Abstract
DNA double strand breaks (DSBs) are potential lethal lesions but can also lead to chromosome rearrangements, a step promoting carcinogenesis. DNA non-homologous end-joining (NHEJ) is the major DSB rejoining process and occurs in all cell cycle stages. Homologous recombination (HR) can additionally function to repair irradiation-induced two-ended DSBs in G2 phase. In mammalian cells, HR predominantly uses a sister chromatid as a template for DSB repair; thus HR functions only in late S/G2 phase. Here, we review current insight into the interplay between HR and NHEJ in G2 phase. We argue that NHEJ represents the first choice pathway, repairing approximately 80% of X-ray-induced DSBs with rapid kinetics. However, a subset of DSBs undergoes end resection and repair by HR. 53BP1 restricts resection, thereby promoting NHEJ. During the switch from NHEJ to HR, 53BP1 is repositioned to the periphery of enlarged irradiation-induced foci (IRIF) via a BRCA1-dependent process. K63-linked ubiquitin chains, which also form at IRIF, are also repositioned as well as receptor-associated protein 80 (RAP80), a ubiquitin binding protein. RAP80 repositioning requires POH1, a proteasome component. Thus, the interfacing barriers to HR, 53BP1 and RAP80 are relieved by POH1 and BRCA1, respectively. Removal of RAP80 from the IRIF core is required for loss of the ubiquitin chains and 53BP1, and for efficient replication protein A foci formation. We propose that NHEJ is used preferentially to HR because it is a compact process that does not necessitate extensive chromatin changes in the DSB vicinity.
The notion that DNA represents the hereditary component of the cell necessitates that it maintains stability. Yet, pioneering work by Thomas Lindahl revealed that DNA incurs substantial damage, including base and sugar damage, DNA–DNA and DNA-protein cross links, single strand breaks and double strand breaks (DSBs).1,2 Given such extensive damage, it became evident that cells must have efficient DNA repair mechanisms if the DNA sequence represents the stably inherited determinant of cellular phenotype, a notion strengthened by the finding that DNA repair defective mutants in lower organisms are genetically unstable.3 The evolutionary conservation of DNA repair pathways further supports a critical role in maintaining genetic stability. The study of model organisms has substantially contributed to our understanding of DNA repair mechanisms, particularly DNA DSB repair, which is our focus here. Such studies have shown that mutants in lower organisms deficient in homologous recombination (HR) are exquisitely radiosensitive owing to the important role of HR in repairing DNA DSBs, the major lethal lesion induced by radiation.4–8 By contrast, mammalian mutants deficient in HR show only modest radiosensitivity. Further, studies examining plasmid rejoining in mammalian cells and DNA integration events revealed a distinct process, initially called illegitimate recombination, which does not require extensive homology.9,10 The concept of a non-homologous end-joining (NHEJ) pathway for DSB repair was further substantiated by the study of radiosensitive mammalian mutants and consolidated by the identification of NHEJ genes.8,11 Indeed, mammalian cell lines, mice and patients with marked radiosensitivity have proved to display deficiency in NHEJ rather than HR (excepting ataxia telangiectasia, arguably the most radiosensitive human disorder, which is predominantly proficient in both pathways). HR does function in mammalian cells, however, and can contribute to DSB repair. Having gained a deep understanding of NHEJ and HR in mammalian cells, we can now evaluate the pathway interplay, and why one pathway dominates. This will be the focus of this review.
An overview of non-homologous end-joining and homologous recombination
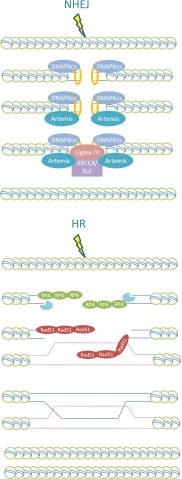
Both, NHEJ and HR, have been well reviewed; only a brief overview encompassing points relevant to the current topic will be given.12–16 NHEJ is initiated by the binding of the Ku heterodimer to double stranded DNA ends, an exceptionally rapid and efficient process, owing to the avid end-binding capacity of Ku and its high abundance. DNA-bound Ku protects ends from nuclease digestion but does not impede ataxia telangiectasia mutated (ATM) activation or signalling.17 DNA-bound Ku recruits the DNA-protein kinase catalytic subunit (DNA-PKcs), generating the DNA-PK complex, which activates the activity of DNA-PK.18 This kinase activity predominantly regulates end-processing and NHEJ through autophosphorylation and also facilitates recruitment of a ligation complex, which encompasses DNA ligase IV (LigIV), X-ray cross complementing Group 4 (XRCC4) and XRCC4-like factor/cernunnos.19 Additional proteins also contribute to end processing, including polynucleotide kinase 3′ phosphatase.20 The structure-specific nuclease, Artemis, is also required for rejoining a subset of DNA ends, which appear to represent those that incur some level of resection, possibly owing to their increased complexity.21 Overall, NHEJ represents a compact process, with current evidence suggesting that only a single Ku molecule binds to each end (Figure 1).22
Figure 1.
Non-homologous end-joining (NHEJ) and homologous recombination (HR) demand different degrees of chromatin remodelling. NHEJ is a compact process that most likely requires little change to the chromatin in the double strand break (DSB) vicinity. HR requires extensive resection, repositioning of damage response proteins and engagement of the sister chromatid. For simplicity, we have shown histone loss in the DSB vicinity. However, the steps in HR may not lead to full histone loss but could involve histone repositioning or modifications to histone proteins. There is extensive evidence that epigenetic changes to histones in the DSB vicinity occur during HR. DNA-PKcs, DNA-protein kinase catalytic subunit; RPA, replication protein A; XLF, XRCC4-like factor; XRCC4, X-ray cross complementing Group 4.
