Abstract
To systematically investigate the impact of immune stimulation upon regulatory variant activity, we exposed primary monocytes from 432 healthy Europeans to interferon-γ (IFN-γ) or differing durations of lipopolysaccharide and mapped expression quantitative trait loci (eQTLs). More than half of cis-eQTLs identified, involving hundreds of genes and associated pathways, are detected specifically in stimulated monocytes. Induced innate immune activity reveals multiple master regulatory trans-eQTLs including the major histocompatibility complex (MHC), coding variants altering enzyme and receptor function, an IFN-β cytokine network showing temporal specificity, and an interferon regulatory factor 2 (IRF2) transcription factor–modulated network. Induced eQTL are significantly enriched for genome-wide association study loci, identifying context-specific associations to putative causal genes including CARD9, ATM, and IRF8. Thus, applying pathophysiologically relevant immune stimuli assists resolution of functional genetic variants.
Inappropriate immune activity and associated inflammation are involved in the pathogenesis of a broad range of common diseases including inflammatory bowel disease, atherosclerosis, rheumatoid arthritis, and cancer. Moreover, a significant proportion of common disease risk loci identified with genome-wide association studies (GWAS) implicate immune genes (1). Most GWAS loci consist of single-nucleotide polymorphisms (SNPs) within noncoding, putatively regulatory DNA, often at a distance from any gene coding regions (2). The identification of functional regulatory variants and associated modulated genes is key to interpreting GWAS findings and establishing how genes are associated with disease. This can be explored by mapping gene expression as a quantitative trait (eQTL mapping) (3–6).
The effect of a regulatory variant on gene expression is frequently dependent on the cell and tissue type (7–9). The extent to which specific cellular pathway activation by environmental agents can modulate regulatory variant effects is relatively unexplored. Monocytes are cardinal innate immune cells that constitutively express receptors responsive to pathogens and cytokines including Toll-like receptors (TLRs) (10), with activity being intricately regulated by the cellular environment and interactions with other immune cell types. Upon stimulation, monocytes exhibit large-scale gene transcription with concomitant cytokine production, enhanced antigen processing, and migration (11, 12). Context-specific genetic variants that directly modulate innate immune responses or gain regulatory capacity upon pathway activation have been subject to selective pressure and may affect disease susceptibility (13). It is probable that such variants are widespread, although they are functionally unidentifiable in resting cells. Supporting this, macrophages derived from different mouse strains demonstrate multiple de novo trans-regulatory loci upon inflammatory stimuli (14). We hypothesized that the transformation in monocyte transcriptional activity accompanying innate immune responses would allow us to detect the effect of context-specific regulatory genetic variation upon gene expression in humans.
Results
Innate Immune Stimuli–Induced Transcriptomes
Interindividual variation in the early (2 hours) and late (24 hours) inflammatory responses to lipopolysaccharide (LPS), a component of gram-negative bacterial cell walls that triggers TLR4 signaling (10), and interferon-γ (IFN-γ) (24 hours), a cytokine acting through the JAK-STAT (Janus kinase–signal transducer and activator of transcription) signaling pathway, important in mycobacterial and viral infections (15, 16), was characterized in highly purified primary CD14+ human monocytes from 432 individuals of European ancestry. Genome-wide gene expression profiling and genotyping were performed with HumanHT-12 v4 BeadChip (Illumina) and HumanOmniExpress-12v1.0 BeadChip (Illumina). We tested 609,704 SNPs [minor allele frequency (MAF) > 0.04] and expression data for 15,421 probes for 414 individuals in the naïve state, 367 individuals after exposure to IFN-γ, 322 individuals after 24-hour LPS, and 261 individuals after 2-hour LPS. To allow comparison with naïve monocytes from the same individuals, expression data from untreated, incubated samples from a subset of 59 individuals were used to regress out expression changes attributable to incubation from the treated samples. Data across all four conditions were available from 228 individuals, permitting cross-treatment comparison. As anticipated, treatment induced significant changes in monocyte expression profiles (table S1) with hierarchical cluster analysis demonstrating that group membership was defined by treatment (fig. S1). Analysis of differential gene expression was consistent with biological expectations in terms of identified canonical pathways, the most significant upstream regulators, and the transcriptomic differences between early and late responses to LPS (table S1 and fig. S1) (17).
Identifying Context-Specific Local Genetic Regulatory Variants
Local, likely cis-acting eQTL (referred to as cis-eQTL) were defined as SNPs showing association with gene expression that were located within a 1-Mb window of the associated probe. We tested for association with gene expression using an additive linear model (18). We included surrogate variables consisting of major principal components (PC) of the expression data as covariates to limit the effect of confounding factors. Similar to others (19, 20), we find this enhanced detection of cis- and trans-eQTLs (fig. S2). Using this approach, we found that the majority of all cis-eQTLs observed affected gene expression only upon cell stimulation, and a significant proportion of constitutive eQTL present in naïve cells were no longer present after treatment (Fig. 1, A and B, and table S2). Across all four data sets, most genes show evidence of eQTL, with 83.7% of probes tested (12,905/15,421 probes) showing association under at least one condition [false discovery rate (FDR) < 0.05].
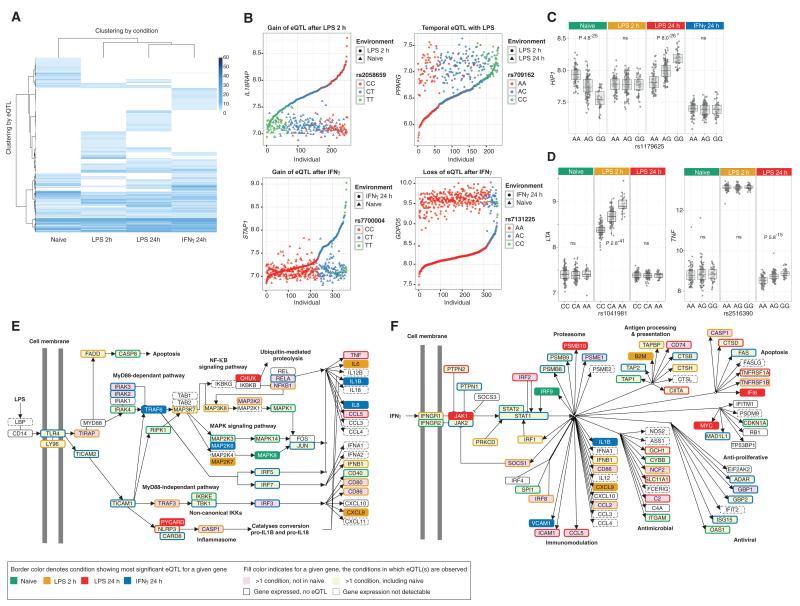
Fig. 1. Genotype modulates the gene expression response to innate immune stimuli in monocytes.
(A) Cis-eQTL mapping of 228 individuals with matched expression data available across all four treatments shows that the majority of eQTL are context-dependent. Genes with significant eQTL (FDR < 0.05) are hierarchically clustered along the y axis according to the t statistic of the most significant eQTL present across treatments (along the x axis). (B) Gene expression values before and after treatment plotted pairwise by individual. In clockwise order: 2-hour LPS induces differential expression of IL18RAP dependent on rs2058659 genotype; 2-hour LPS treatment results in uniform PPARG up-regulation, whereas continued LPS exposure over 24 hours is associated with reduction in expression in carriers of the A allele of rs709162; an eQTL tagged by rs7131225 found in untreated cells (naïve state) for GDPD5 transcription could not be detected after gene induction with IFN-γ; and IFN-γ stimulation leads to induction of STAP1 expression in carriers of the T allele of rs7700004, relative to the C allele. (C) Cis-eQTL displays context-specific directional effects. rs1179625 is the SNP with the greatest effect on expression of HIP1 in the naïve state and after 24-hour LPS but exhibits opposing direction of effect. ns, not significant. (D) Induction by LPS reveals stimulus-specific cis-eQTL for LTA and TNF. (E and F) Genes within the (E) TLR4 signaling and (F) IFN-γ signaling pathways highlighted for those with eQTL for shared data set illustrating context specificity.
For the 228 individuals for whom we had expression data for all four conditions, we observed 21,516 independent cis SNP-probe associations across data sets (defined as the most significantly associated SNP with expression for a given probe per data set), 24.6% of which had the lowest P value for association in naïve cells, 21.6% after 2-hour LPS, 25.4% after 24-hour LPS, and 28.3% after IFN-γ. A total of 11,476 probes had at least one eQTL, of which 43.4% (4977 of 11,476) only had eQTL after monocyte stimulation. We find that eQTL are similarly specific to the unstimulated state, with 54.1% of naïve eQTL (2866 of 5299) not found after treatment (Fig. 1A, fig. S3, and table S2). One explanation for many context-specific eQTL is that treatment increases expression to quantifiable levels, thus allowing eQTL detection. To investigate the extent to which this contributed to the observed stimulus specificity of eQTL, we restricted our analysis to probes with detectable expression (Illumina detection score P < 0.01) in more than 90% of samples from each data set. We found that 33.4% (1702 of 5082) of these probes only had an eQTL after treatment, indicating that context-specific eQTL are identified because of both treatment-induced regulatory effects and treatment-inducing gene expression to detectable levels. We observed that the range of effect sizes (all eQTL, 6.0 to 94.7%; median, 11.3%) was markedly similar across the different conditions, although IFN-γ–associated eQTL had slightly larger effect sizes than all others (fig. S4).
Cell-specific eQTL more often involve enhancer elements distal to the transcription start site (TSS) when compared to eQTL shared across cell types (7). It is unclear whether eQTL induced by innate immune stimuli show a similar distribution. By analyzing all expression-associated SNPs (eSNPs), we found that the more conditions an eSNP was observed across, the more proximal it was to the TSS (P < 2.2 × 10−16). Even when assessing only significant eSNPs with association below a P < 5 × 10−8 threshold, a similar effect was apparent (P < 2.9 × 10−11). We find that the effect size for an eSNP increases with proximity to the TSS (fig. S5) and that eSNPs that could only be observed after treatment were significantly more distal than those observed in the naïve state (P < 2.2 × 10−16) (fig. S5), consistent with the pattern previously observed with cell type–specific eQTLs.
