Abstract
It is hypothesized that the use of targeted drug delivery systems can significantly improve the therapeutic index of small molecule chemotherapies by enhancing accumulation of the drugs at the site of disease. Phage display offers a high-throughput approach for selection of the targeting ligands. We have successfully isolated phage fusion proteins selective and specific for PANC-1 pancreatic cancer cells. Doxorubicin liposomes (Lipodox) modified with tumor-specific phage fusion proteins enhanced doxorubicin uptake specifically in PANC-1 cells as compared with unmodified Lipodox and also compared with normal breast epithelial cells. Phage protein-targeted Lipodox substantially increased the concentration of doxorubicin in the nuclei of PANC-1 cells in spite of P-glycoprotein-mediated drug efflux. The in vitro cytotoxic activity obtained with pancreatic cell-targeted Lipodox was greater than that of unmodified Lipodox. We present a novel and straightforward method for preparing pancreatic tumor-targeted nanomedicines by anchoring pancreatic cancer-specific phage proteins within the liposome bilayer.
Keywords: drug delivery, landscape phage, liposome, major coat protein pVIII, pancreatic cancer, phage display
Introduction
Adenocarcinoma of the exocrine pancreas is the fourth leading cause of cancer deaths in the USA (Jemal et al., 2002). The prognosis of this disease is very poor with a median patient survival for all stages of pancreatic cancer rarely exceeding 3–5 months after initial diagnosis (Molinari et al., 2001). A poor prognosis and low survival rate of patients diagnosed with pancreatic cancer can be attributed to asymptomatic patients at early stages leading to late stage detection and a highly aggressive phenotype with significant metastatic potential often resulting in resistance to conventional chemotherapy. Of patients with resectable disease, only 9–15% are suitable candidates for surgery (Molinari et al., 2001).
Despite a minimal impact on mean survival, chemotherapy remains the best treatment option for patients with metastatic pancreatic cancer (El Kamar et al., 2003). However, conventional chemotherapy demonstrates limited antitumor activity due to multiple factors including: dose-limiting toxicity of antitumor drugs, poor efficacy on accumulating in tumor tissues and intrinsic resistance of cancer cells to common chemotherapy drugs. Effective delivery is a key issue in pancreatic cancer treatment to attain maximum efficiency of chemotherapy. Encapsulation of anticancer drugs into PEGylated liposomes has shown beneficial effects for various types of cancers by minimizing potential side effects while increasing the concentration of encapsulated drug to the tumor site due to the enhanced permeability and retention (EPR) effect (Maeda, 2001). PEGylation of liposomes has minimized non-specific uptake by the reticuloendothelial system and has significantly prolonged their circulation time (Vlerken et al., 2007); however, the therapeutic efficiency of liposomal drugs is suboptimal to exert antitumor activity. It is believed that decoration of therapeutic, long-circulating (PEGylated) liposomes with tumor-targeting moieties, such as antibodies, small peptides or other molecules that specifically bind cancer cell receptors can increase the therapeutic efficiency of drug-loaded liposomes, as they enhance accumulation of the drug at the tumor site (Sofou, 2007; Alexis et al., 2008; Noble et al., 2004; Krumpe and Mori 2006). This concept of targeted drug delivery, though realized in many in vitro and in vivo experiments, is still not applicable in clinical trials and practice. One of the main hurdles towards targeted nanomedicines in clinical applications is the cost ineffectiveness of conjugating nanoparticles with appropriate targeting monoclonal antibodies, antibody fragments or peptides. There now exists an urgent need to develop a simple, cost-effective technology that relies on self-assembly to produce stable, physiologically active targeted nanomedicines.
The integration of phage display technology with nanocarrier-based drug delivery platforms is emerging as a new approach for targeting nanomedicines (Petrenko and Jayanna, 2014). This phage technique evolved as a result of advances in combinatorial chemistry and phage display has allowed identification of tumor-specific peptides in a high-throughput fashion (Mori, 2004; Sergeeva et al., 2006; Aina et al., 2007). Tumor-specific phage can be affinity selected from multibillion-clone libraries (Brigati et al., 2008; Petrenko, 2008) by their ability to interact specifically with cancer cell surface receptors. Using well-established biopanning protocols, a number of phage-borne peptides specific to a broad array of tumors have been identified (Oyama et al., 2003; Krumpe and Mori, 2006; Gray and Brown, 2014).
Recently, we proposed using landscape phage fusion coat proteins—easy to produce ‘substitute antibodies’—as targeting ligands for drug-loaded pharmaceutical nanocarriers, like liposomes (Jayanna et al., 2009, 2014), to overcome the drawbacks associated with chemical modification of nanocarrierswith cancer-selective peptides. This approach is based on the ability of the phage major coat protein to spontaneously insert into bacterial membranes (Broome-Smith et al., 1994; Kuhn, 1995) and lipid bilayers of liposomes (Soekarjo et al., 1996). The ability of the major coat protein pVIII to incorporate into micelles and liposomes emerges from its intrinsic function as a membrane protein, as judged by its biological, chemical and structural properties (Lee et al., 2003). Spontaneous insertion of the gene 9 minor coat protein of bacteriophage M13 in liposomes has also been studied (Houbiers et al., 2001), concluding that protein is able to insert spontaneously into membranes without the need of any machinery or transmembrane gradient. We have shown that the hybrid phage pVIII coat protein fused to the tumor-specific peptides (fpVIII) spontaneously incorporates into the liposome membrane via its C-terminal hydrophobic segment, while its water-exposed N-terminal binding segment is exposed on the surface of drug-loaded liposomes to serve as a targeting moiety. Liposome-incorporated pVIII demonstrates identical binding specificity as a parental phage, indicating that functional activity of the selected phage belongs to their foreign fusion peptides and remains in a functionally active conformation.
In contrast to sophisticated and poorly controllable conjugation procedures used for coupling of synthetic peptides to drug nanovesicles, the phage-based approach relies on precise mechanisms of selection, biosynthesis and self-assembly. When landscape phage serve as a reservoir of the targeted membrane proteins, one of the most troublesome steps of the conjugation technology is bypassed. Furthermore, it does not require idiosyncratic reactions specific for each new shell-decorating polymer or targeting ligand and may be easily adapted to a new nanoparticulate composition and a new addressed target. No reengineering of the selected phage is required at all: the phage themselves serve as the source of the final product—coat protein genetically fused to the targeting peptide. The major coat protein constitutes 98% of the total protein mass of the virion—a level of purity hardly attainable in normal synthetic or bioengineering procedures. A culture of phage-secreting cells is an efficient, convenient and discontinuous protein production system. The yield of phage particles reaches 20 mg/l for the engineered landscape phage and they are secreted from the cell nearly free of intracellular components. Purification of the phage is easily accomplished by simple, routine steps that do not differ from one clone to another. As normal intestinal parasites, phage and their components are not toxic and have already been tested for safety in pre-clinical and clinical trials.
Previously, phage technology has been successfully used to enhance delivery and therapeutic effect of drug-loaded liposomes, targeted to breast and prostate cancer cells (Jayanna et al., 2010; Wang et al., 2010, 2014). In this study, we identified pancreatic cancer-specific phage proteins to prepare drug-loaded liposomes specifically interacting with pancreatic cancer cells. A panel of the pancreatic cell-binding phages was selected from landscape library f8/8 (Petrenko et al., 1996) using an advanced selection procedure in vitro and pancreatic cancer-specific fusion proteins were isolated by size-exclusion chromatography. Doxorubicin-loaded PEGylated liposomes (Lipodox) modified with phage fusion proteins specific towards PANC-1 pancreatic cancer cells demonstrated strong, specific binding with target cells and increased cytotoxicity in vitro.
