Abstract
The quest to understand why and how we age has led to numerous lines of investigation that have gradually converged to consider mitochondrial metabolism as a major player. During mitochondrial respiration a small and variable amount of the consumed oxygen is converted to reactive species of oxygen (ROS). For many years, these ROS have been perceived as harmful by-products of respiration. However, evidence from recent years indicates that ROS fulfill important roles as cellular messengers. Results obtained using model organisms suggest that ROS-dependent signalling may even activate beneficial cellular stress responses, which eventually may lead to increased lifespan. Nevertheless, when an overload of ROS cannot be properly disposed of, its accumulation generates oxidative stress, which plays a major part in the ageing process. Comparative studies about the rates of ROS production and oxidative damage accumulation, have led to the idea that the lower rate of mitochondrial oxygen radical generation of long-lived animals with respect to that of their short-lived counterpart, could be a primary cause of their slow ageing rate. A hitherto largely under-appreciated alternative view is that such lower rate of ROS production, rather than a cause may be a consequence of the metabolic constraints imposed for the large body sizes that accompany high lifespans. To help understanding the logical underpinning of this rather heterodox view, herein I review the current literature regarding the mechanisms of ROS formation, with particular emphasis on evolutionary aspects.
Keywords: Ageing, Lifespan, Mitochondria, OXPHOS, Oxygen, ROS.
OXYGEN, A BIOENERGETIC AND BIOSYNTHETIC NECESSITY FOR METAZOAN EVOLUTION
In the energy metabolism, due to thermodynamic constraints, there is always a trade-off between efficiency and rate [1, 2]. This argument is well illustrated by the comparison of anaerobic glycolysis with oxidative phosphorylation (OXPHOS). Although the yield of ATP produced by mol of glucose consumed is 18-19 times higher in OXPHOS, the rate of ATP production by glycolysis can be up to 100 times faster than that of OXPHOS. In line with these facts, thermodynamic analyses have concluded that a dissipative use of substrates may help cells to grow fast [3, 4]. However, if the strategy is to grow big in size rather than grow fast, then an efficient metabolism fits the bill.
Although the origin of undifferentiated multicellularity might have been rooted in an anoxygenic Earth [5], the complex multicellular forms of life that evolved in the Ediacaran and Cambrian periods, were only possible after oxygen was abundant enough as to support the high-energy demands of these forms of life. Indeed, when we examine life in the biosphere, we find that only aerobic organisms grow large. Anaerobic forms of life, either prokaryotes or eukaryotes, remain unicellular or, at best, uniseriate filaments [6].
The availability of molecular oxygen in the primitive atmosphere opened the way for the efficient harnessing of highly exergonic reactions based on O2 as terminal electron acceptor. Several reasons make this gas to be exceptionally well suited to serve as electron acceptor in the oxidation of carbon-base fuels. With the exception of fluorine and chlorine, the reduction of oxygen provides the largest free energy release per electron transfer. However, unlike the halogen gases, oxygen forms chemical bonds sufficiently stable to allow its accumulation in the atmosphere first and then to be harnessed as a biological oxidant. In contrast, fluorine is useless as biological oxidant because it generates an explosion upon contact with organic matter. Fluorine vigorously attacks R-H bonds and reacts with every other element apart from a few of the noble gases. Similarly, in water solutions, chlorine forms highly reactive hypochlorous acid, which is reactive with a range of organic molecules including thiols, thioesteres, amino acids, amines, unsaturated fatty acids and Fe-S centers [7].
An additional advantage of oxygen is that it is a gas in the pressure-temperature range of liquid water. A solid terminal oxidant (e.g., sulphur) would have to be consumed as food and would limit possible habitats. No such restriction applies with oxygen, which can be freed into an atmosphere and is sufficiently soluble to be distributed throughout an ocean. At the same time, O2 is sufficiently insoluble so that it partitions between the atmosphere and ocean in a 140:1 ratio, allowing the existence of complex life on land. Furthermore, the ability of O2 to diffuse across biological membranes and to bind transporter proteins (eg., hemoglobin) facilitates its delivery to systemic organs. Finally, the biochemical symmetry of oxygenic photosynthesis and aerobic respiration (the former produces oxygen from water and the latter reduces oxygen back to water) maintains homeostasis at the biosphere level.
The importance of oxygen in metazoan development is not limited to its bionergetic role as a final electron acceptor in mitochondrial respiration. Several biochemical networks analyses demonstrate that O2 is involved in a myriad of biosynthetic pathways [8], many of which are essential for specialized cell functions found exclusively in aerobic metazoans [9]. Thus, oxygen is an essential reactive for the biosynthesis of hydroxyproline and hydroxylysine (components of the connective tissue proteins) as well as for the synthesis of the visual pigment retinol, just to mention a few examples among a long list.
MOLECULAR OXYGEN, A DOUBLE-EDGED SWORD
It is not exaggerated to say that the appearance of mammals would be unthinkable in a planet without molecular oxygen. However, the efficient metabolism and the biosynthetic wealth that is sustained by O2, did not come without a price paid in terms of oxidative stress. During the course of cellular life a variable small fraction of the O2 consumed gets converted to potentially harmful reactive species of oxygen (ROS).
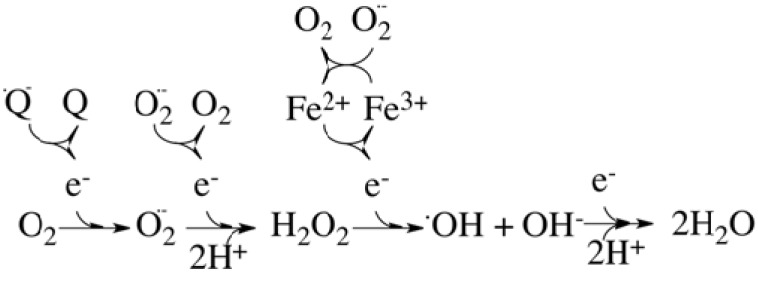
The first step in the production of ROS is the one-electron reduction of molecular oxygen (Fig. 1) to yield superoxide anion (O2.-), which, in spite of being a free radical, is not highly reactive by itself. Superoxide lacks the ability to penetrate lipid membranes and it is therefore enclosed in the compartment where it was produced. Two molecules of superoxide dismutate to hydrogen peroxide (H2O2) and molecular oxygen in a reaction catalysed by superoxide dismutase (SOD). Hydrogen peroxide is not a free radical and it is relatively unreactive. Nevertheless, H2O2 is an important radical forming molecule that can move through biological membranes. In the presence of Fe(II) it forms the very reactive and harmful hydroxyl radical (.OH).
Fig. (1).
Stepwise reduction of molecular oxygen. During cellular respiration, most of the oxygen consumed by the mitochondria is converted to the harmless by-product water at complex IV, where O2 accepts 4 electrons from the reduced form of cytochrome c. However, a variable small fraction of the consumed oxygen can accept a single electron directly from sites others than complex IV (for instance, from the strongly reducing ubisemiquinone anion, .Q-). This side reaction will produce superoxide anion, O2, which can dismutate to produce hydrogen peroxide (H2O2) and molecular oxygen. Hydrogen peroxide can subsequently lead to the formation of the very reactive hydroxyl radical (.OH), in a reaction catalyzed by metal ions (Fe2+ or Cu2+) that is known as the Fenton reaction. Superoxide also plays an important role in connection with the Fenton reaction by recycling the metal ions.
These reactive species are generated in diverse compartments and by different enzymes within the cell [10]. Thus, cytosolic enzymes such as cyclooxygenases [11], lypoxygenases [12] and flavoenzymes like xanthine oxidase [13] are able to produce ROS. Also, NADPH oxidases within the plasma membrane [14] and enzymes involved in lipid metabolism within the peroxisomes, generate ROS. Although all these sources contribute to the overall oxidative burden, the vast majority of the ROS produced in the cell (around 90 %) can be traced back to mitochondria [15].
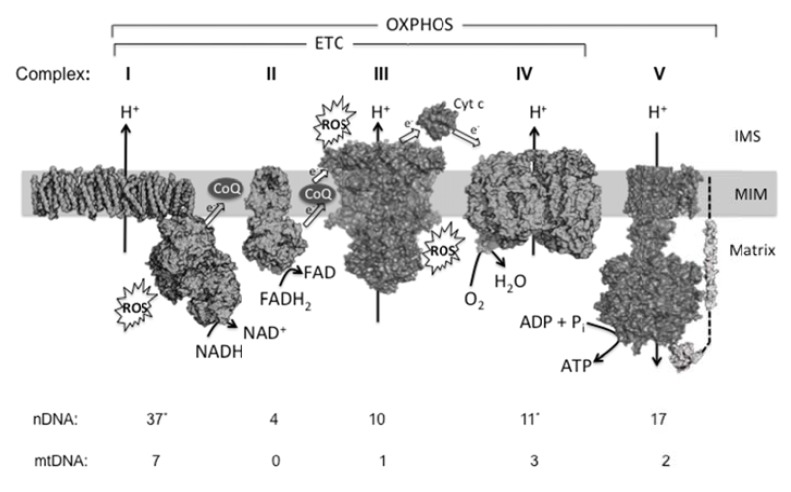
The mitochondrial electron transport chain (ETC) consists of four multi-subunit complexes that mediate the transfer of electrons from NADH or FADH to molecular oxygen (Fig. 2). The energy that is released as the electrons traverse complexes I, III and IV is used to pump protons out of the mitochondrial matrix across the inner mitochondrial membrane, resulting in an electrochemical gradient that will drive the phosphorylation of ADP by the ATP synthase (complex V). Ubiquinone (CoQ), a lipid-soluble molecule, and cytochrome c, a small heme protein, are two mobile elements that act as electron carriers connecting the different macromolecular complexes [16]. During OXPHOS, electrons removed from substrates ultimately reduce oxygen to the harmless by-product water through a four-electron transfer process at complex IV. The oxygen molecule bound to complex IV is not released until all four electrons are transferred. However, oxygen can interact with other centers in the mitochondrial ETC, and, in these cases, oxygen usually receives only a single electron yielding superoxide, which subsequently will lead to other forms of ROS (Fig. 1).
Fig. (2).
Overview of mitochondrial oxidative phosphorylation. The mitochondrial OXPHOS system consists of five multisubunit complexes (I-V) that reside in the mitochondrial inner membrane (MIM). The subunits of these complexes are encoded by the mitochondrial (mtDNA) and nuclear DNA (nDNA). The number of polypeptides being encoded by each genome in mammals is indicated at the bottom. Although organisms other than mammals may exhibit a different pattern of subunits, almost all aerobic organisms carry out OXPHOS according the pathway sketched in this figure. In mammals, complex I is the largest OXPHOS enzyme proposed to consist of 44 different subunits. A simpler bacterial complex I is shown here (3m9s), which is composed of 14 protein chains (corresponding to the “core subunits” from mammals). (*) Recent evidence suggests that the “supernumerary” subunit NDUFA4 hitherto classified as a complex I constituent appears to be a component of complex IV [158], which in this case would have 11 nDNA-encoded subunits. At complex I and complex II (3aee), NADH and FADH2 are oxidized, respectively, and the released electrons are transported to complex III (1be3) via Coenzyme Q10 (CoQ, ubiquinone/ubiquinol). From thereon, electrons are transported to complex IV (2occ) via cytochrome c (Cyt c, 1hrc) and donate to oxygen. Together, complexes I-IV constitute the electron transport chain (ETC). The energy derived from the electron transport is used to expel protons from the mitochondrial matrix across the MIM. This establishes an electrochemical proton-motive force. The backflow of protons is used by complex V (1c17, 1e79) to drive the phosphorylation of ADP and produce ATP. Complexes I and III are thought to be the main ROS production sites.
This side reaction will produce superoxide anion, , which can dismutate to produce hydrogen peroxide (H2O2) and molecular oxygen. Hydrogen peroxide can subsequently lead to the formation of the very reactive hydroxyl radical (.OH), in a reaction catalyzed by metal ions (Fe2+ or Cu2+) that is known as the Fenton reaction. Superoxide also plays an important role in connection with the Fenton reaction by recycling the metal ions.
