Abstract
Eukaryotic cells have evolved an intricate system to resolve DNA damage to prevent its transmission to daughter cells. This system, collectively known as the DNA damage response (DDR) network, includes a large number of proteins responsible for detection of DNA damage, promotion of repair, and coordination with cell cycle progression. Because defects in this network can lead to cancer, this network constitutes a barrier against tumorigenesis. The BRCT domain is a modular protein domain critical for relaying signals in the DDR. We performed a systematic analysis of protein-protein interactions involving tandem BRCT domains (tBRCT) in the DDR by combining literature curation, yeast two hybrid (Y2H) screens, and tandem affinity purification coupled to mass spectrometry (TAP-MS). We identified one previously unrecognized BRCT protein and generated human protein-protein interaction network for this type of modular domain. This study also reveals several novel components in DNA damage signaling such as COMMD1 and mTORC2. Additionally, integration of tBRCT domain interactions with DDR phosphoprotein studies and analysis of kinase-substrate interactions revealed signaling subnetworks that may aid in understanding the involvement of tBRCT in disease and DNA repair.
INTRODUCTION
Cells are constantly subjected to DNA damage from external, as well as internal, causes. The chemical and physical changes associated with DNA damage compromise the faithful transmission of genetic information to daughter cells and, thus, cells have evolved an intricate system to coordinate damage sensing, signal transduction, repair processes, and cell cycle progression (1). Defects in the DNA damage response (DDR) caused by germline or somatic changes have important implications for disease, and the DDR has been proposed to constitute an early barrier to tumorigenesis (2, 3). Current cancer therapy regimens exploit weaknesses in this system to selectively kill cancer cells (4, 5). Thus, the establishment of a platform for the identification of potential sensitizers of therapy should accelerate the development of new treatment strategies.
Transmission of intracellular signals often relies on the coordinated interactions between protein modular domains and linear peptide motifs (6). A large number of modular domains, defined as units of ~100 amino acids that can independently fold in isolation, have been identified, and several of them, such as Src homology 2 (SH2) domain and the BRCA1 C-terminal (BRCT) domain, recognize phosphorylated linear motifs (7–11). The orchestrated recognition of linear motifs by 14-3-3 proteins, proteins with the BRCT domain, and proteins with the forkhead-associated (FHA) domain is at the core of DNA damage signaling (12). Modular domains that recognize phosphorylated linear motifs are components of three-part signaling toolkits that also include protein kinases and phosphatases (10). Thus, in order to achieve a systems view of the DDR it is important to understand not only which modular domains, linear motifs, kinases, and phosphatases participate in conveying DNA damage signals, but also how their dynamic interactions orchestrate the response.
BRCT domains were initially recognized in the C-terminal region of BRCA1, a protein encoded by the major breast and ovarian susceptibility gene with pleiotropic roles in DNA damage repair (13). BRCT domains are present in a large superfamily of ~40 nonorthologous proteins that participate in cell cycle checkpoints and in the DDR (14, 15). BRCT domains are versatile modules that mediate protein-protein and protein-nucleic acid interactions (16). Several BRCT domains recognize linear motifs phosphorylated by kinases that are activated by DNA damage (8, 9, 17). Therefore, proteins found to interact with BRCT domains are likely to have roles in the cellular response to DNA damage.
In order to gain further insight into the determinants of DDR signal transduction, we generated a protein-protein interaction network (PIN) centered on interactions mediated by the BRCT domain with human proteins. This network contains kinases, phosphatases, and other potential BRCT targets previously unknown to participate in the DDR. In addition, bioinformatics analysis of the constituents of the network enabled the identification of biological processes and protein complexes that integrate the DDR with other cellular activities, such as cell cycle regulation and transcription. This interaction network has implications for cancer therapy for which understanding the cellular response to DDR-inducing chemotherapy and radiation therapy can be used to improve patient survival and quality of life during treatment.
RESULTS
Analysis of the minimal complement of human genes encoding BRCT-containing proteins
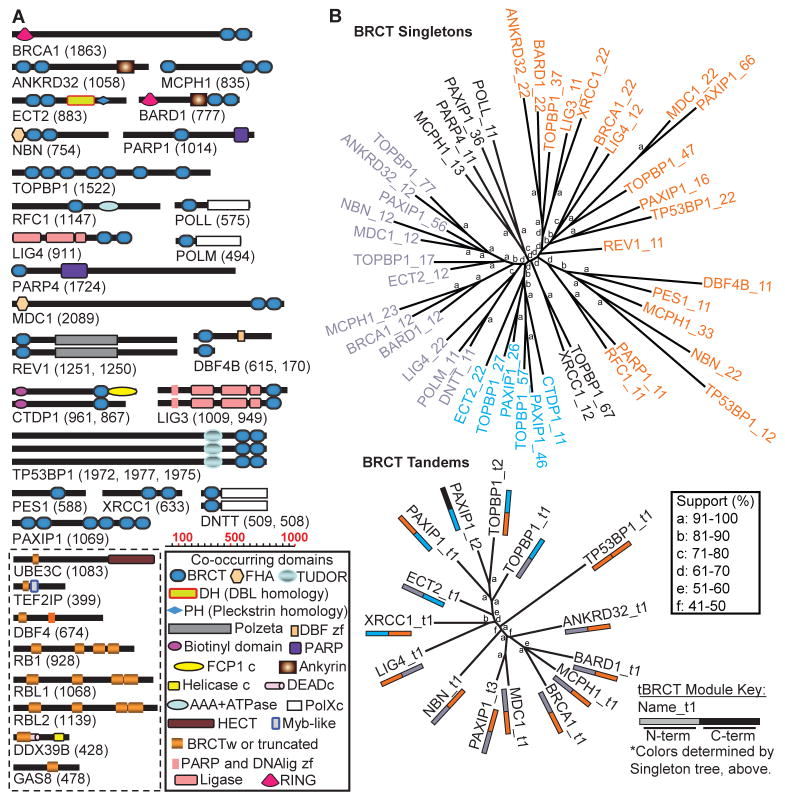
Using a combination of search strategies (Materials and Methods), we identified 23 human genes encoding BRCT domain-containing proteins (Fig. 1A; table S1). Of those, only DBF4B had not been previously recognized as encoding a BRCT-containing protein. Because DBF4B has some, but not all, the structural hallmarks of BRCT domains, final determination of the classification of this protein as a BRCT-containing one requires structural and functional confirmation. We also identified a group of eight genes that encode proteins with truncated or degenerated (lacking conserved residues or structural motifs) BRCT domains (Fig. 1A; table S1). BRCT-encoding genes were located scattered along different autosomal chromosomes; none was located in sex chromosomes (table S1). Several of the genes encode for BRCT domain-containing proteins involved in DNA metabolism, such as DNA polymerases and polymerase-associated proteins (encoded by POLL, POLM, REV1, RFC1, DNTT) and DNA ligases (encoded by LIG4, LIG3). Unlike some other modular domains that preferentially co-occur with other specific modules, BRCT domains co-occurred with many different domains (Fig. 1A).
Fig 1. The BRCT superfamily.
(A) Depiction of the minimal complement of all human proteins containing BRCT domains. Individual diagrams represent the predominant splice forms in the published literature with the number of amino acid residues in parenthesis. BRCT domains (blue boxes) are illustrated with other co-occurring domains. Proteins at the bottom left (dashed line box) represent those with truncated or degenerate BRCT domains (tan boxes). The proteins are drawn according to the scale. (B) Human sequences for the isolated BRCT domains were used to cluster singleton and tBRCT domains on the basis of sequence alignments using neighbor-joining with neighbor method to generate unrooted trees. Top (BRCT Singletons): Unrooted tree illustrating amino acid sequence conservation between individual BRCT singleton domains with bootstrapping scores represented by lower case letters (see key for percent support). Numbers shown after underscore in protein names refer to BRCT relative position (for example, TOPBP1_57 refers to the fifth BRCT in TOPBP1, which contains seven BRCTs, starting from the N-terminus). Bottom (BRCT Tandems): Unrooted tree illustrating amino acid sequence conservation between tBRCT domains. Numbers shown after underscore in protein names refer to tBRCT’s relative position (for example, PAXIP1_t3 refers to the third tBRCT in PAXIP1). Colored bars below protein names represent the position of the individual BRCT domains in different branches from the BRCT singletons tree above.
Editor Questions and Comments:
In the PDF, the colored text labels and the labels on the trees in panel B tend to pixelate. This could create a problem with the files when processed for the print PDF. Ideally, provide vector images if possible, or figures that meet the resolution requirements as specified in the instructions for authors.
In panel B top, is there a difference in the color between the groups containing TOPBP1_67 and POLL_11?
BRCT in Tandem: A different class of BRCT domains
BRCT domains in several proteins recognize and bind phosphorylated serine residues (phosphoserines)(8, 9). BRCT domains found in tandem (Fig. 1A) can behave as a single structural unit (18, 19) that binds phosphopeptides using recognition pockets formed by both BRCT units (20–23). The structural basis for the recognition of phosphopeptide by single BRCTs is presently unknown (9, 24). Therefore, we propose that these tandem BRCT (tBRCT) domains represent a distinct class of BRCT domain. An unrooted tree for all of the individual human BRCT units revealed that they group in several distinct branches (Fig. 1B, Top). To explore the relationship between tBRCT domains, we generated an unrooted tree in which the units were not the individual BRCTs but rather the tBRCT domains (defined as two singleton BRCTs connected by a linker region) (Fig. 1B, Bottom). Interestingly, the two singletons from a tandem frequently originated from different branches of the unrooted BRCT singleton tree (Fig. 1B). Indeed, only in the tandem domains for TP53BP1 did the two singletons come from the same branch. Thus, tBRCT domains present a sequence homology asymmetry reflecting the structural determinants for phosphopeptide recognition and specificity (20–23).
To probe the biology of tBRCT domains, we generated a comprehensive PIN centered on tBRCT domain-mediated interactions. We focused our search on proteins for which there was structural or functional evidence in the literature that the tBRCT functioned as a single unit (Material and Methods). Because these domains might display phosphorylation-independent modes of binding, we did not initially restrict the analysis to phosphorylation-dependent interactions. Our final list contained seven tBRCT domains from ECT2, LIG4, MDC1, BARD1, TP53BP1, BRCA1, and the C-terminal tandem from PAXIP1.
Confirmation of the role of the BRCT domain in the DDR
To determine whether these 23 BRCT-containing proteins were functionally related, we generated a network map of interactions derived from a detailed literature curation, which produced a BRCT-centered PIN with 297 nodes and 420 interactions (fig. S1; table S2). The main network contained 21 BRCT-containing proteins and two isolated small networks were centered on DBF4B and PARP4, respectively (fig. S1). Analysis of the network components using Gene Ontology (GO) (25) confirmed that most BRCT proteins and their interactors were classified in processes related to the DDR and cell cycle checkpoints, and supported a critical role for BRCTs in DNA damage signaling (fig. S2; table S3). We annotated the interactions into three categories: BRCT-mediated, BRCT-independent, and unknown (fig. S1; table S2).
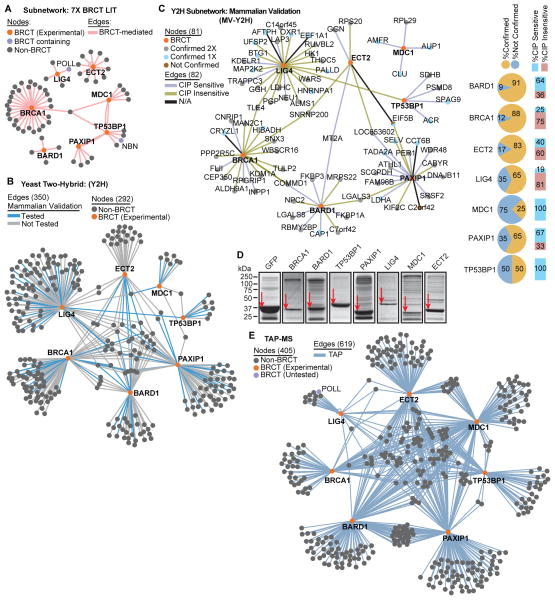
We extracted a subnetwork containing only the seven tBRCT domain-containing proteins that have been explored experimentally and their known BRCT-dependent interactors (referred to as 7X BRCT LIT; Fig. 2A, fig. S3; table S4). GO enrichment analysis of this subnetwork revealed similar results to the parent network with functional roles in the DDR and cell cycle regulation (fig. S4; table S5).
Fig 2. Generating the tBRCT PIN.
(A) Network with BRCT-mediated interactions with the 7 experimentally tested tBRCT domains; orange nodes are derived from the literature (subnetwork from fig. S1). See fig. S3 for an enlarged view of the network with all nodes labeled. (B) Network of the interactions discovered by Y2H screens. Blue edges indicate interactions that were evaluated in the mammalian validation system. See fig. S6 for an enlarged view of the network with all nodes labeled. (C) Orthogonal validation of interactions discovered in (B) using coexpression (TAP-tagged BRCT and GST-tagged non-BRCT proteins) and copurification in mammalian 293FT cells with either streptavidin or glutathione resins. Phosphatase sensitivity was also evaluated in the glutathione affinity purifications. For each BRCT protein, percentages of confirmed interactions in both streptavidin and glutathione pull-down experiments are represented by the pie charts; percentages of interactions sensitive to phosphatase treatment are represented by the bar graphs. (D) Coomassie stain of purified TAP-tagged tBRCT constructs from irradiated 293FT cells. Red arrows indicate the corresponding TAP-tagged bait protein. (E) Network derived from TAP-MS experiments. POLL is the only protein with BRCT domains that was not tested; all others listed as BRCT (Experimental) were tested in the TAP-MS experiments. See fig. S10 for an enlarged view of the network with all nodes labeled.
