Abstract
Caenorhabditis elegans mtf-1 encodes matefin, which has a predicted SUN domain, a coiled-coil region, an anti-erbB-2 IgG domain, and two hydrophobic regions. We show that matefin is a nuclear membrane protein that colocalizes in vivo with Ce-lamin, the single nuclear lamin protein in C. elegans, and binds Ce-lamin in vitro but does not require Ce-lamin for its localization. Matefin is detected in all embryonic cells until midembryogenesis and thereafter only in germ-line cells. Embryonic matefin is maternally deposited, and matefin is the first nuclear membrane protein known to have germ line-restricted expression. Animals homozygous for an mtf-1 deletion allele show that matefin is essential for germ line maturation and survival. However, matefin is also required for embryogenesis because mtf-1 (RNAi) embryos die around the ≈300-cell stage with defects in nuclear structure, DNA content, and chromatin morphology. Down-regulating matefin in mes-3 animals only slightly enhances embryonic lethality, and elimination of UNC-84, the only other SUN-domain gene in C. elegans, has no affect on mtf-1 (RNAi) animals. Thus, mtf-1 mediates a previously uncharacterized pathway(s) required for embryogenesis as well as germ line proliferation or survival.
Lamins are nuclear intermediate filament proteins found in metazoan cells at the nuclear periphery and in the nucleoplasm (1). Lamins interact with most known inner nuclear membrane proteins as well as with several nucleoplasmic proteins (2). Nuclear architecture, cell cycle progression, DNA replication, and RNA transcription and splicing all depend on lamins (3, 4). Consistent with such roles, many of these lamin-binding proteins also bind transcription repressors and chromatin proteins.
To understand nuclear lamins and lamin-associated protein functions in vivo we turned to Caenorhabditis elegans, studying its single lamin protein, Ce-lamin (5), and three inner nuclear membrane proteins, Ce-emerin, Ce-MAN1 (5, 6), and UNC-84 (7). UNC-84 contains an ≈120-residue SUN (Sad1p-UNC-84 homology) domain (8) with an unknown function. The SUN domain is also found in four human proteins, two of which localize at the nuclear envelope (9). UNC-84 is expressed in most C. elegans cells, and it depends on Ce-lamin for its nuclear-envelope localization (7). However, mutations in unc-84 cause nuclear migration or nuclear anchoring defects in only a subset of cells, leading to uncoordinated movement (8). At least two nuclear-envelope proteins, UNC-83 and ANC-1, require the SUN domain of UNC-84 for their nuclear-envelope localization and ability to regulate nuclear position (10). To explain their nuclear-envelope anchoring, a “bridging model” was proposed in which the transmembrane domains of UNC-83 and ANC-1 cross the outer nuclear membrane (ONM), and their luminal domains interact with the luminal domain of UNC-84 embedded in the inner nuclear membrane (7, 10).
Based on the hypothesis that the SUN domain defined a new family of nuclear-envelope proteins, we searched the C. elegans genome for other SUN domains, and found one: an ORF on chromosome V, designated F57B1.2. We report that this gene (mtf-1) encodes matefin, a nuclear membrane protein that binds Ce-lamin and has essential embryonic and germ line-specific functions.
Materials and Methods
C. elegans Strains. C. elegans strains N2, PD4793, ced-3(n717), and unc-84(n369) were obtained from the C. elegans genetic center. Strain PD4793 has an integrated multiconstruct array comprising myo-2::GFP, pes-10::GFP, and gut::GFP, which lead to GFP expression in four-cell embryos, pharyngeal muscle, and gut (11). Strain VC292 containing the gk199 deletion allele of mtf-1 [+/nT1 IV; mtf-1(gk99)/nT1 V] was prepared by the C. elegans Gene Knockout Consortium by using TMP/UV as mutagen. This strain was outcrossed once before balancing. The gk199 allele on chromosome V was outcrossed again with strain PD4793 (11). mes-2 (ss186), mes-3 (ss313), and mes-6 (ss553) were kindly provided by S. Strome (Indiana University, Bloomington). The AZ212 strain expressing H2B::GFP was kindly provided by J. Austin (University of Chicago, Chicago).
Antibodies, Indirect Immunofluorescence Staining of C. elegans, and Time-Lapse Microscopy. C. elegans embryos, larva, and adults were fixed and prepared for indirect immunofluorescence exactly as described (12, 13). Covance (Princeton) produced all antisera against matefin. Rat 3663 was immunized with KLH-conjugated peptide CRHTISPQFSNRHSP (matefin residues 3–16 plus one Cys for conjugation). Rat 3665 and mouse 3666 were immunized with KLH-conjugated peptide PMTDNGTESKLESAC (matefin residues 451–465 plus one Cys for conjugation). Rabbit anti-PGL-1, Rat anti-Mes-2, rabbit anti-Mes-6, and rat anti-Mes-3 were kindly provided by S. Strome. Immunogold transmission electron microscopy (TEM) with matefin serum 3663 (diluted 1:30) and thin-section TEM were performed as described (14). For time-lapse microscopy, F1 progeny of hermaphrodites fed with mtf-1 (RNAi) (see below) were mounted on an agar pad with egg salt buffer, sealed with vacuum grease, and viewed by using a Zeiss Axioplan II microscope equipped for fluorescence. An Axiocam CCD camera and the AxioVision Image Analysis package were used to record time-lapse data every 2 min.
