Abstract
Increasing evidence from mouse models of Alzheimer's disease shows that overexpression of a mutant form of the amyloid precursor protein (APP) and its product, β-amyloid peptide, initiate pathological changes before amyloid deposition. To evaluate the cytological basis for one of these early changes, namely reduced volume of the dentate gyrus (DG), we have used high-throughput diOlistic cell loading and 3D neuronal reconstruction to investigate potential dendritic pathology of granule cells (GCs) in 90-day-old PDAPP mice. Labeled GCs from fixed hippocampal slices were selected randomly and imaged digitally by using confocal laser-scanning microscopy. The dendritic complexity of GCs was quantified according to subordinate morphological parameters, including soma position within the granule cell layer (superficial versus deep) and topographic location within the DG (dorsal versus ventral blade) along the anterior-posterior hippocampal axis. Initial analysis, which included all sampled GC types, revealed a 12% reduction of total dendritic length in PDAPP mice compared with littermate controls. Further analysis, performed with refined subgroups, found that superficially located GCs in the dorsal blade were profoundly altered, exhibiting a 23% loss in total dendritic length, whereas neurons in the ventral blade were unaffected. Superficial GCs were particularly vulnerable (a 32% reduction) in the posterior region of the DG. Furthermore, the dendritic reductions of this select group were uniformly localized within middle-to-outer portions of the dentate molecular layer. We conclude that substantial dendritic pathology is evident in 90-day-old PDAPP mice for a spatially defined subset of GCs well before amyloid accumulation occurs.
The classical neuropathological hallmarks of Alzheimer's disease (AD) include the presence of neurofibrillary tangles within neurons and extracellular cerebrovascular, diffuse, and neuritic plaques (1). The key molecular constituent of the plaque is β-amyloid (Aβ), a 39- to 43-amino acid amyloidogenic peptide, which is a product of the proteolytic processing of the amyloid precursor protein (APP) (2). It has been shown that familial APP mutations may facilitate AD-like neuropathological changes by accelerating aberrant APP proteolytic processing. This hypothesis has been reinforced by evidence from transgenic (Tg) mouse models of AD in which overexpression of mutant APP results in amyloid deposition (3–10). Neuritic dystrophy associated with the deposits of Aβ in AD brains is evidenced by progressive dendritic dystrophy within the hippocampal complex (11–17). Although the assumption has been that Aβ deposition leads to neuritic dystrophy that would impair hippocampal function (18, 19), the causal links between the deposition and pathology have been largely inferential, with minimal data available on the time course of these events.
Tg mouse models have provided a unique opportunity to characterize Aβ-induced neuropathology, including the nature and time of onset of the physiological and morphological alterations of hippocampal neurons characteristic of human AD (20, 21). One such Tg mouse line, PDAPP mice, overexpresses human APP with the 717V→F Indian mutation, and exhibits extensive Aβ deposition in an age- and brain region-specific manner, analogous to that seen in human AD patients, in particular, in the hippocampus (3, 6–8, 16). Recent evidence indicates that PDAPP mice exhibit early behavioral (10, 22–24), biochemical (25, 26), electrophysiological (27, 28), and morphological (29) changes in the hippocampus before the onset of amyloid deposition. The findings from these mouse models support the conclusion that, whereas AD is a progressive human disease culminating in dementia, pathological changes and more subtle functional deficits may appear well in advance of obvious memory loss and cognitive impairment. Given that soluble forms of Aβ have been suggested to lead to neuropathological changes well in advance of its deposition as plaques (23, 24, 26, 30–33), the early detection of any pathological conditions induced by mutant APP overexpression could provide a basis for evaluation of potential early therapeutic intervention (3, 5, 34).
Recently, 100-day-old PDAPP mice were shown to exhibit a 12% overall reduction in hippocampal volume when compared with wild-type mice of the same age (35). Further stereological analysis of hippocampal subfields localized the volumetric reduction (28%) to the molecular layer (ML) and the granule cell layer (GCL) of the dentate gyrus (DG) (35). The present study sought to define the cytological correlate(s) of this early-onset volumetric reduction found within the DG of young PDAPP mice. A high-throughput diOlistic cell loading method (36) combined with computer-assisted 3D reconstruction and morphometric analysis was used to compare the dendritic morphology of granule cells (GCs). The results reveal that overexpression of APP induces selective dendritic pathology in a specific neuronal population within the DG before Aβ deposition.
Materials and Methods
Animals and Tissue Preparation. Ninety-day-old (90-d) heterozygous male PDAPP Tg with the APPV717F mutation (37) and nontransgenic (nTg) littermate mice (n = 10 per group, Taconic Farms) were used with their genotypes blinded throughout the course of the study. Mice were anesthetized with avertin (0.5 mg/g of body weight) and transcardially perfused with normal saline followed by 4% paraformaldehyde (PFA) in 0.1 M phosphate buffer. Brains were dissected, fixed (4% PFA) overnight, coronally sectioned (250 μm) with a vibratome (Leica VT1000S), and then stored in 4% PFA at 4°C before diOlistic loading.
Gene Gun Bullet Preparation. The current optimized protocol has been described in detail elsewhere (36). In brief, 15 mg of 1.6-μm gold particles (Bio-Rad) were thoroughly precipitated with 5 mg of lipophilic dye, DiI (Molecular Probes), and dissolved in 500 μl of methylene chloride. After drying on a glass slide, the DiI-coated gold particles were collected and sonicated in 3 ml of distilled water for 5 min. The solution was vortexed for 15 s and then injected into a tubing preparation station (Bio-Rad). After 30 min of rotation, the solution was slowly withdrawn, and the particle-coated tube was then rotated and air-dried under constant nitrogen flow (0.4 liter/min) for 1 h. The tube was then cut into small bullets and stored in a desiccated environment at room temperature.
