Abstract
Cleft lip and palate (CL/P) is a common disfiguring birth defect with complex, poorly understood etiology. Mice carrying a spontaneous mutation, Dancer (Dc), exhibit CL/P in homozygotes and show significantly increased susceptibility to CL/P in heterozygotes [Deol, M. S. & Lane, P. W. (1966) J. Embryol. Exp. Morphol. 16, 543–558 and Trasler, D. G., Kemp, D. & Trasler, T. A. (1984) Teratology 29, 101–104], providing an animal model for understanding the molecular pathogenesis of CL/P. We genetically mapped Dc to within a 1-cM region near the centromere of chromosome 19. In situ hybridization analysis showed that one positional candidate gene, Tbx10, is ectopically expressed in Dc mutant embryos. Positional cloning of the Dc locus revealed an insertion of a 3.3-kb sequence containing the 5′ region of the p23 gene into the first intron of Tbx10, which causes ectopic expression of a p23-Tbx10 chimeric transcript encoding a protein product identical to a normal variant of the Tbx10 protein. Furthermore, we show that ectopic expression of Tbx10 in transgenic mice recapitulates the Dc mutant phenotype, indicating that CL/Pin Dc mutant mice results from the p23 insertion-induced ectopic Tbx10 expression. These results identify gain of function of a T-box transcription factor gene as a mechanism underlying CL/P pathogenesis.
Cleft lip with or without cleft palate (CL/P) is a common birth defect that affects ≈1 in 700 live births worldwide (1–3). CL/P patients are born with unilateral or bilateral gaps in the upper lip that often extend through the upper jaw into the nostrils. Approximately 50% of CL/P patients exhibit other major developmental defects, for which >300 syndromes, including a number of chromosomal or Mendelian disorders, have been documented (4). The other 50% of CL/P cases are nonsyndromic and do not exhibit simple Mendelian inheritance patterns (5, 6). Nevertheless, extensive epidemiological studies have provided strong evidence, such as familial aggregation, recurrence risks, and high concordance rates in monozygotic twins, that indicate genetic predisposition to CL/P (7). Due to the genetic heterogeneity and paucity of multiplex families for genetic linkage mapping, many studies have attempted to investigate the genetic basis of CL/P by association studies of genes involved in craniofacial development in animal models (6, 7). Despite intense efforts, however, this approach has been largely unsuccessful and the major genes responsible for CL/P pathogenesis remain elusive.
Although a few human disease syndromes have either CL/P or cleft secondary palate without cleft lip as features (8, 9), genetic and embryological studies indicate that CL/P and isolated cleft palate are etiologically distinct (10, 11). Whereas cleft secondary palate is seen in many mutant mouse strains, only four mouse cleft lip mutations, including Dancer (Dc) (12), Twirler (13, 14), Brachyphalangy (15), and legless (16), have been reported. Dc and Twirler are distinct spontaneous mutations in unknown genes and each results in nearly complete penetrance of CL/P in homozygous mutants (12–14). Brachyphalangy was a radiation-induced mutation allelic to the Gli3 gene (15), but the developmental basis of CL/P in the homozygous mutants is not known because several other Gli3 mutations do not have a CL/P phenotype (17, 18). The legless mutation arose from a transgenic insertion that disrupted the Sp8 gene, resulting in gross limb and brain developmental defects with a low frequency of CL/P apparently as a secondary consequence (19). In addition to these Mendelian mutations, CL/P occurs spontaneously at low but stable frequencies in one family of inbred mouse strains, the “A” strains (20). A genome-wide screen for cleft susceptibility loci in the A/WySn strain mapped two interacting recessive loci, clf1 and clf2, that contribute to cleft lip formation (21–23). Understanding the molecular basis of CL/P formation in these mutant mouse strains will provide new insights into the molecular mechanisms of CL/P pathogenesis in humans.
We report here the characterization and positional cloning of the Dc gene. Dc heterozygous mice exhibit head tossing and circling behavior whereas Dc homozygous mutants have CL/P (12). When outcrossed to the C57BL/6 or A/J inbred strains, up to 40% of Dc heterozygous mutants also exhibited CL/P (24). Moreover, Dc heterozygous mutants showed significantly increased susceptibility to 6-aminonicotinamide-induced CL/P (25). These data indicate that Dc predisposes embryos to CL/P and provides a model for investigating gene–gene and gene–environment interactions in CL/P pathogenesis. Dc was previously loosely mapped to proximal chromosome 19 (12). Interestingly, a recent genome-wide screen for CL/P susceptibility loci in humans identified the centromeric region of human chromosome 11, which is syntenic to mouse proximal chromosome 19, as having strong linkage to CL/P (26). Here, we show that the Dc locus contains a translocation insertion in the Tbx10 gene, which encodes a member of the T-box family of DNA-binding transcription factors (27, 28). We show that Dc causes ectopic expression of a variant Tbx10 transcript and that ectopic expression of Tbx10 mRNA results in CL/P in transgenic mice. These data identify a molecular mechanism underlying CL/P pathogenesis.