RNA Interference (RNAi) Experiments. mtf-1 cDNA nucleotides 311–731 (exon 3) were subcloned into feeding vector L4440 (15) and used for RNAi feeding as described (6). Control animals were fed with bacteria carrying an empty L4440 construct. Worms were either examined live to determine viability or fixed and prepared for indirect immunofluorescence staining.
Synthesis of 35S-Met-Labeled Proteins and Blot Overlay Assays. cDNAs encoding Ce-emerin (residues 1–130; lacking only the short C-terminal transmembrane domain), Ce-lamin globular domain (residues 440–566), Ce-mtf-1-N (residues 1–111), and full-length Ce-lamin were cloned into pET20b expression vector. cDNAs encoding Ce-mtf-1-C (residues 129–473) and full-length matefin were cloned into the pET28a expression vector. Ce-MAN1 was cloned into the pET15b expression vector. Mtf-1 cDNAs yk703g5 and yk729e11 were kindly provided by Yuji Kohara (Genome Biology Laboratory, Mishima, Japan). Constructs were transformed into Escherichia coli BL21 (DE3) and proteins were purified via Ni-NTA agarose (Qiagen, Valencia, CA). Equal amounts of protein (determined by a Bradford assay) were resolved by 15% SDS/PAGE and transferred to nitrocellulose membrane. Blot overlay assays were done essentially as described (13).
Results
Matefin Is a Previously Uncharacterized C. elegans Nuclear Envelope Protein with a SUN Domain and Two Putative Transmembrane Domains. Homologs for UNC-84 are known to exist in the nuclear envelope of a wide variety of species and are thought to control the position of the nucleus in the cell (10). UNC-84 contains an evolutionarily conserved SUN domain, which we hypothesized might define a new family of nuclear-envelope proteins. We therefore searched for new SUN-domain genes in C. elegans and found one ORF on chromosome V (F57B1.2). We named this gene mtf-1 and its product matefin (“envelope” in Hebrew), based on its localization. Matefin was predicted to have a 20-residue hydrophobic region, a 36-residue coiled-coil region, a second 21-residue hydrophobic region, one SUN domain, and a 101-residue “anti-erbB-2 Ig” domain overlapping both the second hydrophobic region and the SUN domain (Fig. 1A). Hydropathy programs suggested that the first and possibly both hydrophobic regions were transmembrane domains. Matefin is conserved (57% identical) in the nematode Caenorhabditis briggsae. The SUN domain of matefin is 31%, 28%, and 29% identical and 44%, 37%, and 39% similar to the SUN domains in human UNC-84, UNC-84a, and SUN2, respectively (8, 9). The anti-erbB-2 Ig domain is 29% identical and 45% similar to the human anti-erbB-2 Ig domain (16). However, outside these domains matefin appears to be unique, with no detectable homology to vertebrate, Drosophila, yeast, or plant genes.
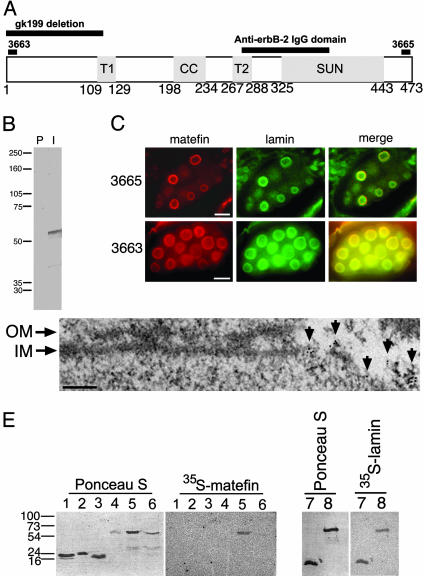
Fig. 1.
Matefin in C. elegans and its interaction with Ce-lamin. (A) A schematic diagram of full-length matefin, drawn approximately to scale. Bars on top indicate the positions of the gk199 deletion, the peptides used to prepare serum 3663 and 3665 antibodies against matefin, and the anti-erbB-2 IgG domain. T1 and T2, putative transmembrane regions 1 and 2, respectively; CC, coiled-coil domain; SUN, SUN domain. (B) Western blot of crude protein extract of mixed stage C. elegans animals probed with preimmune (P) or immune (I) serum 3665. (C) Indirect immunofluorescence double-staining of embryos by using antibodies specific for Ce-lamin plus either the N terminus (serum 3663, Lower) or C terminus (serum 3665, Upper) of endogenous matefin. Please note that although both embryonic and adult nuclei are stained with Ce-lamin, only the embryonic nuclei are stained with matefin (Upper). (Bars, 10 μm.) (D) Immunogold TEM localization of the matefin N terminus (serum 3663) on peripheral chromatin in detergent-extracted nuclei. Fragments of remaining nuclear outer membrane (OM) and inner membrane (IM) are indicated. Arrows indicate the position of the 6-nm gold labeling. (Bar, 100 nm.) (E) Equal amounts of protein, as determined by Bradford assay, were resolved by SDS/PAGE, transferred to nitrocellulose, stained with Ponceau S, and probed with either 35S-matefin or 35S-Ce-lamin. Please note that Ponceau S staining does not always correspond to Bradford assay results. Numbered gel lanes contain the following polypeptides: 1, Ce-emerin (residues 1–130); 2, the C-terminal globular domain of Ce-lamin (residues 440–566); 3, Ce-MAN1 (residues 400–500); 4, BSA; 5, Ce-lamin; 6, matefin; 7, matefin N-terminal domain (residues 1–111); 8, matefin C-terminal domain (residues 129–473).
