Abstract
NO propagates a number of antiatherogenic effects in the endothelium, and diminished availability has been associated with vascular disease. Recently it has been reported that phosphorylation of endothelial NO synthase (eNOS) at Ser-1179 is required to increase activity in response to stimuli, including high-density lipoprotein (HDL). The current study was undertaken to further examine the mechanism by which HDL activates eNOS and to specifically determine the role of the major apolipoprotein of HDL, apolipoprotein AI (ApoAI). Phosphorylation of eNOS residues Ser-116, Ser-617, Ser-635, Ser-1179, and Thr-497 after incubation with ApoAI and HDL was examined. There were significant increases in phosphorylation at Ser-116 in response to both HDL and ApoAI and similar magnitudes of dephosphorylation at Thr-497. Ser-1179 phosphorylation increased transiently but returned to basal level after 2.5 min. Data demonstrating activation of AMP activated protein kinase (AMPK) during HDL and ApoAI incubation suggests that AMPK may play a role in activation of eNOS. NO release in response to HDL and ApoAI stimulation in endothelial cells paralleled the time frames of phosphorylation, suggesting a causal relationship. Furthermore, ApoAI was found to associate with eNOS in endothelial cells and bind transfected eNOS in Chinese hamster ovary cells, whereas confocal data demonstrates colocalization of ApoAI and eNOS in the perinuclear region, suggesting a protein–protein interaction. Collectively, the results indicate that HDL and ApoAI increase eNOS activity by multisite phosphorylation changes, involving AMPK activation after protein association between ApoAI and eNOS.
Decreased bioavailability of endothelium-derived NO is an important antecedent to atherosclerosis (1). NO inhibits events that promote atherosclerotic progression, including vasoconstriction, monocyte adhesion, and smooth muscle cell proliferation (2). The bulk of endothelium-derived NO is generated from l-arginine conversion by endothelial NO synthase (eNOS, NOS III) (3). Activity of eNOS is modulated by complex mechanisms including phosphorylation, protein–protein interactions, substrate availability, and intracellular Ca2+ flux. Numerous biological agents have been associated with changes in eNOS activity, including caveolin (4), Ca2+ calmodulin (5), HSP90 (6), Dynamin-2 (7), bradykinin (8), and more recently high-density lipoprotein (HDL) (9). HDL plays a major role in reversing and preventing progression of vascular disease through its role in reverse cholesterol transport and its involvement in signaling/receptor pathways of cholesterol metabolism (10). Possibly, some cardiovascular protective effects of HDL are mediated via activation of eNOS, although the precise nature of this interaction remains unclear. The current study was undertaken to examine the mechanism by which HDL activates eNOS and to determine whether the major apolipoprotein of HDL, apolipoprotein AI (ApoAI), mediates the response.
Endothelial cells incubated with HDL exhibit an increase in eNOS activity and NO production, most likely involving a receptor-mediated effect through scavenger receptor class B type I (SR-BI) (9, 11). Subsequently, Li et al. (12) have reported that HDL binding to SR-BI activates eNOS in transfected Chinese hamster ovary (CHO) cells in an Akt-independent manner, possibly involving ceramide, whereas Mineo et al. (13) reported that HDL caused eNOS activation through phosphorylation at Ser-1179 in both endothelial cells and COS M6 cells transfected with eNOS. Regulation of eNOS activity by phosphorylation is complex and involves an intricate interaction between multiple sites (Ser-116, Thr-497, Ser-617, Ser-635, and Ser-1179) and the activities of numerous kinases and phosphatases, including AMP activated protein kinase (AMPK) (14), PKA (15), Akt/PKB (16), PKC (17), PP1 (18), and PP2A (19). In the current study, activation of eNOS was characterized by studying five phosphorylation sites within the eNOS enzyme. These included the previously characterized Ser-1179 and Thr-497 but also included three other sites, Ser-116, Ser-617, and Ser-635, which have not yet been studied in relation to HDL activation.
We demonstrated that HDL and ApoAI, but not low-density lipoprotein (LDL), up-regulated the activity of eNOS through specific downstream phosphorylation of a number of these sites and also activated AMPK, suggesting that AMPK plays a role in phosphorylating eNOS. NO release was measured by using diaminofluorescein-2-diacetate (DAF2-DA) fluorescence, which increased over the same time course as phosphorylation occurred. Crosslinking, coimmunoprecipitation, and colocalization studies show that the specific changes in phosphorylation of eNOS involve an interaction between ApoAI with eNOS to increase activity. These findings contribute further understanding of the pivotal roles of both HDL and NO in cardiovascular protection.
Methods
Antibodies. Anti-eNOS monoclonal antibody and monoclonal anti-caveolin-1 antibody were from BD Transduction Laboratories (San Diego). Anti-human ApoAI monoclonal and polyclonal antibodies were created in our laboratory as previously described (20). All anti-phosphorylated eNOS and phospho-AMPK/acetyl-CoA carboxylase (ACC) polyclonal antibodies were created at St. Vincent's Institute of Medical Research as previously described (14). Both anti-mouse and anti-rabbit horseradish peroxidase-conjugated secondary antibodies were from Bio-Rad.
Cell Lines and Culture. Human umbilical vein endothelial cells (HUVEC) and bovine aortic endothelial cells (BAEC) were isolated as previously described (21) from fresh tissue. Endothelial cells were screened by immunohistochemistry for endothelial cell marker von Willebrand factor VIII and smooth muscle cell α-actin to exclude smooth muscle cell cocultures.
Isolation of Lipoproteins and ApoAI. Human HDL3 (1.12–1.21 g/ml) and LDL (1.019–1.063 g/ml) were isolated from whole, pooled plasma by ultracentrifugation as previously described (22). Lipid-free ApoAI was isolated according to the method described previously (23, 24). Presence of phospholipid in the isolated ApoAI was not detectable by using the Bartlett phospholipid assay (25).