Methods
Genetic Mapping of the Dc Mutation. Dc stock mice were obtained from The Jackson Laboratory through rederivation from frozen embryos and were maintained within the stock background by sibling intercrosses. Dc/+ stock females were crossed with CAST/Ei males and their heterozygous male progeny were then crossed to either wild-type females from the Dc stock or C3H/HeJ wild-type females. N2 mice were evaluated for heterozygous phenotype and genomic DNA was isolated from tail biopsy samples for PCR genotyping with microsatellite markers. Molecular markers used for mapping include D19Mit29, D19Mit32, D19Mit44, D19Mit59, D19Mit68, D19Mit69, D19Mit78, D19Mit94, and FosL1. We confirmed recombinations between Dc and D19Mit32 at the centromeric end with a (CA) repeat microsatellite, D19UR1, located ≈8.4 kb proximal to D19Mit32. The PCR primers for D19UR1 were 5′-CATACTTCATCAGGACTTTCATGC-3′ and 5′-GTGCTTCTGGCAGTTCCTCAG-3′, which amplified a 136-bp fragment from C3H/HeJ and Dc stock genomic DNA and an ≈126-bp fragment from CAST/Ei DNA. PCR primers for D19UR2, a marker in the 5′ region of the Tbx10 gene were 5′-CATGTAGACATGTGATCTAGCATG-3′ and 5′-CAGCCCAGATTCTCAGAAGTG-3′, which amplified a 186-bp fragment from C3H/HeJ and Dc stock genomic DNA and an ≈170-bp fragment from CAST/Ei genomic DNA.
Histology and in Situ Hybridization. Embryos were dissected and DNA was prepared from yolk sac or tail samples for genotyping by PCR. Embryos for histology were fixed in Bouin's fixative, dehydrated through graded alcohols, embedded in paraffin, and sectioned and stained with hematoxylin/eosin. Embryos for whole-mount in situ hybridization were fixed in 4% paraformaldehyde in PBS overnight at 4°C. The whole-mount in situ hybridization protocol and the Tbx10 probe have been described (28, 29). For comparison of wild-type and Dc mutant expression patterns of Tbx10, embryos were processed as litters in the same vial, photographs were taken, and embryos were subsequently lysed for PCR genotyping. For negative control, sense RNA probes were used, which did not detect any signal in wild-type or mutant embryos.
RACE and RT-PCR. RACE analysis was carried out by using the GeneRacer kit (Invitrogen) with total RNA isolated from embryonic day (E)11.5 wild-type or Dc/Dc embryos. The sequences of 5′ RACE primers were 5′-GCCCGCTTTGGTGACAATCATCTCTGT-3′ and 5′-GACTCCTGCCCACCTCTGCCTTGTG-3′. RT-PCR amplification of Tbx10 wild-type cDNA was achieved with total RNA from E11.5 mouse embryos by using primers 5′-AATCAGAGGCAGTTTGAGACACC-3′ and 5′-GACATTCGAAGCAGGATTTAGAG-3′. Dc-specific p23-Tbx10 cDNA was amplified by using primers 5′-CACCCGTTTGTCTGGCCCTCT-3′ and 5′-CAGGGCATGATTCAGGGCTTTTG-3′.
Southern Hybridization. Genomic DNA derived from tail samples of Dc/+ and +/+ mice was digested with a panel of restriction enzymes individually. After electrophoresis through 1% agarose gels, DNA was transferred onto Zetaprobe GT membranes (BioRad) and hybridized according to the manufacturer's instructions. The Tbx10 exon 1 probe was an 800-bp HindIII/SphI fragment of the exon 1 region and the exon 2 probe was a 460-bp HindIII/NcoI fragment from the intron 1/exon 2 junction of the Tbx10 gene.
Quantitative Real-Time RT-PCR Analysis of Tbx10 Expression. Template cDNA was synthesized from total RNA isolated independently from Dc mutant and wild-type E11.5 embryos by using the Invitrogen first-strand synthesis kit. Template cDNA was then PCR-amplified in the presence of the fluorogenic DNA dye SYBR Green I by using the Bio-Rad iCycler. Standard curves were generated by a series of 1:10 dilutions of a reference cDNA for each primer pair examined and the expression of each target was then calculated relative to this standard curve. Total Tbx10 transcripts were quantified by amplifying a 165-bp fragment by using a forward primer (5′-TGCGGCAGATTGTGTCCTTG-3′) corresponding to the exon 4 sequence and a reverse primer (5′-AGTTTTCCTGGGCATAGCGGG-3′) complementary to the exon 5 sequence of Tbx10. Wild-type Tbx10 exon 1-containing transcripts were quantified by using a forward primer (5′-TGCTTGAGAGTGAGGTCTGCTGC-3′) corresponding to the exon 1 sequence and a reverse primer (5′-AGCTGGTGCTGGTCTCGGG-3′) complementary to the exon 2 sequence to amplify a 200-bp fragment. These transcripts were quantified relative to the expression of hypoxanthine phosphoribosyl-transferase I (Hprt) and β-actin mRNAs by using the standard curve method. Hprt primers are 5′-TGCTGGTGAAAAGGACCTCTCG-3′ and 5′-CTGGCAACATCAACAGGACTCC-3′. β-actin primers are 5′-GGCCGCCCTAGGCACCAG-3′ and 5′-GGGTCATCTTTTCACGGTTGGC-3′.