We generated rat polyclonal sera against an N-terminal peptide (serum 3663) and a C-terminal peptide (serum 3665; Fig. 1 A; also see Materials and Methods). Western blot analysis of whole worm lysate revealed a major band of 55 kDa, the predicted mass of matefin (Fig. 1B). By indirect immunofluorescence, both antisera specifically stained the nuclear envelope of C. elegans embryos (Fig. 1C). Thin-section immunogold TEM analysis of preembedded stained embryos further showed that the N-terminal matefin epitope (serum 3663) remained associated with peripheral chromatin after cells were extracted with Triton X-100 (Fig. 1D), consistent with the behavior of nuclear membrane proteins that bind lamins (13). Double immunofluorescent labeling of C. elegans embryos with anti-lamin and anti-matefin antibodies showed that endogenous Ce-lamin and matefin colocalized at the nuclear envelope (Fig. 1C).
To determine whether matefin bound directly to Ce-lamin, we used 35S-matefin to probe various recombinant proteins immobilized on blots (Fig. 1E Left, lanes 1–6). There was a strong signal for 35S-matefin binding to the complete Ce-lamin, but not to its isolated tail domain. Matefin also bound weakly to itself (Fig. 1E, lanes 5 and 6). The converse experiment with 35S-Celamin confirmed direct binding to matefin, and further showed that Ce-lamin binds two regions of matefin: residues 1–111 (up to the first putative transmembrane domain) and residues 129–473, comprising all residues after this first transmembrane domain (Fig. 1E, lanes 7 and 8). Pull-down experiments verified the blot overlay results (data not shown). We concluded that matefin is a lamin-binding protein of the inner nuclear membrane. Furthermore, Ce-lamin binding to both “halves” of matefin suggests (but does not prove) that matefin is anchored by two transmembrane domains, with its N and C termini both available to bind Ce-lamin in the nucleoplasm.
To our surprise, RNAi-mediated down-regulation of Ce-lamin had no obvious affect on the nuclear-envelope localization of matefin (Fig. 2A). Matefin is thus the first known C. elegans nuclear membrane protein that remains nuclear-envelope-localized in the absence of Ce-lamin. Matefin localization was also normal in unc-84(n369) null embryos and gonads (Figs. 2C and 6, which is published as supporting information on the PNAS web site). Similarly, RNAi-mediated down-regulation of matefin had no effect on the embryonic nuclear-envelope localization of Ce-lamin, Ce-emerin, Ce-MAN1, FG-repeat nucleoporins (Fig. 2B), or UNC-84 (data not shown).
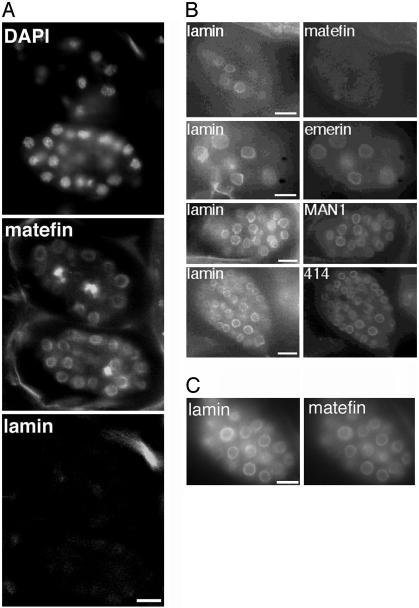
Fig. 2.
Nuclear-envelope localization of matefin does not require Ce-lamin or UNC-84, and matefin is not required to localize other known nuclear-envelope proteins. (A) Ce-lamin-knockdown [lmn-1 (RNAi)] embryos triple-stained for DNA [with 4′,6-diamidino-2-phenylindole (DAPI)] and for endogenous matefin and Ce-lamin by using specific antibodies and indirect immunofluorescence. (B) Double-staining of mtf-1 (RNAi) embryos by indirect immunofluorescence for endogenous Ce-lamin (Left), plus endogenous matefin, Ce-emerin, Ce-MAN1, or FG-nucleoporins (mAb414). (C) Double-staining of unc-84(n369) null embryos by indirect immunofluorescence for endogenous Ce-lamin and endogenous matefin. (Bars, 10 μm.) Wild-type embryos (100–300 cells) were stained with DAPI for DNA and with rat anti-matefin antibodies (serum 3663) by indirect immunofluorescence (IF).
Immunofluorescence studies of early embryos also showed that matefin remained nuclear-envelope-localized through early anaphase and then completely disassembled during mid/late anaphase, similar to Ce-lamin and other known nuclear membrane proteins (refs. 7 and 12 and data not shown).
