Abstract
Duchenne muscular dystrophy (DMD) is the most common lethal muscle disorder in children. It is caused by mutations of the dystrophin gene. Adeno-associated virus (AAV)-mediated gene replacement therapy has been actively pursued to treat DMD. However, this promising therapeutic modality has been challenged by the small packaging capacity of the AAV vector. The size of the full-length dystrophin cDNA is >11 kb, while an AAV virus can carry only a 5 kb genome. Innovative high-capacity AAV vectors may offer an opportunity to express the full-length dystrophin coding sequence. Here we describe several sets of tri-AAV vectors for full-length human dystrophin delivery. In each set, the full-length human dystrophin cDNA was split into three fragments and independently packaged into separate recombinant AAV vectors. Each vector was engineered with unique recombination signals for directional recombination. Tri-AAV vectors were coinjected into the tibialis anterior muscle of dystrophin-deficient mdx4cv mice. Thirty-five days after injection, dystrophin expression was examined by immunofluorescence staining. Despite low reconstitution efficiency, full-length human dystrophin was successfully expressed from the tri-AAV vectors. Our results suggest that AAV can be engineered to express an extra-large (up to 15 kb) gene that is approximately three times the size of the wild-type AAV genome. Further optimization of the trivector strategy may expand the utility of AAV for human gene therapy.
Introduction
Duchenne muscular dystrophy (DMD) is caused by mutations in the dystrophin gene. Currently, there is no cure for this relentless muscle disease. Restoring dystrophin expression by gene replacement therapy holds great promise in changing the clinical course and improving the life quality of DMD patients. Among various viral and nonviral gene delivery vectors, adeno-associated virus (AAV) is particularly attractive (Atchison et al., 1965). Wild-type AAV is nonpathogenic and recombinant AAV vector is weakly immunogenic (Mays and Wilson, 2011; Mingozzi and High, 2011a). Further, AAV is the only gene transfer vector that can effectively transduce all striated muscles in the body in small and large mammals (Bostick et al., 2007; Yue et al., 2008, 2011; Pan et al., 2013). Despite all these advantages, there is a big hurdle for AAV-based DMD gene therapy. The full-length dystrophin coding sequence is approximately 11.2 kb (Koenig et al., 1989). This is far beyond the 5 kb packaging capacity of a single AAV vector (Srivastava et al., 1983; Dong et al., 2010; Lai et al., 2010b). For this reason, the current AAV gene therapy is forced to use the massively truncated micro- or mini-dystrophin genes (Duan, 2011; Lai and Duan, 2012). While these abbreviated genes may offer some help, they are unlikely to accomplish all the functions of the full-length gene. Innovative approaches have to be developed in order to deliver the full-length dystrophin coding sequence with AAV.
We have previously developed a number of dual-vector strategies to double AAV carrying capacity to 10 kb (reviewed by Lai et al., 2010a). However, these vectors still do not have enough room for the full-length dystrophin coding sequence. In dual AAV vectors, an intact expression cassette is split into two parts and each part is separately delivered to the cell by an AAV virion. Gene expression is achieved after intergenome recombination of the two incoming AAV viruses. On the basis of the same logic, one may envision splitting the full-length dystrophin expression cassette into three AAV vectors in the hope of reconstituting the full-length protein upon coinfection and recombination. To explore this possibility, we engineered several independent sets of full-length dystrophin tri-AAV vectors. Basically, the entire full-length human dystrophin expression cassette was split into three fragments and each piece was packaged into an AAV vector. Three AAV vectors were then coinjected into the tibialis anterior (TA) muscle of dystrophin-deficient mdx4cv mice, a murine DMD model with minimal revertant myofibers (Danko et al., 1992). Thirty-five days after injection, we examined dystrophin expression by immunofluorescence staining. Despite a low efficiency of reconstitution, we detected full-length human dystrophin expression in tri-AAV coinfected mdx4cv TA muscles. Our results have provided the proof of principle that AAV can be used to express a super large gene. With additional optimization, we may further improve tri-AAV transduction efficiency and apply it for gene therapy.
Materials and Methods
Animals
All animal experiments were approved by the Animal Care and Use Committee of the University of Missouri and were in accordance with National Institutes of Health guidelines. The original breeding pairs of B6Ros.CgDmdmdx-4Cv/J (mdx4cv) mice were purchased from The Jackson Laboratory. Experimental mice were generated by in-house breeding. Approximately 3-month-old male mdx4cv mice were used in the study. All mice were housed in specific pathogen-free animal care facilities and kept under a 12 hr light (25 lux)/12 hr dark cycle with free access to food and water.
Generation of proviral plasmids
A total of 14 different cis proviral plasmids were used for AAV production (Tables 1 and 2). Eight plasmids were used for generating mini-dystrophin dual AAV vectors, and six were used for generating full-length dystrophin tri-AAV vectors. All the cis plasmids were flanked by the AAV-2 inverted terminal repeats (ITR). In the expression cassette, transcriptional regulation is controlled by the cytomegalovirus (CMV) promoter and the simian virus 40 polyadenylation signal (SV40 pA). The full-length human dystrophin cDNA was used as the template for cloning.
Table 1.
Dual-AAV Constructs Used in the Study
| Name | Description | Reference |
|---|---|---|
| YZ27 | The upstream vector of the R20 overlapping dual-vector set. It contains the CMV promoter, 5′ half of the GFP-fused mini-dystrophin gene (till the end of R20). | Zhang and Duan (2012) and Zhang et al. (2013) |
| YZ22 | The downstream vector of the R20 overlapping dual-vector set. It contains the 3′ half of the GFP-fused mini-dystrophin gene (starting from R20) and the pA signal. | Zhang and Duan (2012) and Zhang et al. (2013) |
| WL40 | The upstream vector of the R7 overlapping dual-vector set. It is similar to YZ27 except that the R20 is replaced by R7. | Fig. 1 (this study) |
| WL43 | The downstream vector of the R7 overlapping dual-vector set. It is similar to YZ22 except that the R20 is replaced by R7. | Fig. 1 (this study) |
| WL41 | The upstream vector of the R8 overlapping dual-vector set. It is similar to YZ27 except that the R20 is replaced by R8. | Fig. 1 (this study) |
| WL44 | The downstream vector of the R8 overlapping dual-vector set. It is similar to YZ22 except that the R20 is replaced by R8. | Fig. 1 (this study) |
| WL42 | The upstream vector of the R9 overlapping dual-vector set. It is similar to YZ27 except that the R20 is replaced by R9. | Fig. 1 (this study) |
| WL45 | The downstream vector of the R9 overlapping dual-vector set. It is similar to YZ22 except that the R20 is replaced by R9. | Fig. 1 (this study) |
AAV, adeno-associated virus; CMV, cytomegalovirus.
All vectors are flanked by AAV inverted terminal repeat.
Table 2.
Tri-AAV Constructs Used in the Study
| Name | Description | Reference |
|---|---|---|
| WL30 | The head vector of the tri-AAV vector set. It contains the CMV promoter, human dystrophin exons 1–26, a splicing donor from Ad41E1a intron, and the highly recombinogenic AP1 sequence. | Figs. 2A, 2B, and 3A (this study) |
| WL34 | The body vector of the hybrid-overlapping (HO) tri-AAV vector set. It contains the AP1 sequence, a splicing acceptor from Ad41E1A intron, human dystrophin exon 27 to the middle of exon 53). | Fig. 2A (this study) |
| WL37 | The tail vector of the HO tri-AAV vector set. It contains exons 50–79, and the pA signal. | Fig. 2A (this study) |
| WL33 | The body vector of the hybrid–hybrid (HH) tri-AAV vector set. It contains the AP1 sequence, a splicing acceptor from Ad41E1A intron, human dystrophin exons 27–48, a synthetic splicing donor from pCI intron, and the highly recombinogenic AP2 sequence. | Figs. 2B and 3A (this study) |
| WL35 | The tail vector of the HH tri-AAV vector set. It contains the AP2 sequence, a synthetic splicing acceptor from pCI intron, human dystrophin exons 49–79, and the pA signal. | Fig. 2B (this study) |
| WL38 | The tail vector of the flagged HH tri-AAV vector set. It is identical to WL35 except for a flag tag fused at the C-terminal end. | Fig. 3A (this study) |
The constructs are listed in the order they appear in the figures. All vectors are flanked by AAV inverted terminal repeat.
