Abstract
In many embryos specification toward one cell fate can be diverted to a different cell fate through a reprogramming process. Understanding how that process works will reveal insights into the developmental regulatory logic that emerged from evolution. In the sea urchin embryo, cells at gastrulation were found to reprogram and replace missing cell types after surgical dissections of the embryo. Non-skeletogenic mesoderm (NSM) cells reprogrammed to replace missing skeletogenic mesoderm cells and animal caps reprogrammed to replace all endomesoderm. In both cases evidence of reprogramming onset was first observed at the early gastrula stage, even if the cells to be replaced were removed earlier in development. Once started however, the reprogramming occurred with compressed gene expression dynamics. The NSM did not require early contact with the skeletogenic cells to reprogram, but the animal cap cells gained the ability to reprogram early in gastrulation only after extended contact with the vegetal halves prior to that time. If the entire vegetal half was removed at early gastrula, the animal caps reprogrammed and replaced the vegetal half endomesoderm. If the animal caps carried morpholinos to either hox11/13b or foxA (endomesoderm specification genes), the isolated animal caps failed to reprogram. Together these data reveal that the emergence of a reprogramming capability occurs at early gastrulation in the sea urchin embryo and requires activation of early specification components of the target tissues.
Keywords: Reprogramming, regulative development, gene regulatory network, sea urchin embryo, cell fate, differentiation, cell fate specification
Introduction
It is widely thought that the differentiation potential of a cell decreases as the cell becomes increasingly specified (Cherry and Daley, 2012). However, this view has been challenged by recent findings that combinations of defined factors can revert the fate of a differentiated cell to a pluripotent state in mammals, leading to induced pluripotent stem cells (iPSCs) (Liu et al., 2008; Park et al., 2008; Takahashi et al., 2007; Takahashi and Yamanaka, 2006; Yu et al., 2007). Thus, studying the reprogramming process will yield insights into how the gene regulatory system buffers perturbations and faithfully carries out the developmental program, and in addition provide useful information for regenerative medicine and disease modeling (Cherry and Daley, 2012).
The sea urchin embryo provides an excellent platform for studying reprogramming. It exhibits high developmental plasticity in that an isolated blastomere from the 4-cell stage embryo develops into a complete pluteus larva, a phenomenon that led to the original concept of “regulative” development (reviewed by Hörstadius, 1973). Later in development, when cells specified toward one fate were surgically removed, cells of other fates reprogrammed to replace the missing cells, a process referred to at the time as “transfating” (Ettensohn and McClay, 1988; McClay and Logan, 1996; Ettensohn et al., 2007). Those cell fate switches during sea urchin embryonic development altered the gene regulatory networks (GRNs) governing specification (Davidson, 2002; Davidson et al., 2006), and resulted in a dynamic and qualitative transition to a different GRN state. The remarkable advance in knowledge of the sea urchin GRN provides an excellent platform for a systems analysis of this reprogramming mechanism.
In the sea urchin, the skeletogenic lineage arises from four large micromeres formed at the vegetal pole of the embryo as a result of two consecutive asymmetric cleavages at the 16-cell and 32-cell stages. At the mesenchyme blastula stage, descendants of large micromeres undergo an epithelial-mesenchymal transition (EMT) and ingress into the blastocoel to form the primary mesenchyme cells (PMCs). The PMCs migrate to specific locations in the blastocoel, fuse via syncitial cables, and secrete the calcium carbonate skeleton of the larva. When skeletogenic cells were removed, either at the 16-cell stage as micromeres or at the mesenchyme blastula stage as PMCs, non-skeletogenic mesoderm cells (NSM) reprogrammed to assume a skeletogenic fate (Ettensohn and McClay, 1988; Sweet et al., 1999). If the PMCs and the archenteron tip (all mesoderm) were removed, the remaining presumptive endoderm reprogrammed to assume skeletal and other mesodermal fates (McClay and Logan, 1996; Sharma and Ettensohn, 2011). From these studies it was clear that the remaining mesoderm or even endoderm had the capacity to reprogram and replace missing skeletogenic cells.
McClay and Logan (1996) showed that if parts of the gut were removed, it too was replaced through reprogramming. In the literature this cell fate change is sometimes referred to classically as “morphallactic regeneration” (Reddien and Sanchez Alvarado, 2004) where one cell type changes to a different cell type. Here we use the term “reprogram”, for the process to indicate that cells normally headed toward one fate demonstrate the capacity to move to a different fate. We distinguish this reprogramming from “regulative” development because, as shown in the experiments below, the regulative changes classically reported in the literature and in textbooks are largely restricted to very early cleavage, before cell specification has progressed very far. Here we show that the regulative capacity ends long before a reprogramming ability can be demonstrated.
Cells of the animal half of the embryo normally are fated to become ectoderm and neural ectoderm. Studies by Ho rstadius demonstrated that these animal cap cells could give rise to vegetal tissues (including endoderm) if isolated and supplied with micromeres at the 16-cell stage, but that inductive capacity could no longer be demonstrated if micromeres were added to animal caps isolated after the 32-cell stage. The animal cap cells lost responsiveness to micromere induction since neither younger or older micromeres induced regulative change if added to animal caps from 32-cell stage embryos or older (Horstadius, 1939; Horstadius, 1973). Thus, a capacity for large inductive and/or regulative change could be demonstrated experimentally between 2 and 32 cell stages, but after that any changes were much more modest, if present at all. These data were in stark contrast to the transfating experiments that demonstrated a later plasticity in reprogramming.
Here we confirm that those regulative capacities are lost early in cleavage, and after that, isolated embryo fragments appear to be largely refractory to an alteration in fate until the beginning of gastrulation at which time the remaining cells of the embryo demonstrate a remarkable ability to reprogram. We returned to the reprogramming question in an attempt to begin to understand the molecular events behind the process in two cases of reprogramming. Work on Primary Mesenchyme Cell-depleted embryos (henceforth referred to as PMC(−) embryos, see Figure 1A), earlier had revealed that about 2-3 hours after removal of PMCs, non-skeletogenic mesenchyme (NSM) cells began to express PMC markers indicating that reprogramming had begun (Sharma and Ettensohn, 2011). In the present study, we began by asking if removal of the PMC predecessors, the micromeres, was followed by a similar 2-3hr delay before replacement was launched. To our surprise there was a much longer delay before there was any evidence of micromere/PMC replacement. Once the reprogramming began, however, the skeletogenic temporal gene expression program was substantially compressed, occurring in about 1/2 of the time normally taken by micromeres during their specification sequence. The NSMs that reprogrammed transiently co-expressed both skeletal and NSM marker genes as the reprogramming progressed.
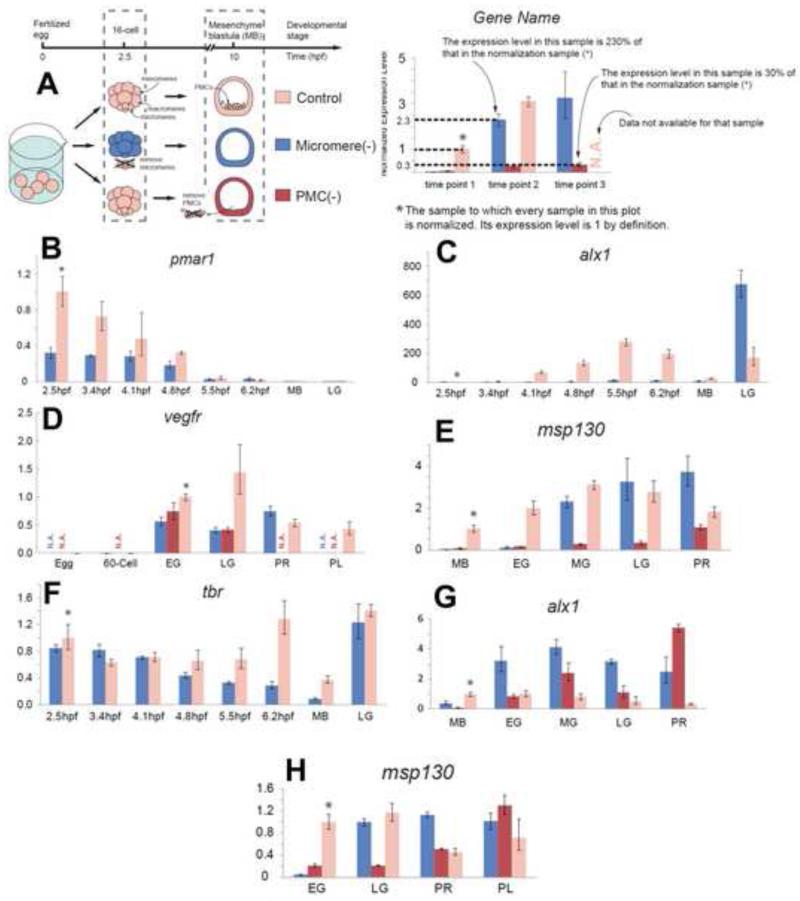
Figure 1.
