Abstract
Genome-wide microarray analysis (Affymetrix array) was used (i) to determine whether only one gene, the cytochrome P450 enzyme Cyp6g1, is differentially transcribed in dichlorodiphenyltrichloroethane (DDT)-resistant vs. -susceptible Drosophila; and (ii) to profile common genes differentially transcribed across a DDT-resistant field isolate [Rst(2)DDTWisconsin] and a laboratory DDT-selected population [Rst(2)DDT91-R]. Statistical analysis (ANOVA model) identified 158 probe sets that were differentially transcribed among Rst(2)DDT91-R, Rst(2)DDTWisconsin, and the DDT-susceptible genotype Canton-S (P < 0.01). The cytochrome P450 Cyp6a2 and the diazepam-binding inhibitor gene (Dbi) were over transcribed in the two DDT-resistant genotypes when compared to the wild-type Drosophila, and this difference was significant at the most stringent statistical level, a Bonferroni correction. The list of potential candidates differentially transcribed also includes 63 probe sets for which molecular function ontology annotation of the probe sets did not exist. A total of four genes (Cyp6a2, Dbi, Uhg1, and CG11176) were significantly different (P < 5.6 e-06) between Rst(2)DDT91-R and Canton-S. Additionally, two probe sets encoding Cyp12d1 and Dbi were significantly different between Rst(2)DDTWisconsin and Canton-S after a Bonferroni correction. Fifty-two probe sets, including those associated with pesticide detoxification, ion transport, signal transduction, RNA transcription, and lipid metabolism, were commonly expressed in both resistant lines but were differentially transcribed in Canton-S. Our results suggest that more than Cyp6g1 is overtranscribed in field and laboratory DDT-resistant genotypes, and the number of commonalities suggests that similar resistance mechanisms may exist between laboratory- and field-selected DDT-resistant fly lines.
The evolution of insecticide resistance is often, but not always, based on major effect alleles (1–4). It has been hypothesized that high selection pressure in the field will favor monogenic forms of pesticide resistance, and that selection for resistance in the laboratory will favor polygenic resistance (5–7). In early genetic studies in Drosophila, dichlorodiphenyltrichloroethane (DDT) resistance was mapped to multiple locations on chromosomes II and III (8–14). Subsequently, low-level DDT resistance was mapped to 64.5 ± 2 centiMorgans on the second chromosome (15), a locus (loci) known as Rst(2)DDT.
Recently, Daborn et al. (16) suggested that resistance to DDT in the field is monogenic and is due to the overexpression of a single P450 gene, Cyp6g1. Le Goff et al. (17) suggested that resistance in field isolates of both Drosophila melanogaster and Drosophila simulans is associated with overtranscription of Cyp6g1, whereas prolonged laboratory selection with DDT apparently coselects additional genes such as Cyp12d1 (18) and Cyp6a8 (19). In contrast to the hypothesis presented by Le Goff et al. (17), Brandt et al. (18) observed that both Cyp6g1 and Cyp12d1 are overexpressed in the field-selected isochromosomal line Rst(2)DDTWisconsin.
Daborn et al. (16) provided a transcriptional profile of a DDT-resistant Drosophila field isolate and suggested that DDT resistance found in many field strains of Drosophila was due to a specific P450 enzyme (Cyp6g1). Daborn et al. (16) used custom-made microarrays comprised of all known members of Drosophila cytochrome P450 genes and metabolic enzymes such as esterases and GSTs in addition to housekeeping genes. Overexpression of Cyp6g1 in transgenic Drosophila showed only slight increases in resistance, suggesting that there is much more to resistance than a single gene.
To date, no genome-wide expression profile has been evaluated to investigate the extent to which gene transcription varies between genotypes that are resistant and susceptible to DDT. This comparison would allow for the testing of the hypothesis that only a single detoxification enzyme, Cyp6g1, is being overexpressed in DDT-resistant strains (16, 17). Additionally, expression profiles of (i) a field-collected isolate and (ii) a laboratory-selected isolate would allow for testing of the hypothesis that different sets of genes are overexpressed in field-collected and laboratory-selected strains. In the following study, we used a laboratory DDT-selected population, Rst(2)DDT91-R, and a field-collected DDT-resistant isolate, Rst(2)DDTWisconsin, to test both of these aforementioned hypotheses. Our transcriptome approach identified a manageable number of genes for further investigation of the molecular basis of pesticide resistance. This analysis also strongly suggests that DDT resistance in Drosophila is more complex than previously described.
Methods
Drosophila Strains. Three D. melanogaster lines were used in the following experiments: (i) the DDT-susceptible Canton-S line; (ii) the DDT-resistant lines Rst(2)DDT91-R; and (iii) Rst(2)DDTWisconsin. The Canton-S strain was obtained from the Drosophila Stock Center (Bloomington, IN) in January 2000 and was used as a standard susceptible strain. Field collected flies from Door County in Wisconsin were exposed to 200 μg of DDT per vial for 24 h, and a single male survivor was mated with the Drosophila balancer line w*;T(2,3)apXa/TM3, Sb1 (18). A single DDT-resistant progeny was again crossed with the balancer line, and the resultant progeny were used to establish an isochromosomal line, Rst(2)DDTWisconsin. The Drosophila populations were reared in an environmentally controlled chamber at 28°C, 80% humidity, and 14 h of light per day. Experimental adult flies were collected as virgins (3 h after eclosion) by using light CO2 anesthesia. For expression analysis, flies were sorted in a 1:1 male/female ratio, flash frozen in liquid nitrogen, and transferred to -80°C.