Pair-wise interactions of the tandem BRCT identified by yeast two-hybrid analysis
To define pair-wise interactions mediated by tBRCT domains, we conducted seven parallel Y2H screens using only the tBRCT regions as bait (Material and Methods). The screens with tBRCTs from MDC1 and TP53BP1 failed to yield any positive colonies, most likely due to their ability to trigger a powerful DNA damage signal when tethered to the DNA (26) (Material and Methods; fig. S5A). To alleviate this problem, we conducted these screens using a low expression vector (Material and Methods).
The Y2H screens generated a PIN with 292 proteins and 350 bait-prey interactions (Fig. 2B, fig. S5B, fig. S6; table S4). To verify the interactions, we subcloned the tBRCT baits into a vector containing a tandem affinity purification (TAP) epitope tag containing streptavidin binding peptide (SBP) and calmodulin-binding peptide (CBP) tags. Preys were cloned into a vector containing a glutathione S-transferase (GST) epitope tag. We tested eighty-two bait-prey interactions (23.4% of the Y2H network) with two complementary pull-down experiments from extracts of 293FT human cells overexpressing individual bait and prey combinations.. In one pull-down experiment, we used the SBP tag to isolate bait complexes and the interacting prey proteins were revealed by probing Western blots with antibody recognizing GST (fig. S7A). In the other, we used the GST tag to isolate the bait complexes and the interactions were revealed by probing with an antibody recognizing CBP (fig. S7B). We defined as true positives only interactions that were replicated in both affinity purification experiments. The application of this stringent criterion resulted in a false positive rate of 23 out of 82 (~28%) (fig. S5B; table S6), which is consistent with previous high-throughput Y2H screens (27–31). In addition, we combined the mammalian validation assay with the complexes isolated with the GST tag with analysis of the effects of phosphatase treatment to determine whether the interactions were dependent on phosphorylation (Fig. 2C, fig. S7B–C; table S6). In the network shown in Fig. 2C, the color of the interactions (edges) indicated whether they were sensitive (increased or decreased) or insensitive to phosphatase treatment. These results suggested a diverse range of potential interaction mechanisms mediated by tBRCT domains. However, these tests were performed using ectopic overexpression of the baits and preys and thus specific interactions should be validated further with endogenous proteins. GO enrichment analysis of the Y2H network suggested that the Y2H method identified a subset of proteins participating in processes different from the network obtained by literature curation (fig. S8; table S7). For example, the literature-curated network was enriched for proteins classified as involved in DNA damage and DNA repair; whereas the Y2H network was enriched in proteins classified as involved in regulation of translational initiation and energy production.
The identification of binary and complex protein interactions for tandem BRCT domains by tandem affinity purification and mass spectrometry
To identify tBRCT domain-mediated protein interactions, we performed tandem affinity purification coupled to mass spectrometry (TAP-MS) using only the tBRCT regions as baits. We decided to focus on the interactions identified under DNA damage conditions because we did not notice a change in the subcellular distribution of ectopically expressed tBRCT fragments tagged with green fluorescent protein (GFP) in response to DNA damage conditions (fig. S9A) and TAP-MS experiments with the PAXIP1 tBRCT indicated that there was substantial overlap in the proteins interacting with this domain under both control and DNA damage conditions (fig. S9B–C), which is consistent with previous studies (32)). These GFP-tBRCT fusions showed that ectopic tBRCTs were predominantly nuclear with some protein also detected in the cytoplasm (fig. S9A). The predominantly nuclear localization and the similarity of the interacting protein profiles may reflect the presence of basal DNA damage incurred in cells from oxidative or replication stresses.
Cells expressing the 7 epitope-tagged tBRCTs were subjected to 20 Gy of ionizing radiation (IR) and protein complexes were isolated from lysates (Fig. 2D) (Material and Methods). These experiments generated a network with 405 proteins and 619 interactions between tBRCT bait and prey partners (Fig. 2E, fig. S10; tables S4, S8). GO enrichment analysis of this network suggested an overrepresentation of proteins involved in RNA processing, cell cycle, and DNA damage (fig. S11; table S9).
We confirmed several PAXIP1 tBRCT interactions discovered by TAP-MS of reciprocal pull-down experiments. We cloned three PAXIP1 tBRCT interactors, MDC1, THRAP3, and Abraxas (also known as FAM175A), into a vector containing a TAP epitope tag comprised of SBP or CBP tags. Each construct was co-expressed with His-tagged PAXIP1 tBRCT in 293FT cells, the SBP tag was used to isolate protein complexes, and the interactions with PAXIP1-tBRCT were confirmed by Western blotting with an antibody recognizing His (fig. S12A–B). Cells exposed to irradiation appeared to have an increased interaction between PAXIP1 and MDC1 (fig. S12A).
An analysis of linear motifs using data from our Y2H screens identified candidate motifs predicted to be recognized by the PAXIP1 tBRCT (8, 33). Visual inspection identified one such motif in THRAP3 (S24RSFS) and two in Abraxas (S188TGFS and S406PTF). To determine whether these motifs mediate the tBRCT interaction, we introduced mutations in these motifs and assessed their effects on binding (fig. S12B). Binding between the PAXIP1 tBRCT and THRAP3 was not impaired by mutations in Ser24 or in Ser28, whereas deletion of the SPTF motif resulted in inhibition of binding between Abraxas and the PAXIP1 tBRCT (fig. S12B). Thus, the tBRCT domains interactions can be either dependent or independent of the SXXF motif.
Functional clustering in the merged tandem BRCT interaction network
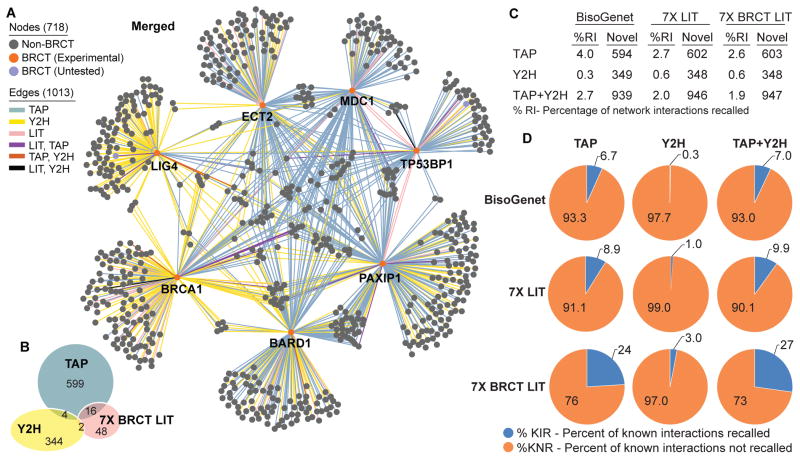
We combined the 7X BRCT LIT (Fig. 2A), Y2H (Fig. 2B), and TAP-MS (Fig. 2E) networks to generate the Merged Network containing 718 nodes with 1013 interactions between tBRCT bait and prey partners (Fig. 3A; table S4) and performed GO enrichment analysis (figs. S13, S14A; table S10). The overlap of identified protein interactions among the different methods, 7X BRCT LIT, Y2H, and TAP-MS was minimal (34, 35)(Fig. 3B), demonstrating the value of utilizing multiple screening approaches to achieve appropriate coverage of protein-protein interactions. However, it is also important to note that the false positive rate for the Y2H dataset is estimated at 28%.
Fig 3. tBRCT Merged Network.
(A) Network compiled from 7X BRCT LIT, Y2H, and TAP-MS networks. Edge color indicates method(s) in which the interaction was identified. Node color denotes presence or absence of the BRCT domain-containing proteins. (B) Venn diagram of overlapping interactions identified in each of the methods used to generate the Merged Network. (C) Table summarizing the % of the interactions within the Merged Network that had been previously identified, represented as % recalled interactions (RI). Datasets: BisoGenet, 7X LIT, and 7X BRCT LIT. The 7X LIT contains all interactions to BRCT-containing proteins and not only those mediated by the BRCT domains. (D) Pie charts indicating the percentage of interactions from database (BisoGenet) or literature curation (7X LIT, 7X BRCT LIT) sources that were also identified in the Y2H and TAP-MS experiments.
The combined experimental TAP-MS and Y2H dataset of tBRCT-mediated interactions is composed of 1.9% previously reported tBRCT-mediated associations (Fig. 3C), yet these interactions constitute approximately 27% of previously reported BRCT-mediated interactions found in the 7X BRCT LIT network (Fig. 3D). When compared to the database derived protein-protein interactions found in BisoGenet (36), which is an application for visualization and analysis of biomolecular relationships, the screenings performed here identified at least 939 previously unreported candidate DDR-implicated protein-protein interactions mediated through association with tBRCT domains (Fig. 3C), suggesting that our knowledge of the DDR components is still rudimentary.
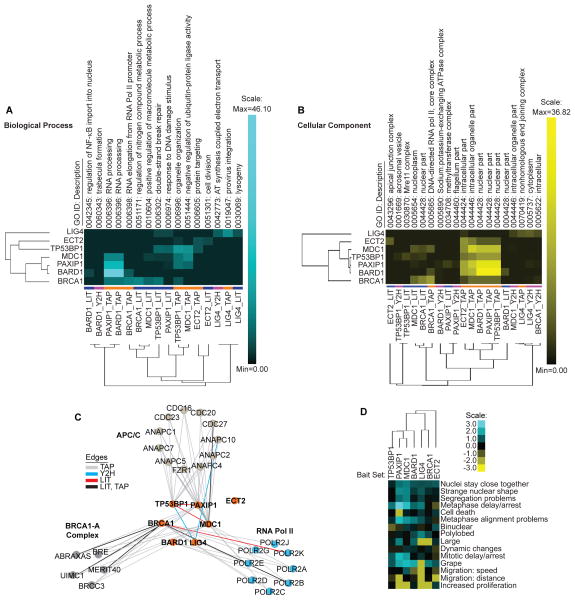
To further define the composition and contribution of the individual screening methods to the network, we conducted GO enrichment analysis for each of the tBRCT bait’s three datasets (7X BRCT LIT, TAP, and Y2H), excluding the bait tBRCT proteins to identify significantly enriched terms for Biological Processes and Cellular Component. We then performed cluster analysis with these enriched terms to (i) assess the functional overlaps between bait-specific datasets and (ii) assess whether the different methods retrieved proteins with similar function or cellular localization or both (Fig. 4A–B). For example, this type of analysis can be used to investigate whether proteins that interact with the BRCA1 tBRCT functionally resemble those that interact with the LIG4 tBRCT.
Fig 4. tBRCT Merged Network Analysis.
(A) Clustering of individual bait sets (columns; excluding the bait) and of each tBRCT bait’s individual screening method (LIT, TAP, or Y2H; rows) according to the top significantly enriched GO terms for Biological Process. (B) Same analysis as in (A) for the top significantly enriched GO terms for Cellular Component. In (A and B), the colors next to the row represent the type of screening method. (C) Subnetwork of highest scoring clusters of proteins from the cell cycle, RNA processing and double-strand break repair GO terms of fig. S14C–E, and the interactions to the tBRCT domains from Fig. 3A. (D) tBRCT Phenoclusters: Clustering of individual bait sets (rows) according to the enrichment of mitotic phenotypes (columns) using the Mitocheck database. Values are represented as the negative log2 transformed enrichment ratio. Positive values indicate enrichment and negative indicate impoverishment.
Clustering of protein sets that interact with the tBRCTs of BRCA1, BARD1, PAXIP1, and MDC1 showed functional overlap of biological processes (Fig. 4A, rows). The functional similarities among these tBRCTs might be a consequence of their structural similarities as illustrated in the unrooted tBRCT tree (Fig. 1B, lower panel). When we analyzed interaction datasets for each individual tBRCT based on screening method, we found that those of BARD1, BRCA1, ECT2, and LIG4 cluster together based on biological processes regardless of screening method (Fig, 4A, columns). However, cellular localization of proteins in each dataset was partially influenced by screening methods, particularly the preferential association of TAP-MS with nuclear proteins (Fig. 4B, rows). Comparison of GO terms enriched in each dataset used in Figures 4A and B to GO terms associated with the corresponding bait proteins in the NCBI Gene database that contain the individual bait tBRCT revealed minimal overlaps between the two (table S11). These results showed that the Y2H and TAP-MS, despite the differences in the types of interactions that they detect (pair-wise versus potentially complex interactions, respectively) retrieve proteins of similar function suggesting that both methods are targeting the same network of interactions, that is the DDR. These results also illustrate the value of using complementary methods and approaches to obtain comprehensive coverage of different cellular compartments.
To determine whether the Merged Network contained clusters of protein-protein interactions enriched for specific biological processes, we collapsed the GO terms present in the Merged Network (fig. S13) into the highest significance common term, which revealed three main biological processes that were overrepresented in the network: RNA Processing, Cell Cycle, and Double-Strand Break Repair (fig. S14B; table S12). We extracted the genes (205 unique genes) encoding proteins associated with these individual processes and we retrieved the known protein-protein interactions among them from publicly available databases using Cytoscape plug-in BisoGenet (36) (Material and Methods). Then, we generated one subnetwork for each of the three processes (fig. S14C–E) and determined the clusters of interacting proteins representing signaling complexes using the Cytoscape plug-in, ClusterOne (Material and Methods). The top-ranking clusters were components of the RNA Polymerase II holoenzyme (RNA Processing network) (fig. S14C), the Anaphase Promoting Complex or Cyclosome (APC/C) (Cell Cycle network) (fig. S14D), and the BRCA1 A-complex (Double-Strand Break Repair network) (fig. S14E) (37–39).