Skeletogenic reprogramming in micromere (−) embryos occurs after a long delay. (A) (Left panel) Illustration of the experiment. At the 16-cell stage micromeres were removed (micromere(−), blue), or at the mesenchyme blastula stage PMCs were removed (PMC(−), red). (Right panel) Explanation of the qPCR plots. (B-H) Analysis of (B) pmar1, (C) alx1, (D) tbr, (E) alx1, late, (F) vegfr, (G) msp130, early, and (H) msp130 late (H). Horizontal axes give hours post fertilization (hpf) until morphogenesis: MB, mesenchyme blastula; EG, early gastrula; MG, mid-gastrula; LG, late gastrula; PR, prism; PL, pluteus. In each plot, the normalization standard sample is indicated by a * sign, and has the relative expression value of 1.
We then asked whether other parts of the embryo reprogrammed with a delayed response. Ho rstadius’ result showing that animal caps quickly lost their inductive response capacity (by the 32-cell stage) was confirmed, and we then found that animal cap descendants obtained an ability to reprogram but only when they reached the beginning of gastrulation, at roughly the same time reprogramming to replace PMCs was initiated. Unlike reprogramming of PMCs, however, the animal caps ability to reprogram required extended contact with the vegetal half embryo until gastrulation. Thus the embryo has the ability to reprogram cells of one germ layer to another and this capacity apparently includes much of the embryo. The reprogramming ability does not appear until early gastrulation, and the reprogramming requires expression of transcription factors used in early specification of the target tissues.
Materials and Methods
Animals
Adult Lytechinus variegatus were obtained from Duke Marine Laboratory (Beaufort, NC, USA) and Reeftopia Inc. (Key West, FL, USA). The embryos were cultured at 23 °C.
Quantitative Polymerase Chain Reaction (qPCR) and Whole Mount In Situ Hybridization (WMISH) analyses
Total RNA was extracted from 15-25 live Lytechinus variegatus embryos using the RNeasy Plus Micro Kit (QIAGEN) and eluted in nuclease-free water. For the 2.5hpf (hours post fertilization) time point, micromere(−) embryos were lysed and homogenized in Buffer RLT Plus (QIAGEN) with 2-Mercaptoethanol added within 5 minutes of micromere removal. cDNA synthesis was performed using the iScript cDNA Synthesis Kit (Bio-Rad). Quantitative PCR reactions were performed using an Eppendorf Mastercycler ep realplex system and Power SYBR Green PCR Master Mix (ABI). QPCR results were analyzed following the 2−ΔΔCT method described by Livak and Schmittgen (2001), using ubiquitin as the normalization gene (Rho and McClay, 2011). Chromogenic and fluorescent WMISH using published anti-sense RNA probes (Croce and McClay, 2010) followed procedures described earlier (Rho and McClay, 2011).
Microsurgery and microinjections
The micromere and PMC removal procedure has been described previously (Sweet et al., 2004). The micromeres were removed at 16-cell to 32-cell stage, approximately 2.5 to 3hpf, resulting in micromere(−) embryos. PMCs were removed at mid to late mesenchyme blastula stage, resulting in PMC(−) embryos. Fluorescent dye FITC (green) or Rhodamine dextran (red) was injected into one-cell stage embryos shortly after fertilization. Animal half embryos were isolated from chimeras produced earlier by combining green fluorescent animal halves and red fluorescent vegetal halves at the 16-32-cell stage. Vegetal half separations were performed under fluorescent light to provide the cleanest possible separation of animal and vegetal halves. As controls for these experiments recombined animal and vegetal halves, if allowed to develop, produced normal pluteus larvae with green ectoderm and red mesoderm and endoderm. Also, most micromere(−) and PMC(−) embryos eventually produced normally patterned larvae if allowed to develop. Morpholinos used were previously published, including extensive controls to show that knockdowns did not include off-target consequences (Oliveri, et al., 2006; McIntyre, et al., 2012).
Results
Skeletogenic reprogramming of non-skeletogenic mesoderm occurs with a delay in micromere-depleted embryos
In a Lytechinus variegatus embryo grown at 23°C, the micromeres appear at 2.5hpf (hours post fertilization), become primary mesenchyme cells (PMCs) at 9.5hpf, and begin synthesizing the larval skeleton at about 14hpf, shortly after archenteron invagination begins. Earlier it was shown that if the PMCs were surgically removed, non-skeletogenic mesoderm (NSM) cells, a subpopulation derived from macromeres of the 16-cell stage embryo, quantitatively replaced the missing PMCs by reprogramming to a skeletogenic fate (Ettensohn and McClay, 1988). Evidence of replacement was first seen about 3 hours after PMCs were removed from mesenchyme blastula stage embryos. At that time transcription factors specific for the skeletogenic fate were expressed by NSM indicating that reprogramming had been initiated (Ettensohn et al., 2007; Sharma and Ettensohn, 2011). These data suggested the possibility that reprogramming of the NSM was triggered quickly after cell loss. If that were true, we predicted that reprogramming also would initiate quickly if the PMC precursors, the micromeres, were removed at 2.5hpf (micromere(−) embryos). Unexpectedly, the micromere(−) embryos did not express PMC lineage-specific markers until early gastrulation, at about 12.5hpf, or at the same time the PMC(−) embryos had expressed the skeletogenic markers. Alx1 is a transcription factor expressed exclusively by the skeletogenic lineage and normally is expressed, and required, early in the specification of that lineage (Ettensohn et al., 2003; Ettensohn et al., 2007)(Fig. 1). The experiments revealed that did not matter whether skeletogenic precursors were removed at 2.5hpf or at 10hpf, the reprogramming to replace them, as defined by appearance of alx1 expression, did not appear until about 12.5-13hpf. To learn more about the extended delay in onset of reprogramming of micromeres we began by quantitatively assessing the expression of markers, starting with the most upstream skeletogenic factor, pmar1 (Oliveri et al., 2002). In control embryos, expression of pmar1 begins shortly after micromere formation at 2.5hpf in Lytechinus variegatus, and is followed by expression of alx1 (Ettensohn et al., 2003) and tbr (Croce and McClay, 2010) at 4.5hpf, vegfr (Duloquin et al., 2007) at about 8.5hpf and msp130 (Leaf et al., 1987) at about 8-9hpf. Each of these genes is expressed exclusively in the skeletogenic lineage downstream of pmar1 and here they are used as markers of the skeletogenic cell specification temporal sequence. At the 16-cell stage micromeres were removed, or PMCs were removed at the mesenchyme blastula stage, and in both cases, embryos were immediately processed for qPCR analysis, or grown in seawater for a period of time and then processed for qPCR analysis. pmar1 mRNA was expressed in embryos lacking micromeres at the time of micromere removal (embryos were processed for RNA no later than 5 min after micromeres were removed), albeit at a much reduced level compared to the sibling controls, suggesting that pmar1 expression is not initially restricted to micromeres (Fig.1B). Initially we thought this might represent an early activation of a reprogramming capacity. However, alx1 and tbr expression did not follow in this low level of pmar1 expression in the micromere(−) embryos as it normally does in micromeres (Oliveri, et al., 2003). There was no evidence of any of the downstream micromere or PMC markers in micromere(−) embryos when the sibling controls reached mesenchyme blastula (MB) stage (about 9.5hpf). Alx1 and tbr markers finally were expressed at early gastrula stage in both micromere(−) and PMC(−) embryos, indicating that the NSM cells apparently did not initiate reprogramming to a PMC fate until that time (Figures 1C and 1D). We hypothesized that in embryos lacking presumptive skeletogenic lineage, that the sequence of reprogramming would follow a similar trajectory as it had earlier in micromeres. We observed that Pmar1 was not expressed in micromere(−) embryos by the NSM, as also reported previously in PMC(−) experiments (Ettensohn, et al., 2007). We examined the expression of alx1, vegfr, and msp130 in both micromere(−) and PMC(−) embryos. Alx1 was expressed at levels equal to, or higher than the controls in both micromere(−) and PMC(−) embryos at early gastrula (EG) stage (Fig. 1E). Expression of vegfr, and msp130 also followed indicating that reprogramming at least roughly followed the skeletogenic specification sequence in both micromere(−) and PMC(−) embryos (Fig. 1F,G). Micromere(−) embryos left in culture produced normally patterned skeletons indicating that reprogramming reached completion.
