Abstract
The mammalian β-globin loci each contain a family of developmentally expressed genes, and a far upstream regulatory element, the locus control region (LCR). In adult murine erythroid cells, the LCR and the transcribed β-globin genes exist within domains of histone acetylation and RNA polymerase II (pol II) is associated with them. In contrast, the silent embryonic genes lie between these domains within hypoacetylated chromatin, and pol II is not found there. We used chromatin immunoprecipitation and real-time PCR to analyze histone modification and pol II recruitment to the globin locus in human erythroid K562 cells that express the embryonic ε-globin gene but not the adult β-globin gene. H3 and H4 acetylation and H3 K4 methylation were continuous over a 17-kb region including the LCR and the active ε-globin gene. The level of modification varied directly with the transcription of the ε-globin gene. In contrast, this region in nonerythroid HeLa cells lacked these modifications and displayed instead widespread H3 K9 methylation. pol II was also detected continuously from the LCR to the ε-globin gene. These studies reveal several aspects of chromatin structure and pol II distribution that distinguish the globin locus at embryonic and adult stages and suggest that both enhancer looping and tracking mechanisms may contribute to LCR–promoter communication at different developmental stages.
The human β-globin locus consists of a family of erythroid specific genes and a far upstream regulatory element termed the locus control region (LCR) (1). The genes are expressed sequentially during development, beginning with the ε-globin gene, which is closest to the LCR, and proceeding to the more distant genes. This organization is mirrored in the murine globin locus. Homologous recombination studies that deleted the LCR in its natural chromosomal context show that the LCR is required for high-level expression of all of the genes, thus fulfilling at a minimum the definition of an enhancer (2–4). There is a different spatial arrangement of the genes in the chicken globin locus, with the adult β-globin genes flanked by the two embryonically expressed genes and a strong bidirectional enhancer located internally within the locus between the adult β-globin gene and the embryonic ε-globin gene (5, 6).
Current views of gene regulation incorporate the concept that two types of complexes participate in the establishment of a chromatin structure which is accessible for transcription factors and the RNA polymerase II (pol II) transcriptional machinery (7, 8). Nucleosome remodeling complexes of the SWI/SNF type use the energy of ATP hydrolysis to alter nucleosome structure and/or move nucleosomes along the chromatin fiber (9). Other enzymatic complexes covalently modify the N-terminal tails of histones by acetylation, methylation, phosphorylation, and ubiquitinylation (10). Core histone H3 and H4 acetylation and H3 K4 methylation are modifications strongly associated with active chromatin (11–13). Conversely, H3 K9 methylation and the presence of the linker histone H1 are associated with condensed and inactive chromatin (14, 15).
The mechanism by which the mammalian globin LCR/enhancers participate in recruitment of chromatin remodeling complexes and pol II, which eventually manifest their activity at the distant promoters, is unclear. Models include (i) tracking along the chromatin between the enhancer and promoter (16) and (ii) direct contact between enhancer and promoter with looping out or linking of the intervening DNA (17, 18). Facilitated tracking is an intermediate view in which components tracking along chromatin toward a promoter retain contact with the enhancer, eventually resulting in loop formation (19). Developmental stage specific domains of intergenic transcription and chromatin remodeling in the human globin locus are supportive of a processive LCR activation mechanism (20). Recent experiments have provided strong evidence that the LCR DNase I hypersensitive sites (HSs) and an active globin promoter are held in close physical proximity when transcription is active, compatible with looping or facilitated tracking (21–23).
Histone modifications have been studied extensively within vertebrate globin loci in erythroid cells expressing the adult β-globin gene (24, 25). There is widespread H3 and H4 acetylation across the loci compared to the pattern in nonerythroid tissues, with peaks of acetylation at the LCR and at the active β-globin genes, although such peaks at the active genes were not notable in the chicken locus (4, 26–30). In chicken embryo erythrocytes, both the active β-globin gene and the developmentally silenced embryonic globin genes exist within the hyperacetylated domain (28). In contrast, in MEL cells and murine fetal liver that express the adult β-globin genes, the globin locus contains a hypoacetylated subdomain, within which reside the silenced embryonic genes (27, 30). This unmodified subdomain makes it unlikely that histone modifications are propagated from the LCR to the adult genes. Interestingly, at the yolk sac stage of murine erythropoiesis, when the embryonic genes are expressed and the adult genes are silenced, high levels of acetylation were observed over both the active and inactive genes, but the subdomain structure was not investigated (27).
In addition to comprising foci of histone hyperacetylation, LCR/enhancers recruit pol II, which is associated with the LCR HSs and the active β-globin gene in MEL cells and in mouse erythroid cells, but absent both at the inactive embryonic εy-globin gene and at sequences between it and the LCR (31–34). The LCR sites of pol II detection are among the nongenic globin sequences that are transcribed at a low rate by pol II in MEL cells and human erythroid K562 cells and within a human globin locus transgene (20, 34–36). Such transcription could potentially deliver pol II to a distant promoter via a tracking mechanism. However, the absence of detectable pol II within the interposed hypoacetylated embryonic subdomain is more consistent with remote transfer of pol II (LPT, long-range polymerase transfer) from the LCR to the β-globin promoter (31, 34). In other enhancer-dependent loci, activators, pol II, and histone acetylation have been detected at both the gene promoter and the enhancer (37, 38), and sometimes in sequences between, consistent with a tracking mechanism of enhancer action (39, 40). In one instance, investigators demonstrated that an activator/HAT/SWI/SNF complex is detected at the 6.5-kb distant enhancer of the HNF-4α gene, subsequently at the intervening sequences between the enhancer and promoter, and finally at both the enhancer and promoter presumably as part of a looped structure, providing an example of facilitated tracking (39).