For a minority of eQTL, associations were observed across conditions, but with the direction of effect switching between those conditions. We mapped the most significant association per condition (17) and identified 20 genes, including HIP1 and STEAP4, with shared peak eQTLs mapping to a single locus but with opposing directions across conditions (Fig. 1C, fig. S6, and table S2).
A diverse set of immune-related genes were found to show eQTL after induction with LPS or IFN-γ, ranging from transcription factor genes such as ETS2, JUN, and IRF2 (interferon regulatory factor 2) to key cytokines and receptors such as IL8 (interleukin-8), IL32, IL18R1, and FAS (table S2). For LTA, an eQTL was observed after 2-hour LPS, whereas no eQTL was apparent for TNF on induction at 2 hours but was seen on down-regulation after 24-hour LPS (Fig. 1D). We investigated the relationship of specific treatments with observed eQTLs in terms of canonical gene pathways. This resolved contiguous pathways of induced eQTL activity (fig. S7). For the TLR4 pathway, eQTL specific to treatment were revealed for nodal genes including TIRAP, TRAF6, FADD, and MAP (mitogen-activated protein) kinase genes, whereas genes encoding key components of the inflammasome such as CASP1 and PYCARD, as well as downstream cytokines including IL6, CXCL9, and IFNB1, showed eQTL specific to early and late responses to LPS (Fig. 1E). Similarly, treatment revealed eQTL in nodal IFN-γ signaling pathway genes and many downstream genes involved in the proteasome, apoptosis, antigen processing, and presentation (Fig. 1F).
Trans-eQTL and Stimulus-Specific Hotspots
Associations involving distant genes (trans-eQTL, defined as SNPs >1 Mb from the probe) may define gene networks regulated by specific SNPs and can provide unbiased insights into pathway identity and underlying biological processes. It is unclear whether the relative paucity of trans-eQTL observed in human eQTL studies, usually attributable to limited power, may also reflect that many trans networks are undetectable in baseline states (21). Our genome-wide eQTL mapping across stimuli in the full data sets, correcting for multiple testing, resolved a total of 1838 trans-associating genes at FDR <0.05 (394 genes at P < 5 × 10−12, approximate to Bonferroni-corrected P = 0.05). Trans-associating loci showed a high degree of context specificity—most notably after IFN-γ treatment when multiple novel trans-eQTLs were identified that were frequently unobservable in the naïve state (Fig. 2, fig. S8, and table S3). Several loci, such as the previously described 12q15 locus at LYZ (8, 22) and the major histocompatibility complex (MHC), showed significant trans associations across treatments, whereas other master regulatory SNPs were resolved only in LPS or IFNγ–induced cells and putatively driven by cis-eQTL modulating cytokine release, enzymes, and transcription factors (Fig. 2).
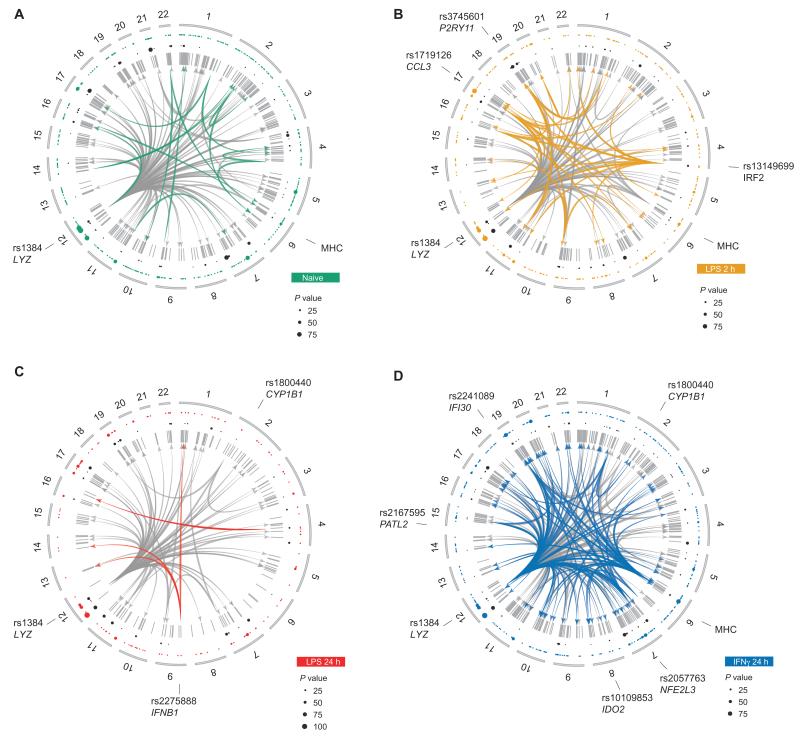
Fig. 2. Trans-eQTL demonstrate context specificity and identify master regulatory loci after treatment.
(A to D) Circular plots demonstrating chromosomal location of trans-acting eQTL of naïve (A), 2-hour LPS treatment (B), 24-hour LPS treatment (C), and IFN treatment (D). From outside to inside: examples of major trans hubs are indicated by rs number of lead eSNP and for that eSNP the name of any cis gene, the nearest gene or genomic region; chromosome number; colored dots are significant trans-eQTL (color denotes treatment, size relative −log10 P value as per legend); black dots are significant cis-eQTL to the same SNP in these treatments, possibly identifying driver genes; gray lines indicate probes significantly regulated in trans; innermost joining lines illustrate start and end points of multiloci trans-eQTL (unique to a treatment by colored lines, observed in a minimum of two data sets by gray lines)—only multiloci eQTL mapping to more than one probe with an FDR <0.05 in the shared data set (n = 228) are illustrated.
Stimulus Specificity for MHC Trans-eQTL
The MHC has been previously associated with trans-eQTL in primary tissues (8, 19). Trans associations to the MHC predominantly map to SNPs in the class II region and demonstrate plasticity across stimuli (Fig. 2 and fig. S9), with 45 genes mapping in trans (FDR < 0.05) to the class II region after IFN-γ treatment in the paired 228 data set, whereas after 24 hours of exposure to LPS, only one observation is made. Moreover, after IFN-γ treatment, we see the same trans-associating genes as in the naïve state, but almost all show increased effect sizes. The number of trans-associated genes mirrors class II expression, where IFN-γ and chronic LPS are associated with robust increases and suppression of class II gene expression, respectively (23) (fig. S9), suggesting that relative expression of class II genes may control trans effects.
Temporal Resolution of Cis- and Trans-eQTL Involving IFNβ Signaling
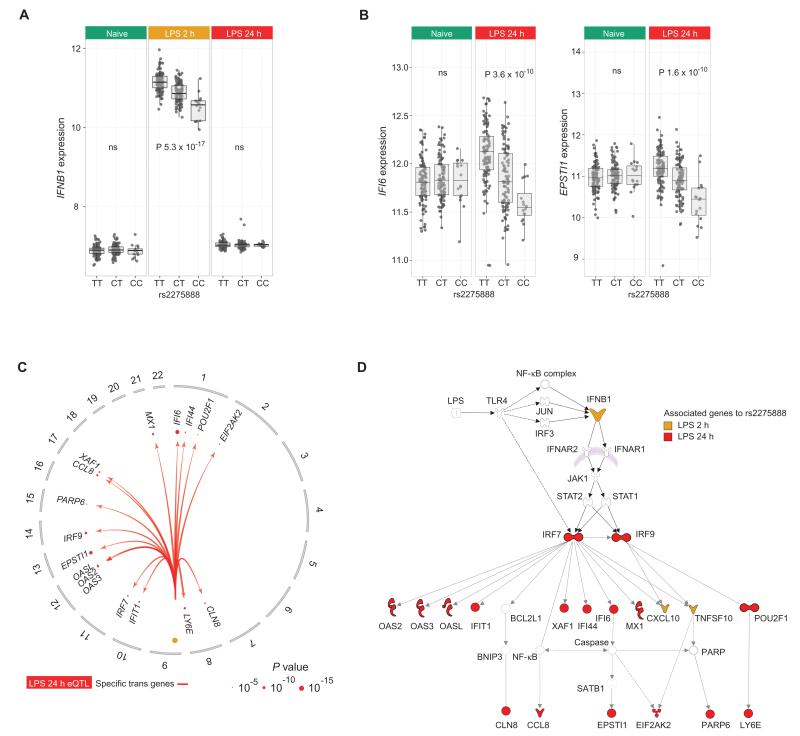
Cis-eQTL regulating cytokine release could be envisaged to affect expression of cytokine-modulated genes in the inflammatory cascade resulting in dose response–invoked eQTL in trans over time. An example of such temporal effects involves IFNB1, encoding the cytokine IFN-β, which exhibits a temporal relationship between genetic modulators and gene networks (Fig. 3). After 2-hour LPS (Fig. 3A), we observe a local cis-eQTL for IFNB1 at rs2275888; the same SNP associated in trans after 24-hour LPS with IRF9, IFI6, EPSTI1, LY6E, and MX1 expression.
Fig. 3. Temporal effects for a stimulus-specific trans-eQTL.
(A) A stimulus-specific local cis-acting eQTL for IFNB1 in monocytes after 2-hour LPS treatment tagged by rs2275888, which is in complete linkage disequilibrium (LD) (r2 = 1) with 15 other SNPs located ~60 kb telomeric to IFNB1 within the intronic sequence of PTPLAD2. (B) rs2275888 is significantly associated in trans with expression of known interferon-induced genes including IFI6 and ESTI1 only after 24-hour LPS. (C) Single SNP analysis at rs2275888 to resolve further genes in trans defined 17 genes showing significant trans association (FDR < 0.05) to rs2275888 after 24-hour LPS stimulation as illustrated in this circular plot. (D) IPA network analysis of trans-associated genes after LPS induction revealed a major network containing 19 genes (P = 1 × 10−44) based on the fit of the trans genes and biological functions. The IFNβ signaling cascade is shown with trans-associated genes to rs2275888 highlighted supporting the associations arising through cis-eQTL–driven differential expression of IFNB1.