Traditionally, these mitochondrial oxidants have been thought of as an inevitable, useless and harmful by-product of respiratory metabolism. That notion has slowly given way to a more nuanced view of ROS as important signalling molecules, reviewed in [17-19]. It has been observed that under hypoxic conditions, complex III-derived ROS lead to the stabilization of the hypoxia-inducible factor-1 alpha, HIF-1α, [20-22], pointing to ROS as key mediators in the cellular response to hypoxia. Recent evidence suggests that starvation triggers a rise in ROS, promoting the oxidation of a critical cysteine residue of the ATG4 gene product (an essential gene involved in autophagy), which, in turn, would lead to the induction of autophagy [23]. Another area where ROS seem to play fundamental roles as signalling elements is in the innate immunity. Thus, pharmacological agents that block complex I or III result in the increased release of ROS and produce an increase in secreted IL-1β, an important mediator of the inflammatory response [24]. Furthermore, antioxidants specifically targeted to the mitochondria appeared to prevent inflammation [24, 25]. Evidences also point to a connection between mitochondrial ROS and stem cell function [26, 27], insulin signalling [28, 29], senescence and tumour suppression [30], transcription factor function [31, 32], growth and development [19], and so on.
WHEN THE THREAT BECOMES A FACT: AGEING AND AGE-RELATED DISEASES
Because of the presence of unpaired valence shell electrons, ROS are highly reactive and can attack biomolecules such as DNA [33], proteins [34] and lipids [35], causing damage to these cellular components. Why, then, has evolution allowed mitochondrial ROS production to persist? Although we do not know the answer, it may well be that ROS production is an inevitable consequence of aerobic metabolism. In this sense, Martin Brand has suggested that historical and structural constraints on the ETC, may have closed the way to retain functionality in the absence of electron leaks [36]. If this view is correct, then the beneficial roles played by ROS as second messengers make sense of Shakespeare’s words in Richard II: “there is no virtue like necessity”. Nevertheless, such a virtue could not be possible without developing antioxidant defenses to limit the magnitude of the oxidative load and its action along the time. However, antioxidant defenses are not infallible. Whenever large changes in tissue ROS levels overwhelm the capacity of an organism to cope with such a burden, that organism will face oxidative stress. Oxidative stress is therefore a state in which cells and tissues experience an abnormal predominance of oxidant over anti-oxidant activity.
There are at least two different mechanisms by which oxidative stress exert cellular damage [37]. First, an imbalanced redox state can cause disruptions in normal mechanisms of cellular signalling where ROS are involved [38]. The resulting imbalance seems to contribute to the pathogenesis of age-related maladies such as cancer [39-41], atherosclerosis [42], neurodegenerative diseases [43], rheumatoid arthritits [44], diabetes [29, 45, 46], etc. On the other hand, an excess of ROS mediates nonspecific irreversible damage to tissues, including cellular and matrix components, that results in a progressive organs dysfunction in a stochastic but nevertheless cumulative manner. According to the current free radical theory of ageing, this oxidative damage to tissues contributes, together with other causes [47], to the ageing process [48]. At this point, it may be convenient to review some recent data that question the ROS theory of ageing, at least in the naïve way that it has been formulated until now.
MITOHORMESIS
During the last few years, a number of key findings have challenged the classical view of the free radical theory of ageing. For instance, cells from mice deficient in mitochondrial DNA polymerase γ exhibit aspects of premature aging and reduced lifespan in association with the accumulation of random point mutations and deletions in mtDNA [49]. However, in these cells accumulation of mtDNA mutations is not associated with increased markers of oxidative stress [50]. Interestingly, in a very recent work Itsara and coworkers report that oxidative stress is not a major contributor to somatic mtDNA mutations [51]. A different line of evidence forcing a re-evaluation of the free radical theory of aging comes from experiments with the model organism Caenorhabditis elegans. Thus, Van Raamsdonk and Hekimi generated animals that completely lacked SOD activity. Although these worms showed an increased sensitivity to multiple stresses, they were viable and exhibited a normal lifespan [52]. On the other hand, although overexpression of the Cu/Zn-SOD isoform in C. elegans leads to an increased lifespan, this effect does not seem to be caused by decreased oxidative damage [53]. Furthermore, this lifespan extension was partially suppressed by the inactivation of the ER stress response, suggesting that the accumulation of the overexpressed protein might have challenged protein-folding homeostasis, triggering a stress response that eventually extended lifespan.
In mammals, exhaustive studies on the effect of under- or overexpressing a large number and a wide variety of genes coding for antioxidant enzymes, also call into question the hypothesis of a simple link between oxidative stress and longevity [54]. Even more, in recent years the view that ROS may mediate lifespan extension has gained popularity. Thus, low doses of the superoxide generating compound paraquat have been shown to extend lifespan in C. elegans [55]. These and additional data in worms, along with complementary evidence obtained from diverse model organisms, has led to the mitohormesis model (reviewed in [56]).
In general, hormesis is the term for favourable biological responses to low exposure to stressors. According to the mitohormesis paradigm, mild mitochondrial stress results in a broad and diverse cytosolic and nuclear response that promotes cytoprotective states. Thus, low levels of potentially damaging ROS may activate beneficial cellular stress responses, while higher levels are obviously detrimental. Inhibition of glycolysis, impairment of insulin-like signalling and certain mutations in mitochondrial ETC components, have been pointed as conditions that promote longevity via ROS-dependent mitohormesis [57]. Indeed, a mild inhibition of mitochondrial respiration extends the lifespan of organisms as diverse as yeast, worms, flies and mice [58-61]. Although the underlying mechanisms are not well understood, it has been proposed that the mild increase in ROS observed in respiration mutants, may stimulate HIF-1 to activate gene expression and promote longevity [62].
In response to mitochondrial mild insults, there exists a stress response that is communicated to the nucleus to increase the expression of mitochondrial chaperones such as HSP-6 and HSP-60. This mitochondrial unfolded protein response (UPRmt) has been suggested as causal for lifespan extension due to inhibition of the ETC in the nematode C. elegans [63]. However, Bennett and coworkers have recently reported that deletion of atfs-1, encoding for a transcription factor required for the induction of the UPRmt, does not avoid lifespan extension after inhibition of the ETC. Furthermore, constitutive activation of the UPRmt by gain of function mutations in atfs-1 fails to extend lifespan [64]. On the other hand, a recent study showed that deficiencies in prohibitin, a protein that influence mitochondrial respiration, mitochondrial fusion and mitochondrial protein quality [65], in addition to shortening lifespan also causes enrichment of several UPRmt components in mitochondria of yeast cells [66]. In summary, although it is still unclear whether the UPRmt is either necessary or sufficient to account for lifespan extension in response to mitochondrial perturbations, it seems clear that, regardless of the signalling pathway(s) involved, ROS can mediate a hermetic response.
COPING WITH OXIDATIVE STRESS: AN EVOLUTIONARY PERSPECTIVE
Mitochondrial function has a profound impact on the aging process [67, 68], not just because mitochondria are the major source of cellular ROS, as we have reviewed above, but because they are also the main target for damage [69, 70]. Consistent with this theory, a number of mitochondrial adaptations to oxidative stress related to longevity have been described in the literature, some of which are reasonably well understood [71, 72], while others are matter of hot discussion [73-77]. In an interesting study, Yu and colleagues used RNA sequencing to compare liver gene expression profiles between naked mole-rats (an extremely long-lived rodent) and wild-derived mice. These authors found that genes associated with oxidoreduction and mitochondria were expressed at higher relative levels in naked mole-rats [78]. These results, thus, point towards a putative role for mitochondrial alterations in the long lifespan of the naked mole-rat.
In an attempt to organize and systematize the current knowledge regarding antioxidant defenses, we can distinguish three different, although non-excluding, general strategies that animals adopt to mitigate the deleterious effects of ROS. A first line of defense against ROS-related damage consists in ameliorating the rate of ROS production. As a second line of defense, organisms have evolved complex antioxidant mechanisms involving low molecular weight compounds as well as antioxidant enzymes. A third strategy that animals seem to have adopted during the course of evolution consists of favouring macromolecular constituents less susceptible to oxidative modification. In the remaining of the current review, I will focus on the first line of defense: control of the rate of ROS production. I will try to dissect facts from interpretations and, whenever possible, stressing current uncertainties and identifying future issues to be addressed to gain a better understanding.
AVOIDING PROBLEMS IS BETTER THAN RESOLVING THEM: RATES OF ROS PRODUCTION
Endogenous antioxidants have been intensively studied in the past in relation to ageing, while much less attention has been paid to the regulation of ROS production, despite the fact that prevention would appear to be the most logical way to reduce oxidative damage. In the current section we will start outlining what we have learnt during the last years about mitochondrial ROS production and the factors affecting it. Afterwards, the relationship between oxygen consumption and ROS production will be examined to conclude that the more oxygen consumed does not necessarily mean the more ROS produced. Finally, two different strategies aimed to reduce the rate of ROS generation will be presented.
What Factors Influence Mitochondrial ROS Production?
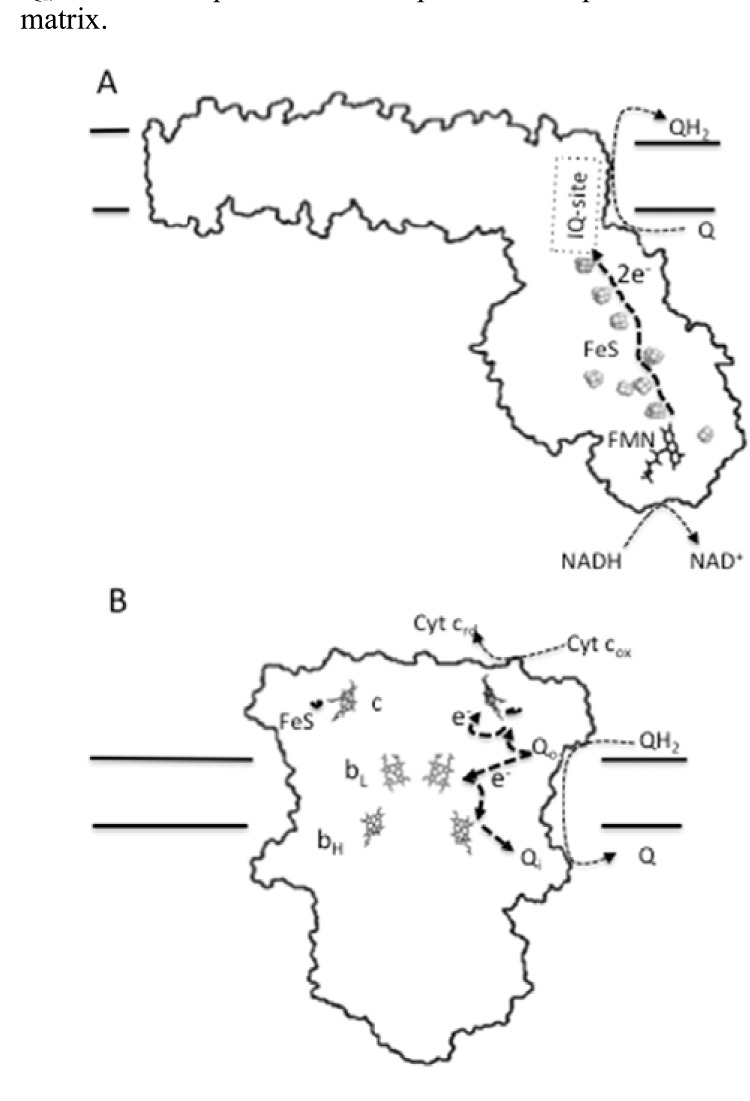
To understand how some animal species have achieved reduced rates of ROS production without compromising their bioenergetic functions, we need previous knowledge of how mitochondria produce ROS and what factors govern mitochondrial ROS generation. There are currently seven separate sites of mammalian mitochondrial ROS production that have been identified [79]. Nevertheless, it is widely accepted that the greatest maximum capacities to generate ROS are at complex I and complex III [80-82]. Complex I remains the least understood of the ETC complexes. In mammals, complex I is composed of around 44 different subunits resulting in a molecular mass of about 1 MDa [83]. The complex contains one flavin mononucleotide (FMN) and nine iron-sulfur clusters as redox prosthetic groups (Fig. 3). This supramolecular complex has a hydrophobic arm, involved in proton pumping [84], and a matrix-protruding hydrophilic arm, which contains the prosthetic groups. A third redox center of complex I is formed by the ubiquinone binding site (IQ-site) at the junction between the arms (Fig. 3), which lacks covalently bound prosthetic groups [85, 86]. Because of the importance of identifying the sites of superoxide production within this large multi-subunit complex, much effort has been devoted to this task. Despite all this effort, the controversy about the ROS generating site(s) dominates the field. Thus, it has been proposed that the iron-sulfur groups [87], the reduced flavin site (IF-site) alone [88] and the IF-site together with the IQ-site [89] are the relevant sites where one-electron reduction of oxygen takes place within complex I. At this point it is pertinent to quote Francis Crick, who used to say that “if you want to understand function, study structure”. In this sense, the structure of the water-soluble arm of complex I from Thermus thermophilus, which contains the FMN and the FeS centers, has been elucidated and is likely to be very similar to that in mammals [90, 91]. This structure indicates that most of the FeS centers in the hydrophilic arm are well shielded from oxygen, so that O2 is most likely to access electrons at the IF and IQ sites [79, 89, 92], yielding superoxide exclusively in the matrix side [79].