Crosslinking and Immunoprecipitation of ApoAI and eNOS. HUVEC were grown to 70% confluence, and cultures were washed twice with serum-free medium and incubated in ApoAI (50 μg/ml) for 1 h. Cells were placed on ice and washed twice before incubation in dithiobis(succinimidyl propionate) (DSP; Pierce) (1 mg/ml) for 30 min. The DSP was aspirated and cells were lysed with RIPA buffer (0.05 M Tris, pH 7.5/0.1 M NaCl/0.05 M NaF/0.1% SDS/0.5% deoxycholate/1% Triton X-100/0.1 M EDTA) for 1 h at 4°C. Cells were then scraped and lysates were centrifuged at 16,000 × g for 30 min. Supernatants were incubated with Protein A agarose and either anti-ApoAI or anti-eNOS antibodies overnight. After immunoprecipitation, beads were washed thoroughly in gradient salt buffers to remove nonspecific binding.
Ligand Blot. CHO cells were transfected with pIRES2-EGFP (BD Biosciences CLONTECH) vector alone or pIRES2-EGFP-eNOS by using Lipofectamine 2000 (Invitrogen Australia). Cells were allowed to recover for 48 h and were then harvested in RIPA buffer, and lysates were separated by SDS/PAGE. Proteins were transferred by Western blot onto nitrocellulose membranes. Membranes were washed, incubated in 20 mM sodium deoxycholate in TBS for 30 min, washed three times in TBS, and incubated in 50 μg/ml ApoAI in LB buffer (20 mM Tris·HCl, pH 7.4/0.15 M NaCl/1 mM EDTA) at room temperature for 1 h before washing and blotting of membranes with anti-ApoAI antibodies.
eNOS Activity Through DAF2-DA Imaging. BAEC were cultured on glass coverslips and serum deprived for 4 h. Coverslips were washed twice before a 15-min incubation in 1 μM DAF2-DA (Calbiochem) at 37°C. Coverslips were washed and placed in the confocal chamber (37°C) with 1 ml of Hanks' balanced salt solution (pH 7.4). Cells were viewed for 30 s before treatment with the following agents: 50 μg/ml HDL, 40 μg/ml ApoAI, 10 μM ionomycin (Sigma-Aldrich), 1 mM NG-monomethyl-l-arginine (l-NMMA) (Calbiochem), or 5 μM S-nitroso-N-acetyl-d,l-penicillamine (SNAP) (Calbiochem). Cells were then viewed for a further 4.5 min, and fluorescence intensity was recorded at 1-s intervals by using confocal microscopy (Olympus IX70 inverted microscope using Perkin-Elmer Wallac Ultraview and Zeiss META 510 systems).
Isolation of Phosphorylated eNOS and AMPK. AMPK was isolated in separate experiments, by using a protocol similar to isolation of phosphorylated eNOS as previously described (26). Briefly, BAEC were grown to 90% confluence before being serum deprived for 4 h. Buffer was aspirated and replaced with serum-free DMEM containing HDL (50 μg/ml), ApoAI (40 μg/ml), or LDL (50 μg/ml), and was incubated for 0.5, 2.5, 5, and 15 min. Cells were harvested on ice in P-Lysis buffer (20 mM Hepes, pH 7.4/2 mM EDTA/50 mM NaF/5 mM sodium pyrophosphate/1 mM DTT/10 μg/ml aprotinin/10 μg/ml pepstatin A/10 μg/ml leupeptin/1 mM PMSF/1% Nonidet P-40; Sigma-Aldrich). Cells were disrupted by snap freezing and cleared by centrifugation. eNOS was isolated by affinity chromatography by using 2′5′ ADP Sepharose (Amersham Pharmacia) at 4°C for 2 h and quantified by Western blotting. For AMPK quantification, cell lysates were loaded directly onto gels. Phosphorylated ACC was isolated from the remainder of these lysates with streptavidin beads overnight at 4°C.
Western Blots. Crosslinked proteins were dissociated from agarose beads in Laemmli buffer containing 10 mM DTT at 37°C for 90 min, separated by SDS/PAGE, and transferred onto nitrocellulose. Phosphorylated eNOS, AMPK lysates, and phosphorylated ACC streptavidin beads were incubated at 100°C in Laemmli buffer containing 10 mM DTT for 5 min, separated by SDS/PAGE, and transferred to PVDF membranes. Immunoblotting was performed with appropriate antibodies. Peroxidase-conjugated secondary anti-rabbit or anti-mouse antibodies were added and visualized by using the enhanced chemiluminescence (ECL) technique (Amersham Pharmacia). Protein was quantitated from digitized films as the product of band density and area by using optimas 6.1 software (Media Cybernetics, LP, Silver Spring, MD).
Statistics. Changes in phosphorylation state from basal were examined by t test. Phosphorylation time course data were analyzed by repeated-measures ANOVA. All data were analyzed by spss (version 10.0, SPSS, Chicago). Results are expressed as mean ± SEM. P < 0.01 was considered significant. Post hoc testing was performed by using Fisher's least significant difference test.