We wondered if the reduced level of pmar1 in micromere(−) embryos, presumably expressed in macromeres and their progeny, was simply insufficient to activate skeletogenesis in these cells. To test this, we examined expression of alx1 in micromere(−) embryos after misexpressing pmar1 in the macromeres (Figure 2A-B). In situ analysis indicated that alx1 indeed was expressed in these embryos at 8.5hpf (Fig. 2C), much earlier than observed in uninjected micromere(−) embryos (Figures 1C and E). These embryos later produced excessive numbers of ingressed cells that, by in situ, stained positively for tbr (Figures 2D-G), a marker of the skeletogenic fate. These data show that the remaining embryo is able to reprogram early but only if artificially activated by over-expression of key transcription factors. In the absence of an artificial stimulus micromere(−) embryos reprogram cells to the skeletogenic fate but with an initiation delay until the early gastrula stage. The low level of pmar1 observed in macromeres apparently occurs normally but is quantitatively insufficient to activate skeletogenesis if micromeres are removed early. The reprogramming at early gastrula bypasses a need for Pmar1. Thus, both micromere(−) and PMC(−) embryos activated skeletogenic reprogramming at early gastrulation. The NSM cells that replaced the PMCs showed no evidence of reprogramming prior to early gastrula, no matter when the micromere/PMCs were removed.
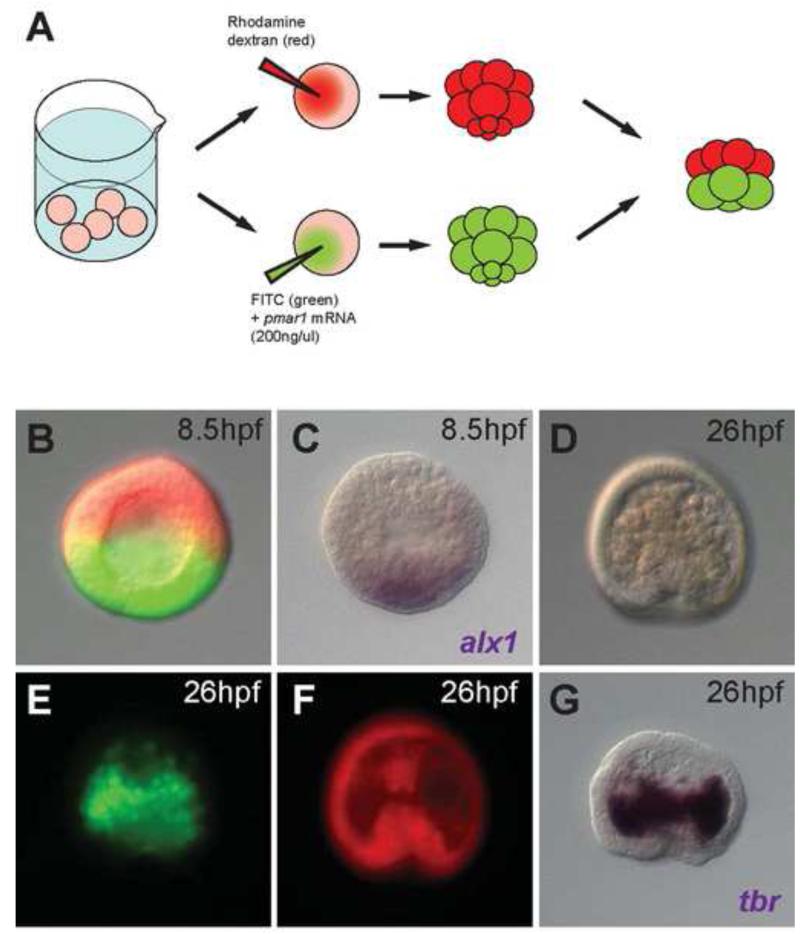
Figure 2.
(A) Experimental procedure for obtaining micromere(−) embryos overexpressing pmar1 in macromeres. (B) A micromere(−) embryo overexpressing pmar1 in its macromeres, with the animal cap cells labeled by rhodamine dextran (red) and progeny of pmar1-injected macromeres labeled by FITC (green). (C) alx1 in situ hybridization for the same embryo in (B) indicating the activation of alx1 in half of the embryo. (D-G) A sibling embryo of (B) developed to 26hpf. (D) The gastrulated embryo showed increased number of ingressed cells in the blastocoel. (E) The ingressed cells derived almost entirely from pmar1-injected macromere labeled with FITC (green) and started to aggregate on the sides of the archenteron. (F) Animal cap cells labeled with rhodamine dextran (red) contributed to the archenteron. (G) In situ hybridization of tbr for the same embryo in (D-F) showing that the FITC-labeled cells (green) were tbr expressing skeletogenic cells.
Once initiated, reprogramming of non-skeletogenic mesoderm to PMCs occurs with compressed timing
A recent study reported that the sequence of skeletogenic gene activation during NSM reprogramming in PMC(−) embryos was similar to that expected of normal development (Sharma and Ettensohn, 2011), although it bypassed expression of pmar1 (Ettensohn et al., 2007). Activation of alx1 normally is followed by vegfr expression after about a 4-5 hour interval. Our qPCR analyses of micromere(−) embryos tended to agree with that earlier finding except it indicated that vegfr was activated with less delay relative to alx1 expression onset than is observed in the normal timing of expression of these markers. This suggested the possibility that reprogramming to a skeletogenic fate, once started, might occur with dynamics different from normal embryos. To more precisely examine the dynamic expression pattern of the factors used as temporal markers, we performed in situ analyses on micromere(−) embryos at frequent intervals from 6.5hpf to 20.5hpf. Consistent with our qPCR data, alx1,vegfr and msp130 were expressed in micromere(−) embryos by 12.5hpf (Fig. 3) indicating only a short delay between alx1 activation as a sign of reprogramming, and expression of downstream genes (in fact vegfr was expressed prior to alx1 in some micromere (−) embryos). The interval was reduced to as short as 1-2hrs (for msp130) relative to the roughly 4hr interval seen during normal micromere specification, and vegfr expression was even earlier. The timing of gene expression obtained from this experiment suggests that once NSM reprogramming is initiated the reprogramming sequence is significantly compressed temporally relative to the endogenous timing of the micromere specification sequence.