Experimental Design and Statistical Analysis. Twelve independent vials from each genotype were established; four vials were pooled for each replicate for a total of three replicates for each genotype and nine samples overall. This strategy was intended to reduce between-vial variations. Total RNA was isolated by using the TRI Reagent extraction protocol (Molecular Research Center, Cincinnati). Messenger RNA, cRNA synthesis, and labeling reactions were performed independently for each replicate following the recommendations of the GeneChip Expression Analysis technical manual (Affymetrix, Santa Clara, CA). The nine Affymetrix Drosophila genome chips were hybridized to the fragmented cRNA, stained, and washed according to the recommendations of the GeneChip Expression Analysis technical manual (Affymetrix) at the Purdue Genomics and Microarray Core Facility. Image data were quantified by using genechip analysis suite/microarray suite 5.0 (MAS 5.0). The identification of informative probe sets was performed by using default settings (α1, 0.04; α2, 0.06; δ, 0.015; scale factor, 1.0; norm factor, 1.0). If all nine replicates for a particular probe set were deemed “absent,” the probe set was removed from further consideration. The remaining probe sets (8,974) were analyzed, and transcript levels were normalized to the chip median and log transformed. For each probe set, which represents the combined expression data from all relevant probe pairs on the chip, the generalized linear model Yi = μ + B1Li + εij was fit (20–22). In each ANOVA, Yi is a log-normalized transcript for the ith line and jth replicate, μ is the overall mean expression for the probe sets, and the Li is the i line represented (Canton-S, Rst(2)DDT91-R, and Rst(2)DDTWisconsin). An F test of the effect of genotype for each gene was conducted as the ratio of the mean squares for line over the mean squares for error. The P value for the test of the null hypothesis λ2 = λ1 = λ0 (i.e., mean expression not different among the three genotypes) was also calculated. We examined the model for conformation to the assumption of normality of the residuals testing the null hypothesis that the residuals for each gene were normally distributed by using the Shapiro–Wilkes Test. All analyses were performed in sas (SAS Institute, Cary, NC). A Bonferroni significance level was used as an initial criterion for rejecting the null hypothesis of a significant treatment effect (0.05/8974), corresponding to a false discovery rate (FDR) of 0.045. We used a second arbitrary nominal threshold of α < 0.01, because type I and II errors are inversely related, with decreases in false positives (type I) being associated with increases in false negatives (type II), and because the Bonferroni correction is overly conservative as tests are correlated (20–22). This threshold corresponded to a FDR of 0.56. We also considered the test for normality of the residuals. If the test of the null hypothesis of difference across genotypes was rejected at the Bonferroni level, and we had no evidence for departure from normality of the residuals, we declared the gene differentially expressed across genotypes and examined additional contrasts comparing the effect of the genotypes [Canton vs. Rst(2)DDT91-R and Canton vs. Rst(2)DDTWisconsin]. If the P value for the test of differences over genotypes was ≤0.01 (56% FDR) but larger than the Bonferroni level, and we had no evidence for departure from normality of the residuals, we considered a gene as being differentially transcribed.
Gene Ontology Analysis. We used blast to link Affymetrix probes to a FlyBase annotation 2, to determine the molecular function, biological process, and cellular component of each gene (www.geneontology.org). We described the transcripts using the molecular function ontology, because it serves well as a foundation for unifying the growing amount of expression information related to genome-wide pesticide studies in different organisms, and it is designed to facilitate the transfer of gene/protein function information among other organisms. However, the three separate ontologies are available which is published as supporting information on the PNAS web site.
Results and Discussion
Multiple Genes Are Differentially Transcribed Between DDT-Resistant and -Susceptible Lines. High-density Affymetrix oligoarrays were used to identify transcripts differentially expressed among (i) the laboratory DDT-selected Rst(2)DDT91-R, (ii) the isochromosomal DDT-resistant field isolate Rst(2)DDTWisconsin, and (iii) the DDT-susceptible Canton-S line. The GeneChip contained 13,966 probe sets, of which 4,992 were uninformative (absent for gene expression in all nine replicates). We identified 158 probe sets that were significantly different among the genotypes (P < 0.01; false discovery rate of 0.56) and the probe sets 143127_at [Cyp6a2 (cytochrome P450)] and 143608_at [Dbi (Diazepambinding inhibitor gene)] were differentially transcribed at the Bonferroni corrected level.
We then investigated the individual contrasts for Rst(2)DDT91-R and Canton-S and Rst(2)DDTWisconsin and Canton-S. A total of four probe sets (Cyp6a2, Dbi, Uhg1, and CG11176) were significantly different between Rst(2)DDT91-R and Canton-S (Tables 1 and 2) at the 5.6 e-06. The P values of two probe sets encoding for Cyp12d1 and Dbi were significantly different between Rst(2)DDTWisconsin and Canton-S after a Bonferroni correction (Table 1). Two cytochrome P450 genes (Cyp6g1 and Cyp12d1) were previously identified in our laboratory (18) and elsewhere (16, 17) as putative transcripts associated with DDT resistance in both Rst(2)DDT91-R and Rst(2)DDTWisconsin.
