Abstract
Differentiation is a highly regulated process whereby cells become specialized to perform specific functions and lose the ability to perform others. In contrast, the question of whether dedifferentiation is a genetically determined process, or merely an unregulated loss of the differentiated state, has not been resolved. We show here that dedifferentiation in the social amoeba Dictyostelium discoideum relies on a sequence of events that is independent of the original developmental state and involves the coordinated expression of a specific set of genes. A defect in one of these genes, the histidine kinase dhkA, alters the kinetics of dedifferentiation and uncouples the progression of dedifferentiation events. These observations establish dedifferentiation as a genetically determined process and suggest the existence of a developmental checkpoint that ensures a return path to the undifferentiated state.
Dedifferentiation is the progression of cells from a more differentiated to a less differentiated state. It is observed in a variety of processes such as cancer, organ regeneration, and stem cell renewal, but it has been difficult to study because there are few experimentally tractable systems that dedifferentiate (1–4). Dedifferentiation in Dictyostelium is an experimentally tractable process. During development, starving cells aggregate and undergo synchronous morphological transitions until they form fruiting bodies after 24 hr (5). If the multicellular structures are disaggregated and incubated in nutrient medium, the cells dedifferentiate. Dedifferentiation is characterized by a loss of developmental markers and a subsequent gain of proliferative capacity (6, 7, 45), but these characteristics do not prove that dedifferentiation is a regulated process. The most convincing evidence in support of regulation has been the observation that a mutant strain (HI4) was defective in dedifferentiation (8, 9).
An argument against the idea that dedifferentiation is a regulated process comes from the observation that dedifferentiation occurs at different rates, depending on the developmental stage at which the cells were disaggregated (6, 10–13). This dependence may indicate that each developmental stage has a dedicated dedifferentiation program that is executed at a different rate, or that dedifferentiation is a stochastic event whereby cells lose developmental markers and regain growth markers.
The purpose of this work was to test whether dedifferentiation is a regulated process by comparing the molecular progression of cells from different developmental stages. We propose that if we found a common set of molecular changes, which is independent of the initial developmental stage, that finding would support the regulated process hypothesis. We used microarray transcriptional profiling to detect changes in the pattern of global gene expression during dedifferentiation. These transcriptional changes reveal physiological changes without prejudice as to what processes are involved (14–18), making them suitable for testing whether dedifferentiation from different developmental stages occurs in a regulated way. We followed the physiological changes that occur in cells during dedifferentiation from three developmental stages: aggregation, finger, and Mexican hat. These stages are quite different from each other in physiology and in morphology (5), yet we found that the dedifferentiating cells exhibited common transcriptional profiles. This finding indicates that dedifferentiation is a regulated process. Examination of the coordinately regulated transcripts revealed genes that are also induced during development, raising the possibility that development and dedifferentiation are regulated by some common genes. We tested that possibility and found that one of the genes, dhkA, regulates both development and dedifferentiation.
Materials and Methods
Growth, Development, and Dedifferentiation. Wild-type Dictyostelium discoideum strains AX2 (19) and dhkA- (20) were grown in HL5 and developed as described (21). At each stage (aggregation, finger, or Mexican hat) structures were harvested by filtration through 77-μm nylon mesh, resuspended in 20 mM potassium phosphate, pH 6.4 (KK2)/20 mM EDTA and dissociated by repeated pipetting. Cells were passed through a 32-μm nylon mesh, resuspended in HL5 at 1–2 × 106 cells per ml, and shaken at 200 rpm at 22°C.
Viability. Cells were counted microscopically, 200 cells were plated in association with Klebsiella pneumoniae (also known as Klebsiella aerogenes) on nutrient agar, and plaques were counted after 3–5 days.
Redifferentiation. Redifferentiation was tested as described (6) with a minor modification: after incubation in HL5, cells were washed in KK2 and developed on filters at 22°C. Aggregation was monitored with a dissecting microscope.
BrdUrd Incorporation. Cells were shaken in HL5 supplemented with 0.5 mM BrdUrd, harvested, and washed in KK2. Genomic DNA was purified as described (22). DNA (10–50 ng) was blotted on Hybond-N+ membranes (Amersham Pharmacia), incubated with a 1:5,000 dilution of anti-BrdUrd antibody (Roche) followed by incubation with 1:40,000 dilution of horseradish peroxidase-conjugated goat anti-mouse IgG antibody (Jackson ImmunoResearch) and developed with SuperSignal West Pico Chemiluminescent Substrate (Pierce). Signals were visualized (BAS system, Fuji) and their intensity was quantified (imagegauge, Fuji).