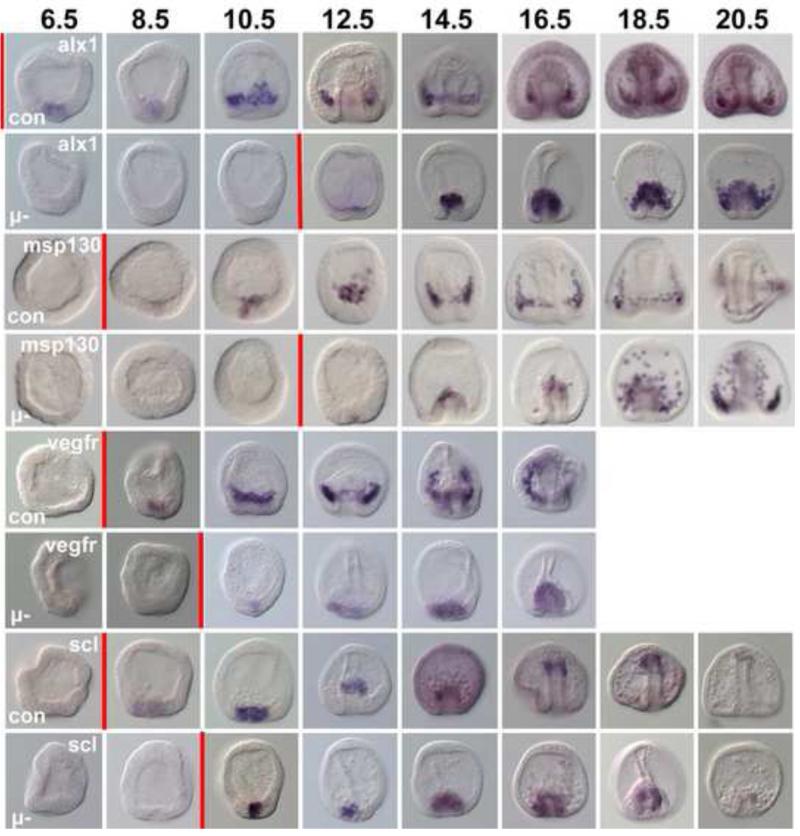
Figure 3.
Reprogramming occurs with compressed temporal dynamics. WMISH analysis of alx1, msp130, vegfr, and scl expression in control embryos (Con) and in micromere(−) (μ-) embryos over time. Numbers at top are hours post fertilization (hpf). Red vertical bars mark the approximate time each row demonstrated expression of the gene in question. First expression of alx1 in controls actually occurs at 4.5hpf (see Fig. 1C). As indicated by the red bars, in controls there is a 4 hr interval between expression of alx1 and msp130 and vegfr. In the reprogramming NSM cells alx1 is seen at 12.5hpf while msp130 is seen within an hour of that, and vegfr actually precedes expression of alx1 suggesting temporal compression relative to the sequence in normal development.
In PMC(−) embryos, NSMs undergoing reprogramming co-express alx1 and an NSM cell marker scl (Sharma and Ettensohn, 2011). We wondered if this hybrid regulatory state occurred in micromere(−) embryos during the reprogramming phase as well. Cells in the vegetal plate of micromere(−) embryos indeed expressed scl and alx1 at 12.5hpf (Fig. 3), and the expression of both became strong in cells covering the cap of the archenteron during gastrulation (Fig. 3, Fig. 4A). Fluorescent two-color in situ hybridization showed that expression of alx1 and scl was by the same cells (Fig. 4A). Unlike PMC(−) embryos where vegfr was only expressed when the alx1/scl co-expressing cells acquired mesenchymal character and left the tip of the archenteron (Sharma and Ettensohn, 2011), in micromere(−) embryos vegfr and scl were also co-expressed with both genes in the same spatial domain (Fig. 4B) at 14.5hpf and prior to the departure of the reprogrammed PMCs from the archenteron. Control embryos at a similar developmental stage showed completely distinct patterns of expression (Fig. 4C,D). These observations indicate an intermediate state when both sets of genes are expressed. Thus, the data suggest that the transition is not a simple flipping of a switch, but that to reprogram there is an extended influence on the transiting cell so it continues its reprogramming trajectory. In this case the NSM is the cell type that reprograms in the absence of PMCs or micromeres. The influence that triggers the reprogramming is unknown but could be a lack of reception of a signal normally produced by PMCs.
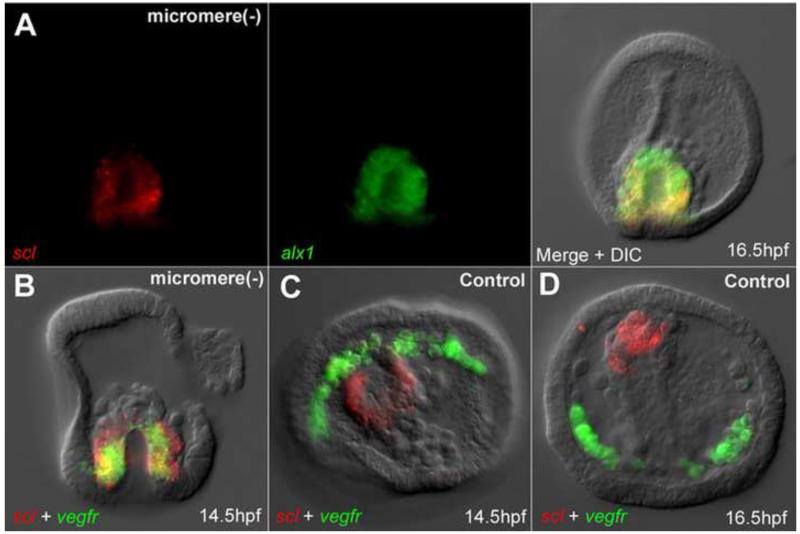
Figure 4.
During reprogramming PMC and NSM markers are co-expressed. (A) Expression of scl and alx1 in reprogramming NSMs at 16.5hpf. (B) Expression of alx1 and vegfr in reprogramming NSMs at 14.5hpf. (C-D) scl and vegfr expressed in separate lineages in control embryos at 14.5hpf (C), and 16.5hpf (D).
Reprogramming of animal cap cells occurs after extended specification by vegetal tissue
We next looked at animal cap cells to ask whether the micromere-PMC-NSM cells reprogramming transition was unique to that group of cells, or whether other cells of the embryo also could also reprogram with a similar delayed onset.