Matefin Is Present in all Early Embryonic Nuclei but Is Restricted to Germ Cells from Midembryogenesis to Adulthood. To address when and where matefin functions, we used the anti-matefin antibodies to investigate its subcellular localization during development. To control for antibody penetration into C. elegans tissues, we always double-labeled the samples with rabbit antibodies against Ce-lamin, which is expressed in all C. elegans cells except mature sperm (5), and used DAPI to label DNA. Matefin was detected at the nuclear envelope of all early embryonic cells (Fig. 1C). In late embryos (after the comma stage) matefin staining decreased in all somatic cells but intensified in the nuclear envelopes of the two primordial germ cells, Z2 and Z3 (Fig. 3A, arrow). The identity of Z2 and Z3 cells was verified by double labeling with antibodies against PGL-1, which is specific to germ cells (data not shown). Throughout larva stages L1–L4 and in adults, matefin was present only in germ cells (Fig. 3B shows staining in L2 larva, and Fig. 3 C and D shows staining in adults). Matefin signal declined during spermatogenesis and was undetectable in sperm (Fig. 3C, arrowhead). These results suggest that matefin functions in the germ cell lineage.
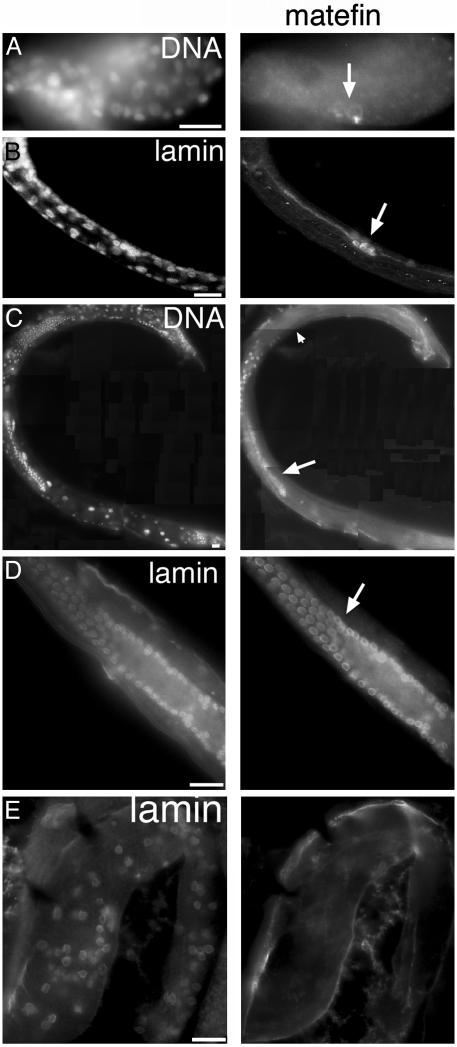
Fig. 3.
Matefin expression is confined to the germ line from midembryogenesis to adulthood. Indirect immunofluorescence images of WT late embryo (A), WT early L2 larva (B), WT adult male (montage shown in C), WT adult hermaphrodites (D), and gonad of an adult hermaphrodite homozygous for the gk199 allele (E), all tripled-stained with DAPI (for DNA), antibodies specific for matefin (serum 3665 for A–C or 3663 for D–E), and Ce-lamin. (A–D) The arrow indicates germ cells. Matefin did not stain mature sperm (arrowhead in C) or gk199 homozygous gonads. Left shows DAPI staining (A and C) or Ce-lamin staining (B, D, and E). Right shows staining results for endogenous matefin. (Bars, 10 μm.)
To test the possibility that matefin protein in early embryos is deposited maternally, we double-stained F1 embryos of strain VC292 by using antibodies against endogenous matefin and Ce-lamin. Strain VC292 contains the gk199 allele, a 435-bp deletion in mtf-1 that removes 105 bp 5′ of the ORF plus the first 110 residues of matefin, including the epitope recognized by serum 3663 (Fig. 1 A).
Among self-fertilizing hermaphrodites heterozygous for the gk199 allele, 25% of the embryos should be homozygous for gk199 and 50% heterozygous for the mtf-1 deletion and the nT1 balancer chromosome V, which expresses wild-type matefin (see below). The remaining 25% were predicted to be homozygous for the nT1 balancer chromosome. We found that ≈97% of embryos (n = 103) stained positively for both matefin (serum 3663) and Ce-lamin antibodies, with roughly similar fluorescence signals for matefin (data not shown). The remaining 3% of embryos (n = 3) stained weakly for both matefin and Ce-lamin, probably because of inefficient antibody penetration into these embryos. In contrast, 100% of L4 larva or adult hermaphrodites stained positive for Ce-lamin, whereas only ≈75% were positive for matefin and had normal germ line (see below). We concluded that matefin is maternally deposited in the egg and persists in somatic cells through early embryonic development. However, in later embryos, larvae and adults matefin expression is limited to germ cells.
Matefin Is Essential for Viability of the Germ Line. Matefin function was analyzed in animals heterozygous or homozygous for the gk199 allele. Heterozygote worms were indistinguishable from wild-type worms, with similar brood sizes. Homozygous gk199/gk199 progeny all developed into adult animals that were sterile but otherwise healthy. We conclude that the maternally supplied matefin is sufficient for embryos to survive development (see below) but insufficient for its role in germ-line cells. Thus, matefin gene expression in embryonic germ cells appeared to be specifically required for the germ line.