The mini-dystrophin dual AAV vectors were used to screen for highly recombinogenic regions in the dystrophin rod domain. Four sets of cis plasmids were used in this study. The first pair of cis plasmids (pYZ27 and pYZ22) has been published before (Zhang and Duan, 2012; Zhang et al., 2013). This set of vectors was originally generated to express a mini-dystrophin gene that is approximately half the size of the full-length dystrophin cDNA. In this pair of vectors, dystrophin spectrin-like repeat 20 (R20) was used to reconstitute mini-dystrophin expression via homologous recombination (Zhang and Duan, 2012). To identify additional highly recombinogenic regions, we replaced R20 with R7, R8, or R9 in pYZ27 and pYZ22 (Table 1). The resulting plasmid sets were (1) pWL40 and pWL43 (in this set of plasmids, R7 is shared as the common region); (2) pWL41 and pWL44 (these two plasmids share R8 as the common region); and (3) pWL42 and pWL45 (this set of plasmids uses R9 for homologous recombination) (Table 1).
A total of six cis plasmids were used to generate split vectors to express the full-length dystrophin coding sequence (Table 2). The head vector (pWL30) carries the 5′ one-third of the full-length cDNA (exons 1–26) (Table 2). Briefly, a 3,548 bp DNA fragment was amplified by polymerase chain reaction (PCR) using the full-length human dystrophin cDNA (a gift from Dr. Jeffrey Chamberlain at the University of Washington) as the template. The forward primer is WL12 (5′-TTTCTCGAGATGCTTTGGTGGGAAGAAGTAGAGG). The underlined nucleotides represent the XhoI site. The reverse primer is WL13 (5′-TTTGCGGCCGCTTTCCGCGGACATCCAGCAGTATAAAACAAAAAAAAAAATCTCTACTTACCTTCATCTCTTCAACTGCTTTCTGTAATTCATCTGG). The underlined nucleotides mark the NotI and SacII sites. The nucleotides in bold font represent the intronic splicing donor signal derived from adenovirus 41 E1A (Ad41E1A) intron. In addition to the dystrophin sequence, pWL30 also contains the AP1 sequence, a ∼0.3 kb highly recombinogenic DNA fragment from the first one-third of the middle part of the human placental alkaline phosphatase (AP) gene cDNA (Supplementary Fig. S1; Supplementary Data are available online at www.liebertpub.com/hum) (Ghosh et al., 2011). The AP1 fragment was generated by PCR using the forward primer WL14 (5′-TTTCTCGAGTTTCCGCGGTCGACCCCCGGGTGCGCGGCGTCGG). The underlined nucleotides mark the XhoI and SacII sites. The reverse primer is WL15 (5′-TTTGCGGCCGCGAAACGGTCCAGGCTATGTGCTCAAGGACGGCGCCCGGCCGG). The underlined nucleotides stand for the NotI site. The SacII/NotI double-digested fragment was then cloned between the CMV promoter and the 3′-ITR of pDD2, a multiple cloning site cis plasmid we published before (Yue and Duan, 2002).
The body vector (pWL33) of the hybrid–hybrid (HH) strategy trivectors was generated by amplifying a 3,678 bp DNA (exons 27–48) fragment from the full-length human dystrophin cDNA (Table 2). The forward primer is WL18 (5′-TTTATGCATTTTGCTAGCCCTAGGTGTGTGGGGGTTAACGTGGCTTTTTTGTGCTTACTAGAGAGAGCTAAAGAAGAGGCCCAACAAAAAGAAGCG). The underlined nucleotides mark the NsiI and NheI sites. The nucleotides in bold font represent intronic acceptor signal from Ad41E1A intron. The reverse primer is WL19 (5′-TTTGGATCCTTTCTCGAGCCTCAGAAACGCAAGAGTCTTCTCTGTCTCGACAAGCCCAGTTTCTATTGGTCTCCTTAAACCTGTCTTGTAACCTTGATACTTACCTGAACGTCAAATGGTCCTTCTTGGTTTGGTTGG). The underlined nucleotides represent the BamHI and XhoI site, respectively, while the nucleotides in bold font represent the intronic donor signal of a synthetic intron from a commercial plasmid pCI (Promega, Fitchburg, WI) (Duan et al., 2001). pWL33 also contains two DNA fragments from different regions of the AP gene. The ∼0.3 kb AP1 fragment is identical to that of pWL30. However, in pWL33 this AP1 fragment was amplified using the forward primer WL16 (5′-TTTATGCATTCGACCCCCGGGTGCGCGGCGTCGG; the underlined nucleotides represent the NsiI site) and the reverse primer WL17 (5′-TTTGGATCCTTTCTCGAGTTTGCTAGCGAAACGGTCCAGGCTATGTGCTCAAGGACGGCGCCCGGCCGG; the underlined nucleotides represent the BamHI, XhoI, and NheI site, respectively). A second highly recombinogenic AP fragment (AP2) was obtained from the last one-third of the middle part of the AP gene (Supplementary Fig. S1) (Ghosh et al., 2011). The AP2 fragment was placed near the 3′-end of pWL33. The AP2 fragment is ∼0.3 kb long, and it was amplified using the forward primer WL20 (5′-TTTATGCATTTTCTCGAGTCGACCGCAGGGCAGCCTCTGTCATCTCCATCAGGGAGGGG; the underlined nucleotides represent the NsiI and XhoI site, respectively) and the reverse primer WL21 (5′-TTTGGATCCATCCTGGAGGCCGAAAGTACATGTTTCGCATGGG; the underlined nucleotides mark the BamHI site). The PCR products including the dystrophin fragment, and two AP fragments were then cloned between the 5′- and 3′-ITR of pDD4 (Yue and Duan, 2002).
The tail vector (pWL35) of the (HH) strategy trivectors was generated by amplifying a 3,864 bp DNA fragment (exons 49–79) using the full-length human dystrophin cDNA as the template (Table 2). The forward primer is WL24 (5′-TTTATGCATTTTTCTAGATAGGCACCTATTGGTCTTACTGACATCCACTTTGCCTTTCTCTCCACAGGAAACTGAAATAGCAGTTCAAGCTAAACAACCGG; the underlined nucleotides represent the XbaI and NsiI sites, respectively; the nucleotides in bold represent the intronic acceptor sequence). The reverse primer is WL25 (5′-TTTGCGGCCGCCTACATTGTGTCCTCTCTCATTGGCTTTCCAGGGG; the underlined nucleotides represent the NotI site). This PCR fragment contains the splicing acceptor signal derived from the pCI intron and the 3′-part of the human dystrophin gene. The 5′-end of the pWL35 plasmid also contains the ∼0.3 kb AP2 sequence. In pWL35, the AP2 sequence was generated by PCR using the forward primer WL22 (5′-TTTATGCATTGCATGCCGCAGGGCAGCCTCTGTCATCTCCATCAGGGAGGGG; the underlined nucleotides represent the NsiI site) and the reverse primer WL23 (5′-TTTGCGGCCGCTTTTCTAGATGGAGGCCGAAAGTACATGTTTCGCATGGGAACCCC; the underlined nucleotides represent NotI and XbaI site, respectively). Finally, the NsiI/NotI double-digested fragment was cloned between 3′-ITR and SV40 pA signal in pDD4 (Yue and Duan, 2002).