Results
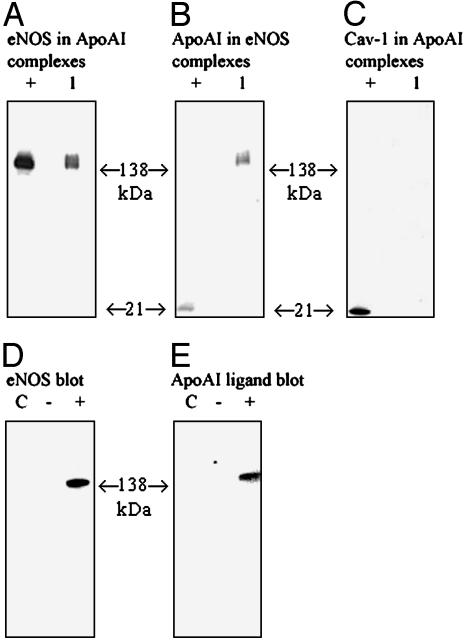
Protein Association Between ApoAI and eNOS. Coimmunoprecipitation data demonstrate that ApoAI associates with eNOS in endothelial cells. HUVEC were incubated with ApoAI, and membrane and cytoplasmic proteins were crosslinked. Upon immunoprecipitation of ApoAI, it was apparent from Western blots that eNOS was present in associated crosslinked complexes (Fig. 1A). Furthermore, this process was reproducible in reverse, in that eNOS was immunoprecipitated and associated complexes contained ApoAI (Fig. 1B). Because eNOS and ApoAI were the major proteins identified in Fig. 1 A or B, data suggest a strong association between these two proteins. However, to determine whether the ApoAI/eNOS binding was an artifact of a primary complex formation involving other known ligands of NOS, we repeated the experiments with caveolin-1. Previous studies have shown that eNOS can directly bind caveolin-1, a protein also thought to be related to the metabolism of HDL (4). However, as shown in Fig. 1C, caveolin-1 was not detected in immunoprecipitated ApoAI complexes. Stronger support for a direct specific interaction between eNOS and ApoAI was observed from binding data by using ligand blots coupled to Western blots. Cell lysates from eNOS- and mock-transfected CHO cells were either Western blotted for eNOS (Fig. 1D) or ligand blotted after incubation with ApoAI. Blotting for ApoAI revealed bands only in the eNOS-transfected lane (Fig. 1E). These data strongly support the hypothesis that ApoAI binds eNOS.
Fig. 1.
Association of ApoAI and eNOS. HUVEC were incubated in serum-free medium containing ApoAI (50 μg/ml) for 1 h. After crosslinking, cells were lysed and harvested before immunoprecipitation with anti-ApoAI antibodies probed by Western blotting for eNOS (A), anti-eNOS antibodies probed by Western blotting for ApoAI (B), and anti-ApoAI antibodies probed by Western blotting for caveolin-1 (cav-1) (C). +, Positive control for immunoblot protein; 1, crosslinked immunoprecipitated complexes. eNOS- and mock-transfected CHO cell lysates were Western blotted for eNOS expression (D), and ligand blotting with ApoAI of the same experiment shows immunoblot for ApoAI (E), where lanes are control (C), mock (-), and eNOS (+). Results are indicative of three separate experiments.
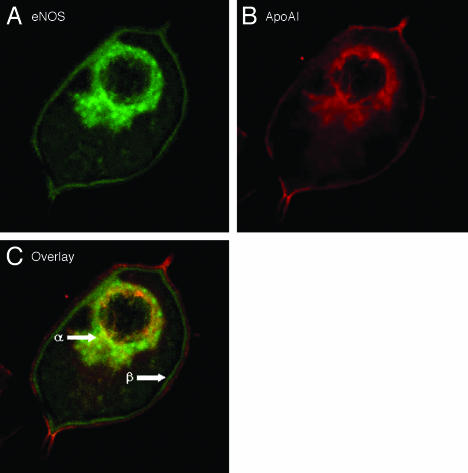
After demonstrating an association between ApoAI and eNOS in vitro, we further demonstrated in situ where specific interactions occurred. Endothelial cells were incubated with ApoAI and fixed on glass slides. After staining for eNOS and ApoAI, colocalization was seen in the perinuclear region as depicted in Fig. 2C (α arrow), which is further highlighted in the perinuclear region of each single labeled image (Fig. 2 A and B). Despite occupying the plasma membrane region, eNOS and ApoAI did not colocalize at the plasma membrane as clearly seen in the overlay image (Fig. 2C, β arrow), where a distinct gap is seen between them. It appears that eNOS resides within the inner membrane, but ApoAI is in the outer leaflet. These results further confirm an association between eNOS and ApoAI within endothelial cells, suggesting an interaction in the perinuclear region.
Fig. 2.
Colocalization of eNOS and ApoAI in endothelial cells. BAEC were serum deprived and incubated in ApoAI for 15 min before cells were fixed and immunostained for eNOS (A) and ApoAI (B); C displays overlay and colocalization of probes. Cells were viewed with confocal microscopy where images were captured by using previously optimized conditions for colocalization. (×400.)
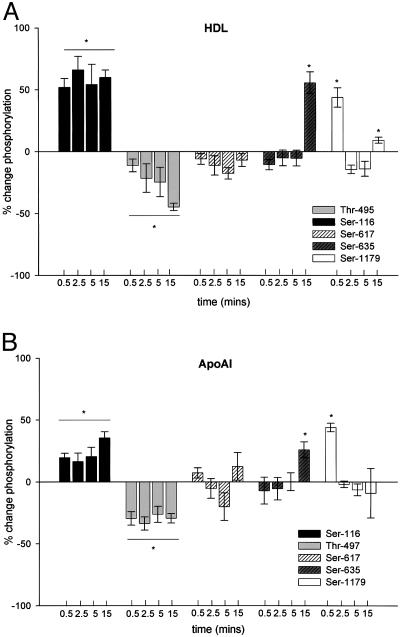
Lipoprotein-Induced Changes in eNOS Phosphorylation. Five known phosphorylation sites within the eNOS amino acid sequence (Ser-116, Thr-497, Ser-617, Ser-635, and Ser-1179) were investigated. In phosphorylation time course studies (0.5, 2.5, 5.0, and 15 min, n = 4; Fig. 3), results are displayed as percent change compared to basal phosphorylation. HDL and ApoAI incubation significantly increased phosphorylation at Ser-116 by 56.5 ± 7.8% and 32.2 ± 6.9% (P < 0.001), respectively, after 15 min. Furthermore, there was a rapid increase in phosphorylation at Ser-116 after 30 s in response to both HDL (Fig. 3A) and ApoAI (Fig. 3B), which was sustained for up to 15 min (P < 0.001). In contrast, time course studies of the Thr-497 site revealed significant dephosphorylation after only 30 s, which was sustained for 15 min in response to ApoAI (Fig. 3B) and increased linearly up to 15 min in response to HDL (P ≤ 0.001; Fig. 3A). The Ser-116 and Thr-497 sites were unaffected by LDL. Interestingly, there were significant (P ≤ 0.01) increases in phosphorylation at site Ser-635 with all three lipoprotein treatments after 15 min, but time course data reveal no significant increase before that time point (Fig. 3 A and B; LDL data not shown). LDL increased phosphorylation at Ser-617 (42.5 ± 8.9%, P ≤ 0.001; data not shown) after 15 min despite neither HDL nor ApoAI having an effect on phosphorylation during the time course. Time course studies carried out on the Ser-1179 site revealed a significant transient increase in phosphorylation of 43 ± 7.8% and 44 ± 3.4% (P < 0.001) after 30 s with HDL and ApoAI, respectively (Fig. 3 A and B). Phosphorylation was returned to basal level after 2.5 min and remained relatively low for the remaining time points.
