from chicken NCAM 120 (21). For expression in eukaryotic cells in vitro and subsequent purification, NCAM constructs are fused at their C-terminal ends with a histidine tag (a two-glycine spacer followed by 10 histidines) and inserted into pcDNA3–1-zeo (Invitrogen). The numbering of amino acids is in accordance with Cunningham et al. (22). All PCR-generated DNAs were sequenced.
NCAM Ig1–5 (amino acids 20–482, lacking the two fibronectin domains, or amino acids 1–463 without the signal peptide) was obtained by PCR amplification with a 5′ primer bearing a Kozak sequence (23) and a 3′ primer containing the histidine tag.
Full-length NCAM extracellular domain (amino acids 20–701, or amino acids 1–682 without signal peptide) was reconstituted from NCAM Ig1–5 by removing the last Ig domain (HindIII site) and reinserting this domain with the following two fibronectin domains isolated from NCAM 120.
NCAM ΔIg 3 (deleted amino acids 227–326, or amino acids 208–307 without signal peptide) was constructed by isolating the NCAM EcoRI/HindIII fragment in pUC19. We then introduced by PCR two BamHI sites at each side of the Ig3 domain (amino acids 226 and 327) and re-ligated the BamHI-digested DNA.
NCAM ΔIg1–2 (deleted amino acids 28–219) was made by isolating the first four Ig domains of NCAM in pUC19 (NCAM DNA to HindIII site). We introduced by PCR two SalI sites flanking Ig domains 1 and 2 (amino acids 27 and 220) and re-ligated the SalI digested DNA.
Cell Transformation. Chinese hamster ovary (CHO)-K1 cells are grown in DMEM with 10% FCS with antibiotics. Stable transfections were carried out by using the Lipofectamine 2000 reagent (Invitrogen) as described by the manufacturer. In brief, CHO cells were plated in a 24-well culture plate and grown to 90% confluence in 0.5 ml of culture medium containing 10% serum. Transfections were carried out at 37°C by directly adding 0.1 ml of OPTI-MEM medium (Life Technologies, Grand Island, NY) mixed with 2.5 μl of Lipofectamine and 1 μg of DNA to each 24-well plate. After 24 h, cells were detached in trypsin/EDTA and seeded in 150-mm dishes in normal medium. The following day, 400 μg/ml zeocin (Invitrogen) was added for 2 weeks to select stable transfected clones. Several clones were isolated and grown separately. Their respective expressions for the NCAM proteins were analyzed by rapid histidine tag purification, SDS/PAGE separation, and Western blotting. Detection with NCAM and histidine tag antibodies and molecular size determination were used to assess of the correct protein expression.
Protein Purification and Characterization. The soluble proteins were purified from culture supernatants in a three-step process. The supernatant was affinity purified on a nickel-nitrilotriacetic acid-Sepharose (Qiagen, Valencia, CA) column, and protein was eluted with a buffer of 20 mM Hepes, 100 mM NaCl, and 250 mM imidazole. The protein fraction was concentrated with a Centricon YM-30 filter (Millipore), and further purified by anion exchange chromatography (Amersham Pharmacia) followed by gel filtration (Sephacryl S2000, Amersham Pharmacia). The collected protein fractions were concentrated in 20 mM Hepes buffer at pH 7.
The protein fragments were characterized by CD with a Jasco 720 spectrometer. Solutions of 0.1 mg/ml protein were scanned three times from 200 to 350 nm at a rate of 50 nm/min. The spectra were the average of three scans, and the buffer background was subtracted. The full-length protein was 60% β-sheet, 35% coil, and 5% helix. The ΔIg3 fragment was 65% β-sheet, 35% coil, and 0.44% helix. The ΔIg12 contained 53% β-sheet, 35% coil, and 9% helix. The high β-sheet content indicates proper protein folding. In 8 M urea, there were only small changes in the spectra of each of the proteins. However, the proteins reverted to random coils when the disulfide bonds were reduced with Tris (2-carboxy-ethyl)-phosphine·HCl (TCEP) in 5 Murea. These large structural changes in urea only after reduction are indicative of correct disulfide bond formation.