The human embryonic ε-globin gene is considerably closer, at ≈6 kb, to the LCR than is the adult β-globin gene (60 kb). Here, we investigated histone modification and pol II recruitment to the LCR and ε-globin gene in the human globin locus at the embryonic stage when this gene is active, as exemplified by the well studied K562 cell model. In contrast to murine erythroid cells expressing the adult globin genes, in K562 cells the domain encompassing the LCR and the active ε-globin gene was enriched for acetylated H3 and H4, and the level of histone modification was directly related to the level of ε-globin transcriptional activity. pol II was also detected broadly in the domain. Thus, distinctive differences in chromatin modification characterize the β-globin locus at embryonic (ε-globin gene is active) and adult (β-globin gene is active) stages. The continuity of histone modifications and of pol II detection between the LCR and the relatively nearby ε-globin gene in an embryonic stage milieu, compared to the discontinuous nature of the modified regions in the adult stage locus, suggests that different mechanisms may contribute to the propagation of the modifications and possibly the transfer of pol II to the active promoter at the two stages of development.
Methods
Cell Culture and Hemin Induction. K562 cells were grown in RPMI medium 1640 containing 10% FBS. For induction, cells at a concentration of 5 × 104 cells per ml were incubated with 30 μM hemin for 4 days. HeLa cells were cultured in DMEM, and MEL cells were cultured in RPMI medium 1640 with 10% FBS. Induction of MEL cells was performed by addition of 2% DMSO for 4 days.
RNase Protection Assay. RNA was prepared from 5 × 106 cells by using the PUREscript kit (Gentra). RNase digestion and gel analyses were performed according to the protocol of manufacturer of the reagent (Ambion, Austin, TX, RPA II kit). Actin served as the loading control.
Chromatin Immunoprecipitation (ChIP). Analysis of histone modification was carried out as described (28, 41). Briefly, nuclei from 5 × 107 K562 cells were digested with different MNase concentrations (0.0025 units/μl, 0.01 units/μl, and 0.04 units/μl) for 10 min at 37°C and combined. Soluble chromatin was fractionated on a sucrose gradient (5–30%), and mono- and dinucleosomes were pooled. Chromatin was precleared by incubation with protein A agarose and reacted with antibodies. Unbound chromatin in the “no antibody” sample was used as input. For analysis of histone H1 and pol II, 2 × 107 cells were cross-linked with 0.4% formaldehyde for 10 min at room temperature and sonicated to 200- to 500-bp fragments (41, 42). Antibodies used were anti-di-acetylated (K9, K14) histone H3, anti-tetra-acetylated (K4, K7, K11, K15) histone H4, anti-di-methylated H3 (K4), anti-di-methylated H3 (K9), and anti-H1 (Upstate Biotechnology, Lake Placid, NY), and normal rabbit IgG and anti-pol II (Santa Cruz Biotechnology).
Quantitative Real-Time PCR Analysis, Primers, and TaqMan Probes. Immunoprecipitated DNA was analyzed by quantitative realtime PCR (ABI Prism 7700) using TaqMan probes and primers (Primer Express 1.0, PE Applied Biosystems). DNA was quantified by using picogreen. DNA immunoprecipitated by antihistone antibodies and input DNA (1 ng each) was amplified with 200 nmol of TaqMan probes and 900 nmol of primers in a 25-μl reaction volume (41). Data were collected at the threshold where amplification was linear. The fold difference for each primer pair was determined by comparing the amount of target sequence in immunoprecipitated DNA to the amount of target sequence in input DNA (28). For anti-pol II antibodies, the fold difference was determined by comparing the amount of target sequence in 2.5% of specifically immunoprecipitated DNA to the amount of target sequence DNA precipitated by normal rabbit IgG. Sequences of primers and TaqMan probes are given in Table 1, which is published as supporting information on the PNAS web site and in ref. 41.
Reverse-Transcription Reaction. Two micrograms of RNA was treated with RNase-free DNase I for 15 min at 25°C, and then 1 μg of RNA was reverse transcribed by using superscript II as suggested by the manufacturer (Invitrogen). cDNA was diluted to 400 μl, and 5 μl of cDNA was amplified in a 25-μl reaction volume by real-time PCR. The amount of cDNA was compared with genomic DNA and then corrected by the amount of actin cDNA compared with genomic DNA.