Fig. (3).
Prosthetic groups and redox centers of ROS-producing complexes I and III. A diagram of complex I indicating the pathway of electrons transfer from NADH to ubiquinone (A). The complex has a L-shaped structure, with a hydrophobic arm embedded into the inner mitochondrial membrane and a matrix-protruding hydrophilic arm, which contains three redox centers. The first center is formed by the flavin mononucleotide, FMN, prosthetic group (IF-site). The second center consists of nine FeS clusters. A third redox center of complex I is formed by the ubiquinone binding site (IQ-site). In (B) the arrangement of the redox centers of complex III is shown. This complex, which is a homodimer, passes electrons from ubiquinol to cytochrome c. Each monomer possesses three subunits with prosthetic groups that serve as redox centers. These are cytochrome b (which contains two hemes: bH and bL), cytochrome c1 (which contains one heme c group) and the Rieske iron-sulfur protein (which contains a 2Fe-2S cluster). In addition to these prosthetic groups, complex III has two separated ubiquinone/ubiquinol binding sites: an ubiquinol oxidizing site (Qo) and an ubiquinone reducing site (Qi), which, despite the lack of covalently bound prosthetic groups, are key factors of the redox process.
Beside complex I, the other main source of mitochondrial ROS is complex III. In mammals, this complex is a homodimer with each monomer consisting of 11 different polypeptide subunits [93], of which three have prosthetic groups that serve as redox centers. These are cytochrome b (which contains two hemes: bH and bL), cytochrome c1 (which contains one heme group) and the Rieske iron-sulfur protein (which contains a 2Fe-2S cluster). This multimeric enzyme funnels electrons from the reduced ubiquinol pool (QH2) to cytochrome c, during the so-called Q-cycle (Fig. 3). For this catalytic redox process, complex III has two separated ubiquinone/ubiquinol binding sites: an ubiquinol oxidizing site (Qo) and an ubiquinone reducing site (Qi). Oxidation of the ubiquinol at the Qo site results in the transfer of one electron to the 2Fe-2S cluster of the Rieske protein with the release of two protons to the intermembrane space. The Rieske protein then transfers an electron to cytochrome c1. The strongly reducing ubisemiquinone anion (.Q-) formed at the Qo site after the transfer of the first electron from ubiquinol, rapidly transfer an electron to the low-potential heme bL, which then transfer one electron to the high-potential heme bH. The reduced bH, in turn, transfer this electron to ubiquinone at the Qi site to form a stable ubisemiquinone. To complete the Q-cycle, a second molecule of ubiquinol is oxided at the Qo site with the release of another two protons and transfer of one electron to cytochrome c (after passing through the Rieske and cytochrome c1), while the second electron of the oxided ubiquinol, after traveling through bL and bH, will eventually reach the ubisemiquinone at the site Qi, to form ubiquinol with the uptake of two protons from the matrix.
A diagram of complex I indicating the pathway of electrons transfer from NADH to ubiquinone (A). The complex has a L-shaped structure, with a hydrophobic arm embedded into the inner mitochondrial membrane and a matrix-protruding hydrophilic arm, which contains three redox centers. The first center is formed by the flavin mononucleotide, FMN, prosthetic group (IF-site). The second center consists of nine FeS clusters. A third redox center of complex I is formed by the ubiquinone binding site (IQ-site). In (B) the arrangement of the redox centers of complex III is shown. This complex, which is a homodimer, passes electrons from ubiquinol to cytochrome c. Each monomer possesses three subunits with prosthetic groups that serve as redox centers. These are cytochrome b (which contains two hemes: bH and bL), cytochrome c1 (which contains one heme c group) and the Rieske iron-sulfur protein (which contains a 2Fe-2S cluster). In addition to these prosthetic groups, complex III has two separated ubiquinone/ubiquinol binding sites: an ubiquinol oxidizing site (Qo) and an ubiquinone reducing site (Qi), which, despite the lack of covalently bound prosthetic groups, are key factors of the redox process.
Mitochondria incubated with substrates able to maintain a pool of reduced ubiquinol and in the presence of antimycin, produce large amounts of O2.- from the reaction of molecular oxygen with a ubisemiquinone bound to the Qo site [94, 95]. This superoxide is released from complex III to both sides of the inner membrane [96]. Since antimycin is an inhibitor that blocks electron transfer from bH to the site Qi, causing a small build-up of ubisemiquinone at the center Qo, some authors argue that the Qo-site from complex III (IIIQo-site) plays a minor role in mitochondrial superoxide production in the absence of antimycin [97], while others argue that site IIIQo is the main site under physiological conditions [81].
Although further efforts, and probably new approaches, are needed to disentangle the relative contribution of each site to mitochondrial ROS generation in vivo, studies carried out with isolated mitochondria have led to propose different modes of mitochondrial operation that lead to superoxide production [92]. The exam of these modes helps to outline the conditions that may favour superoxide production in complexes I and III. Thus, one mode involves the IF-site and ROS generation is favoured by a high NADH/NAD+ ratio in the matrix [98]. Others modes gain relevance when there is a considerable pool of reduced ubiquinol in conjunction with a high proton-motive force (pmf) [99].
Relationship Between Oxygen Consumption and ROS Production
A number of studies have compared mitochondrial ROS production between species showing different longevities. The results of such studies showed that the rates of ROS generation of mitochondria isolated from postmitotic tissues were lower in long-lived than in short-lived species [100-102]. On the other hand, the inverse relationship between mass-specific metabolic rate and lifespan has been recognized since the early twentieth century, when Rubner reported that the total amount of energy consumed through a lifetime by six different mammalian species, which exhibited 6-fold differences in lifespan, was approximately constant [103]. That is, it seemed like those species living longer expended their metabolic potential at lower rate. This way of thinking eventually led to the so-called rate-of-living theory of ageing [104]. According to modern formulations of this theory, a high mass-specific metabolic rate would be the cause of a high rate of ROS production, which would explain the observed positive correlation between metabolic rate and ROS production. However, this causal explanation is based on the implicit, although unsupported, assumption that a fixed proportion of oxygen consumed by cells is diverted to ROS generation [105]. As we will see next, such an assumption is a misleading oversimplification.
Despite the intuitive appeal of this idea, the accumulated evidence challenges this simple view. In this sense, the H2O2 production rate during the active state 3 respiration (high oxygen consumption) is actually lower than in the resting state 4 (low oxygen consumption) [106]. Furthermore, decreases in mitochondrial oxygen radical production occur during many other situations under which oxygen consumption does not decrease and even increases, such as aerobic exercise bouts, chronic exercise training and hyperthyroidism [107]. On the other hand, in yeast, genetic mutants such as deletion of TOR1 (target of rapamycin) or MRG19 (a putative transcription factor that regulates carbon and nitrogen metabolism in yeast), which affect to the expression of ETC components, also produce a reduction in ROS production while oxygen consumption is greatly increased [108, 109]. Another line of evidence in support of a complex relationship between respiration and ROS production, comes from dietary restriction (DR) experiments. Although the details of the underlying mechanisms are still being worked out [110], DR extends lifespan in a wide variety of species including mammals. In these experiments, decreases in ROS production were accompanied either by no changes in oxygen consumption [111, 112] or by mitochondrial biogenesis with increased oxygen consumption [112-114]. Finally, the finding that long-lived birds produce less ROS than short-lived mammals of similar body size, regardless their metabolic rates [107, 115, 116], lends strong support to the hypothesis that oxygen consumption and ROS production are not strictly linked. In line with these observations, Speakman and coworkers reported that individual mice with high metabolism had greater mitochondrial uncoupling and lived longer [117]. To reconcile all these observations with the free-radical theory of ageing, Brand has put forward the hypothesis of “uncoupling to survive” [36]. According to this hypothesis, an increase in the proton conductance of the mitochondrial inner membrane leads to an increased oxygen consumption and a decreased ROS production rate. Indeed, a drop in the proton motive force favours electron transport and oxygen consumption, which will lead to a reduction in the steady concentration of ubisemiquinone and to a lower oxygen tension around the mitochondrion. Thus, proton leak has a twofold beneficial effect on the decline of ROS production. In this sense, it has been known since long that mitochondria from short-lived mammals exhibit higher proton conductances when compared to those from their long-lived counterparts [118]. This negative correlation between longevity and degree of mitochondrial uncoupling, once thought to be the cause of higher oxidative stress in short-lived mammals, has been currently reinterpreted, at the light of the ‘uncoupling to survive’ hypothesis, as an adaptive response aimed at limiting further ROS production in those animals that already present high rates of ROS generation.
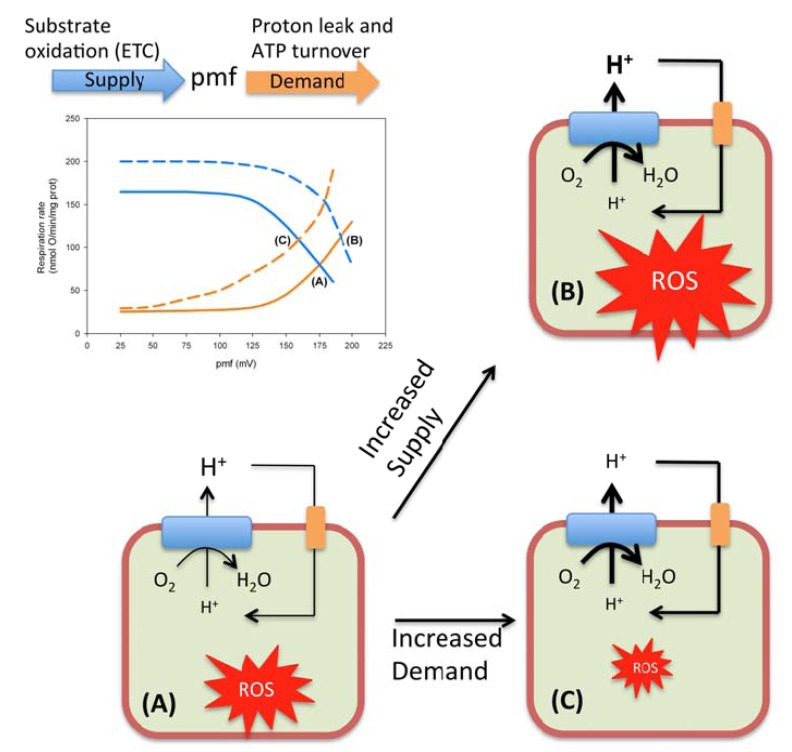
Although until very recently it has been commonly assumed that an increase in oxygen consumption led to an increase in ROS production, we now increasingly appreciate that the rate of oxygen consumption can have opposite effects on the rate of ROS production depending on the force driving this oxygen consumption. Thus, if the rise in O2 consumption is related to a higher tissue pO2 or to an increase in the content of reduced ROS-producing sites, one would expect an enhanced ROS production. In contrast, if the cause of the increased oxygen utilization is a decreased proton motive force, in the setting of constant tissue pO2 and a fixed number of ETC complexes, then the expected output would be a reduced rate of ROS generation (Fig. 4).
Fig. (4).