Table 1. Genes annotated for molecular function with differential transcription among three Drosophila genotypes.
| Molecular function | GO code | Symbol | Probe set | P value | Cytological position | 91-R | Wisconsin |
|---|---|---|---|---|---|---|---|
| Cytochrome P450 activity | GO:0015034 | Cyp6a2* | 143127 | 8.6 e-7 | 42C8 | 2.8 e-7* | 5.9 e-6 |
| Cyp12d1 | 146978 | 7.2 e-6 | 47D4 | 1.0 e-3 | 2.5 e-6* | ||
| Cyp6a17 | 152313 | 4.2 e-5 | 51D1 | 0.43 | 2.5 e-5 | ||
| Cyp6a8 | 142189 | 6.0 e-5 | 51D1 | 7.1 e-5 | 0.17 | ||
| Cyp12d1 | 154692 | 1.5 e-4 | 47D4 | 6.9 e-3 | 5.0 e-5 | ||
| Cyp6w1 | 152494 | 1.8 e-4 | 42A13 | 6.2 e-5 | 0.01 | ||
| Cyp6g1 | 152900 | 6.9 e-4 | 48E8 | 2.9 e-4 | 1.1 e-3 | ||
| Cyp6a14 | 146815 | 7.5 e-4 | 44D3 | 9.4 e-3 | 7.3 e-3 | ||
| Cyp9c1 | 141780 | 7.9 e-4 | 60D10 | 6.5 e-4 | 0.74 | ||
| Cyp4p1 | 143782 | 1.0 e-3 | 45B7 | 4.1 e-4 | 0.11 | ||
| Cyp6a23 | 147225 | 3.3 e-3 | 51D1 | 0.03 | 0.03 | ||
| Glutathione transferase activity | GO:0004364 | CG17530 | 142537 | 2.9 e-5 | 55C7 | 1.9 e-5 | 2.4 e-5 |
| CG17522 | 147434 | 1.8 e-3 | 55C6 | 1.6 e-3 | 9.9 e-4 | ||
| CG1681 | 152675 | 8.3 e-4 | 11F4 | 0.75 | 4.7 e-3 | ||
| Gst3-1 | 141930 | 8.4 e-4 | 55C8 | 0.17 | 3.1 e-3 | ||
| CG6673 | 142740 | 9.3 e-4 | 66D5 | 3.8 e-3 | 0.01 | ||
| Glucuronosyltransferase activity | GO:0015020 | Ugt86Dh | 149669 | 6.6 e-4 | 86D6 | 2.2 e-3 | 0.05 |
| Ugt86Dd | 149663 | 3.5 e-4 | 86D4 | 2.2 e-3 | 0.91 | ||
| Ugt35b | 142271 | 3.6 e-4 | 86D5 | 1.9 e-4 | 3.8 e-4 | ||
| Tyrosine-ester sulfotransferase activity | GO:0017067 | CG5431 | 141306 | 8.7 e-4 | 59F6 | 3.5 e-4 | 0.12 |
| Oxidoreductase activity | GO:0016491 | Pdh | 153433 | 2.6 e-3 | 72E3 | 8.7 e-4 | 0.01 |
| CG30019 | 152569 | 1.1 e-3 | 47C3 | 0.10 | 4.1 e-4 | ||
| Oxidoreductase activity | GO:0016491 | CG3301 | 151819 | 3.5 e-3 | 93D2 | 0.01 | 0.06 |
| CG12224 | 149723 | 4.1 e-3 | 87A4 | 1.3 e-3 | 0.03 | ||
| CG8888 | 152990 | 4.3 e-3 | 48D8 | 0.02 | 0.05 | ||
| CG9360 | 144893 | 4.4 e-3 | 10E2 | 4.1 e-3 | 2.3 e-3 | ||
| CG3603 | 144428 | 4.5 e-3 | 3C3 | 1.5 e-3 | 0.09 | ||
| CG3842 | 144594 | 7.0 e-3 | 5F2 | 0.01 | 2.6 e-3 | ||
| CG15531 | 150879 | 5.9 e-3 | 99E3 | 3.0 e-3 | 5.9 e-3 | ||
| CG9747 | 152137 | 6.4 e-3 | 99E1 | 0.05 | 2.2 e-3 | ||
| CG15093 | 152083 | 7.6 e-3 | 55F2 | 2.7 e-3 | 0.02 | ||
| Calcium channel regulator activity | GO:0005246 | InaF | 143971 | 4.3 e-5 | 10E2 | 2.1 e-5 | 5.5 e-5 |
| Calcium channel activity | GO:0005262 | CG17142 | 147913 | 1.3 e-5 | 61B2 | 0.01 | 4.4 e-4 |
| Calcium ion binding | GO:0005509 | Cpn | 152156 | 2.2 e-4 | 87B1 | 7.3 e-4 | 0.01 |
| CG2185 | 153946 | 4.2 e-3 | 83B8 | 1.9 e-3 | 0.36 | ||
| Voltage-sensitive calcium binding | GO:0005245 | Ca-α1D | 151837 | 3.6 e-3 | 35E5 | 0.09 | 0.01 |
| Calcium-dependent cell adhesion molecule activity | GO:0008014 | Mys | 143507 | 7.0 e-3 | 7D5 | 2.5 e-3 | 0.10 |
| Organic ion transporter activity | GO:0015101 | CG16727 | 150207 | 8.3 e-3 | 92A10 | 3.2 e-3 | 0.22 |
| Ryanodine-sensitive calcium-release channel activity | GO:0005219 | Rya-r44F | 143650 | 3.4 e-3 | 44F1 | 0.63 | 3.1 e-3 |
| Sodium-dependent multivitamin transporter activity | GO:0008523 | CG8932 | 150628 | 8.7 e-4 | 96F8 | 8.1 e-3 | 4.6 e-3 |
| Monosaccharide transporter activity | GO:0015145 | CG15407 | 145701 | 2.4 e-4 | 23E5 | 0.02 | 0.02 |
| Maleylacetoacetate isomerase activity | GO:0016034 | CG9362 | 141293 | 2.7 e-3 | 85D7 | 1.0 e-3 | 5.3 e-3 |
| Long-chain fatty acid transporter activity | GO:0005324 | CG5568 | 141286 | 6.6 e-3 | 64F3 | 4.3 e-3 | 4.6 e-3 |
| Cholesterol O-acyltransferase | GO:0017066 | CG5397 | 151961 | 1.3 e-3 | 21F3 | 1.9 e-3 | 5.5 e-4 |
| Triacylglycerol lipase activity | GO:0004806 | CG17192 | 150700 | 2.0 e-3 | 97D14 | 0.5 | 1.0 e-3 |
| Argininosuccinate lyase activity | GO:0004056 | CG9510 | 141703 | 5.2 e-3 | 29F2 | 2.2 e-3 | 7.7 e-3 |
| Acetyl-CoA C-acyltransferase | GO:0003988 | Yip2 | 153437 | 1.1 e-3 | 30D1 | 3.6 e-4 | 0.03 |
| Diacylglycerol binding | GO:0019992 | CG10737 | 153286 | 9.9 e-3 | 56C1 | 0.89 | 6.2 e-3 |
| Acetylglucosaminyltransferase activity | GO:0008375 | Ext2 | 151844 | 9.9 e-3 | 52F3 | ||