Results and Discussion
Histone Modification Is Continuous Between the LCR and the Active ε-Globin Gene. Histone H3 and H4 acetylation are widespread in adult stage globin loci (24, 25). However, a domain of hypoacetylation encompasses the silent embryonic genes (27, 30). Considerably less is known about histone modification across the loci at earlier developmental stages. We asked in what ways this pattern was altered in human erythroid K562 cells that express the embryonic ε-globin gene but not the adult β-globin gene (Fig. 1). ChIP was performed by using antibodies to acetylated H3 and H4, and to methylated H3 K4 a modification also associated with actively transcribed genes. Chromatin of mono- and dinucleosome size (41) (and see Fig. 6A, which is published as supporting information on the PNAS web site) was immuno-precipitated with antibodies, and the purified DNA was quantitatively analyzed by real-time PCR at 22 sites across the LCR, embryonic ε-globin gene, and adult β-globin gene.
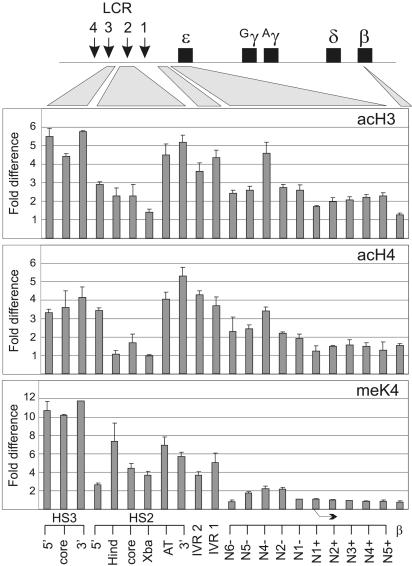
Fig. 1.
Histone modification extends between the LCR and the active ε-globin gene. The human β-globin locus is diagrammed at the top and the positions of amplicons used for real-time PCR are indicated below. IVR denotes positions between the LCR and the gene (IRV 2, -4,801; IVR 1, -3,097). The “N” sites are nucleosome positions relative to the ε-globin transcription start site and extend to -1,708 with respect to the ε-globin transcription start site (56). Mono- and dinucleosomes were prepared from K562 cells by MNase digestion and reacted with antibodies to diacetylated H3, hyperacetylated H4, and H3 dimethylated at K4. Immunoprecipitated DNA was quantitatively analyzed by real-time PCR. The fold difference was determined by comparing the amount of target sequence in immunoprecipitated DNA to the amount of target sequence in input DNA. The results of three chromatin preparations are shown ± SEM. Bent arrow indicates the ε-globin transcription start site.
Within LCR HS3 and HS2, acetylation of H3 and H4 and H3 K4 methylation levels were high (Fig. 1); detection of these modifications was lower at the HS2 core itself, consistent with earlier studies (41). In contrast to what is found in erythroid cells that express adult β-globin genes, H3 and H4 acetylation and H3 K4 methylation continued to be elevated between the LCR and the ε-globin gene. AcH3 and acH4 elevation persisted through the ε-globin gene 5′ flank and coding sequences, whereas H3 K4 methylation levels diminished at these sequences. We conclude that, in an embryonic erythroid milieu, in contrast to the situation in adult stage cells, H3 and H4 acetylation and K4 methylation extend in an uninterrupted fashion from the LCR to the transcribed ε-globin gene.
H3 and H4 acetylation was also detected at the β-globin gene, which is inactive in K562 cells, as was the case for the inactive β-globin gene in 11.5-day mouse fetal yolk sac, suggesting that, insofar as histone modification is concerned, the adult gene exists in a prepared state before its transcription is activated (27). Results in the two systems argue that, at early developmental stages, both the active and inactive globin genes exist in acetylated chromatin, in contrast with adult stages in which the inactive embryonic gene and surrounding chromatin are hypoacetylated.
Histone Modification Levels Are Related to the Transcriptional Activity of the ε-Globin Gene. Histone acetylation at active genes correlates globally with transcriptional activity (43). Very high level H3 and H4 acetylation throughout the chicken β-globin gene, in comparison to more limited hyperacetylation of a housekeeping gene, was most simply interpreted as related to the rate of transcription of each of the genes (44). However, MEL cell induction by DMSO increases β-globin transcription 100-fold, with only an accompanying 2-fold increase in β-globin H3 acetylation (45). Similarly, only modest 2- to 6-fold differences in H3 acetylation accompanied drastic reduction in β-globin transcription in mutant MEL cell lines lacking either the enhancer factor NF-E2 (31) or GATA-1 (46).
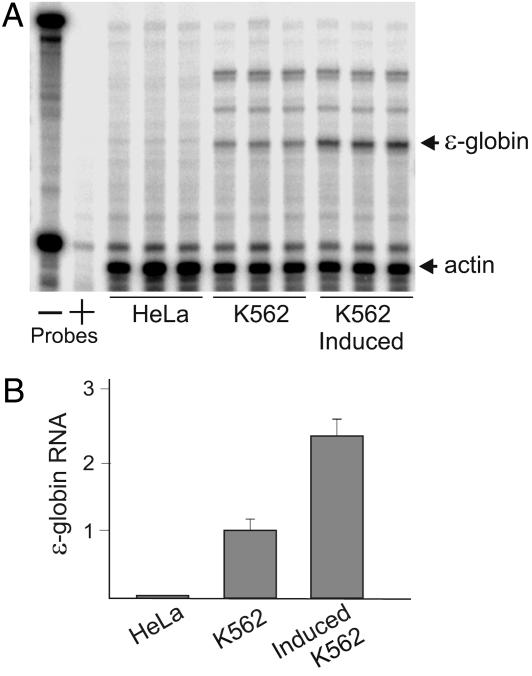
Although the ε-globin gene is transcribed in K562 cells, globin gene expression is much lower than in normal erythroid cells. K562 cells can be induced to higher levels of ε-globin gene transcription, without loss of growth potential, by hemin (47). To address the relationship between ε-globin gene transcription and histone modification, K562 cells were studied with or without induction by 30 μM hemin. HeLa cells served as a nonerythroid control. An RNase protection experiment in which three RNA samples were analyzed for each cell type is shown in Fig. 2A, and the data are depicted graphically in Fig. 2B. Hemin induction resulted in a ≈2.5-fold increase in ε-globin RNA, commensurate with earlier data (48). ε-globin RNA was undetectable in HeLa cells.