Mapping of the proteins in these top-ranking signaling complexes to the individual tBRCT baits (Fig. 4C) revealed an interconnected network to 6 of the 7 tBRCT-containing proteins and members of these complexes. The functional connections revealed in this approach provide a glimpse of overlapping and specific roles of tBRCT domains. Indeed, the ECT2 tBRCT domain was not found to interact with any of these 3 signaling complexes (Fig. 4C).
Because MDC1, PAXIP1, and TP53BP1 were all connected to the APC/C signaling complex, which is a key enzyme complex that controls progression through mitosis, we hypothesized that these three proteins may share common functions in mitosis. We reasoned that if these baits and their interactors (bait sets; from TAP and Y2H) share mitotic functions, then each of these bait sets should contain proteins that when inhibited produce similar mitotic phenotypes. To test this hypothesis, we used the Mitocheck Database (40) (http://www.mitocheck.org/), which contains information about the occurrence of 22 mitosis-related phenotypes when individual genes were targeted by RNAi in human cells. We extracted all genes encoding proteins associated with each bait set and calculated their enrichment ratio for each phenotype scored in Mitocheck. We visualized the results as heat maps for the bait sets clustered according to their functional impact on mitosis (phenocluster) (Fig. 4D). This analysis revealed a clustering pattern for individual bait sets consistent with the interactions found in the APC/C network: TP53BP1, PAXIP1, and MDC1 had the most interactions with the APC/C complex (Fig. 4C) and their corresponding bait sets appeared closely related according to mitotic phenoclusters (Fig. 4D). These data indicate that this type of network and cluster analysis can reveal insight into the function of protein-protein interactions.
Previously unknown players in the DNA damage response
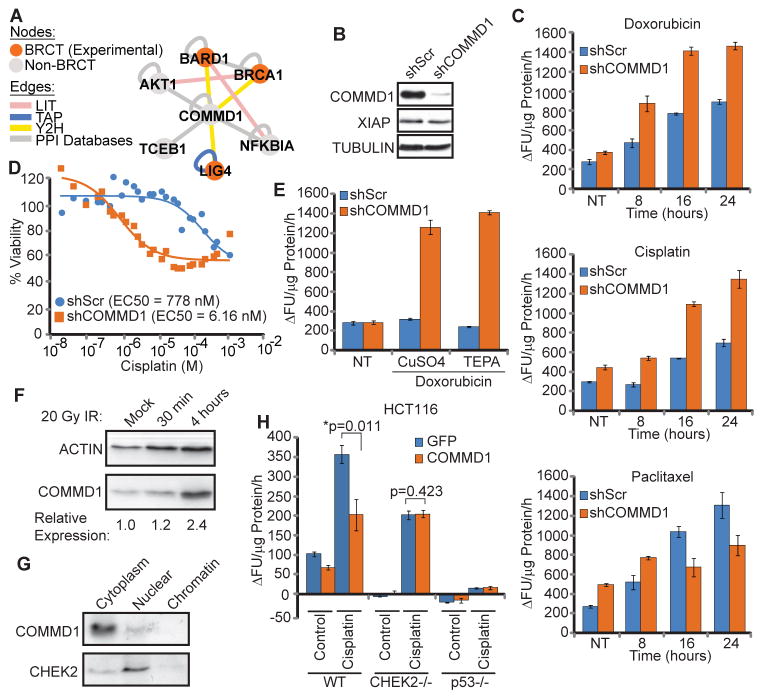
The tBRCT Merged Network also revealed a number of interactions with proteins not previously defined as part of the DDR. We focused on the functional role of COMMD1 because it interacted with multiple tBRCT domains (LIG4, BRCA1, and BARD1) (Fig. 5A). COMMD1 is involved in copper homeostasis, NF-κB signaling, and mouse embryogenesis (41–43). To investigate the role of COMMD1 in response to treatments that can trigger apoptosis, we knocked down COMMD1 using lentivirus-delivered shRNA in breast cancer cells (Fig. 5B, fig. S15A) and examined the response of the cells to either DNA-damaging agents (cisplatin or doxorubicin) (Fig. 5C, fig. S15B) or the microtubule-stabilizing drug paclitaxel (Fig. 5C). Knockdown of COMMD1 sensitized breast cancer cells to treatment with the DNA damaging agents cisplatin and doxorubicin (Fig. 5B–D, fig. S15A,B). The cells exhibited an increase in caspase-3 enzymatic activity, a marker of apoptosis, in response to either cisplatin or doxorubicin (Fig. 5C, fig. S15B), and a decrease in viability in response to cisplatin (Figs. 5D). In contrast, knockdown of COMMD1 resulted in a decrease in caspase 3 activation at later time points in cells treated with microtubule-stabilizing drug, paclitaxel (Fig. 5C, bottom panel). This inverse effect, an increase in sensitivity to DNA-damaging agents but a decrease in sensitivity to drugs targeting microtubules, is similar to the effect of BRCA1 functional impairment (44).
Fig 5. Functional Analysis of COMMD1.
(A) Subnetwork of COMMD1 and interacting proteins from the tBRCT Merged Network including interactions from BisoGenet (PPI Database). Edge color indicates method in which the interaction was identified. (B) Western blot of the indicated proteins in MDA-MB-231 cells expressing control non-targeting (shScr) or COMMD1-specific (shCOMMD1) lentivirus-delivered shRNAs. Data shown are representative of 3 experiments. (C) Caspase-3 activity of lysates from MDA-MB-231 cells expressing the indicated shRNAs and subjected to the indicated drug treatments. Units are defined as the change in fluorescent units from the background per μg protein from whole cell lysates per hour of reaction time (ΔFU/μg Protein/h). Cells were either untreated (NT) or treated for the indicated times with 50 μM cisplatin, 10 μM doxorubicin, or 1 μM paclitaxel (mean + S.D., n=3). (D) Dose response curves using the MTS viability assay of cells expressing the indicated shRNAs and treated with the indicated concentrations of cisplatin. EC50 values (n=3) are also shown. (E) Caspase-3 assay from MDA-MB-231 cells with and without COMMD1 knockdown, either untreated (NT) or treated with 10 μM doxorubicin in the presence of either 100 μM CuSO4 or 50 μM of the copper chelator tetraethylenepentamine (TEPA) (mean + S.D., n=3). (F) Abundance of endogenous COMMD1 in HeLa cells either mock treated, 30 minutes or 4 hours after exposure to 20 Gy IR. Relative abundance normalized to actin control is shown beneath each lane. Data shown are representative of 2 experiments. (G) Cellular localization of COMMD1 and CHEK2 in the nucleus and cytoplasm determined by Western blot from Hela cells. Data shown are representative of 2 experiments. (H) Caspase 3 activity in cells overexpressing COMMD1 in HCT116 wild-type (WT), CHEK2−/−, or p53−/− cells treated with 50 μM cisplatin (mean + S.D., n=3) (*; significant difference using the one-tailed, paired Student’s T-test).
One mechanism through which cells can be sensitized to apoptosis-inducing stimuli is through a decrease in the abundance of antiapoptotic proteins, such as X-linked inhibitor of apoptosis (XIAP). Because COMMD1 has been implicated in copper homeostasis (42) and increased intracellular copper concentration decreases the stability of the inhibitor of apoptosis protein XIAP (45), we assessed if knockdown of COMMD1 altered XIAP abundance or if changing the cellular copper concentration affected the sensitivity of cells in which COMMD1 was knocked down to DNA damage induced by doxorubicin. However, knockdown of COMMD1 had no effect on XIAP abundance, indicating that the increased sensitivity to apoptosis induced by DNA-damaging agents was independent of an effect on XIAP stability (Fig. 5B). Furthermore, the increase in caspase 3 activation in response to doxorubicin treatment of the breast cancer cell line MDA-MB-231 in which COMMD1 was knocked down was unaffected by either increasing or decreasing cellular copper concentration (Fig. 5E).
In line with the role of COMMD1 in the DDR, an independent Y2H assay using the DDR kinase CHEK2 as bait (46) also identified a full-length clone of COMMD1 as an interacting prey. Deletion analysis with the Y2H assay showed that residues 126–190 of COMMD1 were required to interact with CHEK2 and that residues 115–165 (FHA domain) and 225–543 (kinase domain) in CHEK2 were important for binding COMMD1 (fig. S15C). The abundance of COMMD1 in Hela cells increased following ionizing radiation (Fig. 5F) and we detected both CHEK2 and COMMD1 in the cytosol and nucleus (Fig. 5G). Ectopic expression of COMMD1 inhibited cisplatin-induced apoptosis and this effect is abrogated in CHEK2−/− HCT116 cells (Fig. 5H).
Although we were not able to demonstrate the interactions between COMMD1 and BRCT-containing proteins using endogenous proteins, several lines of evidence suggested that the interactions are biological relevant. We detected the interaction in parallel Y2H screens using LIG4, BRCA1, or BARD1 tBRCT as baits. Parallel screens using the same pre-transformed library did not detect an interaction between COMMD1 and MDC1, TP53BP1, ECT2, or PAXIP1, suggesting that COMMD1 is not a spurious “sticky” protein. We confirmed the interaction between COMMD1 and BRCA1 or BARD1 in mammalian cells using two independent pull-down strategies (fig. S7A–B). In pull-down experiments, phosphatase treatment of the lysates decreased the interaction between COMMD1 and BARD1 but did not change in the interaction with BRCA1, suggesting that the interaction with these distinct tBRCTs is mediated by different mechanisms. Modulation of the abundance of COMMD1 altered the response to DNA-damaging agents in a CHEK2-dependent manner, but did not alter the sensitivity to an agent that targets microtubules. These results suggested that COMMD1 has a previously unknown function in the response to DNA damage.
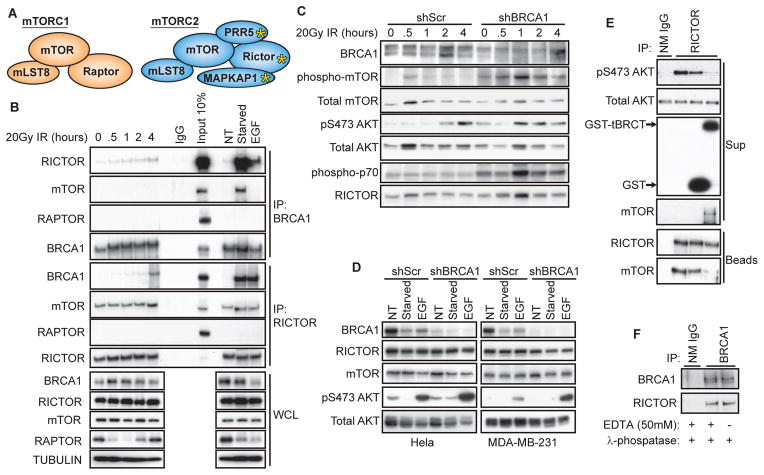
From the TAP dataset, another newly identified interaction with potential clinical implications was found between the BRCA1 tBRCT and members of the mammalian (or mechanistic) target of rapamycin complex 2 (mTORC2) comprised of RICTOR, MAPKAP1, and PRR5 (Fig. 6A), which is involved in cell proliferation and survival through the stimulation of phosphorylation of AKT at Ser473 (pS473) (47). BRCA1 coimmunoprecipitated with RICTOR, but not with mTOR, the catalytic subunit of mTORC2, or RAPTOR, a component of mTORC1 by coimmunoprecipitation of proteins from 239FT cells using low stringency conditions conducive to RICTOR-mTOR interactions (48, 49) (Fig. 6B). The interaction between BRCA1 and RICTOR appeared dynamic: It appeared increased in response to DNA damage and serum starvation and repressed by stimulation of the cells with epidermal growth factor in the BRCA1 IP (Fig. 6B). Although mTOR only coimmunoprecipitated with in BRCA1 under conditions of serum starvation, RICTOR coimmunoprecipitated mTOR under all treatment conditions (Fig. 6B). These data suggested that BRCA1 interacts with RICTOR but not with a functional mTORC2.
Fig 6. BRCA1 regulation of mTORC2.
(A) Schematic of mTORC1 and mTORC2 complexes. Yellow asterisk denotes proteins identified in the TAP-MS network that bound to the tBRCT of BRCA1. (B) Endogenous protein interactions from 293FT cell lysates collected at the indicated times after exposure to 20 Gy IR (0 = No IR), exposed to medium with no FBS for 16 hours (starved), starved then stimulated with 10 ng/mL EGF for 5 minutes or untreated (NT). Coimmunoprecipitations were performed with 0.5% CHAPS lysis buffer. Data are representative of 3 experiments. (IP, antibody recognizing the indicated protein was used for the immunoprecipitation; WCL, whole cell lysate. (C) AKT and downstream mTOR signaling events determined at the indicated times after exposure to 20 Gy IR in MCF-10A cells expressing either control non-targeting (shScr) or BRCA1-targeted (shBRCA1) shRNA. Data are representative of 2 experiments. (D) Western blot analysis of AKT activation and total mTOR and RICTOR abundance in Hela and MDA-MB-231 cells expressing either shScr or BRCA1-targeted shRNAs. Data are representative of 2 experiments. NT, starved, and EGF conditions were the same as those described in panel (B). (E) In vitro kinase assay from HCC1937 cell lysates. RICTOR immunoprecipitates were incubated with either purified GST or GST-BRCA1 tBRCT proteins using AKT as substrate and phosphorylation of Ser473 on AKT (pS473 AKT) was used as a measure of mTORC2 activity. mTORC2 complex integrity after addition of GST or GST-BRCA1 tBRCT was analyzed by Western blot for mTOR retention in the bead fraction with RICTOR. Data are representative of 3 experiments. (F) Immunoprecipitated BRCA1 complexes from 293FT lysates were incubated with λ-phosphatase for 30 minutes at 30° C in the presence and absence of 50 mM EDTA then immunoblotted for RICTOR. Data are representative of 2 experiments.