Generation of Transgenic Mice. Transgenic mice were generated by microinjection of gel-purified, transgenic DNA constructs into the pronuclei of fertilized eggs of B6SJLF2 mice. The CMVβ-Tbx10 transgenic construct was generated by cloning the RT-PCR-isolated p23-Tbx10 cDNA (described above) downstream of the previously described CMV-β-actin promoter (30). Transgenic mice were genotyped by using PCR with a β-actin forward primer (5′-AGCCTCTGCTAACCAT-3′) and Tbx10 exon 2 reverse primer (5′-GCCCGCTTTGGTGACAATCATCTCTGT-3′).
Results and Discussion
Genetic Mapping of Dc and Identification of Tbx10 as a Candidate Gene. Dc mutant mice were removed from breeding colonies many years ago and the stock embryos were cryopreserved at The Jackson Laboratory. We obtained two presumed Dc heterozygous mice with head-tossing behavior rederived from frozen Dc stock embryos and established a breeding colony. We found that nearly 25% of newborn progeny from heterozygous intercrosses exhibited cleft lip at birth. Histological analysis of newborn and late gestation embryos showed that cleft lip is always accompanied by cleft palate in the homozygous mutants (Fig. 1 B and D). In addition, some homozygous mutants exhibited open eyelids at birth (Fig. 1D).
Fig. 1.
Dc/Dc homozygous mutant mice have cleft lip and palate. (A–D) Frontal sections of E16.5 embryonic heads show that Dc/Dc homozygotes have bilateral cleft lip (B) and cleft palate (D). Whereas bilateral secondary palatal shelves have elevated to above the tongue and fused to each other at the midline in wild-type littermates (C), palatal shelves failed to elevate in Dc/Dc embryos (D). Dc/Dc homozygotes also exhibited delayed eyelid closure (arrows in C and D). (E) Dc was mapped to the centromeric end of chromosome 19. Informative microsatellite markers are shown. The Dc locus is mapped between D19Mit32 and FosL1. Markers D19Mit68, D19Mit29, and D19UR2 showed no recombination with Dc in >500 mice genotyped. e, eye; mb, mandible; mn, medial nasal process; mx, maxilla; t, tongue.
To refine the chromosomal location of the Dc mutation, we used an intraspecific backcross mapping strategy and genotyped >500 phenotypically heterozygous N2 mutant mouse progeny from [wild-type × (Dc/+ × CAST/Ei)F1] crosses for microsatellite markers located on proximal chromosome 19. Eleven mice carried informative recombination and were further genotyped with more closely linked markers, which localized the Dc mutation to within a 1-cM region near the centromere, between markers D19Mit32 and FosL1 (Fig. 1E).
Searching the mouse genome databases revealed that the genomic region between D19Mit32 and FosL1 spans ≈2.2 Mb of DNA. This region contains >90 predicted transcription units, one of which encodes a member of the T-box family of DNA-binding transcription factors, Tbx10 (27, 28). T-box genes are evolutionarily conserved and play essential roles in many developmental processes throughout metazoans (31, 32). Tbx10 is closely related to TBX1 and TBX22, two human T-box genes of which loss of function causes cleft palate associated with Di-George Syndrome and X-linked cleft palate with ankyloglossia, respectively (33, 34). Targeted disruption of Tbx1 also caused cleft palate in mice (35). To verify the close linkage between Tbx10 and Dc, we identified a polymorphic microsatellite sequence, D19UR2, located in the 5′ region of the Tbx10 gene and genotyped all DNA samples of our Dc-mapping mice with this marker. No recombination between D19UR2 and Dc was observed in >500 mice genotyped, suggesting that Tbx10 is a strong candidate for the Dc gene.
We investigated the Tbx10 expression pattern during normal craniofacial development. Extensive in situ hybridization analysis showed that Tbx10 mRNA is expressed in a highly restricted pattern in the developing hindbrain and is normally not expressed in the developing facial region (Fig. 2 A and E and ref. 28), suggesting that loss of function of Tbx10 was unlikely to result in CL/P. Moreover, we sequenced PCR fragments covering all eight exons and exon/intron boundaries of the Tbx10 gene amplified from Dc/Dc mutant DNA but did not find any mutation.
Fig. 2.
