Abstract
The completion of the Human Genome Project aroused renewed interest in alternative splicing, an efficient and widespread mechanism that generates multiple protein isoforms from individual genes. Although our knowledge about alternative splicing is growing exponentially, its real impact on cellular life is still to be clarified. Connecting all splicing features (genes, splice transcripts, isoforms, and relative functions) may be useful to resolve this tangle. Herein, we will start from the case of a single gene, Parkinson protein 2, E3 ubiquitin protein ligase (PARK2), one of the largest in our genome. This gene is implicated in the pathogenesis of autosomal recessive juvenile Parkinsonism and it has been recently linked to cancer, leprosy, autism, type 2 diabetes mellitus and Alzheimer’s disease. PARK2 primary transcript undergoes an extensive alternative splicing, which enhances transcriptomic diversification and protein diversity in tissues and cells. This review will provide an update of all human PARK2 alternative splice transcripts and isoforms presently known, and correlate them to those in rat and mouse, two common animal models for studying human disease genes. Alternative splicing relies upon a complex process that could be easily altered by both cis and trans-acting mutations. Although the contribution of PARK2 splicing in human disease remains to be fully explored, some evidences show disruption of this versatile form of genetic regulation may have pathological consequences.
Keywords: Alternative splicing, mRNA, PARK2, Protein isoforms, Splice variants, Splice expression patterns.
INTRODUCTION
With the completion of the Human Genome Project, it came as a surprise to discover that human genome contains only a fraction of genes than originally predicted. It was clear that the small number of human genes could not account for the complexity of the proteome. Therefore, the biological paradigm “one gene to one protein” was groundless. Among several proposed mechanisms, alternative splicing is considered the major driving force for transcriptome and proteome diversity. Because of its ability in increasing the coding potential of a genome, alternative splicing represents a cheap and powerful tool that allows cells to expand their proteome, producing multiple protein products from a single gene. Although our knowledge about alternative splicing is growing exponentially, its real impact on cellular life is still under debate. Connecting each gene to its splice transcripts, corresponding isoforms and relative functions may be useful to resolve this tangle and decipher how splicing acts in physiological and pathological conditions. Indeed, this is not an easy task. In this review, we will represent thesplicing features of a single gene, Parkinson protein 2, E3 ubiquitin protein ligase gene (PARK2), one of the largest genes in human genome [1].
Mutations in PARK2 gene are responsible for the development of a form of autosomal recessive juvenile Parkinsonism (AR-JP) characterized by all the classical symptoms of Parkinson disease (PD), such as tremor, rigidity and bradykinesia [2]. In addition to AR-JP, PARK2 has been recently linked to cancer [3, 4], leprosy [5], autism [6], type 2 diabetes mellitus [7] and Alzheimer’s disease [8].
PARK2 gene spans more than 1.38 Mb of genomic DNA in the long arm of chromosome 6 (6q25.2-q27) [1, 9]. To date, homologous PARK2 genes have been characterized in twelve different organisms, including rat [10], mouse [11, 12], fruit fly [13], zebrafish [14] and worm [15].
The first isolated human PARK2 transcript was of 2,960 bases with an open reading frame (1,395 bases) encoding a protein of 465 amino acids [1]. Based on this transcript, the genomic organization and exon/intron boundary sequences of PARK2 consisted of 12 exons [1]. In the last fifteen years, these 12 exons have been the focus of hundreds of different screenings. The Parkinson Disease Mutation Database (http://www.molgen.vib-ua.be/PDmutDB) [16] currently lists 214 PARK2 mutations: exon rearrangements (deletions, duplications) or, more often, point mutations.
While many studies concentrated on the genetic variations present in the 12 originally established PARK2 exons and in their exon/intron boundaries, there is now enough evidence that additional exonic sequences exist in human and other species, and that they can be alternatively spliced to produce different variants [11, 17-22]. These transcripts show different patterns of expression and encode proteins with different functions [11, 17-22].
This review will provide an update of all human PARK2 alternative splice variants presently known and correlate them to those in rat and mouse, two common animal models for studying human disease genes. Before describing PARK2 splice variants, the next paragraph will briefly introduce the process of alternative splicing.
ALTERNATIVE SPLICING, BASIC CONCEPTS
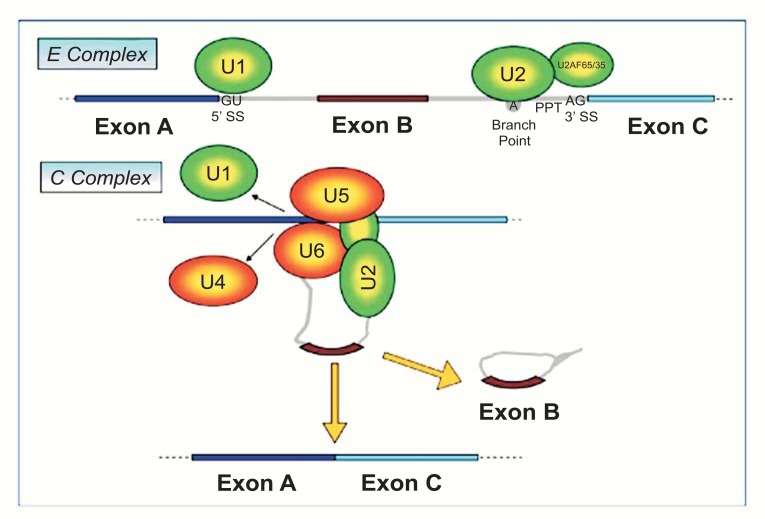
A crucial regulatory stage in the pathway of gene expression is splicing of precursor mRNA (pre-mRNA). In this process, introns are removed and exons are joined to form a mature mRNA, which is then polyadenylated, exported to cytoplasm, and translated into protein. Four conserved regions are important for the splicing process: the 5’ (GU) and the 3’ (AG) splice sites, the branch point placed upstream of the 3’ splice site and the polypyrimidine tract (PPT) placed between the 3’ splice site and the branch point (Fig. 1). Splicing regions are recognized by a large macromolecular complex, the spliceosome, which models the pre-mRNA while it is synthesized by RNA polymerase II in the nucleus (Fig. 1). The splicing machinery is composed of five small nuclear RNAs (U1, U2, U4, U5 and U6) that tie with proteins to form small nuclear ribonucleoproteins (snRNPs) [23]. During the first step of spliceosome assembly (E complex), U1 base-pairs with the 5’-splice site, whereas U2 base-pairs with the branch-point. Then, the tri-snRNP complex U4, U5 and U6 associates with the forming spliceosome (now called B complex) and U4 is ejected. This allows U6 to replace U1 at the 5’ splice site (C complex) and leads to a U6–U2 interaction that gets close together to the 5’-splice site and the branch point, allowing for a transesterification step. At the end, U5 brings near the two exons and allows for the second step of splicing, joining the two exons [24].
Fig. (1).
The alternative splicing mechanism.The spliceosome machinery (U1, U2, U4, U5 and U6) assembles on the nascent pre-mRNA. The conserved sequences that enable recognition of the mRNA by the spliceosome are: the 5’ splice site (GU), the 3’ splice site (AG), the branch point and the polypyrimidine tract (PPT). In E complex, U1 forms a base-pairing interaction with the 5’-splice site, whereas U2 base-pairs with the branch-point. Then, a tri-snRNP complex containing U4, U5 and U6 associates with the forming spliceosome, removing U1 and U4 (C complex). These steps allow the two transesterification reactions and join the exons.
Splicing of exons does not always proceed in the same manner and different combinations of exons can be joined by a process known as alternative splicing. Alternative splicing of coding exons may generate protein isoforms with different biological properties, protein-protein interactions, subcellular localization, signaling pathway or catalytic ability [25]. Alternative splicing in non-coding sequences, instead, can affect the efficiency of mRNA translation, stability or localization [26]. By modifying the reading frame or adding premature stop codons, some splicing events lead to truncated proteins or “Nonsense-Mediated mRNA Degradation” (NMD mechanism) [27]. In this manner, splicing works as an on–off switch in gene expression. Alternative splicing is regulated in time and space, allowing a particular mRNA to be expressed in a specific cell and physiological condition [28]. This process, therefore, represents an extremely economical mean of increasing protein diversity, which can finely tune genomic information to meet the unique needs of each cell. The process of alternative splicing increases the number of the mRNA expressed and explains the divergence between the estimated 24,000 human protein-coding genes and the 100,000 different proteins that are supposed to be generated [29].
ALTERNATIVE SPLICING OF PARK2
In the following paragraphs we will first describe all PARK2 alternative splice variants presently known in human, followed by those in rat and mouse, two common animal models for studying human disease genes.
Human PARK2 Alternative Splice Variants
PARK2 is one of the largest genes in the human genome and spans more than 1.38 Mb of genomic DNA in the long arm of chromosome 6 (6q25.2-q27).