The sterile phenotype of gk199/gk199 animals was further analyzed by staining with DAPI and PGL-1 antibodies, as a germ cell marker. During the L3 stage, gk199/gk199 animals contained an average total of 30 germ cells (n = 20), similar to heterozygote and wild-type animals. During early L4 the average number of germ cells in gk199/gk199 gonads was <80 (n = 20), compared to >250 germ cells in gk199/GFP heterozygous and wild-type animals (n = 20). Both the number of germ cells and gonad size were significantly reduced in adult gk199/gk199 animals (Fig. 4A), relative to heterozygous or wild-type animals. Interestingly, the two gonads in each animal sometimes had nonequal numbers of germ cells (Fig. 4A, arrows indicate gonads of different size), and in many cases germ cells were clustered in separate places in the same gonad (Fig. 4A, arrowhead). The reduced number of germ cells in adult gk199/gk199 gonads suggested germ-line cell proliferation defects, germ cell degeneration, or both. Gametes were never seen in gk199/gk199 animals (Fig. 4). The gk199/gk199 germ cells stained positively for both SYTO 11 and SYTO 12 (Molecular Probes), markers for apoptosis, but staining was detected only when there were already reduced numbers of germ cells (data not shown). We conclude that germ cell reduction and lack of mature gametes probably results from both reduced proliferation and apoptosis. We speculate that apoptosis might be secondary to a primary defect in proliferation. Indirect immunofluorescence staining of adult gk199/gk199 animals showed that surviving germ cells did express the germ-line marker protein PGL-1 (Fig. 3D). We concluded that matefin is not needed to establish germ line fate, but is required for germ line proliferation or survival.
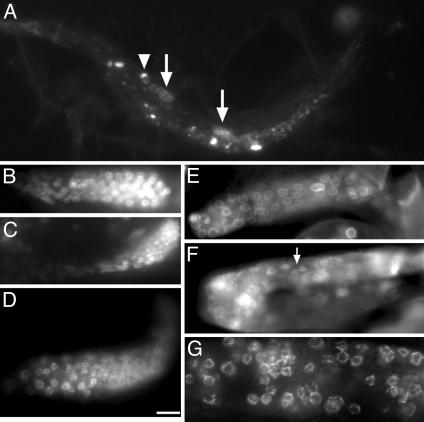
Fig. 4.
Mtf-1 deletion (gk199/gk199) homozygous animals have reduced numbers of germ cells and lack mature gametes. (A) gk199/gk199 animals, which are homozygous for the mtf-1 deletion allele, have germ cells, but their gonads were often of unequal size; arrows in A show a pair of gonads of different size. The arrowhead shows a group of germ cells separated from the main cluster of germ cells. (B and C) Two gk199 homozygous gonads at higher resolution. No gametes were seen in any gk199 gonads. (D–G) Indirect immunofluorescent staining of gk199/gk199 gonads showed positive staining for PGL-1 (D) and normal nuclear-envelope localizations of endogenous Ce-lamin (E), endogenous UNC-84 (F), and endogenous FG-nucleoporins (G). (Bar: 50μm for A; 10 μm for C; 7.5 μm for B and D–F; and 5 μm for G.)
Other inner nuclear membrane/lamina proteins including Ce-lamin, Ce-emerin, Ce-MAN1, UNC-84, and nucleoporins are all normally expressed in the nuclear envelope of germ cells (6, 7, 13). Loss of matefin, verified by indirect immunofluorescence and immunoblotting with 3663 and 3666 sera (Fig. 3E and data not shown), had no effect on the localization of these proteins in germ-line cells, as shown by immunostaining gk199/gk199 gonads with antibodies specific for Ce-lamin (Fig. 3E), UNC-84 (Fig. 4F, arrow), FG-nucleoporins (Fig. 4G), Ce-emerin, or Ce-MAN1 (data not shown), consistent with our RNAi findings in embryonic somatic cells (Fig. 2B).
mtf-1 Is Essential for Early Embryos. To understand the embryonic functions of mtf-1 in vivo, we further examined embryos down-regulated for mtf-1 expression by feeding wild-type (Bristol N2) worms with bacteria expressing mtf-1 dsRNA [mtf-1 (RNAi)]. As noted above, matefin protein was significantly reduced in most mtf-1 (RNAi) embryos (Fig. 2B). Loss of matefin was lethal to embryos, because 59 ± 10% of mtf-1 (RNAi) embryos died (n > 10 independent experiments). Differential interference contrast microscopy and DNA staining showed that most mtf-1 (RNAi) embryos died before the ≈300-cell stage (Fig. 5 A and B). All arrested mtf-1 (RNAi) embryos were abnormal with nuclei that varied in size and DNA content (Fig. 5B). We conclude that matefin is an essential component of the nuclear envelope in early embryos. Embryos that escaped lethality had no visible phenotypes; they developed into normal larva and fertile adults with brood sizes similar to wild-type worms (data not shown). The gonads of these mtf-1 (RNAi) “escapers” stained positive for matefin, indicating that the mtf-1 dsRNA had not affected these animals (data not shown).
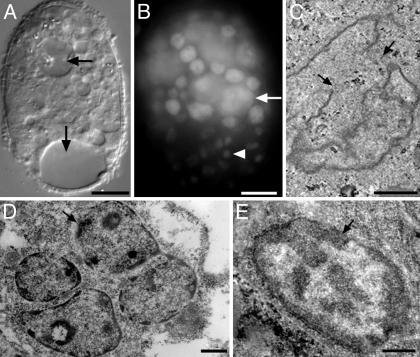
Fig. 5.