Materials and methods
Cells
Cell lines were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA). The target pancreatic adenocarcinoma cells PANC-1 (ATCC, CRL-1469™) were derived from the ductal region of a pancreatic epithelioid carcinoma. Breast adenocarcinoma cells, MCF-7 (ATCC, HTB-22™), were derived from a metastatic pleural effusion of breast adenocarcinoma. Phenotypically normal pancreatic cells, hTERT-HPNE (ATCC, CRL-4023™), derived from cells isolated from the ductal region of the pancreas were used for depletion of the phage library. Phenotypically normal breast epithelial cells, MCF-10A (ATCC, CRL-10317™), were derived from cells of a breast mammary gland. All cells were grown and incubated at 37°C with 5% CO2 as recommended by ATCC. Lipodox (2 mg/ml PEGylated liposomal doxorubicin HCl, containing 1,2-distearoyl-sn-glycero-3-phosphocholine, cholesterol, 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[amino(polyethylene glycol)-2000] (ammonium salt) (DSPE-PEG2000) in molar ratio 56 : 39 : 5 was purchased from Sun Pharma (Mumbai, India).
Landscape phage display library
Multibillion-clone landscape phage display library f8/8 was constructed using the type 8 phage display vector f8-1 (Petrenko et al., 1996; Petrenko and Smith, 2000). In this system, random oligonucleotides were inserted in-frame in the gpVIII, which codes for the major coat protein pVIII resulting in the display of 4000 guest peptides units on the surface of each phage particle. Common phage methods including phage isolation, propagation, purification, titering and sequencing of phage DNA were described in previous protocols (Brigati et al., 2008). Bacterial endotoxins were removed from liter-scale propagations of identified phage clones by three rounds of treatment with 1% Triton X-114 at 4°C for 5 min followed by phase separation of the aqueous and organic phases at 37°C (Aida and Pabst, 1990). Remaining concentrations of endotoxins were quantified by a chromogenic Limulus Amebocyte Lysate endotoxin assay (ToxinSensor, GenScript). Identified phage clones are designated by the structure of their foreign fusion peptides.
Selection of pancreatic cancer cell PANC-1 specific phages
Biased protocol for selection of landscape phages was employed as described (Fagbohun et al., 2012) with some modifications. The phage library (f8/8) was depleted against an empty cell culture flask, a serum-treated flask and normal pancreatic cells (hTERT-HPNE). Unbound phage particles recovered from library depletion were incubated with confluent PANC-1 cells in complete media containing 10% fetal bovine serum (FBS) at room temperature (RT) for 1 h. Based on our previous experiments, we expected to collect in this condition all cell-associated phages, both penetrating and non-penetrating. Unbound phages were removed by washing and cell-associated phages were eluted with elution buffer (200 mM glycine-HCl, pH 2.2, 1 mg/ml bovine serum albumin (BSA), 0.1 mg/ml phenol red) for 10 min on ice. The eluate fraction was neutralized with 376 µl of 1 M Tris-HCl, pH 9.1. Internalized phage was recovered with lysis buffer (2% deoxycholate, 10 mM Tris-HCl, pH 8.0, 0.2 mM ethylenediaminetetraacetic acid (EDTA)) after two additional wash steps. The eluate and two post-elution wash (PEW) fractions were concentrated to a final volume of 80 µl with a 100 kDa Amicon centrifugal unit at 3000 g (Allegra 21R S4180, Beckman Coulter). Phage input and output solutions were titered in bacteria as described previously (Brigati et al., 2008). The results were expressed as a percentage of the ratio of output to input phage. The concentrated elute phage and cell-internalized phage were amplified separately in Escherichia coli K91BluKan bacteria and used in subsequent rounds of selection. Additional rounds of selection were performed similarly to the first round, without the depletion steps. In the subsequent rounds, phage was incubated with PANC-1 cells at 37°C instead of RT to enrich for phage with cell-penetrating properties. Segments of phage gpVIII were amplified by polymerase chain reaction (PCR) and individual phage DNA sequences were identified.
Specificity and selectivity of phage towards PANC-1 pancreatic cancer cells
Individual phage clones were characterized for their selectivity towards target pancreatic cancer cells, PANC-1, in comparison with control cells, hTERT-HPNE (non-neoplastic pancreatic epithelia), MCF-7 (breast adenocarcinoma) and serum in a phage capture assay (Brigati et al., 2008). Briefly, target cells PANC-1, control normal cells hTERT-HPNE and control breast cancer cells MCF-7 were cultivated in triplicate to confluence in separate wells of 96-well cell culture plates. Medium containing 10% FBS was incubated in separate wells in triplicate as a control for serum binding phage. Cells were incubated with phage (1 × 106 cfu) at 37°C for 1 h. Unbound phages were carefully removed and cells were washed with 100 µl washing buffer eight times for 5 min intervals to remove non-specifically interacting phages. Cells were lysed with 25 µl of lysis buffer (2.5% CHAPS) for 10 min with gentle shaking on a rocker. The lysate containing cell-interacting phages was titered in E.coli K91BlueKan starved cells. Phage recovery was calculated as a ratio of output to input phage. An unrelated phage with a non-relevant guest peptide VPEGAFSSD was used as a negative control.
Fusion phage protein-modified Lipodox
A landscape phage bearing pancreatic cancer cell-specific peptide EPSQSWSM was selected from the 8-mer landscape library f8/8 (Petrenko et al., 1996) using biopanning against PANC-1 cells at 37°C, as described above. Phage fusion 55-mer coat proteins AEPSQSWSMDPAKAAFDSLQASATEYIGYAWAMVVVIVGATIGIKLFKKFTSKAS and AETPPSWGGDPAKAAFDSLQASATEYIGYAWAMVVVIVGATIGIKLFKKFTSKAS were isolated by solubilizing phage protein in cholate buffer, as described (Jayanna et al., 2009). Briefly, 6.5 × 1013 virions (∼2.5 mg of pVIII protein) are solubilized to in 120 mM cholate/10 mM Tris-HCl, pH 8.0/0.2 mM EDTA in the presence of 2.5% chloroform before incubation overnight at 37°C. Solubilized phage protein is recovered by size-exclusion chromatography with average recoveries of >50%. Phage protein-modified Lipodox was prepared by incubating Lipodox with the cholate-stabilized fusion phage coat protein fpVIII at a lipid-to-protein weight ratio of 200 : 1. The presence and orientation of the fusion coat protein in phage protein-modified Lipodox was confirmed by western blotting as described previously (Jayanna et al., 2009; Fagbohun et al., 2012). The size distribution and zeta potential of the nanoparticle were measured by dynamic light scattering in distilled water obtained with a Malvern ZetaSizer ZS90 (Malvern, Suwanee, GA).
Confocal immunofluorescence study of subcellular localization of selected phages
PANC-1 cells were seeded in four-well chamber overnight at a density of 2 × 105 cells/ml. The following day, cells were fed with fresh medium. Cell Light reagents (Molecular Probes) for labeling endosome were added in each well and incubated overnight at 37°C. The third day, cells were fed with new fresh medium. Phage (∼109 virions) were diluted in fresh medium, added to cells, and incubated at 37°C for 1h. After removing the unbound phages, cells were washed three times with 1X phosphate-buffered saline (PBS), pH 7.4/0.1% Tween 20 washing buffer and then fixed with 4% formaldehyde for 15 min at 37°C. Cell plasma membrane staining dye (wheat germ agglutinin conjugated with Alexa Fluor 555) was incubated with cells for 10 min at RT. When cell membrane labeling was complete, the labeling solution was removed and the cells were washed twice in 1X HBSS. Cells were permeabilized with 0.2% Triton X-100 at RT for 10 min. Reagent was removed and cells were washed three times with 1X TBS, pH 7.6. Before incubation with antiphage antibodies, cells were treated with blocking buffer for 30 min at RT. Cells were incubated with affinity-purified rabbit anti-fd IgG described previously (Smith et al., 1998) for 1 h, followed by washing and incubation with goat anti-rabbit IgG conjugated with Alexa Flour 488 (Molecular Probes) at a dilution of 1 : 500 in 1X PBS, pH 7.4/1% BSA for 45 min at RT. Cells were washed three times after secondary antibody treatment. TOTO-3 was used for nucleus staining according to the manufacturer's instructions. Cells were covered with cover slides with Prolong Gold Antifade Reagent and sealed with nail polish around the edges of the slide. Digital micrographs were taken using a Nikon Eclipse TE2000-E confocal microscope and analyzed using NIS-Elements software (Nikon).