| Lipid binding | GO:0008289 | Dbi* | 143608 | 1.2 e-6 | 65E8 | 7.9 e-7* | 1.2 e-6 |
| Drug binding | GO:0008144 | CG14715 | 142488 | 4.2 e-3 | 86E19 | 4.9 e-3 | 1.9 e-3 |
| ATP-binding cassette transporter activity | GO:0004009 | CG9892 | 141801 | 8.8 e-3 | 23A2 | 0.01 | 3.7 e-3 |
| Protein binding | GO:0005515 | Arr1 | 152756 | 5.1 e-4 | 36D3 | 2.3 e-4 | 6.6 e-4 |
| Arr2 | 143078 | 1.7 e-3 | 66D9 | 9.1 e-4 | 1.7 e-3 | ||
| Dia | 143636 | 1.0 e-3 | 38E5 | 1.1 e-3 | 5.5 e-4 | ||
| Ank2 | 151478 | 2.1 e-3 | 66A10 | 7.3 e-4 | 7.5 e-3 | ||
| Map205 | 153771 | 3.7 e-5 | 100F5 | 1.6 e-5 | 1.5 e-5 | ||
| DNA binding | GO:0003677 | Sox100B | 151112 | 2.3 e-3 | 100B1 | 1.2 e-3 | 2.2 e-3 |
| RNA polymerase II transcription factor activity | GO:0003702 | Cf2 | 143101 | 5.1 e-3 | 25B1 | 1.8 e-3 | 0.09 |
| NFAT | 153719 | 2.8 e-3 | 12A9 | 3.7 e-3 | 1.2 e-3 | ||
| Odd | 143286 | 2.2 e-3 | 24A1 | 0.71 | 1.3 e-3 | ||
| Transcription coactivator activity | GO:0003713 | Nut2 | 148854 | 7.8 e-3 | 72D10 | 3.5 e-3 | 0.38* |
| Vitamin binding | GO:0019842 | CG3091 | 142199 | 5.1 e-3 | 2F2 | 0.42 | 2.4 e-3 |
| Galactose-binding lectin | GO:0005531 | Lectin-galC1 | 143876 | 7.2 e-4 | 37D1 | 0.05 | 2.0 e-3 |
| Mannose-binding lectin | GO:0005532 | CG11211 | 146673 | 3.0 e-3 | 42A14 | 0.08 | 1.1 e-3 |
| UV-sensitive opsin | GO:0015064 | Rh4 | 143321 | 1.3 e-3 | 73D2 | 5.4 e-4 | 2.2 e-3 |
| Rh3 | 151860 | 2.8 e-3 | 92C5 | 1.5 e-3 | 2.7 e-3 | ||
| Ligand binding or carrier | GO:0005488 | Glob1 | 152233 | 6.9 e-4 | 89A8 | 3.8 e-4 | 6.5 e-4 |
| Blue-sensitive opsin | GO:0015059 | Nina E | 143283 | 3.6 e-3 | 92B4 | 1.8 e-3 | 3.7 e-3 |
| Protein serine/threonine kinase | GO:0004674 | Nina C | 152330 | 8.1 e-3 | 27F3 | 3.7 e-3 | 9.4 e-3 |
| CDP-alcohol phosphotransferase | GO:0008414 | CG10355 | 141512 | 3.7 e-3 | 37D3 | 3.7 e-3 | 0.52 |
| Chaperone activity | GO:0003754 | CG7409 | 148360 | 4.3 e-3 | 66A12 | 0.43 | 5.0 e-3 |
| Aldose 1-epimerase activity | GO:0004034 | CG10467 | 148265 | 4.4 e-3 | 65A6 | 3.7 e-3 | 2.5 e-3 |
| Structural constituent of cuticle (sensu Insecta) activity | GO:0005214 | CG8505 | 141213 | 5.4 e-3 | 49A2 | 2.1 e-3 | 0.19 |
| Peptidase activity | GO:0008233 | CG1304 | 145450 | 5.1 e-4 | 19E6 | 3.2 e-4 | 0.78 |
| CG10477 | 148251 | 9.7 e-4 | 65A3 | 0.07 | 3.6 e-4 | ||
| CG11034 | 145847 | 1.0 e-3 | 25A5 | 0.3 | 1.2 e-3 | ||
| CG9897 | 147730 | 1.4 e-3 | 59C1 | 6.5 e-3 | 4.7 e-4 | ||
| Ser12 | 143698 | 3.0 e-3 | 22D1 | 1.4 e-3 | 3.3 e-3 | ||
| BG:BACR44L 22 | 144331 | 4.7 e-3 | 35D3 | 1.6 e-3 | 0.02 | ||
| Nitrophenylphosphatase activity | GO:0003869 | EG:100G10.4 | 144046 | 8.2 e-3 | 3B3 | 4.3 e-3 | 7.7 e-3 |
| Protein serine/threonine kinase activity | GO:0004674 | InaC | 152951 | 5.7 e-4 | 53E2 | 2.8 e-4 | 6.3 e-4 |
| Signal transducer activity | GO:0004871 | Ggamma30A | 146078 | 1.9 e-4 | 30A2 | 6.9 e-5 | 5.9 e-4 |
| Gbeta76C | 153427 | 5.6 e-3 | 76C1 | 0.21 | 5.8 e-3 | ||
| Sr-Cl | 143747 | 8.6 e-3 | 24D6 | 2.9 e-3 | 0.08 | ||
| Or92a | 151722 | 9.6 e-3 | 92E14 | 0.78 | 5.6 e-3 | ||
| Defense/immunity protein activity | GO:0003793 | LysD | 143466 | 1.9 e-4 | 61F3 | 0.06 | 3.8 e-4 |
| LysB | 143464 | 6.4 e-4 | 61F3 | 0.13 | 1.1 e-3 | ||
| LysC | 143465 | 2.4 e-4 | 61F3 | 0.05 | 5.3 e-4 | ||
| LysE | 143467 | 1.7 e-3 | 61F3 | 0.08 | 4.6 e-3 | ||
| LysP | 143468 | 1.8 e-3 | 61F4 | 8.3 e-3 | 0.03 | ||
| Hydrogen-translocating F-type ATPase activity | GO:0016467 | I(2)06225 | 146216 | 1.5 e-3 | 32B3 | 0.82 | 1.1 e-3 |
| Structural molecule activity | GO:0005198 | InaD | 143203 | 2.7 e-3 | 59B3 | 1.1 e-3 | 5.5 e-3 |
| Farnesyltranstransferase activity | GO:0004311 | Qm | 143901 | 8.8 e-3 | 65F4 | 0.7 | 7.6 e-3 |
| Phosphatidylinositol 3-kinase activity | GO:0016303 | Pi3K59F | 151517 | 4.6 e-4 | 59E4 | 1.6 e-3 | 0.02 |