Microarrays. Test RNA (10 μg) was labeled by reverse transcription using Cy5-conjugated (dT)18 primers, and reference RNA was labeled with Cy3-conjugated (dT)18 (18). Labeled cDNA was purified, resuspended in distilled deionized water, mixed with PerfectHyb Plus hybridization buffer (Sigma), and hybridized to arrays containing nearly 8,000 targets by using a GeneTAC hybridization station (Genomic Solutions, Ann Arbor, MI). Arrays were scanned (ScanArray5000, GSI Lumonics, Billerica, MA) and images were processed with gleams (NuTec Sciences, Atlanta). Data were processed as described (18) and combined into multiexperiment sets for all subsequent analysis (23) as detailed in the Appendix, which is published as supporting information on the PNAS web site.
Cluster Analysis and Generation of Gene Lists. The data analysis procedure is described in the Appendix. Briefly, an analysis of covariance (ANCOVA) model, which contains a categorical term for hybridization batch effects and continuous terms for representing each gene as a time function, was fitted to each time course. Clustering was performed on the smooth time fit coefficients from the three experiments. The three sets of coefficients were concatenated into a 15-element vector of coefficients for each gene and clustering was performed recursively by k-means while varying k from 2 to 10.
A directed filtering method was used to find genes that have a pattern in dedifferentiation and not in development. The filter selected for genes that: (i) have a poor fit to the developmental consensus pattern; (ii) follow the pattern Tmax(Mex) > Tmax(Finger) > Tmax(Agg), where Tmax is the time of maximal expression during dedifferentiation and Mex, Finger, and Agg represent the respective developmental stages; and (iii) whose expression level at the Tmax(Agg) in the dedifferentiation process is greater than their level of expression at the aggregation stage of development. To test for nonrandomness we generated 10,000 random filters and measured group size and variance. Our group was both unusually large and tight (P < 0.05 for both characteristics).
Gene Annotation. To annotate the putative function of selected genes, we used Gene Ontology (GO), which is a controlled vocabulary for describing gene function (24). Near-full-length cDNA sequences of each microarray target were matched to the sequences in the GO database (www.godatabase.org/dev/database). The relevant identifiers were mapped to the GO data structure. Groups with significantly high representation were identified by comparing the number of genes in the experimental group with a common GO category to the total number of genes from that category on the entire array. The data are presented graphically, with bar lengths representing the ratio between the list frequency (no. of genes in list/no. of genes at GO level) and the array group frequency (no. of genes with specific GO annotation on array/no. of all array genes at particular GO level). The x axis is the scale for that ratio.
Results
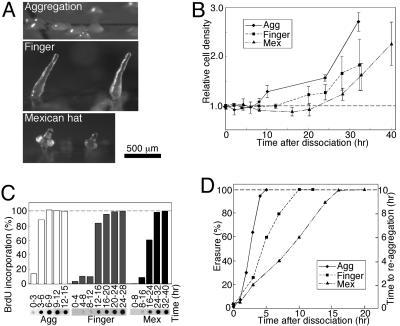
Dedifferentiation from Various Developmental Stages Occurs at Different Rates. We first established that dedifferentiation from different developmental stages occurs at different rates. Cells that developed for 10 hr (aggregates), 13 hr (fingers), and 16 hr (Mexican hats) exhibited different morphologies and physiologies (Fig. 1A). These cells were dedifferentiated by disaggregation and incubation in growth medium. Dedifferentiation was measured by using three parameters: cell division, DNA synthesis, and erasure (Fig. 1). The time of cell division after dedifferentiation was directly proportional to the initial developmental time (Fig. 1B). Aggregation stage cells started to divide after 6 hr, finger cells after 12 hr, and Mexican hat cells after 20 hr. DNA synthesis was monitored by following the incorporation of BrdUrd into nuclear DNA [differentiating Dictyostelium cells do not replicate nuclear DNA (22)]. The aggregation stage cells began to synthesize DNA after 3–6 hr, finger stage cells after 12–16 hr, and Mexican hat stage cells after 16–24 hr of dedifferentiation (Fig. 1C). Finally, we monitored the time required for dedifferentiated cells to reaggregate after dissociation. Disaggregated cells can reaggregate rapidly, and that property is lost during dedifferentiation in a process called erasure (6). We found that erasure was also directly proportional to the time the cells had been developing: aggregation, finger, and Mexican hat stage cells erased after 5, 10, and 16 hr of dedifferentiation, respectively (Fig. 1D).