HR, by contrast, uses an undamaged template to restore any sequence information lost at the DSB site. The initiating step of HR is 5′ to 3′ end resection, generating a 3′ ended single-stranded region.23 Resection can be subdivided into an initiation step involving CtIP/MRE11-RAD50-NBS1 (MRN) followed by a process that extends the length of resected DNA.23 The latter process will be the major focus here. Replication protein A (RPA) rapidly binds to the single stranded DNA (ssDNA) tail, preventing the formation of secondary structures. Subsequently, RPA is displaced by RAD51 via a Breast Cancer Associated Gene 2 (BRCA2)–dependent process.24 RAD51 loading promotes invasion onto the undamaged template and strand displacement, generating D-loop formation, which is necessary to generate a Holliday junction and a heteroduplex molecule (Figure 1). Repair ensues using the undamaged strand as a template, followed by ligation of the DNA ends. Frequently, there is a second Holliday junction formed. Finally, resolution of the Holliday junctions completes the process, giving either cross-over or non–cross-over products, depending on the direction of resolution.
Distinct but overlapping roles of non-homologous end-joining and homologous recombination
Although mammalian cells are diploids, HR rarely uses the homologous chromosome as a template for DSB repair.25 Consequently, HR only functions in late S/G2 phase when a sister chromatid is available. One mechanism underlying this regulation of HR is the control of resection by cyclin-dependent kinases.26,27 Consequently, NHEJ is the major DSB repair pathway in G0/G1 phase cells. Conversely, HR exerts its major role in promoting recovery from replication fork stalling, where the lesion activating HR can be an ssDNA region or a one-ended DSB formed by a collapsed replication fork, as opposed to a two-ended DSB induced by agents such as ionizing radiation (IR).28 Indeed, NHEJ is not a suitable process for rejoining one-ended DSBs generated by replication fork collapse/stalling, as rejoining distant one-ended DSBs can lead to genetic rearrangements. Despite these distinct roles, HR can function to repair two-ended DSBs in late S/G2 phase that arise following IR. Here, we consider the interface between NHEJ and HR at such two-ended DSBs in G2 phase. To avoid roles of HR during replication, we uniquely evaluate DSB repair in G2 cells using cell cycle markers to identify G2 cells rather than exploiting synchrony procedures, which mostly induce DNA damage. Additionally, we frequently prevent S to G2 progression to allow specific analysis of DSBs that arise and undergo repair in G2. Appropriate controls have been undertaken to demonstrate that these procedures do not influence the results; such controls will not be discussed here. Thus, here, by using cell cycle markers, we uniquely evaluate and discuss pathway choice at DSBs that arise and undergo repair in G2 phase.
Non-homologous end-joining is the major and first choice double strand break repair pathway in G2 as in G1 phase
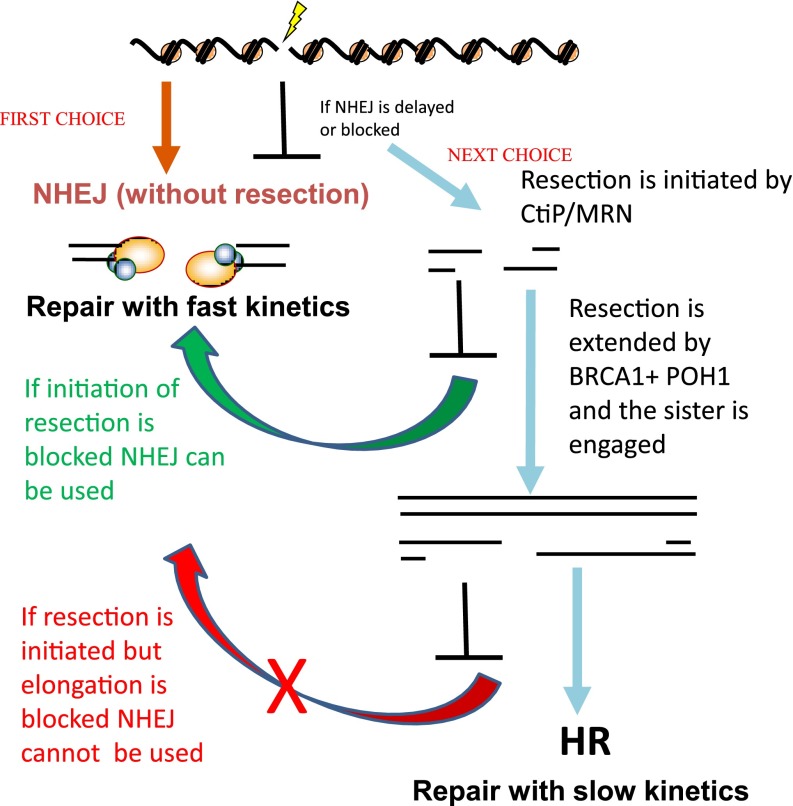
Although G2 phase normally represents only a small fraction of the cell population, efficient repair in G2 is essential to limit chromosome breakage and mitotic cell death. The elegance of HR is its capacity to restore sequence information lost at the DSB using the undamaged sister chromosome as a template. By contrast, there is no plausible mechanism by which NHEJ can restore sequence information lost from both strands. Given this and the abundant usage of HR in lower organisms, it was widely assumed and has became a dogma that HR plays the major role in repairing DSBs in G2, where both pathways can function. Surprisingly, however, cells lacking essential NHEJ proteins, such as DNA LigIV, display a marked G2 phase DSB repair defect, similar to that observed in G1 phase.29,30 Conversely, cells lacking HR proteins show a subtle G2 phase repair defect, failing to repair only 15–20% of the X-ray-induced DSBs. As anticipated, the HR-defective mutants are repair proficient in G1 phase. Additionally, expression of a DNA-PKcs construct with mutations in the DNA-PKcs ABCDE autophosphorylaton cluster (DNA-PKcs 6A) confers a delay in undergoing resection.30 This is significant because ABCDE autophosphorylation promotes release of DNA-PKcs from DNA ends.31 This observation, coupled with the kinetics of repair in HR and NHEJ mutants, supports the notion that NHEJ functions prior to HR30 and that the switch to HR is regulated, at least partly, by DNA-PK activity.32 Thus, we propose that NHEJ makes the first attempt to repair DSBs in G2, but if rejoining is delayed then resection and repair by HR occurs (Figure 2).