Transcripts differentially expressed among three Drosophila genotypes (P < 0.01). “Molecular function” and “GO code” represent the molecular function category and the gene ontology number according to the Gene Ontology annotation. “Symbol” and “Cytological position” columns are given based on Flybase information, release 2 (http://flybase.bio.indiana.edu). “Cytological position” is the gene location in the Drosophila cytological map. A single asterisk indicates a highly significant association between the probe set and the DDT-resistant phenotype (P < 5.6 e-6). “Probe set” is the Affymetrix name for the probe set on the chip. “P value” shows the statistical P value for the test of null hypothesis λ2 = λ1 = λ0 (i.e., mean expression not different among the three genotypes), as described in Methods. “91-R” and “Wisconsin” indicate the P value for t tests when the mean differences of each DDT-resistant Drosophila genotype is compared to the wild-type Canton-S.
Of the 158 probe sets differentially transcribed, 63 were categorized in this group that were not annotated for molecular function using the Gene Ontology (GO) database and that are not discussed hereafter (Table 2). Ninety-five probe sets were identified as belonging to a particular GO molecular function. A large group of detoxification enzymes were differentially expressed (Table 1). Also, transcripts involved in neuronal function were identified. Four transcription factors were differentially expressed (Cf2, NFAT, Odd, and Nut2). The transcript NFAT (Misexpression Suppressor of Ras 1) was overtranscribed in both DDT-resistant lines and may represent extracellular signals that can modulate DDT resistance.
Differential transcription was found in six probe sets coding for peptidase activity. This finding is in keeping with Saleem et al. (23) and Ahmed et al. (24), who observed that proteases have higher enzymatic activities in DDT-resistant houseflies as compared with susceptible ones. The increased proteolytic activity may serve a role to meet energy demands during stress, thus balancing protein degradation and synthesis. Intracellular proteases may play a role in protein biosynthesis or in modification of the conformation of enzymes as part of this induction process (23, 24). Further investigation of protease expression should help to determine the possible role of proteases in pesticide-resistant insects. The following groups of genes were also overexpressed in the resistant lines: (i) transcripts of nearly all forms of molecule binding were overtranscribed in the resistant lines, including DNA, protein, carbohydrate, vitamin, lipid, drug, and ATP binding; (ii) genes involved in perception of abiotic stimuli and oxidoreductase activity; (iii) genes associated with immune defense and signal transduction; as well as (iv) phosphatases, kinases, and structural molecules.
To examine whether the positions of genes overexpressed in DDT-resistant Drosophila were random or clustered together in a region, we checked the cytological position of each differentially transcribed probe set. With the exception of a moderate representation of transcripts in the right arm of the second chromosome, transcripts appear to be widely distributed across all chromosomes [Tables 1 (“Cytological position”) and 2]. These results are consistent with the findings of Dapkus and Merrel (12), who reported that Rst(2)DDT91-R has multifactorial resistance to DDT and is associated with all three of the major chromosomes in Drosophila.
Transcripts Coding for Detoxification Enzymes. We identified 19 differentially transcribed probe sets representing cytochrome P450s, GSTs, or glucuronosyltransferases (Table 1). Just over half (11) of these 19 probe sets were annotated as cytochrome P450 genes. Some of these P450 genes, such as Cyp6g1 and Cyp12d1 (16–18) and Cyp6a2 and Cyp6a8 (19, 25), were expected to show differential transcription. Other P450 genes are potentially interesting for further study, because they have not yet been shown to be overtranscribed in resistant genotypes. The gene Cyp12d2 has been recently described in the literature as differing by only three nucleotide substitutions from Cyp12d1 (http://P450.antibes.inra.fr); although this gene sequence was deposited neither in National Center for Biotechnology Information (www.ncbi.nlm.nih.gov) nor in FlyBase (http://flybase.bio.indiana.edu). Le Goff et al. (17) were not able to separate overtranscription of either Cyp12d1 or Cyp12d2 using a cDNA spotted array.