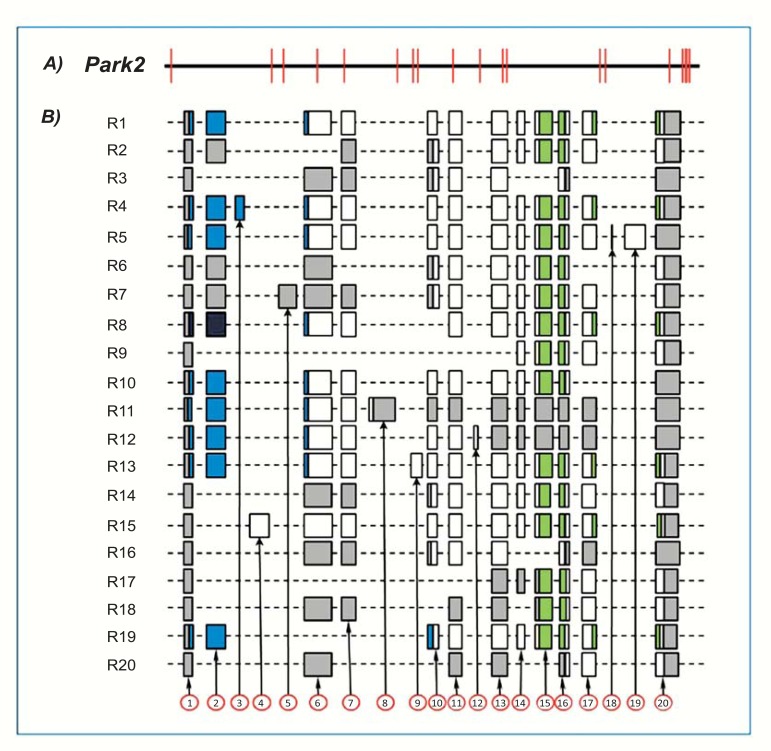
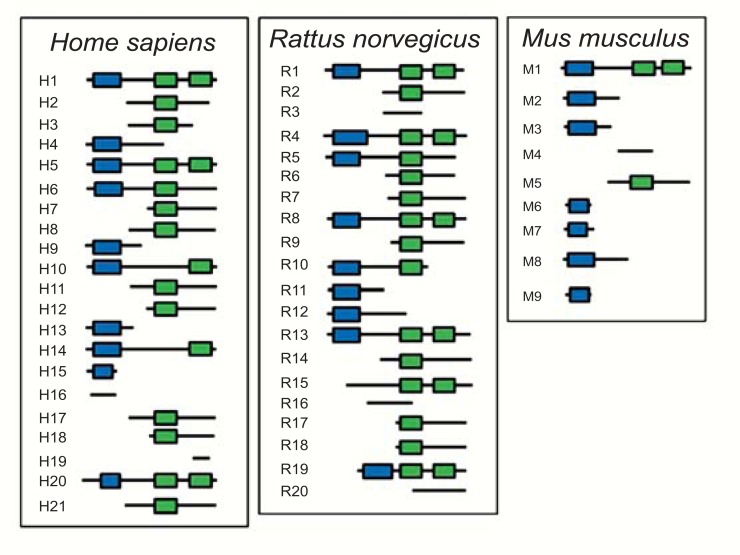
To date, GenBank (Unigene cluster Hs.132954) currently lists 26 human PARK2 transcripts corresponding to 21 different alternative splice variants. Each of these alternative splice variants is shown in (Table 1). The alignment of genomic and transcript sequences (Fig. 2) indicates these alternative splice variants are composed of 17 exons, whose exact length and specific coordinates on PARK2 gene are indicated in (Table 2). With the exception of few exons (9, 11, and 14), each exclusively expressed in a single splice variant, the others are not associated to specific transcripts (Fig. 2). The joining of different exons does not seem to follow any specific order. Indeed, no exonic cluster (i.e. exonic sequences lying close in gene and spliced always, in or out, together) is evident.
Table 1.
Homo sapiens PARK2 splice variants.
| GI PARK2 mRNAs | Code Identifier | Corresponding Homologs |
|---|---|---|
| 3063387; 121308969; 125630744; 158258616; 169790968* | H1 | R1 - M1 |
| 20385797 | H2 | R2 - M7 |
| 20385801 | H3 | R6 |
| 34191069 | H4 | |
| 169790970*; 284468410 | H5 | R8 |
| 169790972* | H6 | |
| 194378189 | H7 | |
| 284468407 | H8 | R14 - M5 |
| 284468408 | H9 | |
| 284468412 | H10 | |
| 284516981 | H11 | |
| 284516982 | H12 | |
| 284516983 | H13 | |
| 284516985 | H14 | |
| 284516987 | H15 | |
| 284516989 | H16 | |
| 284516991 | H17 | |
| 284516993 | H18 | |
| 469609974 | H19 | |
| 469609976 | H20 | |
| 520845529 | H21 |
Gene identifiers corresponding to human (H) PARK2 splice variants currently known are reported. For convenience, a new code identifier based on submission date has been assigned to each variant. In some cases, different submissions (with different Gene Identifiers) exist for the same splice variant. Three records (marked with an asterisk) are Reviewed Reference Sequences (RefSeq) that have been curated by NCBI staff, by assembling transcript and genomic sequences derived from DB023187.1, AK292590.1 and AL32982.12. These RefSeq records include a subset of the publications that are avaible for PARK2 gene. However, we have no certainty of these full lengths, because they are not supported by direct cloning and submitted sequences. Homologous transcripts in rat (R) and mouse (M) are reported in the table; for their Gene Identifiers the reader is referred to Tables 3 and 4.
Fig. (2).
Human PARK2 gene and exonic structure of alternative splice variants. A: Exonic and intronic organization of human PARK2 gene. Exons are represented as red bars. The size of introns (black line) is proportional to their length. B: Exon organization map of the 21 human PARK2 splice variants currently known. Exons are represented by shaded boxes (gray for non coding sequence, white for coding sequence, blue for UBQ domain and green for IBR domains) with a size proportional to their length. The first (1) and last (17) exons are represented entirely, although their sequence is partial in some variants (H1-H5, H7-H21).
Table 2.
Names, gene coordinates and length of human, rat and mouse PARK2 exons.
| Homo Sapiens | Rattus Norvegicus | Mus Musculus | ||||||
|---|---|---|---|---|---|---|---|---|
| Exon | Length | Coordinates on PARK2 Gene | Exon | Length | Coordinates on Park2 Gene | Exon | Length | Coordinates on Park2 Gene |
| 1 | 141 | 5.001-5.141 | 1 | 77 | 178.383-178.459 | 1 | 106 | 183.449-183.554 |
| 2 | 97 | 61.313-61.409 | ||||||
| 3 | 98 | 161.506-161.603 | ||||||
| 4 | 164 | 289.330-289.493 | 2 | 164 | 403.508-403.671 | 2 | 164 | 410.194-410.357 |
| 3 | 72 | 427.602-427.673 | ||||||
| 4 | 154 | 438.685-438.838 | ||||||
| 5 | 156 | 503.452-503.607 | ||||||
| 5 | 241 | 470.038-470.278 | 6 | 241 | 564.217-564.457 | 3 | 241 | 580.527-580.767 |
| 6 | 122 | 531.551-531.672 | 7 | 122 | 627.620-627.741 | 4 | 122 | 646.655-646.776 |
| 8 | 237 | 659.191-659.427 | ||||||
| 9 | 90 | 673.136-673.225 | ||||||
| 7 | 84 | 678.629-678.712 | 10 | 84 | 754.421-754.504 | 5 | 84 | 777.615-777.698 |
| 8 | 116 | 759.386-759.501 | 11 | 116 | 819.899-820.014 | 6 | 116 | 835.744-835.859 |
| 7 | 587 | 899.713-900.299 | ||||||
| 12 | 35 | 950.478-950.512 | ||||||
| 9 | 70 | 895.786-895.855 | ||||||
| 10 | 137 | 946.895-947.031 | 13 | 137 | 953.668-953.804 | 8 | 137 | 978.375-978.511 |
| 11 | 57 | 1016.672-1016.728 | ||||||
| 12 | 62 | 1163.387-1163.448 | 14 | 62 | 1159.331-1159.392 | 9 | 62 | 1181.667-1181.728 |
| 10 | 154 | 1188.275-1188.428 | ||||||
| 13 | 150 | 1183.800-1183.949 | 15 | 150 | 1174.875-1175.024 | 11 | 150 | 1197.792-1197.941 |
| 14 | 326 | 1187.366-1187.691 | ||||||
| 12 | 895 | 1209.731-1210.625 | ||||||
| 15 | 84 | 1345.926-1346.009 | 16 | 84 | 1323.441-1323.524 | 13 | 84 | 1347.110-1347.193 |
| 16 | 118 | 1372.598-1372.715 | 17 | 118 | 1356.481-1356.598 | 14 | 118 | 1393.691-1393.808 |
| 18 | 8 | 1362.100-1362.107 | ||||||
| 19 | 172 | 1362.156-1362.327 | ||||||
| 17 | 2654 | 1382.592-1385.245 | 20 | 209 | 1367.384-1367.592 | 15 | 1821 | 1404.606-1406.426 |
Names, coordinates on human (NG_008289.1), rat (NC_005100.3, selected region from base 49505976 to 51051947) and mouse (NC_000083.6, selected region from base 10656936 to 12246807) genes and length (bp) of PARK2 exons are reported. For convenience, in this work, exons have been renamed consecutively and in ascending order. Homologous exons among different species are reported on the same row.