Fig. 2.
Transcription of the ε-globin gene in K562 cells is induced by hemin. (A) RNase protection analysis was performed by using RNA from uninduced and hemin-induced K562 cells and from nonerythroid HeLa cells. For induction, K562 cells were grown in medium containing 30 μM hemin for 4 days. Three RNA preparations for each type of cell is shown. The ε-globin-protected band is indicated by an arrow on a denaturing gel. RNase protection probes, with (+) or without (-) RNase treatment, are as labeled. Actin served as the loading control. (B) The results are depicted graphically ± SEM.
Hemin induction of K562 cells resulted in elevated levels of acH3 and acH4 compared to uninduced cells in both genic and nongenic regions from the LCR through the ε-globin gene (Fig. 3 A and B). H3 K4 methylation was also increased after hemin induction at most, but not all, positions studied (Fig. 3C). Although hemin induction increased the H3 K4 methylation in ε-globin coding sequences, the levels were never as high as within LCR HS3 and HS2 (but see below). H3 K9 methylation, associated with repressed chromatin, was low at all locations tested in K562 and decreased after hemin induction (Fig. 3D), an inversion of the pattern seen with H3 and H4 acetylation and K4 methylation (49).
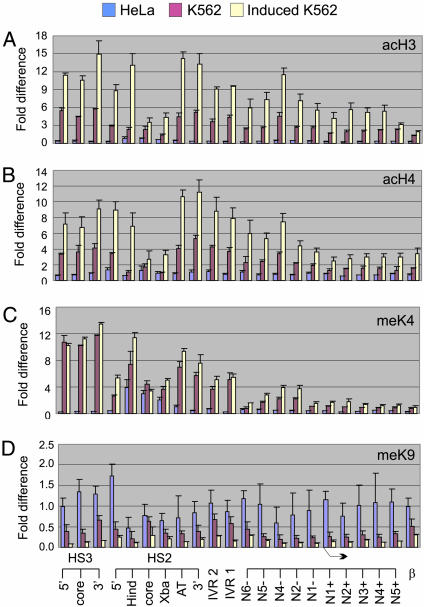
Fig. 3.
Hemin induction of ε-globin transcription in K562 cells increases histone acetylation broadly in the globin locus. ChIP experiments were performed by using HeLa cells and uninduced and hemin-induced K562 cells. Antibodies used included, anti-acH3 (A), anti-acH4 (B), anti-H3-meK4 (C), and anti-H3-meK9 (D). Immunoprecipitated DNA was analyzed by real-time PCR at 22 sites as shown in Fig. 1. The fold difference of immunoprecipitated DNA compared to input DNA is presented for HeLa (blue), uninduced K562 (red), and induced K562 (yellow) cells. The results of three independent experiments ± SEM are depicted. Bent arrow indicates the ε-globin transcription start site.
The β-globin locus in HeLa cells had very low levels of H3 and H4 acetylation and H3 K4 methylation, and elevated levels of H3 K9 methylation, consistent with inactive chromatin. However, some enrichment of H3 K4 methylation was detected at HS2 in HeLa cells. The maintenance of this modification may be related to the moderate sensitivity of HS2 to DNase I in HeLa cells (50). Interestingly, hemin induction resulted in increased H3 and H4 acetylation at the β-globin gene in HeLa cells despite a lack of transcription (see below).
From these results, we conclude that increased H3 and H4 acetylation over a broad region from the LCR to the active ε-globin gene correlates with increased transcriptional activity of the ε-globin gene in K562 cells. These results are strengthened by our observations that, on minichromosomes, where ε-globin transcription is increased a further 2-fold over hemin-induced K562 cells (on a per copy basis), hyperacetylation of H3 from HS2 to the linked ε-globin gene is also increased an additional 2-fold (41). Interestingly, on minichromosomes H3 K4 methylation in the ε-globin coding region is greatly increased, as in highly transcribed yeast genes (43).