Delivery of Particles. DiI-coated gold particles were robustly delivered by using the “Helios gene gun system” (PSD-1000/He, Bio-Rad), which was stabilized with a custom-built adjustable stand. A custom-fabricated centering tool was used to adjust the distance of the gun from the target tissue (3.0 cm) and to focus the landing field of delivered particles onto the region of interest. A membrane filter (BD Falcon, catalog no. 3091) was placed between the gun and the tissue to filter out undesirable large clusters of DiI-coated particles. The particles were accelerated by using inert helium gas at a constant pressure (200 psi) to regulate both the velocity and penetration depth of the particles into the tissue slices. Each side of the tissue slice was shot successively to enhance the yield of labeling. Each shot was performed after completion of confocal image acquisition.
Confocal Imaging Procedure. Labeled neurons were imaged by using a Zeiss Pascal confocal laser-scanning microscope equipped with a ×25 Plan NeoFluor water-immersion objective lens (numerical aperture, 0.8). Labeled structures were excited by using a 1-mW He:Ne laser (543 nm for DiI) with their emissions passed through a 560-nm-long pass filter set. For through-focus imaging, optical sections were collected with 0.5- to 1.2-μm z-axis steps to cover the full depth of the dendritic trees, by using illumination and data acquisition parameters as optimized (36). The 2D stacks were superimposed digitally, and the full 3D data set was generated for visualization of dendritic complexity.
Morphological Criteria for Cell Sampling. The criteria for sampling labeled neurons included (i) the presence of a single DiI-coated gold particle within the soma, (ii) the visualization of complete DiI transport evidenced by tapering in the endpoints of the most distal dendrites, (iii) the visualization of the complete 3D profile of dendritic trees by using the 3D display of the Zeiss Pascal confocal laser-scanning microscope, and (iv) the exclusion of cells with dendrites labeled retrogradely by particles in the surrounding neuropil. Neurons with incomplete transport of dye or severe truncations in their proximal dendrites were excluded.
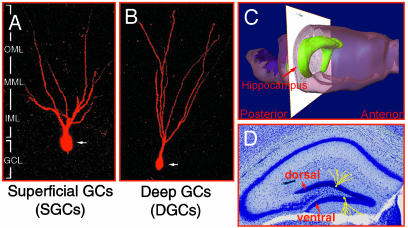
Definition of Subordinate Morphological Parameters. The sampled GCs were classified based on soma position within the GCL across the DG subfields (see Fig. 1). GCs with their somata located in the superficial layer of the GCL were defined as superficial granule cells (SGCs; Fig. 1 A), and those located in the deep layer of the GCL were defined as deep granule cells (DGCs; Fig. 1B). Furthermore, because the hippocampal formation is a 3D anatomical substrate (Fig. 1C), two important topographic contexts were also considered in the quantitative analysis: (i) the location of GCs within the dorsal and ventral blades of the DG (Fig. 1D) and (ii) their locations along the A-P axis of the hippocampus, which was defined based on Hof et al. (38).
Fig. 1.
Morphological parameters of dentate GCs. (A) SGCs. (B) DGCs. Note that the proximal dendrites of SGCs began to bifurcate immediately proximal to the soma, whereas DGCs generally had an unbranched primary dendrite that extended through the GCL without branching until entering the dentate ML. (C and D) DG subfields, dorsal and ventral blades along the anterior-to-posterior (A-P) axis of the hippocampus.
Neuronal Reconstructions and Statistical Analysis. Customized semiautomated 3D reconstruction software was used to contour dendritic processes of digitized 2D optical planes within the 3D image stacks for quantitative analysis of the dendritic complexity of dentate GCs. This software is composed of a group of image-processing algorithms that automate digitized neuron feature extraction and 3D reconstruction for the rapid acquisition of quantitative data. Dendrograms were used to characterize the dendritic morphology of dentate GCs, including the total dendritic length (TDL) of the GCs. Sholl analysis (39) was used to investigate dendritic complexity by examining the number of dendritic intersections and the total dendritic length contained in 20-μm concentric radial intervals from the soma. After analysis of dendritic complexities, the genotypes of the mice were unblinded for statistical analysis. Based on the morphological parameters, a three-way ANOVA (genotype × cell type × topographic location) was used for morphometric comparisons between Tg vs. nTg mice (SPSS, Chicago; P < 0.05, 0.01). For variables with overall significance, comparisons were further analyzed by examining the aforementioned subordinate parameters by using two-way ANOVA, followed by a Tukey honestly significant difference post hoc test (P < 0.05, 0.01).
Results
In the current study, we first validated the efficacy of diOlistic sampling, followed by the internal validation of GC morphology of the animal subjects. The current morphometric comparisons include (i) multiple ANOVA tests to compare the TDL of all sampled GCs between Tg and nTg mice (genotype) and (ii) subsequent analyses with emphasis on subordinate morphological parameters (cell type, topographic location, and A-P axis) to precisely localize the major morphological parameters attributable to any observed dendritic pathology in the 90-d Tg mice.
Nonbiased DiOlistic Sampling of Dentate GCs of Tg vs. nTg Mice. After diOlistic delivery of DiI-coated particles, GCs were randomly sampled across the DG subfields based on the aforementioned standardized morphological criteria (see Materials and Methods). Within the sample pool, an equal number of labeled GCs (n = 81 per group) were sampled from the Tg and nTg groups. Of the 162 sampled GCs, 77 cells were in the dorsal blade and 85 GCs were located in the ventral blade (Table 1). The sampled GCs were also classified into SGCs and DGCs based on their soma positions within the GCL. χ2 tests showed that the distribution of SGCs (n = 97) and DGCs (n = 65) in both blades was statistically similar (χ2 = 2.69, df = 1, P < 0.2), indicating that the current diOlistic protocol provides an objective means to randomly sample both cell types across the DG subfields in Tg and nTg mice.