Uptake of doxorubicin by cells treated with phage protein-modified Lipodox
Uptake of Lipodox and targeted Lipodox by PANC-1 cells was visualized using a Nikon Eclipse TE2000-E confocal microscopy system. PANC-1 cells were seeded in four-well chamber overnight at a density of 2 × 105 cells per well and were grown at 37°C in a 5% CO2 incubator for 24 h. Cells were incubated with 35 µg/ml of doxorubicin contained within unmodified Lipodox or targeted Lipodox in complete media for 4 h. Cells were then washed with 1X PBS, pH 7.4 three times followed by incubation with 5 µg/ml TOTO-3 nuclear stain for 10 min. After washing, a coverslip was placed on the slide over a drop of Prolong Gold Antifade mounting reagent.
Measurement of cell-associated doxorubicin
After cells were counted and pelleted by centrifugation, doxorubicin was extracted by treatment with 0.075 N HCl in 90% isopropyl alcohol at 4°C overnight as described (Goren et al., 2000). Following centrifugation, the supernatant was collected and used for fluorescent doxorubicin determination at an excitation of 470 nm/emission of 590 nm using a Synergy H1 plate reader (Biotek, Vermont, USA). Extracts from untreated cells were used as a blank.
Cellular and nuclear doxorubicin quantitation
PANC-1 cells were exposed to free doxorubicin, Lipodox or phage protein-modified Lipodox for 4 h. Cells were then released by trypsinization as described above, and suspended at a concentration of 5 × 106 cells/ml for 10 min at 4°C in 10 mM Tris-HCl, pH 7.4/100 mM NaCl/1 mM EDTA/1% Triton X-100. The suspension was then centrifuged at 800 g for 15 min and the resulting cell nuclei pellet was separated from the cytosol components found in the supernatant (Goren et al., 2000). Doxorubicin extraction from both nuclear and cytosolic fractions was followed as described previously.
Verapamil blockade of drug efflux
PANC-1 cells grown in monolayer conditions were exposed to 35 µg/ml doxorubicin as free drug, Lipodox or modified Lipodox for 4 h, in the presence or absence of 10 µM verapamil (Sigma). Cells were then washed with 1X PBS, pH 7.4 to remove non-associated drug and then further incubated with fresh medium in the presence or absence of verapamil for 2 h. Cells were released from tissue culture dishes with 0.05% trypsin/0.02% EDTA (Life Technologies, Inc.) and pelleted by centrifugation at 500 g for 7 min. Cell pellets were then washed with 1X PBS, pH 7.4 and centrifuged. Cell pellets were then suspended in fresh culture medium, counted and analyzed for intracellular doxorubicin accumulation.
Cytotoxicity
Modified liposomes
Target PANC-1 cells or non-target MCF-10A cells were seeded into a 96-well microplate at a density of 6 × 104 cells per well. After growth to 90% confluence, cells were treated with varying concentrations of Lipodox, PANC-1-specific Lipodox (L1-Lipodox and P38-Lipodox), irrelevant streptavidin-binding Lipodox (7b1-Lipodox) and doxorubicin in complete Dulbecco's Modified Eagle's medium for 24 h. After 24 h, the medium was gently removed, cells were washed once with 1X PBS, pH 7.4 (the washing step can be omitted to avoid removal of weakly attached cells) and incubated with phenol red-free minimum essential medium (MEM) containing 0.45 mg/ml 3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyltetrazolium Bromide (MTT) reagent for 4 h at 37°C. After 4 h of incubation with MTT, 85 µl was removed from each of the wells and replaced with 50 µl of dimethyl sulfoxide. Solutions were mixed and incubated for 10 min at 37°C to solubilize the formazan byproduct from cell membranes. The absorbance of each well was measured at 540 nm using a Synergy H1 plate reader (BioTek, Vermont). Blank wells containing only culture medium and MTT were subtracted from each sample. Percent viability was expressed as a ratio between the absorbance of treated cells at various concentrations by the average absorbance for a set of untreated cells.
Phage preparations
Ten representative phage clones containing a range of functionally diverse fusion protein sequences was identified for cytotoxicity screening in MCF-7 cells. MCF-7 cells were seeded at an initial density of 5 × 105 cells per well in a 96-well cell culture treated array plate and incubated for 24 h at 37°C. Identified phage clones were diluted in MEM containing 10% FBS and added to the cells for 24 h of treatment. After 24 h of phage incubation, the medium was replaced with fresh MEM for an additional 48 h at 37°C in a 5% CO2 incubator. Medium was removed and incubated with phenol red-free MEM containing 0.45 mg/ml MTT for 4 h at 37°C. After 4 h, 100 μl of 10% sodium dodecyl sulfate in 0.01 N HCl was added to each of the wells and incubated at 37°C for an additional 4 h. The absorbance of each well was measured at 570 nm using a Synergy H1 plate reader (BioTek, Vermont). Calculations were performed as described above.
Statistical analysis
Data from all experiments are expressed as mean ± standard deviation (SD). Differences were determined using Student's independent t-test (P < 0.05).
Results
Selection of phage ligands against PANC-1 cells
One of the primary hypotheses of this project was to determine if targeting nanomedicines with selected phage proteins (i) enhances their cytotoxicity towards pancreatic cancer cells and (ii) enhances their ability to overcome acquired resistance to doxorubicin by delivering a suitable dose to the nucleus. We chose PANC-1 cells as our target as these cells demonstrate a classical multiple drug resistance phenotype often observed after treatment with chemotherapy by virtue of an increased P-glycoprotein (Pgp) efflux pump expression (O'Driscoll et al., 2007). The landscape phage library f8/8 was used to find phage clones that bind with high specificity and selectivity to PANC-1 cells. Extensive depletion of the phage library against plastic, serum and normal pancreatic epithelial cells before enrichment of phage that interact with PANC-1 cancer cells was employed for a robust selection of phage clones specific for cancer cells. To select all cell-associated phages, the first round of biopanning was run at RT. To favor the selection of cell-penetrating phage particles, the second and following rounds of biopanning were run at 37°C. Phage particles associated with cells were eluted sequentially with acid and detergents. The ratio of output to input phage increased from one round to another indicating successful enrichment for phage clones that bind to the target PANC-1 cells (not shown). After the fourth round of selection, 60 phage clones were randomly picked after titering of the eluate, PEW and lysate fractions. Their foreign DNA segments were amplified by PCR, sequenced and translated to reveal the structures of the pVIII fusion peptides. In total, 60 phage clones were isolated and classified into five major families based on their consensus linear peptide motifs (Table I).
Table I.