Erythroid Cell-Specific Depletion of Linker Histone H1 and Localized Nucleosome Eviction at HS2 of the LCR. Linker histone H1 is thought to antagonize transcription and stabilize higher-order chromatin structure. However, the magnitude of H1 depletion in active chromatin is modest. H1 had ≈2-fold lower density in transcribed β-globin chromatin in chicken erythrocytes compared to nontranscribed albumin chromatin (51). Similarly, we have reported a 2- to 3-fold decrease in H1 over the active versus inactive ε-globin gene on minichromosomes (41, 52), but there is no information about H1 in the endogenous human globin locus. We investigated H1 levels by ChIP in both induced and uninduced K562 cells and in HeLa cells. Cells were cross-linked with formaldehyde and the chromatin sonicated to 200- to 500-bp fragments (ref. 41 and see Fig. 6B), immunoprecipitated by H1 antibodies, and analyzed by realtime PCR. Fig. 4A indicates that, in K562 cells, H1 occupancy was 30–80% relative to that seen in HeLa cells, consistent with a transcriptionally active locus. In general, up-regulation of ε-globin transcription by hemin did not affect H1 levels further. However, at a few positions, H1 detection was increased by hemin; notably at HS3 and exon 1(N1+) of ε-globin coding sequence. The significance of these changes with hemin is unclear. Overall, the difference in H1 between K562 cells and HeLa cells is opposite to the pattern of H3 and H4 acetylation; this may relate to the reported activity of H1 as a specific inhibitor of histone acetylation in vitro (53).
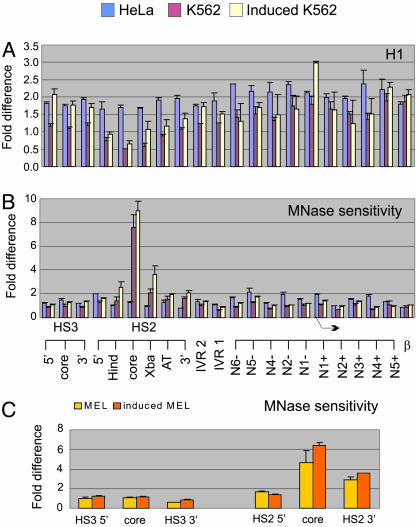
Fig. 4.
Erythroid cell-specific depletion of linker histones and localized nucleosome eviction at HS2. (A) HeLa cells and uninduced and induced K562 cells were cross-linked with formaldehyde and the chromatin sonicated to 200- to 500-bp fragments and immunoprecipitated by H1 antibodies. DNA was analyzed by real-time PCR. Primers and probes were as in Figs. 1 and 3 but without the HS3 3′ and the IVR 2 probes. The fold difference of immunoprecipitated DNA compared to input DNA (results of three independent experiments ± SEM) is graphed. Bent arrow indicates the ε-globin transcription start site. (B) Total genomic DNA from HeLa cells and K562 cells was fragmented with EcoRI. After amplification by real-time PCR, the fold difference was determined by comparing the amount of target sequence in total DNA to the amount of target sequence in DNA prepared by MNase digestion as described in Fig. 1. The results of three independent experiments ± SEM are graphed. (C) MEL cells were uninduced (yellow) or induced (orange) with DMSO. Mono- and dinucleosomal DNA and fragmented genomic DNA were prepared as in B, and amplified with primers at six locations in mouse HS2 and HS3. The results of three independent experiments ± SEM are graphed.
As was the case for acetylated and methylated H3 and H4 (Figs. 1 and 3), low levels of H1 were observed in the HS2 core. We previously reported that HS2 core sequences, but not active globin promoter sequences, were specifically depleted from the input DNA in ChIP experiments of acetylated H3 and H4, consistent with loss of a nucleosome (41). To compare the MNase sensitivities of HS2 and HS3 and to investigate their structure in a nonerythroid cell, we compared input DNA to fragmented total genomic DNA from K562 cells and HeLa cells by using real-time PCR. This analysis revealed that HS2 core sequences, but not HS3 sequences, were depleted in K562 cells (values significantly greater than 1) due to extreme sensitivity to MNase attack (Fig. 4B). Hemin induction had no further effect on HS2 or HS3 structure. In HeLa cells, HS2 sequences were not depleted, consistent with the presence of a nucleosome at HS2.
To ask whether extreme sensitivity to MNase at HS2 is uniquely a characteristic of the embryonic erythroid stage and/or the human globin locus, we studied MEL cells both before and after induction of adult β-globin transcription by DMSO. Similar to K562 cells, HS2 sequences, but not HS3 sequences in MEL cells were highly depleted from input DNA (Fig. 4C), reflecting extreme sensitivity to MNase digestion, even before chemical induction. Neither DMSO nor hemin induction affected the MNase sensitivity of HS2, suggesting, as others have (54), that formation of HS2 is an early step in erythroid differentiation.
pol II Is Widely Distributed from the LCR to the Active ε-Globin Gene. RNA pol II has been detected by ChIP at the murine β-globin LCR HSs and at the active β-globin promoter in adult stage MEL cells (31–34). We investigated the localization of pol II in the embryonic/fetal globin locus in uninduced human K562 cells and after hemin induction. ChIP experiments were carried out by using formaldehyde cross-linked chromatin and antibodies to the large subunit of pol II. Samples were analyzed by quantitative real-time PCR.