Figure 2.
Examination of protein repositioning during homologous recombination. At early and late times post ionizing radiation (IR) in G1 phase and at early times in G2 phase, 53BP1 foci arise in a monomodal distribution around the double strand break. Ubiquitin chains (detected by FK2 antibodies) and receptor-associated protein 80 (RAP80) are similarly positioned. At late times in G2 (8 h post IR), 53BP1 is distributed in a bimodal manner together with ubiquitin chains and RAP80. Replication protein A (RPA) forms in the vacant core. BRCA1 is located between 53BP1 and RPA. This repositioning requires BRCA1 and POH1 to relieve barriers posed by 53BP1 and RAP80, respectively. The figure shows how protein distribution is assessed by monitoring the intensity distribution of the specific proteins along a line drawn through the foci. See reference 37 for details. DDR, DNA damage response; DUB, deubiquitylating enzyme.
Two factors influencing (and enhancing) the switch from NHEJ to HR are the heterochromatic status of DNA and the damage complexity.30,33 It has been shown that heterochromatin (HC) can impede DSB repair in G1 phase and that ATM serves to locally relax HC.34–36 Consistent with such a model, we have observed that, following X-irradiation, DSBs located within HC regions in G2 cells are those that preferentially undergo HR.30,33 Additionally, HR repairs a greater fraction of DSBs following high linear energy transfer radiation, which induces complex DSBs with multiple damages in close proximity.30 This further supports the notion that when NHEJ is impeded (e.g. by the complexity of damage) then repair is switched to HR. These findings, of course, raise the issue of how complex DSBs or DSBs located within the HC regions are repaired in G1 phase, where HR does not ensue. Although an extensive discussion of this issue lies outside the scope of this review, HC-DSBs are repaired with slow kinetics by a process involving NHEJ proteins and the nuclease Artemis in G1 phase.36 Our unpublished findings additionally suggest that complex DSBs are repaired by a similar mechanism. This mechanism appears to represent a process of resection-mediated NHEJ. Thus, in both G1 and G2 phase, NHEJ (most likely without resection) makes the first attempt to repair DSBs, with a resection-mediated process progressing if rapid repair does not ensue. HR represents the resection-dependent repair process in G2 phase; further work is required to define precisely the process in G1 phase. Here, we focus on the switch from NHEJ to HR, since the two processes are well understood.
The initiation of resection by CtIP commits to repair by homologous recombination
Multiple studies, predominantly involving lower organisms, have strongly suggested that resection is initiated by the MRN complex together with CtIP (MRX and Sae2 in yeast).23 In mammalian cells, mutants lacking downstream HR factors, such as BRCA1, BRCA2 or RAD51, have a subtle but distinct G2 phase DSB repair defect assessed by analysis of γH2AX foci, chromosome breakage, pulsed field gel electrophoresis or sister chromatid exchanges (SCEs; all specifically examined in G2 cells).29 This defect is accompanied by impaired formation of RPA and/or RAD51 foci. In striking contrast, G2 phase DSB repair occurs with faster kinetics than control cells following silencing RNA (siRNA)-mediated depletion of CtIP (siCtIP).30 The repair kinetics following CtiP deletion is similar to that observed in G1 cells, and the repair is inhibited when NHEJ is disabled by siRNA-mediated procedures, in NHEJ-defective patient cells, or following DNA-PK inhibition. These findings strongly suggest that CtIP initiates resection, but if it does not occur, then repair can ensue by NHEJ (Figure 2).
Interestingly, siRNA-mediated depletion of BRCA1 (siBRCA1) results in the same subtle repair defect caused by depleting RAD51 or BRCA2.37 However, RPA foci formation is only modestly diminished (reduced to approximately one-third of the control level). A more severe defect in RAD51 foci formation is observed, however, and no IR-induced SCEs arise. Combined siCtIP + siBRCA1 or siCtIP + siBRCA2 restores DSB repair but precludes RPA and RAD51 foci formation.30,37 Collectively, these findings are interpreted to suggest that CtIP regulates an early step that initiates resection, whilst BRCA1 promotes the elongation of resection, i.e. BRCA1 functions downstream of CtIP. If resection is not initiated (owing to loss of CtIP), then DSB repair by NHEJ ensues (Figure 2). However, once HR is initiated, NHEJ cannot be utilized in most instances; hence, siBRCA1 as well as depletion of other downstream HR factors confers a DSB repair defect (Figure 2). The defect is subtle since most DSBs in G2 are repaired by NHEJ and do not undergo resection. Here, we will not consider the initiation step of resection but rather focus on the BRCA1-dependent step that promotes extended resection.