SPR Measurements. The assembly of NCAM monolayers on supported nickel nitrilotriacetic acid–lipid monolayers was monitored by SPR, using a homebuilt instrument described in refs. 24 and 25. Initially, a baseline resonance was established with the bare membrane, and the histidine-tagged protein (1 μM) was injected into the flow cell. After the time-dependent shift in the resonance angle stabilized (≈1 h), the lipid film was washed with 20 mM Hepes buffer to remove nonspecifically bound protein. The difference between the initial and final resonance angles reflected the amount of NCAM immobilized. As a control, an identical experiment was performed with 20 mM EDTA in the buffer. There was negligible NCAM binding in the presence of the EDTA, indicating that nonspecific binding is minimal.
Equilibrium affinity constants were measured between protein covalently immobilized to an amine-reactive surface and soluble NCAM or NCAM fragments, as described (26–28). The immobilization substrates consisted of a monolayer of carboxy-terminated alkanethiol self-assembled onto a thin gold film evaporated on a glass slide (29). The glass slides were cleaned and coated with a 1-nm chromium adhesion layer, followed by a 38-nm gold film. The gold films were coated with a self-assembled monolayer of 11 mercaptoundecanoic acid (29). The terminal carboxyl groups of the latter monolayer were in turn activated by incubating the slides in an aqueous solution containing 3.8 mg/ml 1-ethyl-3-(3-dimethyl-aminopropyl) carbodiimide hydrochloric acid (EDC) and 6.7 mg/ml N-hydroxysulfosuccinimide (NHS) for 15 min at room temperature. The activated substrates were rinsed and subsequently reacted with protein. The specific binding of soluble protein to the immobilized receptor was then monitored by SPR after the injection of protein solution into the flow cell. After equilibration, the total amount of protein bound was recorded from the shift in the plasmon resonance angle, and the solution in the flow cell was exchanged for one with a higher protein concentration. From the concentration dependence of the adsorbed amount, we determined the affinity constant, as documented (26, 28–30).
To ensure that the results were unaffected by the immobilization chemistry, we performed reciprocal experiments in which the soluble protein in the first experiment was immobilized in a second study, and the immobilized protein in the first experiment was then used as the soluble protein.
Force Measurements. The forces, normalized by the geometric average radius of curvature of the supporting disks, F/R were measured as a function of the intersurface distance with a Mark II SFA (31). The SFA quantifies the normalized force F/R between materials on the surfaces of two crossed hemi-cylinders with radii of curvature R = 2 cm (14, 31). From deflections in the spring supporting the lower disk, the total force can be determined with a resolution of ±1 nN, and the normalized force sensitivity ΔF/R of ± 0.02 mN/m (0.02 mJ/m2) allows measurements of weak interactions such as the van der Waals attraction between membranes (12, 14). The absolute separation between the surfaces is determined by multiple beam interferometry with a resolution of ± 0.1 nm (15). The zero separation, D = 0, is defined at contact between the surfaces of the bilayers supporting the proteins (Fig. 2).
This technique was used to measure the interactions during the approach and separation of the opposed extracellular NCAM domains immobilized on lipid bilayers (see Fig. 2A). The supporting lipid layers were prepared by Langmuir–Blodgett deposition. A lipid–chloroform solution containing 75 mol% 6-(9-(2,3-bis(dodecyloxy)propyl)-3,6,9-trioxanonyl-1-oxycarboxylamino)-2-(di(carboxylmethyl)amino)-hexanoic acid (NTA-TRIG-DLGE) and 25 mol % 1,2-ditridecanoyl-sn-glycero-3-phosphocoline (DTPC) was spread at the air–water interface of a Langmuir trough (Nima Coventry, England). The lipid monolayer was compressed to a surface pressure of 36 mN/m (≈65 Å2 per lipid), and then transferred at constant pressure and 25°C onto a hydrophobic 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine (DPPE) monolayer (43 Å2 per lipid) supported on a mica substrate (Fig. 2A) (16, 19). The bilayers were then incubated for 1 h in a solution of 20 mM Hepes, 150 mM NaNO3,20 μMNiSO4, and 1 μM NCAM. The NCAM adsorbed to the NTA head group via its polyhistidine tag (Fig. 2). The samples were then washed with buffer to remove nonspecifically bound protein, and mounted in the surface force apparatus.