Table 1. Nonbiased loading of dentate GCs.
| Dorsal blade | Ventral blade | Total | |
|---|---|---|---|
| SGC | 41 | 56 | 97 |
| DGC | 36 | 29 | 65 |
| Total | 77 | 85 | 162 |
Validation of Interneuronal Variation of Tg and nTg Mice. Before comparative analysis of GC morphology across Tg and nTg mice, interneuronal variations of GCs were quantified within each group for internal validation (Fig. 6, which is published as supporting information on the PNAS web site). A two-way ANOVA revealed no significant difference in TDL between the cell type (P < 0.21) and topographic location (P < 0.25) of GCs in the nTg group (Fig. 6A). The TDL previously found for C57 mice (not shown) is statistically similar to the TDL of nTg mice reported in this study and, hence, serves as an additional source of internal validation of the present morphological analysis. Similarly, no differences in TDLs were found between the cell type (P < 0.78) and topographic location (P < 0.09) in the Tg group (Fig. 6B). These two data sets confirm the homogeneity of dendritic morphology of GCs sampled within each group of mice and merit a comparative analysis between the genotypes.
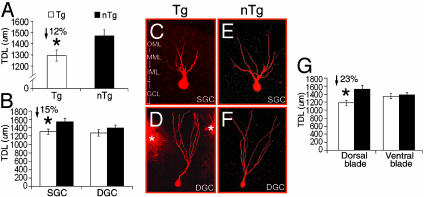
Genotype Effect: Reductions in TDLs of Dentate GCs in Tg Mice. For quantitative analysis, a three-way ANOVA (genotype × cell type × topography) was used to investigate the difference in the TDL of GCs between the 90-d Tg and nTg groups. A statistically significant main effect of genotype (P < 0.02) revealed a 12% reduction of TDL for Tg mice as compared with nTg mice (Fig. 2A) when all GCs studied were included in the analysis. In addition, a trend toward an interaction between topography and genotype was found (P < 0.06) in Tg mice.
Fig. 2.
Comparison of the TDL of dentate GCs between Tg and nTg mice. (A) Genotype. A 12% reduction in TDLs of GCs occurred in Tg mice. (B) Cell type. A 15% reduction in TDLs of SGCs occurred in Tg mice, whereas no significant difference was found in DGCs. (C and D) Dendritic morphology of GCs of Tg mice. (E and F) Dendritic morphology of GCs of nTg mice. Note: SGCs of Tg mice exhibited a significant reduction in dendritic trees in comparison with that of nTg mice (C vs. E), whereas no difference was found for DGCs (D vs. F). Asterisks indicate surrounding DiI clusters. (G) Topographic location. The reduction in TDLs was increased up to 23% in the dorsal blade of Tg mice. No difference was found in the ventral blade of the DG.
This analysis clearly demonstrated a difference in TDL across genotype; however, the possibility that these differences were attributable to a specific subpopulation of GCs was not tested. Accordingly, a series of ANOVA tests was used to further disclose any morphological properties attributable to the reported reduction in TDL of Tg mice. First, cell-type-derived differences in TDL between Tg and nTg mice were examined followed by a separate analysis of topographic-location-derived differences in TDL (independent of cell type). Finally, the two parameters were examined together to elucidate any interaction effects.
Morphological Effects: Subclassification of GCs by Cell Type and Topographic Location. By using the cell type as the only parameter, only the TDLs of SGCs were found to be significantly different (P < 0.03) between Tg and nTg mice. The TDL of SGCs in Tg mice was found to be 15% shorter than that of SGCs in nTg mice (P < 0.03, post hoc, P < 0.05; Fig. 2B), although no significant difference was found for DGCs between the two groups (P < 0.29). These findings demonstrate that the 12% decrease in TDL found when comparing all GCs is principally attributable to SGCs alone. In Fig. 2 C–F, the morphological comparison of GCs between Tg and nTg mice demonstrated a significant shortening of the dendrites of SGCs (Fig. 2 C and E) rather than DGCs of Tg mice (Fig. 2 D and F).
Next, topographic location was used as the only parameter to classify the total GCs for analysis (Fig. 2G). A statistically significant main effect was found for genotype (P < 0.02) and an interaction effect of genotype × topography (P < 0.05). Analysis revealed that Tg mice exhibited a 23% reduction in TDLs of GCs localized to the dorsal blade of the DG (P < 0.001), whereas no changes were seen in the GCs of the ventral blade (P < 0.76).
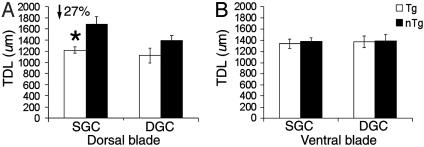
Finally, the interaction effect of both parameters (cell type and topographic location) was examined to further pinpoint the morphological correlates of dendritic reductions in Tg mice. By using ANOVA, a main effect of genotype was found within the dorsal blade (P < 0.001; Fig. 3A). Within the dorsal blade, a statistically significant difference in TDLs was found for SGCs between Tg and nTg mice (P < 0.001), whereas no difference was found in DGCs (P < 0.11). Post hoc tests showed that Tg mice exhibited a 27% reduction in TDLs of SGCs within the dorsal blade as compared with nTg mice (P < 0.05). In contrast, within the ventral blade, no significant main effect was found for genotype (P < 0.81) or for cell type (P < 0.84) between Tg and nTg mice (Fig. 3B). Therefore, the reductions in TDLs of GCs are mainly attributable to the reductions in TDLs exhibited by SGCs restricted to the dorsal blade of the DG.