Studies with cancer cells indicate that BRCA1 inhibits AKT signaling, but the mechanism of action is unclear (50, 51). The amount of pS473 AKT appeared increased in MCF-10A, Hela, or MDA-MB-231 cells in which BRCA1 was knocked down of BRCA1 in increased AKT signaling (Fig. 6C–D). We detected only limited changes in the total abundance of AKT, mTOR, or RICTOR after BRCA1 knockdown, indicating that these proteins were not substrates for the ubiquitin ligase activity of BRCA1 and that this was not how BRCA1 affected AKT phosphorylation (Fig. 6C–D). To examine the effect of the BRCA1 tBRCT domain on the activity of purified mTORC2, we immunoprecipitated mTORC2 with antibodies recognizing RICTOR from HCC1937 breast cancer cells, which lack wild type BRCA1 (it contains a truncated copy of BRCA1 that produces no detectable protein)(52) and thus would not interact with mTORC2 through the BRCT. Addition of bacterially expressed GST-BRCA1 tBRCT to RICTOR immunoprecipitates reduced the amount of mTOR present in the complexes and reduced mTORC2 phosphorylation of AKT at Ser473 (Fig. 6E). The interaction between endogenous BRCA1 and RICTOR was insensitive to λ-phosphatase treatment, suggesting the interaction is phosphorylation-independent (Fig. 6F). These findings suggested that BRCA1 may repress AKT activation by promoting the dissociation of mTOR from mTORC2 complexes.
Phosphorylation events, kinases, and modular domains in the tBRCT network
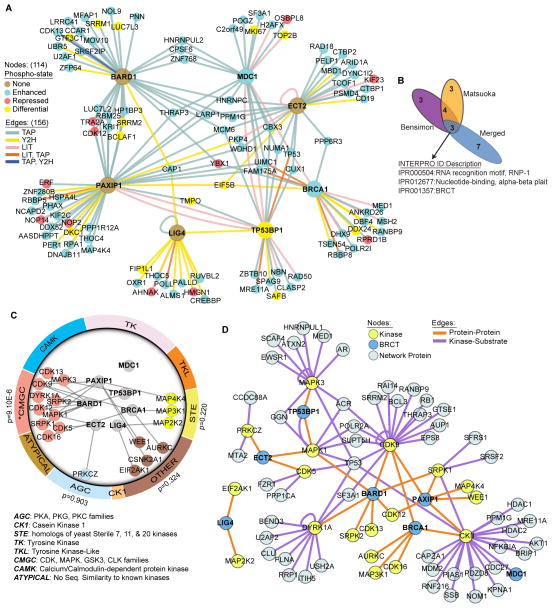
The TAP-MS and Y2H screening approaches were not selective for phosphorylation-specific interactions, yet phosphorylation events, such as those mediated by the kinases ATM and ATR, play a central role in DDR signaling. Therefore, we mapped previously described ATM- or ATR-dependent phosphorylations (53, 54) onto the tBRCT Merged Network (Fig. 7A, fig. S16A). One hundred ten proteins in the tBRCT Merged Network (15.3% of all proteins in the network) were previously documented to be phosphorylated. These 110 proteins represent 12.5% of the proteins reported as modified by ATM- or ATR-dependent phosphorylation in the DDR (53, 54). The tBRCT Merged Network was significantly enriched in proteins phosphorylated in an ATM- or ATR-dependent manner when compared to the expected number of phosphorylated proteins extrapolated from the two large-scale previous studies [Hypergeometric Cumulative Probability, P(X > 110)=9.99 x 10−16; sample size, 718; protein population size, 20000; phosphorylation events described in population, 883; from refs. (53, 54)].
Fig. 7. DDR-dependent phosphorylation events and kinase-substrate relationships in the tBRCT Network.
(A) Integration of tBRCT Merged Network with that of DNA damage-dependent, ATM- and ATR-regulated phosphorylation events using previously reported datasets (53, 54). Node color indicates the change in phosphorylation state in response to DNA damage (None; no change in phosphorylation state, Enhanced; increased phosphorylation, Repressed; decreased phosphorylation, Differential; multiple phosphorylation sites on the same target protein where at least one site differs in its regulation from the others). Edge colors indicate the screening method(s) in which the interaction was observed. (B) Domain enrichment overlap among the three datasets present in Fig. 7A determined using DAVID to compare the top 10 enriched domains from each dataset. The domains enriched in all 3 of the datasets are indicated with the corresponding INTERPRO identification and description. (C) Protein kinases from the TAP and Y2H datasets were mapped to the human kinome. In the outside ring, kinase groups are represented as color segments proportional to total size of the family (93). Inset shows the subnetwork of tBRCT (blue nodes) interactions with the kinases from the network (node color corresponds to the family association). Only the CMGC kinase family is significantly overrepresented in the Y2H and TAP-MS datasets (binomial p-values for individual kinase families are shown). (D) Kinase-substrate associations in the tBRCT Merged Network were determined using PhosphoSitePlus (www.phosphosite.org) to generate lists of known and predicted kinase-substrate interactions on the basis of consensus phosphorylation motifs of the 18 kinases identified in the network. Node color indicates whether the protein is a kinase, BRCT-containing, or other protein from the tBRCT Merged Network. Edge color indicates whether the association between the two network proteins is Protein-Protein or Kinase-Substrate.
To further explore the overlap between the tBRCT Merged Network, the Bensimon et al. (53), and the Matsuoka et al. (54) datasets, we identified which modular domains were enriched in each dataset (Figs. 7B, fig. S16B). In addition to the expected enrichment for BRCT domains, the nucleotide-binding domains RNP-1 and alpha-beta plait were also enriched in each of the three datasets (fig. S16B). Thus, these modular domains may have an essential role in the DDR and should be further explored.
We mapped the 18 protein kinases that interacted with tBRCT domains to their respective kinome families (Fig. 7C). Tyrosine kinases were conspicuously missing and 10 of the 18 kinases belong to the CMGC family, which includes cyclin-dependent kinases, mitogen-activated protein kinases, glycogen synthase kinases, and CDK-like kinases (Fig. 7C). With PhosphoSitePlus (http://www.phosphosite.org) (55) we annotated known and predicted kinase-substrate associations in the tBRCT Merged Network (Fig. 7D), revealing potential targets of phosphorylation by the kinases in the tBRCT Merged Network and may guide future experiments into the regulation of the DDR.
Genes encoding proteins in the tandem BRCT network as common targets of somatic mutation in neoplasia
The tBRCT Merged Network was examined for disease associations common to more than one tBRCT domain dataset (fig. S17). Neoplasias in general were prominent in the tBRCT disease network and “hereditary neoplastic syndromes” was the most highly interconnected term, connected to all of the tBRCTs studied except that of PAXIP1 (fig. S17). Consistent with a prominent role in neoplasia, genes in the tBRCT network represent 4% of all genes but 12% of somatic mutational events in cancer deposited in the COSMIC database (Catalog of Somatic Mutations in Cancer; http://www.sanger.ac.uk/genetics/CGP/cosmic/) (fig. S18A, B). Most of these events can be attributable to mutations in genes in the BRCA1, ECT2, and TP53BP1 bait sets (fig. S18B). We compared the top ten genes and tissues of origin in which these mutations arise between all genes in the human genome and all genes in the tBRCT Network (fig. S18C, D). This analysis indicated distinct distributions of genes disrupted in cancer between all genes in the genome and those in the tBRCT Network. Differences were also found in the tissues in which these mutations arise. Mutational events in genes present in the tBRCT Network are associated with the large intestine, upper areodigestive tract, breast, and ovary.
DISCUSSION
Here, we undertook a systematic analysis of the BRCT domain, which plays a critical role in the DDR. We identified the minimal complement of 23 BRCT-containing proteins in the human proteome and propose that tBRCT domains with their asymmetric organization and phosphopeptide-binding activity constitute a special class of BRCT domains. Although BRCT domains are not as frequent in the human genome as SH2 domains (115 SH2 domains in 105 proteins)(56); the frequency of BRCT domains is comparable to that of PTB domains, which are found in 24 proteins (57).
We established a framework to dissect the molecular mechanisms involved in signaling DNA damage using three independent approaches to build a PIN centered on BRCT domains. Literature curation, TAP-MS, and Y2H methods generated a merged network containing 718 nodes and 1013 edges (Fig. 3A). Our experimental methods detected 27.2% (18/66) of the known domain-mediated interactions for the seven tBRCT tested (Fig. 3D) and we uncovered 947 previously unknown BRCT-mediated interactions in the DDR (Fig. 3C).
Each method used to generate PIN has specific limitations. For example, Y2H screens are prone to false positives and independent screenings may not be comparable. To address these problems with Y2H screens, we used multiple reporters and performed the screenings using a pre-transformed library in parallel matings. We also conducted large-scale validation by affinity purification in mammalian cells. TAP-MS methods provide no information about direct interactions and may miss transient interactions. These limitations can be compensated by the parallel use of the Y2H, which provides direct interaction data as well as more sensitive detection of transient interactions. Thus, the TAP-MS and Y2H provide complementarity to define a comprehensive PIN (58).
Gene Ontology clustering of Biological Process top terms for each bait and screening method indicated that although the specific proteins identified can differ, datasets for the same bait clustered together regardless of the screening method used (Fig. 4A). Analysis of protein clusters in the Merged Network revealed that BRCT domains participated in unique and overlapping interactions with large protein complexes. For example, the tBRCT of TP53BP1, PAXIP1, and MDC1 (and to a lesser extent LIG4) had multiple interactions with components of the APC/C complex; whereas the tBRCT of BRCA1, ECT2, and BARD1 did not (Fig. 4C). This pattern of specificity is also reflected in an independent dataset (Mitocheck), indicating that biologically relevant information can be derived from the PIN. It also provides confirmation for the notion that seemingly distinct DDR pathways might be physically linked (59). Interestingly, ECT2 lacks interactions with the APC/C, BRCA1-A, and RNA polymerase II signaling complexes (Fig. 4C), despite having interactions with 4 proteins involved in double strand break repair (a process associated with BRCA1-A), 9 proteins involved in RNA processing (a process that could be considered associated with RNA polymerase II), and 22 proteins involved in the cell cycle (a process associated with APC/C) (fig. S14B). This suggests that ECT2 participates in processes, such as the cell cycle through control of cytokinesis (60), independently of these complexes.
Our analysis of two newly discovered potential players in the DDR (COMMD1 and RICTOR) also supported the notion that PIN analysis can yield relevant biological information with important implications for cancer therapy (61, 62). For example, it may reveal proteins that could be targeted as sensitizers for chemotherapy and, in some instances, these targets may already have drugs available that have unrecognized utility in cancer therapy. These newly discovered players, such as COMMD1, may also constitute potential biomarkers for drug response. Furthermore, the interaction between tBRCT of BRCA1 and RICTOR may represent a potential mode of integration between the DDR and cell proliferation and viability.
The data presented here represent a framework on which to build a more comprehensive map of the components and interactions involved in the DDR. The snapshot provided by the tBRCT Merged Network suggest that BRCT domains act in the DDR using different modes of interaction including phosphorylation-dependent and -independent binding. Next steps in the identification of determinants of signal transduction include identifying linear motifs (33, 63–65) and kinases involved in regulating the information flow through this network. Phosphorylation-independent interactions may be constitutive interactions that maintain the DDR in a poised state to quickly respond to DNA damage. Phosphorylation-independent interactions may also include inducible interactions mediated by changes in protein abundance or other posttranslational modifications. Future experiments will be designed to add to the existing data and complete the interaction mapping of modular domains (e.g. BRCT, FHA, and 14-3-3) that participate in the DDR using an “interactome walking” (66) approach.
Several screens and integrative methods to define PIN have been performed in the context of the whole proteome, pathway-specific, or disease-specific networks of different model organisms (29, 31, 59, 61, 67, 68). In particular, our study builds on previous approaches to address domain-specific networks for the SH3 domains (69, 70), 14-3-3 proteins (71–73), Polo-box domains (74), and a Caenorhabditis elegans network using 100-aa stretches (the size of an average protein domain)(75). The data presented here correspond to the first comprehensive human study of the tBRCT domain interactome.