Assessing DNA end resection
Prior to discussing factors regulating the progression of resection, we consider limitations of the frequently employed assays. Since ssDNA occurs during replication, genome-wide approaches to monitor resection (such as Western blotting or chromatin binding of RPA) have high backgrounds, since they do not specifically monitor G2 cells. Microscopy-based approaches allow specific analysis of G2 cells using cell cycle markers. Enumeration of RPA foci in G2 cells represents one such approach. siBRCA1 confers a subtle impact on radiation-induced RPA foci numbers in G2, although quantitative assessment at irradiation-induced foci (IRIF) suggests a more marked impairment.37 We propose that RPA loading requires a defined threshold length of ssDNA. Above this threshold, RPA foci form even if the length of resection is diminished; below the threshold, RPA IRIF do not form. siCtIP substantially impairs RPA foci numbers, suggesting dramatically impaired resection. By contrast, depletion of BRCA1 has a more subtle impact. A similar subtle effect is observed following depletion of BLM/Exo1, which also affects the extension of resection.30 In both cases, there is a marked reduction in RAD51 foci numbers and a complete loss of IR-induced SCEs (a late step in HR). Although the precise requirements for RPA IRIF remain unclear, a consideration of these limitations is important when evaluating the results.
BRCA1 repositions 53BP1 to promote resection
Increasing evidence suggests that p53 binding protein (53BP1) together with RAP1-interacting factor (RIF1) and Pax2 transactivation domain-interacting protein functions to promote NHEJ and restrict HR.38 Mice harbouring BRCA1 homozygous mutations display genomic instability, sensitivity to agents that cause replication stalling/collapse and a failure to complete HR.39 Strikingly, mice deficient for both BRCA1 and 53BP1 regain genomic stability and the ability to undergo resection and HR.40–42 Collectively, these findings suggested that BRCA1 relieves a barrier that 53BP1 (via RIF1) creates for resection.38 Exploitation of subdiffraction-limit resolution microscopy revealed that 53BP1 is repositioned to the periphery of enlarged IRIF during HR via a BRCA1-dependent process.43 Extending this, we have demonstrated that 53BP1 IRIF that form in irradiated G2 cells undergo a two-fold enlargement in volume from 0.5 to 8.0 h post IR via a BRCA1-dependent process37 (note that irradiated G2 cells remain in G2 for 8 h owing to cell cycle checkpoint arrest after exposure to 3 Gy. Thus, the IRIF enlargement occurs in cells irradiated and maintained in G2 phase). Such an enlargement does not take place in irradiated G1 phase cells; indeed, although some enlargement of 53BP1 with time can be observed in G1 phase, it is not detected via the measurement of volume. 53BP1 enlargement in G2 phase is accompanied with the generation of a devoid core where RPA foci form. γH2AX is not repositioned in parallel and is localized internally to 53BP1 at 8 h post IR in G2 cells, overlapping with BRCA1. These dramatic changes can also be visualized by quantifying protein intensity along an axis through the IRIF, revealing a change from a monomodal to a bimodal distribution of 53BP1 between 0.5 and 8.0 h post IR, with RPA forming in the 53BP1-devoid core (Figure 3). This process requires BRCA1 BRCT domains but not the ring finger motif. How does BRCA1 promote such a redistribution of 53BP1?
Figure 3.
Model showing how BRCA1 and POH1 regulate 53BP1 repositioning. Resection is initiated by CtIP/MRE11 without the need for BRCA1. BRCA1 allows POH1 to access receptor-associated protein 80 (RAP80) by an undefined mechanism. POH1 facilitates proteasome-dependent degradation of RAP80, allowing an undefined deubiquitylating enzyme (DUB) to degrade the ubiquitin chains. 53BP1 is then no longer held in its original position, causing concomitant loss of RIF1 (RAP1 interacting factor) from the irradiation-induced foci core. Ubiquitin chains, RAP80 and 53BP1 become repositioned at the periphery of enlarged foci.