Results and Discussion
Surface force measurements were carried out with NCAM ectodomains selectively bound and oriented on supported lipid bilayers via engineered C-terminal histidine tags (Fig. 2). The forces were measured between NCAM-decorated membranes as a function of the distance, D, between the surfaces of the lipids supporting the proteins (Fig. 2) (12, 17, 20, 32).
The proteins investigated include the full-length ectodomain, domains Ig1–5 only (ΔFn), NCAM lacking domain 3 (ΔIg3), and NCAM lacking domains 1 and 2 (ΔIg12). Each contained a C-terminal decahistidine tag for immobilization. These fragments enabled us to explicitly determine the roles of isologous Ig3 interactions, Ig12 interactions, and the putative hinge between Ig5 and the first Fn III domain in homophilic NCAM adhesion.
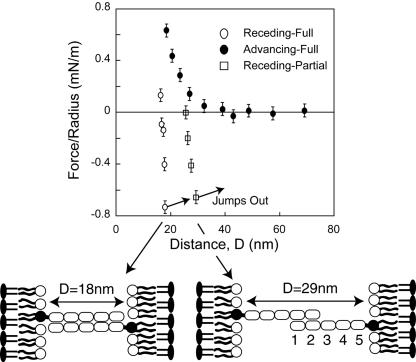
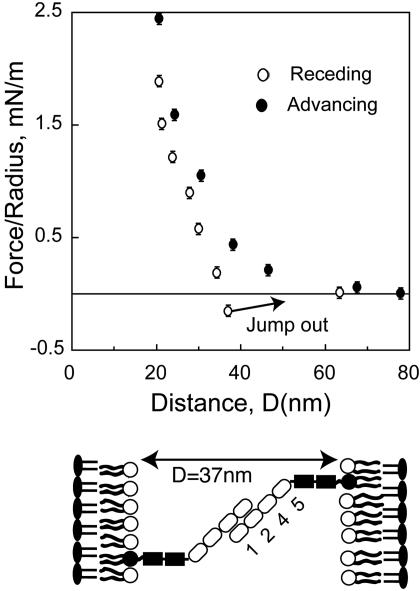
The distance dependence of the force, normalized by the radius, F/R between immobilized monolayers of the Ig1–5(ΔFn) fragment (Fig. 3) shows that the Ig1–5 fragments form two distinct, bound states at two membrane separations. The onset of steric repulsion between the proteins during approach occurs at D < 34 ± 1 nm. From electron micrographs, the end-to-end distance of the Ig1–5 segment of NCAM is 18 nm (3). Thus, the range of repulsion between the two protein monolayers agreed with the expected onset of steric repulsion at ≈37 ± 1 nm between opposing, end-on oriented Ig1–5 proteins. It also indicates that the protein is relatively rigid, as any significant bend would reduce the steric thickness of the Ig1–5 layer below 18 nm.
Fig. 3.
Normalized force versus distance profiles between immobilized Ig1–5 (ΔFn) fragments. Filled circles show forces measured on approach. Open symbols show the force profile measured upon separation. Separation from D <18 nm (open circles) results in adhesion at 18 ± 0.5 nm, consistent with adhesion between fully interdigitated NCAMs (cartoon). Upon separation from 30 nm > D > 20 nm (open squares), the proteins adhere at 29 ± 0.5 nm.
Upon separation, protein adhesion caused the receding curve to drop below the advancing profile. At the minimum in the normalized force profile at 18 ± 0.5 nm the adhesive bonds ruptured, and the surfaces jumped out of contact (Fig. 3). The adhesion at 18 nm agrees with a geometric model in which opposing Ig1–5 domains adhere in a fully overlapping, antiparallel configuration. This is consistent with the formation of isologous bonds between antiparallel Ig3 domains (Fig. 2A). Within the limits of error, this distance is also close to the span of the putative complex formed by antiparallel Ig2–Ig3 contacts (Fig. 2D).