Isolated pancreatic cancer cell-interacting phages are classified into families based on their consensus motifs (shown in bold text), whereas phage clones with no identified consensus motif were regarded as an orphan family of clones.
| Families of selected peptides | ||
|---|---|---|
| E/A– – P– W– G | YL | |
| ADAPAWSG | – – – – –EYL | AP2–4 |
| ADSPTWTG | AGSGQEYL | APPPPST |
| ADTPGWSG | APSNGEYL | APPPPTTA |
| ASSPAWSG | APPPMPSS | |
| EDNPQWSG | – – – – –MYL | APPSNSNT |
| EGGPSWSG | EQSSQMYL | |
| ESNPSWSG | DPSSGMYL | – – – – EPGQ |
| ETAPQWTG | GPDTAMYL | VIMEPGQ |
| VTLTEPGQ | ||
| D/E– – – –W–G | E/DYL | GGPWEPGQ |
| EAMSNWSG | DYL | |
| EPSNTWSG | GDYL | Others |
| E… QTWSG | EYL | ATNM |
| EPTQSWTG | ATSAPELF | |
| DPQTGWSG | – – – – –LYL | DASEYASH |
| DSPQTWAG | ESNNGLYL | DKMSSEHA |
| ETPPSWGG | GSSEQLYL | DLSRDSTM |
| ELPPSWGG | DPDGRLML | |
| EPPAQWQG | – – – – – –YL | DPPMYAAQ |
| GTSNANYL | DQMRLEYP | |
| E-SQSWSM | AAGDTAYL | DSAVTQSE |
| EASQSWSM | ETYNQPYL | DSNAPHSM |
| EPSQSWSM | GQSDTSYL | DTIQSTEA |
| DGQYLGSQ | DTSSQEFL | |
| DTNSAWST | DVAIFLK | |
| VDTTGWSS | – – RGD– – – | |
| DVRGDGLQ | ||
Selectivity of phage towards PANC-1 cancer cells
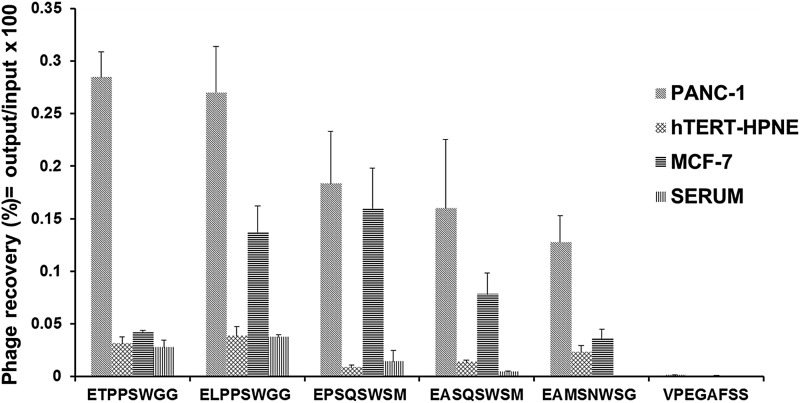
Phage clones obtained by screening of the f8/8 landscape library against pancreatic cancer cells were tested for selectivity in a phage capture assay. Some phages demonstrated high selectivity towards PANC-1 cells only, while some phage showed selectivity for both pancreatic and breast cancer cells. Phages were considered selective if their relative binding to PANC-1 and MCF-7 were at least five times higher than those of hTERT-HPNE and serum. Using these criteria, 30 phage clones were found to be selective for both pancreatic and breast cancer cells. Selectivity of five prominent phage clones is illustrated in Fig. 1. Phage displaying the fusion protein EPSQSWSM and ETPSWGG were isolated from the third round of selection from the lysate and PEW fractions, respectively. Phage clones were propagated in liter scale, endotoxin purified, and phage proteins were isolated from these phages using cholate solubilization followed by size-exclusion chromatography and chosen as a source of protein ligands for construction of targeted nanomedicines.
Fig. 1.
Selectivity of phages towards target cells PANC-1 in comparison with other cells (non-neoplastic pancreatic epithelial cells hTERT-HPNE, breast cancer cells, MCF-7) and serum. Selectivity of the phages was estimated as their recovery (%) = output (cell-associated) phage/input phage. Unrelated phage bearing the peptide VPEGAFSS was used as a control.
Cytotoxicity of phage
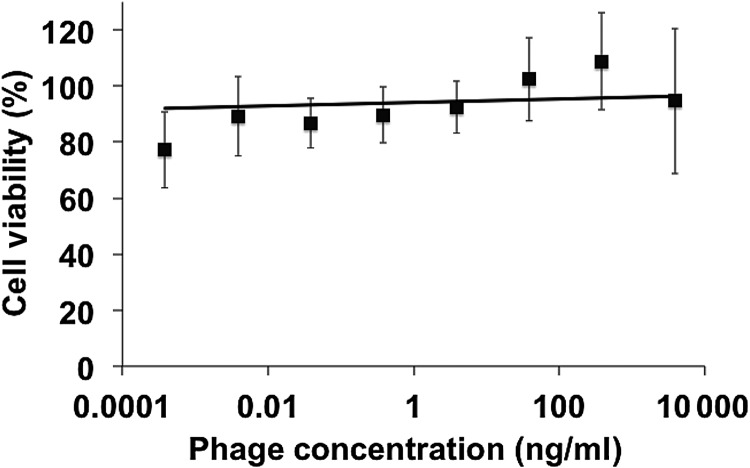
Ten representative phage clones were identified that showed specific interaction with various types of cancer cells and also contained diverse amino acid sequences within the selected fusion peptide region. We sought to determine if identified cancer-specific phage were toxic or produced altered growth kinetics to a chemotherapy-sensitive cancer cell line, MCF-7, to avoid any convoluting effects of Pgp pumps. Intact phage particles were incubated with MCF-7 cells for 24 h followed by an additional 48 h to allow all cells to pass through at least one complete cell cycle. As seen in Fig. 2, no significant dose-dependent effects on cell viability were observed over a wide concentration range. Concentrations of phage in the range of 1–10 μg/ml correspond to the same amount of protein used to target liposome products. This evidence suggests that there is a minimal effect of phage fusion proteins on the viability of cancer cells and any observed effect of toxicity can be translated to the delivery of the liposome itself. Ff class bacteriophage vectors, such as fd and M13, are known to release a significant amount of bacterial endotoxins into the culture medium during routine propagation. A double polyethylene glycol precipitation of phage preparations removes a significant portion of these endotoxins below a physiologically relevant level in vitro; however, treatment with Triton X-114 can extract a significant portion of remaining endotoxins that may become physiologically relevant with nanoparticle preparation or in vivo experiments. We therefore removed residual endotoxins from phage preparations prior to nanoparticle synthesis.
Fig. 2.
Viability of MCF-7 cells determined by MTT assay after treatment with 10 representative phage clones for 24 h followed by 48 h of fresh culture medium. The viability of each preparation was expressed as a percentage of the ratio between viable cells after treatment and viable cells in untreated conditions assumed to represent 100% viability. Data are presented as the mean ± SD, N = 10.
Evaluation of binding and internalization of PANC-1-specific phages to cancer cells
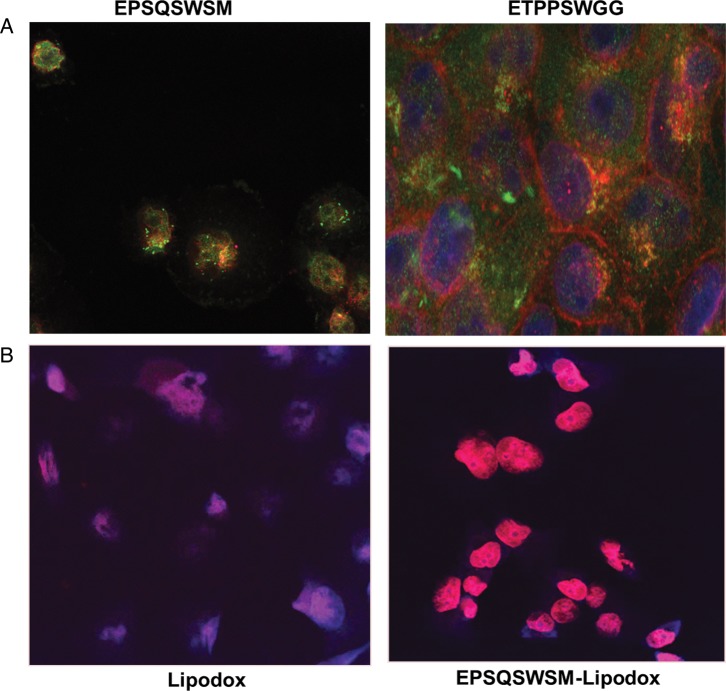
To determine the subcellular localization of phage clones, confocal microscopy of PANC-1 cells treated with intact phage particles (∼109 virions) for 1 h at 37°C was used. Cells were incubated with an affinity-purified rabbit anti-fd IgG for 1 h, washed and incubated with a goat anti-rabbit IgG conjugated with Alexa Flour 488 (Molecular Probes). As seen in Fig. 3A, phages EPSQSWSM and ETPPSWGG readily internalize into PANC-1 cells as seen by the diffuse green fluorescence within the cytoplasm. There was some overlap with labeled endosomes indicating that phage particles internalize via receptor-mediated endocytosis and enters using an endosomal pathway but also can escape endosomes, as seen by cytoplasmic fluorescence.