We further interrogated trans associations to rs2275888 on a single SNP basis. This revealed that most associations occur after 24-hour LPS, with 17 genes in trans, many of which were archetypal early interferon response genes (Fig. 3, B and C, and table S4). Ingenuity pathway analysis (IPA) of trans-associated genes after LPS stimulation resolved an IFN-β–driven network and identified a key role for the upstream regulator IRF7 (P = 9.5 × 10−26) (Fig. 3D). Anti-viral response was the most significant disease and biological annotation (P = 1 × 10−16), whereas significant canonical pathways included interferon signaling (P = 5.8 × 10−8) and pattern recognition receptors (recognition of bacteria and viruses) (P = 6.7 × 10−08). This temporal relationship between a cis-eQTL and trans-associated gene network is consistent with the downstream consequences of differential IFN-β expression after LPS induction modulated by rs2275888.
Coding Polymorphisms Associate with Stimulus-Specific Trans Networks
We identified stimulus-specific trans networks putatively driven by coding variants. One such example is a trans hub observed at rs1800440, which codes for an amino acid substitution (p.Asn453Ser) in the first exon of CYP1B1, encoding a monocyte-expressed cytochrome P450 enzyme (24). This polymorphism is associated with a reduction in the half-life of CYP1B1 to one-third that of wild type (25) and is a cis-eQTL for CYP1B1 (table S2), consistent with reduced half-life driving a compensatory increase in transcription of the variant allele. Single SNP analysis of rs1800440 identified 43 genes, predominantly implicated in cancer and the inflammatory response, associating in trans after IFN-γ treatment (table S4). This variant shows significant variation in interpopulation allele frequency, the minor allele being very rare in African and Asian populations [fixation index (FST) of 0.288 (P = 0.03) and integrated haplotype score (iHS) of −2.126 for the CEU (Caucasian) HapMap population], suggesting possible selection pressure.
Similar examples of structural variants driving trans networks were identified after IFN-γ up-regulation of IDO2 involving a p.Arg248Trp substitution in exon 9 (rs10109853), which results in a 90% reduction in tryptophan catalytic activity (26), and after LPS induction to a p.Ala87Thr variant (rs3745601) in P2RY11 modulating ligand binding (27) (fig. S8 and table S3).
Gene Targets of the Transcription Factor IRF2 Modulated by a Stimulus-Specific Cis-eQTL
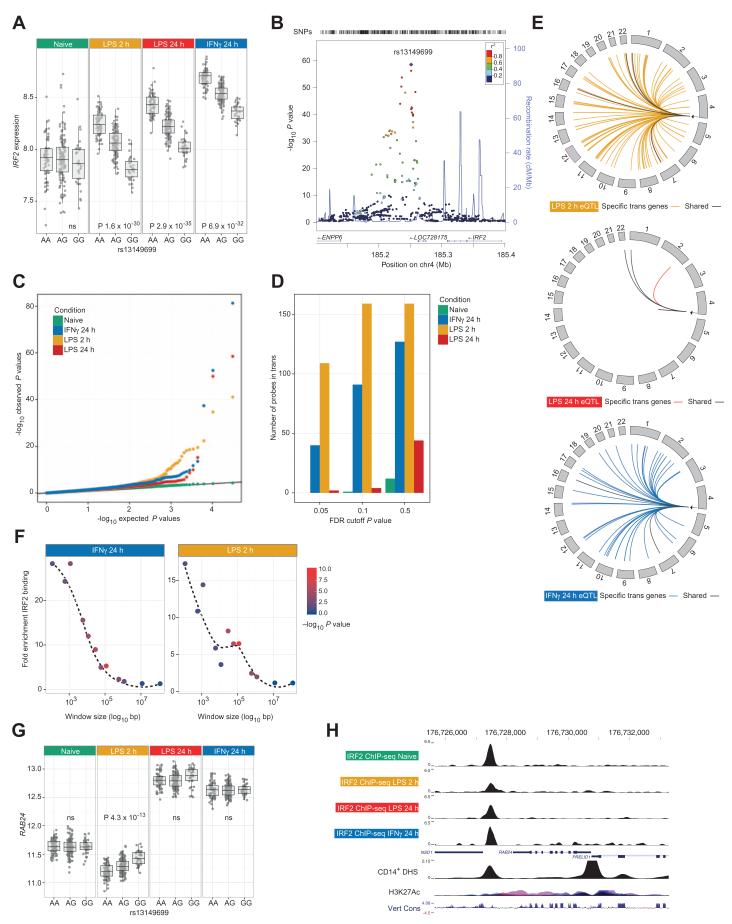
Cis-eQTL that regulate transcription factor abundance may generate associations in trans to transcription factor target genes (8, 19, 22, 28, 29). We report a significant context-specific trans gene hub to chromosome 4q35 tagged by rs13149699. This SNP displayed a strong stimulus-specific cis association with IRF2, encoding the transcription factor IRF2. Carriage of the A allele of rs13149699 resulted in LPS-induced up-regulation of IRF2 expression, whereas the G allele was associated with coincident suppression of expression. IFN-γ stimulation caused up-regulation of both IRF2 alleles, although more markedly so in A allele carriers (P = 6.9 × 10−32) (Fig. 4A). The ENCODE data set (30) shows that rs13149699, which is located in an intergenic region 57 kb telomeric to IRF2 (Fig. 4B), is spanned by a deoxyribonuclease I (DNase I) hypersensitive site in primary CD14+ monocytes and binds a cluster of transcription factors including REL-A (nuclear factor κB p65) (fig. S10). In the stimulated cells, this SNP is associated with a major trans network, and on single SNP analysis, more than 300 trans genes were defined in the complete data sets (Fig. 4, C to E, and table S4). Analysis of paired samples showed no probes associating in the naïve cells, implicating treatment-induced differential expression of IRF2 at rs13149699 as the driver of the trans network. IPA of rs13149699 trans-associated genes supported their modulation by IRF2 and interferon signaling, with overrepresentation of interferon-associated pathways and IRF2 identified as an upstream regulator (P = 7.9 × 10−11).
Fig. 4. Cis regulation of IRF2 at rs13149699 has profound transcriptional consequences in trans.
(A) A significant cis-eQTL to the transcription factor IRF2 seen only after treatment (P values shown for shared data set; in the full data sets, PLPS2 = 2.1 × 10−35, PLPS24 = 6.4 × 10−55 , PIFNγ = 2.6 × 10−59, and Pnaïve = ns). (B) Local association plot for 367 individuals after IFN-γ treatment including imputed genotypes. The peak eQTL for IRF2 remains rs13149699, 57-kb 3′ to the gene. (C) eQTL analysis was performed on rs13149699 alone in the shared (n = 228) data set to identify associated genes missed due to correction for multiple testing across all SNPs—qq plots demonstrating number of probes with expression associated with rs13149699 across treatments. Treatment results in significant numbers of probes deviating from expected, with greatest deviation after 2-hour LPS. (D) Bar charts demonstrating number of significantly associated probes at different FDR thresholds. Note relative absence of associations in untreated state despite the same number of tests being performed. (E) Circular plots demonstrating location of transeQTL (FDR < 0.05) after analysis on rs13149699 in the complete data sets. (F) Windows flanking probes tested for eQTL were defined, and the number of IRF2 ChIP-seq binding sites per treatment was counted. Probes in trans to rs13149699 were significantly enriched for IRF2 binding in both treatments (Fisher’s exact test). (G) Box plot showing allelic expression of RAB24 by rs13149699. (H) ChIP-seq for IRF2 from primary monocytes for different treatments illustrating differential binding 0.5-kb 3′ to RAB24. Also shown are ENCODE tracks for CD14+ DNase I hypersensitivity, H3K27Ac (seven cell lines), and vertebrate conservation (Multiz Alignment & Conservation 100 Species).
To investigate whether these trans-associated genes were bona fide IRF2 targets in monocytes, we performed chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-seq) for primary monocytes from two healthy volunteers using the same experimental conditions to define IRF2 binding sites (table S5). We found that genes in trans to rs13149699 were significantly enriched for IRF2 binding in corresponding treatments (Fig. 4, F to H) with 8.0- and 8.7-fold relative increase in trans-associated genes residing in a 25-kb window around IRF2 binding sites after 2-hour LPS or IFN-γ, respectively, compared to those not in trans to rs13149699 (LPS2 P = 3.5 × 10−6, IFN-γ P = 1.9 × 10−6; Fisher’s exact test). It was apparent that not only were genes in trans to rs13149699 dependent on stimulation, but also that different stimuli elicited different genes in trans, implying that IRF2 regulation is similarly context-dependent. IRF2 is induced by IFN-γ and has both transcriptionally repressive and activating properties, antagonizing the action of IRF1 and modulating the IFN-induced immune response, notably to viral infection (31–33). Consistent with this, whereas most genes in trans to rs13149699 showed opposite allelic association to the cis-eQTL for IRF2, indicating inhibition by IRF2 expression, a significant proportion of trans genes showed the same allelic association consistent with induction by higher IRF2 expression (fig. S10). This analysis highlights how ChIP-seq can confirm putative mechanisms of trans associations and reveal context-specific transcription factor targeting of genes.