Relationship between oxygen consumption and ROS production rates. Modular kinetic analysis has been revealed as a useful approach to investigate the regulation of OXPHOS [159]. OXPHOS is a process that can be divided conceptually into events that either generate (substrate oxidation and electron transport from NADH to O2) or dissipate (ATP turnover and proton leak) the mitochondrial proton-motive force (pmf). The steady-state respiratory rate and pmf, result from the kinetic interplay between these two modules as it is illustrated in the plot. Thus, when the rate of pmf-production (continuous blue curve) matches the rate of pmf-consumption (continuous orange curve), a steady state is established at the respiration rate and pmf given by the coordinates of the point labeled as (A) in the plot. An increased oxygen consumption (respiration rate) can be obtained in two different ways. For instance, an activation of the activities encompassed in the supply module (discontinuous blue curve) will drive the system to a new steady state (B), accompanied by both an increased oxygen consumption and an increased pmf. Since the rate of ROS production is highly sensitive to the pmf, this increase in oxygen consumption is expected to be paralleled by an increase in ROS production. On the other hand, a similar increase in O2 consumption can be imposed by activation of the demand block (discontinuous orange curve). In this case, the new steady state (C) is set at a lower pmf, which will have a reducing effect on the rate of ROS production.
STRATEGIES TO DECREASE MITOCHONDRIAL ROS PRODUCTION
From the exposed above, it seems that natural selection may have two obvious targets to act on. On one side, those interventions leading to a decreased stationary proton motive force should favour a lower basal ROS production. On the other side, reducing electron stalling at the ETC by controlling the entrance and/or the exit of electrons, also may favour a decreased ROS production. In this section, we will review the current evidence suggesting that both strategies might have been successfully adopted by different animal taxa.
Matching the Electron Entrance and Exit Gates
On average, the maximum lifespan potential of birds is twice that of similar-sized mammals [119, 120], while their metabolic rates can be even higher than those exhibited by mammals [121]. In the early nineties, the group of Barja and, independently, the group of Sohal were responsible for a number of seminal rat-pigeon studies that pointed to a decreased rate of ROS production as a potential cause of the superior lifespan of pigeons [115, 122]. Posterior work extended these observations to other species of birds such as canaries and parakeets [116]. Nevertheless, caution is required when generalizing these findings. Although ample evidence supports a negative correlation between rate of ROS generation and lifespan, a number of recent contributions have questioned its general validity [121, 123, 124]. Thus, the long-lived house sparrow, despite of presenting a lower percentage free radical leak than mouse, releases mitochondrial H2O2 at higher rates [125]. The comparison between quails and parrots indicates that despite the 5-fold longevity difference, their mitochondrial ROS production rates were similar [123]. Another example questioning the universal validity of the negative relationship between lifespan and ROS production, generally observed among metazoans, comes from experiments with flies. Miwa and coworkers obtained Drosophila melanogaster mutants overexpressing the mitochondrial adenine nucleotide translocase (ANT). These mutants had significantly lower ROS production due to their lower membrane potential, however their lifespan was not extended compared to the wild type controls [124]. Nevertheless, since lifespan is a complex trait influenced by many variables [126-130], the above described exceptions should not take anyone by surprise.
In any event, work from different laboratories has established that, whenever long-lived species produce less ROS than long-lived animals, differences in complex I biology may be responsible for this differential behaviour [111, 116, 131]. Thus, in a recent and rigorous study, Lambert and coworkers concluded that mitochondria from the long-lived pigeon display low rates of hydrogen peroxide production because they have low levels of complex I [131]. Indeed, complex I represents an entrance gate for electrons, which are incorporated to the ETC. Different lines of evidences suggest that reducing the flux through this gate may mitigate the rate of ROS generation. In this sense, besides the reduced levels of complex I reported for pigeons [131], dietary restriction seems to exert its beneficial effects through a reduction of ROS production at complex I [111]. On the other hand, expression of the yeast Ndi1, which encodes for an alternative NADH dehydrogenase able to bypass the proton-translocating complex I, in Drosophila conferred increased lifespan to the flies [132]. This effect was associated with a significant decrease of several key markers of oxidative damage, indicating that the mechanism by which NDI1 extended lifespan was by protecting flies against oxidative stress emanating from excess ROS production at mitochondrial complex I. These findings are in line with the observation that deletion of MRG19, a putative transcription factor, leads to up-regulation of genes encoding for alternative NADH dehydrogenases such as Ndi and Nde [109], which is accompanied by increases of both chronological and replicative lifespan of yeast [133]. In addition, this mutant showed lower rates of ROS production and higher respiration rates, with respect to its wild type counterpart [109].
Having established that down-regulation of complex I can ameliorate ROS production, questions emerge regarding how these cells manage to maintain proper ATP supply when facing a diminished NADH-ubiquinone oxidoreductase activity. Clues that help to answer this question come from two fronts. First, calorie restriction has been shown to induce mitochondrial biogenesis [113, 134]. Thus, cells from mice under DR showed an increased number of mitochondria per cell [113, 135]. These mitochondria exhibited reduced membrane potential and generated less ROS than controls, but remarkably they were able to maintain their critical ATP production [134]. These observations support the view that a larger amount of low-potential mitochondria would maintain the ability to supply ATP to match the energy demand. A completely different line of evidence, which also argues for a relevant link between mitochondrial biogenesis and low ROS production, comes from deletion experiments using yeast. Thus, deletion of either the TOR1 gene or the MRG19 gene has been reported to up-regulate the biosynthesis of mtDNA-encoded proteins leading to increased respiration accompanied by decreased ROS production and extension of lifespan [108, 109]. Taken together, these results suggests that signalling pathways leading to OXPHOS activation at downstream complex-I sites, would decrease the reduction status of ETC components and, subsequently, ROS release rate. In other words, electron stalling occurs when the rate of entry of electrons via complex I exceeds the rate of transit through the slowest step of the ETC. Since complex IV (cytochrome c oxidase) is thought to be the rate-limiting complex of the ETC [136], rapid entry of electrons through complex I could trigger electron stalling at complexes I and III as well as leading to a high pool of ubisemiquinone, conditions under which superoxide anion radical production is well known to be favoured [95]. Therefore, it can be summarized that complex I seems to act as a pacemaker of the ageing process and therefore as a potential target of natural selection.
The Uncoupling to Survive Hypothesis
Mechanisms that allow protons to bypass the ATP synthase while re-entering the matrix, basically short-circuit the coupling of electron transport (oxygen consumption) to ADP phosphorylation. We know that the coupling of OXPHOS not only can be expected to change with substrate availability but also reflects an ontogenetic response of mitochondria to fit specific organ roles [137]. However, the relevant question we try to address in the current section is, might natural selection have acted on the coupling degree to influence ROS production?
In 1993, Sohal’s group published a seminal article where rates of mitochondrial O2.- and H2O2 production, as well as of O2 consumption, were compared among different mammalian species whose lifespan varied in one order of magnitude [100]. They found that the rates of ROS generation were inversely correlated to lifespan, and directly related to state 4 mitochondrial respiration. State 4 respiration is indicative of the proton leak across the inner mitochondrial membrane, as it was corroborated the same year by Porter and Brand, who reported a negative correlation between proton leak and body size, and consequently lifespan [118]. Nearly twenty years ago, the conclusion to be withdrawn from these observations seemed to be clear: Species showing a higher degree of uncoupling exhibit shorter lifespans because proton-leak drives oxygen consumption, which was thought, in turn, to drive ROS production leading to cellular damage. However, two decades later we have learnt that mild mitochondrial uncoupling lowers ROS by decreasing pmf [138], which may impact cellular ageing, including human tissues [139].
In this context, those genes encoding for proteins that mediate, in a regulated fashion, proton leak, are obvious candidates for selection. The uncoupling proteins (UCPs) are a subfamily of the mitochondrial anion carrier family, which transport substrates across the mitochondrial inner membrane [138]. The canonical UCP1 was the first identified uncoupling protein [140], and mediates non-shivering thermogenesis in eutherian mammals. Subsequently, UCP2 and UCP3 were described as paralogous proteins [141-144]. Although the repertory of functions fulfilled by these other members of the UCP protein is not fully established, available data point to a general role in protection against oxidative stress. Thus, it has been shown that superoxide increases mitochondrial proton conductance through effects on UCP1, UCP2 and UCP3, suggesting that these proteins may form part of a regulated mechanisms for decreasing the concentrations of ROS inside mitochondria [145]. Even the thermogenic UCP1 can mediate decreases in superoxide production in brown adipose tissue mitochondria [146]. This report is in line with the observation that orthologous of UCP1 are present in vertebrates with and without non-shivering thermogenesis [147-149], and with the suggestion that ancestral UCP1 probably had a role in protection against oxidative stress [150].
Thus, a large body of data support a role for the UCPs in defense against oxidations [151, 152]. On the other hand, we know that short-lived animals produce ROS at higher rates than their long-lived counterparts (reviewed above). Since ROS production has a negative feedback mechanism that functions increasing the activity of UCPs [145, 153] it seems reasonable to relate the observed positive correlation between longevity and the degree of mitochondrial coupling with an adaptive mechanism evolved to cope with oxidative stress. Because the regulation of UCPs appears to occur at numerous steps, including transcription, translation, degradation and modulation of protein activity [152], it would be interesting to address whether each of these regulatory steps exhibits species-specific particularities. For instance, how the UCPs from a long-lived species would respond in the oxidative context of a short-lived organism is not addressed in the literature, and it remains unknown whether the higher selective pressure imposed by a higher oxidative stress might have led to positive selection of UCPs in the short-lived lineages, allowing the acquisition of enhanced responsiveness to ROS. Phylogenetic analyses searching for accelerated evolution of UCPs in short-lived lineages may shed light on this issue.
DIFFERENT TARGETS FOR DIFFERENT STRATEGIES
Ageing is an almost universal feature of multicellular organisms. However, different species vary dramatically in their lifespan [120]. For long-lived species, fitness is limited by the time investment and resource acquisition demands of offspring production and selection is expected to promote low-risk, low-wear-and-tear strategies with moderate rates of returns over extended time periods. This life-history strategy relies on a rather slow but efficient metabolism. In contrast, short-lived organisms are expected to pursue a high risk, “live fast, die young” life-history strategy that have the potential to yield high fitness returns over a short time frame; such a strategy demands high metabolic rates at the expense of a reduction in the energetic efficiency [1, 2]. Although from an anthropocentric point of view, we could think that living long lives may represent an appealing goal, it should be pointed that in terms of biological fitness, both strategies (short- and long-lived features) can perform equally well, each within its own ecological niche [4]. Interestingly, it has been suggested that at least in bacteria, ageing may be a conditional strategic choice and not an inevitable outcome [154].
Hitherto, the question of whether these different life-histories impose distinct constraints with regard to the evolution of anti-oxidant defenses, has been an issue largely ignored in the literature. In the remaining of this section I will discuss some ideas that, although rather speculative, pretend to provide a working hypothesis that may guide future research.
Bigger animals live longer [155]. Because of their size they need to make an efficient use of the energetic resources, even if it is at the expense of lower metabolic (growth) rates [156, 157]. An efficient thermodynamic regime requires a rather tight coupling between oxygen consumption and phosphorylation. That is congruent with the observation that proton leak through the inner mitochondrial membrane decreases with increasing body mass in mammals [118]. Therefore, the constraints imposed for a large body size may disfavour UCPs as target of natural selection with regard to the evolution of anti-oxidant defense. Under this scenario, an increased selective pressure on complex I would fit the bill, accounting for the negative relationship observed between lifespan and levels of complex I [131]. In contrast, small animals, which exhibit short lifespan, trade energetic efficiency off for increased rates. Such thermodynamic constraint, would penalize any reduction in complex I activity, while favouring high degree of uncoupling to allow high metabolic fluxes and keeping ROS production rates within tolerable ranges (Fig. 5). In summary, slower rates of ROS formation may be a consequence of a life strategy that favours growth yield (efficiency) over growth rate. This tradeoff between growth rate and growth yield, linked to ROS production is reminiscent of some observations carried out in bacteria. Briefly, in prokaryotes, asymmetric division accompanied by ageing and death of some cells, results in a higher growth rate but a reduced growth yield. In contrast, symmetric division with gradual replacement of the old components, slows down the growth rate but may increase growth yield [154]. These results suggest that ageing may be worth when growing fast is the best strategy for the given conditions.
Fig. (5).