The latter finding supports the model of Ranheim et al. (5), which postulates that the Ig3 domain primarily mediates NCAM adhesion (model 1, Fig. 2A). However, we also measured a second, slightly weaker adhesive interaction at 29 ± 0.5 nm (Fig. 3). We measured the latter bound state by controlling the membrane separation, D, and hence the degree of protein overlap, before separating the protein layers. The position of this second adhesive bond also agrees quantitatively with model 2 (Fig. 2B), in which the outer Ig12 segments from opposing proteins adhere to form a partially inter-digitated, antiparallel complex (Fig. 3) (8, 9, 11, 33). This result supports the role of the Ig1–Ig2 domains in trans binding, and contradicts the view that these domains only form cis bonds (Fig. 2C). No additional bonds were detected, and there was no adhesion at the distance of antiparallel Ig1–Ig3 contact (Fig. 2E).
The magnitude of both adhesive states depended on the time the surfaces were in contact. This is attributed to lateral protein diffusion, which increases the number of intersurface bonds formed (32). The adhesion, measured within 5 min of contact, was -0.8 ± 0.2 mN/m and -0.6 ± 0.2 mN/m for the inner and outer bonds, respectively. However, the adhesion increased to its maximum value within 20 min in both cases. After 20 min, the adhesion was -1.1 ± 0.3 mN/m at 18 nm and -0.7 ± 0.1 mN/m at 29 nm.
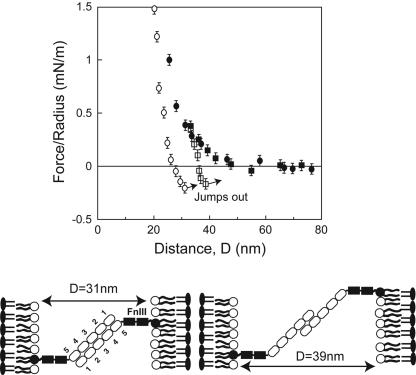
The positions of the two binding interactions and the range of the steric repulsion between full-length NCAM ectodomains (Fig. 4), which contain both Ig1–5 and two fibronectin domains, differed from the ΔFn (Ig1–5) interaction potential. The onset of the steric repulsion was at ≈48 nm, which is short of the 54 nm expected if the seven domains formed a straight rod (see Fig. 1 B and C). Also, the steep, repulsive barrier is farther out at 22 nm. The range of the latter steric barrier is also well below the calculated 27 nm expected for steric contact between rigid NCAM rods and the opposed bilayer. Both of the latter features, together with the ΔFn data, support the hypothesis that NCAM has a flexible hinge C-terminal to Ig5 that tilts the protein relative to the surface. The steric thickness (23 nm = 47 ± 1 nm/2) of the NCAM layer, together with the ≈18 nm and 9 nm lengths of the Ig1–5 (refs. 4 and 6–8, and Protein Data Bank ID 1LWR) and Fn III (ref. 34 and Protein Data Bank ID 1LWR) segments, respectively, suggest that the unstressed protein is bent at the Ig5–Fn junction with an angle of ≈145° [=90° + asin((23 nm-9 nm)/18 nm)] (see Fig. 1A). The most extended proteins determine this value because they define the initial onset of steric repulsion between the two layers.
Fig. 4.
Normalized force versus distance profiles between full-length NCAM extracellular domains. On approach (filled circles), the surfaces repel at D <48 nm, and a steep, steric repulsion occurs at D < 22 nm. On separation from D < 31 nm (open circles), adhesive failure occurs at 31 ± 0.5 nm. At partial protein overlap (48 nm > D > 32 nm, open squares), the proteins form a second adhesive bond at 39 ± 0.5 nm. The cartoon illustrates the proposed models for the protein alignments at each of the two minima, if the hinge angle for both alignments at adhesive failure is 135°.
Upon separation, the full-length ectodomains adhered at 31 ± 0.5 nm and at 39 ± 0.5 nm (Fig. 4). In addition, unlike the previous measurements (Fig. 3), the receding force curves exhibit hysteresis over a larger distance, and the slope is slightly shallower. This feature suggests hinge flexibility. Based on the 18- and 9-nm lengths of the Ig1–5 and two Fn III segments, respectively (refs. 4, 6–8, and 34, and Protein Data Bank ID 1LWR), the data in Figs. 3 and 4, and the distances spanned by the adhesive complexes, the estimated hinge angle at the points of bond failure at 31 nm and at 39 nm is 135 ± 3°, in both cases. This agrees qualitatively with the maximum hinge angle seen in the electron microscopy studies (4, 6). It suggests that, at this point, the force required to further open the hinge exceeds the bond rupture force, and the protein–protein bonds fail. The range and the magnitude of the adhesion measured in the experiments with the full-length ectodomain and with the ΔFn variant were reproducible. Consecutive force curves were super-imposable, indicating that the lipid surface was not damaged and that the protein density remained unchanged. Thus, adhesion does not result in lipid pull-out (20, 32, 35).