The cDNA clone (2,960 bp) submitted by Kitada et al. (Accession number AB009973.1) [1] represents the longest transcript sequence present in GenBank, although the same repository contains three records of PARK2 Reference Sequences with a length between 3 and 4 Kbp. These RefSeq sequences have been generated by NCBI staff by assembling transcript and genomic sequences, and their existence remains uncertain (for further details see Table 1). The shortest human PARK2 transcript variant is 454 bp. This means that PARK2 undergoes a complicated pattern of splicing assembling that greatly reduces primary transcript length up to 3000 times.
Rat Park2 Alternative Splice Variants
Rat Park2 gene is located in the long arm of chromosome 1 and spans more than 1.18 Mb. The cloned rat Park2 transcripts range between 1670 and 534 bp, and thus the splicing assembling strongly reduces the length of the primary transcript more than 2200 times.
GenBank (Unigene cluster Rn.207194) currently lists 27 rat Park2 transcripts (Table 3) that correspond to 20 unique alternative splice variants (Fig. 3). These alternative splice variants are composed of 20 exons, which are reported in (Table 2). As in human, some exons are uniquely present in a single splice variant (exons 3, 4, 5, 8, 9, 12, 18, and 19). Based on the exonic composition of transcript variants, no exonic cluster or preferential rearrangement is evident.
Table 3.
Rattus norvegicus Park2 splice variants.
| GI Park2 mRNAs | Code Identifier | Corresponding Homologs |
|---|---|---|
| 7001383; 7229096; 7717034; 11464986*; 11527823 | R1 | H1 - M1 |
| 18478865 | R2 | H2 - M7 |
| 18478869 | R3 | |
| 20385787 | R4 | |
| 20385789 | R5 | |
| 20385791 | R6 | H3 |
| 20385793; 284810436 | R7 | |
| 20385795; 284066979 | R8 | H5 |
| 20385803 | R9 | |
| 284066981 | R10 | |
| 284468403 | R11 | |
| 284468405 | R12 | |
| 284810438 | R13 | |
| 520845525; 520845527 | R14 | H8 - M5 |
| 520845531 | R15 | |
| 520845533 | R16 | |
| 520845535 | R17 | |
| 520845537 | R18 | |
| 520845539 | R19 | |
| 520845541 | R20 |
Gene identifiers corresponding to rat (R) Park2 splice variants currently known are reported. For convenience, a new code identifier based on submission date has been assigned to each variant. In some cases, different submissions (with different Gene Identifiers) exist for the same splice variant. One record (marked with an asterisk) is a Provisional Reference Sequence (RefSeq) identical to 7229096, which has not yet been subjected to NCBI final review. Homologous transcripts in human (H) and mouse (M) are reported in the table; for their Gene Identifiers the reader is referred to Tables 1 and 4.
Fig. (3).
Rat Park2 gene and exonic structure of alternative splice variants. A: Exons and introns organization of rat Park2 gene. B: Exon organization map of the 20 rat Park2 splice variants currently known. For details, see Fig. 2. Exon 1 and exon 20 sequences are partial in some variants (R1-R13).
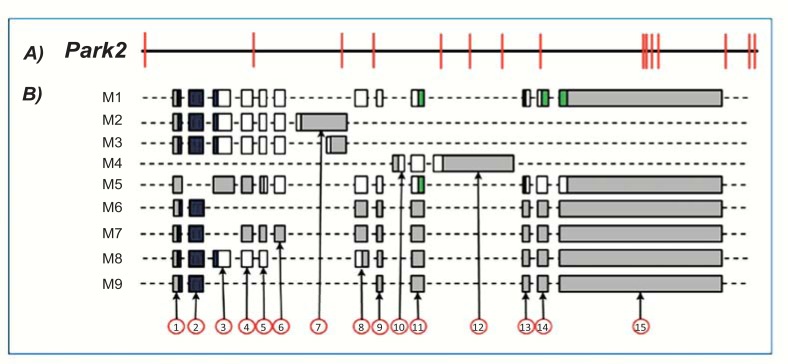
Mouse Park2 Alternative Splice Variants
Park2 splicing has been less investigated in mouse. To date, only 12 transcripts have been cloned and are collected in Unigene cluster Mm.311110 (Table 4). These transcripts arise from splicing of 15 different exons (Table 2) and correspond to 9 unique alternative splice variants (Fig. 4). Among these, a splice variant (M3) is generated by the use of an alternative 5’ donor site inside exon 7 (Fig. 4). Exons 10 and 12 are exclusively expressed in a single splice variant.
Table 4.
Mus musculus Park2 splice variants.
| GI Park2 mRNAs | Code Identifier | Corresponding Homologs |
|---|---|---|
| 5456929; 86577675; 10179808; 118131140* | M1 | H1 - R1 |
| 10179810 | M2 | |
| 10179812 | M3 | |
| 74227131 | M4 | |
| 220961631 | M5 | H8 - R14 |
| 220961633 | M6 | |
| 220961635 | M7 | H2 - R2 |
| 220961637 | M8 | |
| 284829878 | M9 |
Gene identifiers corresponding to mouse (M) Park2 splice variants currently known are reported. For convenience, a new code identifier based on submission date has been assigned to each variant. In some cases, different submissions (with different Gene Identifiers) exist for the same splice variant. One records (marked with an asterisk) is a Provisional Reference Sequence (RefSeq), derived from AC105305.8, AC091254.77, AC091484.8, AC091777.26, AC093450.20, AC122259.2, CT009575.8 and AC163687.5, that has not yet been subjected to NCBI final review. Although this record has been generated by genomic sequence alignments, it perfectly matches to 10179808. Homologous transcripts in human (H) and rat (R) are reported in the table; for their Gene Identifiers the reader is referred to Tables 1 and 3.
Fig. (4).
Mouse Park2 gene and exonic structure of splice variants. A: Exons and introns organization of mouse Park2 gene. B: Exon organization map of the 9 mouse Park2 splice variants currently known. For details, see Fig. 2. Exon 1 and exon 15 sequences are partial in some variants (M5-M9).
Mouse Park2 gene is located in chromosome 17 where it spans 1.22 Mb. Cloned transcripts range from 3226 to 793 bp and, therefore, alternative splicing process reduces primary transcript length by about 1500 times.
SPECIES-SPECIFIC ALTERNATIVE SPLICING OF PARK2 IN HUMAN, RAT AND MOUSE
Alternative splicing is thought to be the major source of phenotypic diversity in higher eukaryotes, especially in mammals. It contributes to enhance transcriptomic diversification, and thus plays an important role in speciation and in the dynamic evolution of genome structure [30].
Inter-species comparison of PARK2 genes is useful to identify the role of alternative cassette exons during evolution. In addition, investigating the analogies and the divergences between species may be fundamental in creating a valid animal model for Parkinson’s disease. To this regard, knocking out Park2 function in mice has been achieved by deletion of exon 2, 3, and 8 in Park2 gene, but no loss of nigrostriatal dopaminergic neurons has been reported [31-34].
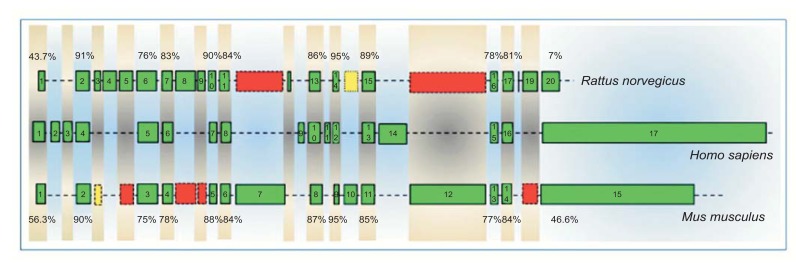
The genomic structures of human PARK2 gene and its homologs in rat and mouse are shown in (Fig. 5). Most of the exons have been conserved during evolution (e.g rat exon 2, human exon 4 and mouse exon 2; rat exon 14, human exon 12 and mouse exon 9), and their sequences have a high degree of homology (up to 95%). Differently, other exons are species-specific (e.g. human exons 2, 3, 9, 11 and 14).
Fig. (5).
Exonic structures of human, rat and mouse PARK2 genes. Homologous sequences are shown in the same column. Green boxes represent the known exons of PARK2 genes. Yellow boxes represent homologous sequences that could be potentially expressed, since they are provided with splice sites (AG/GT). Red boxes represent homologous sequences without splice sites and, therefore, not expressible. Numbers in the top of the figure indicate percentages of homology between human and rat exons, while those at the bottom denote percentages of homology between human and mouse exons.