Fig. 3.
Time course of HDL- and ApoAI-induced changes in phosphorylation of eNOS at sites Ser-116, Thr-497, Ser-617, Ser-635, and Ser-1179. (A and B) HDL-ApoAI-induced changes in the phosphorylation state of Ser-116, Thr-497, Ser-617, Ser-635, and Ser-1179 as percent change from basal phosphorylation. Results are mean ± SEM. *, P < 0.01 by using repeated-measures ANOVA and Fisher's least significant difference test.
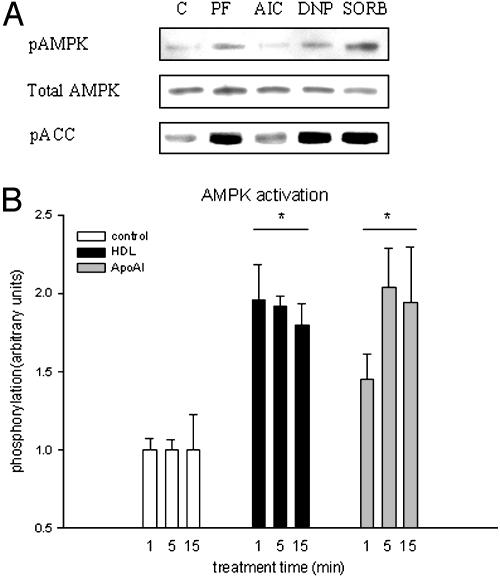
HDL- and ApoAI-Induced Activation of AMPK. That AMPK phosphorylates and associates with eNOS has been previously described (14), and therefore we sought to determine whether AMPK was activated under stimulus from HDL and ApoAI. Endothelial cells incubated with either HDL or ApoAI showed increased activation of AMPK as demonstrated by an increase in phosphorylation of the Thr-172 site of the AMPK-α1 subunit. First we determined phosphorylation of AMPK-α1 to a range of known stimuli to determine whether activation occurred in endothelial cells. Results show an increase in phosphorylation to phenformin, 2,4-dinitrophenol, and sorbitol, but not 5-aminoimidazole-4-carboxamide ribonucleoside (AICAR) (Fig. 4A). To confirm that phosphorylation of the Thr-172 site of AMPK-α1 is in fact increasing kinase activity in endothelial cells, the phosphorylation of the downstream substrate of AMPK, ACC, was determined in response to the same stimuli. Results show an increase in ACC phosphorylation in response to phenformin, 2,4-dinitrophenol, and sorbitol but not to AICAR, suggesting that AMPK is in fact being activated (Fig. 4A). It was therefore assumed from ACC phosphorylation data that any increase in AMPK-α1 phosphorylation was a direct measure of AMPK activity.
Fig. 4.
HDL and ApoAI activation of AMPK in endothelial cells. BAEC were serum-deprived before treatment with positive control for 30 min or HDL/ApoAI over time course of 1, 5, and 15 min. (A) Blots are in order from left to right; control, phenformin, 5-aminoimidazole-4-carboxamide ribonucleoside (AIC), 2,4-dinitrophenol (DNP), and sorbitol. The top row shows Thr-172 phosphorylation of AMPK-α1, the middle row shows total AMPK-α1, and the third row demonstrates Ser-79 ACC phosphorylation. (B) Plot of HDL- and ApoAI-induced Thr-172 AMPK-α1 phosphorylation over time courses of 1, 5, and 15 min as increase compared to control phosphorylation. Results are mean ± SEM. *, P < 0.05 by using repeated-measures ANOVA and Fisher's least significant difference test.
As demonstrated in Fig. 4B, incubations with HDL or ApoAI resulted in rapid activation of AMPK when measured over a 1-, 5-, and 15-min time course. HDL significantly increased phosphorylation by 96% (P < 0.01), 92% (P < 0.001), and 80% (P < 0.01), respectively (Fig. 4B), whereas ApoAI similarly activated AMPK, increasing phosphorylation by 45% (P < 0.05), 104% (P < 0.01), and 95% (P < 0.05), respectively, over the time course (Fig. 4B). All results are percent change of that from control phosphorylation.
Lipoprotein-Induced NO Release in BAEC. A rapid and intense increase in cytoplasmic and perinuclear fluorescence after an average 3-min incubation with SNAP confirmed that the dye was intracellular and quantifiable (see Fig. 6, which is published as supporting information on the PNAS web site). Furthermore, ionomycin, the calcium ionophore positive control, increased NO production and NO release within 2 min in both the cytoplasm and the perinuclear regions (Fig. 6A4). Fluorescence intensity was plotted for positive controls in Fig. 6B, demonstrating NO release in the perinuclear region and cytoplasm separately.