An Inducible Trans Gene Network Involving NFE2L3 and the Proteasome
A further example of a context-specific trans network involving a transcription factor consisted of 23 genes encoding proteins involved in the ubiquitin-proteasome system (34), whose expression was associated with rs2057763 after IFN-γ (fig. S11 and table S4). Twelve of these trans genes encode subunits of the 26S proteasome, including the 20S proteasome core (PSMA1, PSMB2, PSMB4, and PSMB6) together with the 19S regulator complex (PSMC3, PSMC4, PSMC5, PSMD1, PSMD3, PSMD11, PSMD13, and PSMD14), suggesting that this SNP alters the expression of a key proteasomal regulator in response to IFN-γ. IPA revealed that protein ubiquitination was the only significant pathway (P = 9 × 10−20) with two top regulators of this network: NFE2L2 (P = 5.4 × 10−14), encoding Nrf2, a transcription factor that regulates genes containing antioxidant response elements (ARE) in their promoters, and the known Nrf2 inducer molecule 1,2-diothio-3-thione. rs2057763 is an intergenic SNP 36.7 kb from the closest gene on chromosome 7p15 encoding NFE2L3, a relatively poorly characterized NFE2L2 homolog, recently recognized to play a key role in inflammation (35). Probes to NFE2L3 were not analyzed in the primary analysis because of risk of SNP hybridization artifacts. We therefore performed quantitative polymerase chain reaction (qPCR) on both the 3′ untranslated region and exons 1 and 2 of NFE2L3 in monocytes from a randomly selected subset of 84 individuals after IFN-γ and observed a cis-eQTL for rs2057763 (fig. S11), supporting NFE2L3 as the probable driver behind this network.
Informativeness for Disease
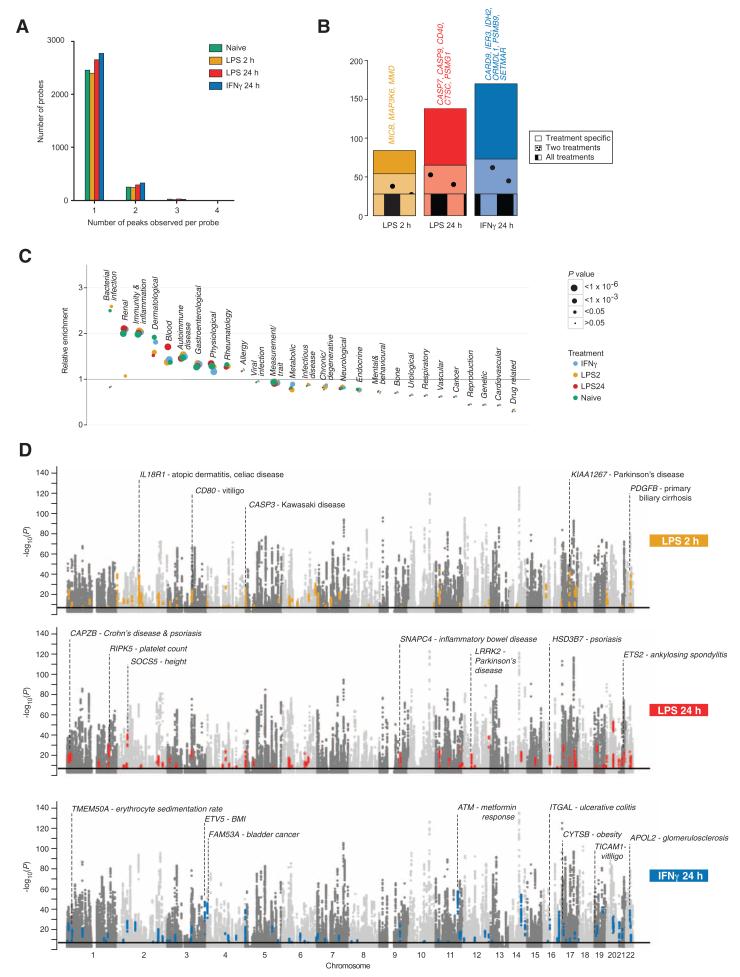
To further investigate the relationship between innate immune stimuli–induced eQTL and human disease risk loci resolved by GWAS, we first defined independent associations to the same probe interrogating gene expression within and between treatments. To do this, we resolved SNPs associated with the same eQTL into independent peaks of association among the shared data set of 228 individuals by conditional analysis (17). This revealed that although the majority of probes associated with a single peak, 9.8% of probes had a second peak in naïve monocytes, with up to three peaks in a small number of instances (Fig. 5A). Similar proportions were observed after treatment, the number of probes with two peaks modestly increasing to 11% after IFN-γ treatment. Considering all probes with a peak in the naïve state, additional peaks specific to stimulation were found, most markedly after IFN-γ treatment (Fig. 5B).
Fig. 5. Stimulus-specific eQTL and GWAS.
(A) Independent associations to the same probe within and between treatments were defined. The frequency distribution of the number of peaks observed per probe is shown by condition. (B) For genes showing an eQTL in naïve cells, the number of such genes with an additional stimulus-specific eQTL is shown according to stimulus and whether the eQTL were specific to that treatment or observed with two or all stimuli. Examples of immune relevant genes with second eQTL specific to a given stimulus are listed. (C) Enrichment analysis of treatment-specific eQTL by GWAS ontology category. (D) Manhattan plots demonstrating eQTL present in denoted treatment. Colored points correspond to eQTL where the primary peak is either a GWAS-identified locus or in r2 > 0.8 with a GWAS locus found in treated, but not naïve, monocytes.
The overlap between primary peaks defined for each condition and disease loci reported in the GWAS catalog (www.genome.gov/gwastudies/) was subsequently assessed. Overall, 467 primary peaks overlapped with a GWAS locus, 248 of which were only observed in a stimulated state (table S6). To ascertain whether specific traits were enriched, we curated GWAS traits for membership of one or more disease categories on an organ-specific and systems approach. Observed cis-eQTL were found to be most significantly overrepresented in traits related to bacterial infection (naïve, 2-hour LPS), renal diseases (all except 2-hour LPS), and inflammation and immunity (all subsets) (Fig. 5C). Correspondingly, we observed underrepresentation in cancer, cardiovascular, and genetic traits.
The observation that many GWAS loci overlapped with eQTL specific to induced cells (Fig. 5D) is consistent with the hypothesis that disease risk alleles may manifest activity in a context-specific manner (36, 37). Noteworthy examples we identified include a unique cis-eQTL to the serine threonine kinase gene ATM after exposure to IFNγ (rs609261, P = 7.8 × 10−89) in complete LD to a GWAS SNP for metformin response in diabetes (38). Notably, IFN-γ has been shown to activate the ATM pathway (39), and ATM itself plays a key role in the IFN-γ response (40). Our data suggest that differential expression of ATM may be driving this association with metformin response in a highly context-specific manner.
For IRF8, a novel treatment-specific cis-eQTL is seen, maximal after induction of gene expression by LPS for 2 hours (P = 9.9 × 10−19), for which the peak SNP, rs17445836, is also a lead GWAS SNP for multiple sclerosis (MS) and associated with up-regulation of interferon response gene sets in peripheral blood mononuclear cells (PBMCs) from MS patients (41). For the chemokine receptor 3 gene (CCR3), we also find evidence of a stimulus-specific cis-eQTL, after 24-hour LPS (P = 2.3 × 10−14), for which the lead eSNP was recently reported as the peak GWAS SNP for Behçet’s disease, a multisystem inflammatory disorder involving severe vasculitis (42). Other observations include an eQTL to IRF1 with the peak SNP (PLPS2 = 6.9 × 10−17) overlapping with a Crohn’s locus (43) and an eQTL observed to the gene LILRA5 only after 24-hour LPS intersecting with a prostate cancer locus (44).
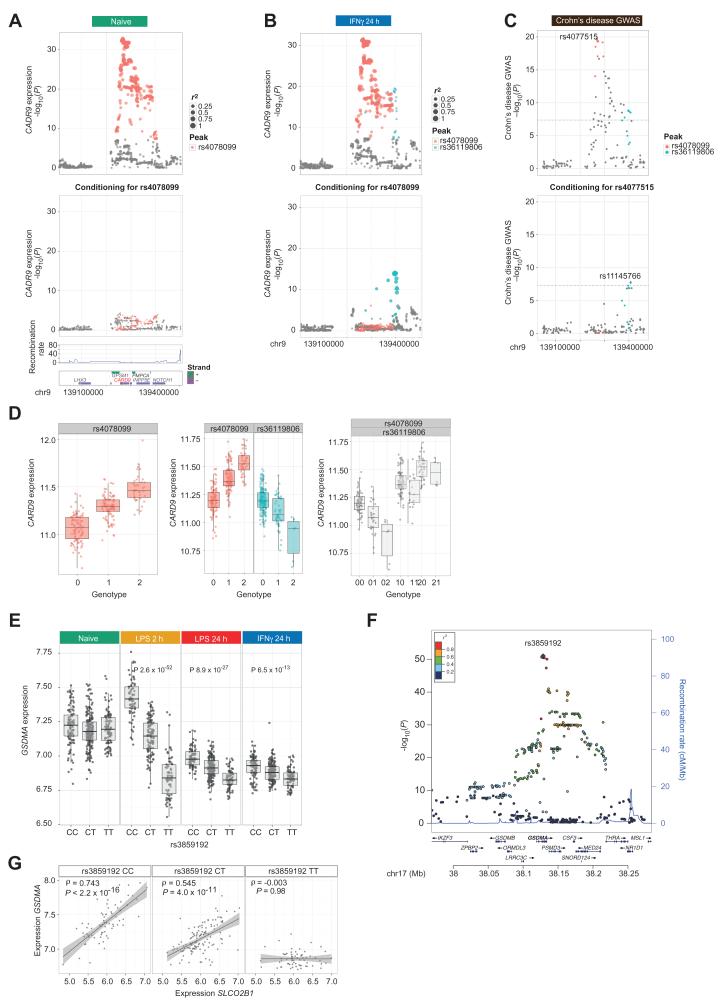
Stimulus-Specific eQTL, CARD9, and Crohn’s Disease Risk
The resolution of context-specific eQTL can also enable the identification and interpretation of new disease associations from existing GWAS. For the autoimmune disease–associated gene CARD9, we identified a second independent stimulus-specific eQTL following IFN-γ in addition to a constitutively active eQTL (Fig. 6, A and B, and fig. S12). The latter is tagged by peak SNPs that define a GWAS locus for inflammatory bowel disease and ankylosing spondylitis (45, 46). We hypothesized that if differential expression of CARD9 is the functional mechanism underlying observed GWAS signals, the independent IFNγ-specific eQTL may additionally show evidence of disease association. A large cohort of Crohn’s disease (CD) patients and controls (46) confirmed that the constitutive CARD9 peak eQTL is associated with a risk allele for CD [rs4077515, PGWAS 2.2 × 10−20 odds ratio (OR) 1.25 (95% confidence interval, 1.20 to 1.30)]. Notably, conditional analysis demonstrated that the induced CARD9 peak eQTL was further associated with an independent signal of protection for CD [rs11145766 shows independent association with reduced risk of CD, P = 3.7 × 10−9 GWAS OR 0.73 (0.63 to 0.84)] (Fig. 6, C and D). These effects were independent of reported associations that involved rare variants of CARD9 (47). Both risk alleles were associated with reduced CARD9 expression, consistent with the key role of CARD9 as an adaptor protein coordinating innate and adaptive immune responses and mounting an appropriate IFNγ gut response to pathogens (48).