Species-specific cassette exons may originate by two different mechanisms: i) exonization of common ancestral intronic sequences, and ii) exon shuffling, in which a new exon is inserted into an existing gene [30]. Rat and mouse specific Park2 exons may have originated by the first mechanism (exonization). Gene-comparison, in fact, reveals that some rat-specific Park2 exons (e.g., rat exon 3, 5, 8, 9 and 19) have a high degree of identity level with intronic sequences of mouse and vice versa (e.g. mouse exon 7, 10 and 12). During evolution, therefore, these sequences may have gained splice sites (AG/GT) and been expressed in one species but not in the other one. Conversely, human-specific PARK2 exons might have originated by exon shuffling, since their sequences are unique in human PARK2 gene and do not match to any of the corresponding Park2 intronic regions of rat and mouse (Fig. 5).
Analyzing the exonic structures of PARK2 alternative splice variants of the three species (Figs. 2, 3 and 4), we can separate exons with a low (e.g. human exons 2, 3, 9, 11 and 14) or high inclusion level (e.g. human exons 1, 4-8, 10, 12, 13, 15-17) in splice variants. Exons with a high inclusion level in splice variants coincide with conserved exons (with the exception of the first and the last exons, whose evolutionary conservation is relatively modest), while those having a low inclusion level coincide with species-specific exons (Fig. 5).
ALTERNATIVE SPLICING OF PARK2 PRODUCES DIVERSITY
The combinatorial arrangement of PARK2 exons generates a large number of alternatively spliced mRNAs that may diverge for untranslated regions, half-life, regulation or degradation modality (e.g. the NMD mechanism seen above). Moreover, their different coding regions may lead the expression of distinct PARK2 protein isoforms, which are different in amino acid sequence, post-translational modifications and functional domain composition (Fig. 6).
Fig. (6).
Predicted molecular architecture of PARK2 isoforms.PARK2 isoforms contain one or more of the following domains: an N-terminal Ubiquitin-like domain (UBQ, in blue) and one or two In Between Ring finger domains (IBR, in green). The code identifier of each isoform, reported on their left, corresponds to that of the encoding splice variants listed in (Tables 1, 3 and 4).
The original (canonical) PARK2 protein (Accession number BAA25751.1) [1] comprises an N-terminal ubiquitin-like (UBQ) domain and two C-terminal in-between ring fingers (IBR) domains, encoded by specific PARK2 exons (Figs. 2, 3 and 4). The UBQ domain targets specific protein substrates for degradation by the proteasome, whereas IBR domains occur between pairs of ring fingers and play a role in protein quality control. PARK2 isoforms, encoded by the alternative splice transcripts currently known, structurally diverge from the canonic one for the presence or absence of the UBQ domain and for one or both IBR domains (Fig. 6). Moreover, when UBQ domain is present, it often differs in length from the canonic one. Interestingly, some isoforms miss some of these domains (Fig. 6).
Alternative splicing also affects intrinsically disordered protein regions (e.g. regions lacking of stable tertiary structure), thus playing a critical role in remodeling protein-protein interactions [35]. Alternative splicing events on intrinsically disordered protein regions could regulate interactions of PARK2 isoforms with specific cellular targets. In addition, PARK2 isoforms generated by different alternative splice transcripts could interact with each other mutually regulating their functions, as it has been reported for RBCK1, a protein with IBR and E3 ubiquitin ligase domains, whose migration in the nucleus is inhibited by interaction with RBCK2, an isoform lacking IBR domain [36].
In addition to molecular architectures, alternative splicing may also influence stability, localization and catalytic efficiency of PARK2 isoforms. Although scientific evidences concerning this are still few, preliminary studies reported the identification of a PARK2 isoform, missing exons 5-8, with a defective degradation activity of Cyclin E and control of cellular cycle [19]. Another study detected a splice variant of pdr-1 (a Caenorhabditis elegans homologous of PARK2) with an in frame deletion, characterized by altered solubility and intracellular localization [15].
Besides the well-known involvement in proteasome-dependent degradation of target proteins [37, 38], PARK2 has been implicated in apoptosis regulation [39], mitochondrial homeostasis, mitophagy [40, 41] and mitochondrial DNA stability [42]. In addition, UBQ proteins such as PARK2, are implicated in endocytosis, cellular trafficking, signal transduction, transcriptional regulation and DNA repair, in ubiquitin degradation independent manner [43]. It may not be excluded that PARK2 alternative splice isoforms (included those missing functional domains) act in a different cellular context, operating in a still, not yet characterized manner.
DIFFERENTIAL EXPRESSION OF PARK2 IN TISSUES AND CELLS
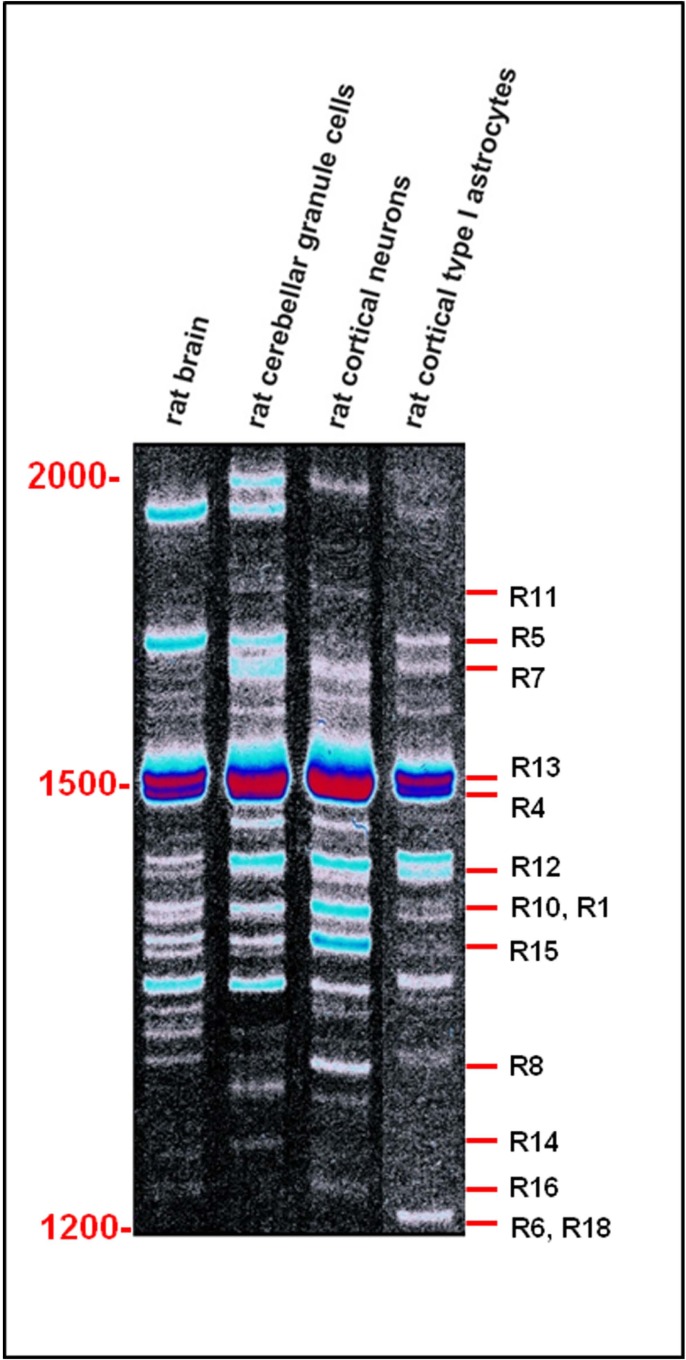
Alternative splicing events are finely regulated in time and space, and thus contribute to cell specialization and tissue definition. Although we are not yet able to define the tissue and cell specific spectrum of expression of PARK2 and its homologs, the few evidences reported to date undoubtedly demonstrate that a regional and cellular differential expression of transcripts and isoforms exists. Distinct transcript-expression profiles occur in human brain regions [44] and leukocytes [17], in rat brain and isolated nerve cells [18] and in a wide variety of mouse tissues including brain, heart, lung, liver, skeletal muscle, kidney, and testis [11]. (Fig. 7), for example, shows the different expression patterns of Park2 splice variants in rat cerebellar granule cells, cortical neurons and type I astrocytes [18].
Fig. (7).
Differential expression of Park2 transcript variants in rat brain and isolated cells. Single-stranded cDNAs from adult rat brain, rat cortical neurons, rat cerebellar granule cells and rat cortical type I astrocytes mRNAs was PCR amplified with primers flanking the start and the stop codon of Park2. The resulting splicing patterns clearly show a regional and cellular differential expression among different rat neuronal cells, producing spatial and functional diversification. Marker length (bp) is shown on the left. Known rat splice variants with a length between 1200 and 2000 bp are shown on the right (for codes, see Table 3). All the other products have not yet been cloned and remain unknown.