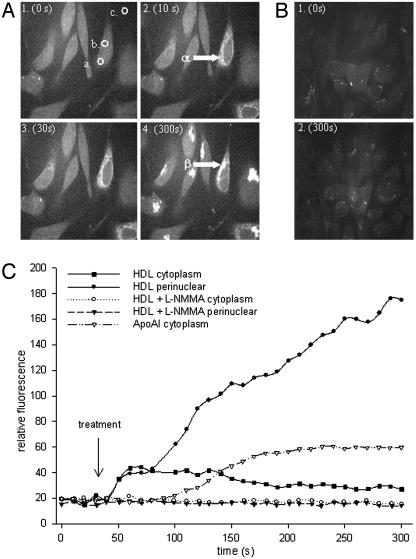
Cells were stimulated with ApoAI, HDL alone, and HDL plus a pretreatment of the specific NOS inhibitor l-NMMA. Responses to HDL (Fig. 5A) were recorded at 0, 10, 30, and 300 s. Images for HDL+l-NMMA are shown at 0 and 300 s (Fig. 5B). HDL incubation rapidly increased cytoplasmic NO release within 10 s (Fig. 5A2, α arrow). Cytoplasmic release peaked at 40 s after treatment and slowly dissipated (Fig. 5C). NO release from the perinuclear region began to increase when cytoplasmic NO release dissipated, and it continued to increase for the experimental duration (300 s) (Fig. 5A4, β arrow). ApoAI-induced NO release, however, was a slower process, where a rise in NO was not seen until 60 s after treatment (Fig. 5C). This response was a more uniform cytosolic and perinuclear NO release. l-NMMA blocked HDL-stimulated NO release from both the cytoplasm and perinuclear regions (Fig. 5 B and C).
Fig. 5.
NO release in cultured BAEC in response to HDL with and without l-NMMA treatment. BAEC were cultured on glass coverslips and serum-deprived before 15-min incubation in DAF2-DA. Cells were viewed by using confocal microscopy, and images were captured for HDL-induced fluorescence at 0 s (A1), 10 s (A2), 30 s (A3), and 300 s (A4) after treatment. (B) HDL+l-NMMA-induced fluorescence at 0 s (B1) and 300 s (B2) after treatment. Arrow α shows cytoplasmic NO release, and arrow β demonstrates perinuclear NO release. (C) HDL- and ApoAI-induced NO release, recorded as increased fluorescence in a designated region of interest, was plotted against time in comparison to control regions. Plots show time-dependent NO release in the perinuclear region and cytoplasm in response to ApoAI and HDL alone and HDL after cells were preincubated with l-NMMA. Treatment commenced at 30 s.
Discussion
HDL-mediated NO release from endothelial cells has been well established (9, 11). This study demonstrates that eNOS activity can be up-regulated in response to incubation with physiological concentrations of HDL and lipid-poor ApoAI through regulation of phosphorylation sites on the eNOS enzyme. In particular, prolonged increase in phosphorylation of Ser-116 and transient increase at Ser-1179, which may be partly due to AMPK activation, together with dephosphorylation of Thr-497 are collectively likely to be responsible for increased eNOS activity. Furthermore, ApoAI and eNOS closely associate in the cell, most likely in the perinuclear region providing further understanding of the mechanisms by which HDL/ApoAI activates eNOS in endothelial cells. Live cell confocal microscopy illustrated real-time NO production consistent with the phosphorylation time course, as well as novel data regarding the spatial release of NO within individual cells.
Although previous studies revealed that HDL increases eNOS activity through a number of proposed mechanisms, including the need for HDL binding to SR-BI and downstream phosphorylation (9, 11–13), the precise mechanism between binding to receptor and downstream signaling is still to be elucidated. Recently, it has been demonstrated that SR-BI mediates uptake of holo-HDL in hepatocytes and CHO cells, providing evidence for internalization of ApoAI in these cells (27). Whether or not a similar mechanism for SR-BI-mediated HDL uptake occurs in endothelial cells has not been investigated. Other pathways for internalization of ApoAI have also been proposed through receptors such as cubilin (28) and a recently revealed ApoAI-binding protein, beta chain of the ATP synthase (29). However this process occurs, it most likely dissociates ApoAI within the cell (28), rendering it available for protein–protein interaction and possible signaling functions. eNOS is known to be activated by proteins such as HSP90 and CaM in a similar fashion. Such interactions highlight the fact that eNOS possesses binding sites for several ligands involved in its activation. The formation of such complexes may recruit kinases and other associated enzymes for downstream signaling through mechanisms that, in the case of HDL, likely include activation of AMPK and phosphorylation.
Previous studies have demonstrated that specific phosphorylation of eNOS at Ser-1179 and dephosphorylation at Thr-497 can increase the activity of the enzyme and increase NO production (14–17). Many of these studies have also shown that multiple kinases and phosphatases are responsible for the regulation of phosphorylation under various stimuli. In this study, we have broadened the growing list of stimuli and shown not only that the sites Ser-1179 and Thr-497 are involved in eNOS activation, but also that phosphorylation of the previously less-characterized site Ser-116, accompanies activation by lipoproteins. The eNOS Ser-116 site has been rarely implicated in agonist-mediated phosphorylation, with shear stress being the sole example of increased phosphorylation (30). Detailed mutagenesis studies indicate that Ser-116 does not affect basal NO release but is a negative regulator of agonist activation.