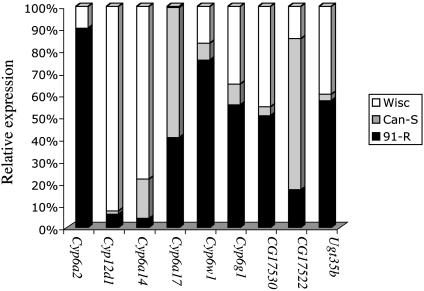
Six detoxification enzyme genes were constitutively over-transcribed in both Rst(2)DDTWisconsin and Rst(2)DDT91-R (Cyp6g1, Cyp12d1, Cyp6a2, Cyp6w1, Ugt35b, and CG17530) (Fig. 1). Cyp6a17 and CG17522 had higher transcript abundance in wild-type flies compared to resistant genotypes. This result suggests potential endogenous functions for these respective gene products. The relative transcript expression of Cyp6a2 in Rst(2)DDT91-R was 255- and 9.1-fold greater than Canton-S and Rst(2)DDTWisconsin. It is important to note that the Rst(2)DDT91-R line is more resistant to DDT than Rst(2)DDTWisconsin and Canton-S (data not shown). The respective LC50s for these three genotypes are 1,304, 89, and 0.71 μg of DDT per vial (with a 24-h bioassay; data not shown). Dunkov et al. (26) reported that Cyp6a2 is able to metabolize xenobiotics, which suggests that Cyp6a2 may play a role in the higher level of DDT resistance observed in Rst(2)DDT91-R as compared to Rst(2)DDTWisconsin. However, we do not rule out the possible occurrence of target site insensitivity in combination with metabolic resistance by Rst(2)DDT91-R. Three cytochrome P450s (Cyp6g1, Cyp12d1, and Cyp6a2) were previously described as being associated with pesticide resistance in laboratory-selected Drosophila (16–19, 25), whereas it has been hypothesized that only Cyp6g1 is overexpressed in DDT-resistant flies collected from the field (16, 17). To the contrary, our report shows that multiple cytochrome P450s are overexpressed and potentially contribute to the DDT resistance phenotype.
Fig. 1.
Relative expression for probe sets derived from detoxification enzyme coding genes among three Drosophila fly lines. Bars show the percentage of each genotype contributing to a total transcriptional level across the probe sets. The wild-type Canton-S is represented by gray (Can-S), and the two DDT resistant genotypes, Rst(2)DDT91-R and Rst(2)DDT Rst(2)DDTWisconsin, are represented by black (91-R) and white (Wisc) columns, respectively. Detoxification enzymes were annotated as cytochrome P450 enzymes (GO: 0015034) (Cyp6a2, Cyp12d1, Cyp6a14, Cyp6a17, Cyp6w1, and Cyp6g1), GSTs (GO, 0004363) (CG17530 and CG17522) and UDP-glucuronosyltransferases (GO, 0015020) (Ugt35b).
GSTs are able to conjugate glutathione to xenobiotics, thus converting the xenobiotics to nonreactive water-soluble conjugates that are easily excreted (27). GSTs have been associated with organochlorine and organophosphorous insecticide resistance both in the malaria mosquito (Anopheles gambiae) and in Drosophila (28–31). We found five GST transcripts overexpressed in the DDT-resistant lines (Fig. 1 and Table 1). Another group of conjugative detoxification enzymes, UDP-glucuronosyltransferases (UGTs), catalyzes the conjugation of glucuronic acid to a wide variety of endobiotics and xenobiotics (32, 33). Okazaki and Katayama (34) have shown that dietary DDT in rats increases enzymatic activity of hepatic 4-nitrophenol-UDP glucuronosyltransferases. We also have identified three UGT transcripts as being differentially transcribed in the resistant genotypes (Fig. 1 and Table 1).
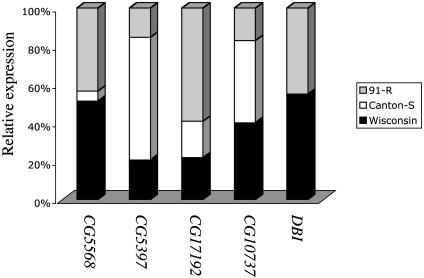
Transcripts Coding for Lipid Metabolism Genes. DDT and its metabolites have been shown to act as estrogen receptor agonists, thereby producing estrogen-like effects (35). We detected differential transcript levels of (i) long-chain fatty acid transporter, (ii) triacylglycerol lipase, (iii) cholesterol O-acyl transferase, and (iv) “diacylglycerol binding” in both resistant genotypes. These findings suggest a possible relationship between DDT resistance and lipid metabolism (Fig. 2). Diazepam-binding inhibitor (DBI) is a polypeptide found in several organisms that has been shown to be involved in benzodiazepine receptor modulation, acyl CoA metabolism, steroidogenesis, insulin secretion, and drug dependence (36, 37). The relative transcript expression of DBI in Rst(2)DDT91-R and Rst(2)DDTWisconsin was 108- and 89-fold greater than Canton-S, suggesting a possible association between DDT resistance and cholesterol metabolism (Fig. 2).
Fig. 2.