Fig. 3.
(A) Cellular localization of phage EPSQSWSM (left) and ETPPSWGG (right) in PANC-1 cells. PANC-1 cells were treated with phage, fixed, permeabilized, treated with anti-fd phage IgG, stained with an Alexa Fluor 488 conjugated goat anti-rabbit IgG and were visualized with a FITC filter under a confocal microscope. (B) Microscopy of doxorubicin uptake by PANC-1 cells delivered by unmodified Lipodox (left) compared with EPSQSWSM-Lipodox (right).
The observed specific binding and internalization of PANC-1-specific phage clones was translated to phage protein-modified Lipodox. PANC-1 cells in monolayer were exposed to 35 µg/ml doxorubicin in Lipodox or in phage protein-modified Lipodox for 4 h. After washing, cells were fixed and visualized for doxorubicin fluorescence. As seen in Fig. 3B, even after 4 h of incubation, little doxorubicin fluorescence was observed with Lipodox while intense nuclear fluorescence was seen with phage protein-modified Lipodox indicating that phage protein navigated Lipodox internalization into PANC-1 cells and facilitated doxorubicin delivery to nucleus in 4 h. Nuclear delivery of free doxorubicin, Lipodox or modified Lipodox was estimated quantitatively by cell fractionation experiments. As illustrated in Table II, a major portion of the drug is found in the nuclear fraction with both free doxorubicin and modified Lipodox after 4 h of incubation as compared with unmodified Lipodox.
Table II.
Mean fluorescence intensity of doxorubicin after subcellular fractionation of nuclei and cytosol compartments of PANC-1 cells after exposure to free doxorubicin, unmodified Lipodox and Lipodox targeted with either phage protein EPSQSWSM or ETPPSWGG at a constant doxorubicin concentration of 35 µg/ml for 4 h.
| Mean intensity fluorescence (%) | Nucleus | Cytoplasm |
|---|---|---|
| Lipodox | 58.38 | 41.52 |
| EPSQSWSM-Lipodox | 81.12 | 18.88 |
| ETPPSWGG-Lipodox | 83.37 | 16.63 |
| Doxorubicin | 82.72 | 17.28 |
Phage protein-Lipodox characterization
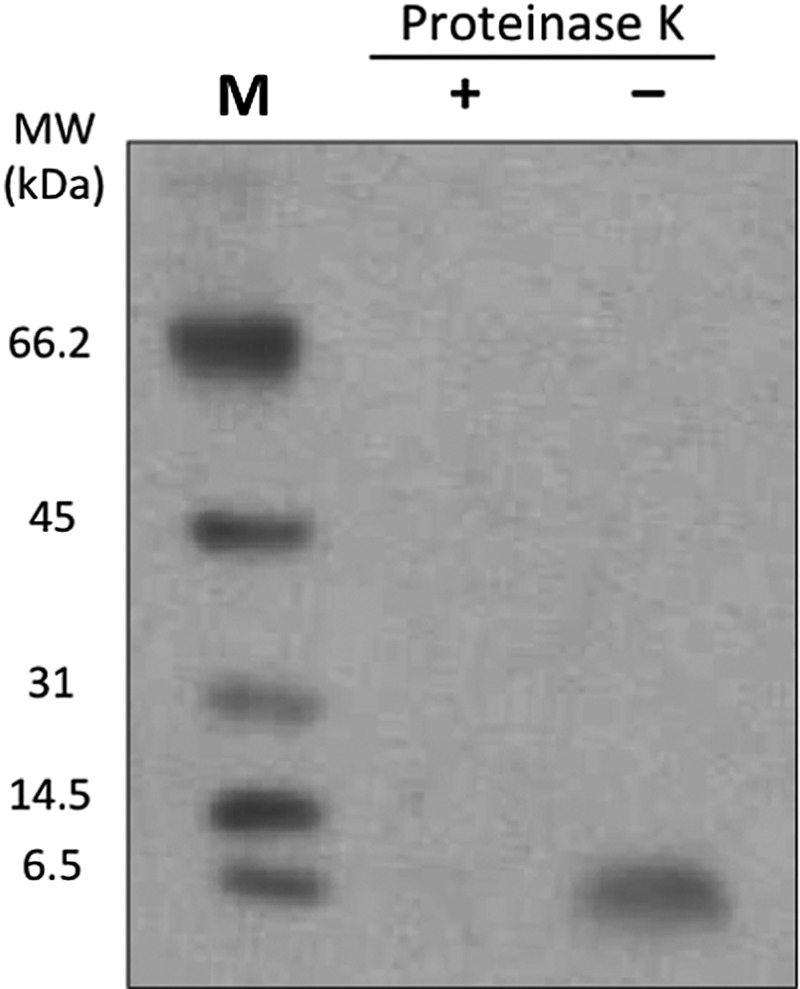
Phage proteins EPSQSWSM and ETPPSWGG were inserted into Lipodox as described (Jayanna et al., 2010; Wang et al., 2010). Lipodox modified with non-relevant phage fusion peptides specific for streptavidin (VPEGAFSSD) was used as a non-specific control of liposome modification. Modification of Lipodox with phage proteins did not alter its surface architecture indicated by a comparative size distribution and zeta potential, though there was a slight increase in Lipodox size distribution after phage protein modification. The average size and zeta potential of unmodified Lipodox was 75.8 nm and −25.6 mV, respectively, and of modified Lipodox was 134 and −23.01 mV, respectively. The presence of phage coat protein and N-terminal orientation of the protein in the liposomal preparations was determined by western blotting after digestion with proteinase K as described previously (Jayanna et al., 2009) (Fig. 4). Insertion of the phage protein into liposomes protects C-terminus of the protein and leaves N-terminus sensitive to proteinase K degradation.
Fig. 4.
Western blotting pattern showing the presence of the modified Lipodox with an N-terminal-out orientation. Liposomal preparations modified with phage proteins were treated with proteinase K and then probed with antibodies specific for N-terminus of the phage coat protein. Lane 1—marker, 2—liposomal preparations treated with proteinase K, lane 3—liposomal preparations untreated with proteinase K.
Selectivity of modified Lipodox towards cancer cells
We explored whether the observed selectivity of phage towards PANC-1 cells was translated further to a specificity of modified Lipodox. PANC-1 cells and normal breast epithelial cells, MCF-10A, were treated with Lipodox, modified Lipodox or free doxorubicin for 4 h at 35 μg/ml and cell-associated doxorubicin was determined quantitatively. As shown in Table III, doxorubicin uptake was markedly increased in PANC-1 cells treated with modified Lipodox as compared with unmodified Lipodox, while doxorubicin uptake was essentially the same in Lipodox- or modified Lipodox-treated MCF-10A cells. Doxorubicin uptake was highest in free doxorubicin-treated PANC-1 or MCF-10A cells as free drug is readily permeable into the cells.