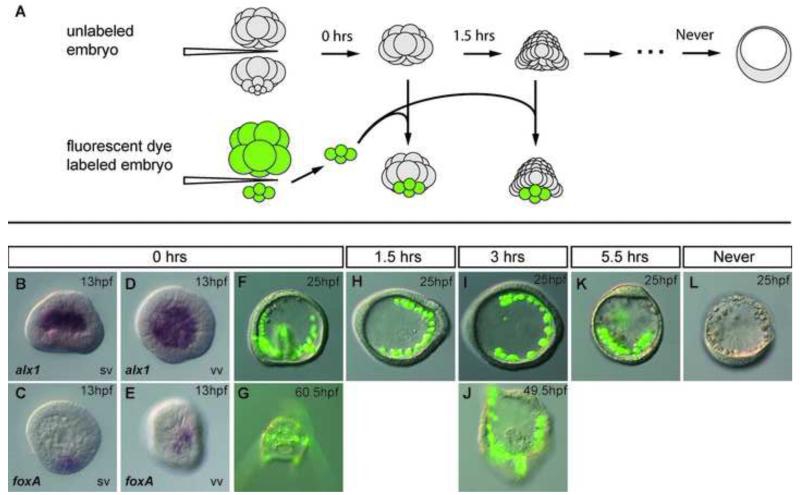
We first asked if and when isolated animal caps acquired an ability to reprogram and replace the endomesoderm. From the classical studies of Horstadius and others animal caps isolated at the 16-cell or 32-cell stage form only dauer blastulae and do not exhibit a reprogramming capacity, though at the 16-32-cell stage animal caps responded to inductions (Horstadius, 1939; Horstadius, 1973; Amemiya, 1996). However, in further study, Horstadius also observed that after the 32-cell stage the animal cap cells also lost the ability to be induced by micromeres, even if the micromeres were younger 16-cell, induction competent cells (Horstadius, 1939). We wondered whether the animal cap cells were permanently locked out of an ability to reprogram or whether they might be like the micromere-NSM replacement cells and reprogram if isolated later in development. We began by repeating the Ho rstadius experiment and assessed both the duration of time animal caps responded to micromere induction, and the response of animal caps if isolated at later and later stages. Gut formation was used as the primary phenotypic indicator of successful induction or reprogramming change. We also examined the expression pattern of gut specific marker genes in recombinant embryos. To assure that we examined only animal cap tissue, animal caps (all 8 mesomeres) were surgically isolated from 32-cell stage embryos and were cultured for 0hrs, 1.5hrs, 3hrs and 5.5hrs before being recombined on the vegetal side with a 16-cell stage micromere quartet labeled with a fluorescent dye (Figure 5A). After the animal cap ring received micromeres at the 32-cell stage, the skeletogenic marker alx1 was expressed later by all the ingressed PMCs (Fig. 5B,D) and the early endomesoderm marker foxA was produced by the responding animal caps and localized to a small region of the blastula (Fig. 5C, E). When observed at 25hpf, a significant number of the 16-cell micromere to 16-32-cell animal cap recombinants (8/14) gastrulated and developed to the pluteus stage (Fig. 5F,G). Thus, consistent with Horstadius’ observations 32-cell stage animal caps were capable of being induced by micromeres to give rise to a gut and correctly pattern the added PMCs. However, after that the animal caps became refractory to an induction response. if animal caps were isolated at 32-cell stage, then incubated for 1.5 hr or more before receiving 16-cell micromeres, the animal caps failed to respond to the micromere induction (Figures 5H-K). These embryos produced an incorrectly patterned calcium carbonate skeleton derived entirely from transplanted micromeres. Animal caps that never received micromeres became dauer blastulae (Fig. 5L) Animal caps isolated between the 32-cell stage and mesenchyme blastula also became dauer blastulae. These observations showed that animal cap cells initially respond to micromere induction if at the 16-32-cell stage, but lose this inductive competence shortly thereafter during cleavage, and by themselves, demonstrated no capacity for reprogramming up to the mesenchyme blastula stage (about 9hpf).
Figure 5.
Animal cap cells lose an ability to regulate during early cleavage stages. (A) Schematic of the experiment. Unlabeled animal cap cells isolated from 32-cell stage embryos were cultured for 0, 1.5, 3, or 5.5hrs before receiving freshly isolated micromere quartets from fluorescently labeled 16-cell stage donor embryos. Numerals at the top of each arrow indicate the times elapsed before isolated animal halves received donor micromere quartets. (B-E) In situ images of embryos developed from an animal half receiving micromere quartet immediately after isolation, fixed at 13hpf. (B,D) in situ hybridization of alx1 in the chimera embryo. (C,E) in situ hybridization of foxA in the chimera embryo. (F-K) Embryos developed from an animal half receiving FITC labeled (green) micromere quartet. (F) micromeres received at 32-cell stage, imaged at 25hpf and (G) 60.5hpf; (H) micromeres received after 1.5hrs of isolated culture, imaged at 25hpf; (I) micromeres received after 3hrs of isolated culture, at 25hpf and (J) 49.5hpf; (K) micromeres received after 5.5hrs and imaged at 25hpf. (L) A dauer blastula developed from an isolated animal half that never received micromeres. SV side view, VV ventral view.
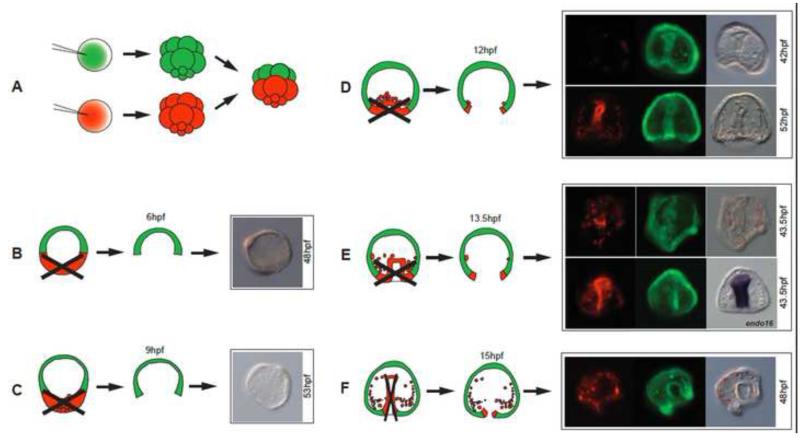
There was a dramatic change as we isolated animal caps at later and later stages. Animal caps isolated from the beginning of gastrulation and beyond reprogrammed and replaced vegetal tissues remarkably well. To clearly distinguish cells with animal or vegetal half origin, we established an assay in which each embryo was a chimera of a normal animal half labeled with a fluorescent dye, and a normal vegetal half labeled with a different fluorescent dye, obtained and recombined at the 16 to 32-cell stage (Fig. 6A). At each isolation time point, best attempts were made to remove all cells of vegetal origin (Figures 6B-F). Most animal-vegetal separations were complete, however at later stages a few PMCs tended to adhere tightly to the basal lamina, so we were unable to remove all PMCs from some of those older embryos. Nevertheless, results of this set of experiments were quite clear. After not responding if isolated between the 60-cell stage (3hpf) and 12hpf (Fig. 6B,C), isolated animal caps began to demonstrate reprogramming abilities and replaced missing germ layers (4/6, and 3/5 in two initial trials when animal caps were isolated at early gastrula, Figure 6D). Many of these animal caps contained correctly patterned PMCs and coelomic pouches in addition to a gut, all from animal cap cells (Fig. 6D, upper panels imaged at 42hpf), and in further experiments where embryos were cultured longer the animal caps became pluteus larvae (see Fig. 7). Animal caps isolated at the mid-gastrula stage (Fig. 6E) or later still reprogrammed to replace missing parts as well, and later expressed the endoderm marker gene endo16 (Romano and Wray, 2003) (Figure 6E lower panels). Thus, the presumptive ectoderm is able to reprogram but only after reaching a certain specification state and only after being in contact with the vegetal halves until gastrulation (because animal caps isolated from embryos between 3-10hpf developed only as dauer blastulae). Perhaps not coincidentally, the timing of the reprogramming onset by ectoderm (12-13hpf) coincided with the time NSMs first demonstrated their capacity to switch to PMCs in both micromere(−) and PMC(−) embryos. Thus the sea urchin embryo has a remarkable ability to completely re-pattern the limits of germ layers using the remaining cell population and to adjust relative proportions, and experiments suggest this ability is first initiated at early gastrula stage.
Figure 6.
Animal caps reprogram. (A) Embryos were produced with animal caps and vegetal halves labeled with different fluorescent dyes and recombined at the 32-cell stage. (B-F) After 6, 9, 12, 13.5 and 15hpf the red and green halves were separated. (B,C) Halves separated at 6 and 9hpf did not develop beyond dauer blastulae. (D-F) Animal halves isolated at 12, 13.5 or 15hpf, contributed to vegetal germ layers (7/11 at 12hpf; 3/4 at 13.5hpf; 4/9 at 15hpf in this experiment). In E, lower right panel, in situ for endo16 of the same embryo seen to its left.
Figure 7.