Fig. 1.
Dedifferentiation markers. Cells were developed on filters to the indicated stage [Agg, aggregation (♦); Finger (▪); Mex, Mexican hat (▴)], dissociated, incubated in HL5, and sampled as indicated.(A) Developmental morphology. Aggregates appear after 10 hr, fingers after 13 hr, and Mexican hats after 16 hr of development. (B) Proliferation. Dedifferentiating cells were counted and the data were plotted relative to the initial density. Data are means and standard deviation of four experiments. (C) Nuclear DNA synthesis. Dedifferentiating cells were labeled with BrdUrd as indicated. Nuclear DNA was dot-blotted and BrdUrd incorporation was detected (photographs). Labeling intensity was plotted relative to the terminal level in each experiment (graphs). (D) Erasure. Dedifferentiating cells were starved on filters and reaggregation was determined microscopically (right y axis). Erasure is the fraction (percent, left y axis) of the time required for vegetative cells to aggregate (10 hr).
In summary, the three measurements of dedifferentiation time were internally consistent and directly proportional to the initial developmental time (Fig. 1). They confirm and extend the observation that dedifferentiation from the three stages occurs at different rates (6, 10–13). We used these three initial conditions to introduce variability into the system. We hypothesize that if dedifferentiation were a regulated process, it would be accompanied by an invariant set of transcriptional changes that is independent of the original differentiated state.
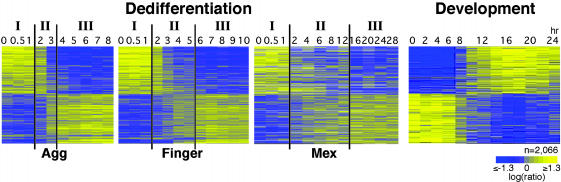
Reversal of Developmental Gene Expression Patterns in Dedifferentiation. Dedifferentiation reverses development, so we expected that the transcriptional profile of dedifferentiating cells would be reversed relative to that of developing cells. We dedifferentiated cells from the three developmental stages and analyzed their RNA with a microarray that represents nearly 6,000 genes. Previously we showed that 2,000 Dictyostelium genes are robustly coregulated during development. Half of them are up-regulated and half are down-regulated between 8 and 12 hr of development (Fig. 2, Development) (18). We found that during dedifferentiation, these genes were coregulated with a reversed pattern (Fig. 2). During the first hour of dedifferentiation, cells from the three stages exhibited similar transcriptional profiles: developmentally induced genes were expressed at a high level and the others at a low level. We define this period as phase I of dedifferentiation (Fig. 2). By the second hour, the distinction between developmentally up- and down-regulated genes became blurred (phase II). The duration of phase II was directly proportional to the length of time the cells had been developing. In aggregation cells, phase II was almost undetected at the 2-hr resolution of the experiment and the genes exhibited a fairly sharp transition. In finger cells, phase II lasted 4 hr and in Mexican hat cells it lasted 10 hr. The difference between the up- and down-regulated genes became clear again during phase III, when proliferation began. Aggregation cells entered phase III after 4 hr, finger cells after 6 hr, and Mexican hat cells after 16 hr of dedifferentiation (Fig. 2).
Fig. 2.
Transcriptional profiles of dedifferentiation. RNA samples were collected from dedifferentiating cells at the indicated times (hr). Color charts (Agg, dedifferentiation from aggregates; Finger, from fingers; Mex, from Mexican hats; Development, normal development) represent the expression pattern of 2,066 developmentally regulated genes (rows) (18). The gene order is identical in all charts. Vertical lines delineate three transcriptional phases (I, II, and III). Colors indicate lower than average (blue), average (gray), and higher than average (yellow) expression as described (18). Scale: log2 of the ratio between sample and standard.
The division into three phases was independent of the length of time the cells had been developing, but the duration of phase II was directly proportional to the initial developmental time (Fig. 2), similar to the duration of dedifferentiation described in Fig. 1. We propose that after disaggregation, the cells remain differentiated for about 1 hr (phase I). They then down-regulate the expression of the developmental genes and degrade their mRNA (phase II), and the duration of this phase is directly proportional to the length of time the cells had developed. Eventually, the cells begin to express growth genes (phase III) at the time of DNA synthesis and cell division. These results were verified by Northern blot analysis with the prespore genes cotA and DP87, the prestalk gene ecmA, and the vegetative genes V14 and V18 (data not shown).