POH1 relieves a barrier that receptor-associated protein 80 creates to 53BP1 relocalization
To understand how BRCA1 repositions 53BP1, it is necessary to evaluate the factors required for 53BP1 IRIF formation.44,45 In brief, ATM activation at DSBs promotes phosphorylation of H2AX (γH2AX) and recruitment of the mediator protein, MDC1. MDC1, like ATM, interacts with MRN, thereby tethering MRN and hence ATM at the DSB and extending the region with bound ATM.46 Thus, the IRIF progressively increase in size. MDC1 also recruits two ubiquitin ligases, RNF8 and RNF168.47,48 Current evidence suggests that RNF8 promotes the recruitment of RNF168, which binds to K63-linked ubiquitin chains.49 This step is required for 53BP1 recruitment to IRIF, but the details remain unclear. 53BP1 can bind to dimethylated histone H4K20 (H4K20Me2) via its tudor domain.50,51 The dimethylases, Jumonji D2A/2B, (JMJD2A/JMJD2B), have a hybrid tandem tudor domain, which also binds H4K20Me2. JMJD2A has a higher affinity for H4K20me2 than 53BP1, and one proposal is that in the absence of DNA damage, JMJD2 out-competes 53BP1 for H4K20Me2 binding. Following DNA damage, RNF8/168 activity promotes degradation or loss of JMJD2 allowing 53BP1 to bind to H4K20Me2.52 More recently, 53BP1 was also shown to interact with H2A ubiquitylated on K15, a modification dependent on RNF168.53 Thus, although the mechanism lacks detail, 53BP1 IRIF requires RNF8-RNF168-dependent K63-linked ubiquitin chain formation.54 We reasoned that repositioning of 53BP1 to the IRIF extremity would necessitate the repositioning of ubiquitin chains. Indeed, using anti-FK2 antibodies to identify conjugated ubiquitin including K63-linked ubiquitin chains, we observed that they are repositioned in a similar manner to 53BP1 during the progression of HR in G2.37 Receptor-associated protein 80 (RAP80) encompasses a tandem ubiquitin interacting motif and has been proposed to bind to and to protect ubiquitin chains assembled on H2A.55 A BRCA1-RAP80 complex has been suggested to inhibit resection with loss of RAP80 leading to unbridled resection.56 Significantly, RAP80 was repositioned concomitantly with 53BP1 at 8 h post IR in G2 via a BRCA1-dependent process.37 POH1, a deubiquitylating enzyme (DUB) and proteasome component, has been proposed to influence 53BP1 IRIF size in G1 by regulating RNF8/168-dependent ubiquitination and JMJD2 chromatin retention.57 Strikingly, we found that depletion of POH1, like siBRCA1, precluded the formation of a 53BP1-devoid core in the IRIF and RPA foci formation and that combined depletion of POH1 and RAP80 restored the bimodal distribution as well as RPA foci.37 These findings demonstrate that 53BP1 and RAP80 provide barriers to resection, which are relieved by BRCA1 and POH1, respectively (Figure 3). Combinatorial analysis showed that both barriers must be relieved to achieve repositioning of 53BP1 and resection. However, the barriers are co-ordinated since 53BP1 is not repositioned in the absence of POH1 and vice versa; RAP80 is not repositioned following siBRCA1. Since RAP80 binds and protects ubiquitin chains, we propose that POH1 promotes degradation of RAP80 allowing an undefined deubiquitinase (DUB) to degrade the ubiquitin chains. Since combined depletion of RAP80 and POH1 allows repositioning of the ubiquitin chains, it appears that POH1 is not essential for ubiquitin chain degradation, suggesting that it may function as a proteasome component to degrade RAP80. BRCA1 appears to initiate the entire process since the ubiquitin chains are not repositioned in the absence of BRCA1, although they are when BRCA1 and 53BP1 are co-depleted. Taken together, we propose a model whereby 53BP1 restricts POH1-dependent degradation of RAP80, thereby stabilizing its own positioning. We suggest that BRCA1 affects 53BP1 in some way to allow POH1-dependent clearance of RAP80. This in turn allows degradation of the ubiquitin chains, promoting full clearance of 53BP1 (Figure 4). Notably, the process does not require BRCA1's ring finger domain.37 How BRCA1 initiates the process remains unclear.
Figure 4.
The initiation of resection by CtIP/MRE11 commits to homologous recombination (HR). Non-homologous end-joining (NHEJ) represents the pathway of first choice. If NHEJ is delayed, resection is initiated by CtIP/MRE11. CtIP silencing RNA (siRNA) allows NHEJ to occur. Resection is elongated by a process involving BRCA1, POH1 and Exo/BLM. If this process is blocked, then neither NHEJ nor homologous recombination can proceed, and there is a double strand break repair defect. MRN, MRE11-RAD50-NBS1.
An orchestrated handover from non-homologous end-joining to homologous recombination
The findings above argue that NHEJ represents the pathway of first choice to repair two-ended DSBs in G2 cells. However, if NHEJ progression is impeded, either because of the chromatin or damage complexity, then resection is initiated by CtIP/MRN, and there is a commitment to repair by HR. This initiation step does not require 53BP1 repositioning since CtIP depletion can rescue the repair defect in siBRCA1 cells, suggesting that BRCA1 functions downstream of CtIP. Instead, BRCA1 promotes downstream steps of HR involving extending the region of resected DNA, and consequently (or as well) RAD51 loading. This role requires removing the barriers posed by 53BP1 and RAP80 to resection via 53BP1 repositioning to the IRIF periphery. Ubiquitination contributes to this process demonstrating its central function in regulating DSB repair pathway usage. NHEJ represents a compact process that is not dependent on IRIF formation (most NHEJ occurs normally in MEFs lacking γH2AX).21 HR, by contrast, requires extensive chromatin modification to allow resection and RAD51 loading, engagement of a sister chromatid and branch migration (Figure 1). These findings, therefore, reveal the progression from a compact process of DSB repair (NHEJ) to HR via a regulated and orchestrated process. Indeed, we suggest that a significant role of damage response signalling is the orchestration and regulation of this pathway handover via extensive chromatin modification.
An important question is whether significant histone loss occurs during HR. Although the 53BP1-devoid core appears to be a substantial region, γH2AX remains at least partially within the core. However, a smaller devoid core region of γH2AX is detectable in the region where RPA is present.37 Although the magnitude of histone loss remains unclear, it is likely that resection, RAD51 loading and ensuing steps of HR do necessitate histone modification, loss or at least histone sliding. This raises the issue of how epigenetic modifications are re-established post HR, and the possibility that in mammalian cells, where epigenetic modifications are a critical aspect regulating transcription or gene silencing, the process of HR could promote changes in epigenetic modifications in G2, where the elaborate mechanisms to re-establish the epigenetic code that exist in S phase may not efficiently operate. We suggest that this could provide an explanation for why the compact process of NHEJ is the preferred mechanism for DSB repair in mammalian cells rather than HR.