MATERIAL AND METHODS
Generating the list of the minimal complement of human BRCT proteins
A dataset of human proteins was obtained using a program developed by our group to extract all proteins identified by the taxonomic index Homo sapiens from the non-redundant protein sequences database (NCBI “nr”). This dataset was searched using hmmsearch (HMMER package) to screen for matches to the Pfam BRCT domain (PF00533 version 17)(76). Selected proteins were extracted and then a BLAST search was conducted against the UniProt database (SWISS-PROT), and conserved domains were identified by local and global alignments using hmmpfam (HMMER package) and the Pfam-A database. We then removed false positive sequences presenting low-quality BRCT hits (Pfam cutoff values were: length smaller than 36AA with score lower than 6.7 and e-value higher than 5.6). The lowest BLAST score and highest e-value retained for an isolated domain was 15.3 and 5 x 10−5, respectively (LIG3). Redundant sequence grouping was based on UniProt database BLAST results and CLUSTALW (77) multiple alignments. BRCT domains were determined in all proteins by comparing results from hmmpfam with results from NCBI (CDD), Pfam, and BLOCKs conserved domain identification web tools. Secondary structure prediction results were also used to confirm divergent BRCTs. This procedure resulted in a list of 23 human proteins, which we refer to as the BRCT superfamily (Fig. 1A; table S1).
We also generated a list of proteins containing degenerated or truncated BRCTs by combining three approaches. First, we performed a psi-blast search with the Retinoblastoma protein family members, RB1, RBL1 (p130), and RBL2 (p107). Second, we performed iterative hmmcreate-hmmsearches starting with Rb BRCT initial alignment (RB1, RBL1, RBL2), creating and improving an Rb BRCT HMM and alignment during the procedure. We also implemented a HMM-HMM search using HHpred, starting with an iterated Rb BRCT alignment. These procedures generated a list of seven proteins (including RB1, RBL1, and RBL2) with degenerated (BRCTw) (78) or truncated BRCT domains (table S1). Finally, after visual inspection of the BAT1 sequence and N-terminal structure (PBD: 1XTK), we concluded that the BAT1 putative BRCT domain lacked some hallmarks of BRCTs and the region coincided with another different domain, DEADc. However, we included it in the list of degenerated BRCT domains pending further characterization.
To identify co-occurring modular domains and motifs, we extracted the protein RefSeq (79) from Entrez Gene database using the official protein name and searched with NCBI CDD (Conserved Domains Database) web tool. The identified specific motifs and domains were extracted from the retrieved records.
To generate the unrooted trees, BRCT domains and tBRCT domains were extracted and aligned with the clustalW and PRALINE web tools, respectively. After inspection and manual editing, protdist (phylip package v4) was used to calculate BRCTs distances with the PMB (Probability Matrix from Blocks)(80) category model. Unrooted trees were created using neighbor-joining with neighbor method (phylip package v4). Support values were obtained using 10,000 replicates created with seqboot (phylip package v4), using Jackknife with 1% deletion. Trees were generated using figtree.
Literature curation
Although there are several databases describing protein interactions, they are prone to errors (70) and the exact region of interaction is seldom reported. Thus, to derive the literature curated network, we searched PubMed for all records containing any of the 23 BRCT genes or proteins. Searches conducted using the official name or the documented aliases (from Entrez Gene) generated 21,652 records. The full text of every paper that reported protein interactions was manually inspected and only information on human genes or proteins was retained. In deciding whether to record an interaction we followed the recommendations from Cusick et al. (81). To ensure that all protein interactions had been extracted, we inspected the interactions listed in the Entrez Gene record for each BRCT-containing protein. The interactions listed were inspected in the original papers and only considered if the interaction was sufficiently documented. We annotated every interaction as (a) mediated by the BRCT domain or (b) not mediated by the BRCT domain. When fragments used for domain mapping were larger than the BRCT domains, we classified the interaction as (a) only if the segment including the BRCT domain was more than half of the length of the fragment. Interactions from cases in which there were no mapping experiments or in which the region mapped was too large to make a determination were classified as (c) unknown. Interactions with nucleic acid were annotated, but not included in the network figure. Cytoscape (82) was used for network visualization. The annotated dataset with references can be found in table S2.
Choice of functional tandem BRCT domains
Twelve proteins contained more than one BRCT domain (Fig. 1) with fifteen possible tandems. We initially focused on tBRCTs for which there was evidence in the literature that they would function as a single unit by binding to phosphopeptides using structural features of both BRCT domains (21, 22, 83). There was compelling evidence for the tandems present in BRCA1, MDC1, BARD1, and LIG4 (17, 21, 22, 83). For the most C-terminally located BRCT tandem of PAXIP1, the functional tandem could be plausibly inferred from phosphopeptide binding data (8, 17). We extended our list by including the tandem BRCTs from ECT2 and TP53BP1 as potential functional tandems given their domain organization. It is important to note that the second BRCT of NBN (84) was only identified after we finalized our bait selection and was not included as one of our baits. To determine domain borders, we used the amino acid residue number given in the hmmpfam analysis and information derived from the literature. In cases in which the domain was not completely identified, Pfam, NCBI (CDD), and SMART web tools were used, in that order, to aid the determination. The following tandem domains (which included flanking amino acids) were used for cloning in Y2H and in TAP-tag vectors: BARD1 (aa 578–802), BRCA1 (aa 1650–1863), ECT2 (aa 111–341), LIG4 (aa 618–912), MDC1 (aa 1895–2095), PAXIP1 (aa 850–1069), TP53BP1 (aa 1779–2031).
Y2H screening
To detect pair-wise interactions, we employed the MATCHMAKER Two-Hybrid 3 System (Clontech, Palo Alto, CA). The tBRCT domains of each protein of interest were cloned into the pGBKT7 vector as a fusion to GAL4 DNA binding domain (DBD) and transformed into the S. cerevisiae strain AH109 [MATa] alone or in combination with empty pGADT7 vector. With the exception of the tBRCT of TP53BP1, which did not generate any transformants, all other fusion proteins were expressed (fig. S5A). We conducted the screens in a stringent selection medium lacking Trp, Leu, His, and Adenine, conditions in which no self-activation was detected (fig. S5A). Use of multiple reporters has been shown to decrease the number of false positives (85).
Expression of the MDC1 tBRCT consistently produced very few colonies suggesting toxicity, albeit to a lesser extent than that found for TP53BP1. Expression of TP53BP1 or MDC1 tBRCTs was not toxic when these domains were fused to GAL4 Activation domain (AD), indicating that toxicity was a result of tethering the fusion protein to the DNA, which is consistent with data from mammalian cells (26). Attempts to conduct screening with tBRCTs of MDC1 in the context of pGBKT7 also failed. To circumvent this toxicity problem, the tBRCT domains of MDC1 and TP53BP1 were subcloned into pGBT9, which has proven to be less toxic in Y2H studies (Clontech). We conducted these two screens in triple drop out medium (SD-Trp-Leu-His). This strategy led to the identification of a few clones with a higher percentage of false positives than those for the tBRCTs tested in more stringent conditions (fig. S5B).
The AH109 transformants containing the bait alone were mated to Y187 [MATα] strain containing a pre-transformed human testes cDNA library (Clontech) and incubated for eight days. We conducted screens with the seven tBRCT domains in parallel to increase the likelihood that the screens would be comparable (each bait was mated to an aliquot of the same pre-transformed library; MDC1 and ECT2 were screened twice). In most of the screens, >1 x 106 transformants were screened (fig. S5B). Yeast miniprep DNA was amplified by PCR and sequenced. The frame of each sequence was identified using the fusion protein sequence as the reference. Each sequence was then analyzed using BLASTp and in-frame clones were then subjected to further bioinformatics analysis.
The screening of a human mammary gland cDNA library using full-length human CHEK2 was previously described (46). We identified a clone corresponding to full-length COMMD1 that was also positive in when isolated plasmids were re-transformed and tested for growth in selective medium. Fine mapping of the interaction site in the two proteins was done by fusing full-length CHEK2 and deletions mutants (aa 1–543; 20–75; 115–165; 225–543; 143–390) to GAL4 DBD in the pGBKT7 vector and full-length human COMMD1 and deletion mutants (aa 1–190; 1–76; 60–140; 126–190) to GAL4AD in the pACT2 vector. CHEK2 constructs were co-transformed with COMMD1 constructs in AH109 in triple drop out medium (SD-Trp-Leu-His).
Antibodies, Cell Lines, and RNAi
Antibody recognizing CBP (GenScript); antibodies recognizing GAL4 DBD, RICTOR, mTOR, phospho(Ser2448)mTOR, -phospho(Thr412/Thr398)-p70S6K I & II, and total AKT (Millipore); antibodies recognizing XIAP, phospho473-AKT, and RAPTOR (Cell Signaling); antibody recognizing COMMD1 (Abnova); antibody recognizing BRCA1 (Ab-1) (Calbiochem); antibody recognizing GST (Pharmacia Biotech); antibody recognizing TUBULIN (Sigma).
HEK 293FT (Invitrogen) and Hela (ATCC) cells were grown in DMEM supplemented with 10% fetal bovine serum, 50 U/mL penicillin, and 50 μg/mL streptomycin (Invitrogen). MDA-MB-231 (ATCC) cells were grown in RPMI supplemented with 10% fetal bovine serum, 50U/mL penicillin, and 50 μg/mL streptomycin. MCF-10A (ATCC) cells were grown in DMEM/F12 supplemented with 5 % horse serum, 20 ng/mL EGF, 0.5 μg/mL hydrocortisone, 100 ng/mL cholera toxin, 10 μg/mL Insulin, 50 U/mL penicillin, and 50 μg/mL streptomycin (86).
Lentiviral shRNA constructs in pLK0.1 targeting BRCA1 and COMMD1, as well as non-targeting scrambled control, were purchased from Open Biosystems. Viral particles were generated in 293FT cells using the ViraPower system (Invitrogen). Target cells were transduced with virus then 48 hours later selected on 1 μg/mL puromycin for 10–14 days followed by the indicated treatments.
Immunoprecipitations, Kinase and Phosphatase Assays
Immunoprecipitations were carried out in CHAPS lysis buffer (0.5% CHAPS, 150 mM NaCl, 10 mM HEPES pH 7.4). To 1 mg whole cell lysates, 5 μg antibody recognizing BRCA1 or 2 μg antibody recognizing RICTOR were added and immune complexes were allowed to form by incubating for 120 min at 4° C, after which 20 μl Protein A/G Sepharose (Santa Cruz) was added as a 50% slurry in CHAPS lysis buffer and then the samples were incubated for another 1 hr with rotation. Immune complexes were washed four times with CHAPS lysis buffer then analyzed by Western blot with the indicated antibodies. Immune complexes used in kinase assays were washed an additional time with RICTOR-mTOR kinase buffer (25 mM HEPES pH 7.5, 100 mM potassium acetate, 1 mM MgCl2), as described previously (87). Briefly, RICTOR immune complexes were incubated in 15 μl kinase buffer containing 500 ng recombinant unmodified AKT (Abcam) and 500 μM ATP for 20 min at 37° C. The reaction was then centrifuged and a 7 μl aliquot was taken and added to 43 μl Laemmli buffer of which 5 μl was analyzed by Western blot. The immune complexes were then washed an additional two times with CHAPS lysis buffer then boiled in 50 μl laemmli buffer of which 5 μl was analyzed by Western blot.
For phosphatase assays, 293FT cells were grown to 75% confluence then treated with 20 Gy IR. 1 mg whole cell lysate (0.5% CHAPS, 150 mM NaCl, 10 mM HEPES pH 7.5) was used in the immunoprecipitation with the Ab1 antibody recognizing BRCA1. The immunoprecipitation was washed 3X in lysis buffer then incubated with 800 U lambda phosphatase in 1X buffer (NEB) both with and without 50 mM EDTA to inhibit the enzyme for 30 min at 30°C. The beads were then washed an additional time with 1mL lysis buffer, boiled in Laemlli buffer, and analyzed by Western blot.
Gene Ontology Clustering
Each tBRCT dataset was reduced to its individual detection method (7X BRCT LIT, TAP-MS, Y2H). These datasets were analyzed using WebGestalt to determine enrichment of GO terms. The top GO term was taken for each screen for all datasets, if there was significant enrichment, and compared to the other tBRCT datasets from the corresponding screen. A term was considered enriched if it was calculated to have a Benjamini and Hochberg adjusted hypergeometric p-value less than 0.05. A p-value of 0.05 was used if no significant enrichment was present. The p-values were then -log10 transformed and clustering analysis was performed with the PermutMatrix software using Pearson distance, McQuitty’s method (WPGMA) for hierarchical clustering, and Multiple-fragment heuristic seriation for both columns and rows (88).
Phenoclusters
A master list with 679 unique experimentally identified genes (from 405 and 292 identified by TAP and Y2H, respectively) including the tBRCT baits but removing redundant genes was converted to Ensembl names using Clone/Gene ID Converter (http://idconverter.bioinfo.cnio.es/). The list was manually curated to resolve inconsistencies and multiple Ensembl entries for the same Entrez Gene name. The Mitocheck phenotype database (20,921 genes), which scores 22 mitosis-related phenotypes in a binary form (presence = 1; absence = 0) from RNA interference screens was downloaded from http://www.mitocheck.org/ as a tab-delimited file in which genes are represented in rows and phenotypes in columns. Only 116 experimentally identified genes (from 679) in which knockdown leads to at least one phenotype were retained.