An alternative form of the pWL35 plasmid was made in which a flag tag was included at the 3′-end (Table 2). This flag-tagged plasmid is called pWL38. Briefly, pWL38 was generated by amplifying a dystrophin fragment from the full-length human dystrophin cDNA using the forward primer WL24 (see above) and a reverse primer WL29 (5′- TTTGCGGCCGCCTACTTGTCATCGTCATCCTTGTAGTCCTTGTCATCGTCATCCTTGTAGTCCTTGTCATCGTCATCCTTGTAGTCCATTGTGTCCTCTCTCATTGGCTTTCCAGGGG; The underlined nucleotides represent the NotI site and the nucleotides in bold stand for the flag tag).
The body vector (pWL34) of the hybrid-overlapping (HO) strategy trivectors was generated by replacing the 3′-end of pWL33 with a PCR-amplified dystrophin fragment (exon 41 to the middle of exon 53) (Table 2). The forward primer is WL01 (5′-TTTAGCGCTGGCGGGAAATTGAGAGC; the underlined nucleotides represent the AfeI site). The reverse primer is WL26 (5′-TTTGGATCCTCTTCCTTAGCTTCCAGCC; the underlined nucleotides represent the BamHI site).
The tail vector (pWL37) of the (HO) strategy trivectors was generated by replacing the 5′-end of pWL35 with a PCR-amplified dystrophin fragment containing exons 50–59 (Table 2). The full-length dystrophin cDNA was used as the template. The forward primer was WL30 (5′-TTTATGCATTGGAGGTACCTGCTCTGGC; the underlined nucleotides represent the NsiI site) and the reverse primer was WL05 (5′-TTTCTCGAGGTGATCTTGGAGAGAG; the underlined nucleotides represent the XhoI site).
Recombinant AAV production and in vivo gene delivery
Y445F AAV-6 and Y731F AAV-9 tyrosine mutant AAV vectors were used in the study. Recombinant AAV vectors were generated by triple-plasmid cotransfection using a cis plasmid, an AAV helper plasmid containing the AAV-2 Rep gene and the Y445F AAV-6 (or Y731F AAV-9) capsid gene (gifts from Dr. Arun Srivastava at the University of Florida) and an adenovirus helper plasmid (Stratagene, La Jolla, CA) (Zhong et al., 2008; Qiao et al., 2010; Zhang et al., 2013). Viral stocks were purified through two rounds of CsCl isopycnic ultracentrifugation according to our published protocol (Qiao et al., 2010; Shin et al., 2012). For dual-AAV coinfection studies, we used Y445F tyrosine mutant AAV-6. Briefly, 2.25×1010 viral genome (vg) particles of each vector were coinjected into the TA muscle of ∼3-month-old male mdx4cv mice. For triple-AAV coinfection studies, we used both Y445F tyrosine mutant AAV-6 and Y731F tyrosine mutant AAV-9. Y445F tyrosine mutant AAV-6 was used to compare the transduction efficiency of two different sets of the full-length dystrophin tri-AAV vectors (the HH vectors and the hybrid-overlapping vectors). In this experiment, 4×1010 vg particles/vector of each vector set were coinjected into the TA muscle of mdx4cv mice. Y731F tyrosine mutant AAV-9 was used to evaluate transduction efficiency of the flag-tagged HH tri-AAV vectors. In this study, 4.7×1011 (low-dose group) or 1.2×1012 (high-dose group) vg particles of each vector were coinjected into the TA muscle of mdx4cv mice. AAV transduction was evaluated at 35 days postinjection.
Cell culture studies
MO59K cell line, a human glioblastoma cell line, was obtained from American Type Culture Collection (No. CRL-2365; Manassas, VA) (Ghosh et al., 2008). Cells were cultured in a 1:1 mixture of Dulbecco's modified Eagle's medium and Ham's F12 medium (Gibco, Carlsbad, CA). Culture medium was supplemented with 10% fetal bovine serum, 100 unit/ml penicillin, 0.1 mg/ml streptmycin, 0.05 mM nonessential amino acids, and 0.5 mM sodium pyruvate (Gibco). Twenty-four hours after plating, cells were infected with Y445F AAV-6 tyrosine mutant dual AAV vectors at the multiplicity of infection of 10,000 vg particles/vector/cell. Cells were fixed in 4% formaldehyde 48 hr later and examined for GFP expression under a confocal microscope.
Immunostaining and GFP visualization in muscle
Freshly dissected muscles were snap-frozen in liquid nitrogen-cooled isopentane in optimal cutting temperature compound (Sakura Finetek Inc., Torrance, CA). Ten-micrometer cryo-tissue sections were used for GFP visualization or immunofluorescence staining. GFP was detected under the FITC channel using a Nikon E800 fluorescence microscope. Photomicrographs were taken with a Qimage REtiga 1300 camera. Dystrophin was examined with two antibodies—a human dystrophin-specific monoclonal antibody Dys 3 (Hum-Dys antibody) (1:20; Novocastra, Newcastle, UK) and a second antibody (Dys 2) that reacts with dystrophin from any species (Pan-Dys antibody) (1:100; Novocastra). The Dys 3 antibody reacts with an epitope located in dystrophin hinge 1 (Yue et al., 2003; Lai et al., 2005, 2009). The Dys 2 antibody recognizes an epitope located at the C-terminal domain of dystrophin. The flag-tag was examined with an anti-flag M2 antibody (1:500; Sigma-Aldrich, St. Louis, MO).
Quantitative reverse transcription PCR
Quantitative reverse transcription PCR (qRT-PCR) analyses were performed as described previously by Bartoli et al. (2006). Briefly, total RNA was extracted from muscles by the Trizol method (Life Technologies, Carlsbad, CA) from the ground muscle lysate. Residual DNA was removed from the samples using the Free DNA kit (Ambion, Austin, TX). One microgram RNA was reverse-transcribed using random hexamers according to the protocol Superscript II first-strand synthesis system for RT-PCR (Life Technologies). TaqMan quantitative PCR was performed using the ABI PRISM 7700 system (Life Technologies) with 0.2 mM of each primer and 0.1 mM of the probe according to the protocol Absolute QPCR Rox Mix (Thermo Scientific, Pittsburgh, PA). The primers and probes are listed in Table 3 (see also Supplementary Fig. S2). The probes were labeled with 6-carboxyfluorescein at the 5′-end and dihydrocyclopyrroloindole tripeptide minor groove binder at the 3′-end. The primer a/b set and the probe WL145 were used to amplify the transcribed junction of the head and body vectors. The primer c/d set and the probe WL146 were used to amplify the transcribed junction of the body and tail vectors. The primer a/d set and the probe WL147 were used to amplify the transcribed junction of the head and tail vectors (Table 3 and Supplementary Fig. S2). The ubiquitous acidic ribosomal phosphoprotein, large (Rplp0), was used to normalize the data across samples. Each experiment was performed in duplicate.
Table 3.