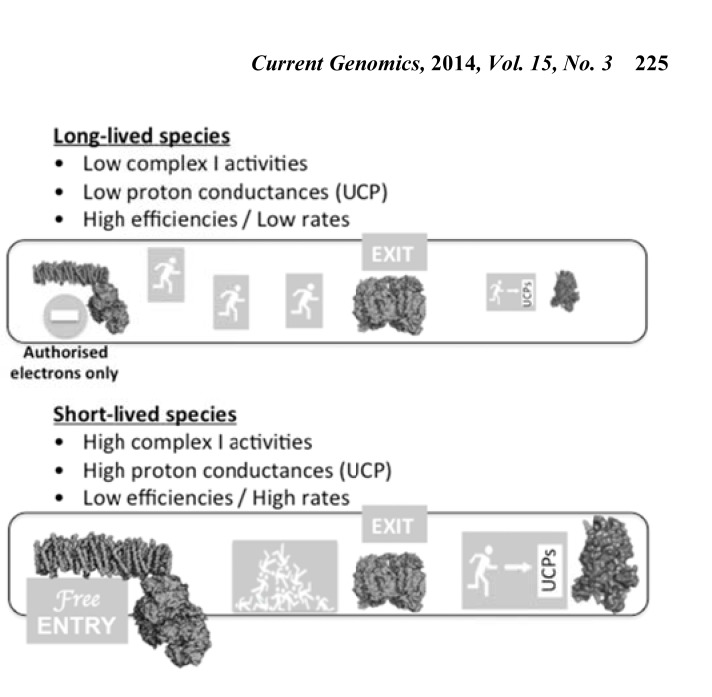
Different selective targets for different life strategies. The electron transport chain can be envisioned as a motorway, where electrons travel from NADH to molecular oxygen. Thus, complex I represents the motorway entrance while complex IV corresponds to the motorway exit. In this metaphor, uncoupling proteins (UCPs) assume the role of emergency exits. Whenever a traffic jam may be a problem, the dissipation of the proton-motive force through UCPs will speed up the flow of electrons and their proper exit through complex IV. Long-lived organisms require an efficient energy metabolism. This requirement, beside a low metabolic rate, imposes a rather tight coupling between substrate oxidation and ADP phosphorylation, which in turn urges for low proton conductances through UCPs. Therefore, in this life-history context, controlling the electron entrance through complex I seems to have been an important target of natural selection. In contrast, short-lived animals demand a high metabolic rate, even if that is at the expense of low efficiencies. In this scenario, reducing the entrance of electron to the ETC does not seem to be an affordable option. Thus, UCPs gain relevance as anti-oxidant devices.
CONCLUDING REMARKS
The appearance of metazoa was possible only after that the unique bioenergetics and biosynthetic demands imposed for these new complex forms of life could be satisfied. Due to its favorable physicochemical properties, O2 was selected during biological evolution to serve both as a terminal electron acceptor during substrate oxidation and as a key metabolite of anabolic pathways. Although aerobic respiration and biosynthetic reactions supported by O2 have significant advantages, they did not come without a price paid in terms of oxidative stress. During cellular respiration, most of the oxygen consumed by the cell is converted to the harmless by-product water in the mitochondria. However, a variable small percentage of this oxygen can receive a single electron yielding superoxide, which subsequently will lead to other forms of ROS, which fulfill important roles as cellular messengers. Nevertheless, any imbalance of ROS homeostasis will cause oxidative stress, which plays a major part in the ageing process as well as in the development of a plethora of chronic and degenerative diseases. Through evolutionary time, animals have developed diverse and complementary strategies aimed at combating oxidative damage. This review is focused on the mechanisms that control the rate of ROS production, with strong emphasis on the evolutionary aspects. Although more than half a dozen of mitochondrial sites have been identified as ROS producing points, the controversy about their relative importance dominates the field. Nevertheless, there is a large consensus on the importance of complex I and III as sources of ROS. Although further efforts, and probably new approaches, are needed to disentangle the mechanisms of mitochondrial ROS generation in vivo, it seems well established that a high NADH/NAD+ ratio, a high steady state level of ubisemiquinone and a high proton-motive force, are conditions that promote ROS production and oxidative damage. Although until very recently it has been commonly assumed that an increase in oxygen consumption led to an increase in ROS production, we now increasingly appreciate that the rate of oxygen consumption can have opposite effects on the rate of ROS production depending on the force driving this oxygen consumption. Thus, when an increase in respiration rate is brought about by an increased NADH dehydrogenase activity, the consequence will be a rise in the proton-motive force accompanied by higher rates of ROS production. In contrast, if the increased respiration rate is caused by either an incremented ATP turnover or an augmented proton leak, then, at the new steady state, the proton-motive force will be lower as well as the generation of ROS. Therefore, complex I and uncoupling proteins seem to be obvious targets of natural selection to reduce the formation of ROS. Further insights into the biology of ROS production come from comparative studies across species. In this sense, longevity has been negatively correlated both with complex I expression and with proton leak. These key observations can be interpreted in the following way. Long-lived animals require an efficient energy metabolism able to support their large body sizes. This requirement imposes a rather tight coupling of oxidative phosphorylation, which, in turn, urges for low proton conductances through uncoupling proteins. Therefore, in this life-history context, controlling the electron entrance through complex I might have been the main way to ameliorate ROS production during the evolution of large, long-lived animals. In contrast, the evolutionary success of small short-lived organisms is highly dependent on a rapid rather than efficient metabolism. This constraint might have precluded downregulation of complex I as an affordable option for this life-history strategy, which had to resort to uncoupling as a mechanism to ameliorate ROS production while keeping high rates of substrate oxidation.
ACKNOWLEDGEMENTS
The author would like to thank Alicia Esteban del Valle and the anonymous reviewers for their valuable comments and suggestions to improve the quality of the paper.
CONFLICT OF INTEREST
The author(s) confirm that this article content has no conflicts of interest.
REFERENCES
- 1.Aledo JC, Esteban delValle A. Glycolysis in wonderland: the importance of energy dissipation in metabolic pathways. J. Chem. Educ. 2002;79:1336–1339. [Google Scholar]
- 2.Aledo J C, Esteban del Valle A. The ATP paradox is the expression of an economizing fuel mechanism. J. Biol. Chem. 2004;279:55372–55375. doi: 10.1074/jbc.M410479200. [DOI] [PubMed] [Google Scholar]
- 3.Aledo JC. Glutamine breakdown in rapidly dividing cells: waste or investment?. BioEssays. 2004;26:778–785. doi: 10.1002/bies.20063. [DOI] [PubMed] [Google Scholar]
- 4.Aledo JC, Pérez-Claros JA, Valle AE. Switching Between Cooperation and Competition in the Use of Extracellular Glucose. J. Mol. Evol. 2007;65:328–339. doi: 10.1007/s00239-007-9014-z. [DOI] [PubMed] [Google Scholar]
- 5.Aledo J C. An Early and Anaerobic Scenario for the Transition to Undifferentiated Multicellularity. J. Mol. Evol. 2008;67:145–153. doi: 10.1007/s00239-008-9128-y. [DOI] [PubMed] [Google Scholar]
- 6.Fenchel T, Finlay B J. The Evolution of Life without Oxygen. Am. Scietist. 1994;82:22–29. [Google Scholar]
- 7.Catling DC, Glein CR, Zahnle KJ, Mckay CP. Why O 2Is Required by Complex Life on Habitable Planets and the Concept of Planetary “Oxygenation Time. Astrobiology. 2005;5:415–438. doi: 10.1089/ast.2005.5.415. [DOI] [PubMed] [Google Scholar]
- 8.Raymond J. The Effect of Oxygen on Biochemical Networks and the Evolution of Complex Life. Science. 2006;311:1764–1767. doi: 10.1126/science.1118439. [DOI] [PubMed] [Google Scholar]
- 9.Goldfine H. The evolution of oxygen as a biosynthetic reagent. J. Gen. Physiol. 1965;49:253–274. doi: 10.1085/jgp.49.1.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chance B, Sies H, Boveris A. Hydroperoxide metabolism in mammalian organs. Physiol. Rev. 1979;59:527–605. doi: 10.1152/physrev.1979.59.3.527. [DOI] [PubMed] [Google Scholar]
- 11.Kontos H A, Wei E P, Ellis E F, Jenkins L W, Povlishock J T, Rowe G T, Hess M L. Appearance of superoxide anion radical in cerebral extrecellular space during increased prostaglandin synthesis in cats. Circ. Res. 1985;57:142–151. doi: 10.1161/01.res.57.1.142. [DOI] [PubMed] [Google Scholar]
- 12.Catalano A, Rodilossi S, Caprari P, Coppola V, Procopio A. 5-Lipoxygenase regulates senescence-like growth arrest by promoting ROS-dependent p53 activation. EMBO J. 2005;24:170–179. doi: 10.1038/sj.emboj.7600502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuppusamy P, Zweier J L. Characterization of free radical generation by xanthine oxidase.Evidence for hydroxyl radical generation. J. Biol. Chem. 1989;264:9880–9884. [PubMed] [Google Scholar]
- 14.Lambeth J D. NOX enzymes and the biology of reactive oxygen. Nat. Rev. Immunol. 2004;4:181–189. doi: 10.1038/nri1312. [DOI] [PubMed] [Google Scholar]
- 15.Balaban R S, Nemoto S, Finkel T. Mitochondria, Oxidants, and Aging. Cell. 2005;120:483–495. doi: 10.1016/j.cell.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 16.Rich P R. The molecular machinery of Keilin's respiratory chain. Biochem. Soc. Trans. 2003;31:1095–1105. doi: 10.1042/bst0311095. [DOI] [PubMed] [Google Scholar]
- 17.Finkel T. Signal Transduction by Mitochondrial Oxidants. J. Biol. Chem. 2012;287:4434–4440. doi: 10.1074/jbc.R111.271999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Forman H J, Maiorino M, Ursini F. Signaling Functions of Reactive Oxygen Species. Biochemistry. 2010;49:835–842. doi: 10.1021/bi9020378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Magalhães JP, Church GM. Cells discover fire: Employing reactive oxygen species in developmentand consequences for aging. Exp. Gerontol. 2006;41:1–10. doi: 10.1016/j.exger.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 20.Mansfield KD, Guzy RD, Pan Y, Young RM, Cash TP, Schumacker PT, Simon MC. Mitochondrial dysfunction resulting from loss of cytochrome c impairs cellular oxygen sensing and hypoxic HIF-a activation. Cell Metab. 2005;1:393–399. doi: 10.1016/j.cmet.2005.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guzy R D, Hoyos B, Robin E, Chen H, Liu L, Mansfield K D, Simon M C, Hammerling U, Schumacker P T. Mitochondrial complex III is required for hypoxia-induced ROS production and cellular oxygen sensing. Cell Metabol. 2005;1:401–408. doi: 10.1016/j.cmet.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 22.Brunelle J K, Bell E L, Quesada N M, Vercauteren K, Tiranti V, Zeviani M, Scarpulla R C, Chandel N S. Oxygen sensing requires mitochondrial ROS but not oxidative phosphorylation. Cell Metabol. 2005;1:409–414. doi: 10.1016/j.cmet.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 23.Scherz-Shouval R, Shvets E, Fass E, Shorer H, Gil L, Elazar Z. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J. 2007;26:1749–1760. doi: 10.1038/sj.emboj.7601623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou R, Yazdi A S, Menu P, Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2010;469:221–225. doi: 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]
- 25.Bulua A C, Simon A, Maddipati R, Pelletier M, Park H, Kim K-Y, Sack M N, Kastner D L, Siegel R M. Mitochondrial reactive oxygen species promote production of proinflammatory cytokines and are elevated in TNFR1-associated periodic syndrome (TRAPS). J. Exp. Med. 2011;208:519–533. doi: 10.1084/jem.20102049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ito K, Hirao A, Arai F, Matsuoka S, Takubo K, Hamaguchi I, Nomiyama K, Hosokawa K, Sakurada K, Nakagata N, Ikeda Y, Mak T W, Suda T. Regulation of oxidative stress by ATM is required for self-renewal of haematopoietic stem cells. Nature. 2004;431:997–1002. doi: 10.1038/nature02989. [DOI] [PubMed] [Google Scholar]