Relative expression for probe sets derived from lipid metabolism coding genes among three Drosophila fly lines. Bars show the percentage of each genotype contributing to a total transcriptional level across the probe sets. The wild-type Canton-S is represented by white (Canton-S), and the two DDT-resistant genotypes, Rst(2)DDT91-R and Rst(2)DDTWisconsin, are represented by gray (91-R) and black (Wisconsin) columns, respectively. Transcripts associated with lipid metabolism were annotated as long chain fatty acid transporter, CG5568 (GO, 0005324); cholesterol O-acyltransferase, CG5397 (GO, 0017066); triacylglycerol lipase, CG17192 (GO, 0004806); diacylglycerol binding, CG10737 (GO, 0019992); and lipid binding, DBI (GO, 0008289).
Resistance appears more complex than the overtranscription of Cyp6g1, as suggested by Daborn and colleagues (16, 17). Also, RNA expression profiles in Rst(2)DDT91-R and Rst(2)DDTWisconsin Drosophila genotypes identified several common metabolic pathways components associated with DDT resistance. The comparison of microarray data among different research groups is difficult. Differences in experimental design of microarrays, techniques used (cDNA spotted array vs. high-density oligoarrays), and statistical analysis make such comparisons challenging. Sexual dimorphism plays a pivotal role in Drosophila gene expression. Studies of the transcriptome of sexually mature males and females have shown clear sex-dependent gene regulation (38–41). Finally, aging is associated with changes in the expression of many genes. Whole genome transcript profiles showed that nearly 23% of the expressed genome changed with age (42). Therefore, discrepancies between our findings and those of Daborn and colleagues (16, 17) may be due to differences in statistical methodologies, array technology used, age, or gender.
Nonetheless, the use of genome-wide microarray technology allows an efficient and quantitative evaluation of transcripts in insecticide-resistant genotypes and has the potential to suggest interesting genes for further study. To confirm the role of any genes described above from DDT-resistant Drosophila, validation studies such as quantitative PCR, Northern blots, RNA interference, P element transformation, and messenger RNA and protein integration are essential. Our group (18) and others (19, 43) have validated, by using Northern blot, that Cyp6g1, Cyp12d1 (Cyp12d2), Cyp6a2, and Cyp6a8 are differentially transcribed in DDT-resistant Drosophila genotypes. Pesticide metabolism studies in Escherichia coli and yeast using recombinant detoxification enzymes should shed more light on DDT metabolism by resistant Drosophila. This study has also allowed for the identification of several other gene transcripts with undefined relationships to detoxification processes. Functional characterizations of these mechanisms will almost certainly yield highly informative findings not previously considered in pesticide resistance research.
Supplementary Material
Acknowledgments
We are grateful to the Purdue Genomics Center MicroArray Core Facility for excellent technical assistance, particularly Dr. Philip San Miguel, Doug Murphy, Rick Westerman, and Fred Rakhshan. We are indebted to Lisa Bono for assisting with data management and analyses. We also thank Drs. Rebecca Doerge, James Fleet, Larry Murdock, and Greg Hunt for excellent comments. This work was supported by grants from the National Institutes of Health (NIH 1R01 AI51513-01, to B.R.P.), the U.S. Agriculture Department (USDA-IFAFS N00149419318, to L.M.M.), the Purdue Research Foundation (to B.R.P.), and the Department of Entomology, Purdue University (to B.R.P.). The Purdue Research Foundation supported J.H.F.P. This is publication no. 17370 of the Purdue University Agricultural Experimental Station (West Lafayette, IN).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviation: DDT, dichlorodiphenyltrichloroethane.
References
- 1.Roush, R. T. & Mackenzie, J. A. (1987) Annu. Rev. Entomol. 32, 361-380. [DOI] [PubMed] [Google Scholar]
- 2.Denholm, I. & Rowland, M. W. (1992) Annu. Rev. Entomol. 37, 91-112. [DOI] [PubMed] [Google Scholar]
- 3.Heckel, D. G., Gahan, L. J., Liu, Y. B. & Tabashnik, B. E. (1999) Proc. Natl. Acad. Sci. USA 96, 8373-8377.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oakeshott, J. G., Home, I., Sutherland, T. D. & Russel, R. J. (2003) Genome Biol. 4, 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McKenzie, J. A. (1996) Ecological and Evolutionary Aspects of Insecticide Resistance (Academic/Landes, Austin, TX).