Summary and perspective
We propose that NHEJ is the first choice DSB repair pathway in mammalian cells in G2 but, if repair is hindered, an orchestrated and regulated switch to resection and HR occurs, providing the more elaborate HR pathway the opportunity to specifically repair more complex DSBs. Since NHEJ cannot accurately repair complex DSBs involving loss of sequence information on both strands, this appears a logical strategy to optimize repair pathway usage in higher organisms, where epigenetic modifications are important. Interestingly, HR also appears to preferentially repair DSBs that arise at HC regions.33,58 Whilst this initially appeared counter intuitive, it could arise as a consequence of the barrier that HC creates to repair by core NHEJ. An important future question is to evaluate the extent of histone loss or modification during G2 phase HR and the mechanisms promoting the maintenance of epigenetic modifications. Interestingly, recent studies have shown that epigenetic silencing can arise as a consequence of HR.59,60 Whilst insight into the process underlying the formation of a 53BP1-devoid core in IRIF has been gained, the process leading to the reformation of ubiquitin chains and 53BP1 remains unaddressed. One possibility is that this process involves RNF168 but not RNF8, a distinction that requires the generation of specific tools or reagents. Finally, a question emerging is whether HR necessitates chromatin modifications on the undamaged template. Indeed, one possibility is that the two-fold enlargement of 53BP1 represents its assimilation onto the undamaged strand.
REFERENCES
- 1.Lindahl T. DNA repair enzymes. Annu Rev Biochem 1982; 51: 61–87. doi: 10.1146/annurev.bi.51.070182.000425 [DOI] [PubMed] [Google Scholar]
- 2.Lindahl T. Instability and decay of the primary structure of DNA. Nature 1993; 362: 709–14. [DOI] [PubMed] [Google Scholar]
- 3.Friedberg EC. Out of the shadows and into the light: the emergence of DNA repair. Trends Biochem Sci 1995; 20: 381. [DOI] [PubMed] [Google Scholar]
- 4.Sargentini NJ, Smith KC. Quantitation of the involvement of the recA, recB, recC, recF, recJ, recN, lexA, radA, radB, uvrD, and umuC genes in Escherichia coli. Radiat Res 1986; 107: 58–72. [PubMed] [Google Scholar]
- 5.Horii ZI, Clark AJ. Genetic analysis of the recF pathway to genetic recombination in Escherichia coli K-12: isolation and characterization of mutants. J Mol Biol 1973; 80: 327–44. [DOI] [PubMed] [Google Scholar]
- 6.Haynes RH, Kunz BA. DNA repair and mutagenesis in yeast. In: Strathern J, Jones EW, Broach JR, eds. The molecular biology of the yeast Saccharomyces. Life cycle and inheritance. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1981. pp. 371–414. [Google Scholar]
- 7.Friedberg EC. Deoxyribonucleic acid repair in the yeast Saccharomyces cerevisiae. Microbiol Rev 1988; 52: 70–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jeggo PA. Studies on mammalian mutants defective in rejoining double-strand breaks in DNA. Mutat Res 1990; 239: 1–16. [DOI] [PubMed] [Google Scholar]
- 9.Wilson JH, Berget PB, Pipas JM. Somatic cells efficiently join unrelated DNA segments end-to-end. Mol Cell Biol 1982; 2: 1258–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brenner DA, Kato S, Anderson RA, Smigocki AC, Camerini-Otero RD. The recombination and integration of DNAs introduced into mouse L cells. Cold Spring Harb Symp Quant Biol 1984; 49: 151–60. [DOI] [PubMed] [Google Scholar]
- 11.Jeggo PA. Pathways of DNA double strand break repair in mammalian cells. Radioprotection 1997; 32: C1–117. [Google Scholar]
- 12.Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem 2010; 79: 181–211. doi: 10.1146/annurev.biochem.052308.093131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neal JA, Meek K. Choosing the right path: does DNA-PK help make the decision? Mutat Res 2011; 711: 73–86. doi: 10.1016/j.mrfmmm.2011.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kass EM, Jasin M. Collaboration and competition between DNA double-strand break repair pathways. FEBS Lett 2010; 584: 3703–08. doi: 10.1016/j.febslet.2010.07.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mazon G, Mimitou EP, Symington LS. SnapShot: homologous recombination in DNA double-strand break repair. Cell 2010; 142: 646.e1. doi: 10.1016/j.cell.2010.08.006 [DOI] [PubMed] [Google Scholar]
- 16.Moynahan ME, Jasin M. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat Rev Mol Cell Biol 2010; 1: 196–207. doi: 10.1038/nrm2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liang F, Jasin M. Ku80-deficient cells exhibit excess degradation of extrachromosomal DNA. J Biol Chem 1996; 271: 14405–11. [DOI] [PubMed] [Google Scholar]
- 18.Gottlieb TM, Jackson SP. The DNA-dependent protein kinase: requirement of DNA ends and association with Ku antigen. Cell 1993; 72: 131–42. [DOI] [PubMed] [Google Scholar]