- 27.Liu J, Cao L, Chen J, Song S, Lee I H, Quijano C, Liu H, Keyvanfar K, Chen H, Cao L-Y, Ahn B-H, Kumar N G, Rovira I I, Xu X-L, van Lohuizen M, Motoyama N, Deng C-X, Finkel T. Bmi1 regulates mitochondrial function and the DNA damage response pathway. Nature. 2009;459:387–392. doi: 10.1038/nature08040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bashan N, Kovsan J, Kachko I, Ovadia H, Rudich A. Positive and Negative Regulation of Insulin Signaling by Reactive Oxygen and Nitrogen Species. Physiol. Rev. 2009;89:27–71. doi: 10.1152/physrev.00014.2008. [DOI] [PubMed] [Google Scholar]
- 29.Houstis N, Rosen E D, Lander E S. Reactive oxygen species have a causal role in multiple forms of insulin resistance. Nature. 2006;440:944–948. doi: 10.1038/nature04634. [DOI] [PubMed] [Google Scholar]
- 30.Takahashi A, Ohtani N, Yamakoshi K, Iida S-I, Tahara H, Nakayama K, Nakayama K I, Ide T, Saya H, Hara E. Mitogenic signalling and the p16INK4a-Rb pathway cooperate to enforce irreversible cellular senescence. Nat. Cell Biol. 2006;8:1291–1297. doi: 10.1038/ncb1491. [DOI] [PubMed] [Google Scholar]
- 31.Schreck R, Rieber P, Baeuerle P A. Reactive oxygen intermediates as apparently widely used messengers in the activation of NF-kB transcription factor and HIV-1. EMBO J. 1991;10:2247–2258. doi: 10.1002/j.1460-2075.1991.tb07761.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dalton T P, Shertzer H G, Puga A. Regulation of gene expression by reactive oxygen. Annu. Rev. Pharmacol. Toxicol. 1999;39:67–101. doi: 10.1146/annurev.pharmtox.39.1.67. [DOI] [PubMed] [Google Scholar]
- 33.Cooke M S, Evans M D, Dizdaroglu M, Lunec J. Oxidative DNA damage: mechanisms, mutation, and disease. FASEB J. 2003;17:1195–1214. doi: 10.1096/fj.02-0752rev. [DOI] [PubMed] [Google Scholar]
- 34.Cabiscol E. Oxidative stress promotes specific protein damage in Saccharomyces cerevisiae. J. Biol. Chem. 2000;275:273893–27398. doi: 10.1074/jbc.M003140200. [DOI] [PubMed] [Google Scholar]
- 35.Pamplona R. Advanced lipoxidation end-products. Chem. Biol. Interact. 2011;192:14–20. doi: 10.1016/j.cbi.2011.01.007. [DOI] [PubMed] [Google Scholar]
- 36.Brand MD. Uncoupling to survive?.The role of mitochondrial inefficiency in ageing. Exp. Gerontol. 2000;35:811–820. doi: 10.1016/s0531-5565(00)00135-2. [DOI] [PubMed] [Google Scholar]
- 37.Thannickal V J. Oxygen in the Evolution of Complex Life and the Price We Pay. Am. J. Respir. Cell Mol. Biol. 2009;40:507–510. doi: 10.1165/rcmb.2008-0360PS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matés J M, Segura J A, Alonso F J, Márquez J. Intracellular redox status and oxidative stress: implications for cell proliferation, apoptosis, and carcinogenesis. Arch. Toxicol. 2008;82:273–299. doi: 10.1007/s00204-008-0304-z. [DOI] [PubMed] [Google Scholar]
- 39.Borek C. Dietary Antioxidants and Human Cancer. Integr. Canc. Ther. 2004;3:333–341. doi: 10.1177/1534735404270578. [DOI] [PubMed] [Google Scholar]
- 40.Schafer Z T, Grassian A R, Song L, Jiang Z, Gerhart-Hines Z, Irie H Y, Gao S, Puigserver P, Brugge J S. Antioxidant and oncogene rescue of metabolic defects caused by loss of matrix attachment. Nature. 2009;461:109–113. doi: 10.1038/nature08268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weigl S, Paradiso A, Tommasi S. Mitochondria and Familial Predisposition to Breast Cancer. Curr. Genomics. 2013;14:195–203. doi: 10.2174/1389202911314030005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stocker R. Role of Oxidative Modifications in Atherosclerosis. Physiol. Rev. 2004;84:1381–1478. doi: 10.1152/physrev.00047.2003. [DOI] [PubMed] [Google Scholar]
- 43.Lebovitz RM, Zhang H, Vogel H, Cartwright J, Dionne L, Lu N, Huang S, Matzuk MM. Neurodegeneration, myocardial injury, and perinatal death in mitochondrial superoxide dismutase-deficient mice. Proc. Natl. Acad. Sci. U S A. 1996;93:9782–9787. doi: 10.1073/pnas.93.18.9782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pham-Huy L A, He H, Pham-Huy C. Free radicals, antioxidants in disease and health. Int. J. Biomed. Sci. 2008;4:89–96. [PMC free article] [PubMed] [Google Scholar]
- 45.Naudi A, Jove M, Ayala V, Cassanye A, Serrano J, Gonzalo H, Boada J, Prat J, Portero-Otín M, Pamplona R. Cellular dysfunction in diabetes as maladaptive response to mitochondrial oxidative stress. Exper. Diabetes Res. 2012;2012:696215. doi: 10.1155/2012/696215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de Magalhães J P. Programmatic features of aging originating in development: aging mechanisms beyond molecular damage?. FASEB J. 2012;26:4821–4826. doi: 10.1096/fj.12-210872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Harman D. Free Radical Theory of Aging: An Update: Increasing the Functional Life Span. Ann. N. Y. Acad. Sci. 2006;1067:10–21. doi: 10.1196/annals.1354.003. [DOI] [PubMed] [Google Scholar]
- 48.Trifunovic A, Wredenberg A, Falkenberg M, Spelbrink J F, Rovio A T, Bruder C E, Bohlooly-Y M, Gidlöf S, Oldfors A, Wibom R, Törnell J, Jacobs H T, Larsson N-G. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429:417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- 49.Kujoth G C, Hiona A, Pugh T D, Someya S, Wohlgemuth S E, Hofer T, Seo A Y, Sullivan R, Jobling W A, Morrow J D, Van Remmen H, Sedivy JM, Yamasoba T, Tanokura M, Weindruch R, Leeuwenburgh C, Prolla TA. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science. 2005;309:481–484. doi: 10.1126/science.1112125. [DOI] [PubMed] [Google Scholar]
- 50.Itsara L S, Kennedy S R, Fox E J, Yu S, Sánchez-Contreras M, Cardozo-Peláez F, Pallanck L J. Oxidative stress is not a major contributor to somatic mitochondrial DNA mutations. PLoS Genet. 2014;10:e1003974. doi: 10.1371/journal.pgen.1003974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Van Raamsdonk J M, Hekimi S. Superoxide dismutase is dispensable for normal animal lifespan. Proc. Natl. Acad. Sci. U S A. 2012;109:5785–5790. doi: 10.1073/pnas.1116158109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cabreiro F, Ackerman D, Doonan R, Araiz C, Back P, Papp D, Braeckman B P, Gems D. Increased life span from overexpression of superoxide dismutase in Caenorhabditis elegans is not caused by decreased oxidative damage. Free Radic. Biol. Med. 2011;51:1575–1582. doi: 10.1016/j.freeradbiomed.2011.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pérez V, Bokov A, Van Remmen H, Mele J, Ran Q, Ikeno Y, Richardson A. Is the oxidative stress theory of aging dead?. Biochim. Biophis. Acta. 2009;1790:1005–1014. doi: 10.1016/j.bbagen.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang W, Hekimi S. A mitochondrial superoxide signal triggers increased longevity in Caenorhabditis elegans. PLoS Biol. 2010;8:e10000556. doi: 10.1371/journal.pbio.1000556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yun J, Finkel T. Mitohormesis. Cell Metabol. 2014;19(5):752–766. doi: 10.1016/j.cmet.2014.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bennett C F, Kaeberlein M. The mitochondrial unfolded protein response and increased longevity: Cause, consequence, or correlation?. Exp. Gerontol. 2014 doi: 10.1016/j.exger.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kenyon CJ. The genetics of ageing. Nature. 2010;464:504–512. doi: 10.1038/nature08980. [DOI] [PubMed] [Google Scholar]
- 58.Feng J, Bussière F, Hekimi S. Mitochondrial electron transport is a key determinant of life span in Caenorhabditis elegans. Develop. Cell. 2001;1:633–644. doi: 10.1016/s1534-5807(01)00071-5. [DOI] [PubMed] [Google Scholar]
- 59.Copeland J M, Cho J, Lo T, Hur H J, Bahadorani S, Arabyan T, Rable J, Soh J, Walker D W. Extension of Drosophila life span by RNAi of the mitochondrial respiratory chain. Curr. Biol. 2009;19:1591–1598. doi: 10.1016/j.cub.2009.08.016. [DOI] [PubMed] [Google Scholar]
- 60.Dell'Agnello C, Leo S, Agostino A, Szabadkai G, Tiveron C, Zulian A, Prelle A, Roubertoux P, Rizzuto R, Zeviani M. Increased longevity and refractoriness to Ca2+-dependent neurodegeneration in Surf1 knockout mice. Hum. Mol. Genet. 2007;16:431–444. doi: 10.1093/hmg/ddl477. [DOI] [PubMed] [Google Scholar]
- 61.Lee S-J, Hwang A B, Kenyon C. Inhibition of respiration extends C.elegans' lifespan via reactive oxygen species that increase HIF-1 activity. Curr. Biol. 2010;20:2131–2136. doi: 10.1016/j.cub.2010.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Durieux J, Wolff S, Dillin A. The cell non-autonomous nature of electron transport chain-mediated longevity. Cell. 2011;144:79–91. doi: 10.1016/j.cell.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bennett C, Wende H V, Simko M, Klum S, Barfield S, Choi H, Pineda VV, Kaeberlein M. Activation of the mitochondrial unfolded protein response does not predict longevity in Caenorhabditis elegans. Nat. Commun. 2014;5:3483. doi: 10.1038/ncomms4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Artal-Sanz M, Tavernarakis N. Prohibitin and mitochondrial biology. Trends Endocrinol. Metab. 2009;20:394–401. doi: 10.1016/j.tem.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 65.Schleit J, Johnson S C, Bennett C F, Simko M, Trongtham N, Castanza A, Hsieh E J, Moller R M, Wasko B M, Delaney J R. Molecular mechanisms underlying genotype-dependent responses to dietary restriction. Aging Cell. 2013;12:1050–1061. doi: 10.1111/acel.12130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.López-Otín C, Blasco M A, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153:1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Leeuwenburgh C, Pamplona R, Sanz A. Mitochondria and aging. J. Aging Res. 2011;2011:782946. doi: 10.4061/2011/782946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li Y, Huang TT, Carlson EJ, Melov S, Ursell P C, Olson J L, Noble L J, Yoshimura M P, Berger C, Chan P H, Wallace DG, Epstein CJ. Dilated cardiomyopathy and neonatal lethality in mutant mice lacking manganese superoxide dismutase. Nature. 1995;11:376–381. doi: 10.1038/ng1295-376. [DOI] [PubMed] [Google Scholar]
- 69.Pamplona R. Mitochondrial DNA Damage and Animal Longevity: Insights from Comparative Studies. J. Aging Res. 2011;2011:1–9. doi: 10.4061/2011/807108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pamplona R, Portero-Otín M, Requena J R, Thorpe S R, Herrero A, Barja G G. A low degree of fatty acid unsaturation leads to lower lipid peroxidation and lipoxidation-derived protein modification in heart mitochondria of the longevous pigeon than in the short-lived rat. Mech. Ageing Dev. 1999;106:283–296. doi: 10.1016/s0047-6374(98)00121-3. [DOI] [PubMed] [Google Scholar]