Fig. 6. Examples of context-specific eQTL informative for disease risk.
(A) Local association plot showing evidence of cis-eQTL for CARD9 in naïve cells. After conditioning for rs4078099, no evidence of association is seen. (B) After induction with IFNγ, in addition to the eQTL tagged by rs4078099, a second stimulus-specific independent peak of association is tagged by rs36119806. (C) Local association plots for CD with SNPs colored as per (A) and (B). The lead SNP rs40771515 is in high LD with rs4078099 (r2 = 0.93); after conditioning, a second independent GWAS peak is seen (lead SNP rs11145766, r2 = 0.84 with rs136119806). (D) Box plots showing expression of CARD9 by allele in naïve cells, and after IFNγ stimulation, showing allelic association by SNP corrected for the effect of the other eQTL or by combination of inherited alleles for rs4078099 and rs36119806. (E) rs3859192 is the peak eQTL for expression of GSDMA, a transforming growth factor–β–regulated proapoptotic gene. (F) Local association plot for GSDMA expression after 2-hour LPS using imputed genotyping. (G) Allele-specific correlation for expression values per genotype demonstrates significant regulation of GSDMA expression in homozygous carriers of the C allele by the solute transporter SLCO2B1 after 2-hour LPS with no association being present in homozygous carriers of the T allele. Similar results were obtained analyzing monocytes after 24-hour LPS, and with both probes to SLCO2B1, with no association observed in the naïve state.
Dissecting GWAS Mechanisms
We wished to explore whether allele-specific correlation analyses can identify gene sets and pathways associated with risk genotypes that are potentially informative as to underlying pathogenic mechanisms. A notable treatment-specific cis-eQTL was observed at rs3859192 to GSDMA, most significantly after 2-hour LPS (P = 2.6 × 10−52) (Fig. 6, E and F). This SNP is associated with white cell count (49) and asthma susceptibility (50), and is a known eQTL in lung resections (51). The CEU genotype frequencies of this SNP are 0.257:0.496:0.248 (CC/CT/TT), making it an ideal candidate for exploring underlying allele-specific regulation.
We performed allele-specific correlation analyses across all probes with GSDMA expression, with probes deemed to be associated if P < 1 × 10−6, equivalent to Bonferroni-corrected P < 0.05. We observed 50 probes (corresponding to 40 genes; table S6) with expression significantly correlated with GSDMA in individuals homozygous for the CC genotype but with no association in the TT genotype (P > 0.01). The most significant probes mapped to SLCO2B1 (Fig. 6G), an organic ion transporter implicated in the transport of large molecules including eicosanoids, leukotrienes, steroids, and certain drugs (52). Pathway analysis of these genes showed the top-upstream regulators to be IL13 (P = 1.5 × 10−8), IL6 (P = 1.6 × 10−7), and CSF1 (P = 2.0 × 10−7), strongly implicated in asthma pathogenesis, inflammatory responses, and white cell count, respectively. These data suggest that certain risk alleles can alter the correlated gene expression profiles of specific genes in a context-specific manner, possibly removing such genes from their normal regulatory influences. More generally, this association illustrates how eQTL defined for innate immune stimuli can potentially further inform understanding of organ-specific disease.
Discussion
Polymorphisms within gene loci implicated in immune responses are associated with a wide range of diseases. Understanding the genetic underpinnings of interindividual variation in immune and inflammatory responses is therefore of fundamental importance. Here, we have assessed the effect of carefully defined innate immune stimuli of high relevance to monocyte activity upon the generation of primary monocyte eQTL, contrasting observations to those in the naïve state. We illustrate how the majority of genes analyzed in monocytes demonstrate an eQTL under at least one condition. The stimuli examined act through well-annotated pathways, causing large-scale changes in gene expression, and are known to affect chromatin remodeling. These two factors together likely underpin the high degree of context specificity observed for induced eQTL. Our data demonstrate that genetic polymorphisms play a significant role in determining the transcriptional response to differing innate immune stimuli in human monocytes and that such effects may be context-specific.
The modulatory effects of genetic variation on gene expression are seen for specific genes dependent on particular stimuli and the kinetics of the response. We uncover that such genetic effects may only be seen in the early response after induction of expression or less commonly in modulating the extent of subsequent down-regulation. Although we only analyzed two time points for LPS, it is plausible that similar temporal effects exist for IFNγ and other stimuli. We frequently observe pathways where multiple genes show individual and specific associations with particular genetic markers. Although we anticipated that eQTL might manifest only after cellular stimulation in highly inducible genes such as cytokines, as observed in IL1B, IL6, and LTA, less expected was the observation of similar induced eQTL in many upstream nodal genes such as TRAF6 and SOCS1.
The resolution of trans-acting genetic factors and associated gene networks is recognized to be highly informative but challenging (19, 28, 53). Treatment with innate immune stimuli reveals a number of marked trans-eQTL including regulatory hubs. This may reflect a synchronizing influence of stimulation on the expression patterns of monocytes, allowing underlying genetic influences to be defined by reducing stochastic variation at the individual cell level. Notably, we observe opposing quantitative effects of chronic LPS and IFNγ on the numbers of trans-associated genes to the MHC class II region that parallel underlying class II expression. These trans-eQTL are likely to arise from cis effects in the MHC and/or to specific human leukocyte antigen (HLA) alleles; however, elucidating the mechanisms underpinning this observation requires further investigation. Context-specific differential cis effects at other loci similarly are perceived to induce consequential trans networks as illustrated by those observed at IFNB1 and IRF2. These data support a model where identifying the genetic effects on a system requires the identification of a pathway, and any relevant effects of variation on upstream activation before the underlying impact of genetic variation and trans associations can be revealed.
The integration of mapping cis- and transacting loci for specific cellular contexts with comparable data sets defining transcription factor binding, chromatin accessibility, and DNA looping across the genome (30) is a key step in resolving the functional genomic landscape on an individual basis. This study demonstrates the importance of considering the cellular context, and by extension a myriad of other stimuli and the developmental state, in the continued effort to understand the nature and functional consequences of genetic variation and translate this into patient benefit through individualized therapy and more rational drug design.
Materials and Methods
Volunteer Collection and PBMC Purification
This study was approved by the Oxfordshire Research Ethics Committee (COREC reference 06/Q1605/55). A total of 432 volunteers of European ancestry were recruited in the Oxfordshire area after written informed consent. The cohort consisted of 194 males and 238 females with a median age of 30 years (mean, 33 years; interquartile range, 19 to 56 years). Whole blood was collected into anticoagulant EDTA-containing blood collection tubes (Vacutainer System, Becton Dickinson). PBMCs, purified using Ficoll-Paque density gradients from 50 ml of whole blood, were washed three times in Hanks’ balanced salt solution without Ca2+ and Mg2+ (Invitrogen), and the number of cells was determined using a hemocytometer.
Monocyte Separation
Magnetic-activated cell sorting (MACS, Miltenyi) was performed to positively separate CD14+ monocytes from 20 million PBMCs, as per the manufacturer’s instructions. This is designed to provide a sample of ~99% purity. Flow cytometry was performed on a subset of samples with similar purities observed. Typical monocyte yields ranged from 1 × 106 to 10 × 106 monocytes per individual, of which 5 × 105 untreated cells were immediately suspended in RLT reagent (Qiagen) to form the naïve subset and snap-frozen for RNA extraction at a later date. All other monocytes were resuspended at a concentration of 1 × 106/ml in 400 μl of prewarmed RPMI 1640 complete medium, supplemented with 20% fetal calf serum, penicillin/streptomycin, and l-glutamine. Cells were rested overnight (16 hours) at 37°C, 5% CO2 in 5-ml nonadherent polypropylene cell culture tubes (BD Biosciences) before stimulation assays.
Stimulation Assays
For stimulation assays, 4 × 105 monocytes were exposed to ultrapure LPS (20 ng/ml) (catalog # tlrl-pelps, Invivogen) for either 2 or 24 hours. Alternatively, monocytes were exposed to IFNγ (catalog # 285-IF, R&D Systems) at a concentration of 20 ng/ml for 24 hours. The number of treated samples per individual depended on the total number of cells purified, and the priority on sample accrual was collation of IFNγ-treated followed by 24-hour LPS and then 2-hour LPS, respectively. Experiments were terminated simultaneously, ensuring identical incubation periods for all samples. A subset of samples was kept in the incubator without stimulation throughout this period to ascertain incubator effects. All experiments were completed within 48 hours of blood sample collection.
Genomic DNA Extraction
Genomic DNA was extracted from whole blood using the Gentra Puregene Blood Kit following the manufacturer’s instruction (Qiagen). The DNA wasquantified by the PicoGreen dsDNA (double-stranded DNA) quantification assay (Invitrogen).