Fig. 3.
Comparison of the TDL of cell type and topographic location between Tg and nTg mice. (A) Cell type within the dorsal blade. A significant reduction (27%) occurred in TDLs of SGCs, whereas no difference was found in DGCs. Note a marginal decrease in the TDL of DGCs of Tg mice. (B) Cell type within the ventral blade. No significant differences were found in either SGCs or DGCs.
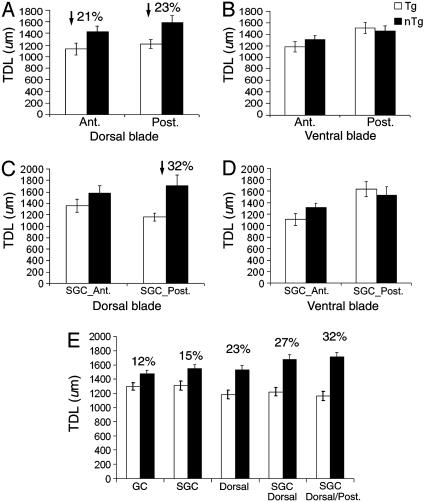
Reduction in TDLs of GCs Across the A-P Axis of the DG in Tg Mice. By considering the 3D structure of the DG within the hippocampus, we have further analyzed dendritic structure by reference to where a given analyzed neuron was positioned along the A-P axis. By using the topographic location as a parameter, a three-way ANOVA (genotype × topographic location × A-P axis) revealed significant main effects of both genotype (P < 0.007) and location along the A-P axis (P < 0.02) and the interaction effect of topography × A-P axis (P < 0.05). In Fig. 4A, dendritic reductions of GCs were consistent along the dorsal blade of the A-P axis in Tg mice (P < 0.007), with a 21% reduction anteriorly and a 23% reduction was found posteriorly (post hoc tests, P < 0.05). In contrast, no significant difference was found between Tg and nTg mice across the A-P axis of the ventral blade (P < 0.75; Fig. 4B).
Fig. 4.
Comparison of the TDL of GCs across the A-P axis between Tg and nTg mice. (A) Consistent reductions in TDLs of GCs within the dorsal blade by A-P axis. A 21% reduction was found anteriorly, whereas a 23% reduction was found posteriorly. Ant., anterior; Post., posterior. (B) No significant differences were found in the ventral blade across the A-P axis. (C) A prominent reduction occurred exclusively in SGCs of the dorsal blade across the A-P axis (DGC data not shown). The analysis has further localized a significant reduction (32%) in TDLs only in the posterior pole. (D) No significant differences were found in SGCs of the ventral blade along the A-P axis. (E) Localization of selective dendritic pathology of GCs in 90-d Tg mice.
In terms of the cell type, ANOVA (genotype × cell type × A-P axis) revealed a significant main effect of both genotype (P < 0.02) and cell type (SGCs) along the A-P axis (P < 0.01). Post hoc tests confirmed that dendritic reductions of SGCs were consistent along the A-P axis (P < 0.05). Moreover, in considering each cell type corresponding to its topographic location along the A-P axis, ANOVA revealed that significant reductions in TDLs of Tg mice within the dorsal blade were seen only for SGCs (P < 0.007) and not for DGCs (P < 0.19). Further analysis has localized a significant reduction (32%) of TDLs to the SGCs within the dorsal blade in the posterior DG of Tg mice (P < 0.02), with no difference found anteriorly (Fig. 4C). No significant difference was found in SGCs of the ventral blade along the A-P axis (P < 0.81; Fig. 4D). Moreover, no significant reductions were found in TDLs of Tg mice for DGCs in either blade along the A-P axis (P < 0.19; not shown). Therefore, the dendritic reductions in Tg mice seem to be completely localized to the SGCs of the dorsal blade in the posterior region of the DG, whereas no loss in TDL exists for either cell type in the ventral blade along the A-P axis.
Based on these results, the progressive refinement in identification of affected GCs for reduction of their TDL is portrayed in Fig. 4E. The percent loss in TDL was found to be mainly attributable to the SGCs within the dorsal blade, with the most pronounced dendritic reduction of these cells in the posterior pole of the DG.
Alterations in Dendritic Complexity of Dentate GCs in Tg Mice. Sholl analysis was used to localize the changes in dendritic complexity within ML of the DG.
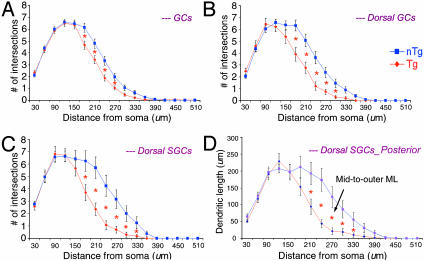
First, the number of dendritic intersections of GCs at 20-μm radial intervals from the soma was compared between Tg and nTg mice (Fig. 5A). Significant differences were found at a range of 180 to 270 μm from the soma (P < 0.05) for GCs within the dorsal blade, whereas no differences were found for GCs in the ventral blade of the DG. When considering topographic location within the dorsal blade, significant differences in dendritic intersections were only found at regions from 180 to 300 μm from the soma (P < 0.05; Fig. 5B). Furthermore, when taking both the cell type (SGCs) and its topographic location (the dorsal blade) into account, significant differences in the number of dendritic intersections were found at regions from 180 to 330 μm from the soma (P < 0.05; Fig. 5C), whereas no differences were found for either cell type within the ventral blade of the DG (data not shown). Note that, similar to the dendrogram analyses above, the difference in the number of dendritic intersections between Tg and nTg mice was linearly resolved by including the analysis of subordinate morphological parameters. Along these lines, by comparing TDLs between Tg and nTg mice for SGCs within the dorsal blade across the A-P axis, the reduction in TDL was found to be most pronounced in the posterior region of the DG. This reduction was localized to the zone 180 to 330 μm from the soma center (P < 0.05), the region that falls approximately in the mid-to-outer ML (Fig. 5D).