Primers and Probes Used for Quantitative Reverse Transcription Polymerase Chain Reaction
| Name | Gene/location | Sequence |
|---|---|---|
| Primer a (WL102) | Dystrophin exon 26 (forward primer) | 5′ gattttgaatataaaactccagatgaa |
| Primer b (WL103) | Dystrophin exon 27 (reverse primer) | 5′ ggagtttcactttcgcttcttt |
| Primer c (WL104) | Dystrophin exon 48 (forward primer) | 5′ aaccaaccaaaccaagaagg |
| Primer d (WL105) | Dystrophin exon 49 (reverse primer) | 5′ gctgccctttagacaaaatctc |
| Rplp0 forward primer | Ribosomal protein large P0 | 5′ ctccaagcagatgcagcaga |
| Rplp0 reverse primer | Ribosomal protein large P0 | 5′ atagccttgcgcatcatggt |
| Probe WL145 | The junction of the dystrophin head and body vectors | 5′ agagatgaagagagctaaagaagaggccc |
| Probe WL146 | The junction of the dystrophin body and tail vectors | 5′ tttgacgttcaggaaactgaaatagcag |
| Probe WL147 | The junction of the dystrophin head and tail vectors | 5′ agagatgaaggaaactgaaatagcagtt |
| Rplp0 Probe | Ribosomal protein large P0 | 5′ ccgtggtgctgatgggcaagaa |
Statistical analysis
The sample size is indicated in the corresponding figure legend. Data are presented as mean±standard error of mean unless otherwise stated. Statistical difference for two-group comparisons (Fig. 2D, between HO vectors and HH vectors; Fig. 3C, between flag antibody and Pan-Dys antibody; Fig. 3D, between WL30/33/38 coinfected and noninfected) was assessed with the Student's t-test. Statistical difference for multiple group comparison (Fig. 1D) was examined by one-way ANOVA followed by Bonferroni post hoc analysis using the IBM SPSS software (IBM Corporation, Armonk, NY). A p<0.05 was considered statistically significant.
FIG. 2.
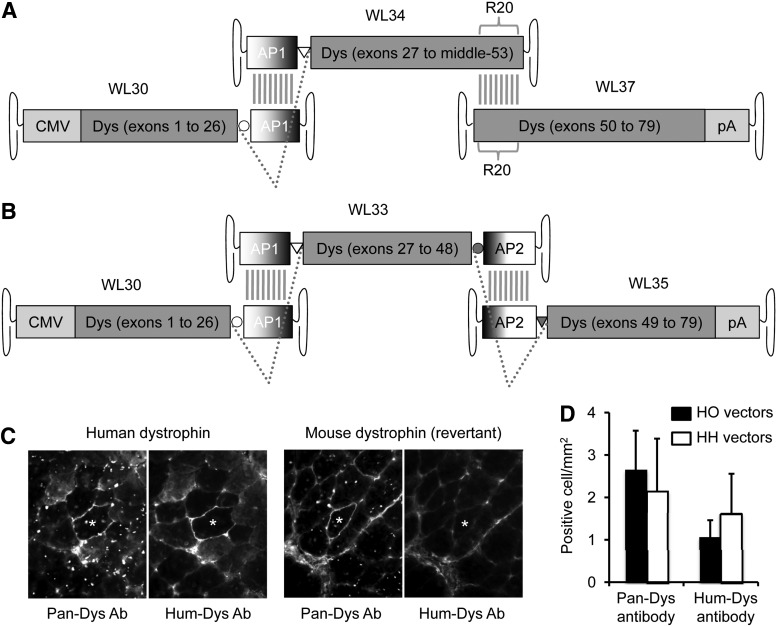
Expression of full-length human dystrophin with two different sets of tri-AAV vectors. (A) Schematic outline of the hybrid-overlapping (HO) tri-AAV vectors. The first (WL30) and the middle (WL34) one-third of the dystrophin coding sequences are split at the junction of exons 26 and 27. A highly recombinogenic alkaline phosphatase gene sequence (AP1) is engineered at the end of the first vector (WL30) and the beginning of the second vector (WL34). Splicing donor (circle) and acceptor (triangle) signals are also engineered in WL30 and WL34, respectively, at the indicated position. Reconstitution of these two vectors can be achieved by either homologous recombination of the AP1 sequence (vertical gray lines) or end-to-end joining of the inverted terminal repeat (ITR). In either case, the open reading frame can be re-established in mRNA after splicing (dotted lines). The recombination between the middle one-third of dystrophin (WL34) and the last one-third of dystrophin (WL37) is mediated by homologous recombination of dystrophin spectrin-like repeat 20 (R20) (vertical gray lines). (B) Schematic outline of the hybrid–hybrid (HH) tri-AAV vectors. The full-length dystrophin expression cassette is carried in three AAV vectors (WL30, WL33, and WL34). The recombination between the first (WL30) and middle (WL33) one-third of dystrophin is identical to that of the HO tri-AAV vectors. A similar approach is used to reconstitute the middle (WL33) and the last (WL35) one-third of dystrophin. The dystrophin coding sequence is split at the junction of exons 48 and 49. A different but also highly recombinogenic region of the AP gene (AP2) and a different set of splicing signals (gray circle and gray triangle) are engineered in WL33 and WL35 to mediate recombination. (C) Representative immunofluorescence staining photomicrographs of serial muscle sections that are stained by two different dystrophin antibodies. The Pan-Dys antibody recognizes dystrophin from any species. The Hum-Dys antibody reacts only with human dystrophin. The two panels at the left show human dystrophin expression in a myofiber (positive for both Pan-Dys and Hum-Dys antibodies). The two panels at the right show expression of mouse dystrophin in a revertant myofiber (positive with the Pan-Dys antibody but negative with the Hum-Dys antibody). *The same myofiber in serial sections. (D) Quantitative evaluation of dystrophin expression in muscles infected by the HO or HH tri-AAV vectors. N=4 for each group.
FIG. 3.
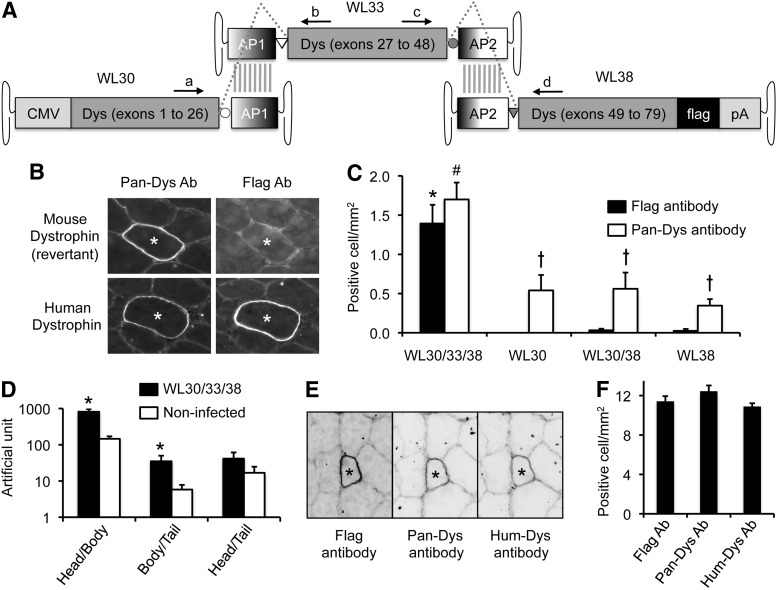
Quantitative examination of the flag-tagged HH tri-AAV vectors in mdx4cv mouse muscle. (A) Schematic outline of the flag-tagged HH tri-AAV vectors. The structure is identical to the original HH tri-AAV vectors except the addition of a flag tag at the C-terminal end of the human dystrophin coding sequence. Locations of the primers used for quantitative RT-PCR are marked (see Table 3 for details). (B) Representative Pan-Dys and flag antibody immunofluorescence staining photomicrographs. Top two panels show a revertant myofiber (positive for Pan-Dys staining but negative for flag staining). *The same myofiber in serial sections. (C) Quantification of human dystrophin reconstitution in mdx4cv mice infected by one, two, or all three vectors of the flag-tagged HH tri-AAV vectors. N=14 for WL30/WL33/WL38 coinfected mice; N=6 for WL30 infected mice; N=8 for WL30/WL38 coinfected mice; N=8 for WL38 infected mice. *Significantly higher than other groups by flag antibody staining. #Significantly higher than other groups by Pan-Dys antibody staining. Cross, significantly higher than that of flag antibody staining in the same group. (D) Quantitative RT-PCR results. The primer set used in each qRT-PCR is indicated in the x-axis. N=14 for tri-AAV infected mice; N=6 for noninfected control mice. *Significantly higher than that of uninfected. (E) Representative flag, Pan-Dys, and Hum-Dys antibody immunofluorescence staining photomicrographs. *The same myofiber in serial sections. (F) Quantification of human dystrophin reconstitution in mdx4cv mice infected with high-dose flag-tagged HH tri-AAV vectors. N=2 mice for each group. The error bar stands for the difference between the mean value and each individual value. RT-PCR, reverse transcription polymerase chain reaction.