RNA Extraction
Total RNA was extracted in batches using the RNeasy Mini Kit from cells collected in the RLT reagent following the manufacturer’s instruction using an additional DNase step to minimize contamination with genomic DNA (Qiagen). Total RNA was quantified by NanoDrop and Bioanalyzer for a subset of samples (Bioanalyzer RNA 6000 Nano Kit, Agilent).
Genotyping
Genotyping was performed with Illumina HumanOmniExpress-12v1.0 BeadChip with coverage of 733,202 separate markers. PLINK (54) was used for initial genotyping quality control (QC), including identity by descent (IBD) analysis to ensure individuals were unrelated, and heterozygosity testing and multidimensional scaling to identify population outliers compared to HapMap populations. For SNP filtering, we used sample and SNP call rates >98%. We used a MAF of 4% and a Hardy-Weinberg equilibrium threshold set at 1 × 10−6. We removed 11 individuals because of failure in one or more QC criteria, resulting in 421 individuals for further analysis with genotyping data available at 609,704 SNPs. Genotyping files were subsequently converted from PLINK output for use in MatrixEQTL.
Gene Expression Analysis
Total RNA from naïve and, where available, treated monocytes from each individual was quantified using the Illumina HumanHT-12 v4 BeadChip gene expression array platform with 47,231 probes. Of these, 28,688 probes correspond to coding transcripts with well-established annotations (RefSeq content NM), 11,121 to coding transcript with provisional annotation (XM), 1752 to noncoding transcript with well-established annotation (NR), 2209 to noncoding transcript with provisional annotation (XR), and 3461 to experimentally confirmed mRNA sequences that align to expressed sequence tag (EST) clusters from UniGene. Analysis of 1488 arrays was performed in three separate batches, the first consisting of 288 naïve monocyte samples as previously described (8). The second batch consisted of 636 samples from the same individuals randomized to individual and treatment across array. A third batch consisted of 564 samples composed of 144 further naïve monocyte samples, as well as 420 treated samples that were similarly randomized to individual and treatment across arrays. Complementary RNA synthesis, cleanup, and hybridization were as per the manufacturer’s instructions. Arrays were read, and data were exported using BeadStudio software (Illumina). Samples were subsequently processed in R using the R packages lumi, limma, and ComBat (55–57). Principal component analysis (PCA) was performed to identify samples within each treatment cohort with outlier expression for removal from further analyses. After QC and removal of duplicates and sample outliers, 1438 independent samples were suitable for further analysis. Raw data were transformed and normalized using robust spline normalization within lumi. Samples were then corrected for batch effects using ComBat. A linear model incorporating data from 59 untreated, incubated samples and 421 untreated, nonincubated samples was used to estimate the effect of incubation on gene expression. This was then regressed from all treated samples to minimize the influence of incubator effects on the analysis. Differential gene expression analysis was performed with the limma package. Pathway analysis was performed with IPA (Ingenuity Systems) to define gene networks, significant upstream regulators, and relationships with canonical signaling pathways. For network analysis of trans gene lists, a global molecular network based on the IP knowledge base (IPKB) was used with a P value based on the fit of the trans genes and list of biological functions within IPKB.
Probe Filtering
Normalization and correction for batch and incubator effects were carried out on all probes. Probes were subsequently selected for eQTL analysis. As previously described (8), probe sequences mapping to more than one genomic location using Basic Local Alignment Search Tool (BLAST) were excluded from eQTL analysis. Additionally, 6137 probes found to anneal in regions with SNPs present at a MAF of at least 1% in the European population of the 1000 Genomes Project were excluded from eQTL analysis to minimize potential confounding effects, as were probes mapping to non-autosomal locations. Remaining probes were included in eQTL analysis if they were present (Illumina detection P < 0.01) in >2.5% of samples from a specific treatment or, alternatively, >5% of all samples. This resulted in 15,421 probes for analysis, mapping to 11,168 independent Entrez ID.
eQTL Analysis
To account for confounding variation in the gene expression data, we carried out PCA for each of the four conditions. For each set of eQTL analyses, the dominant PC were chosen for inclusion as covariates in the model such that the number of significant associations was maximized across all conditions. This resulted in the use of 30 (cis analysis of 228 shared samples), 16 (trans analysis of 228 shared samples), 40 (cis analysis of full data sets), and 20 PC (trans analysis of full data sets) as shown in fig. S2. The eQTL analysis was performed with the R package MatrixEQTL (18) using an additive linear model. SNPs were included in the cis analysis if they were located within 1 Mb of the gene expression probe under consideration. Correspondingly, associations between SNPs and probes outside this window were deemed trans. FDRs reported for the resulting associations are as reported by MatrixEQTL. For single SNP eQTL analysis, the same procedure was carried out, but with all analysis performed on a single SNP.
ChIP-seq Analysis
Monocytes were purified from donor blood cones from two healthy volunteers, and 5 × 106 cells were used per stimulation assay (n = 6). Cells were either untreated or treated as per LPS2 and IFNγ stimulations for the eQTL analysis. At the end of the experiment, cells were cross-linked and pellets were frozen. ChIP was performed with cross-linked cells lysed on ice in a lysis buffer [140 mM NaCl, 50 mM Hepes (pH 7.5), 1 mM EDTA (pH 8), 0.5 mM EGTA (pH 8), 0.5% (v/v) NP-40, 0.25% (v/v) Triton X, and 10% glycerol]. Lysis buffer was supplemented with 1× Complete Protease Inhibitor Cocktail (Roche), 0.5 mM phenylmethylsulfonyl fluoride (Sigma), pepstatin (10 μg/μl) (Sigma), and leupeptin hemisulfate (10 μg/μl) (Sigma) before usage. Nuclear pellets were collected after brief centrifugation and washed using a buffer [10 mM tris (pH 8), 1 mM EDTA (pH 8), 2% (v/v) Triton X, 0.2% (v/v) Na deoxycholate, 10% glycerol] similarly supplemented as described above. Sonication to fragment cross-linked chromatin was carried out using the Bioruptor (Diagenode) at high power over 12 cycles of 30 s each. Fragmented chromatin was incubated overnight with Dynabeads (Life Technologies) precoated with 10 μg of anti-IRF2 antibody [IRF-2 (C-19): sc-498, Santa Cruz Biotechnology]. Progressive washing of the chromatin-Dynabead mixture was then carried out in 10 mM tris (pH 8), 1 mM EDTA (pH 8), 1% (v/v) NP-40, 0.7% (v/v) Na deoxycholate, 0.5 M LiCl, supplemented as previously but using only 0.5× Complete Protease Inhibitor. Cross-links were reversed after an overnight incubation in elution buffer [10 mM tris (pH 8), 1 mM EDTA (pH 8), and 1% SDS]. Immunoprecipitated DNA was obtained using phenol-chloroform extraction and purified after precipitation in ethanol. DNA was quantified using the High Sensitivity Qubit system (Life Technologies), and the fragmentation profile was assessed using a DNA HS 2200 TapeStation chip (Agilent).
ChIP libraries were prepared using the NEBNext DNA Sample Prep Master Mix Set 1 according to the manufacturer’s specifications, but with the following amendments: end repair was carried out using 5 μl of NEBNext End Repair Reaction Buffer and 0.5 μl of NEBNext End Repair Enzyme Mix. A-tail was carried out using 5 μl of NEBNext A-tail Reaction Buffer and 0.3 μl of NEBNext Klenow Fragment (3′→5′ exo-) Enzyme Mix. Ligation was carried out using 10 μl of NEBNext Quick Ligation Reaction Buffer and 0.5 μl of NEBNext Quick T4 DNA Ligase Enzyme Mix. The NEBNext adapters were diluted 100-fold, and the amount added was adjusted if less than 5 ng of ChIP material was available. No size selection was carried out. PCR was done using the following reaction mix with 18 cycles of PCR: DNA (36 μl), 5× Phusion buffer (10 μl), custom paired end (PE) primer 1 (1 μl), custom PE primer 2 (1 μl), dNTP (deoxynucleotide triphosphate) mix (1.5 μl), Phusion polymerase (0.5 μl). The cleanup after amplification was done at 1:0.85 (AMPure beads/DNA). The concentration of each library was determined by real-time PCR using Agilent qPCR Library Quantification Kit and a MX3005P instrument (Agilent). Sequencing was performed on an Illumina HiSeq 2000 using 50–base pair paired-end reads.
Reads were mapped using Stampy (58), and peaks were called with MACS2 (59) using non-immunoprecipitated DNA as a control for enrichment. Analysis was performed with BEDTools (60). Only peaks found in both samples were used for comparison with trans-associated probes to rs13149699. We observed 481 independent peaks that overlapped by a minimum of 10% in the two biological replicates in the untreated state: 170 after 2-hour LPS2 and 154 after IFNγ stimulation. The number of peaks found within defined distances (as per Fig. 4F) of trans-associated probes was counted and compared to those found within the same distance of non–trans-associated probes that had been similarly tested for eQTL. A Fisher’s exact test was performed to test significance of number of occurrences per genomic window flanking the trans-associated probes versus non-associated probes.
Imputation of Genotypes
Genotypes were prephased with SHAPEIT2 (61), and missing genotypes were imputed with IMPUTE2 (62) in 5-Mb chunks against the 1000 Genomes reference panel (63). Summary statistics for each chunk were obtained using SNPTEST (https://mathgen.stats.ox.ac.uk/genetics_software/snptest/snptest.html), and sites with an information score of less than 0.9 or significant departure from Hardy-Weinberg equilibrium (P < 1 × 10−3) were excluded from further analysis. Genotype probabilities for all remaining sites were converted into dosage estimates and formatted for use with MatrixEQTL.