Fig. 5.
Comparison of the dendritic complexity of GCs at 20-μm radius intervals from the soma between Tg and nTg mice. (A) Dendritic intersections of total GCs. Significant differences occurred in a region 180 to 270 μm from the soma. (B) Dendritic intersections of topographic location (dorsal blade). Significant differences occurred in a region 180 to 300 um from the soma. (C) Dendritic intersection of cell type (SGC) × topography (dorsal blade). Significant differences were found at a region 180 to 330 μm from the soma of dorsal SGCs. Note: an increase in the difference of two graphs as compared with that of A and B. (D) Prominent reductions (32%) in TDLs of SGCs within the dorsal blade in the posterior portion of the DG of Tg mice. Note: the difference falls into the mid-to-outer ML.
Discussion
The present study used a cell-loading method combined with 3D reconstruction to analyze quantitatively the morphological differences in GCs between 90-d Tg and nTg mice. With systematic analysis focused on subordinate parameters, the major morphological features attributable to transgene-induced alterations in dendritic complexity of GCs were revealed. Dendritic abnormalities were consistently restricted to a defined subpopulation of dentate GCs. Specifically, SGCs located within the dorsal blade of the posterior pole of the DG exhibited transgene-induced selective reductions in dendritic length and branching before the onset of Aβ accumulation. Several experimentally testable mechanisms to account for these changes merit discussion.
Transgene-Induced Dendritic Pathology Is Cell- and Region-Specific at an Early Age. In the present study, the reduction of TDL was found to be linearly augmented when a breakdown analysis was used with emphasis on subordinate parameters. The incremental losses detected were found to be attributable to several morphological properties of GC location, within which one subset of GCs appeared to have the highest vulnerability. The percent loss of dendritic length for Tg mice compared with their wild-type controls was found to increase from 12% when comparing all labeled GCs, to 15% when comparing SGCs, and to a 23% loss of dendritic length when comparing only GCs of the dorsal blade of the DG. Selective vulnerability was further refined when the cell classes being compared were further restricted to dorsal blade SGCs (a 27% loss of dendritic length), and finally to a 32% loss of dendritic length when comparing dorsal blade SGCs within the posterior region of the DG (see Fig. 4E). In concert with selective pathology reported in AD brains (14, 15, 40–42), these data strongly support a transgene-induced selective vulnerability of a specific group of dentate GCs before any amyloid deposition. This trend in percent loss of dendritic length appears quite similar to the DG volume loss reported in the same Tg mice by Redwine et al. (35), in which a 12% volumetric reduction of the whole hippocampus was in fact attributable to a 28% volumetric reduction of the DG. Thus, these findings suggest that the volumetric reduction is a consequence of the extensive loss of dendrites demonstrated in this analysis.
Two recent studies support the transgene-induced selective dendritic pathology of the 90-d Tg mice reported in the present study (43, 44). First, a recent birth-dating study (43) demonstrated that, within the DG, the dorsal blade is more active in neurogenesis than the ventral blade. Moreover, BrdUrd+ cells were found mostly in the inner two thirds of the GCL (i.e., the DGCs that showed no changes with genotype in our studies), whereas no evidence of the cell proliferation labeling was found in the outer one-third portion of the GCL (i.e., the SGCs where all of the dendritic alterations were seen). Given that the neurogenesis of dentate GCs follows an “outside-in” fashion (45–47), early generated “older” neurons migrate to reside in superficial portions of the GCL (defined here as SGCs) with more recently generated neurons in the deep portions of the GCL (defined here as DGCs). Also, the dorsal blade develops relatively earlier then the ventral blade, and the posterior pole develops relatively earlier than the anterior pole of the DG (46, 47). Accordingly, we reason that SGCs within the dorsal blade along the posterior region are relatively more mature and are less actively undergoing neurogenesis than DGCs of either blade and, hence, are more susceptible to the consequences of the overexpression of mutant APP or toxicity of soluble Aβ species. This result suggests the possibility of a transgene-induced premature aging process of SGCs located in the dorsal blade of the DG in 90-d Tg mice. In contrast, DGCs, which are located in the inner two thirds of the GCL of either blade of the DG, are perhaps more likely to be replaced by newly generated neurons. Early susceptibility of SGCs in the dorsal blade of the DG does not, however, rule out the possibility of additional later-onset transgene effects on dendritic morphology of DGCs. The premature aging process of GCs in 90-d Tg mice is further supported by the fact that there is a significant reduction in TDLs of GCs in 15-month-old nTg mice (C.-C. Wu, unpublished data). In addition, although the dorsal blade of the DG is particularly vulnerable to overexpressed APP at 90 days, preliminary data suggest that the dendritic retraction of both Tg and nTg mice affects both dorsal and ventral blades at 15 months of age. Future studies are thus necessary to investigate the progression of dendritic pathology of DGCs of Tg mice ranging from 90 days to the onset age of Aβ deposition (3, 7).