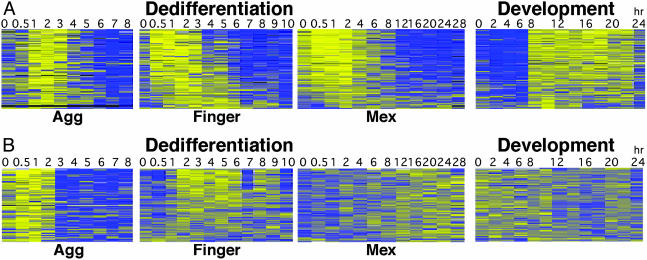
Invariant Gene Expression During Dedifferentiation. Fig. 2 showed that developmentally coregulated genes are also coregulated during dedifferentiation. To find dedifferentiation-specific genes, we performed cluster analysis and selected genes that were up-regulated during phase II, regardless of the length of time the cells had developed (Fig. 3A). Expression of these 272 genes was observed at 1–3 hr of dedifferentiation in the aggregation cells, 0.5–5 hr in the finger cells, and 0–8 hr in the Mexican hat cells. Therefore, the down-regulation time was directly proportional to the initial developmental time, but the time of induction was inversely proportional. This observation suggested that the dedifferentiation genes have already been expressed during late development. We therefore examined the expression of these genes during development and found them to be sharply induced after 8 hr (Fig. 3A, Development). This finding accounts for the early expression during dedifferentiation of finger (13-hr) and Mexican hat (16-hr) cells and suggests that some dedifferentiation genes have a role in development as well.
Fig. 3.
Dedifferentiation-specific gene expression. Data were collected as in Fig. 2. Time (hr) is indicated above and the developmental stage below each chart (Agg, dedifferentiation from aggregates; Finger, from fingers; Mex, from Mexican hats; Development, normal development). (A) The 259 genes up-regulated in phase II. The gene order is identical in all charts (the genes are represented by 272 targets). (B) The 122 genes coregulated during dedifferentiation but not during normal development.
The results in Fig. 3A raised the possibility that all dedifferentiation genes are also developmentally regulated. To test that possibility, we used a filtering method to find genes that are induced only during dedifferentiation; we found 122 genes that were coordinately regulated during dedifferentiation but not during development (Fig. 3B). The grouping of these genes was independent of the original developmental stage, but their time of maximal expression was directly proportional to the original developmental time. Aggregation cells expressed these genes as soon as dedifferentiation began and turned them down after 3 hr of dedifferentiation; finger cells expressed them between 2 and 6 hr; and Mexican hat cells, from 8 hr on (Fig. 3B). The gene group was found to be highly statistically significant for its large size and its low variance (P < 0.05 for both characteristics).
The data in Fig. 3 show that dedifferentiation is accompanied by the regulation of an invariant group of genes. This finding strongly supports the idea that dedifferentiation is a regulated process, because a random process would not be expected to induce consistent gene expression profiles from cells that begin the process from different developmental stages.
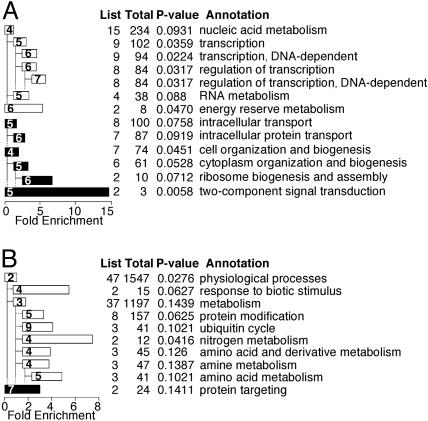
Biological Processes Implicated by Dedifferentiation-Specific Gene Expression. Genes that may have important roles in biological processes can be found by annotation of gene groups discovered by microarray experiments (15), despite the fact that there is little correlation between function and expression of individual genes (25, 26). Because the annotation of the Dictyostelium genome is incomplete, we compared the gene sequences with sequences of several well annotated genomes. We then assigned the GO classification of the closest homologue to the respective Dictyostelium gene and used the classifications to annotate the coregulated genes in Fig. 3. The chart in Fig. 4A describes the GO annotation of the group described in Fig. 3A. This group contains genes that participate in transcriptional regulation and RNA metabolism (Fig. 4A, white bars). Interestingly, the maximal expression of these RNA metabolism genes (Fig. 3A) coincides with the time of major changes in gene expression (Fig. 2), suggesting a true functional correlation. Other significant groups of genes suggest a role for protein transport and ribosome biogenesis (Fig. 4A, black bars). The most significant group is the two-component signal transduction system (Fig. 4A, bottom black bar), suggesting the intriguing possibility that signaling is involved in the regulation of dedifferentiation.