Phenotypes of mtf-1 (RNAi) embryos, which became arrested by the ≈300-cell stage. Arrested embryos were viewed by differential interference contrast microscopy (A), stained for DNA with DAPI (B), or viewed by thin section transmission EM (C–E). Arrows in A indicate vacuoles. The arrow in B indicates a large nucleus, and the arrowhead indicates a condensed nucleus. Nuclei in C and D have abnormal shapes. The nucleus in C also has a discontinuous nuclear envelope (arrows) and membrane blebs. The cell shown in D contains four nuclei. Cells in D and E have abnormally condensed chromatin (arrows). (Bars: A and B, 10 μm; C–E, 1 μm.)
The cellular phenotypes of mtf-1 (RNAi) embryos were examined by differential interference contrast, DAPI staining, and thin-section TEM. Arrested embryos had large vacuoles, which are commonly observed in dying embryos (Fig. 5A, arrows), aberrant nuclei with aneuploid DNA content (Fig. 5B, arrow shows a giant nucleus), or abnormally condensed chromatin (Fig. 5B, arrowhead). To further analyze these nuclear and chromatin phenotypes, mtf-1 (RNAi) embryos were fixed, embedded, sectioned, and viewed by TEM. Control embryos were derived from hermaphrodites fed an empty L4440 vector. We saw abnormally shaped nuclei, discontinuous and “blebbed” nuclear envelopes (Fig. 4C), and clumped chromatin (arrows in Fig. 5 D and E) in ≈60% of embryos (n = 24), corresponding to the percent embryonic lethality. A subset of mtf-1 (RNAi) embryos had cells with multiple (2–4) nuclei (Fig. 5D), a phenomenon never seen in control embryos.
To determine when these nuclear and chromatin abnormalities appeared, we analyzed living mtf-1 (RNAi) embryos by time-lapse microscopy in wild-type worms or strain AZ212, which expresses GFP-fused histone H2B (H2B-GFP) (17). The mtf-1 (RNAi) nuclei had morphologically normal structure and chromosome segregation for most cell divisions and showed the above defects late, just before embryonic arrest [Movie 1, which is published as supporting information on the PNAS web site, shows a normal mitosis in mtf-1 (RNAi) embryos].
The mes Mutant Backgrounds Do Not Significantly Affect Embryonic Lethality of mtf-1 (RNAi). The above expression pattern for matefin overlapped with maternal effect sterile (Mes) proteins (18, 19), suggesting that the Mes protein complex and matefin might interact. Mes proteins are required for germ line survival, and their absence causes the germ line to degenerate starting at the L3/L4 larva stages (20). Mes proteins are also important for chromatin silencing in the germ line (21) and for regulating Hox gene expression in the soma of C. elegans (22). To determine whether mes mutations affect matefin expression, we immunostained for endogenous matefin in F1 embryos and gonads of mes-2 (ss186), mes-3 (ss313), or mes-6 (ss553) homozygous animals. Matefin expression was normal in all cases (see Fig. 7A, which is published as supporting information on the PNAS web site, for staining in mes-3 and mes-6 embryos). Similarly, immunostaining showed that Mes-2, Mes-3, and Mes-6 proteins localized normally in mtf-1 (RNAi) embryos (data not shown).
The lethality of mtf-1 (RNAi) embryos was only slightly increased (P < 0.01) in the mes-3 (ss313) or mes-6 (ss553) genetic background (Fig. 7B). If mtf-1 and mes genes directly interacted or were components of the same pathway, we would expect more extensive (“synthetic”) lethality. Thus, the slight increase in lethality probably reflects the additive effects of two independently essential genes.
UNC-84 Activity Does Not Overlap with Matefin. The only other gene in C. elegans with a SUN domain is UNC-84, which like matefin is localized to the nuclear envelope (7, 8). Animals homozygous for unc-84 mutations are viable (8). We therefore tested the possibility that matefin and UNC-84 have overlapping functions by feeding unc-84 homozygous null animals (n369 allele) with mtf-1 dsRNA. The average percentage of dead embryos (58.6%) was similar to that of wild-type embryos fed with mtf-1 dsRNA (60.5%; n = 20). Furthermore, UNC-84 did not affect matefin expression or localization, because matefin expression was normal in unc-84(n369) animals (Figs. 2C and 6).
Discussion
We characterized matefin, a previously uncharacterized SUN-domain protein and the second of two SUN-domain proteins in C. elegans. Outside its SUN and “anti-erbB-2 Ig” domains, matefin is not related to other SUN-domain proteins or other proteins in the database. Our findings suggest that matefin is anchored at the nuclear inner membrane by two putative transmembrane domains, with exposed N- and C-terminal domains that both bind Ce-lamin in vitro. These findings predicted lamin-dependent localization of matefin at the nuclear envelope. However, we found no requirement for Ce-lamin in vivo, suggesting a lamin-independent retention mechanism for matefin at the nuclear membrane. This mechanism might involve binding to chromatin or other stable (lamin-independent) partners at the inner membrane. The lack of binding of matefin to the isolated globular tail domain of Ce-lamin suggests that matefin might instead bind the rod domain, as seen previously for LAP2, otefin, and DNA (23). It is unlikely that matefin binds the globular head domain of Ce-lamin because the head domain mediates polymerization (1), and no other lamin-binding proteins are known to bind there.