The strength of the two adhesive bonds between the full ectodomains is lower than between the ΔFn monolayers. This is not unexpected if the hinge increases the protein's flexibility, as suggested by the range of hinge angles (3, 4). This would increase its configurational entropy, thereby increasing the entropic repulsion between the protein monolayers and reducing the net attraction. Similar behavior was observed with streptavidin and polymer-tethered biotin. The adhesion measured with flexibly tethered biotin was three to four times lower than that measured with surface anchored ligand at comparable surface densities (20, 32).
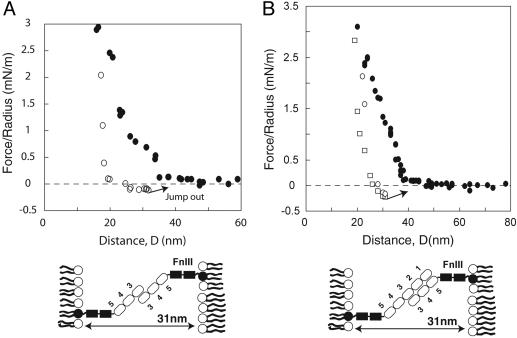
Measurements with domain deletions validated the models in Figs. 3 and 4, and identified the domains primarily responsible for the two observed bound states. The inner adhesive bond was absent in force profiles between NCAM ectodomains lacking the Ig3 domain (ΔIg3), but the fragments adhered at 37 ± 0.5 nm (Fig. 5). This is expected for antiparallel Ig12 contacts (model 2, Fig. 5). These data are, however, incompatible with model 3, in which the two putative trans bonds require double-reciprocal binding between antiparallel Ig1 and Ig3 (Fig. 2D) and antiparallel Ig2 and Ig3 (Fig. 2E). Our data show that the proteins adhere despite the absence of Ig3. As with the full-length extracellular NCAM, the calculated hinge angle is 136 ± 3°, based on the point of failure at 37 ± 0.5 nm and on the protein dimensions.
Fig. 5.
Normalized force versus distance profiles between ΔIg3 NCAM ectodomains. Removing Ig3 abrogates the inner bond, but a single bound state at 37 ± 0.5 nm survives. The inward shift of the minimum to 37 nm is as expected, given the length of the two deleted Ig domains (one from each side) and assuming a 135 ± 3° bend in the hinge, as illustrated.
Conversely, the ΔIg12 fragments lacked the outer adhesive minimum, but retained the adhesive bond at 31 ± 0.5 nm (Fig. 6). This supports the formation of antiparallel, isologous Ig3 bonds (Fig. 2A), and confirms that Ig12 forms the outer, trans adhesive bond. Adhesion between ΔIg12 fragments was weak at -0.15 ± 0.07 (Fig. 6A). However, ΔIg12 similarly bound the NCAM at 31 ± 0.5 nm, but with an adhesion of -0.31 ± 0.05 mN/m (Fig. 6B).
Fig. 6.
Normalized force versus the distance between homotypic ΔIg12 fragments (A) and between ΔIg12 and the full NCAM ectodomain (B). The filled circles were measured upon approach, and open circles indicate detachment. The single adhesive bond formed in both cases is at 31 ± 1 nm (arrows).
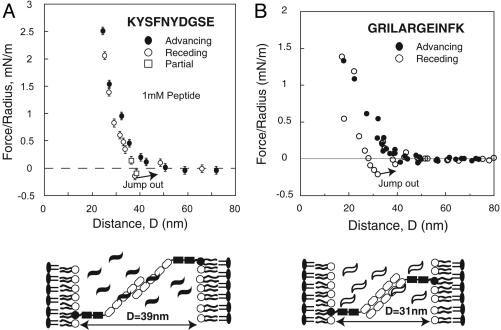
Force measurements between the full ectodomains in the presence of a soluble decapeptide KYSFNYDGSE further tested the involvement of Ig3 (6). This sequence matches an endogonous sequence in Ig3, and was shown to inhibit NCAM-mediated cell adhesion (6). Force measurements with the full-length NCAM ectodomains in the presence of 1 mM decapeptide showed that the peptide blocked adhesion at 31 nm, but had no effect on the outer bond at 39 nm (Fig. 7A). This interference further supports Ig3 involvement in the formation of the inner, slightly stronger homophilic NCAM adhesive bond.