- 71.Pamplona R, Portero-Otín M, Sanz A, Ayala V, Vasileva E, Barja G G. Protein and lipid oxidative damage and complex I content are lower in the brain of budgerigar and canaries than in mice. Relation to aging rate. AGE. 2005;27:267–280. doi: 10.1007/s11357-005-4562-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Moosmann B, Behl C. Mitochondrially encoded cysteine predicts animal lifespan. Aging Cell. 2008;7:32–46. doi: 10.1111/j.1474-9726.2007.00349.x. [DOI] [PubMed] [Google Scholar]
- 73.Bender A, Hajieva P, Moosmann B. Adaptive antioxidant methionine accumulation in respiratory chain complexes explains the use of a deviant genetic code in mitochondria. Proc. Natl. Acad. Sci. U S A. 2008;105:16496–16501. doi: 10.1073/pnas.0802779105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kitazoe Y, Kishino H, Hasegawa M, Nakajima N, Thorne J L, Tanaka M. Adaptive Threonine Increase in Transmembrane Regions of Mitochondrial Proteins in Higher Primates. PLoS ONE. 2008;3:e3343. doi: 10.1371/journal.pone.0003343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jobson R W, Dehne-Garcia A, Galtier N. Apparent longevity-related adaptation of mitochondrial amino acid content is due to nucleotide compositional shifts. Mitochondrion. 2010;10:540–547. doi: 10.1016/j.mito.2010.05.013. [DOI] [PubMed] [Google Scholar]
- 76.Aledo J C, Li Y, de Magalhães J P, Ruíz-Camacho M, Pérez-Claros J A. Mitochondrially encoded methionine is inversely related to longevity in mammals. Aging Cell. 2011;10:198–207. doi: 10.1111/j.1474-9726.2010.00657.x. [DOI] [PubMed] [Google Scholar]
- 77.Yu C, Li Y, Holmes A, Szafranski K, Faulkes C G, Coen C W, Buffenstein R, Platzer M, de Magalhães J P, Church G M. RNA Sequencing Reveals Differential Expression of Mitochondrial and Oxidation Reduction Genes in the Long-Lived Naked Mole-Rat When Compared to Mice. PLoS ONE. 2011;6:e26729. doi: 10.1371/journal.pone.0026729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Brand M D. The sites and topology of mitochondrial superoxide production. Exp. Gerontol. 2010;45:466–472. doi: 10.1016/j.exger.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Barja G. Mitochondrial Oxygen Radical Generation and Leak: Sites of Production in States 4 and 3, Organ Specificity, and Relation to Aging and Longevity. J. Bioenerg. Biomembr. 1999;31:347–366. doi: 10.1023/a:1005427919188. [DOI] [PubMed] [Google Scholar]
- 80.Chen Q. Production of Reactive Oxygen Species by Mitochondria: central role of complex III. J. Biol. Chem. 2003;278:36027–36031. doi: 10.1074/jbc.M304854200. [DOI] [PubMed] [Google Scholar]
- 81.Kudin A P. Characterization of Superoxide-producing Sites in Isolated Brain Mitochondria. J. Biol. Chem. 2003;279:4127–4135. doi: 10.1074/jbc.M310341200. [DOI] [PubMed] [Google Scholar]
- 82.Carroll J, Fearnley I M, Shannon R J, Hirst J, Walker J E. Bovine complex I is a complex of 45 different subunits. J. Biol. Chem. 2006;281:32724–32727. doi: 10.1074/jbc.M607135200. [DOI] [PubMed] [Google Scholar]
- 83.Mourier A, Larsson N-G. Tracing the Trail of Protons through Complex I of the Mitochondrial Respiratory Chain. PLoS Biol. 2011;9:e1001129. doi: 10.1371/journal.pbio.1001129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Efremov R G, Baradaran R, Sazanov L A. The architecture of respiratory complex I. Nature. 2010;465:441–445. doi: 10.1038/nature09066. [DOI] [PubMed] [Google Scholar]
- 85.Efremov R G, Sazanov L A. Structure of the membrane domain of respiratory complex I. Nature. 2011;476:414–420. doi: 10.1038/nature10330. [DOI] [PubMed] [Google Scholar]
- 86.Genova M L, Ventura B, Giuliano G, Bovina C, Formiggini G, Castelli G P, Lenaz G. The site of production of superoxide radical in mitochondrial comples I is not a bound ubisemiquinone but presumably iron-sulfur cluster N2. FEBS Lett. 2001;505:364–368. doi: 10.1016/s0014-5793(01)02850-2. [DOI] [PubMed] [Google Scholar]
- 87.Pryde K R, Hirst J. Superoxide Is Produced by the Reduced Flavin in Mitochondrial Complex I: a single, unified mechanism that applies during both forward and reverse electron transfer. J. Biol. Chem. 2011;286:18056–18065. doi: 10.1074/jbc.M110.186841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Treberg J R, Quinlan C L. Evidence for Two Sites of Superoxide Production by Mitochondrial NADH-Ubiquinone Oxidoreductase (Complex I). J. Biol. Chem. 2011;286:27103–27110. doi: 10.1074/jbc.M111.252502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sazanov L A. Structure of the Hydrophilic Domain of Respiratory Complex I from Thermus thermophilus. Science. 2006;311:1430–1436. doi: 10.1126/science.1123809. [DOI] [PubMed] [Google Scholar]
- 90.Sazanov L A. Respiratory Complex I: Mechanistic and Structural Insights Provided by the Crystal Structure of the Hydrophilic Domain. Biochemistry. 2007;46:2275–2288. doi: 10.1021/bi602508x. [DOI] [PubMed] [Google Scholar]
- 91.Murphy M P. How mitochondria produce reactive oxygen species. Biochem. J. 2009;417:1. doi: 10.1042/BJ20081386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Iwata S, Lee J W, Okada K, Lee J K, Iwata M, Rasmussen B, Link T A, Ramaswamy S, Jap B K. Complete structure of the 11-subunit bovine mitochondrial cytochrome bc1 complex. Science. 1998;281:64–71. doi: 10.1126/science.281.5373.64. [DOI] [PubMed] [Google Scholar]
- 93.Zhang L, Yu L, Yu C-A. Generation of superoxide anion by succinate-cytochrome c reductase from bovine heart mitochondria. J. Biol. Chem. 1998;273:33972–33976. doi: 10.1074/jbc.273.51.33972. [DOI] [PubMed] [Google Scholar]
- 94.Turrens J F, Alexandre A, Lehninger A L. Ubisemiquinone is the electron donor for superoxide formation by complex III of heart mitochondria. Arch. Biochem. Biophys. 1985;237:408–414. doi: 10.1016/0003-9861(85)90293-0. [DOI] [PubMed] [Google Scholar]
- 95.Muller FL. Complex III Releases Superoxide to Both Sides of the Inner Mitochondrial Membrane. J. Biol. Chem. 2004;279:49064–49073. doi: 10.1074/jbc.M407715200. [DOI] [PubMed] [Google Scholar]
- 96.Liu Y, Fiskum G, Schubert D. Generation of reactive oxygen species by the mitochondrial electron transport chain. J. Neurochem. 2002;80:780–787. doi: 10.1046/j.0022-3042.2002.00744.x. [DOI] [PubMed] [Google Scholar]
- 97.Starkov A A, Fiskum G. Regulation of brain mitochondrial H2O2 production by membrane potential and NAD(P)H redox state. J. Neurochem. 2003;86:1101–1107. doi: 10.1046/j.1471-4159.2003.01908.x. [DOI] [PubMed] [Google Scholar]
- 98.Lambert A J, Brand M D. Superoxide production by NADH:ubiquinone oxidoreductase (complex I) depends on the pH gradient across the mitochondrial inner membrane. Biochem. J. 2004;382:511–517. doi: 10.1042/BJ20040485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ku H-H, Brunk U T, Sohal R S. Relationship between mitochondrial superoxide and hydrogen peroxide production and longevity of mammalian species. Free Radic. Biol. Med. 1993;15:621–627. doi: 10.1016/0891-5849(93)90165-q. [DOI] [PubMed] [Google Scholar]
- 100.Lambert A J, Boysen H MM, Buckingham J A, Yang T, Podlutsky A, Austad S N, Kunz T H, Buffenstein R, Brand M D. Low rates of hydrogen peroxide production by isolated heart mitochondria associate with long maximum lifespan in vertebrate homeotherms. Aging Cell. 2007;6:607–618. doi: 10.1111/j.1474-9726.2007.00312.x. [DOI] [PubMed] [Google Scholar]
- 101.Barja G. Mitochondrial Free Radical Production and Aging in Mammals and Birds. Ann. N.Y. Acad. Sci. 1998;854:224–238. doi: 10.1111/j.1749-6632.1998.tb09905.x. [DOI] [PubMed] [Google Scholar]
- 102.Rubner M. Das problem der lebensdauer und seine beziehunger zum watchstum und ernarhnung. Oldenberg Munich. 1908 [Google Scholar]
- 103.Pearl R. The rate of living (University of London Press LTD. 1928 [Google Scholar]
- 104.Kirkinezos IG, Moraes CT. Reactives oxygen species and mitochondrial diseases. Sem. Cell Dev. Biol. 2001;12:449–457. doi: 10.1006/scdb.2001.0282. [DOI] [PubMed] [Google Scholar]
- 105.Boveris A, Chance B. The mitochondrial generation of hydrogen peroxide.General properties and effect of hyperbaric oxygen. Biochem. J. 1973;134:707–716. doi: 10.1042/bj1340707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Barja G. Mitochondrial oxygen consumption and reactive oxygen species production are independently modulated: implications for aging studies. Rejuvenation Res. 2007;10:215–224. doi: 10.1089/rej.2006.0516. [DOI] [PubMed] [Google Scholar]
- 107.Bonawitz N D, Chatenay-Lapointe M, Pan Y, Shadel G S. Reduced TOR Signaling Extends Chronological Life Span via Increased Respiration and Upregulation of Mitochondrial Gene Expression. Cell Metab. 2007;5:265–277. doi: 10.1016/j.cmet.2007.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mittal N, Babu MM, Roy N. The efficiency of mitochondrial electron transport chain is increased in the long-lived mrg19 Saccharomyces cerevisiae. Aging Cell. 2009;8:643–653. doi: 10.1111/j.1474-9726.2009.00518.x. [DOI] [PubMed] [Google Scholar]
- 109.Heestand B N, Shen Y, Liu W, Magner D B, Storm N, Meharg C, Habermann B, Antebi A. Dietary restriction indiced longevity is mediated by nuclear receptor NHR-62 in Caenorhabditis elegans. PLoS Genet. 2013;9:e1003651. doi: 10.1371/journal.pgen.1003651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gredilla R, Sanz A, Lopez-Torres M, Barja G. Caloric restriction decreases mitochondrial free radical generation at complex I and lowers oxidative damage to mitochondrial DNA in the rat heart. FASEB J. 2001;15:1589–1591. doi: 10.1096/fj.00-0764fje. [DOI] [PubMed] [Google Scholar]
- 111.Sanz A. Methionine restriction decreases mitochondrial oxygen radical generation and leak as well as oxidative damage to mitochondrial DNA and proteins. FASEB J. 2006;20:1064–1073. doi: 10.1096/fj.05-5568com. [DOI] [PubMed] [Google Scholar]
- 112.Nisoli E. Calorie Restriction Promotes Mitochondrial Biogenesis by Inducing the Expression of eNOS. Science. 2005;310:314–317. doi: 10.1126/science.1117728. [DOI] [PubMed] [Google Scholar]
- 113.Guarente L. Mitochondria—A Nexus for Aging, Calorie Restriction, and Sirtuins?. Cell. 2008;132:171–176. doi: 10.1016/j.cell.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ku H-H, Sohal R S. Comparison of mitochondrial pro-oxidant generation and anti-oxidant defenses between rat and pigeon: possible basis of variation in longevity and metabolic potential. Mech. Ageing Dev. 1993;72:67–76. doi: 10.1016/0047-6374(93)90132-b. [DOI] [PubMed] [Google Scholar]
- 115.Herrero A, Barja GG. H2O2 production of heart mitochondria and aging rate are slower in canaries and parakeets than in mice: sites of free radical generation and mechanisms involved. Mech. Ageing Dev. 1998;103:133–146. doi: 10.1016/s0047-6374(98)00035-9. [DOI] [PubMed] [Google Scholar]
- 116.Speakman J R, Talbot D A, Selman C, Snart S, McLaren J S, Redman P, Krol E, Jackson D M, Johnson M S, Brand M D. Uncoupled and surviving: individual mice with high metabolism have greater mitochondrial uncoupling and live longer. Aging Cell. 2004;3:87–95. doi: 10.1111/j.1474-9728.2004.00097.x. [DOI] [PubMed] [Google Scholar]