Table III.
Mean fluorescence intensity of doxorubicin in PANC-1 and MCF-10A cells after exposure of cancer cells to free doxorubicin, unmodified Lipodox, EPSQSWSM-Lipodox and ETPPSWGG-Lipodox at a constant doxorubicin concentration of 35 µg/ml for 4 h.
| Mean fluorescence intensity | PANC-1 | MCF-10A |
|---|---|---|
| Lipodox | 1000 | 2100 |
| EPSQSWSM-Lipodox | 4810 | 3095 |
| ETPPSWGG-Lipodox | 2655 | 2111 |
| Doxorubicin | 4925 | 12 675 |
Phage protein-modified Lipodox partially overcomes Pgp-mediated drug efflux
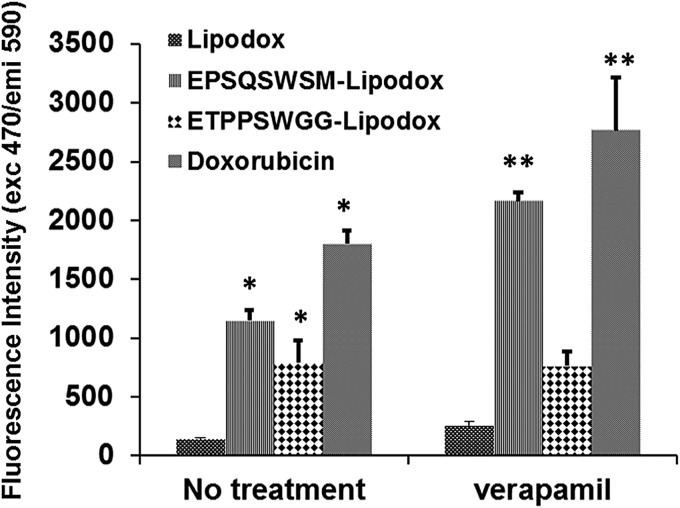
Pancreatic cancer PANC-1 cells overexpress the Pgp efflux pump which is sensitive to blockade by verapamil (O'Driscoll et al., 2007). We sought to evaluate if modified Lipodox can overcome Pgp-mediated drug efflux and increase intracellular concentration of doxorubicin. Figure 5 shows that the level of retention of doxorubicin after treatment of target cells by free drug or modified Lipodox is substantially higher than unmodified Lipodox even after 2 h of post-treatment efflux. The intracellular retention of free doxorubicin within cells treated with doxorubicin and ETPSQWSM-Lipodox increased by 50% in the presence of verapamil, a Pgp pump inhibitor (Lamy et al., 1995), whereas identical drug levels accumulated in cells exposed to ETPSWGG-Lipodox in the absence or presence of verapamil suggesting that internalization of ETPSWGG-Lipodox bypasses multiple drug resistance (MDR) efflux machinery.
Fig. 5.
Fluorescence intensity of doxorubicin in PANC-1 cells after exposure of cancer cells to free doxorubicin, non-targeted Lipodox and Lipodox targeted with phage proteins EPSQSWSM and ETPPSWGG at a concentration of 35 µg/ml doxorubicin for 4 h in the presence or absence of verapamil. *P<0.05, Student's t-test. ** P < 0.05, Student's t-test vs no treatment.
In vitro cytotoxic activity of doxorubicin delivered by phage protein-modified Lipodox
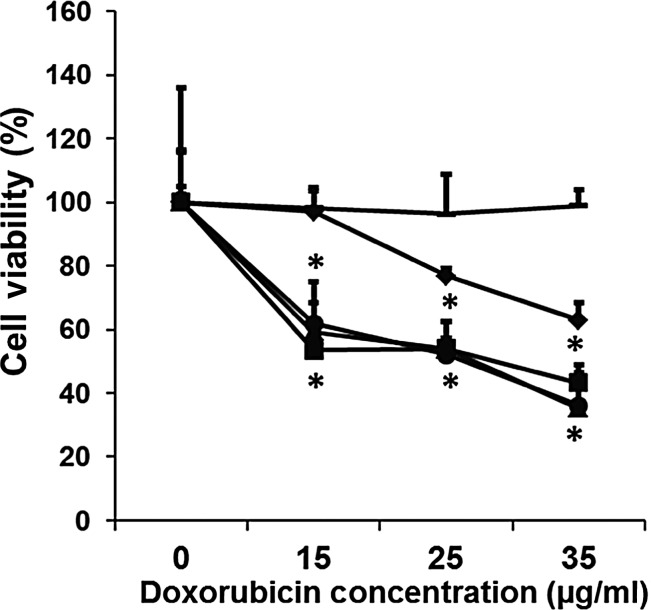
We then explored whether the observed enhanced nuclear delivery of doxorubicin via modified Lipodox would increase doxorubicin cytotoxicity against PANC-1. PANC-1 cells were exposed to Lipodox, modified Lipodox or free doxorubicin for 24 h at various concentrations. After treatment, PANC-1 cell viability was assessed by MTT assay. As seen in Fig. 6, a growth inhibition curve of doxorubicin in modified Lipodox was clearly superior (10-fold drop in IC50) to that of free doxorubicin, unmodified Lipodox or non-specifically modified Lipodox, stressing a key role of liposome binding and internalization in enhancement of cytotoxic activity.
Fig. 6.
Toxicity towards PANC-1 cells after 24 h treatment with Lipodox (diamond), free doxorubicin (square), EPSQSWSM-Lipodox (triangle), ETPPSWGG-Lipodox (circle) and non-specific phage-Lipodox (line), (mean ± SD, n = 3). The cytotoxicity of each preparation was expressed as percent survival compared with untreated cells in which survival was taken as 100%. *P < 0.05, Student's t-test.
Discussion
Pancreatic cancer remains the fourth leading cause of death in the USA (Jemal et al., 2002). Most patients are diagnosed at a clinically advanced stage, when the cancer has been metastasized to distal organs and surgery stops being a viable treatment option. A majority of patients are identified at surgically unresectable advanced stages such as metastatic or locally advanced disease, and have a median survival of 3–6 months for metastatic disease and 6–10 months for locally advanced disease (Molinari et al., 2001). Gemcitabine either alone or in combination with erlotinib are the only approved chemotherapy treatments for patients with advanced pancreatic cancer, of whom the overall survival time is generally around 6 months (Cleary et al., 2004; Moore et al., 2007; Oettle and Neuhaus, 2007). However, treatment with multiple combination chemotherapy in patients with advanced pancreatic cancer is often hindered by their systemic toxicity leading to dose-limiting toxicities and also the performance status of patients. New treatment strategies are mandatory to improve the therapeutic outcomes of patients with advanced, unresectable pancreatic cancer. The concept of using encapsulated drugs within nanoparticles to create nanomedicines has a 2-fold effect that is continuing to play a significant role in the next wave of chemotherapy options: (i) to passively target the drug to the site of disease primarily by leaky vasculature created by the EPR effect and (ii) to modulate the pharmacokinetics and the corresponding therapeutic index of existing chemotherapeutic agents approved for pancreatic cancer treatment (Peer et al., 2007). Several non-targeted nanomedicines have been explored for the treatment of pancreatic cancer including: albumin-based paclitaxel (Nab-paclitaxel), liposomal doxorubicin (Doxil, Lipodox), liposomal cisplatin (Lipoplatin), cationic liposomes encapsulating paclitaxel (EndoTAGTM-1) and polymeric micelles (Tsai et al., 2011). Though non-targeted nanomedicine therapy has been successful in reducing overall systemic toxicity of the free drug, targeting of nanomedicines can further increase their therapeutic index.