- 6.McKenzie, J. A. (2000) Bull. Entomol. Res. 90, 3-7. [DOI] [PubMed] [Google Scholar]
- 7.McKenzie, J. A. & Batterdam, P. (1994) Trends Ecol. Evol. 9, 166-169. [DOI] [PubMed] [Google Scholar]
- 8.Crow, J. F. (1954) J. Econ. Entomol. 47, 393-398. [Google Scholar]
- 9.King, J. C. & Somme, L. (1958) Genetics 43, 577-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ogita, Z. (1960) Botyu-Kagagu 26, 7-18. [Google Scholar]
- 11.Ogita, Z. (1961) Botyu-Kagagu 26, 88-93. [Google Scholar]
- 12.Dapkus, D. C. & Merrel, D. J. (1977) Genetics 87, 685-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shepanski, M. M. C., Glover, T. J. & Kuhr, R. J. (1977) J. Econ. Entomol. 70, 539-543. [DOI] [PubMed] [Google Scholar]
- 14.Dapkus, D. (1992) J. Econ. Entomol. 85, 340-377. [DOI] [PubMed] [Google Scholar]
- 15.Daborn, P., Boundy, S., Yen, J., Pittendrigh, B.& Ffrench-Constant, R. (2001) Mol. Genet. Genom. 266, 556-563. [DOI] [PubMed] [Google Scholar]
- 16.Daborn, P., Yen, J. L., Bogwitz, M., Le Goff, G., Feil, E., Jeffers, S., Tijet., N., Perry, T., Heckel., D., Batterham., P., et al. (2002) Science 297, 2253-2256. [DOI] [PubMed] [Google Scholar]
- 17.Le Goff, G., Boundy, S., Daborn, P. J., Yean, J. L., Sofer, L., Lind, R., Sabourault, C., Madi-Ravazzi, L. & Ffrench-Constant, R. H. (2003) Insect Biochem. Mol. Biol. 33, 701-708. [DOI] [PubMed] [Google Scholar]
- 18.Brandt, A., Scharf, M., Pedra, J. H. F., Holmes, G., Dean, A., Kreitman, M. & Pittendrigh, B. R. (2002) Insect Mol. Biol. 11, 337-341. [DOI] [PubMed] [Google Scholar]
- 19.Maitra, S., Dombrowiski, S. M., Waters, L. C. & Ganguly, R. (1996) Gene 180, 165-171. [DOI] [PubMed] [Google Scholar]
- 20.Kerr, M. K. & Churchill, G. A. (2001) Proc. Natl. Acad. Sci. USA 98, 8961-8965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wolfinger, R. D., Gibson, G., Wolfinger, E. D., Bennet, L., Hamadeh, H., Bushel, P., Afshari, C. & Paules, R. S. (2001) J. Comput. Biol. 8, 625-637. [DOI] [PubMed] [Google Scholar]
- 22.Wayne, M. L. & McIntyre, L. M. (2002) Proc. Natl. Acad. Sci. USA 99, 14903-14906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saleem, A. M., Shakoori, A. R., Wilkins, R. M. & Mantle, D. (1994) Pakistan J. Zool. 26, 327-333. [Google Scholar]
- 24.Ahmed, S., Wilkins, R. M. & Mantle, D. (1998) Insect Biochem. Mol. Biol. 28, 629-639. [DOI] [PubMed] [Google Scholar]
- 25.Maitra, S., Dombrowiski, S. M., Basu, M., Raustol, O., Waters, L. C. & Ganguly, R. (2000) Gene 248, 147-156. [DOI] [PubMed] [Google Scholar]
- 26.Dunkov, B. C., Mocelin, G., Shotkoski, F., Ffrench-Constant, R. H. & Feyereisen, R. (1997) DNA Cell Biol. 16, 1345-1356. [DOI] [PubMed] [Google Scholar]
- 27.Hayes, J. D. & Pulford, D. J. (1995) Crit. Rev. Biochem. Mol. Biol. 30, 445-600. [DOI] [PubMed] [Google Scholar]
- 28.Wang, J. Y., McCommas, S. & Syvanen, M. (1991) Mol. Gen. Genet. 227, 260-266. [DOI] [PubMed] [Google Scholar]
- 29.Ranson, H., Prapanthadara, L. A. & Hemingway, J. (1997) Biochem. J. 324, 97-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ranson, H., Jensen, B., Wang, X., Prapanthadara, L., Hemingway, J. & Collins, F. H. (2000) Insect Mol. Biol. 9, 499-507. [DOI] [PubMed] [Google Scholar]
- 31.Ranson, H., Rossiter, L., Ortelli, F., Jensen, B., Wang, X., Roth, C. W., Collins, F. H. & Hemingway, J. (2001) Biochem. J. 359, 295-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McGurk, K. A., Brierley, C. H. & Burchell, B. (1998) Biochem. Pharmacol. 55, 1005-1012. [DOI] [PubMed] [Google Scholar]
- 33.Naydenova, Z., Krauss, G.-J, Golovinsky, E. & Grancharov, K. (1999) Pesticide Sci. 55, 825-830 [Google Scholar]
- 34.Okazaki, Y. & Katayama, T. (2003) J. Nutr. Biochem. 14, 81-89. [DOI] [PubMed] [Google Scholar]
- 35.Diel, P., Schulz, T., Smolnikar, K., Strunck, E., Vollmer, G. & Michna, H. (2000) J. Steroid Biochem. Mol. Biol. 73, 1-10. [DOI] [PubMed] [Google Scholar]
- 36.Kolmer, M., Roos, C., Tirronen, M., Myohanen, S. & Alho, H. (1994) Mol. Cell. Biol. 14, 6983-6995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Papadopolous, V. & Brown, A. S. (1995) J. Steroid Biochem. Mol. Biol. 53, 103-110. [DOI] [PubMed] [Google Scholar]
- 38.Jin, W., Riley, R. M., Wolfinger, R. D., White, K. P., Passador-Gurgel, G. & Gobson, G. (2001) Nat. Genet. 29, 389-395. [DOI] [PubMed] [Google Scholar]
- 39.Arbeitmann, M. N., Furlong, E. E. M., Iman, F., Johnson, E., Null, B. H., Baker, B. S., Krasnow, M. A., Scott, M. P., Davis, R. W. & White, K. P. (2002) Science 297, 2270-2275. [DOI] [PubMed] [Google Scholar]
- 40.Ranz, J. M., Castillo-Davis, C. I., Meiklejohn, C. D. & Hartl, D. L. (2003) Science 300, 1742-1745. [DOI] [PubMed] [Google Scholar]
- 41.Parisi, M., Nuttal, R., Naiman, D., Boufard, G., Malley, J., Andrews, J., Eastman, S. & Oliver, B. (2003) Science 299, 697-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pletcher, S. D., MacDonald, S. J., Marguerie, R., Certa, U., Sterns, S. C., Goldstein, D. B. & Partridge, L. (2002) Curr. Biol. 12, 712-723. [DOI] [PubMed] [Google Scholar]
- 43.Waters, L. C., Zelhof, A. C., Shaw, B. J. & Cha'ng, L.-Y. (1992) Proc. Natl. Acad. Sci. USA 89, 4855-4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.