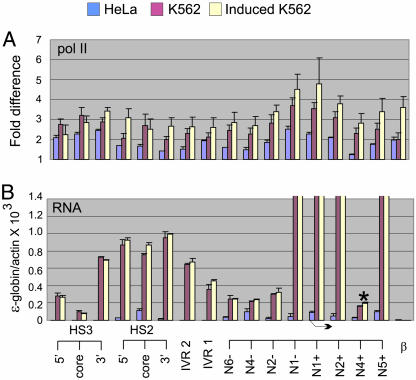
pol II was detected at HS2 and HS3 in K562 cells (Fig. 5A), consistent with studies in the murine locus (33, 34), and at the active ε-globin gene. Strikingly, however, pol II was associated with sequences in the region intervening between the LCR and the ε-globin gene as well as throughout the gene itself. This result provides a counterpoint to the pattern in the murine adult globin locus where pol II was not detected between the LCR and the inactive εy-globin gene, and these sequences were part of a hypoacetylated chromatin domain (27, 34). Hemin induction resulted in increased association of pol II with HS2, intervening sequences, and the ε-globin gene, most notably within coding sequences, consistent with the increase in transcription. Surprisingly, pol II was detected at the inactive β-globin gene in K562 cells and at higher levels after hemin induction, even in the absence of transcription of the gene (see below). Similarly, others have found continued association of pol II with the inactive β-globin gene when the LCR is deleted from the mouse globin locus (33). In HeLa cells, pol II detection was much reduced throughout the globin locus, albeit with low-level association at HS3, and at the two globin genes.
Fig. 5.
Widespread distribution of pol II between the LCR and ε-globin gene in embryonic/fetal stage K562 cells. (A) Formaldehyde cross-linked chromatin was prepared from HeLa cells and uninduced and induced K562 cells and immunoprecipitated with anti-pol II antibodies or with control normal rabbit IgG. Aliquots were amplified by real-time PCR using the primers and probes indicated in Fig. 1, without the Hind, Xba, and AT HS2 probes, or the N5- and N3+ ε-globin probes. The fold difference was determined by comparing the amount of target sequence in immunoprecipitated DNA to the amount of target sequence in DNA precipitated by normal rabbit IgG. The results of three independent experiments ± SEM are graphed. (B) cDNA was reverse transcribed from 1 μg of RNA from HeLa cells and uninduced and induced K562 cells and then amplified by real-time PCR. The amount of cDNA was compared with genomic DNA and then corrected by the amount of actin cDNA. Probes and primers were as in A. Genic transcription is beyond the scale presented in the figure to visualize low level intergenic transcription. The results of three independent experiments ± SEM are graphed. Bent arrow indicates the ε-globin transcription start site.
Transcription of β-globin locus intergenic regions on the same strand as that of the globin genes occurs in K562 cells (35, 36). To investigate whether RNA transcripts characterize both the intergenic and genic regions in which pol II is detected, RNA was isolated from untreated and hemin-induced K562 cells and from HeLa cells. Reverse-transcribed cDNA was analyzed by realtime PCR (Fig. 5B). In ε-globin coding sequences, RNA transcripts were abundant (beyond the scale of the graph in Fig. 5B). A reduced signal at the second intron of the ε-globin gene (asterisk) reflects the level of unspliced transcripts. In the LCR and the intervening sequences to the ε-globin gene, transcripts were detected at a level greater than that seen in the second intron, and at ≈5% of the level of genic transcripts. Importantly, even though pol II was detected at the inactive β-globin gene, transcripts were not detected there, suggesting a block to elongation similar to that seen in a mouse globin locus from which the LCR had been deleted (33).
Long intergenic pol II transcripts in the globin locus are suggestive of a tracking process for opening chromatin domains during development (20). Our results support the possibility that some pol II may track from the LCR to the embryonic ε-globin gene, the most proximal gene of the locus. Furthermore, the region between the LCR and the ε-globin gene also contains acetylated histones, which suggests that acetylation could be established by the tracking of HATs associated with elongating polymerases (55). In adult stage MEL cells, the patterns of histone acetylation and pol II distribution in the LCR are not greatly altered when pol II elongation is inhibited by DRB, arguing against this mechanism (34), but this may reflect conditions peculiar to the adult, and not the embryonic, stage locus. Alternatively, histone acetylation and detection of pol II over a broad region could result from a local high concentration of HATs and pol II in the vicinity of an LCR and a proximal transcribing gene (23).
Our results indicate that, in an embryonic/fetal β-globin milieu, as exemplified in K562 cells, histone modifications and pol II are detected over a broad region including HS3 and HS2 and extending to the active ε-globin gene 6 kb distant. In contrast, in the adult stage globin locus in MEL cells and in mouse erythroid cells, histone acetylation and pol II association are discontinuous, detected at the LCR and at the active β-globin gene, but not between (27, 30). We propose that widespread histone modification and the presence of pol II in intervening sequences between the LCR and the active ε-globin gene reflect activation of this embryonic globin gene by the LCR. Further, translocation of histone modifying complexes and pol II could participate in establishment of LCR-gene proximity (21, 23), consistent with facilitated tracking (19). Such a processive component of enhancer–promoter communication at an early erythroid developmental stage may relate to the 6-kb relative proximity of the ε-globin gene to the LCR, in contrast with the 60-kb-distant β-globin gene. However, it is also possible, and consistent with the model proposed by Gribnau et al. (20), that a processive mechanism underlies sequential histone modification and opening of chromatin domains in the globin locus, and establishment of close contacts between the LCR and the later expressed globin genes during development. To fully evaluate these possibilities, it will be necessary and important to extend our observations to human globin loci which accurately reflect normal hemoglobin switching.