Phosphorylation of Ser-116 increased by 56.5% and 32.2% during a 15-min incubation with HDL and ApoAI, respectively. Phosphorylation occurred within 30 s of addition of the stimulus and was maintained for at least 15 min, suggesting a prolonged activation of the enzyme rather than a transient stimulation. It should be noted that ApoAI-induced phosphorylation of Ser-116, although significant, is ≈50% less than that of HDL-induced phosphorylation, which may partially explain the difference between NO release to each treatment. Moreover, the kinase responsible for phosphorylation at Ser-116 in response to ApoAI is not known; however, other studies have outlined kinases involved in phosphorylation of eNOS, revealing crucial roles for AMPK, Akt, PKA, PKC, and CaM II kinase at other sites (13–17). The AMPK results obtained in this study demonstrate that activation is strong during incubation with HDL after 1 min which persisted for 15 min, paralleling the phosphorylation of Ser-116 to HDL. This relationship suggests that AMPK may be playing a role in the phosphorylation of Ser-116. Further supporting evidence of AMPK phosphorylation at Ser-116 is the fact that ApoAI induces a less dramatic activation of AMPK, being slight activation after 1 min, but an equivalent activation as HDL after 5 min. These data also parallel phosphorylation trends of Ser-116 in response to ApoAI. One possible explanation for the differences between responses to HDL and “free” ApoAI may lie with differences in ligand/receptor kinetics and subsequent post receptor processing that influences the rate of ApoAI transport to activation sites. A previous study on AMPK coprecipitation with and phosphorylation of eNOS by Chen et al. (14) outlines the ability of AMPK to phosphorylate eNOS at Ser-1179 and Thr-497, but it does not present data on the phosphorylation of other sites, such as Ser-116. Therefore, it does not rule out the possibility that AMPK is phosphorylating Ser-116, and such data require much broader investigation, clearly outside the scope of this article. Tyrosine kinase, src, and MAP kinase have also been implicated in the activation of eNOS, although the action of these kinases is uncertain (13).
Although Ser-1179 is well characterized, its effect on eNOS activation in response to lipoproteins remains controversial. A study by Li et al. (12) suggested activation of eNOS in eNOS- or SR-BI-transfected CHO cells occurred through downstream ceramide signaling after binding of HDL to SR-BI, rather than Akt-mediated phosphorylation at Ser-1179. In contrast, Mineo et al. (13) have reported that HDL phosphorylation of Ser-1179 in endothelial cells does regulate eNOS activity through the actions of kinases, as does this study. However, current data demonstrate a rapid transient phosphorylation of Ser-1179 within 30 s of HDL and ApoAI stimulation, which then returned to basal within 150 s. This result is in contrast to previous findings, where phosphorylation from other stimuli is more prolonged, lasting up to 20 min (13). Our results are consistent with the hypothesis that phosphorylation of this site increases NO release, and that disparity of phosphorylation of this site may relate to the origin of the cell system studied. Collectively, these data indicate eNOS regulation at this site is emerging as an extremely complex scheme, probably involving a number of different pathways. The results reported by Chen et al. (14), who observed increased phosphorylation of Ser-1179 through the action of AMPK, support this suggestion. Our data suggest a rapid transient phosphorylation of Ser-1179 after 30 s, being returned to basal after 5 min. This finding appears unrelated to AMPK activity, which in contrast increased from the earliest time point and persists throughout the time course. However, this does not completely rule out the involvement of AMPK on this site but may simply highlight the actions of phosphatases, which act more readily on eNOS dephosphorylation than on AMPK inactivation.
eNOS Thr-497 is a regulatory site, where increased phosphorylation prevents CaM binding by increasing steric hindrance. Michel et al. (17) have shown that PKA-mediated PP1 phosphatase activation dephosphorylated Thr-497 and increased eNOS activity. In this study, we show a significant decrease in phosphorylation of this site compared to basal, during incubations of both HDL and ApoAI. Thr-497 was dephosphorylated rapidly and the response persisted for at least 15 min. The dephosphorylation in response to HDL and ApoAI likely contributes to enzyme activation, as previously demonstrated (17). Similar results have been demonstrated in cells stimulated with bradykinin (18, 31), IBMX, forskolin, and VEGF (17).
To date, the Ser-617 and Ser-635 sites have not been extensively explored, although these sites are both phosphorylated in BAEC under VEGF, bradykinin, and ATP stimulation (32). Ser-617 appears to be important for regulating protein interaction with HSP90 and Akt, and it is also likely to negatively regulate basal NO release but not stimulated release. Ser-617 was unaffected by both HDL and ApoAI, but phosphorylation was significantly increased by LDL. Because oxLDL inhibits activation of eNOS (9), our results seem consistent with mutation studies (33) demonstrating that increased phosphorylation at this site decreases eNOS activity by preventing interaction with stimulatory proteins. Ser-635, however, has been proposed as a regulator of both basal and stimulated release of NO. This study demonstrates an increase in phosphorylation at Ser-635 after 15 min to all three lipoprotein stimuli, making it unlikely to be part of the activation mechanism for stimulatory NO release.
Measurement of NO release in living cells was used to determine time-related changes in eNOS activity. The results clearly demonstrate that HDL-induced NO release was diminished by l-NMMA, confirming that NO release occurred through eNOS. That the mechanism of eNOS activation was mediated through phosphorylation is supported by the parallel time frames for these events. There was an initial release of NO in a uniform whole cytoplasmic response, most likely linked to membrane-bound eNOS. This is consistent with HDL binding to the cell membrane, allowing fast receptor-mediated access to eNOS. After ≈60 s, a new intracellular pool of NO becomes apparent. The perinuclear compartment is most likely that of the Golgi apparatus, and by 300 s most NO is produced at this site. Previous studies have suggested Golgi participation in the eNOS pathway. In particular, Fulton et al. (26) present the appearance of two distinct pools of eNOS, one being of membrane origin and the other in a perinuclear compartment, later thought to be the Golgi. Further to this hypothesis, this study's colocalization data reveal that the probable interaction between ApoAI and eNOS most likely occurs in the perinuclear region and not at the membrane. Therefore, the fact that HDL produced an initial release of NO within 10 s of stimulation and ApoAI does not suggests that ApoAI present in holo HDL either targets activation sites more rapidly or that other nonprotein moieties in HDL influence other pathways that initiate eNOS, perhaps through the estrogen- or ceramide-mediated activation suggested by Gong et al. (34) and Li et al. (12), respectively. But after the initial response by HDL, it seems that ApoAI, either in a pure lipid-free form or recycled from HDL, is actively recruited to the Golgi, where it binds eNOS and initiates a delayed NO release.