Fig. 4.
Annotation of dedifferentiation genes. Genes from Fig. 3 were GO-annotated and the “biological process” annotation of significantly enriched groups is shown (P values are indicated below). The GO tree levels of the “biological process” annotation are shown as numbers inside bars (ranging from 2 to 9). The table on the right indicates the number of genes in each group (List), genes with that annotation on the entire array (Total), the P value, and the annotation. Bar lengths represent the fold enrichment (scale, x axis). Indented bars are subgroups of bars immediately above, as indicated by the branching pattern. Bar colors represent the group annotation at GO level 2: white, physiological process; black, cellular process. (A) Genes regulated during phase II of dedifferentiation and during development as shown in Fig. 3A (P < 0.1). (B) Genes regulated during phase II of dedifferentiation but not during development as shown in Fig. 3B (P < 0.15).
Fig. 4B shows a similar analysis of the genes from Fig. 3B. It implicates genes that participate in protein modification (probably protein degradation due to the ubiquitin cycle annotation) and amino acid metabolism (Fig. 4B, white bars) as well as protein targeting (Fig. 4B, black bar). These findings may reflect an adaptation of the cells to the exogenous source of nutrients as they cease the developmental utilization of endogenous proteins as an energy source (5).
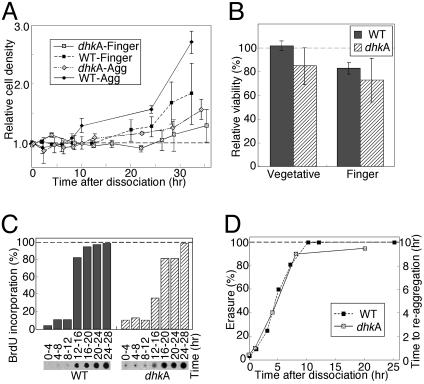
A Dedifferentiation-Induced Gene Is Required for Dedifferentiation. The above analysis implicated several groups of genes as being involved in dedifferentiation, but it only provided reasonable correlations. To begin to test whether dedifferentiation-induced genes have a causative relationship to the process, we tested the effect of one gene on dedifferentiation. We selected dhkA because it is a two-component signal transduction system gene up-regulated during phase II (Fig. 3) and because it has a known role in signal transduction (20, 27). dhkA- cells were developed to the finger stage, disaggregated, and incubated in nutrient medium to induce dedifferentiation. Monitoring cell number revealed that the mutant cells dedifferentiated more slowly than the wild type (Fig. 5A, WT). dhkA- cells started to proliferate after 25–30 hr, whereas stage-matched wild-type cells divided after 12 hr. To exclude the possibility that the difference between the wild type and the dhkA- mutant was due to cell death, we measured viability by testing plating efficiency. We found that the two strains were nearly indistinguishable in viability during growth (vegetative) and at the finger stage of development (Fig. 5B). The delayed dedifferentiation of the mutant strain suggests that dhkA has a role in dedifferentiation.
Fig. 5.
Dedifferentiation of dhkA- cells. Wild-type and mutant aggregate (WT, solid diamonds; dhkA-, hatched diamonds) and finger cells (WT, solid squares; dhkA-, hatched squares) were dedifferentiated and sampled as indicated. (A) Growth. Cells were counted and the data are plotted relative to the initial density. Data are means and standard deviation of three or four experiments. The WT data are from Fig. 1 A.(B) Viability. Plating efficiency of vegetative and dissociated finger cells was measured. Viability is the percentage of microscopically visible cells that formed plaques. Results are means and standard deviations of three experiments. WT, gray bars; dhkA-, hatched bars. (C) Nuclear DNA synthesis. Cells were labeled with BrdUrd; incorporation into nuclear DNA was determined and quantified as in Fig. 1 (photographs). WT, gray bars; dhkA-, hatched bars. (D) Erasure. Dedifferentiating cells were starved on filters and reaggregation time was monitored microscopically. Erasure is the fraction (percent, left x axis) of the time required for vegetative cells to aggregate (10 hr).