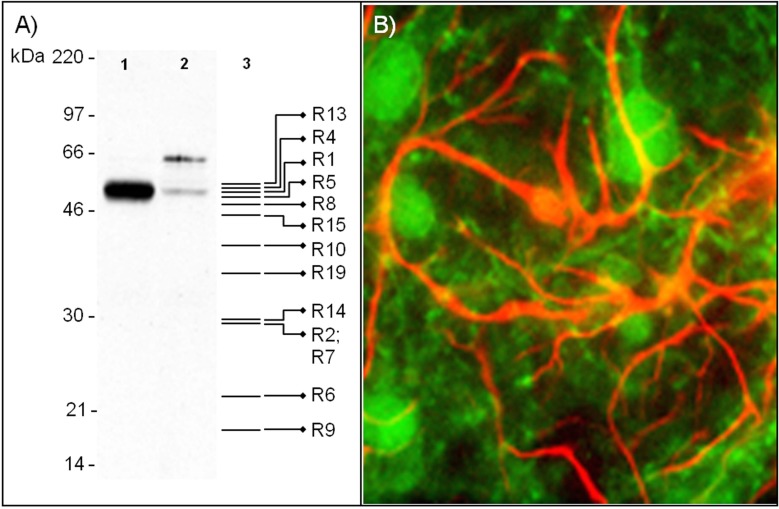
Differential expression of PARK2 transcripts is also mirrored at the protein level. In mice, Park2 protein isoforms show a differential distribution in different nervous system areas (cerebral cortex/diencephalons, hippocampus, cerebellum, brainstem, striatum, spinal cord), peripheral tissues (heart, liver, spleen, pancreas, kidney), and developmental stages [45, 46]. Distinct expression patterns occur also in rat brain regions (substantia nigra and cerebellum) [46-48] and isolated nervous cells [49]. To this regard, a representative example is shown in (Fig. 8), where Park2 isoforms are clearly differentially expressed in rat cortical neurons and type I astrocytes. Both western blot and immunofluorescence assays demonstrate not only a quantitative difference in Park2 expression levels between these cell types, but also a different expression pattern of splice isoforms. The expression of PARK2 has been recently assessed in human blood [50]. This study revealed the presence of different splice variants and protein isoforms.
Fig. (8).
Differential expression of Park2 isoforms in rat cortical neurons and type I astrocytes. A: Western Blot analysis of Park2 isoforms in rat neurons (lane 1) and type I astrocytes (lane 2). The assay was performed using a rabbit anti-Park2 polyclonal antibody (AB5112, Millipore) as previously described ADDIN EN.CITE ADDIN EN.CITE.DATA [49]. Expected molecular weights of Park2 isoforms potentially recognized by this antibody are drawn on lane 3. The very intense immunoreactive band of about 51 kDa in cortical neurons overlaps with the expected molecular weight of several isoforms (R1, R2, R4-R10, R13-R15, and R19). Instead, type I astrocytes express a faint band of 51 kDa which may correspond to R1 isoform, and a 65 kDa band that corresponds to a still uncharacterized variant. It should be noted that the antibody used in this western blot analysis recognizes only one epitope, which is not present in all isoforms. Other isoforms, therefore, may be expressed in these cell types and not be immunoreactive to the antibody used. B: Park2 and GFAP immunoreactivity in mixed cortical cultures. Primary cultures of cortical neurons and type I astrocytes were double labeled with antibodies against Park2 protein (green signal) and GFAP (red signal), as previously described ADDIN EN.CITE ADDIN EN.CITE.DATA [49], and observed by fluorescence microscopy.
When the extensive PARK2 alternative splicing was still unknown, correlation between mRNA and protein expression patterns was not an easy task. The presence of unexpected immunoreactive bands on western blot, for example, was very often explained as the result of partially translated forms of PARK2 full-length, post-translational modifications [46, 47] or cross reactivity with other proteins [51, 52]. Interestingly, the datasheet of many commercially available PARK2 antibodies (see below for further details) shows the presence of multiple immunoreactive bands without providing sufficient explanations. Unlike the past, splicing of PARK2 can now be investigated at the protein level in more details by the use of different antibodies. To date more than 160 PARK2 antibodies are commercially available. They are generally raised from rabbit or mouse and commercialized by various companies. (Table 5) list 35 commercially available PARK2 antibodies whose immunogens used are known. Some of them have been raised against the same immunogen, and thus recognize common epitopes. These 35 antibodies may allow recognizing 15 different PARK2 epitopes. Although no epitope is probably isoform specific, the combinatorial use of antibodies targeting different protein regions, together with the use of different techniques such as two dimensional gel assays, may provide a precious aid to decode the exact spectrum of PARK2 isoforms expressed in tissues and cells.
Table 5.
List of commercially available antibodies against human, rat and mouse PARK2.
| Product Number | Provider | Immunogen (aa) | Recognized Isoforms |
|---|---|---|---|
| H00005071-B01P | Abnova | 1-387 | H4 |
| H00005071-D01P | Abnova | ||
| H00005071-D01 | Abnova | ||
| OASA06385 | Aviva Systems biology | 83-97 | H1, H4, H5, H8, H9, H10, H13, H14, H17, H20, H21 |
| AHP495 | AbD Serotec | ||
| MD-19-0144 | Raybiotech | ||
| DS-PB-01562 | Raybiotech | ||
| PAB14022 | Abnova | ||
| MCA3315Z | AbD Serotec | 288-388 | H4 |
| H00005071-M01 | Abnova | ||
| PAB1105 | Abnova | 62-80 | H1, H4, H5, H9, H10, H13, H14, H20 |
| 70R-PR059 | Fitzgerald | ||
| PAB0714 | Abnova | 305-323 | H1-H6, H8, H11, H17, H20, H21, R1, R2, R4-R10, R13-R15, R19 |
| AB5112 | Millipore | ||
| R-113-100 | Novus biologicals | ||
| P5748 | Sigma | 298-313 | H1-H6, H8, H11, H17, H20, H21, R1, R2, R4-R10, R13-R15, R19, M1, M5 |
| GTX25667 | GeneTex | ||
| ABIN122870 | Antibodies online | ||
| PA1-751 | Thermo Scientific | ||
| R-114-100 | Novus biologicals | 295-311 | H1-H6, H8, H10, H11, H14, H17, H20, H21, R1, R2, R4-R10, R13-R15, R19, M1, M5 |
| AB5978 | Millipore | ||
| MAB5512 | Millipore | 399-465 | H1, H2, H5-H8, H10-H12, H14, H17-H21 |
| 05-882 | Millipore | ||
| sc-32282 | Santa Cruz | ||
| sc-30130 | Santa Cruz | 61-360 | H1-H3, H6, H9, H11-H13 |
| sc-133167 | Santa Cruz | ||
| sc-136989 | Santa Cruz | ||
| EB07439 | Everest Biotech | 394-409 | H2, H6, H7, H11, H12, H18 |
| GTX89242 | Gene Tex | ||
| NB100-53798 | Novus biologicals | ||
| GTX113239 | GeneTex | 28-258 | H1 |
| 10R-3061 | Fitzgerald | 390-406 | H1, H2, H5-H8, H10-H12, H14, H17, H18, H20, H21 |
| A01250-40 | GenScript | 300-350 | H1-H6, H8, H11, H17, H20, H21 |
| NB600-1540 | Novus biologicals | 399-412 | H1, H2, H5-H7, H10-H12, H18, H20 |
| ARP43038_P050 | Aviva Systems biology | 311-360 | H2, H3, H6, H7, H11, H12, H18, M1, M5 |
Some PARK2 antibodies have been raised against the same immunogen. Amino acids positions refer to PARK2 isoform NP_004553.2. Human (H), rat (R), and mouse (M) recognized isoforms are indicated in the right column.
ALTERNATIVE SPLICING OF PARK2 AND PATHOLOGY
Alternative splicing process is a key element in PARK2 gene expression and could be easily disrupted through multiple errors. An aberrant alternative splicing may arise from changes of regulatory sequences required for correct pre-mRNA processing, such as splice sites, branch point, polypyrimidine tract, exonic splicing enhancers/silencers and intronic enhancers/silencers, which are called cis-acting mutations. To this regard, a number of PARK2 cis-acting mutations identified in patients with Parkinson’s disease have been collected in Parkinson Disease Mutations database. To date, point mutations localized in splice acceptor or donor sites of PARK2 introns 1, 6, 7, 10, 12, 13 and 16 have been investigated [53-59], while cis-acting mutations in splice sites of exons 2-5, 8, 9, 11, 14, 15, 17 and in the other splicing regulatory regions have not yet been explored and need to be assessed. Moreover, deregulation of alternative splicing may be the result of changes in the components of spliceosome machinery (trans-acting mutations). Further studies are required to discover their possible pathogenic influence on PARK2 alternative splicing, and to elucidate the exact splice pattern/phenotype correlations. Disruption of PARK2 alternative splicing by both cis- and trans-acting mutations, in fact, could result in a functionally harmful PARK2 expression pattern, creating aberrant events with pathological consequences that may provide an explanation for the broad spectrum of phenotypic abnormalities observed in patients with PARK2 mutations [60-62].