The magnitude and spatial and temporal pattern of ApoAI-induced NO release differed from the response induced by HDL. First, phosphorylation of eNOS in response to ApoAI incubation at nearly all sites investigated was less than HDL-induced phosphorylation. Second, NO release was detected after 10 s during HDL incubation but was detected only after 60 s with ApoAI. Finally, NO release in response to ApoAI incubation was uniform in the cell cytoplasm and perinuclear region. These effects may relate to differential effects of free or lipidated ApoAI and other potential signaling effects of the lipid component of HDL as mentioned above.
The pathway of activation of eNOS is clearly complex and involves multiple phosphorylation sites. One possible interpretation of the pathways involved is that HDL/ApoAI binding to a cell surface receptor (SR-BI or other HDL receptor) triggers endocytosis and then separation of lipid and protein moieties, which initiates an initial lipid-/estrogen-based response. The ApoAI then moves to the Golgi and binds eNOS, a process that actively recruits the necessary initiating enzymes, such as AMPK, which phosphorylates and activates the enzyme. A similar in vivo mechanism (of eNOS activation by ApoAI) is supported by human clinical studies in which it was reported that acute infusions of ApoAI phospholipid discs (reconstituted HDL) can completely normalize the decreased vasomoter response in vessels of the forearm of hypocholesterolemic men (35, 36). These outcomes were a result of direct restoration of the NO pathway. Such studies highlight the necessity for treatments that restore the NO-mediated response during diseased states of low HDL/ApoAI, and this study reveals possible therapeutic targets that may lead to such an outcome.
Supplementary Material
Acknowledgments
We thank Dr. Ian Harper (Monash Micro Imaging, Monash University, Victoria, Australia) for technical expertise with confocal imaging, Dr. Belinda Michell (St. Vincent's Institute of Medical Research, Victoria, Australia) for expertise with eNOS phosphorylation antibodies, and Belinda Ahlers (Baker Heart Research Institute, Victoria, Australia) for help with endothelial cell cultures and DAF2-DA techniques. This work was supported by grants from National Health and Medical Research Council of Australia, the National Heart Foundation of Australia, and the Australian Reseach Council. B.E.K. is a Federation Fellow.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ACC, acetyl-CoA carboxylase; AMPK, AMP activated protein kinase; ApoAI, apolipoprotein AI; BAEC, bovine aortic endothelial cells; CHO, Chinese hamster ovary; DAF2-DA, diaminofluorescein-2-diacetate; eNOS, endothelial NO synthase; HDL, highdensity lipoprotein; HUVEC, human umbilical vein endothelial cells; LDL, low-density lipoprotein; l-NMMA, NG-monomethyl-l-arginine; SNAP, S-nitroso-N-acetyl-d,l-penicillamine; SR-BI, scavenger receptor class B type I.
References
- 1.Cannon, R. O., 3rd. (1998) Clin. Chem. 44, 1809-1819. [PubMed] [Google Scholar]
- 2.Channon, K. M., Qian, H. & George, S. E. (2000) Arterioscler. Thromb. Vasc. Biol. 20, 1873-1881. [DOI] [PubMed] [Google Scholar]
- 3.Mayer, B. & Hemmens, B. (1997) Trends Biochem. Sci. 22, 477-481. [DOI] [PubMed] [Google Scholar]
- 4.Gratton, J. P., Fontana, J., O'Connor, D. S., Garcia-Cardena, G., McCabe, T. J. & Sessa, W. C. (2000) J. Biol. Chem. 275, 22268-22272. [DOI] [PubMed] [Google Scholar]
- 5.Pollock, J. S., Forstermann, U., Mitchell, J. A., Warner, T. D., Schmidt, H. H., Nakane, M. & Murad, F. (1991) Proc. Natl. Acad. Sci. USA 88, 10480-10484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shah, V., Wiest, R., Garcia-Cardena, G., Cadelina, G., Groszmann, R. J. & Sessa, W. C. (1999) Am. J. Physiol. 277, G463-G468. [DOI] [PubMed] [Google Scholar]
- 7.Cao, S., Yao, J., McCabe, T. J., Yao, Q., Katusic, Z. S., Sessa, W. C. & Shah, V. (2001) J. Biol. Chem. 276, 14249-14256. [DOI] [PubMed] [Google Scholar]
- 8.Cargnoni, A., Comini, L., Bernocchi, P., Bachetti, T., Ceconi, C., Curello, S. & Ferrari, R. (2001) Br. J. Pharmacol. 133, 145-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Uittenbogaard, A., Shaul, P. W., Yuhanna, I. S., Blair, A. & Smart, E. J. (2000) J. Biol. Chem. 275, 11278-11283. [DOI] [PubMed] [Google Scholar]
- 10.Fielding, C. J. & Fielding, P. E. (1995) J. Lipid Res. 36, 211-228. [PubMed] [Google Scholar]