FIG. 1.
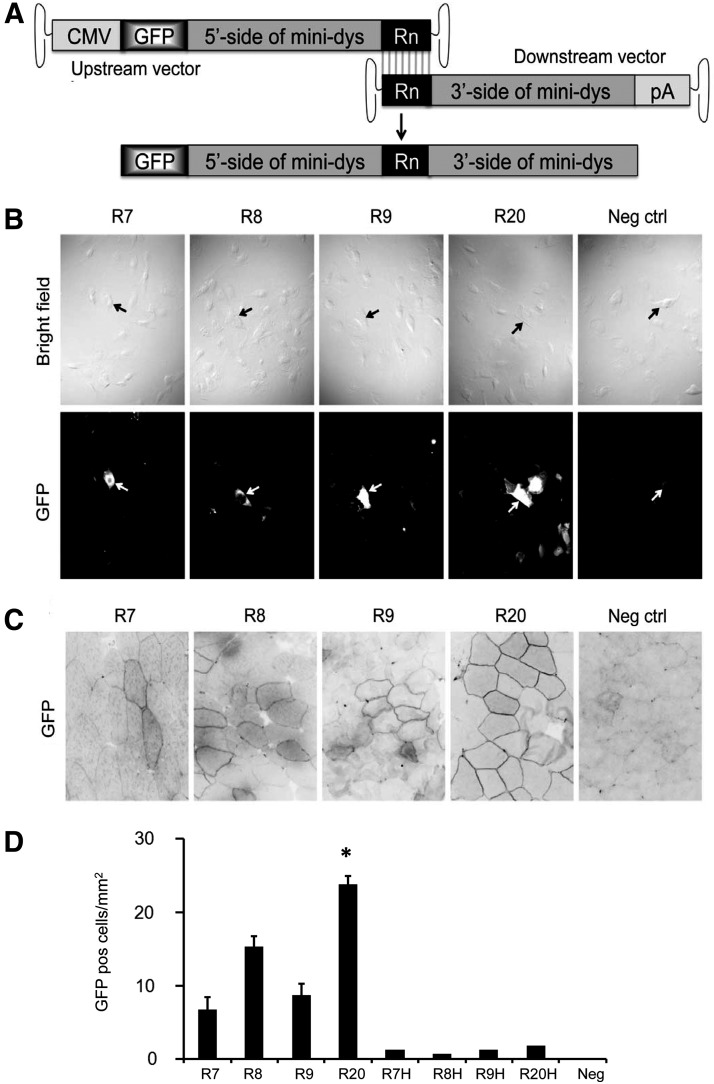
Evaluation of homologous recombination efficiency of dystrophin R7, R8, and R9 in overlapping dual AAV vectors. (A) Schematic outline of the constructs and the overlapping strategy. A 7 kb GFP-fused mini-dystrophin gene is split into two AAV vectors. The upstream vector contains the CMV promoter, the GFP coding sequence, and the 5′-half of the mini-dystrophin gene. The downstream vector contains the 3′-half of the mini-dystrophin gene and the polyadenylation signal. The region shared by both constructs is marked as Rn, which can be R7, R8, R9, or R20. Homologous recombination between the Rn regions of the two vectors reconstitutes the expression of the GFP-fused mini-dystrophin gene. (B) In vitro evaluation of the reconstitution efficiency in MO59K cells. Representative photomicrographs of the bright-field and GFP images are shown in the top and bottom panels, respectively. The dual vectors used in transduction are marked by the homologous region they shared (R7, R8, R9, or R20). Neg ctrl, uninfected negative control. Arrow marks the same cell in the bright-field image and the corresponding GFP fluorescence image. (C) In vivo evaluation of the reconstitution efficiency in mdx4cv mice. Representative photomicrographs of the GFP images from muscles infected by the indicated set of the dual vectors are shown. Neg ctrl, un-infected negative control. (D) Quantitative evaluation of the reconstitution efficiency in mdx4cv mice. N=3–5 mice/group. R7H, R8H, R9H, and R20H refer to the muscles that were injected only with the indicated head vector instead of the paired dual vectors. Neg, the muscle from uninfected mice. *Significantly higher than all other groups. AAV, adeno-associated virus; CMV, cytomegalovirus.
Results
Screening for highly recombinogenic regions in the dystrophin gene
One approach to deliver the full-length dystrophin coding sequence is through homologous recombination (Duan et al., 2001). In this approach, the dystrophin cDNA is split into three overlapping fragments, including the head, body, and tail part. The 3′-end of the head fragment shares a common region with the 5′-end of the body fragment and the 3′-end of the body fragment shares a common region with the 5′-end of the tail fragment. Each fragment is packaged into an AAV vector. After coinfection, recombination at the overlapping regions reconstitutes full-length dystrophin. The success of this approach depends on efficient recombination of the overlapping sequences (Ghosh et al., 2006). Recent studies suggest that dystrophin R20 mediates robust homologous recombination (Odom et al., 2011; Zhang and Duan, 2012; Zhang et al., 2013). We have thus selected R20 for body and tail fragment reconstitution. To reconstitute the head fragment and the body fragment, another highly recombinogenic region is needed. Considering the packaging limitation of a single AAV vector, three candidate regions (R7, R8, and R9) were chosen for further evaluation.
We have previously described a set of mini-dystrophin dual AAV vectors (Zhang and Duan, 2012; Zhang et al., 2013). In this set of the vectors, R20 is used to reconstitute a 7 kb GFP-fused ΔR2–15/ΔR18–19 minigene (Fig. 1A). To determine homologous recombination efficiency of R7, R8, and R9, we swapped each of these regions into the ΔR2–15/ΔR18–19 minigene dual AAV vectors (Fig. 1A and Table 1). We first tested reconstitution efficiency in MO59K cells (Ghosh et al., 2006). Transduction with either upstream or downstream vector alone did not yield GFP expression (data not shown). GFP-positive cells were observed only after coinfection (Fig. 1B). Among four dual-vector sets, the original R20-based dual vectors appeared to be more efficient than the other three sets (Fig. 1B). To determine in vivo reconstitution efficiency, dual vectors were delivered to the TA muscle of adult mdx4cv mice (Fig. 1C and D). At 35 days after gene transfer, we detected GFP expression in muscles coinjected with both the upstream and downstream vectors, but not in muscle injected with a single vector (Fig. 1C and data not shown). Consistent with the in vitro results, R20-based dual vectors yielded significantly more GFP-positive cells (Fig. 1D). None of the candidate regions (R7, R8, and R9) resulted in efficient homologous recombination. Our results suggest that it may not be ideal to reconstitute the head and body fragments using the overlapping strategy.
Design and construction of the full-length dystrophin tri-AAV vectors
The trans-splicing and hybrid methods are two other strategies that are often used to reconstitute a split gene using AAV. The efficiency of the trans-splicing approach is limited by the selection of the splicing site (Lai et al., 2005, 2006). Only the hybrid strategy can mediate efficient reconstitution in a transgene-independent manner (Ghosh et al., 2008, 2011).