Additional supportive evidence for early dendritic pathology suggests that a correlation exists between the reductions in calbindin (CB) and c-Fos within the DG and cognitive deficits in Tg mice (44). Although not directly stated in their study, the reported data of Palop et al. (44) clearly indicate that young Tg mice exhibit a significant reduction in CB immunoreactivity along the dorsal blade of the DG, which corresponds well with the present findings that the dorsal blade exhibited selective vulnerability with respect to dendritic pathology in young Tg mice.
Possible Mechanisms Underlying Dendritic Atrophy of GCs of PDAPP Mice. Although the observed dendritic reductions in dentate GCs support the previous reports of early-onset transgene-induced pathologies before plaque formation (22, 23, 25–29, 35), the mechanisms underlying these early changes remain to be determined. Several possibilities exist for explaining the observed dendritic atrophy of GCs in 90-d Tg mice.
It has been shown that Aβ deposits are localized to the entorhinal cortex and to the DG of the hippocampus (40, 48, 49), arguing for circuit-specific degeneration observed in AD brains (14, 15, 40, 41). It is reasoned, therefore, that the dendritic pathology in Tg mice is most likely due to the cumulative buildup of APP-derived fragments or products by anterograde transport of APP from the entorhinal cortex to the terminal fields of the DG (50, 51). In the terminal fields, APP undergoes proteolysis to generate Aβ fragments, subsequently inducing neuritic pathology of the dentate ML. Although this hypothesis corresponds well with our Sholl analysis findings of dendritic shortening of dorsal blade SGCs found mainly within the mid-to-outer ML of the DG, it remains an inadequate explanation for the present finding because of the lack of Aβ deposition in 90-d Tg mice.
Alternatively, early-onset selective dendritic pathology may be due to an elevated level of soluble forms of Aβ, induced by the overexpression of APP (33, 42, 52, 53). This hypothesis seems reasonable because increased soluble Aβ has been demonstrated to exhibit neurotoxicity (33, 42, 54), which is likely to induce early-onset neuropathology reminiscent of AD (23, 24, 26, 29–32, 42). The contribution of soluble Aβ to observed dendritic pathology was further supported by the fact that diffuse deposits of Aβ were undetectable with 3D6 immunohistochemistry in the DG of 90-d Tg mice (J. F. Reilly, personal communication; see ref. 7). Moreover, the fact that the impact of soluble Aβ induces dendritic alterations of the GCs in the absence of Aβ deposits is likely to play a major role in cognitive deficits of 90-d Tg mice (10, 24). Although the mechanisms of APP overexpression that induce early-onset dendritic pathology require further investigation, e.g., the intracellular accumulation and/or aberrant proteolytic processing of APP (55–59), the present study has confirmed the presence of Aβ toxicity before the deposition of plaques and has specifically localized it to the dorsal blade SGCs of the posterior portion of the DG, providing a morphological correlate to the pathological deficits observed in young Tg mice (22–28, 35). Future studies are necessary to investigate the A-P levels of soluble Aβ within the subfields of the DG, which can then be correlated to the selective dendritic pathology of GCs reported in the current study. Also, the follow-up study should focus on the quantitative analysis of spine morphology that would likely predate robust changes in the dendrites of dorsal blade SGCs of the posterior DG of 90-d or younger Tg mice (29).
Conclusion
The present study used a series of systematic analyses with emphasis on precise cellular locations within the DG to localize what appears to be a spatially restricted decrease in GC dendritic complexity in mice overexpressing mutant APP. The selective vulnerability of particular dentate GCs to the mutant APP at an early age, before plaque formation, suggests a neurotoxic effect of soluble Aβ and/or an accelerated cell-restricted aging process. The basis for this accelerated sensitivity to mutant APP may provide additional opportunities for developing early therapeutic interventions.
Supplementary Material
Acknowledgments
We thank Drs. J. M. Redwine and J. F. Reilly for scientific advice and Dr. J. Polich (The Scripps Research Institute, La Jolla, CA) for statistical analysis.
Abbreviations: DG, dentate gyrus; GC, granule cell; Aβ, β-amyloid; APP, amyloid precursor protein; ML, dentate molecular layer; GCL, GC layer; SGC, superficial GC; DGC, deep GC; Tg, transgenic; nTg, nontransgenic; TDL, total dendritic length; A-P, anterior-to-posterior.