Conditional Analysis
To investigate the presence of local associations in addition to the most significant SNP reported for each probe, we carried out a conditional analysis with imputed genotypes. To this end, the cis-eQTL analysis was repeated for all gene expression probes that had produced an association with a P value of no more than 5 × 10−8, conditioning on the initial association. If no additional independent eQTL are present, it is expected that this will not identify any additional significant associations. Conversely, any associations that are independent of the first one are expected to remain and may increase in significance. This process was iterated until no further significant associations remained (P < 5 × 10−8). To confirm the presence of an independent signal, this process was repeated for the second and third most significantly associated SNP for a given probe. Only examples where the secondary signal was present in all instances are reported.
The resulting eQTL were further characterized by identifying the list of SNPs associated with each of the peak eSNPs. To this end, all SNPs that are showing significant association with gene expression are assigned to the eSNP with which they have the highest r2. Peaks identified in this way were matched across conditions, and the list of SNPs observed in all treatments in which the peak is present is reported.
Overlap Between eQTL Signals and GWAS Catalog
All traits listed in the catalog of published GWAS (www.genome.gov/gwastudies/; date accessed 10 January 2013) were extracted and expertly curated into phenotypic groupings blind to GWAS or eSNP data. Groups were defined on the basis of organ specificity of a particular trait, disease process, or type. These included cardiovascular, respiratory, gastroenterological, urological, rheumatological, neurological, renal, endocrine, hematological, dermatological, bone, cancer, immunity and inflammation, autoimmune, allergy, genetic, viral infection, bacterial infection, parasitic disease, measurement, physiological, metabolic, chronic or degenerative disease, reproduction, and drug-related. We recognized that classification of a given trait was possible into multiple groups, and to capture this diversity, for each trait, we assigned trait membership into three possible groups.
A reported GWAS SNP was considered to coincide with an eQTL identified during the conditional analysis of cis associations if the GWAS SNP itself or any SNPs with r2 > 0.8 with this SNP were part of one of the eQTL peaks (P < 5 × 10−8). Enrichment of peak eQTLs for individual GWAS categories, as well as overall enrichment of GWAS SNPs, was tested with Fisher’s exact test by comparing the overlap obtained for peak eQTLs to that observed for all SNPs tested for cis associations. Publicly available summary statistics for a 200-kb window around CARD9 from the latest CD GWAS meta-analysis (46) were extracted using the Ricopili tool (www.broadinstitute.org/mpg/ricopili/). LD calculations were performed with PLINK (54) and the phase 1 1000 Genomes data (63). Approximate conditional analysis, based on summary statistics and LD properties, was carried out using the method described in (46).
Flow Cytometry
To validate the changes in class II HLA observed at transcriptomic level with IFN-γ and LPS treatment, we assessed class II expression on primary monocytes by multiparametric flow cytometry. Monocytes were stimulated, as described, before being washed and stained with violet viability dye (Invitrogen) as per the manufacturer’s instructions. To reduce nonspecific staining, a three-step procedure adapted from a protocol shared by R. Apps (National Cancer Institute, National Institutes of Health) was used. Human immunoglobulin G (IgG) was added to each tube, followed by mouse anti-human antibodies directed against HLA-DP (clone B7/21, Abcam), HLA-DQ (clone SPV-L3, Abcam), HLA-DR (clone L243, BD Biosciences), or all class II alleles (clone Tu39 and SK10/SPV-L3 combined) or an isotype control. After 20 min, cells were washed and stained with sheep anti-mouse PE-labeled antibody (Sigma) and sheep IgG.
After a further 20 min, cells were washed and stained with mouse anti-human CD14–FITC (fluorescein isothiocyanate) (clone 61D3, eBioscience) and mouse IgG (Sigma). Cells were fixed with the paraformaldehyde containing Reagent A (Life Technologies), and data were acquired immediately on a FACSCanto (BD Biosciences) machine. A minimum of 10,000 gated monocytes were acquired for each sample. Fluorescence-minus-one controls were used to set gate thresholds.
Data Presentation
Graphs were generated using ggplot2 (64) and R base packages. Box plots of gene expression data show values corrected for confounding variance using gene expression PC as per eQTL analysis. Circular plots were created using the ggbio R package (65), and local association plots were generated with LocusZoom (66).
Supplementary Material
Acknowledgments
Gene expression and ChIP-seq data are available through ArrayExpress. Genotyping data have been deposited at the European Genome-phenome Archive (EGA) and are available on request (EGAS00000000109). We are very grateful to all the volunteers who participated in this study together with members of the Knight laboratory for their support. We also thank G. McVean and D. Kwiatkowski for support of this study and helpful suggestions. This work was supported by the Wellcome Trust [grants 074318 (J.C.K.), 088891 (B.P.F.), and 090532/Z/09/Z (core facilities Wellcome Trust Centre for Human Genetics including High-Throughput Genomics Group)], the European Research Council (ERC) under the European Union’s Seventh Framework Programme (FP7/2007-2013)/ERC grant agreement no. 281824 ( J.C.K.), the Medical Research Council (98082, J.C.K.), and the National Institute for Health and Research Oxford Biomedical Research Centre.
Footnotes
Supplementary Materials www.sciencemag.org/content/343/6175/1246949/suppl/DC1
Materials and Methods
Figs. S1 to S12
Database S1
Tables S1 to S7
References (67-83)
References and Notes
- 1.Parkes M, Cortes A, van Heel DA, Brown MA. Genetic insights into common pathways and complex relationships among immune-mediated diseases. Nat. Rev. Genet. 2013;14:661–673. doi: 10.1038/nrg3502. doi: 10.1038/nrg3502. [DOI] [PubMed] [Google Scholar]
- 2.Maurano MT, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nica AC, et al. Candidate causal regulatory effects by integration of expression QTLs with complex trait genetic associations. PLOS Genet. 2010;6:e1000895. doi: 10.1371/journal.pgen.1000895. doi: 10.1371/journal.pgen.1000895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Emilsson V, et al. Genetics of gene expression and its effect on disease. Nature. 2008;452:423–428. doi: 10.1038/nature06758. doi: 10.1038/nature06758. [DOI] [PubMed] [Google Scholar]
- 5.Moffatt MF, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448:470–473. doi: 10.1038/nature06014. doi: 10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
- 6.Cookson W, Liang L, Abecasis G, Moffatt M, Lathrop M. Mapping complex disease traits with global gene expression. Nat. Rev. Genet. 2009;10:184–194. doi: 10.1038/nrg2537. doi: 10.1038/nrg2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dimas AS, et al. Common regulatory variation impacts gene expression in a cell type–dependent manner. Science. 2009;325:1246–1250. doi: 10.1126/science.1174148. doi: 10.1126/science.1174148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fairfax BP, et al. Genetics of gene expression in primary immune cells identifies cell type–specific master regulators and roles of HLA alleles. Nat. Genet. 2012;44:502–510. doi: 10.1038/ng.2205. doi: 10.1038/ng.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nica AC, et al. The architecture of gene regulatory variation across multiple human tissues: The MuTHER study. PLOS Genet. 2011;7:e1002003. doi: 10.1371/journal.pgen.1002003. doi: 10.1371/journal.pgen.1002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Medzhitov R. Toll-like receptors and innate immunity. Nat. Rev. Immunol. 2001;1:135–145. doi: 10.1038/35100529. doi: 10.1038/35100529. [DOI] [PubMed] [Google Scholar]
- 11.Serbina NV, Jia T, Hohl TM, Pamer EG. Monocyte-mediated defense against microbial pathogens. Annu. Rev. Immunol. 2008;26:421–452. doi: 10.1146/annurev.immunol.26.021607.090326. doi: 10.1146/annurev.immunol.26.021607.090326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Escoubet-Lozach L, et al. Mechanisms establishing TLR4-responsive activation states of inflammatory response genes. PLOS Genet. 2011;7:e1002401. doi: 10.1371/journal.pgen.1002401. doi: 10.1371/journal.pgen.1002401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grossman SR, et al. Identifying recent adaptations in large-scale genomic data. Cell. 2013;152:703–713. doi: 10.1016/j.cell.2013.01.035. doi: 10.1016/j.cell.2013.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Orozco LD, et al. Unraveling inflammatory responses using systems genetics and gene-environment interactions in macrophages. Cell. 2012;151:658–670. doi: 10.1016/j.cell.2012.08.043. doi: 10.1016/j.cell.2012.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Manry J, et al. Evolutionary genetic dissection of human interferons. J. Exp. Med. 2011;208:2747–2759. doi: 10.1084/jem.20111680. doi: 10.1084/jem.20111680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu X, Ivashkiv LB. Cross-regulation of signaling pathways by interferon-γ: Implications for immune responses and autoimmune diseases. Immunity. 2009;31:539–550. doi: 10.1016/j.immuni.2009.09.002. doi: 10.1016/j.immuni.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.See supporting materials and methods in Science Online.