Monitoring nuclear DNA synthesis revealed that the mutant began to synthesize DNA at 12–16 hr of dedifferentiation, much like the wild type (Fig. 5C). Moreover, in reaggregation experiments, dhkA- cells erased after 8–10 hr, just like the wild type (Fig. 5D). These results show that dhkA does not affect dedifferentiation by regulating DNA replication or erasure. Therefore, dhkA has a specific effect on dedifferentiation because it uncouples cell division from DNA synthesis and erasure. This finding further supports the idea that dedifferentiation is a regulated, multistep process because the steps can be separated by a specific mutation.
Discussion
Our data support the hypothesis that dedifferentiation is a regulated process because cells induced to dedifferentiate from three developmental stages underwent an identical set of transcriptional changes. In the first hour of dedifferentiation, developmental genes were still abundant in the cells and some dedifferentiation genes began to be expressed. During the second phase of dedifferentiation, the abundance of the developmental transcripts was greatly reduced, indicating a massive down-regulation of gene expression and a degradation of many transcripts. This process was also accompanied by the induction of 381 genes. Annotation of these genes revealed that some are probably related to mRNA metabolism and regulation of gene expression, likely relations in light of the massive changes in transcript abundance during phase II. Finally, in phase III, the cells began to express vegetative genes. The compelling finding is that the sequence of events and the specific regulation of several hundred dedifferentiation genes were independent of the initial developmental stage and the consequential variable rates of dedifferentiation. In addition, all of the measured parameters were coordinately regulated: the microarray profiles, DNA synthesis, cell division, and erasure. It is unlikely that such coordination could result from a stochastic unregulated process.
Annotation of the dedifferentiation genes revealed the possible involvement of several processes, including signal transduction, ubiquitin-mediated proteolysis, and regulation of transcription. The signal transduction genes are from the two-component system family (28, 29), including the histidine kinase gene dhkA, a hybrid histidine kinase essential for proper terminal differentiation (20, 27).
It is tempting to speculate about the function of a gene in dedifferentiation based on its annotation, but a functional test is better. We found that the signal transduction gene dhkA was essential for dedifferentiation, supporting the idea that dedifferentiation is a genetically regulated process because it is mutable. This finding also indicates that signal transduction is involved, and it adds confidence to the interpretation that the other gene groups may regulate dedifferentiation. The promise of manipulating the process genetically is supported by previous studies of a strain (HI4) defective in dedifferentiation (8, 9, 13). Unfortunately, the gene mutated in HI4 could not be cloned at the time and the strain has been lost. The dedifferentiation characteristics of dhkA- cells also indicate that regulation of dedifferentiation is a stepwise process because dhkA is required only for cell division, not for DNA replication or for erasure.
Finally, our findings suggest that dedifferentiation and development have coevolved while using common genes, because dhkA is a regulator of both processes and several hundred genes are regulated during both processes. We propose that development consists of checkpoints that ensure a return path to the undifferentiated state in case development fails. The checkpoint could condition developmental progression on the accumulation of a protein, such as DhkA, that is essential for dedifferentiation. If development cannot proceed, the cells may attempt to reinitiate development as some strains undergo several cycles of development and dedifferentiation due to mutations in gene of the lagC pathway (30). However, dedifferentiation is not merely a reversal of development because it involves the coordinate regulation of 122 genes in patterns not seen in development.
The selective advantage of dedifferentiation is obvious, but the need for an intricately regulated process is intriguing. In vertebrate wound healing and regeneration, regulation may attenuate dedifferentiation to preserve information about tissue proportioning (31, 32). The lack of regulation may be a causative factor in cancer and other diseases (1–3, 33–41). Dictyostelium is a soil amoeba, and the sheath that surrounds the multicellular organism cannot protect it from all of the mechanical insults that the soil environment may inflict. Therefore, Dictyostelium dedifferentiation probably evolved under the pressure of occasional mechanical disaggregation during development. Regulating dedifferentiation allows the cells to reaggregate rapidly and continue to develop if starvation persists. It also provides an alternative: in the presence of food, cells can return to the more advantageous process of growth. Because Dictyostelium and higher eukaryotes share many signal transduction mechanisms (42–44), further investigation of dedifferentiation in this model organism may help identify molecules that regulate dedifferentiation in vertebrates.
Supplementary Material
Acknowledgments
We are grateful to N. Whitehouse for assistance with the GO annotation and to A. Kuspa, V. Lundblad, P. Hastings, and C. Thompson for critique and discussion. The Japanese cDNA project team is grateful to Prof. N. Ogasawara of the Nara Institute of Science and Technology for supporting the project. This work was supported by National Institute of Child Health and Human Development Grant P01 HD39691-01. Work in Japan was supported by the Japan Society for the Promotion of Science–Research for the Future Program (96L00105 and 00L01412) and a Grant-in-Aid for Scientific Research on Priority Area C (12206001).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviation: GO, Gene Ontology.