For each bait set (all proteins that interact with each BRCT bait), the percentage of all genes in the set that displayed each phenotype was obtained. The percentage was then divided by the percentage of genes in the complete Mitocheck database with that phenotype to obtain an enrichment ratio. Enrichment ratios that had a 0.0 value were replaced by 0.2 (half of the lowest non-zero value, 0.4) in the complete set. To assess the significance of this approach, we conducted heterogeneity analysis on the raw counts collapsing the data (TP53BP1, MDC1, and PAXIP1 clustering together and ETC2, BARD1, BRCA1, and LIG4 clustering together with ETC2 as the ‘outlier’ of the second cluster). Collapsing the data, the test for independence yields p=0.073, which we considered suggestive evidence for the validity of this approach. Bait sets were clustered with Cluster 3.0 using Correlation (centered) metric of similarity with no filtering but log2-transformed to depict increase and decrease changes as numerically equal but with opposite sign. The clustering method chosen was complete linkage. It was visualized using Java TreeView v 1.1.rr2.
Network Generation and Analysis
Network graphics were generated with Cytoscape version 2.7 (82). Where indicated, protein-protein interactions were imported using BisoGenet version 1.41 from the SysBiomics database, which integrates data from BIOGRID, INTACT, MINT, DIP, BIND, and HPRD (36). The ClusterONE version 0.91 Cytoscape plug-in was used with the indicated parameters to determine molecular complexes based on imported protein-protein interactions. DDR-regulated phosphoproteins described in two previously reported publications (53, 54) were investigated for overlap between proteins identified in this study’s Merged Network with those of Table S1 from Matsuoka et al. (54) and Table S11 from Bensimon et al. (53)
To create the overlapping disease enrichment network, gene sets for each of the individual tBRCT domains derived from the Merged Network were generated using Metacore (http://portal.genego.com) to determine enriched disease associations (by biomarkers). P-value cutoff was 0.001 (binomial). Disease terms enriched in 2 or more tBRCT gene sets were kept for the analysis.
To identify the association of the tBRCT network with somatic mutations, the complete Cosmic (Catalog of Somatic Mutations in Cancer) database not including fusions (CosmicCompleteExport_v56_151111.tsv) was downloaded from Cosmic FTP server (ftp://ftp.sanger.ac.uk/pub/CGP/cosmic/data_export/). Every analysis was performed on three datasets: unfiltered, filter A (removing unknown and silent mutations), and filter B (removing unknown, silent, and missense mutations).
Validation of Y2H interactions
To estimate the rate of false positives, DNAs from yeast containing positive interactions were isolated by minipreps and a PCR reaction was performed using the following oligonucleotide primers targeting pACT2 vector sequences, which contain the attb sites: 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTCTTACCCATACGATGTTCCAGA-3′ and 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCGGGGTTTTTCAGTATCTACGA-3′. PCR products were cloned into pDONR221 then subsequently into pDEST27, producing an N-terminal GST-fusion, using Gateway recombination cloning (Invitrogen). 293FT cells were cotransfected with the indicated constructs using the calcium phosphate method. For the Streptavidin pull-down panels, cell lysates were collected 24 hours post transfection in 1% CHAPS lysis buffer (1% CHAPS, 150 mM NaCl, 10 mM HEPES pH 7.4 with protease and phosphatase inhibitors), followed by affinity purification of the TAP-tagged tBRCT constructs using streptavidin-conjugated agarose beads, which were washed 4 times with 1% CHAPS lysis buffer then analyzed by Western blot with an antibody recognizing GST to assess the interaction.
For the glutathione pull-down panels, cells were transfected as above in Phosphatase Lysis Buffer (PLB) (1% CHAPS, 100 mM NaCl, 10 mM MgCl2, 10 mM HEPES pH 7.0, 1 mM DTT, and protease inhibitors). GST-tagged proteins were pulled down from 2 mg whole cell lysates using 40 μL Glutathione Sepharose 4B beads (GE Bioscience) and washed 3 times with 500 μL PLB. On the last washing step, the beads were equally aliquoted into 2 tubes, spun down, and supernatant removed. The beads were then incubated in 20 μL PLB containing 10 Units calf intestinal phosphatase (CIP) (New England BioLabs) with and without 50 mM EDTA and incubated at 37°C for 15 minutes. The supernatant was then removed and the beads were washed once more with 500 μL PLB followed by Western blot analysis of tBRCT protein binding using an antibody recognizing the CBP tag. As a control for phosphatase activity and the effects of EDTA inhibition, 3 mg of 293FT lysate stimulated with 20 ng/mL EGF was incubated with 3 μg an antibody recognizing AKT and 45 μL protein A/G beads to pull down total AKT. The beads were equally divided between 3 tubes and either left untreated or incubated with phosphatase in the presence and absence of EDTA.
Caspase-3 activity and MTS assays
Breast cancer cell lines were treated as indicated then lysed in 1% CHAPS lysis buffer (1% CHAPS, 150 mM NaCl, 10 mM HEPES pH 7.4). Caspase-3 enzymatic activity assays measure cellular induction of apoptosis represented by hydrolysis of the peptide substrate acetyl-Asp-Glu-Val-Asp-7-amido-4-methylcoumarin (Ac-DEVD-AMC) by caspase-3, resulting in the release of fluorescent 7-amino-4-methylcoumarin (AMC) using excitation and emission wavelengths of 360 nm and 460 nM, respectively. 50 μg whole cell lysates were used to determine caspase-3 induction measured as DEVDase enzymatic activity with the caspase-3 fluorescence assay kit (Sigma). Cell viability assays were performed using the MTS [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium] assay in 96-well format (Promega).
Cellular Localization and Tandem Affinity Purification
To confirm nuclear expression of the tBRCTs for further TAP experiments, tBRCT domain inserts were subcloned into the pEGFP-C2 vector. The pEGFP-C2 vectors containing the subcloned inserts were transiently transfected into Hela cells using Fugene 6 (Roche). Twenty four hours post transfection the cells were treated with gamma irradiation (10 Gy) or mock treated. Subcellular compartment localization was then assessed 1 hour post-irradiation. Fluorescence microscopy was performed with a Nikon Eclipse TE2000-U inverted microscope.
The tBRCT constructs were subcloned into pNTAP. Tandem affinity purification was used to guarantee high efficiency and specificity (89). Log growing 293FT cells were transfected using the calcium phosphate method with the TAP-tagged tBRCTs or control TAP-tagged GFP vectors (Fig. 2D). Approximately 1 X 108 cells were used for the purification of TAP-tagged complexes using the InterPlay TAP purification kit (Stratagene) with the exception of the lysis buffer, which was changed to NETN buffer (0.5% (v/v) Nonidet P-40, 20 mM Tris (pH 8.0), 50 mM NaCl, 50 mM NaF, 100 μM Na3VO4, 1 mM DTT, and 50 μg/ml PMSF). Lysates were cleared by centrifugation at 13,000g for 5 min at 4° C. All wash steps were performed with 1 mL of NETN three times. After the final elution, the supernatant was boiled for 5 min at 95° C with Laemmli buffer then run on a SDS-PAGE (Bio-Rad).
Mass Spectrometry
Protein fractions (n = 5) created by SDS-PAGE were excised, destained, reduced with tris-carboxyethylphosphine, alkylated with iodoacetamide, and digested overnight with sequencing grade trypsin (Promega, Madison, WI). Tryptic peptides were eluted from the gel and concentrated to 20 ml using vacuum centrifugation. A nanoflow liquid chromatograph (U3000, Dionex, Sunnyvale, CA) coupled to an electrospray ion trap mass spectrometer (LTQ or LTQ-Orbitrap, Thermo, San Jose, CA) was used for tandem mass spectrometry peptide sequencing experiments. Samples were first loaded onto a pre-column (5 mm x 300 mm ID, C18 PepMap100, Dionex) and washed for 8 min with aqueous 2% acetonitrile and 0.04% trifluoroacetic acid. Using a flow rate of 300 nl/min, the trapped peptides were eluted onto the analytical column (75 μm ID x 15 cm, C18 PepMap 100, Dionex). The 60-minute gradient program began at 95% solvent A (aqueous 2% acetonitrile/0.1% formic acid) for 8 min; solvent B (aqueous 90% acetonitrile/0.1% formic acid) was ramped from 5% to 50% over 35 min. Then solvent B was ramped from 50% to 90% B over 5 min, followed by washing and re-equilibration of the column. The mass spectrometer cycled through a survey scan and 5 tandem mass spectra collected in a data-dependent manner in the linear ion trap using 60-second exclusion for previously sampled peptide peaks.
Database searches were conducted against human entries in the SwissProt database (v.20090505) using Mascot (Matrix Science, London, UK; version 2.2.04) (90), assuming the digestion enzyme trypsin and allowing as many as 2 missed cleavages. Tandem mass spectra were matched to peptide sequences with a peptide ion mass tolerance of 1.2 Da and fragment ion mass tolerance of 0.80 Da. Oxidation of methionine and carbamidomethylation of cysteine were specified as variable modifications. Assignments were manually verified by inspection of the tandem mass spectra and coalesced into Scaffold reports (v.2.0, available at www.proteomesoftware.com) for statistical analysis and data presentation.
Scaffold (version Scaffold_2_04_00, Proteome Software Inc., Portland, OR) was used to validate MS-MS based peptide and protein identifications. Peptide identifications were accepted if they could be established at greater than 95.0% probability as specified by the Peptide Prophet algorithm (91). Protein identifications were accepted if they could be established at greater than 50.0% probability and contained at least 2 identified peptides. Protein probabilities were assigned by the Protein Prophet algorithm (92). Proteins that contained similar peptides and could not be differentiated based on MS-MS analysis alone were grouped to satisfy the principles of parsimony. Proteins appearing in control GFP purification and in the tBRCT samples were removed from the final datasets.
Supplementary Material
Figure S1. Literature curation PIN of BRCT-containing proteins
Figure S2. GO enrichment for the LIT Network
Figure S3. Enlarged and annotated 7X BRCT LIT Subnetwork
Figure S4. GO enrichment for 7X BRCT LIT Subnetwork
Figure S5. Y2H controls
Figure S6. Enlarged and annotated Y2H Network
Figure S7. Mammalian-based validation of Y2H interactions
Figure S8. GO enrichment for the Y2H Network
Figure S9. The effect of irradiation on the localization of tBRCT domain-containing proteins and protein - protein interactions
Figure S10. Enlarged and annotated TAP-MS Network
Figure S11. GO enrichment for the TAP-MS Network
Figure S12. Validation of interactions by reverse tagging
Figure S13. GO enrichment for the Merged Network
Figure S14. GO enrichment for the Merged Network and PIN clusters
Figure S15. COMMD1 knock down enhances the apoptotic response of breast cancer cells treated with DNA-damaging agents
Figure S16. Comparison of the experimental designs and the top 10 enriched domains in datasets of this study and two others
Figure S17. The tBRCT disease network
Figure S18. Mutational analysis of tBRCT datasets using the COSMIC database
Table S1. Human BRCT domains
Table S2. Literature-curated interactions with BRCT domain-containing proteins
Table S3. GO enrichment for the LIT network
Table S4. tBRCT-mediated interactions
Table S5. GO enrichment for the 7X BRCT LIT subnetwork
Table S6. Mammalian-based validation of interactions detected byY2H assay and sensitivity of the interactions to phosphatase treatment
Table S7. GO enrichment for the Y2H network
Table S8. TAP-MS number of assigned spectra
Table S9. GO enrichment for the TAP-MS network
Table S10. GO enrichment for the Merged Network
Table S11. GO enrichments infrom TAP, Y2H and LIT interaction datasets compared to the BRCT bait alone
Table S12. Genes from the Merged Network associated with enriched GO terms for Biological Processes
Acknowledgments
We thank Sarah Repasky for help with constructs, and members of the Protein Modules Consortium (http://www.proteinmodules.org/) and, Eric Haura, Tom Sellers, Michael Schell, Eric Welsh and the Monteiro Lab for helpful suggestions.
Funding: This work was supported by US Army Medical Research and Materiel Command, National Functional Genomics Center project (W81XWH-08-2-0101; opinions, interpretations, conclusions and recommendations are those of the author(s) and are not necessarily endorsed by the US Army) and an NIH Lung SPORE (P50-CA119997); and supported in part by the Proteomics and the Molecular Genomics cores at the H. Lee Moffitt Cancer Center & Research Institute. N.W. is supported by a Florida Breast Cancer Foundation fellowship. Y.L. is supported by a fellowship from Agency for Science, Technology, and Research (A*STAR), Singapore.
Footnotes
Competing interests: none.
Data and materials availability: The mass spectrometry data associated with this manuscript may be downloaded from ProteomeCommons.org Tranche using the following hash: A3Qfib3l0qvDJLz/Djwgx9MWVj4GX/fi4hRjlJ4QHGBfDrkvDPzZauxF0zowksIjUSSA mfqbMZ4sL4J27YAIbnJmarMAAAAAAAAdsA==.