Both cis- and trans-acting mutations have been previously associated to human diseases. For example, cis-acting mutations have been found in genes involved in Alzheimer’s Disease (PSEN1, MAPT, GRN) [63], while trans-acting mutations have been detected in Retinitis pigmentosa (U4/U5/ U6 protein complex) [64], in Spinal Muscle Atrophy [65], and in Dystrophia Myotonica [66].
Emerging evidences support the importance of PARK2 splice variants expression changes in disease development. When compared to control patients, distinct PARK2 splice patterns have been identified in frontal cortex of Parkinson’s disease, pure dementia with Lewy bodies, common Lewy body disease and Alzheimer’s disease patients [20-22]. The disease-specific expression profiles of PARK2 isoforms suggest a role for splicing deregulation in the development of neurodegenerative disorders. In addition, aberrant PARK2 transcripts have been reported in some human cancers (ovarian and colorectal cancer, chronic myeloid leukemia and other cancer-derived cell lines) [67]. Based on these evidences, the possibility that PARK2 aberrant alternative splicing may be involved in PD and other human diseases represents an interesting hypothesis that needs to be further investigated.
CONCLUSION
PARK2 gene provides a fascinating example of the use of alternative splicing to create different variants within single cell types. Disruption of this versatile form of genetic regulation may alter the fine-tuning of the encoded proteins to suit specific cellular needs. Investigating the full spectrum of PARK2 alternative spliced mRNAs, studying the complete pattern of expression of alternative splicing transcripts and isoforms in tissue and cell-types, and determining each of their functions will require additional studies. Furthermore, investigating cis- and trans-acting mutations may provide novel insights into the pathogenesis of human diseases associated to PARK2 and their cure.
ACKNOWLEDGEMENTS
We gratefully acknowledge Cristina Calì, Alfia Corsino, Maria Patrizia D’Angelo and Francesco Marino for their administrative and technical support.
CONFLICT OF INTEREST
The authors confirm that this article content has no conflicts of interest.
REFERENCES
- 1.Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, Yokochi M, Mizuno Y, Shimizu N. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392(6676):605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi H, Ohama E, Suzuki S, Horikawa Y, Ishikawa A, Morita T, Tsuji S, Ikuta F. Familial juvenile parkinsonism: clinical and pathologic study in a family. Neurology. 1994;44(3 Pt 1):437–41. doi: 10.1212/wnl.44.3_part_1.437. [DOI] [PubMed] [Google Scholar]
- 3.Cesari R, Martin ES, Calin GA, Pentimalli F, Bichi R, McAdams H, Trapasso F, Drusco A, Shimizu M, Masciullo V, D'Andrilli G, Scambia G, Picchio MC, Alder H, Godwin AK, Croce CM. Parkin, a gene implicated in autosomal recessive juvenile parkinsonism, is a candidate tumor suppressor gene on chromosome 6q25-q27. Proc. Natl. Acad. Sci. U S A. 2003;100(10):5956–61. doi: 10.1073/pnas.0931262100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Veeriah S, Taylor B S, Meng S, Fang F, Yilmaz E, Vivanco I, Janakiraman M, Schultz N, Hanrahan A J, Pao W, Ladanyi M, Sander C, Heguy A, Holland E C, Paty P B, Mischel P S, Liau L, Cloughesy T F, Melling-hoff I K, Solit D B, Chan T A. Somatic mutations of the Parkinson's disease-associated gene PARK2 in glioblastoma and other human malignancies. Nat. Genet. 2010;42(1):77–82. doi: 10.1038/ng.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mira M T, Alcais A, Nguyen V T, Moraes M O, Di Flumeri C, Vu H T, Mai C P, Nguyen T H, Nguyen N B, Pham X K, Sarno E N, Alter A, Montpetit A, Moraes M E, Moraes J R, Dore C, Gallant C J, Lepage P, Verner A, Van De Vosse E, Hudson T J, Abel L, Schurr E. Susceptibility to leprosy is associated with PARK2 and PACRG. Nature. 2004;427(6975):636–40. doi: 10.1038/nature02326. [DOI] [PubMed] [Google Scholar]
- 6.Glessner JT, Wang K, Cai G, Korvatska O, Kim CE, Wood S, Zhang H, Estes A, Brune CW, Bradfield JP, Imielinski M, Frackelton EC, Reichert J, Crawford EL, Munson J, Sleiman PM, Chiavacci R, Annaiah K, Tho-mas K, Hou C, Glaberson W, Flory J, Otieno F, Garris M, Soorya L, Klei L, Piven J, Meyer KJ, Anagnostou E, Sakurai T, Game R M, Rudd DS, Zurawiecki D, McDougle CJ, Davis LK, Miller J, Posey DJ, Michaels S, Kolevzon A, Silverman J M, Bernier R, Levy SE, Schultz RT, Dawson G, Owley T, McMahon WM, Wassink TH, Sweeney JA, Nurnberger JI, Coon H, Sutcliffe JS, Minshew NJ, Grant SF, Bucan M, Cook EH, Buxbaum JD, Devlin B, Schellenberg GD, Hakonarson H. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature. 2009;459(7246):569–73. doi: 10.1038/nature07953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wongseree W, Assawamakin A, Piroonratana T, Sinsomros S, Limwongse C, Chaiyaratana N. Detecting purely epistatic multi-locus interactions by an omnibus permutation test on ensembles of two-locus analyses. BMC Bioinform. 2009;10:294. doi: 10.1186/1471-2105-10-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burns M P, Zhang L, Rebeck G W, Querfurth H W, Moussa C E. Parkin promotes intracellular Abeta1-42 clearance. Hum. Mol. Genet. 2009;18(17):3206–16. doi: 10.1093/hmg/ddp258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matsumine H, Saito M, Shimoda-Matsubayashi S, Tanaka H, Ishikawa A, Nakagawa-Hattori Y, Yokochi M, Kobayashi T, Igarashi S, Takano H, Sanpei K, Koike R, Mori H, Kondo T, Mizutani Y, Schaffer AA, Yamamura Y, Nakamura S, Kuzuhara S, Tsuji S, Mizuno Y. Localization of a gene for an autosomal recessive form of juvenile Parkinsonism to chromosome 6q25.-27. . Am. J. Hum. Genet. 1997;60(3):588–96. [PMC free article] [PubMed] [Google Scholar]
- 10.D'Agata V, Zhao W, Cavallaro S. Cloning and distribution of the rat parkin mRNA. Brain Res. Mol. Brain Res. 2000;75(2):345–9. doi: 10.1016/s0169-328x(99)00286-7. [DOI] [PubMed] [Google Scholar]
- 11.Kitada T, Asakawa S, Minoshima S, Mizuno Y, Shimizu N. Molecular cloning, gene expression, and identification of a splicing variant of the mouse parkin gene. Mamm. Genome. 2000;11(6):417–21. doi: 10.1007/s003350010080. [DOI] [PubMed] [Google Scholar]
- 12.Tomac AC, Hoffer BJ. Assignment of the mouse Park2 (PARKIN): the homologue to a new human Parkinson candidate gene, to the telomeric region of mouse 17A3.-3.3 by in situ hybridization. Cytogenet. Cell Genet. 2001;95(1-2):120–1. doi: 10.1159/000057032. [DOI] [PubMed] [Google Scholar]
- 13.Bae Y J, Park K S, Kang S J. Genomic organization and expression of parkin in Drosophila melanogaster. Exp. Mol. Med. 2003;35(5):393–402. doi: 10.1038/emm.2003.52. [DOI] [PubMed] [Google Scholar]
- 14.Fett M E, Pilsl A, Paquet D, van Bebber F, Haass C, Tatzelt J, Schmid B, Winklhofer K F. Parkin is protective against proteotoxic stress in a transgenic zebrafish model. PloS One. 2010;5(7):e11783. doi: 10.1371/journal.pone.0011783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Springer W, Hoppe T, Schmidt E, Baumeister R. A Caenorhabditis elegans Parkin mutant with altered solubility couples alpha-synuclein aggregation to proteotoxic stress. Hum. Mol. Gen. 2005;14(22):3407–23. doi: 10.1093/hmg/ddi371. [DOI] [PubMed] [Google Scholar]