To take the advantage of the hybrid vector strategy, we designed two different sets of tri-AAV vectors (Fig. 2A and B, and Table 2). In both sets of the trivectors (Fig. 2A and B), the head vector (WL30) contains the CMV promoter, dystrophin exons 1–26, the splicing donor from Ad41E1a intron, and the highly recombinogenic AP1 sequence (Ghosh et al., 2011). In the hybrid-overlapping (HO) vector set, the body vector (WL34) contains the AP1 sequence, the splicing acceptor from Ad41E1a intron, and dystrophin exon 27 to the middle of exon 53. The tail vector (WL37) of the HO vector set contains dystrophin exons 50–79 and the SV40 polyA signal (Fig. 2A). In the HH vector set, the body vector (WL33) contains (starting from the 5′-end) the AP1 sequence, the splicing acceptor from Ad41E1a intron, dystrophin exons 27–48, the splicing donor of synthetic pCI intron, and the highly recombinogenic AP2 sequence. The tail vector (WL35) of the HH vector set contains the AP2 sequence, the splicing donor of synthetic pCI intron, and the SV40 polyA signal (Fig. 2B).
To evaluate full-length dystrophin reconstitution, we used two antibodies: a Hum-Dys Ab that only recognizes the hinge 1 region at the N-terminal end of human dystrophin, and a Pan-Dys Ab that reacts with the C-terminal domain of both human and mouse dystrophin. The HO and HH tri-AAV vectors were packaged in Y445F AAV-6 and injected into the TA muscle of mdx4cv mice at the dose of 4×1010 vg particles per vector. Dystrophin expression was examined 35 days later. Reconstituted human dystrophin was positive by both Pan-Dys and Hum-Dys antibody immunostaining (Fig. 2C, left two panels). In contrast, endogenous mouse dystrophin revertant fibers were positive by Pan-Dys antibody immunostaining only (Fig. 2C, right two panels). Hum-Dys-positive myofibers were not detected in muscles injected with single vector (WL30, WL33, WL34, WL35, or WL37 alone) or various dual-vector combinations (WL30+WL34, WL34+WL37, WL30+WL37, WL30+WL33, WL33+WL35, or WL30+WL35) (data not shown). As expected, human dystrophin reconstitution was achieved in muscles coinjected with all three vectors (WL30+WL34+WL37 for the HO trivectors and WL30+WL33+WL35 for the HH trivectors) (Fig. 2C). However, only a few full-length human dystrophin-positive cells were detected. Further, there was no significant difference between the HO and HH trivectors (Fig. 2D).
Characterization of the flag-tagged tri-AAV vectors
In the original HH trivector set, the C-terminal end of human dystrophin was detected by the Pan-Dys antibody. This antibody cross-reacts with endogenous mouse dystrophin (Fig. 2B and C). To further increase the detection specificity, we engineered a flag tag at the C-terminal end of human dystrophin and generated the flag-tagged HH tri-AAV vectors (Fig. 3A). This set of vectors is identical to the original HH trivectors except for the flag tag added to the 3-end of the tail vector (Fig. 3A). The flag-tagged HH trivectors were packaged in Y731F AAV-9 and injected into the TA muscle of mdx4cv mice at the dose of 4.7×1011 vg particles per vector (Fig. 3B and C). Dystrophin expression was examined 35 days after injection. As expected, human dystrophin was detected by both the Pan-Dys and flag antibodies, while revertant mouse dystrophin was only recognized by the Pan-Dys antibody (Fig. 3B). Importantly, coinfection with WL30, WL33, and WL38 resulted in significantly more flag and Pan-Dys double-positive myofibers (50-fold higher than that of single- or dual-vector transduction) (Fig. 3C).
To confirm full-length dystrophin reconstitution at the mRNA level, we performed quantitative RT-PCR using total RNA (Fig. 3D). Compared with that of uninfected muscles, tri-AAV (WL30+WL33+WL38) coinfected muscles yielded significantly more transcripts with the primer/probe sets specific for the head/body and body/tail junctions (Fig. 3D and Supplementary Fig. S2). Importantly, similar levels of transcripts were detected in uninfected and tri-AAV coinfected muscles when the primer/probe set that is specific to the head/tail junction was used.
High-dose coinfection enhances tri-AAV reconstitution
The proof-of-principle studies with the HO, HH, and flagged HH vectors demonstrated the feasibility of trivector-mediated full-length dystrophin expression. However, reconstitution efficiency was low (≤2 positive myofibers/mm2) (Figs. 2D and 3C). To determine whether increasing AAV dose can improve recombination, high-dose flag-tagged HH trivectors (1.2×1012 vg particles/vector, Y731F AAV-9) were delivered to the TA muscles of two mdx4cv mice. We examined dystrophin expression 35 days later using the Pan-Dys, Hum-Dys, and flag antibodies (Fig. 3E). Positive staining with all three antibodies unambiguously confirmed successful reconstitution of full-length flag-tagged human dystrophin (Fig. 3E). Surprisingly, doubling the dose resulted in a five-fold increase in the number of full-dystrophin-transduced myofibers (∼11 positive myofibers/mm2) (Fig. 3F).
Discussion
In this study, we showed successful expression of full-length human dystrophin with the tri-AAV vectors. Specifically, the full-length human dystrophin cDNA expression cassette was divided into three fragments and each fragment engineered into an AAV vector. The complete expression cassette was reconstituted in muscle after coinfection with all three vectors.
Over the last few years, AAV-based gene therapy has shown indisputable clinical benefit in several diseases (Carter, 2005; Mingozzi and High, 2011b; Buning, 2013). A common feature in these successes is that they all have a relatively small therapeutic gene that can fit into a single AAV virion. The wild-type AAV genome is ∼4.7 kb (Srivastava et al., 1983). Recombinant AAV virus can accommodate a slightly larger genome (up to 5 kb). The packaging efficiency drops sharply when the genome size exceeds 5 kb (Dong et al., 1996). The small packaging capacity creates a significant hurdle for larger therapeutic genes.
Several innovative dual-vector strategies have been explored to deliver a large therapeutic gene with AAV. The basic idea is to break the intact expression cassette into two pieces so that each piece can fit into a single AAV vector. Subsequently, the fragmented cassette is restored through intermolecular recombination between independent vector genomes. The trans-splicing and the overlapping vectors are the two prototypes (Duan et al., 2001). In the trans-splicing vectors, recombination is achieved via end-to-end joining of AAV ITRs. After removing the junction by the engineered splicing signals, transgene expression is restored. In the overlapping vectors, two split fragments share a common region. Homologous recombination between the common regions reconstitutes the intact expression cassette. Theoretically, both vector systems can deliver a large gene. However, there are dramatic variations in the efficiency depending on the unique properties of the target gene. The trans-splicing vectors require an optimal gene-splitting site, while the overlapping vectors require a highly recombinogenic sequence (Xu et al., 2004; Lai et al., 2005; Ghosh et al., 2006). The hybrid vectors are developed to overcome these limitations (Ghosh et al., 2008). In the hybrid vectors, reconstitution can be achieved either via ITR joining or homologous recombination. The development of dual-AAV vectors has partially addressed the size barrier by doubling the packaging capacity to 10 kb. Yet, this still cannot meet the need of the 11.2 kb full-length dystrophin coding sequence.