Supplementary Material
Acknowledgments
We thank Gary Felsenfeld and Michael Litt for real-time PCR expertise and helpful discussions, and Gerd Blobel, Jane Little, and Cecilia Trainor for critical comments on the manuscript.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: LCR, locus control region; pol II, RNA polymerase II; ChIP, chromatin immunoprecipitation; HS, DNase I hypersensitive site.
References
- 1.Stamatoyannopoulos, G. & Grosveld, F. (2001) in The Molecular Basis of Blood Diseases, eds. Stamatoyannopoulos, G., Majerus, P. W., Perlmutter, R. M. & Varmus, H. (Saunders, Philadelphia), pp. 135-182.
- 2.Epner, E., Reik, A., Cimbora, D., Telling, A., Bender, M. A., Fiering, S., Enver, T., Martin, D. I., Kennedy, M., Keller, G., et al. (1998) Mol. Cell 2, 447-455. [DOI] [PubMed] [Google Scholar]
- 3.Bender, M. A., Bulger, M., Close, J. & Groudine, M. (2000) Mol. Cell 5, 387-393. [DOI] [PubMed] [Google Scholar]
- 4.Schubeler, D., Groudine, M. & Bender, M. A. (2001) Proc. Natl. Acad. Sci. USA 98, 11432-11437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi, O.-R. & Engel, J. D. (1986) Nature 323, 731-734. [DOI] [PubMed] [Google Scholar]
- 6.Nickol, J. M. & Felsenfeld, G. (1988) Proc. Natl. Acad. Sci. USA 85, 2548-2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peterson, C. L. & Workman, J. L. (2000) Curr. Opin. Genet. Dev. 10, 187-192. [DOI] [PubMed] [Google Scholar]
- 8.Aalfs, J. D. & Kingston, R. E. (2000) Trends Biochem. Sci. 25, 548-555. [DOI] [PubMed] [Google Scholar]
- 9.Vignali, M., Hassan, A. H., Neely, K. E. & Workman, J. L. (2000) Mol. Cell. Biol. 20, 1899-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berger, S. L. (2002) Curr. Opin. Genet. Dev. 12, 142-148. [DOI] [PubMed] [Google Scholar]
- 11.Strahl, B. D., Ohba, R., Cook, R. G. & Allis, C. D. (1999) Proc. Natl. Acad. Sci. USA 96, 14967-14972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Strahl, B. D. & Allis, C. D. (2000) Nature 403, 41-45. [DOI] [PubMed] [Google Scholar]
- 13.Noma, K. & Grewal, S. I. (2002) Proc. Natl. Acad. Sci. USA 99, 16438-16445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laybourn, P. J. & Kadonaga, J. T. (1991) Science 254, 238-245. [DOI] [PubMed] [Google Scholar]
- 15.Kouzarides, T. (2002) Curr. Opin. Genet. Dev. 12, 198-209. [DOI] [PubMed] [Google Scholar]
- 16.Travers, A. (1999) Proc. Natl. Acad. Sci. USA 96, 13634-13637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dorsett, D. (1999) Curr. Opin. Genet. Dev. 9, 505-514. [DOI] [PubMed] [Google Scholar]
- 18.Bulger, M. & Groudine, M. (1999) Genes Dev. 13, 2465-2477. [DOI] [PubMed] [Google Scholar]
- 19.Blackwood, E. M. & Kadonaga, J. T. (1998) Science 281, 61-63. [DOI] [PubMed] [Google Scholar]
- 20.Gribnau, J., Diderich, K., Pruzina, S., Calzolari, R. & Fraser, P. (2000) Mol. Cell 5, 377-386. [DOI] [PubMed] [Google Scholar]
- 21.Carter, D., Chakalova, L., Osborne, C. S., Dai, Y. & Fraser, P. (2002) Nat. Genet. 32, 623-626. [DOI] [PubMed] [Google Scholar]
- 22.Tolhuis, B., Palstra, R. J., Splinter, E., Grosveld, F. & de Laat, W. (2002) Mol. Cell 10, 1453-1465. [DOI] [PubMed] [Google Scholar]
- 23.Palstra, R. J., Tolhuis, B., Splinter, E., Nijmeijer, R., Grosveld, F. & de Laat, W. (2003) Nat. Genet. 35, 190-194. [DOI] [PubMed] [Google Scholar]
- 24.Forsberg, E. C. & Bresnick, E. H. (2001) BioEssays 23, 820-830. [DOI] [PubMed] [Google Scholar]
- 25.Bulger, M., Sawado, T., Schubeler, D. & Groudine, M. (2002) Curr. Opin. Genet. Dev. 12, 170-177. [DOI] [PubMed] [Google Scholar]
- 26.Schubeler, D., Francastel, C., Cimbora, D. M., Reik, A., Martin, D. I. & Groudine, M. (2000) Genes Dev. 14, 940-950. [PMC free article] [PubMed] [Google Scholar]
- 27.Forsberg, E. C., Downs, K. M., Christensen, H. M., Im, H., Nuzzi, P. A. & Bresnick, E. H. (2000) Proc. Natl. Acad. Sci. USA 97, 14494-14499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Litt, M. D., Simpson, M., Recillas-Targa, F., Prioleau, M. N. & Felsenfeld, G. (2001) EMBO J. 20, 2224-2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kiekhaefer, C. M., Grass, J. A., Johnson, K. D., Boyer, M. E. & Bresnick, E. H. (2002) Proc. Natl. Acad. Sci. USA 99, 14309-14314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bulger, M., Schubeler, D., Bender, M. A., Hamilton, J., Farrell, C. M., Hardison, R. C. & Groudine, M. (2003) Mol. Cell. Biol. 23, 5234-5244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson, K. D., Christensen, H. M., Zhao, B. & Bresnick, E. H. (2001) Mol. Cell 8, 465-471. [DOI] [PubMed] [Google Scholar]