Fig. 7.
Normalized force versus distance profiles between full-length NCAM ectodomains in the presence of soluble peptides. Filled circles indicate the profile measured during approach, and open symbols were measured during separation. (A) Measurements with 1 mM of the decapeptide KYSFNYDGSE. The proteins only adhered at 39 ± 0.5 nm. The cartoon illustrates the proposed NCAM alignment at 39 nm. (B) The force curves measured with the peptide GRILAR-GEINFK. The proteins adhered at 31 ± 0.5 nm, and adhesion at 39 nm was abolished. The cartoon illustrates the proposed NCAM alignment at D = 31 nm.
The binding surface and specificity of the decapeptide is unknown, so we carried out a second set of experiments with a different peptide GRILARGEINFK, which inhibits the Ig12 interaction (11). In contrast, this peptide had no effect on the inner bond, but abrogated the outer putative Ig12 bond (Fig. 7B).
The finding that the decapeptide inhibits Ig3-dependent adhesion contrasts with NMR studies where the peptide did not bind soluble Ig3 domains. However, the latter NMR study also did not detect Ig3 association (7). In this work, the difference in adhesive behavior between the full-length and ΔIg3 variants clearly demonstrates that Ig3 is required for the inner adhesive bond between fully overlapping Ig1–5 domains. Whereas these force measurements were carried out with the full Ig1–5 segments, the NMR studies were performed with recombinant Ig3 domains (7). The N-terminal Ig3 domain boundary in our ΔIg3 and ΔIg12 variants differs from that of Atkins et al. (7), which had 14 additional N-terminal residues. Nevertheless, ΔIg12, which has a shorter Ig3 sequence is functional, adheres at the predicted distance for isologous Ig3 contacts, and CD shows that it is folded. Although we don't know the origin of this difference in behavior (7, 8), it is important to point out that our findings agree with nearly all of the published literature on NCAM adhesion.
The model suggested by the force data is supported by equilibrium binding measurements, which are summarized in Table 1. The affinity constant Ka between full NCAM ectodomains is 4 × 107 M-1. The models in Figs. 2 A and B, 3, and 4 predict that the NCAM ectodomain should bind both ΔIg12 and ΔIg3 fragments. These proteins do bind with the affinities given in Table 1. Similarly, previous work (5, 6) and our force measurements predict that ΔIg12 fragments will bind homophylically, whereas the recent model of Soroka et al. (10) predicts no homophylic binding. This fragment binds homophylically with an affinity of 1.7 ± 0.3 × 107 M-1 (Table 1). Likewise, Fig. 1A and our force data predict that ΔIg12 would bind ΔIg3 weakly or not at all, but Fig. 2D predicts binding. Both SPR (Table 1) and force data (not shown) show that they do not adhere.
Table 1. Determined affinity constants (× 10-7 M-1).
| Protein | NCAM | ΔIg3 | ΔIg12 |
|---|---|---|---|
| NCAM | 4 ± 1 | 1.1 ± 0.3 | 3.4 ± 0.4 |
| ΔIg3 | 1.0 ± 0.6 | - | |
| ΔIg12 | 1.7 ± 0.3 |
-, not detected.
The adhesive strengths of the fragments differ from that of the full ectodomain. There are two possible reasons for this. First is the inherent risk in cutting and pasting different protein regions, which, despite careful design, could impair function. Although the CD measurements indicate that the proteins are folded, subtle changes caused by structural alterations can compromise function. Alternatively, this could result from lost secondary interactions by flanking domains in the deletion mutants. This could explain, for example, the weaker homotypic ΔIg12 binding relative to adhesion between NCAM, or between ΔIg12 and NCAM. Nevertheless, these differences in adhesive strength in no way detract from the findings that the fragments are functional, and they adhere at the distances predicted by the model in Figs. 3 and 4.