Dc mutant embryos have ectopic Tbx10 mRNA expression. In situ hybridization analysis revealed widespread ectopic overexpression of Tbx10 at E9.5 (B and C) and E10.5 (F and G)in Dc mutants compared with the wild-type (A and E) littermates. Because of the short detection time used due to overexpression of Tbx10 in mutant embryos, the restricted wild-type Tbx10 expression pattern in the developing hindbrain is barely visible. (D and H) The pattern of p23 mRNA expression in wild-type embryos is very similar to that of the ectopic Tbx10 expression in Dc mutant embryos.
Because the Dc mutation is semidominant, we hypothesized that Dc might be a neomorphic regulatory mutation in the Tbx10 gene. T-box genes are known to be dose-sensitive and overexpression of several T-box transcription factors resulted in various developmental consequences (36, 37). In particular, transgenic mice carrying a BAC construct containing the human TBX1 gene displayed hyperactive head-tilting behavior, which resembled the Dc/+ heterozygous phenotype (38). We therefore investigated whether Tbx10 mRNA expression was altered in Dc mutant embryos. In situ hybridization analysis revealed widespread ectopic Tbx10 mRNA expression in approximately three-quarters of Dc/+ intercross progeny from E9 through E12, with strong ectopic expression in the first pharyngeal arches and the limb buds, whereas the remaining embryos expressed Tbx10 in the highly restricted wild-type pattern as reported (28). Genotyping the embryos confirmed that ectopic Tbx10 mRNA expression is specific to the Dc/+ and Dc/Dc mutant embryos (Fig. 2), indicating that Dc causes misregulation of the Tbx10 gene.
The Dc Locus Contains an Insertion in the First Intron of Tbx10. To investigate the cause of ectopic Tbx10 expression in Dc mutants, we carried out 5′ RACE analysis by using reverse primers corresponding to either the exon 2 or exon 3 sequences of the Tbx10 gene (Fig. 3). Sequence analysis of the 5′ RACE products indicated that the majority of Tbx10 transcripts in Dc mutant embryos have the Tbx10 exon 1 sequence replaced by a 99-nt sequence identical to the 5′ UTR of the p23 mRNA (Fig. 3A). RT-PCR analysis of E11.5 embryonic RNA confirmed expression of p23-Tbx10 chimeric transcripts specifically in Dc/+ and Dc/Dc mutant embryos (Fig. 3 B and C). Because the p23 sequence in the chimeric transcript does not contain an AUG codon, translation of the p23-Tbx10 chimeric transcript is predicted to initiate within the Tbx10 exon 2 sequence and to produce a protein with an intact T-box domain but lacking the N-terminal 41-aa residues of the previously reported Tbx10 protein (Fig. 3C). Interestingly, RT-PCR analysis showed that wild-type embryos express Tbx10 transcripts of two different sizes (Fig. 3B). Sequence analysis showed that the longer RTPCR product corresponds to the previously reported Tbx10 cDNA (37), whereas the shorter product corresponds to an alternatively spliced Tbx10 mRNA that lacks sequences from the 3′ half of exon 1 (GenBank accession no. AY542280). This alternatively spliced shorter Tbx10 mRNA is predicted to encode a protein product identical to that of the p23-Tbx10 chimeric transcripts (Fig. 3C).
Fig. 3.
Dc mutant embryos express a p23-Tbx10 chimeric transcript. (A) Sequence analysis of 5′ RACE products showed that the majority of Tbx10 transcripts in Dc mutants have their exon 1 sequence replaced by 99 nucleotides from the first exon of the p23 gene. The p23 nucleotide sequence is shown in lowercase and the Tbx10 exon 2 sequence in uppercase. Translation of the chimeric transcript is predicted to initiate in the Tbx10 exon 2 sequence. (B) RT-PCR analysis confirmed expression of p23-Tbx10 chimeric transcripts in Dc/+ and Dc/Dc mutant embryos and revealed expression of two different-sized wild-type Tbx10 transcripts. (C) Schematic representation of wild-type and Dc Tbx10 cDNAs. Numbers in the boxes indicate exons. The positions of predicted translation start (ATG) and stop (TGA) codons are marked. Polyadenylation signals (ATTAAA) are also indicated. Positions of the primers used for RT-PCR are marked. Sequence analysis of RT-PCR products shown in B showed that the two different wild-type Tbx10 transcripts result from alternative splicing in exon 1. The mutant p23-Tbx10 chimeric transcript is predicted to encode a protein truncated by 41 amino acids in the N terminus compared with the full-length protein predicted from the long isoform of wild-type Tbx10 mRNA, wt Tbx10 (I), but identical to that encoded by the shorter wild-type splice variant, wt Tbx10 (II).