- 32.Johnson, K. D., Grass, J. A., Boyer, M. E., Kiekhaefer, C. M., Blobel, G. A., Weiss, M. J. & Bresnick, E. H. (2002) Proc. Natl. Acad. Sci. USA 99, 11760-11765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sawado, T., Halow, J., Bender, M. A. & Groudine, M. (2003) Genes Dev. 17, 1009-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson, K. D., Grass, J. A., Park, C., Im, H., Choi, K. & Bresnick, E. H. (2003) Mol. Cell. Biol. 23, 6484-6493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ashe, H. L., Monks, J., Wijgerde, M., Fraser, P. & Proudfoot, N. J. (1997) Genes Dev. 11, 2494-2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Plant, K. E., Routledge, S. J. & Proudfoot, N. J. (2001) Mol. Cell. Biol. 21, 6507-6514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shang, Y., Myers, M. & Brown, M. (2002) Mol. Cell 9, 601-610. [DOI] [PubMed] [Google Scholar]
- 38.Spicuglia, S., Kumar, S., Yeh, J. H., Vachez, E., Chasson, L., Gorbatch, S., Cautres, J. & Ferrier, P. (2002) Mol. Cell 10, 1479-1487. [DOI] [PubMed] [Google Scholar]
- 39.Hatzis, P. & Talianidis, I. (2002) Mol. Cell 10, 1467-1477. [DOI] [PubMed] [Google Scholar]
- 40.Louie, M. C., Yang, H. Q., Ma, A. H., Xu, W., Zou, J. X., Kung, H. J. & Chen, H. W. (2003) Proc. Natl. Acad. Sci. USA 100, 2226-2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim, A. & Dean, A. (2003) Mol. Cell. Biol. 23, 8099-8109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Johnson, K. D. & Bresnick, E. H. (2002) Methods 26, 27-36. [DOI] [PubMed] [Google Scholar]
- 43.Bernstein, B. E., Humphrey, E. L., Erlich, R. L., Schneider, R., Bouman, P., Liu, J. S., Kouzarides, T. & Schreiber, S. L. (2002) Proc. Natl. Acad. Sci. USA 99, 8695-8700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Myers, F. A., Evans, D. R., Clayton, A. L., Thorne, A. W. & Crane-Robinson, C. (2001) J. Biol. Chem. 276, 20197-20205. [DOI] [PubMed] [Google Scholar]
- 45.Sawado, T., Igarashi, K. & Groudine, M. (2001) Proc. Natl. Acad. Sci. USA 98, 10226-10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Letting, D. L., Rakowski, C., Weiss, M. J. & Blobel, G. A. (2003) Mol. Cell. Biol. 23, 1334-1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dean, A., Erard, F., Schneider, A. P. & Schechter, A. N. (1981) Science 212, 459-461. [DOI] [PubMed] [Google Scholar]
- 48.Dean, A., Ley, T. J., Humphries, R. K., Fordis, M. & Schechter, A. N. (1983) Proc. Natl. Acad. Sci. USA 80, 5515-5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Litt, M. D., Simpson, M., Gaszner, M., Allis, C. D. & Felsenfeld, G. (2001) Science 293, 2453-2455. [DOI] [PubMed] [Google Scholar]
- 50.Dhar, V., Nandi, A., Schildkraut, C. L. & Skoultchi, A. I. (1990) Mol. Cell. Biol. 10, 4324-4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Postnikov, Y. V., Shick, V. V., Belyavsky, A. V., Khrapko, K. R., Brodolin, K. L., Nikolskaya, T. A. & Mirzabekov, A. D. (1991) Nucleic Acids Res. 19, 717-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gui, C. Y. & Dean, A. (2001) Mol. Cell. Biol. 21, 1155-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Herrera, J. E., West, K. L., Schiltz, R. L., Nakatani, Y. & Bustin, M. (2000) Mol. Cell. Biol. 20, 523-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jimenez, G., Griffiths, S. D., Ford, A. M., Greaves, M. F. & Enver, T. (1992) Proc. Natl. Acad. Sci. USA 89, 10618-10622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wittschieben, B. O., Otero, G., de Bizemont, T., Fellows, J., Erdjument-Bromage, H., Ohba, R., Li, Y., Allis, C. D., Tempst, P. & Svejstrup, J. Q. (1999) Mol. Cell 4, 123-128. [DOI] [PubMed] [Google Scholar]
- 56.Gong, Q. H., McDowell, J. C. & Dean, A. (1996) Mol. Cell. Biol. 16, 6055-6064. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.