In conclusion, these direct force measurements show that NCAM forms either of two adhesive complexes, which require different Ig domains and span different membrane separations. Approaches used in previous studies of NCAM binding could detect only one or the other complex, thereby creating an apparent contradiction. However, with the ability to measure the absolute membrane separations at which opposed NCAM ectodomains adhere we directly demonstrated that NCAM adheres both in a fully, antiparallel alignment requiring Ig3, and in a separate anti-parallel interaction between the Ig12 domains. By contrast, methods such as x-ray crystallography reflect a single configuration and are thus less well suited to describe the complexities of the type of molecular interaction exhibited by NCAM.
These results contribute to the increasing body of evidence that multidomain adhesion molecules can use multiple domains to adhere at different relative alignments (16–18). As found for C-cadherin (16–18), the strongest homophilic NCAM bond occurs between extensively overlapping ectodomains. Although Ig3 is required for cell adhesion (6), the biological significance of the Ig12-mediated binding has yet to be established. Nevertheless, it is interesting to speculate that cell–cell adhesion occurs through a sequence of protein interactions in which the Ig12 binding provides an initial, weaker, and perhaps unstable “capture” mode at a relatively long membrane–membrane distance. This, in turn, could lead to the stronger, more stable and intimate interaction afforded by the fully overlapping configuration. In this vein, it is also reasonable to view the flexibility afforded by the NCAM hinge as a mechanism that further reduces the constraints of membrane-membrane distance in the formation of adhesions between cells. Conversely, it could also allow adherent cells to adjust their proximity to serve other cell surface functions.
Acknowledgments
We thank Nancy Harmon for her excellent technical assistance. This work was supported by National Institutes of Health Grant RO1 GM63536.
Abbreviations: NCAM, neural cell adhesion molecule; Fn, fibronectin; SPR, surface plasmon resonance; SFA, surface force apparatus.
References
- 1.Walsh, F. & Doherty, P. (1997) Annu. Rev. Cell. Biol. 13, 425-456. [DOI] [PubMed] [Google Scholar]
- 2.Cremer, H., Lange, R., Christof, A., Plomann, M., Vopper, G., Roes, J., Brown, R., Baldwin, S., Kraemer, P. & Scheff, S. (1994) Nature 367, 455-459. [DOI] [PubMed] [Google Scholar]
- 3.Hall, A. & Rutishauser, U. (1987) J. Cell Biol. 104, 1579-1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Becker, J. W., Erickson, H. P., Hoffmann, S., Cunningham, B. A. & Edelman, G. M. (1989) Proc. Natl. Acad. Sci. USA 86, 1088-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ranheim, T. S., Edelman, G. M. & Cunningham, B. A. (1996) Proc. Natl. Acad. Sci. USA 93, 4071-4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rao, Y., Wu, X.-F., Gariepy, J., Rutishauser, U. & Siu, C.-H. (1992) J. Cell Biol. 118, 937-949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Atkins, A. R., Chung, J., Songpon, D., Little, E., Edelman, G. M., Wright, P. E., Cunningham, B. A. & Dyson, H. J. (2001) J. Mol. Biol. 311, 161-172. [DOI] [PubMed] [Google Scholar]
- 8.Kiselyov, V., Berezin, V., Maar, T. E., Soroka, V., Edvardsen, K., Schousboe, A. & Bock, E. (1997) J. Biol. Chem. 272, 10125-10134. [DOI] [PubMed] [Google Scholar]