- 117.Porter R K, Brand M D. Body mass dependence of H+ leak in mitochondria and its relevance to metabolic rate. Nature. 1993;362:628–630. doi: 10.1038/362628a0. [DOI] [PubMed] [Google Scholar]
- 118.Hulbert A J, Pamplona R, Buffenstein R, Buttemer W A. Life and Death: Metabolic Rate, Membrane Composition, and Life Span of Animals. Physiol. Rev. 2007;87:1175–1213. doi: 10.1152/physrev.00047.2006. [DOI] [PubMed] [Google Scholar]
- 119.de Magalhães JP, Costa J. A database of vertebrate longevity records and their relation to other life-history traits. J. Evol. Biol. 2009;22:1770–1774. doi: 10.1111/j.1420-9101.2009.01783.x. [DOI] [PubMed] [Google Scholar]
- 120.Montgomery MK, Hulbert AJ, Buttemer WA. The long life of birds: the rat-pigeon comparison revisited. PLoS One. 2011;6(8):e24138 1–15. doi: 10.1371/journal.pone.0024138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Barja G, Cadenas S, Rojas C, Lopez-Torres M, Pérez-Campos R. A decrease of free radical production near critical targets as a cause of maximum longevity in animals. Comp. Biochem. Physiol. 1994;108B:1501–1512. doi: 10.1016/0305-0491(94)90103-1. [DOI] [PubMed] [Google Scholar]
- 122.Montgomery M K, Hulbert A J, Buttemer WA. Does the oxidative stress theory of aging explain longevity differences in birds?. I. Mitochondrial ROS production. Exp. Gerontol. 2012;47:203–210. doi: 10.1016/j.exger.2011.11.006. [DOI] [PubMed] [Google Scholar]
- 123.Miwa S, Riyahi K, Partridge L, Brand MD. Lack of correlation between mitochondrial reactive oxygen species production and life span in Drosophila. Ann. N. Y. Acad. Sci. 2004;1019:388–391. doi: 10.1196/annals.1297.069. [DOI] [PubMed] [Google Scholar]
- 124.Brown JCL, McClelland GB, Faure PA, Klaiman JM, Staples JF. Examining the mechanisms responsible for lower ROS release rates in liver mitochondria from the long-lived house sparrow (Passer domesticus) and big brown bat (Eptesicus fuscus) compared to the short-lived mouse (Mus musculus). Mech. Ageing Dev. 2009;130:467–476. doi: 10.1016/j.mad.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 125.Samuels D C. Mitochondrial DNA repeats constrain the life span of mammals. Trends Genet. 2004;20:226–229. doi: 10.1016/j.tig.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 126.Bender A, Hajieva P, Moosmann B. From the Cover: Adaptive antioxidant methionine accumulation in respiratory chain complexes explains the use of a deviant genetic code in mitochondria. Proc. Natl. Acad. Sci. U S A. 2008;105:16496–16501. doi: 10.1073/pnas.0802779105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Samuels D C. Life span is related to the free energy of mitochondrial DNA. Mech. Ageing Dev. 2005;126:1123–1129. doi: 10.1016/j.mad.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 128.Aledo JC, Li Y, de Magalhães JP, Ruíz-Camacho M, Pérez-Claros JA. Mitochondrially encoded methionine is inversely related to longevity in mammals. Aging Cell. 2010;10:198–207. doi: 10.1111/j.1474-9726.2010.00657.x. [DOI] [PubMed] [Google Scholar]
- 129.Aledo JC, Valverde H, Magalhães JP. Mutational Bias Plays an Important Role in Shaping Longevity-Related Amino Acid Content in Mammalian mtDNA-Encoded Proteins. J. Mol. Evol. 2012;74:332–341. doi: 10.1007/s00239-012-9510-7. [DOI] [PubMed] [Google Scholar]
- 130.Lambert AJ, Buckingham JA, Boysen HM, Brand MD. Low complex I content explains the low hydrogen peroxide production rate of heart mitochondria from the long-lived pigeon, Columba livia. Aging Cell. 2010;9:78–91. doi: 10.1111/j.1474-9726.2009.00538.x. [DOI] [PubMed] [Google Scholar]
- 131.Sanz A, Soikkeli M, Portero-Otín M, Wilson A, Kemppainen E, McIlroy G, Ellila S, Kemppainen K K, Tuomela T, Lakanmaa M, Kiviranta E, Stefanatos R, Dufour E, Hutz B, Naudí A, Jové M, Zeb A, Vartiainen S, Matsuno-Yagi A, Yagi T, Rustin P, Pamplona R, Ja-cobs HT. Expression of the yeast NADH dehydrogenase Ndi1 in Drosophila confers increased lifespan independently of dietary restriction. Proc. Natl. Acad. Sci. U S A. 2010;107:9105–9110. doi: 10.1073/pnas.0911539107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kharade S V, Mittal N, Das S P, Sinha P, Roy N. Mrg19 depletion increases S.cerevisiae lifespan by augmenting ROS defence. FEBS Lett. 2005;579:6809–6813. doi: 10.1016/j.febslet.2005.11.017. [DOI] [PubMed] [Google Scholar]
- 133.López-Lluch G, Hunt N, Jones B, Zhu M, Jamieson H, Hilmer S, Cascajo M V, Ingram D K, Navas P, de Cabo R. Calorie restriction induces mitochondrial biogenesis and bioenergetic efficiency. Proc. Natl. Acad. Sci. U S A. 2006;103:1767–1773. doi: 10.1073/pnas.0510452103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Cerletti M, Jang Y C, Finley L W S, Haigis M C, Wagers A J. Short-term calorie restriction enhances skeletal muscle stem cell function. Cell Stem Cell. 2012;10:515–519. doi: 10.1016/j.stem.2012.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Chen Z X, Velaithan R, Pervaiz S. mitoEnergetics and cancer cell fate. Biochim. Biophys. Act. (BBA) - Bioenergetics. 2009;1787:462–467. doi: 10.1016/j.bbabio.2008.12.009. [DOI] [PubMed] [Google Scholar]
- 136.Cairns C B, Walther J, Harken AH, Banerjee A. Mitochondrial oxidative phosphorylation thermodynamic efficiencies reflect physiological organ roles. Am. J. Physiol. 1998;274:1–8. doi: 10.1152/ajpregu.1998.274.5.R1376. [DOI] [PubMed] [Google Scholar]
- 137.Mookerjee S A, Divakaruni A S, Jastroch M, Brand M D. Mitochondrial uncoupling and lifespan. Mech. Ageing Dev. 2010;131:463–472. doi: 10.1016/j.mad.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Amara C E, Shakland E G, Jubrias S A, Marcinek D J, Kushmerick M J, Conley K E. Mild mitochondrial uncoupling impacts cellular aging in human muscle in vivo. Proc. Natl. Acad. Sci. U S A. 2007;104:1057–1062. doi: 10.1073/pnas.0610131104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Nicholls DG, Rial E. A History of the First Uncoupling Protein, UCP1. J. Bioenerg. Biomembr. 1999;31:399–406. doi: 10.1023/a:1005436121005. [DOI] [PubMed] [Google Scholar]
- 140.Boss O, Samec S, Paoloni-Giacobino A, Rossier C, Dulloo A, Seydoux J, Muzzin P, Giacobino J-P. Uncoupling protein-3: a new member of the mitochondrial carrier family with tissue-specific expression. FEBS Lett. 1997;408:39–42. doi: 10.1016/s0014-5793(97)00384-0. [DOI] [PubMed] [Google Scholar]
- 141.Fleury C, Neverova M, Collins S, Raibault S, Champigny O, Levi-Meyrueis C, Bouillaud F, Selding M F, Surwit R S, Ricquier D, Warden G H. Uncoupling protein-2: a novel gene linked to obesity and hyperinsulinemia. Nat. Genet. 1997;15:269–272. doi: 10.1038/ng0397-269. [DOI] [PubMed] [Google Scholar]
- 142.Gong D-W, He Y, Karas M, Reitman M. Uncoupling protein-3 is a mediator of thermogenesis regulated by thyroid hormone, beta3-adrenergic agonist, and leptin. J. Biol. Chem. 1997;272:24129–24132. doi: 10.1074/jbc.272.39.24129. [DOI] [PubMed] [Google Scholar]
- 143.Vidal-Puig A, Solanes G, Grujic D, Flier JS, Lowell BB. UCP3: An Uncoupling Protein Homologue Expressed Preferentially and Abundantly in Skeletal Muscle and Brown Adipose Tissue. Biochem. Biophys. Res. Commun. 1997;235:79–82. doi: 10.1006/bbrc.1997.6740. [DOI] [PubMed] [Google Scholar]
- 144.Echtay KS, Roussel D, St-Pierre J, Jekabsons MB, Cadenas S, Stuart JA, Harper JA, Roebuck SJ, Morrison A, Pickering S, Clapham JC, Brand MD. Superoxide activates mitochondrial uncoupling proteins. Nautre. 2002;415:96–99. doi: 10.1038/415096a. [DOI] [PubMed] [Google Scholar]
- 145.Oelkrug R, Kutschke M, Meyer CW, Heldmaier G, Jastroch M. Uncoupling Protein 1 Decreases Superoxide Production in Brown Adipose Tissue Mitochondria. J. Biol. Chem. 2010;285:21961–21968. doi: 10.1074/jbc.M110.122861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Hughes J, Criscuolo F. Evolutionary history of the UCP gene family: gene duplication and selection. BMC Evol. Biol. 2008;8:306. doi: 10.1186/1471-2148-8-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Saito S, Saito C T, Shingai R. Adaptive evolution of the uncoupling protein 1 gene contributed to the acquisition of novel nonshivering thermogenesis in ancestral eutherian mammals. Gene. 2008;408:37–44. doi: 10.1016/j.gene.2007.10.018. [DOI] [PubMed] [Google Scholar]
- 148.Jastroch M, Withers K W, Taudien S, Frappell P B, Helwig M, Fromme T, Hirschberg V, Heldmaier G, McAllan B M, Firth B T, Burmester T, Platzer M, Klingenspor M. Marsupial uncoupling protein 1 sheds light on the evolution of mammalian nonshivering thermogenesis. Physiol. Genom. 2007;32:161–169. doi: 10.1152/physiolgenomics.00183.2007. [DOI] [PubMed] [Google Scholar]
- 149.Rial E, Zardoya R. Oxidative stress, thermogenesis and evolution of uncoupling proteins. J. Biol. 2009;8:58. doi: 10.1186/jbiol155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Rousset S, Alves-Guerra M-C, Mozo J, Miroux B, Cassard-Doulcier A-M, Bouillaud F, Ricquier D. The biology of mitochondrial uncoupling proteins. Diabetes. 2004;53:S130–S135. doi: 10.2337/diabetes.53.2007.s130. [DOI] [PubMed] [Google Scholar]
- 151.Azzu V, Brand M D. The on-off switches of the mitochondrial uncoupling proteins. Trends Biochem. Sci. 2010;35:298–307. doi: 10.1016/j.tibs.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Echtay K S. Superoxide Activates Mitochondrial Uncoupling Protein 2 from the Matrix Side.Studies using targeted antioxidants. J. Biol. Chem. 2002;277:47129–47135. doi: 10.1074/jbc.M208262200. [DOI] [PubMed] [Google Scholar]
- 153.Watve M, Parab S, Jogdand P, Keni S. Aging may be a conditional strategic choice and not an inevitable outcome for bacteria. Proc. Natl. Acad. Sci. U S A. 2006;103:14831–14835. doi: 10.1073/pnas.0606499103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Speakman J R. Body size, energy metabolism and lifespan. J. Exper. Biol. 2005;208:1717–1730. doi: 10.1242/jeb.01556. [DOI] [PubMed] [Google Scholar]
- 155.Pfeiffer T, Schuster S, Bonhoeffer S. Cooperation and competition in the evolution of ATP-producing pathways. Science. 2001;292:504–507. doi: 10.1126/science.1058079. [DOI] [PubMed] [Google Scholar]
- 156.del Valle A E, Aledo J C. What Process Is Glycolytic Stoichiometry Optimal For?. J. Mol. Evol. 2006;62:488–495. doi: 10.1007/s00239-005-0135-y. [DOI] [PubMed] [Google Scholar]
- 157.Balsa E, Marco R, Perales-Clemente E, Szklarczyk R, Calvo E, Landázuri M O, Enríquez JA. NDUFA4 Is a Subunit of Complex IV of the Mammalian Electron Transport Chain. Cell Metabol. 2012;16:378–386. doi: 10.1016/j.cmet.2012.07.015. [DOI] [PubMed] [Google Scholar]
- 158.Affourtit C, Quinlan C L, Brand M D. Measurement of Proton Leak and Electron Leak in Isolated Mitochondria. In: Methods Mol. Biol. (Totowa NJ: Humana Press) 2011:165–182. doi: 10.1007/978-1-61779-382-0_11. [DOI] [PubMed] [Google Scholar]