Searching the mouse genome databases revealed that the p23 gene is located on mouse chromosome 10 and the p23 sequence in the p23-Tbx10 chimeric transcript corresponds to the exon 1 sequence of the p23 gene. Thus, the Dc mutation resulted most likely from a translocation insertion of a chromosome 10 segment containing at least part of the p23 gene into the vicinity of the Tbx10 gene on chromosome 19. In situ hybridization analysis showed that p23 mRNA expression patterns in wild-type embryos matched the ectopic Tbx10 expression patterns in Dc mutant embryos (Fig. 2), suggesting that expression of the p23-Tbx10 chimeric mRNA in Dc mutant embryos is driven by the p23 gene promoter.
To identify the insertion breakpoints at the Dc locus, we carried out Southern hybridization analyses by using genomic DNA fragments corresponding to the exon 1 and exon 2 regions of the Tbx10 gene as probes. Both probes detected distinct-sized mutant-specific EcoRI fragments that were smaller than the wild-type EcoRI fragment (Fig. 4B), indicating that the p23 fragment is inserted into the first intron. Hybridization of the two probes to StuI-digested wild-type and Dc/+ DNA samples further localized the insertion site to within a 1.2-kb region between the StuI site in intron 1 and exon 2 (Fig. 4).
Fig. 4.
The Dc locus contains an insertion of the 5′ region of the p23 gene in the first intron of Tbx10. (A) Schematic of the Dc mutant locus indicating insertion of a 3.3-kb fragment containing the p23 exon 1 in the first intron of Tbx10. The Tbx10 exons are labeled E1–E8. The ATG codons in E1 and E2 correspond to the translation start sites of the alternatively spliced wild-type Tbx10 mRNAs, respectively. Recognition sites for the restriction enzymes BglII, EcoRI, and StuI are marked. Positions of PCR primers used to amplify the mutant genomic fragments are labeled as B1–B4. The genomic regions corresponding to the exon 1 and exon 2 probes used for Southern hybridization are also indicated. (B) Southern hybridization analysis of genomic DNA isolated from +/+ and Dc/+ mutant mice. The sizes of the hybridized fragments are marked to the right of each image.
According to the mouse genome database annotation, the p23 gene is composed of eight exons spanning ≈22 kb of genomic DNA, with the first intron being ≈9.4 kb in size. Because the p23-Tbx10 chimeric transcript only contained the exon 1 sequences of the p23 gene, we investigated the possibility that the 3′ end of the insertion may lie in intron 1 of the p23 gene. We paired forward PCR primers corresponding to sequences in intron 1 of the p23 gene with reverse primers from the exon 2 region of the Tbx10 gene and attempted PCR amplification across the 3′ insertion break point in Dc/Dc homozygous mutant DNA. Sequence analysis of PCR-amplified fragments indicated that the insertion contained exon 1 and the first 2,995 bp of intron 1 of the p23 gene and was inserted at 917 bp 5′ to exon 2 of Tbx10 (Fig. 4A).
Southern hybridization analysis also provided information for estimating the size of the insertion. As shown in Fig. 4B, the exon 1 and exon 2 probes both detected an ≈13-kb mutant-specific BglII fragment, which is ≈3.3 kb larger than the wild-type BglII fragment. Analysis of the p23 gene region indicated the presence of a BglII site ≈270 bp upstream of the 99-bp p23 exon 1 sequence. If the Dc locus contained this BglII site, however, the exon 2 probe would be expected to detect a mutant-specific BglII fragment of ≈8 kb in size. The absence of this predicted BglII site at the Dc locus suggested that the 5′ end of the insertion is probably downstream of this BglII site in the p23 promoter region. We tested this possibility by PCR amplification across the predicted 5′ end of the insertion. Sequence analysis of PCR products showed that the entire insertion contained a 3,328-bp fragment of the p23 gene region, from 234 bp upstream of the 99-bp exon 1 to 2,995 bp into intron 1 (Fig. 4A). This insertion did not cause any deletion of Tbx10 sequence (data not shown).
The Dc mutation, while causing ectopic expression of the p23-Tbx10 chimeric transcripts, did not alter the expression levels of the full-length Tbx10 transcripts (Fig. 5B). The highly restricted expression pattern of the full-length Tbx10 mRNA was also maintained in Dc/+ and Dc/Dc mutant embryos (Fig. 5 C and D). We performed quantitative real-time RT-PCR analyses of mRNA expression of four other genes in the vicinity of Tbx10 but did not find any alterations in Dc/Dc mutant embryos (data not shown), indicating that the p23 insertion did not disrupt other genes at the Dc locus.
Fig. 5.
Full-length Tbx10 mRNA is expressed normally in Dc mutant embryos. (A) Quantitative real-time RT-PCR by using Tbx10 primers corresponding to sequences of the fourth and fifth exons showed a dramatic overexpression of Tbx10 transcripts in E11.5 Dc/+ and Dc/Dc mutant embryos compared with wild-type (+/+) littermates. (B) Quantitative real-time RT-PCR by using primers corresponding to sequences of the first and second exons showed no significant difference in expression levels of full-length Tbx10 mRNA between +/+, Dc/+, and Dc/Dc embryos. Error bars represent SD. (C and D) In situ hybridization analysis by using an cRNA probe specific to the Tbx10 exon 1 sequence showed that full-length Tbx10 mRNA expression is highly restricted to the fourth rhombomere in Dc/+ (C) and Dc/Dc (D) embryos at E10.5, which is indistinguishable from the expression pattern in +/+ embryos. ov, otic vesicle; r4, fourth rhombomere.