References and Notes
- 1.Harper JW, Elledge SJ. The DNA damage response: ten years after. Mol Cell. 2007;28:739–745. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 2.Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, Zieger K, Guldberg P, Sehested M, Nesland JM, Lukas C, Orntoft T, Lukas J, Bartek J. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- 3.Gorgoulis VG, Vassiliou LV, Karakaidos P, Zacharatos P, Kotsinas A, Liloglou T, Venere M, Ditullio RA, Jr, Kastrinakis NG, Levy B, Kletsas D, Yoneta A, Herlyn M, Kittas C, Halazonetis TD. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434:907–913. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- 4.Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, Mortimer P, Swaisland H, Lau A, O’Connor MJ, Ashworth A, Carmichael J, Kaye SB, Schellens JH, de Bono JS. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. New England Journal of Medicine. 2009;361:123–134. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 5.Helleday T, Petermann E, Lundin C, Hodgson B, Sharma RA. DNA repair pathways as targets for cancer therapy. Nat Rev Cancer. 2008;8:193–204. doi: 10.1038/nrc2342. [DOI] [PubMed] [Google Scholar]
- 6.Pawson T, Nash P. Assembly of cell regulatory systems through protein interaction domains. Science. 2003;300:445–452. doi: 10.1126/science.1083653. [DOI] [PubMed] [Google Scholar]
- 7.Bhattacharyya RP, Remenyi A, Yeh BJ, Lim WA. Domains, motifs, and scaffolds: the role of modular interactions in the evolution and wiring of cell signaling circuits. Annu Rev Biochem. 2006;75:655–680. doi: 10.1146/annurev.biochem.75.103004.142710. [DOI] [PubMed] [Google Scholar]
- 8.Manke IA, Lowery DM, Nguyen A, Yaffe MB. BRCT repeats as phosphopeptide-binding modules involved in protein targeting. Science. 2003;302:636–639. doi: 10.1126/science.1088877. [DOI] [PubMed] [Google Scholar]
- 9.Yu X, Chini CC, He M, Mer G, Chen J. The BRCT domain is a phospho-protein binding domain. Science. 2003;302:639–642. doi: 10.1126/science.1088753. [DOI] [PubMed] [Google Scholar]
- 10.Lim WA, Pawson T. Phosphotyrosine signaling: evolving a new cellular communication system. Cell. 2010;142:661–667. doi: 10.1016/j.cell.2010.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Birge RB, Hanafusa H. Closing in on SH2 specificity. Science. 1993;262:1522–1524. doi: 10.1126/science.7504323. [DOI] [PubMed] [Google Scholar]
- 12.Mohammad DH, Yaffe MB. 14-3-3 proteins, FHA domains and BRCT domains in the DNA damage response. DNA Repair (Amst) 2009;8:1009–1017. doi: 10.1016/j.dnarep.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koonin EV, Altschul SF, Bork P. BRCA1 protein products … Functional motifs … [letter] Nat Genet. 1996;13:266–268. doi: 10.1038/ng0796-266. [DOI] [PubMed] [Google Scholar]
- 14.Callebaut I, Mornon JP. From BRCA1 to RAP1: a widespread BRCT module closely associated with DNA repair. FEBS Letters. 1997;400:25–30. doi: 10.1016/s0014-5793(96)01312-9. [DOI] [PubMed] [Google Scholar]
- 15.Bork P, Hofmann K, Bucher P, Neuwald AF, Altschul SF, Koonin EV. A superfamily of conserved domains in DNA damage-responsive cell cycle checkpoint proteins. FASEB J. 1997;11:68–76. [PubMed] [Google Scholar]
- 16.Mesquita RD, Woods NT, Seabra-Junior ES, Monteiro AN. Tandem BRCT domains: DNA’s Praetorian Guard. Genes Cancer. 2010;1:1140–1146. doi: 10.1177/1947601910392988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodriguez M, Yu X, Chen J, Songyang Z. Phosphopeptide binding specificities of BRCA1 COOH-terminal (BRCT) domains. Journal of Biological Chemistry. 2003;278:52914–52918. doi: 10.1074/jbc.C300407200. [DOI] [PubMed] [Google Scholar]
- 18.Mirkovic N, Marti-Renom MA, Weber BL, Sali A, Monteiro AN. Structure-based assessment of missense mutations in human BRCA1: implications for breast and ovarian cancer predisposition. Cancer Research. 2004;64:3790–3797. doi: 10.1158/0008-5472.CAN-03-3009. [DOI] [PubMed] [Google Scholar]
- 19.Joo WS, Jeffrey PD, Cantor SB, Finnin MS, Livingston DM, Pavletich NP. Structure of the 53BP1 BRCT region bound to p53 and its comparison to the Brca1 BRCT structure. Genes and Development. 2002;16:583–593. doi: 10.1101/gad.959202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Botuyan MV, Nomine Y, Yu X, Juranic N, Macura S, Chen J, Mer G. Structural basis of BACH1 phosphopeptide recognition by BRCA1 tandem BRCT domains. Structure (Camb) 2004;12:1137–1146. doi: 10.1016/j.str.2004.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Williams RS, Lee MS, Hau DD, Glover JN. Structural basis of phosphopeptide recognition by the BRCT domain of BRCA1. Nat Struct Mol Biol. 2004;11:519–525. doi: 10.1038/nsmb776. [DOI] [PubMed] [Google Scholar]
- 22.Clapperton JA, Manke IA, Lowery DM, Ho T, Haire LF, Yaffe MB, Smerdon SJ. Structure and mechanism of BRCA1 BRCT domain recognition of phosphorylated BACH1 with implications for cancer. Nat Struct Mol Biol. 2004;11:512–518. doi: 10.1038/nsmb775. [DOI] [PubMed] [Google Scholar]
- 23.Shiozaki EN, Gu L, Yan N, Shi Y. Structure of the BRCT repeats of BRCA1 bound to a BACH1 phosphopeptide: implications for signaling. Mol Cell. 2004;14:405–412. doi: 10.1016/s1097-2765(04)00238-2. [DOI] [PubMed] [Google Scholar]
- 24.Ghosh A, Shuman S, Lima CD. The structure of Fcp1, an essential RNA polymerase II CTD phosphatase. Mol Cell. 2008;32:478–490. doi: 10.1016/j.molcel.2008.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. published online EpubMay. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soutoglou E, Misteli T. Activation of the cellular DNA damage response in the absence of DNA lesions. Science. 2008;320:1507–1510. doi: 10.1126/science.1159051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sprinzak E, Sattath S, Margalit H. How reliable are experimental protein-protein interaction data? J Mol Biol. 2003;327:919–923. doi: 10.1016/s0022-2836(03)00239-0. [DOI] [PubMed] [Google Scholar]
- 28.Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci USA. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giot L, Bader JS, Brouwer C, Chaudhuri A, Kuang B, Li Y, Hao YL, Ooi CE, Godwin B, Vitols E, Vijayadamodar G, Pochart P, Machineni H, Welsh M, Kong Y, Zerhusen B, Malcolm R, Varrone Z, Collis A, Minto M, Burgess S, McDaniel L, Stimpson E, Spriggs F, Williams J, Neurath K, Ioime N, Agee M, Voss E, Furtak K, Renzulli R, Aanensen N, Carrolla S, Bickelhaupt E, Lazovatsky Y, DaSilva A, Zhong J, Stanyon CA, Finley RL, Jr, White KP, Braverman M, Jarvie T, Gold S, Leach M, Knight J, Shimkets RA, McKenna MP, Chant J, Rothberg JM. A Protein Interaction Map of Drosophila melanogaster. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- 30.Goehler H, Lalowski M, Stelzl U, Waelter S, Stroedicke M, Worm U, Droege A, Lindenberg KS, Knoblich M, Haenig C. A Protein Interaction Network Links GIT1, an Enhancer of Huntingtin Aggregation, to Huntington’s Disease. Molecular Cell. 2004;15:853–865. doi: 10.1016/j.molcel.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 31.Li S, Armstrong CM, Bertin N, Ge H, Milstein S, Boxem M, Vidalain PO, Han JD, Chesneau A, Hao T, Goldberg DS, Li N, Martinez M, Rual JF, Lamesch P, Xu L, Tewari M, Wong SL, Zhang LV, Berriz GF, Jacotot L, Vaglio P, Reboul J, Hirozane-Kishikawa T, Li Q, Gabel HW, Elewa A, Baumgartner B, Rose DJ, Yu H, Bosak S, Sequerra R, Fraser A, Mango SE, Saxton WM, Strome S, Van Den HS, Piano F, Vandenhaute J, Sardet C, Gerstein M, Doucette-Stamm L, Gunsalus KC, Harper JW, Cusick ME, Roth FP, Hill DE, Vidal M. A map of the interactome network of the metazoan C. elegans. Science. 2004;303:540–543. doi: 10.1126/science.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutilier K, Yang L, Wolting C, Donaldson I, Schandorff S, Shewnarane J, Vo M, Taggart J, Goudreault M, Muskat B, Alfarano C, Dewar D, Lin Z, Michalickova K, Willems AR, Sassi H, Nielsen PA, Rasmussen KJ, Andersen JR, Johansen LE, Hansen LH, Jespersen H, Podtelejnikov A, Nielsen E, Crawford J, Poulsen V, Sorensen BD, Matthiesen J, Hendrickson RC, Gleeson F, Pawson T, Moran MF, Durocher D, Mann M, Hogue CW, Figeys D, Tyers M. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 33.Liu Y, Woods NT, Kim D, Sweet M, Monteiro AN, Karchin R. Yeast two-hybrid junk sequences contain selected linear motifs. Nucleic Acids Res. 2011;39:e128. doi: 10.1093/nar/gkr600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aloy P, Russell RB. The third dimension for protein interactions and complexes. Trends Biochem Sci. 2002;27:633–638. doi: 10.1016/s0968-0004(02)02204-1. [DOI] [PubMed] [Google Scholar]
- 35.von Mering C, Krause R, Snel B, Cornell M, Oliver SG, Fields S, Bork P. Comparative assessment of large-scale data sets of protein-protein interactions. Nature. 2002;417:399–403. doi: 10.1038/nature750. [DOI] [PubMed] [Google Scholar]
- 36.Martin A, Ochagavia ME, Rabasa LC, Miranda J, Fernandez-de-Cossio J, Bringas R. BisoGenet: a new tool for gene network building, visualization and analysis. BMC Bioinformatics. 2010;11:91. doi: 10.1186/1471-2105-11-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang B, Matsuoka S, Ballif BA, Zhang D, Smogorzewska A, Gygi SP, Elledge SJ. Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science. 2007;316:1194–1198. doi: 10.1126/science.1139476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Greenberg RA, Sobhian B, Pathania S, Cantor SB, Nakatani Y, Livingston DM. Multifactorial contributions to an acute DNA damage response by BRCA1/BARD1-containing complexes. Genes and Development. 2006;20:34–46. doi: 10.1101/gad.1381306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu X, Chen J. DNA damage-induced cell cycle checkpoint control requires CtIP, a phosphorylation-dependent binding partner of BRCA1 C-terminal domains. Mol Cell Biol. 2004;24:9478–9486. doi: 10.1128/MCB.24.21.9478-9486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Neumann B, Walter T, Heriche JK, Bulkescher J, Erfle H, Conrad C, Rogers P, Poser I, Held M, Liebel U, Cetin C, Sieckmann F, Pau G, Kabbe R, Wunsche A, Satagopam V, Schmitz MH, Chapuis C, Gerlich DW, Schneider R, Eils R, Huber W, Peters JM, Hyman AA, Durbin R, Pepperkok R, Ellenberg J. Phenotypic profiling of the human genome by time-lapse microscopy reveals cell division genes. Nature. 2010;464:721–727. doi: 10.1038/nature08869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Burstein E, Ganesh L, Dick RD, van De Sluis B, Wilkinson JC, Klomp LW, Wijmenga C, Brewer GJ, Nabel GJ, Duckett CS. A novel role for XIAP in copper homeostasis through regulation of MURR1. EMBO J. 2004;23:244–254. doi: 10.1038/sj.emboj.7600031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van de Sluis B, Muller P, Duran K, Chen A, Groot AJ, Klomp LW, Liu PP, Wijmenga C. Increased Activity of Hypoxia-Inducible Factor 1 Is Associated with Early Embryonic Lethality in Commd1 Null Mice. Molecular and Cellular Biology. 2007;27:4142–4156. doi: 10.1128/MCB.01932-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maine GN, Mao X, Komarck CM, Burstein E. COMMD1 promotes the ubiquitination of NF-kappaB subunits through a cullin-containing ubiquitin ligase. EMBO J. 2007;26:436–447. doi: 10.1038/sj.emboj.7601489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kennedy RD, Quinn JE, Mullan PB, Johnston PG, Harkin DP. The role of BRCA1 in the cellular response to chemotherapy. JNCI Cancer Spectrum. 2004;96:1659–1668. doi: 10.1093/jnci/djh312. [DOI] [PubMed] [Google Scholar]
- 45.Mufti AR, Burstein E, Csomos RA, Graf PC, Wilkinson JC, Dick RD, Challa M, Son JK, Bratton SB, Su GL, Brewer GJ, Jakob U, Duckett CS. XIAP Is a copper binding protein deregulated in Wilson’s disease and other copper toxicosis disorders. Mol Cell. 2006;21:775–785. doi: 10.1016/j.molcel.2006.01.033. [DOI] [PubMed] [Google Scholar]
- 46.Freeman AK, Dapic V, Monteiro AN. Negative regulation of CHK2 activity by protein phosphatase 2A is modulated by DNA damage. Cell Cycle. 2010;9:736–747. doi: 10.4161/cc.9.4.10613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 48.Sarbassov DD, Ali SM, Kim DH, Guertin DA, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol. 2004;14:1296–1302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- 49.Pearce LR, Huang X, Boudeau J, Pawlowski R, Wullschleger S, Deak M, Ibrahim AF, Gourlay R, Magnuson MA, Alessi DR. Identification of Protor as a novel Rictor-binding component of mTOR complex-2. Biochem J. 2007;405:513–522. doi: 10.1042/BJ20070540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ma Y, Hu C, Riegel AT, Fan S, Rosen EM. Growth factor signaling pathways modulate BRCA1 repression of estrogen receptor-alpha activity. Mol Endocrinol. 2007;21:1905–1923. doi: 10.1210/me.2006-0397. [DOI] [PubMed] [Google Scholar]