Matefin and UNC-84 are both SUN-domain proteins at the nuclear envelope, but their specific localizations and functions appear to differ significantly. Much evidence localizes matefin to the nuclear inner membrane: its direct binding to Ce-lamin, colocalization with Ce-lamin in embryos, and association with peripheral chromatin in detergent-extracted nuclei (shown in this work). In contrast, UNC-84 requires Ce-lamin for its nuclear localization but may not bind directly to lamins (7). SUN domains are likely to have a conserved folded backbone structure. However, this structure is unknown, and whether different SUN domains have conserved surface residues or partners remains open to question.
Matefin is maternally deposited in eggs, but the mtf-1 gene is expressed exclusively in germ-line cells. All other characterized C. elegans nuclear membrane proteins are expressed in both the soma and germ-line cells (5–7, 12). Matefin and UNC-83 in C. elegans, as well as Young Arrest in Drosophila (24) and various vertebrate lamin isoforms (25) comprise a growing list of nuclear-envelope proteins with cell- or tissue-specific expression and function. This theme is likely to resound in human cells, with implications for tissue-specific diseases (23).
Matefin is required for germ line and embryonic development. Animals homozygous for an mtf-1 deletion had normal somatic development and produced germ cells and gonads until larval L3 stage, when the number of germ cells in mtf-1 deletion animals was still similar to wild type. After L3/early L4, the number of germ cells declined, the remaining germ cells made no gametes, and adult animals were sterile. It remains open to question whether matefin is required for germ line proliferation at late stages, germ line maintenance, or both. This germ-line phenotype per se was not surprising, because matefin gene expression is restricted to germ cells. Matefin is not required to specify germ cell fate, but its actual role(s) in germ cells versus embryonic cells remain unclear.
The mtf-1 and mes genes have similar germ-line expression patterns (20). However, the mtf-1 phenotype is seen in homozygous F1 animals, whereas the mes phenotype is seen only in F2 homozygous animals (grandchildless) (26). The number of germ cells present at each stage of development is similar in gk199/gk199 homozygote and mes F2 animals. Mes proteins regulate gene expression, and so might nuclear-envelope proteins (3). However, the nature of matefin's function is unknown, except that matefin and Mes proteins do not regulate each other's expression or localization. Further work is needed to determine whether mtf-1 functions in a known germ line pathway [e.g., nos, puf, glh, pgl, glp-1,or glp-4 (27)] or whether matefin defines a new nuclear-membrane-dependent pathway.
Matefin also appears to be functionally independent of UNC-84, the other SUN-domain protein in C. elegans (7), because UNC-84 and matefin did not affect each other's expression and down-regulating matefin in unc-84 null embryos did not increase lethality. These results suggest that matefin has unique biological role(s) in embryos and germ cells.
In addition to its presence in the germ line, matefin was present in all nuclei of early embryos. The role of matefin in early embryonic soma could not be analyzed in mtf-1 homozygous animals, because they are sterile and mtf-1 heterozygous animals provided sufficient matefin to their embryos. RNAi-induced down-regulation of matefin in early embryos was lethal to embryos, which became arrested before the comma stage with <300 cells. However, in marked contrast to the loss of Ce-lamin, LEM-domain proteins, or BAF (28), which cause anaphase-bridged chromatin and nuclear structure defects at very early stages, the cell cycle in mtf-1 (RNAi) embryos looked normal. Thus, matefin is unlikely to have fundamental roles in nuclear assembly per se, and we can only speculate that it has a previously uncharacterized role late in embryogenesis and/or a specialized role in the germ line or meiosis. The idea of novel specialized roles is supported by the large number (>70) of proposed nuclear membrane proteins identified in a proteomic analysis of rat liver (29).
It was recently suggested that matefin bind ZYG-12, a protein required for centrosome attachment to the nucleus (30). Our data showing that matefin is present only in early embryos and germ cells would limit this hypothetical interaction to a small number of cell types and would further suggest that in most cell types, centrosome positioning by ZYG-12 would require partners other than matefin.
Supplementary Material
Acknowledgments
We thank Susan Strome for PGL-1 and Mes antibodies, mes strains, and critical discussions; the C. elegans Gene Knockout Consortium and C. elegans Reverse Genetics Core Facility at the University of British Columbia for providing the mtf-1 deletion allele (gk199); Merav Gazit and Orly Barshalev for their help in establishing a protocol for matefin down-regulation; Bill Kelly for providing PBK 48.5; Yuji Kohara for providing mtf-1 cDNAs; and Kelly Liu for critical review of the manuscript. This work was supported by grants from the Israel Science Foundation and by National Institutes of Health Grant GM64535 (to K.L.W. and Y.G.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: DAPI, 4′,6-diamidino-2-phenylindole; RNAi, RNA interference; TEM, transmission electron microscopy.