References
- 1.Gimm, O. (2001) Cancer Lett. 163, 143-156. [DOI] [PubMed] [Google Scholar]
- 2.Dominguez-Malagon, H. & Gaytan-Graham, S. (2001) Ultrastruct. Pathol. 25, 497-516. [DOI] [PubMed] [Google Scholar]
- 3.Matias-Guiu, X., Catasus, L., Bussaglia, E., Lagarda, H., Garcia, A., Pons, C., Munoz, J., Arguelles, R., Machin, P. & Prat, J. (2001) Hum. Pathol. 32, 569-577. [DOI] [PubMed] [Google Scholar]
- 4.Tsonis, P. A. (2000) Dev. Biol. 221, 273-284. [DOI] [PubMed] [Google Scholar]
- 5.Loomis, W. F. (1975) Dictyostelium discoideum: A Developmental System (Academic, New York).
- 6.Waddell, D. R. & Soll, D. R. (1977) Dev. Biol. 60, 83-92. [DOI] [PubMed] [Google Scholar]
- 7.Soll, D. R. & Waddell, D. R. (1975) Dev. Biol. 47, 292-302. [DOI] [PubMed] [Google Scholar]
- 8.Soll, D. R., Mitchell, L. H., Hedberg, C. & Varnum, B. (1984) Dev. Genet. 4, 167-184. [Google Scholar]
- 9.Kraft, B., Chandrasekhar, A., Rotman, M., Klein, C. & Soll, D. R. (1989) Dev. Biol. 136, 363-371. [DOI] [PubMed] [Google Scholar]
- 10.Finney, R., Varnum, B. & Soll, D. R. (1979) Dev. Biol. 73, 290-303. [DOI] [PubMed] [Google Scholar]
- 11.Finney, R. E., Mitchell, L. H., Soll, D. R., Murray, B. A. & Loomis, W. F. (1983) Dev. Biol. 98, 502-509. [DOI] [PubMed] [Google Scholar]
- 12.Finney, R., Ellis, M., Langtimm, C., Rosen, E., Firtel, R. & Soll, D. R. (1987) Dev. Biol. 120, 561-576. [DOI] [PubMed] [Google Scholar]
- 13.Alexander, S., Cibulsky, A. M., Mitchell, L. & Soll, D. R. (1985) Differentiation 30, 1-6. [DOI] [PubMed] [Google Scholar]
- 14.Alizadeh, A. A., Eisen, M. B., Davis, R. E., Ma, C., Lossos, I. S., Rosenwald, A., Boldrick, J. C., Sabet, H., Tran, T., Yu, X., et al. (2000) Nature 403, 503-511. [DOI] [PubMed] [Google Scholar]
- 15.Hughes, T. R., Marton, M. J., Jones, A. R., Roberts, C. J., Stoughton, R., Armour, C. D., Bennett, H. A., Coffey, E., Dai, H., He, Y. D., et al. (2000) Cell 102, 109-126. [DOI] [PubMed] [Google Scholar]
- 16.Good, J., Cabral, M., Sharma, S., Yang, J., Van Driessche, N., Shaw, C., Shaulsky, G. & Kuspa, A. (2003) Development (Cambridge, U.K.) 130, 2953-2965. [DOI] [PubMed] [Google Scholar]
- 17.Kibler, K., Nguyen, T. L., Svetz, J., Van Driessche, N., Ibarra, M., Thompson, C., Shaw, C. & Shaulsky, G. (2003) Dev. Biol. 259, 193-208. [DOI] [PubMed] [Google Scholar]
- 18.Van Driessche, N., Shaw, C., Katoh, M., Morio, T., Sucgang, R., Ibarra, M., Kuwayama, H., Saito, T., Urushihara, H., Maeda, M., et al. (2002) Development (Cambridge, U.K.) 129, 1543-1552. [DOI] [PubMed] [Google Scholar]
- 19.Watts, D. J. & Ashworth, J. M. (1970) Biochem. J. 119, 171-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang, N., Shaulsky, G., Escalante, R. & Loomis, W. F. (1996) EMBO J. 15, 3890-3898. [PMC free article] [PubMed] [Google Scholar]
- 21.Shaulsky, G. & Loomis, W. F. (1993) Dev. Biol. 160, 85-98. [DOI] [PubMed] [Google Scholar]