- 51.Xiang T, Ohashi A, Huang Y, Pandita TK, Ludwig T, Powell SN, Yang Q. Negative Regulation of AKT Activation by BRCA1. Cancer Res. 2008;68:10040–10044. doi: 10.1158/0008-5472.CAN-08-3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tomlinson GE, Chen TT, Stastny VA, Virmani AK, Spillman MA, Tonk V, Blum JL, Schneider NR, Wistuba II, Shay JW, Minna JD, Gazdar AF. Characterization of a breast cancer cell line derived from a germ-line BRCA1 mutation carrier. Cancer Res. 1998;58:3237–3242. [PubMed] [Google Scholar]
- 53.Bensimon A, Schmidt A, Ziv Y, Elkon R, Wang SY, Chen DJ, Aebersold R, Shiloh Y. ATM-dependent and -independent dynamics of the nuclear phosphoproteome after DNA damage. Sci Signal. 2010;3:rs3. doi: 10.1126/scisignal.2001034. 3/151/rs3. [DOI] [PubMed] [Google Scholar]
- 54.Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, III, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, Shiloh Y, Gygi SP, Elledge SJ. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 55.Hornbeck PV, Kornhauser JM, Tkachev S, Zhang B, Skrzypek E, Murray B, Latham V, Sullivan M. PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 2012;40:D261–270. doi: 10.1093/nar/gkr1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu BA, Jablonowski K, Raina M, Arce M, Pawson T, Nash PD. The human and mouse complement of SH2 domain proteins-establishing the boundaries of phosphotyrosine signaling. Mol Cell. 2006;22:851–868. doi: 10.1016/j.molcel.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 57.Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141:1117–1134. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Scholtens D, Vidal M, Gentleman R. Local modeling of global interactome networks. Bioinformatics. 2005;21:3548–3557. doi: 10.1093/bioinformatics/bti567. [DOI] [PubMed] [Google Scholar]
- 59.Boulton SJ, Gartner A, Reboul J, Vaglio P, Dyson N, Hill DE, Vidal M. Combined Functional Genomic Maps of the C. elegans DNA Damage Response. Science. 2002;295:127–131. doi: 10.1126/science.1065986. [DOI] [PubMed] [Google Scholar]
- 60.Tatsumoto T, Xie X, Blumenthal R, Okamoto I, Miki T. Human ECT2 is an exchange factor for Rho GTPases, phosphorylated in G2/M phases, and involved in cytokinesis. J Cell Biol. 1999;147:921–928. doi: 10.1083/jcb.147.5.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pujana MA, Han JD, Starita LM, Stevens KN, Tewari M, Ahn JS, Rennert G, Moreno V, Kirchhoff T, Gold B, Assmann V, Elshamy WM, Rual JF, Levine D, Rozek LS, Gelman RS, Gunsalus KC, Greenberg RA, Sobhian B, Bertin N, Venkatesan Nyivi-Guedehoussou K, Sole X, Hernandez P, Lazaro C, Nathanson KL, Weber BL, Cusick ME, Hill DE, Offit K, Livingston DM, Gruber SB, Parvin JD, Vidal M. Network modeling links breast cancer susceptibility and centrosome dysfunction. Nat Genet. 2007;39:1338–1349. doi: 10.1038/ng.2007.2. [DOI] [PubMed] [Google Scholar]
- 62.Taylor IW, Linding R, Warde-Farley D, Liu Y, Pesquita C, Faria D, Bull S, Pawson T, Morris Q, Wrana JL. Dynamic modularity in protein interaction networks predicts breast cancer outcome. Nat Biotechnol. 2009;27:199–204. doi: 10.1038/nbt.1522. [DOI] [PubMed] [Google Scholar]
- 63.Diella F, Haslam N, Chica C, Budd A, Michael S, Brown NP, Trave G, Gibson TJ. Understanding eukaryotic linear motifs and their role in cell signaling and regulation. Front Biosci. 2008;13:6580–6603. doi: 10.2741/3175. [DOI] [PubMed] [Google Scholar]
- 64.Yaffe MB, Leparc GG, Lai J, Obata T, Volinia S, Cantley LC. A motif-based profile scanning approach for genome-wide prediction of signaling pathways. Nat Biotechnol. 2001;19:348–353. doi: 10.1038/86737. [DOI] [PubMed] [Google Scholar]
- 65.Miller ML, Jensen LJ, Diella F, Jorgensen C, Tinti M, Li L, Hsiung M, Parker SA, Bordeaux J, Sicheritz-Ponten T, Olhovsky M, Pasculescu A, Alexander J, Knapp S, Blom N, Bork P, Li S, Cesareni G, Pawson T, Turk BE, Yaffe MB, Brunak S, Linding R. Linear motif atlas for phosphorylation-dependent signaling. Sci Signal. 2008;1:ra2. doi: 10.1126/scisignal.1159433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cusick ME, Klitgord N, Vidal M, Hill DE. Interactome: gateway into systems biology. Human Molecular Genetics. 2005:ddi335. doi: 10.1093/hmg/ddi335. [DOI] [PubMed] [Google Scholar]
- 67.Ghavidel A, Cagney G, Emili A. A skeleton of the human protein interactome. Cell. 2005;122:830–832. doi: 10.1016/j.cell.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 68.Lim J, Hao T, Shaw C, Patel AJ, Szabo G, Rual JF, Fisk CJ, Li N, Smolyar A, Hill DE, Barabasi AL, Vidal M, Zoghbi HY. A protein-protein interaction network for human inherited ataxias and disorders of Purkinje cell degeneration. Cell. 2006;125:801–814. doi: 10.1016/j.cell.2006.03.032. [DOI] [PubMed] [Google Scholar]
- 69.Tong AH, Drees B, Nardelli G, Bader GD, Brannetti B, Castagnoli L, Evangelista M, Ferracuti S, Nelson B, Paoluzi S, Quondam M, Zucconi A, Hogue CW, Fields S, Boone C, Cesareni G. A combined experimental and computational strategy to define protein interaction networks for peptide recognition modules. Science. 2002;295:321–324. doi: 10.1126/science.1064987. [DOI] [PubMed] [Google Scholar]
- 70.Carducci M, Perfetto L, Briganti L, Paoluzi S, Costa S, Zerweck J, Schutkowski M, Castagnoli L, Cesareni G. The protein interaction network mediated by human SH3 domains. Biotechnol Adv. 2012;30:4–15. doi: 10.1016/j.biotechadv.2011.06.012. [DOI] [PubMed] [Google Scholar]
- 71.Meek SE, Lane WS, Piwnica-Worms H. Comprehensive proteomic analysis of interphase and mitotic 14-3-3-binding proteins. J Biol Chem. 2004;279:32046–32054. doi: 10.1074/jbc.M403044200. [DOI] [PubMed] [Google Scholar]
- 72.Pozuelo Rubio M, Geraghty KM, Wong BH, Wood NT, Campbell DG, Morrice N, Mackintosh C. 14-3-3-affinity purification of over 200 human phosphoproteins reveals new links to regulation of cellular metabolism, proliferation and trafficking. Biochem J. 2004;379:395–408. doi: 10.1042/BJ20031797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jin J, Smith FD, Stark C, Wells CD, Fawcett JP, Kulkarni S, Metalnikov P, O’Donnell P, Taylor P, Taylor L, Zougman A, Woodgett JR, Langeberg LK, Scott JD, Pawson T. Proteomic, functional, and domain-based analysis of in vivo 14-3-3 binding proteins involved in cytoskeletal regulation and cellular organization. Curr Biol. 2004;14:1436–1450. doi: 10.1016/j.cub.2004.07.051. [DOI] [PubMed] [Google Scholar]
- 74.Lowery DM, Clauser KR, Hjerrild M, Lim D, Alexander J, Kishi K, Ong SE, Gammeltoft S, Carr SA, Yaffe MB. Proteomic screen defines the Polo-box domain interactome and identifies Rock2 as a Plk1 substrate. EMBO J. 2007;26:2262–2273. doi: 10.1038/sj.emboj.7601683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Boxem M, Maliga Z, Klitgord N, Li N, Lemmens I, Mana M, de LL, Mul JD, van de Peut D, Devos M, Simonis N, Yildirim MA, Cokol M, Kao HL, de Smet AS, Wang H, Schlaitz AL, Hao T, Milstein S, Fan C, Tipsword M, Drew K, Galli M, Rhrissorrakrai K, Drechsel D, Koller D, Roth FP, Iakoucheva LM, Dunker AK, Bonneau R, Gunsalus KC, Hill DE, Piano F, Tavernier J, van den Heuvel S, Hyman AA, Vidal M. A protein domain-based interactome network for C. elegans early embryogenesis. Cell. 2008;134:534–545. doi: 10.1016/j.cell.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, Holm L, Sonnhammer EL, Eddy SR, Bateman A. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sibbald PR, Argos P. Weighting aligned protein or nucleic acid sequences to correct for unequal representation. J Mol Biol. 1990;216:813–818. doi: 10.1016/S0022-2836(99)80003-5. [DOI] [PubMed] [Google Scholar]
- 78.Yamane K, Katayama E, Sugasawa K, Tsuruo T. Retinoblastoma susceptibility protein, Rb, possesses multiple BRCT-Ws, BRCA1 carboxyl-terminus-related W regions with DNA break-binding activity. Oncogene. 2000;19:1982–1991. doi: 10.1038/sj.onc.1203528. [DOI] [PubMed] [Google Scholar]
- 79.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Research. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Veerassamy S, Smith A, Tillier ER. A transition probability model for amino acid substitutions from blocks. J Comput Biol. 2003;10:997–1010. doi: 10.1089/106652703322756195. [DOI] [PubMed] [Google Scholar]
- 81.Cusick ME, Yu H, Smolyar A, Venkatesan K, Carvunis AR, Simonis N, Rual JF, Borick H, Braun P, Dreze M, Vandenhaute J, Galli M, Yazaki J, Hill DE, Ecker JR, Roth FP, Vidal M. Literature-curated protein interaction datasets. Nat Methods. 2009;6:39–46. doi: 10.1038/nmeth.1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Stucki M, Clapperton JA, Mohammad D, Yaffe MB, Smerdon SJ, Jackson SP. MDC1 Directly Binds Phosphorylated Histone H2AX to Regulate Cellular Responses to DNA Double-Strand Breaks. Cell. 2005;123:1213–1226. doi: 10.1016/j.cell.2005.09.038. [DOI] [PubMed] [Google Scholar]
- 84.Xu C, Wu L, Cui G, Botuyan MV, Chen J, Mer G. Structure of a Second BRCT Domain Identified in the Nijmegen Breakage Syndrome Protein Nbs1 and its Function in an MDC1-Dependent Localization of Nbs1 to DNA Damage Sites. Journal of Molecular Biology. 2008;381:361–372. doi: 10.1016/j.jmb.2008.05.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256–268. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- 87.Sarbassov DD, Ali SM, Sengupta S, Sheen JH, Hsu PP, Bagley AF, Markhard AL, Sabatini DM. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol Cell. 2006;22:159–168. doi: 10.1016/j.molcel.2006.03.029. [DOI] [PubMed] [Google Scholar]
- 88.Caraux G, Pinloche S. PermutMatrix: a graphical environment to arrange gene expression profiles in optimal linear order. Bioinformatics. 2005;21:1280–1281. doi: 10.1093/bioinformatics/bti141. [DOI] [PubMed] [Google Scholar]
- 89.Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 90.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 91.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 92.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 93.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Literature curation PIN of BRCT-containing proteins
Figure S2. GO enrichment for the LIT Network
Figure S3. Enlarged and annotated 7X BRCT LIT Subnetwork
Figure S4. GO enrichment for 7X BRCT LIT Subnetwork
Figure S5. Y2H controls
Figure S6. Enlarged and annotated Y2H Network
Figure S7. Mammalian-based validation of Y2H interactions
Figure S8. GO enrichment for the Y2H Network
Figure S9. The effect of irradiation on the localization of tBRCT domain-containing proteins and protein - protein interactions
Figure S10. Enlarged and annotated TAP-MS Network
Figure S11. GO enrichment for the TAP-MS Network
Figure S12. Validation of interactions by reverse tagging
Figure S13. GO enrichment for the Merged Network
Figure S14. GO enrichment for the Merged Network and PIN clusters
Figure S15. COMMD1 knock down enhances the apoptotic response of breast cancer cells treated with DNA-damaging agents
Figure S16. Comparison of the experimental designs and the top 10 enriched domains in datasets of this study and two others
Figure S17. The tBRCT disease network
Figure S18. Mutational analysis of tBRCT datasets using the COSMIC database
Table S1. Human BRCT domains
Table S2. Literature-curated interactions with BRCT domain-containing proteins
Table S3. GO enrichment for the LIT network
Table S4. tBRCT-mediated interactions
Table S5. GO enrichment for the 7X BRCT LIT subnetwork
Table S6. Mammalian-based validation of interactions detected byY2H assay and sensitivity of the interactions to phosphatase treatment
Table S7. GO enrichment for the Y2H network
Table S8. TAP-MS number of assigned spectra
Table S9. GO enrichment for the TAP-MS network
Table S10. GO enrichment for the Merged Network
Table S11. GO enrichments infrom TAP, Y2H and LIT interaction datasets compared to the BRCT bait alone
Table S12. Genes from the Merged Network associated with enriched GO terms for Biological Processes