References
- 1.Goldman, R. D., Gruenbaum, Y., Moir, R. D., Shumaker, D. K. & Spann, T. P. (2002) Genes Dev. 16, 533-547. [DOI] [PubMed] [Google Scholar]
- 2.Mattout-Drubezki, A. & Gruenbaum, Y. (2003) Cell Mol. Life Sci. 60, 2053-2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cohen, M., Lee, K. K., Wilson, K. L. & Gruenbaum, Y. (2001) Trends Biochem. Sci. 26, 41-47. [DOI] [PubMed] [Google Scholar]
- 4.Hutchison, C. J., Alvarez-Reyes, M. & Vaughan, O. A. (2001) J. Cell Sci. 114, 9-19. [DOI] [PubMed] [Google Scholar]
- 5.Liu, J., Rolef-Ben Shahar, T., Riemer, D., Spann, P., Treinin, M., Weber, K., Fire, A. & Gruenbaum, Y. (2000) Mol. Biol. Cell 11, 3937-3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gruenbaum, Y., Lee, K. K., Liu, J., Cohen, M. & Wilson, K. L. (2002) J. Cell Sci. 115, 923-929. [DOI] [PubMed] [Google Scholar]
- 7.Lee, K. K., Starr, D., Liu, J., Cohen, M., Han, M., Wilson, K. & Gruenbaum, Y. (2002) Mol. Biol. Cell 13, 892-901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Malone, C. J., Fixsen, W. D., Horvitz, H. R. & Han, M. (1999) Development (Cambridge, U.K.) 126, 3171-3181. [DOI] [PubMed] [Google Scholar]
- 9.Dreger, M., Bengtsson, L., Schoneberg, T., Otto, H. & Hucho, F. (2001) Proc. Natl. Acad. Sci. USA 98, 11943-11948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Starr, D. A. & Han, M. (2003) J. Cell Sci. 116, 211-216. [DOI] [PubMed] [Google Scholar]
- 11.Edgley, M. L., Liu, J., Riddle, D. L. & Fire, A. (1999) Worm Breeder's Gazette 15, 20. [Google Scholar]
- 12.Lee, K. K., Gruenbaum, Y., Spann, P., Liu, J. & Wilson, K. L. (2000) Mol. Biol. Cell 11, 3089-3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu, J., Lee, K. K., Segura-Totten, M., Neufeld, E., Wilson, K. L. & Gruenbaum, Y. (2003) Proc. Natl. Acad. Sci. USA 100, 4598-4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cohen, M., Tzur, Y. B., Neufeld, E., Feinstein, N., Delannoy, M. R., Wilson, K. L. & Gruenbaum, Y. (2002) J. Struct. Biol. 140, 232-240. [DOI] [PubMed] [Google Scholar]
- 15.Timmons, L. & Fire, A. (1998) Nature 395, 854. [DOI] [PubMed] [Google Scholar]
- 16.Schier, R., Marks, J. D., Wolf, E. J., Apell, G., Wong, C., McCartney, J. E., Bookman, M. A., Huston, J. S., Houston, L. L., Weiner, L. M. & Adams, G. P. (1995) Immunotechnology 1, 73-81. [DOI] [PubMed] [Google Scholar]
- 17.Praitis, V., Casey, E., Collar, D. & Austin, J. (2001) Genetics 157, 1217-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holdeman, R., Nehrt, S. & Strome, S. (1998) Development (Cambridge, U.K.) 125, 2457-2467. [DOI] [PubMed] [Google Scholar]
- 19.Xu, L., Fong, Y. & Strome, S. (2001) Proc. Natl. Acad. Sci. USA 98, 5061-5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Capowski, B. & Strome, S. (1991) Worm Breeder's Gazette 11, 87-89. [Google Scholar]
- 21.Kelly, W. G. & Fire, A. (1998) Development (Cambridge, U.K.) 125, 2451-2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ross, J. M. & Zarkower, D. (2003) Dev. Cell 4, 770-772. [DOI] [PubMed] [Google Scholar]
- 23.Gruenbaum, Y., Goldman, R. D., Meyuhas, R., Milles, E., Margalit, A., Fridkin, A., Dayani, Y., Prokocimer, M. & Enosh, A. (2003) Int. Rev. Cytol. 226, 1-62. [DOI] [PubMed] [Google Scholar]
- 24.Liu, J., Song, K. & Wolfner, M. F. (1995) Genetics 141, 1473-1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stuurman, N., Heins, S. & Aebi, U. (1998) J. Struct. Biol. 122, 42-66. [DOI] [PubMed] [Google Scholar]
- 26.Seydoux, G. & Strome, S. (1999) Development (Cambridge, U.K.) 126, 3275-3283. [DOI] [PubMed] [Google Scholar]
- 27.Beanan, M. J. & Strome, S. (1992) Development (Cambridge, U.K.) 116, 755-766. [DOI] [PubMed] [Google Scholar]
- 28.Zheng, R., Ghirlando, R., Lee, M. S., Mizuuchi, K., Krause, M. & Craigie, R. (2000) Proc. Natl. Acad. Sci. USA 97, 8997-9002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schirmer, E. C., Florens, L., Guan, T., Yates, J. R. & Gerace, L. (2003) Science 531, 1380-1382. [DOI] [PubMed] [Google Scholar]
- 30.Malone, C. J., Misner, L., Le Bot, N., Tsai, M. C., Campbell, J. M., Ahringer, J. & White, J. G. (2003) Cell 115, 825-836. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.