Ectopic Expression of Tbx10 in Transgenic Mice Recapitulates the Dc Mutant Phenotypes. We next investigated whether ectopic Tbx10 expression causes the Dc mutant phenotype. We generated transgenic mice expressing the p23-Tbx10 chimeric transcripts by using the CMVβ promoter (30). Three transgenic founder mice survived to adulthood, of which one male founder transmitted the transgene through the germ line. Whole-mount in situ hybridization analysis of embryos collected from wild-type females mated with this transgenic founder male showed that the Tbx10 transgene was expressed throughout hemizygous transgenic embryos (Fig. 6B). Breeding the transgenic founder male with wild-type females of the Dc stock generated six surviving hemizygous transgenic progeny, which all exhibited head-tossing behavior that resembled the Dc/+ mutant mice. Examination of two litters of newborn pups found two hemizygous transgenic progeny with CL/P and open eyelids (data not shown). Crossing F1 transgenic hemizygotes with Dc/+ heterozygous mice resulted in three transheterozygous mutants, which all exhibited CL/P and open eyelids at birth (Fig. 6 D and F), whereas none of the Dc/+ heterozygous littermates that did not carry the transgene exhibited CL/P. In addition, breeding Dc/+ heterozygous stock mice with the nontransmitting transgenic founders yielded >20 Dc/+ heterozygous progeny, none of which had CL/P. That some hemizygous transgenic mice exhibited CL/P, whereas Dc/+ heterozygous mice did not in the transgenic strain background, is likely due to higher levels of Tbx10 mRNA overexpression from the transgenes. The fact that ectopic expression of Tbx10 mRNA by using a heterologous promoter different from that in the Dc locus is sufficient to recapitulate both the heterozygous and homozygous phenotypes of the Dc mutation indicates that the Dc mutant phenotypes result from ectopic Tbx10 expression induced by the p23 insertion.
Fig. 6.
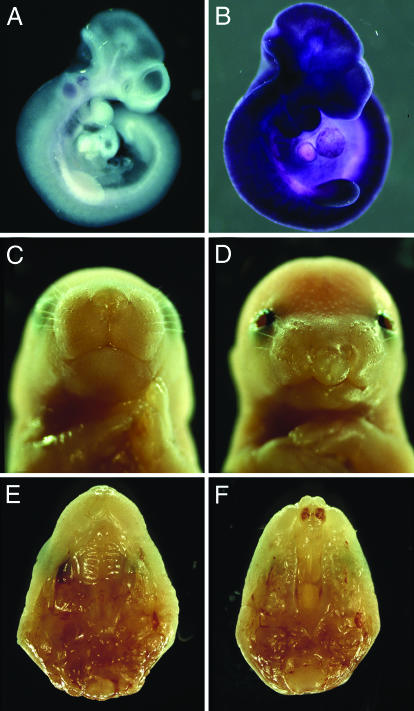
Transgenic Tbx10 overexpression causes CL/P. (A and B) Wholemount in situ hybridization analysis showed ubiquitous overexpression of Tbx10 transcripts in CMVβ-Tbx10 transgenic embryos at E10.5 (B) compared with the highly restricted hindbrain expression in wild-type littermates (A). (C and D) Frontal views of a wild-type E18.5 embryo (C) and of a littermate transheterozygous for Dc and the CMVβ-Tbx10 transgene (D). The transheterozygous embryo exhibited bilateral cleft lip and open eyelids. (E and F) Palatal views of wild-type (E) and transheterozygous mutant (F) upper jaws showed cleft primary and secondary palates in the mutant.
Several possibilities exist for the action of ectopic Tbx10 in Dc embryos. The ectopically expressed Tbx10 protein may directly activate or repress downstream target genes, resulting in disturbance of craniofacial development. In both Dc mutant mice and the CMVβ-Tbx10-transgenic mice, Tbx10 mRNA is expressed nearly ubiquitously, but the developmental defects are highly specific to the craniofacial region. This finding suggests that the downstream effects of transcriptional modulation of any target gene may be highly specific to the developing craniofacial region. Alternatively, ectopic Tbx10 expression may antagonize the function of other closely related T-box factors, such as Tbx1, Tbx15, Tbx18, and Tbx22, which all share high amino acid sequence identity in their DNA-binding domains and are all expressed in partially overlapping patterns during craniofacial development (39–43). Mutations in Tbx1 and TBX22 cause craniofacial defects that include cleft palate in mice and humans, respectively (34, 35). A recent study in zebrafish (44) showed that closely related T-box transcription factors can interact either synergistically or antagonistically to regulate region specific developmental fate in overlapping expression domains. Further investigation of Tbx10 function in Dc mutants will lead to a better understanding of the molecular pathways involved in craniofacial development and CL/P pathogenesis. Interestingly, a recent genome-wide scan of CL/P susceptibility loci in humans identified strong linkage with the centromeric chromosome 11 region where TBX10 resides (26). It is possible that gain of function of TBX10 may underlie a subset of CL/P cases in humans.