- 16.Cruts M, Theuns J, Van Broeckhoven C. Locus-specific mutation databases for neurodegenerative brain diseases. Hum. Mutation. 2012;33(9):1340–4. doi: 10.1002/humu.22117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sunada Y, Saito F, Matsumura K, Shimizu T. Differential expression of the parkin gene in the human brain and peripheral leukocytes. Neurosci. Lett. 1998;254(3):180–2. doi: 10.1016/s0304-3940(98)00697-1. [DOI] [PubMed] [Google Scholar]
- 18.Dagata V, Cavallaro S. Parkin transcript variants in rat and human brain. Neurochem. Res. 2004;29(9):1715–24. doi: 10.1023/b:nere.0000035807.25370.5e. [DOI] [PubMed] [Google Scholar]
- 19.Ikeuchi K, Marusawa H, Fujiwara M, Matsumoto Y, Endo Y, Watanabe T, Iwai A, Sakai Y, Takahashi R, Chiba T. Attenuation of proteolysis-mediated cyclin E regulation by alternatively spliced Parkin in human colorectal cancers. J. Int. du Canc. 2009;125(9):2029–35. doi: 10.1002/ijc.24565. [DOI] [PubMed] [Google Scholar]
- 20.Beyer K, Domingo-Sabat M, Humbert J, Carrato C, Ferrer I, Ariza A. Differential expression of alpha-synuclein, parkin, and synphilin-1 isoforms in Lewy body disease. Neurogenetics. 2008;9(3):163–72. doi: 10.1007/s10048-008-0124-6. [DOI] [PubMed] [Google Scholar]
- 21.Humbert J, Beyer K, Carrato C, Mate J L, Ferrer I, Ariza A. Parkin and synphilin-1 isoform expression changes in Lewy body diseases. Neurobiol. Dis. 2007;26(3):681–7. doi: 10.1016/j.nbd.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 22.Tan E K, Shen H, Tan J M, Lim K L, Fook-Chong S, Hu W P, Paterson M C, Chandran V R, Yew K, Tan C, Yuen Y, Pavanni R, Wong M C, Puvan K, Zhao Y. Differential expression of splice variant and wild-type parkin in sporadic Parkinson's disease. Neurogenetics. 2005;6(4):179–84. doi: 10.1007/s10048-005-0001-5. [DOI] [PubMed] [Google Scholar]
- 23.Behzadnia N, Golas M M, Hartmuth K, Sander B, Kastner B, Deckert J, Dube P, Will C L, Urlaub H, Stark H, Luhrmann R. Composition and three-dimensional EM structure of double affinity-purified, human prespli-ceosomal A complexes. EMBO J. 2007;26(6):1737–48. doi: 10.1038/sj.emboj.7601631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tazi J, Bakkour N, Stamm S. Alternative splicing and disease. Biochimica et Biophysica Acta. 2009;1792(1):14–26. doi: 10.1016/j.bbadis.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stamm S, Ben-Ari S, Rafalska I, Tang Y, Zhang Z, Toiber D, Thanaraj T. A.oreq H. Function of alternative splicing. . Gene . 2005; 344:1–20. doi: 10.1016/j.gene.2004.10.022. [DOI] [PubMed] [Google Scholar]
- 26.Hughes T A. Regulation of gene expression by alternative untranslated regions. Trends Genet. 2006;22(3):119–22. doi: 10.1016/j.tig.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 27.Lejeune F, Maquat L E. Mechanistic links between nonsense-mediated mRNA decay and pre-mRNA splicing in mammalian cells. Curr. Opin. Cell. Biol. 2005;17(3):309–15. doi: 10.1016/j.ceb.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 28.Kelemen O, Convertini P, Zhang Z, Wen Y, Shen M, Falaleeva M, Stamm S. Function of alternative splicing. Gene. 2012;514(1):1–30. doi: 10.1016/j.gene.2012.07.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Modrek B, Lee C. A genomic view of alternative splicing. Nat Genet. 2002;30(1):13–9. doi: 10.1038/ng0102-13. [DOI] [PubMed] [Google Scholar]
- 30.Lev-Maor G, Goren A, Sela N, Kim E, Keren H, Doron-Faigenboim A, Leibman-Barak S, Pupko T, Ast G. The "alternative" choice of constitutive exons throughout evolution. PLoS Genetics. 2007;3(11):e203. doi: 10.1371/journal.pgen.0030203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goldberg M S, Fleming S M, Palacino J J, Cepeda C, Lam H A, Bhatnagar A, Meloni E G, Wu N, Ackerson L C, Klapstein G J, Gajendiran M, Roth B L, Chesselet M F, Maidment N T, Levine M S, Shen J. Parkin-deficient mice exhibit nigrostriatal deficits but not loss of dopaminergic neurons. J. Biol. Chem. 2003;278(44):43628–35. doi: 10.1074/jbc.M308947200. [DOI] [PubMed] [Google Scholar]
- 32.Perez F A, Palmiter R D. Parkin-deficient mice are not a robust model of parkinsonism. Proc. Natl. Acad. Sci. U S A. 2005;102 (6):2174–9. doi: 10.1073/pnas.0409598102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Itier J M, Ibanez P, Mena M A, Abbas N, Cohen-Salmon C, Bohme G A, Laville M, Pratt J, Corti O, Pradier L, Ret G, Joubert C, Periquet M, Araujo F, Negroni J, Casarejos M J, Canals S, Solano R, Serrano A, Gallego E, Sanchez M, Denefle P, Benavides J, Tremp G, Rooney T A, Brice A, Garcia de Yebenes J. Parkin gene inactivation alters behaviour and dopamine neurotransmission in the mouse. Hum. Mol. Genet. 2003;12(18):2277–91. doi: 10.1093/hmg/ddg239. [DOI] [PubMed] [Google Scholar]
- 34.Von Coelln R, Thomas B, Savitt JM, Lim KL, Sasaki M, Hess EJ, Dawson VL, Dawson TM. Loss of locus coeruleus neurons and reduced startle in parkin null mice. Proc. Natl. Acad. Sci. U.S.A. 2004;101(29):10744–9. doi: 10.1073/pnas.0401297101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barbosa-Morais NL, Irimia M, Pan Q, Xiong HY, Gueroussov S, Lee LJ, Slobodeniuc V, Kutter C, Watt S, Colak R, Kim T, Misquitta-Ali CM, Wilson MD, Kim PM, Odom D T, Frey BJ, Blencowe BJ. The evolutionary landscape of alternative splicing in vertebrate species. Science. 2012;338(6114):1587–93. doi: 10.1126/science.1230612. [DOI] [PubMed] [Google Scholar]
- 36.Tatematsu K, Yoshimoto N, Okajima T, Tanizawa K, Kuroda S. Identification of ubiquitin ligase activity of RBCK1 and its inhibition by splice variant RBCK2 and protein kinase Cbeta. J. Biol. Chem. 2008;283(17):11575–85. doi: 10.1074/jbc.M706961200. [DOI] [PubMed] [Google Scholar]
- 37.Jiang H, Ren Y, Zhao J, Feng J. Parkin protects human dopaminergic neuroblastoma cells against dopamine-induced apoptosis. Hum. Mol. Genet. 2004;13(16):1745–54. doi: 10.1093/hmg/ddh180. [DOI] [PubMed] [Google Scholar]
- 38.Huynh DP, Scoles DR, Nguyen D, Pulst SM. The auto-somal recessive juvenile Parkinson disease gene product, parkin, interacts with and ubiquitinates synaptotagmin XI. Hum. Mol. Genet. 2003;12(20):2587–97. doi: 10.1093/hmg/ddg269. [DOI] [PubMed] [Google Scholar]
- 39.da Costa CA, Sunyach C, Giaime E, West A, Corti O, Brice A, Safe S, Abou-Sleiman PM, Wood NW, Takahashi H, Goldberg MS, Shen J, Checler F. Transcriptional repression of p53 by parkin and impairment by mutations associated with autosomal recessive juvenile Parkinson's disease. Nat. Cell Biol. 2009;11(11):1370–5. doi: 10.1038/ncb1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoshii S R, Kishi C, Ishihara N, Mizushima N. Parkin mediates proteasome-dependent protein degradation and rupture of the outer mitochondrial membrane. J. Biol. Chem. 2011;286(22):19630–40. doi: 10.1074/jbc.M110.209338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen Y, Dorn G W. 2nd PINK1-phosphorylated mitofusin 2 is a Parkin receptor for culling damaged mitochondria. Science. 2013;340(6131):471–5. doi: 10.1126/science.1231031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rothfuss O, Fischer H, Hasegawa T, Maisel M, Leitner P, Miesel F, Sharma M, Bornemann A, Berg D, Gasser T, Patenge N. Parkin protects mitochondrial genome integrity and supports mitochondrial DNA repair. Hum. Mol. Genet. 2009;18(20):3832–50. doi: 10.1093/hmg/ddp327. [DOI] [PubMed] [Google Scholar]
- 43.Winklhofer K F, Tatzelt J, Haass C. The two faces of protein misfolding: gain- and loss-of-function in neurodegenerative diseases. EMBO J. 2008;27(2):336–49. doi: 10.1038/sj.emboj.7601930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Solano SM, Miller DW, Augood SJ, Young AB, Penney J BJr. Expression of alpha-synuclein, parkin, and ubiquitin carboxy-terminal hydrolase L1 mRNA in human brain: genes associated with familial Parkinson's disease. Ann. Neurol. 2000;47(2):201–10. [PubMed] [Google Scholar]