References
- 1.Price, D. L. & Sisodia, S. S. (1998) Annu. Rev. Neurosci. 21, 479-505. [DOI] [PubMed] [Google Scholar]
- 2.Selkoe, D. J. & Schenk, D. (2003) Annu. Rev. Pharmacol. Toxicol. 43, 545-584. [DOI] [PubMed] [Google Scholar]
- 3.Games, D., Adams, D., Alessandrini, R., Barbour, R., Berthelette, P., Blackwell, C., Carr, T., Clemens, J., Donaldson, T., Gillespie, F., et al. (1995) Nature 373, 523-527. [DOI] [PubMed] [Google Scholar]
- 4.Chen, K. S., Masliah, E., Grajeda, H., Guido, T., Huang, J., Khan, K., Motter, R., Soriano, F. & Games, D. (1998) Prog. Brain Res. 117, 327-334. [PubMed] [Google Scholar]
- 5.Games, D., Bard, F., Grajeda, H., Guido, T., Khan, K., Soriano, F., Vasquez, N., Wehner, N., Johnson-Wood, K., Yednock, T., et al. (2000) Ann. N.Y. Acad. Sci. 920, 274-284. [DOI] [PubMed] [Google Scholar]
- 6.Irizarry, M. C., Soriano, F., McNamara, M., Page, K. J., Schenk, D., Games, D. & Hyman, B. T. (1997) J. Neurosci. 17, 7053-7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson-Wood, K., Lee, M., Motter, R., Hu, K., Gordon, G., Barbour, R., Khan, K., Gordon, M., Tan, H., Games, D., et al. (1997) Proc. Natl. Acad. Sci. USA 94, 1550-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Masliah, E., Sisk, A., Mallory, M. & Games, D. (2001) J. Neuropathol. Exp. Neurol. 60, 357-368. [DOI] [PubMed] [Google Scholar]
- 9.Wilson, C. A., Doms, R. W. & Lee, V. M. (1999) J. Neuropathol. Exp. Neurol. 58, 787-794. [DOI] [PubMed] [Google Scholar]
- 10.Dodart, J. C., Mathis, C., Saura, J., Bales, K. R., Paul, S. M. & Ungerer, A. (2000) Neurobiol. Dis. 7, 71-85. [DOI] [PubMed] [Google Scholar]
- 11.Flood, D. G., Buell, S. J., Horwitz, G. J. & Coleman, P. D. (1987) Brain Res. 402, 205-216. [DOI] [PubMed] [Google Scholar]
- 12.Einstein, G., Buranosky, R. & Crain, B. J. (1994) J. Neurosci. 14, 5077-5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ji, Y., Gong, Y., Gan, W., Beach, T., Holtzman, D. M. & Wisniewski, T. (2003) Neuroscience 122, 305-315. [DOI] [PubMed] [Google Scholar]
- 14.Smith, T. D., Adams, M. M., Gallagher, M., Morrison, J. H. & Rapp, P. R. (2000) J. Neurosci. 20, 6587-6593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thal, D. R., Rub, U., Schultz, C., Sassin, I., Ghebremedhin, E., Del Tredici, K., Braak, E. & Braak, H. (2000) J. Neuropathol. Exp. Neurol. 59, 733-748. [DOI] [PubMed] [Google Scholar]
- 16.Masliah, E., Sisk, A., Mallory, M., Mucke, L., Schenk, D. & Games, D. (1996) J. Neurosci. 16, 5795-5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Selkoe, D. J. (1991) Neuron 6, 487-498. [DOI] [PubMed] [Google Scholar]
- 18.Knowles, R. B., Wyart, C., Buldyrev, S. V., Cruz, L., Urbanc, B., Hasselmo, M. E., Stanley, H. E. & Hyman, B. T. (1999) Proc. Natl. Acad. Sci. USA 96, 5274-5279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Le, R., Cruz, L., Urbanc, B., Knowles, R. B., Hsiao-Ashe, K., Duff, K., Irizarry, M. C., Stanley, H. E. & Hyman, B. T. (2001) J. Neuropathol. Exp. Neurol. 60, 753-758. [DOI] [PubMed] [Google Scholar]
- 20.Price, D. L., Wong, P. C., Markowska, A. L., Lee, M. K., Thinakaren, G., Cleveland, D. W., Sisodia, S. S. & Borchelt, D. R. (2000) Ann. N.Y. Acad. Sci. 920, 179-191. [DOI] [PubMed] [Google Scholar]
- 21.Morrison, J. H. (2001) Neurobiol. Aging 22, 349-350. [DOI] [PubMed] [Google Scholar]
- 22.Huitron-Resendiz, S., Sanchez-Alavez, M., Gallegos, R., Berg, G., Crawford, E., Giacchino, J. L., Games, D., Henriksen, S. J. & Criado, J. R. (2002) Brain Res. 928, 126-137. [DOI] [PubMed] [Google Scholar]
- 23.Koistinaho, M., Ort, M., Cimadevilla, J. M., Vondrous, R., Cordell, B., Koistinaho, J., Bures, J. & Higgins, L. S. (2001) Proc. Natl. Acad. Sci. USA 98, 14675-14680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.King, D. L., Arendash, G. W., Crawford, F., Sterk, T., Menendez, J. & Mullan, M. J. (1999) Behav. Brain Res. 103, 145-162. [DOI] [PubMed] [Google Scholar]
- 25.Hsia, A. Y., Masliah, E., McConlogue, L., Yu, G. Q., Tatsuno, G., Hu, K., Kholodenko, D., Malenka, R. C., Nicoll, R. A. & Mucke, L. (1999) Proc. Natl. Acad. Sci. USA 96, 3228-3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mucke, L., Masliah, E., Yu, G. Q., Mallory, M., Rockenstein, E. M., Tatsuno, G., Hu, K., Kholodenko, D., Johnson-Wood, K. & McConlogue, L. (2000) J. Neurosci. 20, 4050-4058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanchez-Alavez, M., Gallegos, R. A., Kalafut, M. A., Games, D., Henriksen, S. J. & Criado, J. R. (2002) Neurosci. Lett. 330, 45-48. [DOI] [PubMed] [Google Scholar]
- 28.Larson, J., Lynch, G., Games, D. & Seubert, P. (1999) Brain Res. 840, 23-35. [DOI] [PubMed] [Google Scholar]
- 29.Lanz, T. A., Carter, D. B. & Merchant, K. M. (2003) Neurobiol. Dis. 13, 246-253. [DOI] [PubMed] [Google Scholar]
- 30.Lue, L. F., Kuo, Y. M., Roher, A. E., Brachova, L., Shen, Y., Sue, L., Beach, T., Kurth, J. H., Rydel, R. E. & Rogers, J. (1999) Am. J. Pathol. 155, 853-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gong, Y., Chang, L., Viola, K. L., Lacor, P. N., Lambert, M. P., Finch, C. E., Krafft, G. A. & Klein, W. L. (2003) Proc. Natl. Acad. Sci. USA 100, 10417-10422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McLean, C. A., Cherny, R. A., Fraser, F. W., Fuller, S. J., Smith, M. J., Beyreuther, K., Bush, A. I. & Masters, C. L. (1999) Ann. Neurol. 46, 860-866. [DOI] [PubMed] [Google Scholar]
- 33.Klein, W. L., Krafft, G. A. & Finch, C. E. (2001) Trends Neurosci. 24, 219-224. [DOI] [PubMed] [Google Scholar]
- 34.Schenk, D., Barbour, R., Dunn, W., Gordon, G., Grajeda, H., Guido, T., Hu, K., Huang, J., Johnson-Wood, K., Khan, K., et al. (1999) Nature 400, 173-177. [DOI] [PubMed] [Google Scholar]
- 35.Redwine, J. M., Kosofsky, B., Jacobs, R. E., Games, D., Reilly, J. F., Morrison, J. H., Young, W. G. & Bloom, F. E. (2003) Proc. Natl. Acad. Sci. USA 100, 1381-1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu, C.-C., Reilly, J. F., Morrison, J. H. & Bloom, F. E. (2004) Cereb. Cortex 14, 543-554. [DOI] [PubMed] [Google Scholar]
- 37.Murrell, J., Farlow, M., Ghetti, B. & Benson, M. D. (1991) Science 254, 97-99. [DOI] [PubMed] [Google Scholar]
- 38.Hof, P. R., Young, W. G., Bloom, F. E., Belichenko, P. V. & Celio, M. R. (2000) Comparative Cytoarchitectonic Atlas of the C57BL/6 and 129/Sv Mouse Brains (Elsevier, New York).