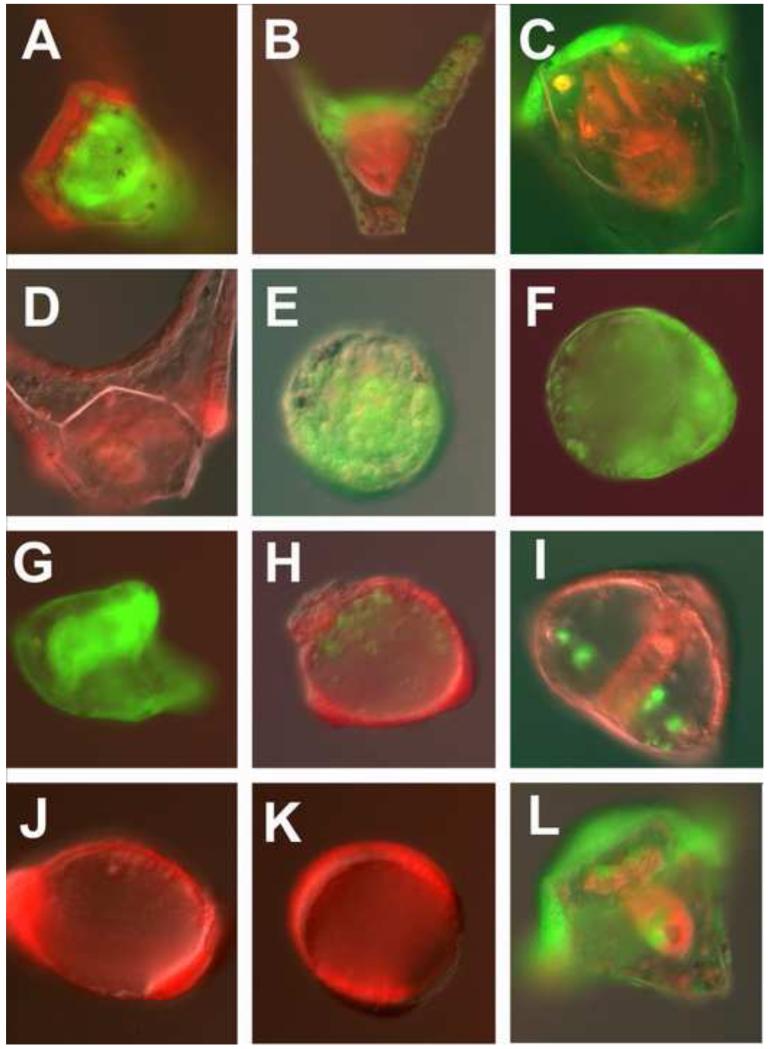
Endoderm GRN necessary for reprogramming. Red/green half embryos were recombined at the 32-cell stage and imaged at 48 hpf. In each case the embryos were imaged in both the red and green channels and merged. (A). Control red animal cap embryo recombined with a green vegetal half embryo. (B). Green Hox11/13b knockdown in the animal cap recombined with a control red vegetal half embryo. (C). Green FoxA knockdown in the animal cap recombined with control red vegetal half. Note: there is no mouth. (D). Control red animal cap after removal of green control vegetal half at EG. (E). Green Hox11/13b knockdown animal cap after removal of control red vegetal half at EG. (F). Green FoxA knockdown animal cap after removal of control red vegetal half at EG. (G). Control green animal cap after removal of control red vegetal half at EG. (H). Control red animal cap after removal of green Hox11/13b knockdown vegetal half at EG. (I). Control red animal cap after removal of green FoxA knockdown vegetal half at EG. (J). Red animal cap isolated at 32-cell stage and imaged at 48hpf. (K). Red animal cap recombined with green control vegetal half at 32-cell stage, isolated at HB, and imaged at 48hpf. (L). Green animal cap recombined with red vegetal half at 32-cell stage and half the red vegetal half removed at HB. HB, hatched blastula (7.5hpf); EG, early gastrula (13-14hpf).
Reprogramming requires activation of endomesoderm specification genes as animal cap cells switch to new cell fates
For animal cap cells to switch cell fates we predicted that it would be necessary to use endomesoderm specification genes to accomplish this transition. We designed an assay to test this prediction using two of early specification genes necessary for endomesoderm. Hox11/13b is an early endomesoderm transcription factor that is activated prior to hatched blastula and is near the top of the endoderm GRN (Peter and Davidson, 2010). FoxA is also an endoderm specification transcription factor that is activated early in endomesoderm specification, is necessary for endoderm specification, and is downstream of Hox11/13b (Oliveri et al., 2006; Peter and Davidson, 2011). Chimeric embryos were produced as before at the 32-cell stage with each half labeled with red or green fluorescent markers. Morpholinos to either Hox11/13b or FoxA were co-injected with one of the fluorescent markers so that the morpholino was present either in the animal caps or the vegetal halves of the experimental chimeras. Then, at various times the animal caps were separated from the vegetal halves and cultured to 48hpf, or control chimeras were never separated.
Control chimeras without morpholinos developed to the pluteus stage with the red-green boundary at predicted locations based on earlier lineage analyses (n=8/9;Fig. 7A). If the animal caps were separated from the vegetal halves of control embryos at early gastrula, 13/13 reprogrammed to produce endoderm and mesoderm (9/13 became plutei; 4/13 were late gastrulae at 48hpf; Fig. 7D,G). If the morpholino for Hox11/13b or FoxA was present only in the animal cap the intact chimeras developed to pluteus larvae also, presumably because neither Hox11/13b nor FoxA are necessary for specification of animal cap-derived tissues (Hox11/13b n=18/18; Fig. 7B; FoxA n=4/6; Fig. 7C). The only exception to this was that the FoxA animal cap knockdowns did not develop a mouth, but since FoxA is known to be necessary for stomodeum development this was not a surprise (Fig. 7C) (Oliveri et al., 2006). On the other hand if the morpholinos were present only in the vegetal halves of chimeras left intact, endomesoderm development was severely compromised (Hox11/13b knockdowns, n=7/16 had partial guts, 9/16 had no recognizable guts; FoxA knockdowns, n=1/7 had a partial gut, 6/7 had no recognizable guts; data not shown).
With the above as controls we now examined animal caps isolated at early gastrula in which the morpholinos were either carried in the animal caps, or in the vegetal halves. If the animal caps carried Hox11/13b morpholino or FoxA morpholino they lost much of their capacity to reprogram (Hox11/13b, n=2/13 had any recognizable gut or mesenchyme cells; Fig. 7E; FoxA, n=0/7 had gut or mesenchyme (Fig. 7F). Thus, although the animal cap cells of intact chimeras participated normally in ectoderm development and patterning while carrying the Hox11/13b or FoxA morpholinos in the animal caps, those same cells were unable to reprogram to replace endomesoderm. This supports the hypothesis that, like the transfating of the NSM cells to replace PMCs, the animal caps must express endomesoderm specification components to accomplish reprogramming.
If the vegetal halves carried the morpholinos there was a distinct outcome. Remember, for the animal caps to reprogram they had to be in contact with the vegetal halves until mesenchyme blastula stage. Isolation of animal caps prior to that resulted in dauer blastula formation (Fig. 7J,K). If, in the chimera experiment, the vegetal halves carried the Hox11/13 morpholino, the isolated animal caps (again, isolated at early gastrula) were severely compromised in their ability to reprogram (4/14 had any guts or skeleton from the animal cap halves, Fig. 7H). If the vegetal halves carried the FoxA morpholino, the isolated animal caps were better able to reprogram (n=6/7 had endoderm and/or mesoderm, Fig. 7I), suggesting that a hypothetical vegetal signal required for animal cap reprogramming capability does not require FoxA for its production. Since the vegetal signal is only hypothetical based on the observations of these experiments, the actual vegetal input to the animal cap that enables reprogramming has yet to be discovered, but it likely is expressed downstream of Hox11/13b.