Evolved as a result of advances in combinatorial chemistry and phage display, phage technique provided a new way of identification of tumor-specific peptide ligands in a high-throughput fashion (Krumpe and Mori, 2006; Aina et al., 2007). The integration of phage display technology with the nanocarrier-based drug delivery platforms emerged recently as a new drug targeting approach (Koivunen et al., 1999; Medina et al., 2001; Lee et al., 2004; Petrenko, 2008). In particular, landscape phage-based nanotechnology (Petrenko and Jayanna, 2014) offers a highly scalable method of protein isolation in which phage can first be propagated by routine microbiological methods at sufficient scale that can then produce milligram quantities of high purity phage major coat protein after isolation by size-exclusion chromatography. Full-length, 55-amino acid phage protein is inserted into preformed liposomes by spontaneous incorporation into the lipid membrane and anchored by the highly hydrophobic core of the protein. Thus, the protein can be seen as a multifunctional protein with two major functional domains: an anchoring domain at the C-terminus of the fusion protein and a targeting domain located the N-terminus of the fusion protein. We hypothesized that the insertion of major coat protein into liposomes is a two-step process involving (i) an interaction between the highly positive C-terminus and the negatively charged phosphate head groups of the lipid bilayer followed by (ii) the interaction of the hydrophobic core of the major coat protein with the lipid bilayer (Petrenko and Jayanna, 2014).
To enhance a potential anticancer efficiency of doxorubicin liposomes in multidrug-resistant pancreatic cancer cells, we specifically targeted them via fusion with preselected phage protein specific for cancer cells PANC-1. The tumor-specific proteins were isolated from the phages that were affinity selected from multibillion-clone landscape phage library f8/8 by their ability to bind very specifically cancer cells. The phages EPSQSWSM and ETPPSWGG used in this work demonstrated high selectivity and specificity towards target cells versus control unrelated cells. The binding and internalization of the phages EPSQSWSM and ETPPSWGG into PANC-1 cells were also confirmed by immunofluorescence microscopy.
Isolated major coat proteins of selected phages were inserted into preformed liposomal doxorubicin (Lipodox) preparations, in which the phage-derived protein spans the width of the lipid bilayer, therefore displaying the tumor-binding peptides on the surface of the liposome vesicles, as described in Petrenko and Jayanna (2014). This topology of the fusion protein in modified liposomes was confirmed by protease digestion experiments. Treatment of the modified liposomes with proteinase K resulted in complete loss of N-terminus signal, as determined by western blotting with an N-terminal-specific antibody, indicating that the N-terminus of the major coat protein is exposed on the surface of the liposomes and able to interact with the local environment.
Phage proteins effectively enhanced the delivery of targeted Lipodox preparations specifically to PANC-1 cells as compared with non-targeted Lipodox, indicating that the selective binding and internalization properties of phage to target cells were efficiently translated to modified Lipodox. The amount of doxorubicin delivered by targeted Lipodox was overall substantially higher than unmodified Lipodox. Subcellular analysis showed that the amount of drug located in the nucleus of PANC-1 cells after treatment with targeted Lipodox was also significantly higher in comparison with non-targeted Lipodox, as visualized by confocal microscopy and quantified by fluorometric assay after doxorubicin extraction. These results indicate a role of enhanced endocytosis and internalization rates achieved by phage protein-targeted Lipodox resulting in a more rapid nuclear delivery of doxorubicin than untargeted.
PANC-1 cells are known to overexpress Pgp efflux pumps, a protein within the ATP-binding cassette (ABC) transporter family of membrane-bound ATP-dependent efflux pumps known to reduce the intracellular drug accumulation by actively removing a wide array of compounds such as doxorubicin (Gottesman et al., 1996). Whereas treatment of cells with verapamil, a known Pgp inhibitor, effectively inhibited doxorubicin efflux by >50% in cells exposed to either Lipodox, EPSQSWSM-Lipodox or free doxorubicin. However, in the absence of verapamil, retention of doxorubicin in cells exposed to EPSQSWSM-Lipodox was significantly higher than in cells treated with Lipodox, demonstrating an efficient delivery of doxorubicin to the desired site of action, the nucleus, with targeted liposomes versus non-targeted liposomes. We therefore show that ETPPSWGG-Lipodox can effectively bypass the efflux pump essentially by allowing more doxorubicin to accumulate in the nucleus faster than non-targeted Lipodox. We observed a higher retention of doxorubicin within cells after treatment with free doxorubicin than targeted Lipodox in the absence of the verapamil inhibitor. It is hypothesized that the Pgp pumps were unable to sufficiently remove the large residual of doxorubicin possibly sequestered and preventing Pgp activity.
Nuclear delivery of doxorubicin is observed to be more effective than either liposome preparation because it is freely permeable to cells in an in vitro setting and accumulate primarily in the nucleus. Once doxorubicin has penetrated the cell, active Pgp pumps can rapidly export the free drug back into the extracellular environment before the doxorubicin molecules are able to accumulate in the nucleus. It can then be hypothesized that this free drug is then able to be taken up into neighboring cells of the tumor microenvironment and have more of an effect within a tumor. This model of drug uptake after being exported from a drug-resistant cell type can aid in deeper drug penetration within a tumor at higher drug doses compared with free drug, which is rapidly eliminated from the circulation by the kidneys and results in minimal drug retention at the site of pathology.
Higher doxorubicin uptake and retention mediated by targeting Lipodox with phage proteins resulted in more effective cell killing compared with non-targeted counterparts. In conclusion, our results support the proposition that phage protein-targeted Lipodox offers an attractive means of delivering doxorubicin into multidrug-resistant tumor cells, which accumulate drug more effectively than non-targeted Lipodox, irrespective of Pgp efflux activity. Phage protein-mediated drug delivery systems have the potential to circumvent the multidrug-resistant phenotype commonly observed in late stage and recurrent cancer therapies. This technology will be especially useful to other drug delivery devices if they inherit stable drug retention and enhanced tumor accumulation in vivo from their PEGylated liposome carrier as expected.
Funding
This project was funded by a National Institute of Health (NIH) NIH U54 grant 5U54 CA151881-0 to Dr Vladimir Torchilin as Project 4 awarded to V.A.P.
Acknowledgements
We gratefully thank Logan Stallings for the excellent quality of phage preparations used in this study. We also thank Dr John Dennis of the Department of Anatomy, Physiology, and Pharmacology at Auburn University's College of Veterinary Medicine for his expertise in confocal microscopy and for equipment use.