- 39.Sholl, D. A. (1953) J. Anat. 87, 387-406. [PMC free article] [PubMed] [Google Scholar]
- 40.Morrison, J. H. & Hof, P. R. (1997) Science 278, 412-419. [DOI] [PubMed] [Google Scholar]
- 41.Morrison, J. H. & Hof, P. R. (2002) Prog. Brain Res. 136, 467-486. [DOI] [PubMed] [Google Scholar]
- 42.Kim, H. J., Chae, S. C., Lee, D. K., Chromy, B., Lee, S. C., Park, Y. C., Klein, W. L., Krafft, G. A. & Hong, S. T. (2003) FASEB J. 17, 118-120. [DOI] [PubMed] [Google Scholar]
- 43.Kempermann, G., Gast, D., Kronenberg, G., Yamaguchi, M. & Gage, F. H. (2003) Development (Cambridge, U.K.) 130, 391-399. [DOI] [PubMed] [Google Scholar]
- 44.Palop, J. J., Jones, B., Kekonius, L., Chin, J., Yu, G. Q., Raber, J., Masliah, E. & Mucke, L. (2003) Proc. Natl. Acad. Sci. USA 100, 9572-9577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bayer, S. A., Yackel, J. W. & Puri, P. S. (1982) Science 216, 890-892. [DOI] [PubMed] [Google Scholar]
- 46.Altman, J. & Bayer, S. A. (1990) J. Comp. Neurol. 301, 365-381. [DOI] [PubMed] [Google Scholar]
- 47.Schlessinger, A. R., Cowan, W. M. & Gottlieb, D. I. (1975) J. Comp. Neurol. 159, 149-175. [DOI] [PubMed] [Google Scholar]
- 48.Su, J. H., Cummings, B. J. & Cotman, C. W. (1998) Acta Neuropathol. (Berlin) 96, 463-471. [DOI] [PubMed] [Google Scholar]
- 49.Reilly, J. F., Games, D., Rydel, R. E., Freedman, S., Schenk, D., Young, W. G., Morrison, J. H. & Bloom, F. E. (2003) Proc. Natl. Acad. Sci. USA 100, 4837-4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sheng, J. G., Price, D. L. & Koliatsos, V. E. (2002) J. Neurosci. 22, 9794-9799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lazarov, O., Lee, M., Peterson, D. A. & Sisodia, S. S. (2002) J. Neurosci. 22, 9785-9793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kuo, Y. M., Beach, T. G., Sue, L. I., Scott, S., Layne, K. J., Kokjohn, T. A., Kalback, W. M., Luehrs, D. C., Vishnivetskaya, T. A., Abramowski, D., et al. (2001) Mol. Med. 7, 609-618. [PMC free article] [PubMed] [Google Scholar]
- 53.Hsiao, K., Chapman, P., Nilsen, S., Eckman, C., Harigaya, Y., Younkin, S., Yang, F. & Cole, G. (1996) Science 274, 99-102. [DOI] [PubMed] [Google Scholar]
- 54.Lambert, M. P., Barlow, A. K., Chromy, B. A., Edwards, C., Freed, R., Liosatos, M., Morgan, T. E., Rozovsky, I., Trommer, B., Viola, K. L., et al. (1998) Proc. Natl. Acad. Sci. USA 95, 6448-6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Benowitz, L. I., Rodriguez, W., Paskevich, P., Mufson, E. J., Schenk, D. & Neve, R. L. (1989) Exp. Neurol. 106, 237-250. [DOI] [PubMed] [Google Scholar]
- 56.Masliah, E., Cole, G. M., Hansen, L. A., Mallory, M., Albright, T., Terry, R. D. & Saitoh, T. (1991) J. Neurosci. 11, 2759-2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Saitoh, T., Horsburgh, K. & Masliah, E. (1993) Ann. N.Y. Acad. Sci. 695, 34-41. [DOI] [PubMed] [Google Scholar]
- 58.Mucke, L., Abraham, C. R. & Masliah, E. (1996) Ann. N.Y. Acad. Sci. 777, 82-88. [DOI] [PubMed] [Google Scholar]
- 59.Rossner, S., Mehlhorn, G., Schliebs, R. & Bigl, V. (2001) Eur. J. Neurosci. 13, 269-278. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.