Finally, we asked what would happen if we separated the animal caps from the vegetal halves but included some of the vegetal tissue on purpose. The embryos adapted quite well, and developed to the pluteus stage (n=5/5; Fig. 7L). In each case they used the remaining vegetal tissue for endomesoderm tissues, but incorporated animal cap tissue to replace some of the missing vegetal tissue. Notice in Fig. 7L a gut formed from the remaining endomesoderm tissue (red in this chimera), but part of the gut was filled in with animal cap tissue (green). Given the outcome of the timing seen in reprogramming to replace the PMC and gut, we assume that when parts of the animal or vegetal tissue were removed earlier than 12hpf, that replacement did not begin until early gastrula.
Discussion
Reprogramming, regulative development and regeneration
Contrary to the common notion that cell fate switching potential declines monotonically during development, evidence here suggests that specified cells in the sea urchin embryo initiate a reprogramming capacity after, and not before they reach a relatively advanced state of specification toward their normal fate. This delayed emergence of reprogramming in response to tissue removal may be a more general feature of development in many organisms rather than specific to sea urchins. Other species in the animal kingdom also exhibit remarkable ability to reprogram cell fates in later stages of development. Asexual budding by free-living larvae has been reported in several echinoderm classes (Eaves and Palmer, 2003; Vaughn and Strathmann, 2008). In lobate ctenophores, although their embryonic development is generally mosaic, the adults exhibit high regenerative ability. Specific populations of cells replace missing body structures but the replacement was demonstrated to happen only after the animal developed into the adult stage, a phenomenon referred to as “post-regeneration” (Henry and Martindale, 2000).
Our results further suggest a distinction between the so-called regulative capacity of sea urchins and the reprogramming activity that emerges later in development. The classical concept of regulative development was defined experimentally as the ability of early blastomeres to produce something other than what they would normally make, a phenomenon also seen in reprogramming. The difference is that all regulative development demonstrations occur before the blastomeres begin or progress deeply into their specification program. Here, by contrast, after a long period during which the cells demonstrated essentially no plasticity, isolated tissues gained the ability to reprogram. The reprogramming appears different from epimorphosis regeneration in that during epimorphosis cells typically de-differentiate from the fully differentiated state, and then are reprogrammed to form the same or a different cell type. In the case of the sea urchin the responding cells transit through intermediate states where both previous and reprogrammed markers are present, and the transit is compressed temporally relative to the control specification sequence for a tissue. The reprogramming is also different from stem cell-mediated replacement of cells because the cells that respond are not a set-aside population of pleuripotent cells. The cells that reprogram presumably are influenced by changes in a signaling environment that causes a change in their fate. Sea urchins thus demonstrate a remarkable ability to reprogram at many stages of their life history. The capacity demonstrated here, as with other stages, aligns most closely with morphallactic regeneration in which existing tissues are repatterned or reprogrammed. Here, we show that reprogramming occurs beginning at gastrulation. It may well be that the capacity for regeneration in adults (Ebert, 1967), is initiated at gastrulation as seen here.
A mechanistic view of reprogramming
Transcription factors necessary for specifying a lineage during normal development were also necessary for reprogramming to that lineage. This suggests, perhaps not surprisingly, that the regulatory logic underlying normal development, (Davidson, 2002; Davidson et al., 2006), is used also to achieve cell fate reprogramming. It was shown earlier that the cell fate transition does not necessarily return to the beginning of a specification sequence (Ettensohn et al., 2007), but the transition includes at least part of the original specification sequence. Thus reprogramming may be understood in terms of the regulatory logic organizing normal development. The key is to figure out how the initial switch is made. In Lytechinus growing at 23° C the step time between successive regulatory transactions is approximately twice as fast as those observed in S. purpuratus which was previously calculated to be about 3hrs (Peter et al., 2012). In the reprogramming sequence seen here, the interval between expression of the first transcription factor (alx1) and differentiation genes (msp130 and vegfr), which had been 4-5hr during normal specification, was less than an hour in the reprogramming. These data suggest that there was time for no more than one or perhaps two steps in that particular reprogramming sequence.
Network theory and experiment suggest that different cell fates correspond to distinct attractors of GRN dynamics (to use the language of that field)(Huang et al., 2005; Kauffman, 1969). By that concept micromere(−), PMC(−) and normal embryos move along different trajectories in gene expression state space, and converge, experimentally, at the skeletal fate attractor. If that is true, the appearance of reprogramming capacity at a rather precise time in the developmental sequence provides an attractive target period for application of recently proposed mechanistic mathematical models of the GRN that attempt to explain the development of normal and perturbed embryos (Peter et al., 2012). In the case of the animal caps, the absence of Hox11/13b in animal caps had no effect on specification of normal animal caps, but that absence prevented the animal caps from reprogramming to replace endomesodermal fates. Reprogramming, as observed here, starts from one attractor (specification state) and transitions into another attractor. In the reprogramming seen in the present experiments, it appears to pass through an intermediate phase that can settle into different attractors depending on the perturbation to the system. What was somewhat surprising to us was that the capacity for reprogramming was only achieved after specification had progressed both in the animal caps, and in the NSM, before each was able to respond to missing tissue and transition to a new state.
Manipulating signaling environment as a possible way of achieving reprogramming
iPSC induction generally requires forced expression of transcription factors (Yamanaka and Blau, 2010) and in general has low rates of successful reprogramming (Hochedlinger and Plath, 2009). In the sea urchin embryo, by contrast, the reprogramming observed here was highly robust and efficient, required no introduction of external molecules into the cells and achieved a high success rate of cell fate conversion. In both the micromere(−) replacement and the animal cap reprogramming signaling had to be involved. The NSM transfating occurred only in the absence of PMCs suggesting that the NSM somehow sensed the absence of the PMCs. The animal caps reprogrammed only after extended contact with the vegetal half embryos, and the Hox11/13b knockdown in the vegetal halves suggested that those halves not only had to undergo specification, but also must have supplied the animal caps with the capacity to reprogram, likely through signaling. The underlying developmental mechanisms that naturally emerged from evolution make these several reprogramming cases attractive models for learning principles of cell fate conversion, which may in turn be instructive for iPSC generation.
Highlights.
Cellular differentiation potential does not decline monotonically in development
A reprogramming capacity begins at gastrulation in the sea urchin embryo.
Gene expression dynamics are compressed during reprogramming.
The cells undergoing reprogramming switch to the GRN of the targeted cell type.