- 19.Cottarel J, Frit P, Bombarde O, Salles B, Negrel A, Bernard S, et al. A noncatalytic function of the ligation complex during nonhomologous end joining. J Cell Biol 2013; 200: 173–86. doi: 10.1083/jcb.201203128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bernstein NK, Hammel M, Mani RS, Weinfeld M, Pelikan M, Tainer JA, et al. Mechanism of DNA substrate recognition by the mammalian DNA repair enzyme, polynucleotide kinase. Nucleic Acids Res 2009; 37: 6161–73. doi: 10.1093/nar/gkp597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Riballo E, Kuhne M, Rief N, Doherty A, Smith GC, Recio MJ, et al. A pathway of double-strand break rejoining dependent upon ATM, Artemis, and proteins locating to gamma-H2AX foci. Mol Cell 2004; 16: 715–24. doi: 10.1016/j.molcel.2004.10.029 [DOI] [PubMed] [Google Scholar]
- 22.Britton S, Coates J, Jackson SP. A new method for high-resolution imaging of Ku foci to decipher mechanisms of DNA double-strand break repair. J Cell Biol 2013; 202: 579–95. doi: 10.1083/jcb.201303073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Symington LS, Gautier J. Double-strand break end resection and repair pathway choice. Annu Rev Genet 2011; 45: 247–71. doi: 10.1146/annurev-genet-110410-132435 [DOI] [PubMed] [Google Scholar]
- 24.Pellegrini L, Yu DS, Lo T, Anand S, Lee M, Blundell TL, et al. Insights into DNA recombination from the structure of a RAD51-BRCA2 complex. Nature 2002; 420: 287–93. doi: 10.1038/nature01230 [DOI] [PubMed] [Google Scholar]
- 25.Johnson RD, Jasin M. Sister chromatid gene conversion is a prominent double-strand break repair pathway in mammalian cells. EMBO J 2000; 19: 3398–407. doi: 10.1093/emboj/19.13.3398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aylon Y, Liefshitz B, Kupiec M. The CDK regulates repair of double-strand breaks by homologous recombination during the cell cycle. EMBO J 2004; 23: 4868–75. doi: 10.1038/sj.emboj.7600469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ira G, Pellicioli A, Balijja A, Wang X, Fiorani S, Carotenuto W, et al. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature 2004; 431: 1011–17. doi: 10.1038/nature02964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Petermann E, Helleday T. Pathways of mammalian replication fork restart. Nat Rev Mol Cell Biol 2010; 1: 683–87. doi: 10.1038/nrm2974 [DOI] [PubMed] [Google Scholar]
- 29.Beucher A, Birraux J, Tchouandong L, Barton O, Shibata A, Conrad S, et al. ATM and Artemis promote homologous recombination of radiation-induced DNA double-strand breaks in G2. EMBO J 2009; 28: 3413–27. doi: 10.1038/emboj.2009.276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shibata A, Conrad S, Birraux J, Geuting V, Barton O, Ismail A, et al. Factors determining DNA double-strand break repair pathway choice in G2 phase. EMBO J 2011; 30: 1079–92. doi: 10.1038/emboj.2011.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chan DW, Lees-Miller SP. The DNA-dependent protein kinase is inactivated by autophosphorylation of the catalytic subunit. J Biol Chem 1996; 271: 8936–41. [DOI] [PubMed] [Google Scholar]
- 32.Neal JA, Dang V, Douglas P, Wold MS, Lees-Miller SP, Meek K. Inhibition of homologous recombination by DNA-dependent protein kinase requires kinase activity, is titratable, and is modulated by autophosphorylation. Mol Cell Biol 2011; 3: 1719–33. doi: 10.1128/MCB.01298-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kakarougkas A, Ismail A, Klement K, Goodarzi AA, Conrad S, Freire R, et al. Opposing roles for 53BP1 during homologous recombination. Nucleic Acids Res 2013; 41: 9719–13. doi: 10.1093/nar/gkt729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goodarzi AA, Jeggo PA. The heterochromatic barrier to DNA double strand break repair: how to get the entry visa. Int J Mol Sci 2012; 13: 11844–60. doi: 10.3390/ijms130911844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goodarzi AA, Kurka T, Jeggo PA. KAP-1 phosphorylation regulates CHD3 nucleosome remodeling during the DNA double-strand break response. Nat Struct Mol Biol 2011; 18: 831–39. doi: 10.1038/nsmb.2077 [DOI] [PubMed] [Google Scholar]
- 36.Goodarzi AA, Noon AT, Deckbar D, Ziv Y, Shiloh Y, Lobrich M, et al. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol Cell 2008; 31: 167–77. doi: 10.1016/j.molcel.2008.05.017 [DOI] [PubMed] [Google Scholar]
- 37.Kakarougkas A, Ismail A, Katsuki Y, Freire R, Shibata A, Jeggo PA. Co-operation of BRCA1 and POH1 relieves the barriers posed by 53BP1 and RAP80 to resection. Nucleic Acids Res 2013; 441: 10298–311. doi: 10.1093/nar/gkt802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zimmermann M, de Lange T. 53BP1: pro choice in DNA repair. Trends Cell Biol Oct 2013. Epub ahead of print. doi: 10.1016/j.tcb.2013.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moynahan ME, Chiu JW, Koller BH, Jasin M. Brca1 controls homology-directed DNA Repair. Mol Cell 1999; 4: 511–18. [DOI] [PubMed] [Google Scholar]