The dystrophin gene is one of the largest genes in the body. Out-of-frame mutations in the dystrophin gene lead to DMD. Early studies suggest that some regions of the dystrophin gene might be dispensable. For example, deletion of a large section of the dystrophin rod domain or the dystrophin C-terminal domain is tolerated in human patients and mice (England et al., 1990; Crawford et al., 2000; Harper et al., 2002). However, recent findings from several groups suggest that the structure–function relationship of the dystrophin gene is more complicated than previously thought (Lai et al., 2009, 2013; Koo et al., 2011; Rumeur et al., 2012; Taghli-Lamallem et al., 2013). Ideally, a full-length dystrophin cDNA should be used for gene replacement therapy because full-length dystrophin offers better protection than abbreviated micro/mini-dystrophins (Phelps et al., 1995). Considering the 5 kb maximal packaging capacity of a single AAV vector, we will need three independent AAV vectors to carry a full-length dystrophin expression cassette. We initially hypothesized that reconstitution can be achieved via any one of the recombination strategies discussed above (the trans-splicing, overlapping or hybrid). Since the cloning of the overlapping vectors is simple and straightforward, we explored this strategy first. Studies from several laboratories have shown that dystrophin R20 can mediate efficient homologous recombination (Odom et al., 2011; Zhang and Duan, 2012; Zhang et al., 2013). R20 is encoded by exons 51–53. Thus, it can be used to reconstitute the body and tail parts. To identify a highly recombinogenic region for the head fragment and body fragment reconstitution, we screened R7, R8, and R9. Unfortunately, none of these regions resulted in robust homologous recombination (Fig. 1). The exact reason why R20 is more recombinogenic is currently not clear. However, in the context of the overlapping vectors, similar differences have been seen between different genes (the AP gene versus the LacZ gene) or among different parts of the same gene (Ghosh et al., 2006, 2011).
Next, we tested the hybrid approach-based tri-AAV vectors. To capitalize on the high recombination efficiency of R20, we designed two different sets of tri-AAV vectors. In both sets of the vectors, reconstitution of the head and body parts is achieved via homologous recombination of a foreign sequence (AP1) and/or ITR-mediated end joining (Fig. 2A and B). To reconstitute the body and tail parts, we used either R20-mediated homologous recombination (for the HO trivectors) or the hybrid method based on a different foreign sequence (AP2) (for the HH trivectors). In vivo studies in mdx4cv mice suggest that both HO and HH trivectors can lead to low-level human dystrophin expression (Fig. 2C and D). To further confirm full-length human dystrophin cDNA reconstitution, we designed a third set of trivectors by attaching a flag tag to the C-terminal end of the tail vector of the HH tri-AAV vectors (Fig. 3A). Consistent with our initial observation with the HH trivector, we detected flag-positive human dystrophin expression in mdx4cv mice infected by the flag-tagged HH trivectors (Fig. 3).
An alternative approach to determine tri-AAV reconstitution is to evaluate mRNA. We designed human dystrophin-specific primers and probes to cover the possible junctions between the head and body vectors, body and tail vectors, and head and tail vectors (Supplementary Fig. S2). Because of high sequence homology between the human and mouse dystrophin mRNA, low-level signals were detected in noninfected mice by qRT-PCR (Fig. 3D). Importantly, transcripts specific to the full-length human dystrophin mRNA (head/body recombination and body/tail recombination) were log-fold higher in trivector-infected mice. On the other hand, no significant difference was seen between trivector-infected and noninfected mice for head/tail qRT-PCR, suggesting that there was nominal recombination between the head and tail vectors (likely because of a lack of homology between the AP1 and AP2 sequences) (Fig. 3D).
An important requirement in DMD gene therapy is to treat sufficient numbers of myofibers (Chamberlain, 1997). The sporadic expression obtained from tri-AAV coinfection is far from enough. To improve transduction efficiency, we doubled the vector dose. Interestingly, we observed a fivefold increase in human dystrophin-positive cells (Fig. 3F). The exact mechanism underlying this disproportional dose–response is unclear. We speculate that this may likely reflect a threshold effect. In other words, by doubling the vector dose we may have surpassed a critical concentration threshold for tri-AAV recombination. Despite the improvement, the overall transduction efficiency remains too low to have a significant impact on muscle disease (Chamberlain, 1997). Future studies are needed to identify the rate-limiting steps and further improve tri-AAV transduction efficiency.
In this study, we observed few Flag-positive cells in muscles that received WL38 alone (tail vector only) or both WL30 and WL38 (head and tail vector together) (Fig. 3C). Our results revealed the possibility of generating toxic and/or immunogenic products from a single vector or the head/tail-recombined genome. Although such events are rare, they still represent important safety and immunological concerns. Future studies are needed to fully characterize these untoward products, and more importantly to develop effective strategies to minimize and/or eliminate such products.
In summary, our results suggest that tri-AAV vectors may be used to expand the total carrying capacity of the AAV vectors to 15 kb. This not only opens the door to deliver a large therapeutic gene (such as the full-length dystrophin cDNA), but also provides more space for engineering various regulatory elements into the expression cassette.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institutes of Health (NIH) NS-62934 (D.D.), HL-91883 (D.D.), and AR-49419 (D.D.), and the Muscular Dystrophy Association (D.D.). We thank Dr. Jeffrey Chamberlain (University of Washington, Seattle, WA) for providing the full-length human dystrophin plasmid. We thank Dr. Arun Srivastava (University of Florida, Gainesville, FL) for providing tyrosine mutant AAV capsid packaging plasmids. We thank Dr. Chady Hakim for the help with statistical analysis.
Author Disclosure Statement
The authors declare no competing financial interests.
During the submission of our manuscript, Koo et al. reported a different set of tri-AAV vectors based on the trans-splicing approach (Koo et al., 2014). Collectively, results from both studies suggest that AAV packaging capacity can be significantly expanded using the tri-vector strategy.