Acknowledgments
This work was supported by NIH R01 HD14483, P01 HD037105, and P050 GM081883. We appreciate the help of Esther Miranda with experiments, the McClay laboratory for reagents, and for critical evaluation of the manuscript. The authors declare no competing financial interests.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amemiya S. Complete regulation of development throughout metamorphosis of sea urchin embryos devoid of macromeres. Dev Growth Differ. 1996;38:465–476. doi: 10.1046/j.1440-169X.1996.t01-4-00003.x. [DOI] [PubMed] [Google Scholar]
- Cherry AB, Daley GQ. Reprogramming cellular identity for regenerative medicine. Cell. 2012;148:1110–1122. doi: 10.1016/j.cell.2012.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croce JC, McClay DR. Dynamics of Delta/Notch signaling on endomesoderm segregation in the sea urchin embryo. Development. 2010;137:83–91. doi: 10.1242/dev.044149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson EH. Academic; Burlington, MA; San Diego: 2006. The regulatory genome: gene regulatory networks in development and evolution. [Google Scholar]
- Davidson EH, Rast JP, Oliveri P, Ransick A, Calestani C, Yuh CH, Minokawa T, Amore G, Hinman V, Arenas-Mena C, Otim O, Brown CT, Livi CB, Lee PY, Revilla R, Rust AG, Pan Z, Schilstra MJ, Clarke PJ, Arnone MI, Rowen L, Cameron RA, McClay DR, Hood L, Bolouri H. A genomic regulatory network for development. Science. 2002;295:1669–1678. doi: 10.1126/science.1069883. [DOI] [PubMed] [Google Scholar]
- Duloquin L, Lhomond G, Gache C. Localized VEGF signaling from ectoderm to mesenchyme cells controls morphogenesis of the sea urchin embryo skeleton. Development. 2007;134:2293–2302. doi: 10.1242/dev.005108. [DOI] [PubMed] [Google Scholar]
- Eaves AA, Palmer AR. Reproduction: widespread cloning in echinoderm larvae. Nature. 2003;425:146. doi: 10.1038/425146a. [DOI] [PubMed] [Google Scholar]
- Ebert TA. Growth and Repair of Spines in Sea Urchin Strongylocentrotus Purpuratus (Stimpson) Biol Bull. 1967;133:141–&. doi: 10.1126/science.157.3788.557. [DOI] [PubMed] [Google Scholar]
- Ettensohn CA, Illies MR, Oliveri P, De Jong DL. Alx1, a member of the Cart1/Alx3/Alx4 subfamily of Paired-class homeodomain proteins, is an essential component of the gene network controlling skeletogenic fate specification in the sea urchin embryo. Development. 2003;130:2917–2928. doi: 10.1242/dev.00511. [DOI] [PubMed] [Google Scholar]
- Ettensohn CA, Kitazawa C, Cheers MS, Leonard JD, Sharma T. Gene regulatory networks and developmental plasticity in the early sea urchin embryo: alternative deployment of the skeletogenic gene regulatory network. Development. 2007;134:3077–3087. doi: 10.1242/dev.009092. [DOI] [PubMed] [Google Scholar]
- Ettensohn CA, McClay DR. Cell lineage conversion in the sea urchin embryo. Dev Biol. 1988;125:396–409. doi: 10.1016/0012-1606(88)90220-5. [DOI] [PubMed] [Google Scholar]
- Henry JJ, Amemiya S, Wray GA, Raff RA. Early inductive interactions are involved in restricting cell fates of mesomeres in sea urchin embryos. Dev Biol. 1989;136:140–153. doi: 10.1016/0012-1606(89)90137-1. [DOI] [PubMed] [Google Scholar]
- Henry JQ, Martindale MQ. Regulation and regeneration in the ctenophore Mnemiopsis leidyi. Dev Biol. 2000;227:720–733. doi: 10.1006/dbio.2000.9903. [DOI] [PubMed] [Google Scholar]
- Hochedlinger K, Plath K. Epigenetic reprogramming and induced pluripotency. Development. 2009;136:509–523. doi: 10.1242/dev.020867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horstadius S. The mechanics of sea urchin development, studied by operative methods. Biol Rev Camb Philos. 1939;14:132–179. [Google Scholar]
- Hörstadius S. Experimental Embryology of Echinoderms. Clarendon Press; Oxford: 1973. [Google Scholar]
- Huang S, Eichler G, Bar-Yam Y, Ingber DE. Cell fates as high-dimensional attractor states of a complex gene regulatory network. Phys Rev Lett. 2005;94:128701. doi: 10.1103/PhysRevLett.94.128701. [DOI] [PubMed] [Google Scholar]
- Kauffman SA. Metabolic stability and epigenesis in randomly constructed genetic nets. J Theor Biol. 1969;22:437–467. doi: 10.1016/0022-5193(69)90015-0. [DOI] [PubMed] [Google Scholar]
- Leaf DS, Anstrom JA, Chin JE, Harkey MA, Showman RM, Raff RA. Antibodies to a fusion protein identify a cDNA clone encoding msp130, a primary mesenchyme-specific cell surface protein of the sea urchin embryo. Developmental biology. 1987;121:29–40. doi: 10.1016/0012-1606(87)90135-7. [DOI] [PubMed] [Google Scholar]
- Liu H, Zhu F, Yong J, Zhang P, Hou P, Li H, Jiang W, Cai J, Liu M, Cui K, Qu X, Xiang T, Lu D, Chi X, Gao G, Ji W, Ding M, Deng H. Generation of induced pluripotent stem cells from adult rhesus monkey fibroblasts. Cell Stem Cell. 2008;3:587–590. doi: 10.1016/j.stem.2008.10.014. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- McClay DR, Logan CY. Regulative capacity of the archenteron during gastrulation in the sea urchin. Development. 1996;122:607–616. doi: 10.1242/dev.122.2.607. [DOI] [PubMed] [Google Scholar]
- Minokawa T, Hamaguchi Y, Amemiya S. Skeletogenic potential of induced secondary mesenchyme cells derived from the presumptive ectoderm in echinoid embryos. Dev Genes Evol. 1997;206:472–476. doi: 10.1007/s004270050077. [DOI] [PubMed] [Google Scholar]
- Oliveri P, Carrick DM, Davidson EH. A regulatory gene network that directs micromere specification in the sea urchin embryo. Dev Biol. 2002;246:209–228. doi: 10.1006/dbio.2002.0627. [DOI] [PubMed] [Google Scholar]
- Oliveri P, Walton KD, Davidson EH, McClay DR. Repression of mesodermal fate by foxa, a key endoderm regulator of the sea urchin embryo. Development. 2006;133:4173–4181. doi: 10.1242/dev.02577. [DOI] [PubMed] [Google Scholar]
- Oliveri P, Tu Q, Davidson EH. Global regulatory logic for specification of an embryonic cell lineage. Proc Natl Acad Sci U S A. 2008;105:5955–5962. doi: 10.1073/pnas.0711220105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park I-H, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, Lerou PH, Lensch MW, Daley GQ. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- Peter IS, Davidson EH. The endoderm gene regulatory network in sea urchin embryos up to mid-blastula stage. Dev Biol. 2010;340:188–199. doi: 10.1016/j.ydbio.2009.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter IS, Davidson EH. A gene regulatory network controlling the embryonic specification of endoderm. Nature. 2011;474:635–639. doi: 10.1038/nature10100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter IS, Faure E, Davidson EH. Predictive computation of genomic logic processing functions in embryonic development. Proc Natl Acad Sci U S A. 2012;109:16434–16442. doi: 10.1073/pnas.1207852109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien PW, Sanchez Alvarado A. Fundamentals of planarian regeneration. Annu Rev Cell Dev Biol. 2004;20:725–757. doi: 10.1146/annurev.cellbio.20.010403.095114. [DOI] [PubMed] [Google Scholar]
- Rho HK, McClay DR. The control of foxN2/3 expression in sea urchin embryos and its function in the skeletogenic gene regulatory network. Development. 2011;138:937–945. doi: 10.1242/dev.058396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano LA, Wray GA. Conservation of Endo16 expression in sea urchins despite evolutionary divergence in both cis and trans-acting components of transcriptional regulation. Development. 2003;130:4187–4199. doi: 10.1242/dev.00611. [DOI] [PubMed] [Google Scholar]
- Sharma T, Ettensohn CA. Regulative deployment of the skeletogenic gene regulatory network during sea urchin development. Development. 2011;138:2581–2590. doi: 10.1242/dev.065193. [DOI] [PubMed] [Google Scholar]
- Sweet H, Amemiya S, Ransick A, Minokawa T, McClay DR, Wikramanayake A, Kuraishi R, Kiyomoto M, Nishida H, Henry J. Blastomere isolation and transplantation. Methods Cell Biol. 2004;74:243–271. doi: 10.1016/s0091-679x(04)74011-x. [DOI] [PubMed] [Google Scholar]
- Sweet HC, Hodor PG, Ettensohn CA. The role of micromere signaling in notch activation and mesoderm specification during sea urchin embryogenesis. Development. 1999;126:5255–5265. doi: 10.1242/dev.126.23.5255. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Vaughn D, Strathmann RR. Predators induce cloning in echinoderm larvae. Science. 2008;319:1503. doi: 10.1126/science.1151995. [DOI] [PubMed] [Google Scholar]
- Yamanaka S, Blau HM. Nuclear reprogramming to a pluripotent state by three approaches. Nature. 2010;465:704–712. doi: 10.1038/nature09229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin II, Thomson JA. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]