- 45.Huynh D P, Dy M, Nguyen D, Kiehl T R, Pulst S M. Differential expression and tissue distribution of parkin isoforms during mouse development. Dev. Brain Res. 2001;130(2):173–81. doi: 10.1016/s0165-3806(01)00234-6. [DOI] [PubMed] [Google Scholar]
- 46.Stichel C C, Augustin M, Kuhn K, Zhu X R, Engels P, Ullmer C, Lubbert H. Parkin expression in the adult mouse brain. Eur. J. Neurosci. 2000;12(12):4181–94. [PubMed] [Google Scholar]
- 47.Horowitz J M, Myers J, Stachowiak M K, Torres G. Identification and distribution of Parkin in rat brain. Neuroreport. 1999;10 (16):3393–7. doi: 10.1097/00001756-199911080-00025. [DOI] [PubMed] [Google Scholar]
- 48.Gu W J, Abbas N, Lagunes M Z, Parent A, Pradier L, Bohme G A, Agid Y, Hirsch E C, Raisman-Vozari R, Brice A. Cloning of rat parkin cDNA and distribution of parkin in rat brain. J. Neurochem. 2000;74(4):1773–6. doi: 10.1046/j.1471-4159.2000.0741773.x. [DOI] [PubMed] [Google Scholar]
- 49.D'Agata V, Grimaldi M, Pascale A, Cavallaro S. Regional and cellular expression of the parkin gene in the rat cerebral cortex. Eur. J. Neurosci. 2000;12(10):3583–8. doi: 10.1046/j.1460-9568.2000.00244.x. [DOI] [PubMed] [Google Scholar]
- 50.Kasap M, Akpinar G, Sazci A, Idrisoglu H A, Vahaboglu H. Evidence for the presence of full-length PARK2 mRNA and Parkin protein in human blood. Neurosci. Lett. 2009;460(3):196–200. doi: 10.1016/j.neulet.2009.05.079. [DOI] [PubMed] [Google Scholar]
- 51.Horowitz JM, Vernace VA, Myers J, Stachowiak MK, Hanlon DW, Fraley G S, Torres G. Immunodetection of Parkin protein in vertebrate and invertebrate brains: a comparative study using specific antibodies. J. Chem. Neuro-anat. 2001;21(1):75–93. doi: 10.1016/s0891-0618(00)00111-3. [DOI] [PubMed] [Google Scholar]
- 52.Pawlyk AC, Giasson BI, Sampathu DM, Perez FA, Lim K L, Dawson V L, Dawson T M, Palmiter R D, Trojanowski J Q, Lee VM. Novel monoclonal antibodies demonstrate biochemical variation of brain parkin with age. J. Biol. Chem. 2003;278(48):48120–8. doi: 10.1074/jbc.M306889200. [DOI] [PubMed] [Google Scholar]
- 53.Illarioshkin SN, Periquet M, Rawal N, Lucking CB, Zagorovskaya TB, Slominsky PA, Miloserdova OV, Markova ED, Limborska SA, Ivanova-Smolenskaya IA, Brice A. Mutation analysis of the parkin gene in Russian families with autosomal recessive juvenile parkinsonism. Mov. Disord. 2003;18(8):914–9. doi: 10.1002/mds.10467. [DOI] [PubMed] [Google Scholar]
- 54.Pigullo S, De Luca A, Barone P, Marchese R, Bellone E, Colosimo A, Scaglione C, Martinelli P, Di Maria E, Pizzuti A, Abbruzzese G, Dallapiccola B, Ajmar F, Mandich P. Mutational analysis of parkin gene by denaturing high-performance liquid chromatography (DHPLC) in essential tremor. Parkinsonism Relat. Disord. 2004;10(6):357–62. doi: 10.1016/j.parkreldis.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 55.Scherfler C, Khan N L, Pavese N, Eunson L, Graham E, Lees A J, Quinn NP, Wood NW, Brooks DJ, Piccini PP. Striatal and cortical pre- and postsynaptic dopaminergic dysfunction in sporadic parkin-linked parkinsonism. Brain. 2004;127(Pt 6):1332–42. doi: 10.1093/brain/awh150. [DOI] [PubMed] [Google Scholar]
- 56.Bertoli-Avella A M, Giroud-Benitez J L, Akyol A, Barbosa E, Schaap O, van der Linde H C, Martignoni E, Lopiano L, Lamberti P, Fincati E, Antonini A, Stocchi F, Montagna P, Squitieri F, Marini P, Abbruzzese G, Fabbrini G, Marconi R, Dalla Libera A, Trianni G, Guidi M, De Gaetano A, Boff Maegawa G, De Leo A, Gallai V, de Rosa G, Vanacore N, Meco G, van Duijn C M, Oostra B A, Heutink P, Bonifati V. Italian Parkinson Genetics, N. Novel parkin mutations detected in patients with early-onset Parkinson's disease. Mov. Disord. 2005;20(4):424–31. doi: 10.1002/mds.20343. [DOI] [PubMed] [Google Scholar]
- 57.Bardien S, Keyser R, Yako Y, Lombard D, Carr J. Molecular analysis of the parkin gene in South African patients diagnosed with Parkinson's disease. Parkinsonism Relat. Disord. 2009;15(2):116–21. doi: 10.1016/j.parkreldis.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 58.Nuytemans K, Meeus B, Crosiers D, Brouwers N, Goossens D, Engelborghs S, Pals P, Pickut B, Van den Broeck M, Corsmit E, Cras P, De Deyn P P, Del-Favero J, Van Broeckhoven C, Theuns J. Relative contribution of simple mutations vs.copy number variations in five Par-kinson disease genes in the Belgian population. Hum. Mut. 2009;30(7):1054–61. doi: 10.1002/humu.21007. [DOI] [PubMed] [Google Scholar]
- 59.Pankratz N, Kissell D K, Pauciulo M W, Halter C A, Rudolph A, Pfeiffer R F, Marder K S, Foroud T, Nichols W C, Parkinson Study Group P I. Parkin dosage mutations have greater pathogenicity in familial PD than simple sequence mutations. Neurology. 2009;73(4):279–86. doi: 10.1212/WNL.0b013e3181af7a33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Abbas N, Lucking C B, Ricard S, Durr A, Bonifati V, De Michele G, Bouley S, Vaughan J R, Gasser T, Marconi R, Broussolle E, Brefel-Courbon C, Harhangi B S, Oostra B A, Fabrizio E, Bohme G A, Pradier L, Wood N W, Filla A, Meco G, Denefle P, Agid Y, Brice A. A wide variety of mutations in the parkin gene are responsible for autosomal recessive parkinsonism in Europe.French Parkinson's Disease Genetics Study Group and the European Consortium on Genetic Susceptibility in Parkinson's Disease. Hum. Mol. Genet. 1999;8(4):567–74. doi: 10.1093/hmg/8.4.567. [DOI] [PubMed] [Google Scholar]
- 61.Hattori N, Kitada T, Matsumine H, Asakawa S, Yamamura Y, Yoshino H, Kobayashi T, Yokochi M, Wang M, Yoritaka A, Kondo T, Kuzuhara S, Nakamura S, Shimizu N, Mizuno Y. Molecular genetic analysis of a novel Parkin gene in Japanese families with autosomal reces-sive juvenile parkinsonism: evidence for variable homozygous deletions in the Parkin gene in affected individuals. Ann. Neurol. 1998;44(6):935–41. doi: 10.1002/ana.410440612. [DOI] [PubMed] [Google Scholar]
- 62.Lucking CB, Durr A, Bonifati V, Vaughan J, De Michele G, Gasser T, Harhangi BS, Meco G, Denefle P, Wood NW, Agid Y, Brice A. French Parkinson's Disease Genetics Study, G. European Consortium on Genetic Susceptibility in Parkinon's D. Association between early-onset Parkinson's disease and mutations in the parkin gene. N. Engl. J. Med. 2000; 342(21):1560–7. doi: 10.1056/NEJM200005253422103. [DOI] [PubMed] [Google Scholar]
- 63.Mills JD, Janitz M. Alternative splicing of mRNA in the molecular pathology of neurodegenerative diseases. Neurobiol. Aging. 2012;33(5):1012 e11–24. doi: 10.1016/j.neurobiolaging.2011.10.030. [DOI] [PubMed] [Google Scholar]
- 64.Mordes D, Luo X, Kar A, Kuo D, Xu L, Fushimi K, Yu G, Sternberg P, Jr., Wu JY. Pre-mRNA splicing and retinitis pigmentosa. Mol. Vis. 2006;12:1259–71. [PMC free article] [PubMed] [Google Scholar]
- 65.Lefebvre S, Burglen L, Reboullet S, Clermont O, Burlet P, Viollet L, Benichou B, Cruaud C, Millasseau P, Zeviani M. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80(1):155–65. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 66.Faustino N A, Cooper T A. Pre-mRNA splicing and human disease. Genes Dev. 2003;17(4):419–37. doi: 10.1101/gad.1048803. [DOI] [PubMed] [Google Scholar]
- 67.Xu L, Lin D C, Yin D, Koeffler H P. An emerging role of PARK2 in cancer. J. Mol. Med. 2014;92(1):31–42. doi: 10.1007/s00109-013-1107-0. [DOI] [PubMed] [Google Scholar]