Acknowledgments
We thank Robert Angerer, Dirk Bohmann, Tom Gridley, and Joanna Olmsted for comments on the manuscript; Lin Gan for the CMVβ promoter vector; and the University of Rochester Transgenic Mouse Facility for generation of transgenic founder mice. This work was supported by National Institutes of Health/National Institute of Dental and Craniofacial Researh Grants DE13681, DE14593, and DE15207 (to R.J.). J.O.B. is supported by National Institutes of Health Training Grant DE07202. All animal protocols were approved by the University of Rochester Committee on Animal Resources in accordance with National Institutes of Health guidelines.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: CL/P, cleft lip with or without cleft palate; Dc, Dancer; En, embryonic day n.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AY542280).
References
- 1.Vanderas, A. P. (1987) Cleft Palate J. 24, 216-225. [PubMed] [Google Scholar]
- 2.Tolarova, M. M. & Cervenka, J. (1998) Am. J. Med. Genet. 75, 126-137. [PubMed] [Google Scholar]
- 3.Croen, L. A., Shaw, G. M., Wasserman, C. R. & Tolarova, M. M. (1998) Am. J. Med. Genet. 79, 42-47. [DOI] [PubMed] [Google Scholar]
- 4.Gorlin, R. J., Cohen, M. M., Jr., & Hennekam, R. C. M. (2001) in Syndromes of the Head and Neck (Oxford Univ. Press, New York), pp. 850-976.
- 5.Shutte, B. C. & Murray, J. C. (1999) Hum. Mol. Genet. 8, 1853-1859. [DOI] [PubMed] [Google Scholar]
- 6.Murray, J. C. (2002) Clin. Genet. 61, 248-256. [DOI] [PubMed] [Google Scholar]
- 7.Prescott, N. J., Winter, R. M. & Malcolm, S. (2001) Ann. Hum. Genet. 65, 505-515. [DOI] [PubMed] [Google Scholar]
- 8.Kondo, S., Shutte, B. C., Richardson, R. J., Bjork, B. C., Knight, A. S., Watanabe, Y., Howard, E., de Lima, R. L., Daack-Hirsch, S., Sander, A., et al. (2002) Nat. Genet. 32, 285-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van den Boogaard, M. J., Dorland, M., Beemer, F. A. & van Amstel, H. K. (2000) Nat. Genet. 24, 342-343. [DOI] [PubMed] [Google Scholar]
- 10.Diehl, S. R. & Erickson, R. P. (1997) Proc. Natl. Acad. Sci. USA 94, 5231-5236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilkie, A. O. & Morriss-Kay, G. M. (2001) Nat. Genet. Rev. 2, 458-468. [DOI] [PubMed] [Google Scholar]
- 12.Deol, M. S. & Lane, P. W. (1966) J. Embryol. Exp. Morphol. 16, 543-558. [PubMed] [Google Scholar]
- 13.Lyon, M. F. (1958) J. Embryol. Exp. Morphol. 6, 105-116. [PubMed] [Google Scholar]
- 14.Gong, S.-G., White, N. J. & Sakasegawa, A. Y. (2000) Arch. Oral Biol. 45, 87-94. [DOI] [PubMed] [Google Scholar]
- 15.Johnson, D. R. (1969) Genet. Res. 13, 275-280. [DOI] [PubMed] [Google Scholar]
- 16.McNeish, J. D., Scott, W. J., Jr., & Potter, S. S. (1988) Science 241, 837-839. [DOI] [PubMed] [Google Scholar]
- 17.Johnson, D. R. (1967) J. Embryol. Exp. Morphol. 17, 543-558. [PubMed] [Google Scholar]
- 18.Vortkamp, A., Franz, T., Gessler, M. & Grzeschik, K. H. (1992) Mamm. Genome 3, 461-463. [DOI] [PubMed] [Google Scholar]
- 19.Bell, S. M., Schreiner, C. M., Waclaw, R. R., Campbell, K., Potter, S. S. & Scott, W. J. (2003) Proc. Natl. Acad. Sci. USA 100, 12195-12200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kalter, H. (1979) Teratology 20, 213-232. [DOI] [PubMed] [Google Scholar]