- 18.Shabalin AA. Matrix eQTL: Ultra fast eQTL analysis via large matrix operations. Bioinformatics. 2012;28:1353–1358. doi: 10.1093/bioinformatics/bts163. doi: 10.1093/bioinformatics/bts163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fehrmann RS, et al. Trans-eQTLs reveal that independent genetic variants associated with a complex phenotype converge on intermediate genes, with a major role for the HLA. PLOS Genet. 2011;7:e1002197. doi: 10.1371/journal.pgen.1002197. doi: 10.1371/journal.pgen.1002197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Biswas S, Storey JD, Akey JM. Mapping gene expression quantitative trait loci by singular value decomposition and independent component analysis. BMC Bioinformatics. 2008;9:244. doi: 10.1186/1471-2105-9-244. doi: 10.1186/1471-2105-9-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gaffney DJ. Global properties and functional complexity of human gene regulatory variation. PLOS Genet. 2013;9:e1003501. doi: 10.1371/journal.pgen.1003501. doi: 10.1371/journal.pgen.1003501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rotival M, et al. Integrating genome-wide genetic variations and monocyte expression data reveals trans-regulated gene modules in humans. PLOS Genet. 2011;7:e1002367. doi: 10.1371/journal.pgen.1002367. doi: 10.1371/journal.pgen.1002367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cullell-Young M, et al. From transcription to cell surface expression, the induction of MHC class II I-Aα by interferon-γ in macrophages is regulated at different levels. Immunogenetics. 2001;53:136–144. doi: 10.1007/s002510100312. doi: 10.1007/s002510100312. [DOI] [PubMed] [Google Scholar]
- 24.Baron JM, et al. Cytochrome P450 1B1: A major P450 isoenzyme in human blood monocytes and macrophage subsets. Biochem. Pharmacol. 1998;56:1105–1110. doi: 10.1016/s0006-2952(98)00105-1. doi: 10.1016/S0006-2952(98)00105-1. [DOI] [PubMed] [Google Scholar]
- 25.Bandiera S, et al. Proteasomal degradation of human CYP1B1: Effect of the Asn453Ser polymorphism on the post-translational regulation of CYP1B1 expression. Mol. Pharmacol. 2005;67:435–443. doi: 10.1124/mol.104.006056. doi: 10.1124/mol.104.006056. [DOI] [PubMed] [Google Scholar]
- 26.Metz R, et al. Novel tryptophan catabolic enzyme IDO2 is the preferred biochemical target of the antitumor indoleamine 2,3-dioxygenase inhibitory compound D-1-methyl-tryptophan. Cancer Res. 2007;67:7082–7087. doi: 10.1158/0008-5472.CAN-07-1872. doi: 10.1158/0008-5472.CAN-07-1872. [DOI] [PubMed] [Google Scholar]
- 27.Amisten S, Melander O, Wihlborg AK, Berglund G, Erlinge D. Increased risk of acute myocardial infarction and elevated levels of C-reactive protein in carriers of the Thr-87 variant of the ATP receptor P2Y11. Eur. Heart J. 2007;28:13–18. doi: 10.1093/eurheartj/ehl410. doi: 10.1093/eurheartj/ehl410. [DOI] [PubMed] [Google Scholar]
- 28.Small KS, et al. Identification of an imprinted master trans regulator at the KLF14 locus related to multiple metabolic phenotypes. Nat. Genet. 2011;43:561–564. doi: 10.1038/ng.833. doi: 10.1038/ng.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Westra HJ, et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat. Genet. 2013;45:1238–1243. doi: 10.1038/ng.2756. doi: 10.1038/ng.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.ENCODE Project Consortium et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matsuyama T, et al. Targeted disruption of IRF-1 or IRF-2 results in abnormal type I IFN gene induction and aberrant lymphocyte development. Cell. 1993;75:83–97. doi: 10.1016/S0092-8674(05)80086-8. [PubMed] [Google Scholar]
- 32.Kröger A, Köster M, Schroeder K, Hauser H, Mueller PP. Activities of IRF-1. J. Interferon Cytokine Res. 2002;22:5–14. doi: 10.1089/107999002753452610. doi: 10.1089/107999002753452610. [DOI] [PubMed] [Google Scholar]
- 33.Harada H, et al. Structurally similar but functionally distinct factors, IRF-1 and IRF-2, bind to the same regulatory elements of IFN and IFN-inducible genes. Cell. 1989;58:729–739. doi: 10.1016/0092-8674(89)90107-4. doi: 10.1016/0092-8674(89)90107-4. [DOI] [PubMed] [Google Scholar]
- 34.Krüger E, Kloetzel PM. Immunoproteasomes at the interface of innate and adaptive immune responses: Two faces of one enzyme. Curr. Opin. Immunol. 2012;24:77–83. doi: 10.1016/j.coi.2012.01.005. doi: 10.1016/j.coi.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 35.Chevillard G, Blank V. NFE2L3 (NRF3): The Cinderella of the Cap‘n’Collar transcription factors. Cell. Mol. Life Sci. 2011;68:3337–3348. doi: 10.1007/s00018-011-0747-x. doi: 10.1007/s00018-011-0747-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gaulton KJ, et al. A map of open chromatin in human pancreatic islets. Nat. Genet. 2010;42:255–259. doi: 10.1038/ng.530. doi: 10.1038/ng.530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gregory AP, et al. TNF receptor 1 genetic risk mirrors outcome of anti-TNF therapy in multiple sclerosis. Nature. 2012;488:508–511. doi: 10.1038/nature11307. doi: 10.1038/nature11307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.GoDARTS and UKPDS Diabetes Pharmacogenetics Study Group et al. Common variants near ATM are associated with glycemic response to metformin in type 2 diabetes. Nat. Genet. 2011;43:117–120. doi: 10.1038/ng.735. doi: 10.1038/ng.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sasaki M, Ikeda H, Sato Y, Nakanuma Y. Proinflammatory cytokine-induced cellular senescence of biliary epithelial cells is mediated via oxidative stress and activation of ATM pathway: A culture study. Free Radic. Res. 2008;42:625–632. doi: 10.1080/10715760802244768. doi: 10.1080/10715760802244768. [DOI] [PubMed] [Google Scholar]
- 40.Watling D, Carmo CR, Kerr IM, Costa-Pereira AP. Multiple kinases in the interferon-γ response. Proc. Natl. Acad. Sci. U.S.A. 2008;105:6051–6056. doi: 10.1073/pnas.0710814105. doi: 10.1073/pnas.0710814105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.De Jager PL, et al. Meta-analysis of genome scans and replication identify CD6, IRF8 and TNFRSF1A as new multiple sclerosis susceptibility loci. Nat. Genet. 2009;41:776–782. doi: 10.1038/ng.401. doi: 10.1038/ng.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kirino Y, et al. Genome-wide association analysis identifies new susceptibility loci for Behçet’s disease and epistasis between HLA-B*51 and ERAP1. Nat. Genet. 2013;45:202–207. doi: 10.1038/ng.2520. doi: 10.1038/ng.2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Franke A, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat. Genet. 2010;42:1118–1125. doi: 10.1038/ng.717. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu J, et al. Genome-wide association study in Chinese men identifies two new prostate cancer risk loci at 9q31.2 and 19q13.4. Nat. Genet. 2012;44:1231–1235. doi: 10.1038/ng.2424. doi: 10.1038/ng.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Evans DM, et al. Interaction between ERAP1 and HLA-B27 in ankylosing spondylitis implicates peptide handling in the mechanism for HLA-B27 in disease susceptibility. Nat. Genet. 2004;43:761–767. doi: 10.1038/ng.873. doi: 10.1038/ng.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jostins L, et al. Host–microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rivas MA, et al. Deep resequencing of GWAS loci identifies independent rare variants associated with inflammatory bowel disease. Nat. Genet. 2011;43:1066–1073. doi: 10.1038/ng.952. doi: 10.1038/ng.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sokol H, et al. Card9 mediates intestinal epithelial cell restitution, T-helper 17 responses, and control of bacterial infection in mice. Gastroenterology. 2013;145:591–601.e593. doi: 10.1053/j.gastro.2013.05.047. doi: 10.1053/j.gastro.2013.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Crosslin DR, et al. Genetic variants associated with the white blood cell count in 13,923 subjects in the eMERGE Network. Hum. Genet. 2012;131:639–652. doi: 10.1007/s00439-011-1103-9. doi: 10.1007/s00439-011-1103-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marinho S, Custovic A, Marsden P, Smith JA, Simpson A. 17q12-21 variants are associated with asthma and interact with active smoking in an adult population from the United Kingdom. Ann. Allergy Asthma Immunol. 2012;108:402–411.e409. doi: 10.1016/j.anai.2012.03.002. doi: 10.1016/j.anai.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 51.Hao K, et al. Lung eQTLs to help reveal the molecular underpinnings of asthma. PLOS Genet. 2012;8:e1003029. doi: 10.1371/journal.pgen.1003029. doi: 10.1371/journal.pgen.1003029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kalliokoski A, Niemi M. Impact of OATP transporters on pharmacokinetics. Br. J. Pharmacol. 2009;158:693–705. doi: 10.1111/j.1476-5381.2009.00430.x. doi: 10.1111/j.1476-5381.2009.00430.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grundberg E, et al. Mapping cis- and trans-regulatory effects across multiple tissues in twins. Nat. Genet. 2012;44:1084–1089. doi: 10.1038/ng.2394. doi: 10.1038/ng.2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Purcell S, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Du P, Kibbe WA, Lin SM. lumi: A pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–1548. doi: 10.1093/bioinformatics/btn224. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 56.Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. ComBat, sva: Surrogate Variable Analysis, R package version 3.6.0.
- 57.Smyth GK. Limma: Linear models for microarray data. In: Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W, editors. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. Springer; New York: 2005. pp. 397–420. [Google Scholar]
- 58.Lunter G, Goodson M. Stampy: A statistical algorithm for sensitive and fast mapping of Illumina sequence reads. Genome Res. 2011;21:936–939. doi: 10.1101/gr.111120.110. doi: 10.1101/gr.111120.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Quinlan AR, Hall IM. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Delaneau O, Zagury JF, Marchini J. Improved whole-chromosome phasing for disease and population genetic studies. Nat. Methods. 2013;10:5–6. doi: 10.1038/nmeth.2307. doi: 10.1038/nmeth.2307. [DOI] [PubMed] [Google Scholar]
- 62.Marchini J, Howie B. Genotype imputation for genome-wide association studies. Nat. Rev. Genet. 2010;11:499–511. doi: 10.1038/nrg2796. doi: 10.1038/nrg2796. [DOI] [PubMed] [Google Scholar]
- 63.1000 Genomes Project Consortium et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wickham H. ggplot2: Elegant Graphics for Data Analysis. Springer; New York: 2009. [Google Scholar]
- 65.Yin T, Cook D, Lawrence M. ggbio: An R package for extending the grammar of graphics for genomic data. Genome Biol. 2012;13:R77. doi: 10.1186/gb-2012-13-8-r77. doi: 10.1186/gb-2012-13-8-r77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pruim RJ, et al. LocusZoom: Regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.