- 40.Bunting SF, Callen E, Wong N, Chen HT, Polato F, Gunn A, et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell 2010; 141: 243–54. doi: 10.1016/j.cell.2010.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bouwman P, Aly A, Escandell JM, Pieterse M, Bartkova J, van der Gulden H, et al. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat Struct Mol Biol 2010; 17: 688–95. doi: 10.1038/nsmb.1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bothmer A, Robbiani DF, Feldhahn N, Gazumyan A, Nussenzweig A, Nussenzweig MC. 53BP1 regulates DNA resection and the choice between classical and alternative end joining during class switch recombination. J Exp Med 2010; 207: 855–65. doi: 10.1084/jem.20100244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chapman JR, Barral P, Vannier JB, Borel V, Steger M, Tomas-Loba A, et al. RIF1 is essential for 53BP1-dependent nonhomologous end joining and suppression of DNA double-strand break resection. Mol Cell 2013; 49: 858–71. doi: 10.1016/j.molcel.2013.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Panier S, Durocher D. Regulatory ubiquitylation in response to DNA double-strand breaks. DNA Repair (Amst) 2009; 8: 436–43. doi: 10.1016/j.dnarep.2009.01.013 [DOI] [PubMed] [Google Scholar]
- 45.Bekker-Jensen S, Mailand N. Assembly and function of DNA double-strand break repair foci in mammalian cells. DNA Repair (Amst) 2010; 9: 1212–28. doi: 10.1016/j.dnarep.2010.09.010 [DOI] [PubMed] [Google Scholar]
- 46.Lukas C, Melander F, Stucki M, Falck J, Bekker-Jensen S, Goldberg M, et al. Mdc1 couples DNA double-strand break recognition by Nbs1 with its H2AX-dependent chromatin retention. EMBO J 2004; 23: 2674–83. doi: 10.1038/sj.emboj.7600269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Doil C, Mailand N, Bekker-Jensen S, Menard P, Larsen DH, Pepperkok R, et al. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell 2009; 136: 435–46. doi: 10.1016/j.cell.2008.12.041 [DOI] [PubMed] [Google Scholar]
- 48.Stewart GS, Panier S, Townsend K, Al-Hakim AK, Kolas NK, Miller ES, et al. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell 2009; 136: 420–34. doi: 10.1016/j.cell.2008.12.042 [DOI] [PubMed] [Google Scholar]
- 49.Mattiroli F, Vissers JH, van Dijk WJ, Ikpa P, Citterio E, Vermeulen W, et al. RNF168 ubiquitinates K13-15 on H2A/H2AX to drive DNA damage signaling. Cell 2012; 150: 1182–95. doi: 10.1016/j.cell.2012.08.005 [DOI] [PubMed] [Google Scholar]
- 50.Sanders SL, Portoso M, Mata J, Bahler J, Allshire RC, Kouzarides T. Methylation of histone H4 lysine 20 controls recruitment of Crb2 to sites of DNA damage. Cell 2004; 119: 603–14. doi: 10.1016/j.cell.2004.11.009 [DOI] [PubMed] [Google Scholar]
- 51.Botuyan MV, Lee J, Ward IM, Kim JE, Thompson JR, Chen J, et al. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell 2006; 127: 1361–73. doi: 10.1016/j.cell.2006.10.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mallette FA, Mattiroli F, Cui G, Young LC, Hendzel MJ, Mer G, et al. RNF8- and RNF168-dependent degradation of KDM4A/JMJD2A triggers 53BP1 recruitment to DNA damage sites. EMBO J 2012; 31: 1865–78. doi: 10.1038/emboj.2012.47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fradet-Turcotte A, Canny MD, Escribano-Diaz C, Orthwein A, Leung CC, Huang H, et al. 53BP1 is a reader of the DNA-damage-induced H2A Lys 15 ubiquitin mark. Nature 2013; 499: 50–54. doi: 10.1038/nature12318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pei H, Zhang L, Luo K, Qin Y, Chesi M, Fei F, et al. MMSET regulates histone H4K20 methylation and 53BP1 accumulation at DNA damage sites. Nature 2011; 470: 124–28. doi: 10.1038/nature09658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim H, Chen J, Yu X. Ubiquitin-binding protein RAP80 mediates BRCA1-dependent DNA damage response. Science 2007; 316: 1202–05. doi: 10.1126/science.1139621 [DOI] [PubMed] [Google Scholar]
- 56.Coleman KA, Greenberg RA. The BRCA1-RAP80 complex regulates DNA repair mechanism utilization by restricting end resection. J Biol Chem 2011; 286: 13669–80. doi: 10.1074/jbc.M110.213728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Butler LR, Densham RM, Jia J, Garvin AJ, Stone HR, Shah V, et al. The proteasomal de-ubiquitinating enzyme POH1 promotes the double-strand DNA break response. EMBO J 2012; 31: 3918–34. doi: 10.1038/emboj.2012.232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Goodarzi AA, Jeggo P, Lobrich M. The influence of heterochromatin on DNA double strand break repair: getting the strong, silent type to relax. DNA Repair (Amst) 2010; 9: 1272–82. doi: 10.1016/j.dnarep.2010.09.013 [DOI] [PubMed] [Google Scholar]
- 59.O'Hagan HM, Mohammad HP, Baylin SB. Double strand breaks can initiate gene silencing and SIRT1-dependent onset of DNA methylation in an exogenous promoter CpG island. PLoS Genet 2008; 4: e1000155. doi: 10.1371/journal.pgen.1000155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee B, Morano A, Porcellini A, Muller MT. GADD45alpha inhibition of DNMT1 dependent DNA methylation during homology directed DNA repair. Nucleic Acids Res 2012; 40: 2481–93. doi: 10.1093/nar/gkr1115 [DOI] [PMC free article] [PubMed] [Google Scholar]