- 9.Kasper, C., Rasmussen, H., Kastrup, J. S., Ikemizu, S., Jones, R. Y., Berezin, V., Bock, E. & Larsen, I. K. (2000) Nat. Struct. Biol. 7, 389-393. [DOI] [PubMed] [Google Scholar]
- 10.Soroka, V., Kolkova, K., Kastrup, J. S., Diederichs, K., Breed, J., Kiselyov, V. V., Poulsen, F. M., Poulsen, F. M., Larsen, I. K., Welte, W., et al. (2003) Structure (London) 10, 1291-1301. [DOI] [PubMed] [Google Scholar]
- 11.Soroka, V., Kiryushko, D., Novitskaya, V., Ronn, C. B., Poulson, F. M., Holm, A., Bock, E. & Berezin, V. (2002) J. Biol. Chem. 277, 24676-24683. [DOI] [PubMed] [Google Scholar]
- 12.Marra, J. & Israelachvili, J. (1985) Biochemistry 24, 4608-4618. [DOI] [PubMed] [Google Scholar]
- 13.Israelachvili, J. (1992) Surface Sci. Rep. 14, 110-159. [Google Scholar]
- 14.Leckband, D. & Israelachvili, J. N. (2001) Q. Rev. Biophys. 34, 105-267. [DOI] [PubMed] [Google Scholar]
- 15.Israelachvili, J. (1973) J. Coll. Int. Sci. 44, 259-272. [Google Scholar]
- 16.Sivasankar, S., Brieher, W., Lavrik, N., Gumbiner, B. & Leckband, D. (1999) Proc. Natl. Acad. Sci. USA 96, 11820-11824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sivasankar, S., Gumbiner, B. M. & Leckband, D. (2001) Biophys. J. 80, 1758-1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu, B., Chappuis-Flament, S., Wong, E., Jensen, I., Gumbiner, B. M. & Leckband, D. E. (2003) Biophys. J. 41, 12163-12170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu, B., Davies, E. A., van der Merwe, A. & Leckband, D. (2002) Biochemistry 42, 12163-12170. [DOI] [PubMed] [Google Scholar]
- 20.Wong, J. Y., Kuhl, T. L., Israelachvili, J. N., Mullah, N. & Zalipsky, S. (1997) Science 275, 820-822. [DOI] [PubMed] [Google Scholar]
- 21.Fujimoto, I., Bruses, J. L. & Rutishauser, U. (2001) J. Biol. Chem. 276, 31745-31751. [DOI] [PubMed] [Google Scholar]
- 22.Cunningham, B. A., Hemperly, J. J., Murray, B. A., Prediger, E. A., Brackenbury, R. & Edelman, G. M. (1987) Science 236, 799-806. [DOI] [PubMed] [Google Scholar]
- 23.Kozak, M. (1987) Nucleic Acids Res. 15, 8125-8148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yeung, C. & Leckband, D. (1997) Langmuir 13, 6746-6754. [Google Scholar]
- 25.Lavrik, N. & Leckband, D. (2000) Langmuir 16, 1842-1851. [Google Scholar]
- 26.Yeung, C., Purves, T., Kloss, A. A., Kuhl, T. L., Sligar, S. & Leckband, D. (1999) Langmuir 15, 6829-6836. [Google Scholar]
- 27.Efremova, N. V., Bondurant, B., O'Brien, D. E. & Leckband, D. (2000) Biochemistry 39, 3441-3456. [DOI] [PubMed] [Google Scholar]
- 28.Johnson, C. P., Jensen, I. E., Prakasam, A., Vijayendran, R. & Leckband, D. (2003) Bioconjugate Chem. 14, 974-978. [DOI] [PubMed] [Google Scholar]
- 29.Vijayendran, R. A., Motsegood, K. M., Beebe, D. J. & Leckband, D. E. (2003) Langmuir 19, 1824-1828. [Google Scholar]
- 30.Vijayendran, R., Hammer, D. & Leckband, D. (1998) J. Chem. Phys. 108, 1162-1169. [Google Scholar]
- 31.Israelachvili, J. N. & Adams, G. E. (1978) J. Chem. Soc. Faraday Trans. I 75, 975-1001. [Google Scholar]
- 32.Leckband, D., Schmitt, F.-J., Israelachvili, J. & Knoll, W. (1994) Biochemistry 33, 4611-4624. [DOI] [PubMed] [Google Scholar]
- 33.Jenson, P., Soroka, V., Thompson, N. K., Ralets, I., Berezin, V., Bock, E. & Poulsen, F. M. (1999) Nat. Struct. Biol. 6, 486-493. [DOI] [PubMed] [Google Scholar]
- 34.Leahy, D. J., Hendrickson, W. A., Aukhil, I. & Erickson, H. P. (1992) Science 258, 987-991. [DOI] [PubMed] [Google Scholar]
- 35.Leckband, D., Müller, W., Schmitt, F.-J. & Ringsdorf, H. (1995) Biophys. J. 69, 1162-1169. [DOI] [PMC free article] [PubMed] [Google Scholar]