- 21.Juriloff, D. M. (1995) J. Craniofac. Genet. Dev. Biol. 15, 1-12. [PubMed] [Google Scholar]
- 22.Juriloff, D. M. & Mah, D. G. (1995) Mamm. Genome 6, 63-69. [DOI] [PubMed] [Google Scholar]
- 23.Juriloff, D. M., Harris, M. J. & Brown, C. J. (2001) Mamm. Genome 12, 426-435. [DOI] [PubMed] [Google Scholar]
- 24.Trasler, D. G. & Leong, S. (1982) Teratology 27, 259-265. [DOI] [PubMed] [Google Scholar]
- 25.Trasler, D. G., Kemp, D. & Trasler, T. A. (1984) Teratology 29, 101-104. [DOI] [PubMed] [Google Scholar]
- 26.Prescott, N. J., Lees, M. M., Winter, R. M. & Malcolm, S. (2000) Hum. Genet. 106, 345-350. [DOI] [PubMed] [Google Scholar]
- 27.Law, D. J., Garvey, N., Agulnik, S. I., Perlroth, V., Hahn, O. M., Rhinehart, R. E., Gebuhr, T. C. & Silver, L. M. (1998) Mamm. Genome 9, 397-399. [DOI] [PubMed] [Google Scholar]
- 28.Bush, J. O., Maltby, K. M., Cho, E.-S. & Jiang, R. (2003) Gene Expr. Patterns 3, 533-538. [DOI] [PubMed] [Google Scholar]
- 29.Jiang, R., Lan, Y., Norton, C. R., Sundberg, J. P. & Gridley, T. (1998) Dev. Biol. 198, 277-285. [PubMed] [Google Scholar]
- 30.Niwa, H., Yamamura, K. & Miyazaki, J. (1991) Gene 108, 193-199. [DOI] [PubMed] [Google Scholar]
- 31.Papaioannou, V. E. & Silver, L. M. (1998) BioEssays 20, 9-19. [DOI] [PubMed] [Google Scholar]
- 32.Papaioannou, V. E. (2001) Int. Rev. Cytol. 207, 1-70. [DOI] [PubMed] [Google Scholar]
- 33.Merscher, S., Funke, B., Epstein, J. A., Heyer, J., Puech, A., Lu, M. M., Xavier, R. J., Demay, M. B., Russell, R. G., Factor, S., et al. (2001) Cell 104, 619-629. [DOI] [PubMed] [Google Scholar]
- 34.Braybrook C., Doudney, K., Marcano, A. C., Arnason, A., Bjornsson, A., Patton, M. A., Goodfellow, P. J., Moore, G. E. & Stanier, P. (2001) Nat. Genet. 29, 179-183. [DOI] [PubMed] [Google Scholar]
- 35.Jerome, L. A. & Papaioannou, V. E. (2001) Nat. Genet. 27, 286-291. [DOI] [PubMed] [Google Scholar]
- 36.Hatcher, C. J. & Basson, C. T. (2001) Nat. Med. 7, 1185-1186. [DOI] [PubMed] [Google Scholar]
- 37.Takeuchi, J. K. (2003) Development (Cambridge, U.K.) 130, 2729-2739. [DOI] [PubMed] [Google Scholar]
- 38.Funke, B., Epstein, J. A., Kochila, L. K., Lu, M. M., Pandita, R. K., Liao, J., Bauerndistel, R., Schuler, T., Schorle, H., Brown, M. C., et al. (2001) Hum. Mol. Genet. 10, 2549-2556. [DOI] [PubMed] [Google Scholar]
- 39.Chapman, D. L., Garvey, N., Hancock, S., Alexiou, M., Agulnik, S. I., Gibson-Brown, J. J., Cebra-Thomas, J., Bollag, R. J., Silver, L. M. & Papaioannou, V. E. (1996) Dev. Dyn. 206, 379-390. [DOI] [PubMed] [Google Scholar]
- 40.Agulnik, S. I., Papaioannou, V. E. & Silver, L. M. (1998) Genomics 51, 68-75. [DOI] [PubMed] [Google Scholar]
- 41.Kraus, F., Haenig, B. & Kispert, A. (2001) Mech. Dev. 100, 83-86. [DOI] [PubMed] [Google Scholar]
- 42.Bush, J. O., Lan, Y., Maltby, K. M. & Jiang, R. (2002) Dev. Dyn. 225, 322-326. [DOI] [PubMed] [Google Scholar]
- 43.Braybrook, C., Lisgo, S., Doudney, K., Henderson, D., Marcano, A. C., Strachan, T. A., Patton, M. A., Villard, L., Moore, G. E., Stanier, P., et al. (2002) Hum. Mol. Genet. 11, 2793-2804. [DOI] [PubMed] [Google Scholar]
- 44.Goering, L. M., Hoshijima, K., Hug, B., Bisgrove, B., Kispert, A. & Grunwald, D. J. (2003) Proc. Natl. Acad. Sci. USA 100, 9410-9415. [DOI] [PMC free article] [PubMed] [Google Scholar]