References
- Atchison R.W., Casto B.C., and Hammon W.M. (1965). Adenovirus-associated defective virus particles. Science 149, 754–756 [DOI] [PubMed] [Google Scholar]
- Bartoli M., Poupiot J., Goyenvalle A., et al. (2006). Noninvasive monitoring of therapeutic gene transfer in animal models of muscular dystrophies. Gene Ther. 13, 20–28 [DOI] [PubMed] [Google Scholar]
- Bostick B., Ghosh A., Yue Y., et al. (2007). Systemic AAV-9 transduction in mice is influenced by animal age but not by the route of administration. Gene Ther. 14, 1605–1609 [DOI] [PubMed] [Google Scholar]
- Buning H. (2013). Gene therapy enters the pharma market: the short story of a long journey. EMBO Mol. Med. 5, 1–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter B.J. (2005). Adeno-associated virus vectors in clinical trials. Hum. Gene Ther. 16, 541–550 [DOI] [PubMed] [Google Scholar]
- Chamberlain J.S. (1997). Dystrophin levels required for correction of Duchenne muscular dystrophy. Basic Appl. Myol. 7, 251–255 [Google Scholar]
- Crawford G.E., Faulkner J.A., Crosbie R.H., et al. (2000). Assembly of the dystrophin-associated protein complex does not require the dystrophin COOH-terminal domain. J. Cell Biol. 150, 1399–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danko I., Chapman V., and Wolff J.A. (1992). The frequency of revertants in mdx mouse genetic models for Duchenne muscular dystrophy. Pediatr. Res. 32, 128–131 [DOI] [PubMed] [Google Scholar]
- Dong J.Y., Fan P.D., and Frizzell R.A. (1996). Quantitative analysis of the packaging capacity of recombinant adeno-associated virus. Hum. Gene Ther. 7, 2101–2112 [DOI] [PubMed] [Google Scholar]
- Dong B., Nakai H., and Xiao W. (2010). Characterization of genome integrity for oversized recombinant AAV vector. Mol. Ther. 18, 87–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan D. (2011). Duchenne muscular dystrophy gene therapy: lost in translation? Res. Rep. Biol. 2, 31–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan D., Yue Y., and Engelhardt J.F. (2001). Expanding AAV packaging capacity with trans-splicing or overlapping vectors: a quantitative comparison. Mol. Ther. 4, 383–391 [DOI] [PubMed] [Google Scholar]
- England S.B., Nicholson L.V., Johnson M.A., et al. (1990). Very mild muscular dystrophy associated with the deletion of 46% of dystrophin. Nature 343, 180–182 [DOI] [PubMed] [Google Scholar]
- Ghosh A., Yue Y., and Duan D. (2006). Viral serotype and the transgene sequence influence overlapping adeno-associated viral (AAV) vector-mediated gene transfer in skeletal muscle. J. Gene Med. 8, 298–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A., Yue Y., Lai Y., and Duan D. (2008). A hybrid vector system expands aden-associated viral vector packaging capacity in a transgene independent manner. Mol. Ther. 16, 124–130 [DOI] [PubMed] [Google Scholar]
- Ghosh A., Yue Y., and Duan D. (2011). Efficient transgene reconstitution with hybrid dual AAV vectors carrying the minimized bridging sequence. Hum. Gene Ther. 22, 77–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper S.Q., Hauser M.A., Dellorusso C., et al. (2002). Modular flexibility of dystrophin: implications for gene therapy of Duchenne muscular dystrophy. Nat. Med. 8, 253–261 [DOI] [PubMed] [Google Scholar]
- Koenig M., Beggs A.H., Moyer M., et al. (1989). The molecular basis for Duchenne versus Becker muscular dystrophy: correlation of severity with type of deletion. Am. J. Hum. Genet. 45, 498–506 [PMC free article] [PubMed] [Google Scholar]
- Koo T., Malerba A., Athanasopoulos T., et al. (2011). Delivery of AAV2/9-microdystrophin genes incorporating helix 1 of the coiled-coil motif in the C-terminal domain of dystrophin improves muscle pathology and restores the level of alpha1-syntrophin and alpha-dystrobrevin in skeletal muscles of mdx mice. Hum. Gene Ther. 22, 1379–1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo T., Popplewell L., Athanasopoulos T., and Dickson G. (2014). Triple trans-splicing adeno-associated virus vectors capable of transferring the coding sequence for full-length dystrophin protein into dystrophic mice. Hum. Gene Ther. 25, 98–108 [DOI] [PubMed] [Google Scholar]
- Lai Y., and Duan D. (2012). Progress in gene therapy of dystrophic heart disease. Gene Ther. 19, 678–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Y., Yue Y., Liu M., et al. (2005). Efficient in vivo gene expression by trans-splicing adeno-associated viral vectors. Nat. Biotechnol. 23, 1435–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Y., Yue Y., Liu M., and Duan D. (2006). Synthetic intron improves transduction efficiency of trans-splicing adeno-associated viral vectors. Hum. Gene Ther. 17, 1036–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Y., Thomas G.D., Yue Y., et al. (2009). Dystrophins carrying spectrin-like repeats 16 and 17 anchor nNOS to the sarcolemma and enhance exercise performance in a mouse model of muscular dystrophy. J. Clin. Invest. 119, 624–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Y., Yue Y., Bostick B., and Duan D. (2010a). Delivering large therapeutic genes for muscle gene therapy. In Muscle Gene Therapy. Duan D., ed. (Springer Science+Business Media, LLC, New York: ) pp 205–218 [Google Scholar]
- Lai Y., Yue Y., and Duan D. (2010b). Evidence for the failure of adeno-associated virus serotype 5 to package a viral genome>or=8.2 kb. Mol. Ther. 18, 75–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Y., Zhao J., Yue Y., and Duan D. (2013). alpha2 and alpha3 helices of dystrophin R16 and R17 frame a microdomain in the alpha1 helix of dystrophin R17 for neuronal NOS binding. Proc. Natl. Acad. Sci. USA 110, 525–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mays L.E., and Wilson J.M. (2011). The complex and evolving story of T cell activation to AAV vector-encoded transgene products. Mol. Ther. 19, 16–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mingozzi F., and High K.A. (2011a). Immune responses to AAV in clinical trials. Curr. Gene Ther. 11, 321–330 [DOI] [PubMed] [Google Scholar]
- Mingozzi F., and High K.A. (2011b). Therapeutic in vivo gene transfer for genetic disease using AAV: progress and challenges. Nat. Rev. Genet. 12, 341–355 [DOI] [PubMed] [Google Scholar]
- Odom G.L., Gregorevic P., Allen J.M., and Chamberlain J.S. (2011). Gene therapy of mdx mice with large truncated dystrophins generated by recombination using rAAV6. Mol. Ther. 19, 36–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X., Yue Y., Zhang K., et al. (2013). Long-term robust myocardial transduction of the dog heart from a peripheral vein by adeno-associated virus serotype-8. Hum. Gene Ther. 24, 584–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelps S.F., Hauser M.A., Cole N.M., et al. (1995). Expression of full-length and truncated dystrophin mini-genes in transgenic mdx mice. Hum. Mol. Genet. 4, 1251–1258 [DOI] [PubMed] [Google Scholar]
- Qiao C., Zhang W., Yuan Z., et al. (2010). AAV6 capsid tyrosine to phenylalanine mutations improve gene transfer to skeletal muscle. Hum. Gene Ther. 21, 1343–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumeur E.L., Hubert J.F., and Winder S.J. (2012). A new twist to coiled coil. FEBS Lett. 586, 2717–2722 [DOI] [PubMed] [Google Scholar]
- Shin J.H., Yue Y., and Duan D. (2012). Recombinant adeno-associated viral vector production and purification. Methods Mol. Biol. 798, 267–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava A., Lusby E.W., and Berns K.I. (1983). Nucleotide sequence and organization of the adeno-associated virus 2 genome. J. Virol. 45, 555–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taghli-Lamallem O., Jagla K., Chamberlainc J.S., and Bodmer R. (2014). Mechanical and non-mechanical functions of Dystrophin can prevent cardiac abnormalities in Drosophila. Exp. Gerontol. 49, 26–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Yue Y., Lai Y., et al. (2004). Trans-splicing adeno-associated viral vector-mediated gene therapy is limited by the accumulation of spliced mRNA but not by dual vector coinfection efficiency. Hum. Gene Ther. 15, 896–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Y., and Duan D. (2002). Development of multiple cloning site cis-vectors for recombinant adeno- associated virus production. Biotechniques 33, 672, 674, 676–678 [DOI] [PubMed] [Google Scholar]
- Yue Y., Li Z., Harper S.Q., et al. (2003). Microdystrophin gene therapy of cardiomyopathy restores dystrophin-glycoprotein complex and improves sarcolemma integrity in the mdx mouse heart. Circulation 108, 1626–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Y., Ghosh A., Long C., et al. (2008). A single intravenous injection of adeno-associated virus serotype-9 leads to whole body skeletal muscle transduction in dogs. Mol. Ther. 16, 1944–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Y., Shin J.-H., and Duan D. (2011). Whole body skeletal muscle transduction in neonatal dogs with AAV-9. Methods Mol. Biol. 709, 313–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., and Duan D. (2012). Novel mini-dystrophin gene dual adeno-associated virus vectors restore neuronal nitric oxide synthase expression at the sarcolemma. Hum. Gene Ther. 23, 98–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Yue Y., Li L., et al. (2013). Dual AAV therapy ameliorates exercise-induced muscle injury and functional ischemia in murine models of Duchenne muscular dystrophy. Hum. Mol. Genet. 22, 3720–3729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong L., Li B., Mah C.S., et al. (2008). Next generation of adeno-associated virus 2 vectors: point mutations in tyrosines lead to high-efficiency transduction at lower doses. Proc. Natl. Acad. Sci. USA 105, 7827–7832 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.