- 22.Shaulsky, G. & Loomis, W. F. (1995) Proc. Natl. Acad. Sci. USA 92, 5660-5663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bolstad, B. M., Irizarry, R. A., Astrand, M. & Speed, T. P. (2003) Bioinformatics 19, 185-193. [DOI] [PubMed] [Google Scholar]
- 24.Asburner, M., Ball, C., Blake, J., Botstein, D., Butler, H., Cherry, J., Davis, A., Dolinski, K., Dwight, S., Eppig, J., et al. (2000) Nat. Genet. 25, 25-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giaever, G., Chu, A. M., Ni, L., Connelly, C., Riles, L., Veronneau, S., Dow, S., Lucau-Danila, A., Anderson, K., Andre, B., et al. (2002) Nature 418, 387-391. [DOI] [PubMed] [Google Scholar]
- 26.Winzeler, E. A., Shoemaker, D. D., Astromoff, A., Liang, H., Anderson, K., Andre, B., Bangham, R., Benito, R., Boeke, J. D., Bussey, H., et al. (1999) Science 285, 901-906. [DOI] [PubMed] [Google Scholar]
- 27.Wang, N., Soderbom, F., Anjard, C., Shaulsky, G. & Loomis, W. F. (1999) Mol. Cell. Biol. 19, 4750-4756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thomason, P. & Kay, R. (2000) J. Cell Sci. 113, 3141-3150. [DOI] [PubMed] [Google Scholar]
- 29.Loomis, W. F., Shaulsky, G. & Wang, N. (1997) J. Cell Sci. 110, 1141-1145. [DOI] [PubMed] [Google Scholar]
- 30.Kibler, K., Svetz, J., Nguyen, T. L., Shaw, C. & Shaulsky, G. (2003) Dev. Biol. 264, 506-521. [DOI] [PubMed] [Google Scholar]
- 31.Echeverri, K. & Tanaka, E. M. (2002) Semin. Cell Dev. Biol. 13, 353-360. [DOI] [PubMed] [Google Scholar]
- 32.Liu, Y. & Rao, M. S. (2003) J. Cell. Biochem. 88, 29-40. [DOI] [PubMed] [Google Scholar]
- 33.Clerc, P., Saillan-Barreau, C., Desbois, C., Pradayrol, L., Fourmy, D. & Dufresne, M. (2002) Pharmacol. Toxicol. 91, 321-326. [DOI] [PubMed] [Google Scholar]
- 34.Aigner, T. (2002) Virchows Arch. 441, 219-230. [DOI] [PubMed] [Google Scholar]
- 35.Dei Tos, A. P. (2000) Ann. Diagn. Pathol. 4, 252-266. [DOI] [PubMed] [Google Scholar]
- 36.Arendt, T. (2000) Neurobiol. Aging 21, 783-796. [DOI] [PubMed] [Google Scholar]
- 37.Arendt, T., Holzer, M., Stobe, A., Gartner, U., Luth, H. J., Bruckner, M. K. & Ueberham, U. (2000) Ann. N.Y. Acad. Sci. 920, 249-255. [DOI] [PubMed] [Google Scholar]
- 38.Heusch, G. & Schulz, R. (2002) Ital. Heart J. 3, 282-284. [PubMed] [Google Scholar]
- 39.Khan, I. A. (2003) Am. Heart J. 145, 787-794. [DOI] [PubMed] [Google Scholar]
- 40.Previtali, M. (2001) Ital. Heart J. 2, 93-99. [PubMed] [Google Scholar]
- 41.Vignola, A. M., Gagliardo, R., Siena, A., Chiappara, G., Bonsignore, M. R., Bousquet, J. & Bonsignore, G. (2001) Curr. Allergy Asthma Rep. 1, 108-115. [DOI] [PubMed] [Google Scholar]
- 42.Chung, C. Y., Funamoto, S. & Firtel, R. A. (2001) Trends Biochem. Sci. 26, 557-566. [DOI] [PubMed] [Google Scholar]
- 43.Parent, C. A. & Devreotes, P. N. (1999) Science 284, 765-770. [DOI] [PubMed] [Google Scholar]
- 44.Coates, J. C. & Harwood, A. J. (2001) J. Cell Sci. 114, 4349-4358. [DOI] [PubMed] [Google Scholar]
- 45.Takeuchi, I. & Sakai, Y. (1971) Devel. Growth Differ. 13, 201-210. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.