- 11.Yuhanna, I. S., Zhu, Y., Cox, B. E., Hahner, L. D., Osborne-Lawrence, S., Lu, P., Marcel, Y. L., Anderson, R. G., Mendelsohn, M. E., Hobbs, H. H. & Shaul, P. W. (2001) Nat. Med. 7, 853-857. [DOI] [PubMed] [Google Scholar]
- 12.Li, X. A., Titlow, W. B., Jackson, B. A., Giltiay, N., Nikolova-Karakashian, M., Uittenbogaard, A. & Smart, E. J. (2002) J. Biol. Chem. 277, 11058-11063. [DOI] [PubMed] [Google Scholar]
- 13.Mineo, C., Yuhanna, I. S., Quon, M. J. & Shaul, P. W. (2003) J. Biol. Chem. 278, 9142-9149. [DOI] [PubMed] [Google Scholar]
- 14.Chen, Z. P., Mitchelhill, K. I., Michell, B. J., Stapleton, D., Rodriguez-Crespo, I., Witters, L. A., Power, D. A., Ortiz de Montellano, P. R. & Kemp, B. E. (1999) FEBS Lett. 443, 285-289. [DOI] [PubMed] [Google Scholar]
- 15.Boo, Y. C., Sorescu, G., Boyd, N., Shiojima, I., Walsh, K., Du, J. & Jo, H. (2002) J. Biol. Chem. 277, 3388-3396. [DOI] [PubMed] [Google Scholar]
- 16.Michell, B. J., Griffiths, J. E., Mitchelhill, K. I., Rodriguez-Crespo, I., Tiganis, T., Bozinovski, S., de Montellano, P. R., Kemp, B. E. & Pearson, R. B. (1999) Curr. Biol. 9, 845-848. [DOI] [PubMed] [Google Scholar]
- 17.Michell, B. J., Chen, Z., Tiganis, T., Stapleton, D., Katsis, F., Power, D. A., Sim, A. T. & Kemp, B. E. (2001) J. Biol. Chem. 276, 17625-17628. [DOI] [PubMed] [Google Scholar]
- 18.Fleming, I., Fisslthaler, B., Dimmeler, S., Kemp, B. E. & Busse, R. (2001) Circ. Res. 88, E68-E75. [DOI] [PubMed] [Google Scholar]
- 19.Cieslik, K., Zembowicz, A., Tang, J. L. & Wu, K. K. (1998) J. Biol. Chem. 273, 14885-14890. [DOI] [PubMed] [Google Scholar]
- 20.Allan, C. M., Fidge, N. H., Morrison, J. R. & Kanellos, J. (1993) Biochem. J. 290 (Pt 2), 449-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jaffe, E. A., Hoyer, L. W. & Nachman, R. L. (1973) J. Clin. Invest. 52, 2757-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fidge, N. H., Nestel, P. J. & Suzuki, N. (1983) Biochim. Biophys. Acta 753, 14-21. [DOI] [PubMed] [Google Scholar]
- 23.Sviridov, D., Fidge, N., Beaumier-Gallon, G. & Fielding, C. (2001) Biochem. J. 358, 79-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morrison, J. R., Fidge, N. H. & Grego, B. (1990) Anal. Biochem. 186, 145-152. [DOI] [PubMed] [Google Scholar]
- 25.Bartlett, G. R. (1959) J. Biol. Chem. 234, 466-468. [PubMed] [Google Scholar]
- 26.Fulton, D., Fontana, J., Sowa, G., Gratton, J. P., Lin, M., Li, K. X., Michell, B., Kemp, B. E., Rodman, D. & Sessa, W. C. (2002) J. Biol. Chem. 277, 4277-4284. [DOI] [PubMed] [Google Scholar]
- 27.Silver, D. L., Wang, N., Xiao, X. & Tall, A. R. (2001) J. Biol. Chem. 276, 25287-25293. [DOI] [PubMed] [Google Scholar]
- 28.Fidge, N. H. (1999) J. Lipid Res. 40, 187-201. [PubMed] [Google Scholar]
- 29.Martinez, L. O., Jacquet, S., Esteve, J. P., Rolland, C., Cabezon, E., Champagne, E., Pineau, T., Georgeaud, V., Walker, J. E., Terce, F., et al. (2003) Nature 421, 75-79. [DOI] [PubMed] [Google Scholar]
- 30.Gallis, B., Corthals, G. L., Goodlett, D. R., Ueba, H., Kim, F., Presnell, S. R., Figeys, D., Harrison, D. G., Berk, B. C., Aebersold, R. & Corson, M. A. (1999) J. Biol. Chem. 274, 30101-30108. [DOI] [PubMed] [Google Scholar]
- 31.Harris, M. B., Ju, H., Venema, V. J., Liang, H., Zou, R., Michell, B. J., Chen, Z. P., Kemp, B. E. & Venema, R. C. (2001) J. Biol. Chem. 276, 16587-16591. [DOI] [PubMed] [Google Scholar]
- 32.Michell, B. J., Harris, M. B., Chen, Z. P., Ju, H., Venema, V. J., Blackstone, M. A., Huang, W., Venema, R. C. & Kemp, B. E. (2002) J. Biol. Chem. 277, 42344-42351. [DOI] [PubMed] [Google Scholar]
- 33.Bauer, P. M., Fulton, D., Boo, Y. C., Sorescu, G. P., Kemp, B. E., Jo, H. & Sessa, W. C. (2003) J. Biol. Chem. 278, 14841-14849. [DOI] [PubMed] [Google Scholar]
- 34.Gong, M., Wilson, M., Kelly, T., Su, W., Dressman, J., Kincer, J., Matveev, S. V., Guo, L., Guerin, T., Li, X. A., et al. (2003) J. Clin. Invest. 111, 1579-1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bisoendial, R. J., Hovingh, G. K., Levels, J. H., Lerch, P. G., Andresen, I., Hayden, M. R., Kastelein, J. J. & Stroes, E. S. (2003) Circulation 107, 2944-2948. [DOI] [PubMed] [Google Scholar]
- 36.Spieker, L. E., Sudano, I., Hurlimann, D., Lerch, P. G., Lang, M. G., Binggeli, C., Corti, R., Ruschitzka, F., Luscher, T. F. & Noll, G. (2002) Circulation 105, 1399-1402. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.