References
- Aida Y., Pabst M.J. J. Immunol. Methods. 1990;132:191–195. doi: 10.1016/0022-1759(90)90029-u. [DOI] [PubMed] [Google Scholar]
- Aina O.H., Liu R., Sutcliffe J.L., Marik J., Pan C.X., Lam K.S. Mol. Pharm. 2007;4:631–651. doi: 10.1021/mp700073y. [DOI] [PubMed] [Google Scholar]
- Alexis F., Rhee J.W., Richie J.P., Radovic-Moreno A.F., Langer R., Farokhzad O.C. Urol. Uncol. 2008;26:74–85. doi: 10.1016/j.urolonc.2007.03.017. [DOI] [PubMed] [Google Scholar]
- Brigati J.R., Samoylova T.I., Jayanna P.K., Petrenko V.A. Curr. Protoc. Protein Sci. 2008;3(Suppl 51):1–27. doi: 10.1002/0471140864.ps1809s51. Chapter 18, Unit 18–9. [DOI] [PubMed] [Google Scholar]
- Broome-Smith J.K., Gnaneshan S., Hunt L.A., Mehraein-Ghomi F., Hashemzadeh-Bonehi L., Tadayyon M., Hennessey E.S. Mol. Membr. Biol. 1994;11:3–8. doi: 10.3109/09687689409161023. [DOI] [PubMed] [Google Scholar]
- Cleary S.P., Gryfe R., Guindi M., et al. J. Am. Coll. Surg. 2004;198:722–731. doi: 10.1016/j.jamcollsurg.2004.01.008. [DOI] [PubMed] [Google Scholar]
- El Kamar F.G., Grossbard M.L., Kozuch P.S. Oncologist. 2003;8:18–34. doi: 10.1634/theoncologist.8-1-18. [DOI] [PubMed] [Google Scholar]
- Fagbohun O.A., Bedi D., Grabchenko N.I., Deinnocentes P.A., Bird R.C., Petrenko V.A. Protein Eng. Des. Sel. 2012;25:271–283. doi: 10.1093/protein/gzs013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goren D., Horowitz A.T., Tzemach D., Tarshish M., Zalipsky S., Gabizon A. Clin. Cancer Res. 2000;6:1949–1957. [PubMed] [Google Scholar]
- Gottesman M.M., Pastan I., Ambudkar S.V. Curr. Opin. Genet. Dev. 1996;6:610–617. doi: 10.1016/s0959-437x(96)80091-8. [DOI] [PubMed] [Google Scholar]
- Gray B.P., Brown K.C. Chem. Rev. 2014;114:1020–1081. doi: 10.1021/cr400166n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbiers M.C., Spruijt R.B., Demel R.A., Hemminga M.A., Wolfs C.J.A.M. Biochim. Biophys. Acta. 2001;1511:309–316. doi: 10.1016/s0005-2736(01)00288-7. [DOI] [PubMed] [Google Scholar]
- Jayanna P.K., Torchilin V.P., Petrenko V.A. Nanomedicine. 2009;5:83–89. doi: 10.1016/j.nano.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayanna P.K., Bedi D., Gillespie J.W., DeInnocentes P., Wang T., Torchilin V.P., Bird R.C., Petrenko V.A. Nanomedicine. 2010;6:538–546. doi: 10.1016/j.nano.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemal A., Thomas A., Murray T., Thun M. CA Cancer J. Clin. 2002;52:23–47. doi: 10.3322/canjclin.52.1.23. [DOI] [PubMed] [Google Scholar]
- Koivunen E., Arap W., Valtanen H., et al. Nat. Biotechnol. 1999;17:768–774. doi: 10.1038/11703. [DOI] [PubMed] [Google Scholar]
- Krumpe L.R., Mori T. Int. J. Pept. Res. Ther. 2006;12:79–91. doi: 10.1007/s10989-005-9002-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn A. FEMS Microbiol. Rev. 1995;17:185–190. doi: 10.1111/j.1574-6976.1995.tb00201.x. [DOI] [PubMed] [Google Scholar]
- Lamy T., Drenou B., Grulois I., et al. Leukemia. 1995;9:1549–1555. [PubMed] [Google Scholar]
- Lee S., Meslsh M.F., Opella S.J. J. Biomol. NMR. 2003;26:327–334. doi: 10.1023/a:1024047805043. [DOI] [PubMed] [Google Scholar]
- Lee T.Y., Wu H.C., Tseng Y.L., Lin C.T. Cancer Res. 2004;64:8002–8008. doi: 10.1158/0008-5472.CAN-04-1948. [DOI] [PubMed] [Google Scholar]
- Maeda H. Adv. Enzyme Regul. 2001;41:189–207. doi: 10.1016/s0065-2571(00)00013-3. [DOI] [PubMed] [Google Scholar]
- Medina O.P., Soderlund T., Laakkonen L.J., Tuominen E.K., Koivunen E., Kinnunen P.K. Cancer Res. 2001;61:3978–3985. [PubMed] [Google Scholar]
- Molinari M., Helton W.S., Espat N.J. Surg. Clin. North Am. 2001;81:651–666. doi: 10.1016/s0039-6109(05)70151-1. [DOI] [PubMed] [Google Scholar]
- Moore M.J., Goldstein D., Hamm J., et al. J. Clin. Oncol. 2007;25:1960–1966. doi: 10.1200/JCO.2006.07.9525. [DOI] [PubMed] [Google Scholar]
- Mori T. Curr. Pharm. Des. 2004;10:2335–2343. doi: 10.2174/1381612043383944. [DOI] [PubMed] [Google Scholar]
- Noble C.O., Kirpotin D.B., Hayes M.E., Mamot C., Hong K., Park J.W., Benz C.C., Marks J.D., Drummond D.C. Expert Opin. Ther. Targets. 2004;8:335–353. doi: 10.1517/14728222.8.4.335. [DOI] [PubMed] [Google Scholar]
- O'Driscoll L., Walsh N., Larkin A., et al. Anticancer Res. 2007;27:2115–2120. [PubMed] [Google Scholar]
- Oettle H., Neuhaus P. Drugs. 2007;67:2293–2310. doi: 10.2165/00003495-200767160-00001. [DOI] [PubMed] [Google Scholar]
- Oyama T., Sykes K.F., Samli K.N., Minna J.D., Johnston S.A., Brown K.C. Cancer Lett. 2003;2:219–230. doi: 10.1016/j.canlet.2003.08.011. [DOI] [PubMed] [Google Scholar]
- Pastorino F., Brignole C., DiPaolo D., et al. Cancer Res. 2006;66:10073–10082. doi: 10.1158/0008-5472.CAN-06-2117. [DOI] [PubMed] [Google Scholar]
- Peer D., Karp J.M., Hong S., Farokhzad O.C., Margalit R., Langer R. Nat. Nanotechnol. 2007;2:751–760. doi: 10.1038/nnano.2007.387. [DOI] [PubMed] [Google Scholar]
- Petrenko V.A. Expert Opin. Drug Deliv. 2008;5:825–836. doi: 10.1517/17425247.5.8.825. [DOI] [PubMed] [Google Scholar]
- Petrenko V.A., Jayanna P.K. FEBS Lett. 2014;2:341–349. doi: 10.1016/j.febslet.2013.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrenko V.A., Smith G.P. Protein Eng. 2000;13:589–592. doi: 10.1093/protein/13.8.589. [DOI] [PubMed] [Google Scholar]
- Petrenko V.A., Smith G.P., Gong X., Quinn T. Protein Eng. 1996;9:797–801. doi: 10.1093/protein/9.9.797. [DOI] [PubMed] [Google Scholar]
- Sergeeva A., Kolonin M.G., Molldrem J.J., Pasqualini R., Arap W. Adv. Drug Deliv. Rev. 2006;58:1622–1654. doi: 10.1016/j.addr.2006.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G.P., Petrenko V.A., Matthews L.J. J. Immunol. Methods. 1998;215:151–161. doi: 10.1016/s0022-1759(98)00087-8. [DOI] [PubMed] [Google Scholar]
- Soekarjo M., Eisenhawer M., Kuhn A., Vogel H. Biochemistry. 1996;35:1232–1241. doi: 10.1021/bi951087h. [DOI] [PubMed] [Google Scholar]
- Sofou S. Nanomedicine. 2007;2:711–724. doi: 10.2217/17435889.2.5.711. [DOI] [PubMed] [Google Scholar]
- Tsai C.S., Park J.W., Chen L.T. J. Gastrointest. Oncol. 2011;2:185–194. doi: 10.3978/j.issn.2078-6891.2011.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlerken V.I.E., Vyas T.K., Amiji M.M. Pharm. Res. 2007;24:1405–1414. doi: 10.1007/s11095-007-9284-6. [DOI] [PubMed] [Google Scholar]
- Wang T., D'Souza G.G., Bedi D., Fagbohun O.A., Potturi L.P., Papahadjopoulos-Sternberg B., Petrenko V.A., Torchilin V.P. Nanomedicine. 2010;5:563–574. doi: 10.2217/nnm.10.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Hartner W.C., Gillespie J.W., Praveen K.P., Yang S., Mei L.A., Petrenko V.A., Torchilin V.P. Nanomedicine. 2014;10:421–430. doi: 10.1016/j.nano.2013.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]