Abstract
The hippocampal formation contains a distinct population of neurons organized into separate anatomical subregions. Each hippocampal subregion expresses a unique molecular profile accounting for their differential vulnerability to mechanisms of memory dysfunction. Nevertheless, it remains unclear which hippocampal subregion is most sensitive to the effects of advancing age. Here we investigate this question by using separate imaging techniques, each assessing different correlates of neuronal function. First, we used MRI to map cerebral blood volume, an established correlate of basal metabolism, in the hippocampal subregions of young and old rhesus monkeys. Second, we used in situ hybridization to map Arc expression in the hippocampal subregions of young and old rats. Arc is an immediate early gene that is activated in a behavior-dependent manner and is correlated with spike activity. Results show that the dentate gyrus is the hippocampal subregion most sensitive to the effects of advancing age, which together with prior studies establishes a cross-species consensus. This pattern isolates the locus of age-related hippocampal dysfunction and differentiates normal aging from Alzheimer's disease.
Cross-species studies have documented that the hippocampal formation, a structure vital for learning new memories, is particularly vulnerable to the aging process (1–3). A complex structure, the hippocampus is divided into separate but inter-connected anatomic subregions: the entorhinal cortex, the dentate gyrus, the CA subfields, and the subiculum (4). Each hippocampal subregion contains a distinct population of neurons that express a unique molecular profile (5). This uniqueness can account for why each hippocampal subregion is differentially vulnerable to mechanisms of memory dysfunction (6). For example, transient hypoperfusion will cause hippocampal-dependent memory deficits by targeting the CA1 subregion, whereas early Alzheimer's disease causes an overlapping memory deficit by targeting the entorhinal cortex.
Although a number of studies suggest some cell loss with age (7, 8), compared to neurodegeneration, aging is characterized by a relative absence of frank cell death and lacks definitive histopathological markers (9). Rather, aging affects hippocampal performance by impairing normal neuronal physiology, expressed as synaptic dysfunction. This physiologic feature of aging accounts, to a large degree, for the difficulty identifying the primary hippocampal subregions most sensitive to normal aging. Not only is quantifying synaptic dysfunction difficult in postmortem tissue (10–12), but because of hippocampal inter-connectivity, dysfunction in one subregion affects physiologic properties in other hippocampal subregions (1) and even throughout the circuit as a whole. Thus, assessing the functional integrity of each hippocampal subregion individually and simultaneously in living subjects is an effective approach for pinpointing the primary site of age-related hippocampal dysfunction.
Most in vivo functional imaging techniques can assess global hippocampal function but do not have sufficient spatial resolution required to visualize individual hippocampal subregions. One study used a high-resolution variant of functional MRI (13) to image individual hippocampal subregions across the human lifespan (14). Although a decline in signal was observed throughout, the dentate gyrus was the dominant hippocampal subregion that had a pattern of decline most consistent with normal aging (14). These results remain inconclusive, however, because of the difficulty in excluding individuals in the earliest stages of Alzheimer's disease, even among nondemented elderly. Furthermore, although the MRI technique used in the study is sensitive to deoxyhemoglobin, a hemodynamic correlate of brain metabolism, it is also sensitive to other tissue elements (13), such as iron, which cannot be excluded as factors influencing the observed effect.
Our first experiment was designed to address both limitations. Rhesus monkeys were chosen as our experimental group, because they, like all nonhuman mammals, develop age-related hippocampal dysfunction but do not develop Alzheimer's disease (9). Animals were imaged with an MRI technique that generates a measure of regional cerebral blood volume (CBV). Among the three correlates of oxidative metabolism that can be measured with MRI, cerebral blood flow, cerebral blood volume, and deoxyhemoglobin content (15, 16), we chose to rely on MRI measures of CBV to assess hippocampal function. CBV can be measured with higher spatial resolution compared with cerebral blood flow (17), and it provides a purer measure of basal metabolism compared to maps sensitive to basal deoxyhemoglobin (13). MRI measures of CBV have been shown to tightly correlate with regional energy metabolism and cerebral blood flow (for example, see refs. 16 and 18), and can successfully map dysfunction in the brain (18–21), including the hippocampal formation.
Any hemodynamic measure is only an indirect correlate of brain metabolism and cannot exclude that an effect is influenced by primary vascular alterations (22). Furthermore, MRI does not have neuronal resolution. Our second experiment was designed to address these issues. Arc is an immediate early gene whose expression is correlated with spike activity. Arc also modulates long-term potentiation (23–25), a mechanism associated with memory and impaired by aging (1). The cellular localization of Arc mRNA can be used to determine which individual neurons participate in a specific behavioral experience. This new methodology, cellular compartment analysis of temporal activity by fluorescence in situ hybridization combined with high resolution confocal microscopy (or catFISH) was used to map the expression of Arc mRNA in single cells of young, middle-aged, and old rats, after exploration of a novel environment. This imaging approach can directly assess behaviorally induced physiological function with single-cell resolution.
Methods
Mapping Regional CBV in Rhesus Monkeys. Fourteen rhesus monkeys (Macaca mulatta), 7–31 years of age, participated in the study and were imaged with a 1.5-T scanner. During preliminary studies, we identified a T1-weighted acquisition protocol that optimized visualization of the hippocampal subregions (Fig. 1). Whole-brain 3D T1-weighted images (time to repeat, 50 ms; time to echo, 5 ms; flip angle, 35°; in plane resolution, 0.62 mm × 0.62 mm; slice thickness, 2 mm) oriented perpendicular to the hippocampal long-axis were acquired before and 4 min after i.v. administration of the contrast agent gadolinium (0.1 mmol/kg). Studies suggest that this slice thickness does not introduce volume average artifacts, which could hinder reliable regional identification (26). As previously described (17, 27), the change in signal intensity was used to calculate absolute steady-state CBV maps, filtering out pixels with a value ≥10% that reflect large blood vessels. For each subject, the precontrast scan was used to identify slices in which the external morphology and the internal architecture of the hippocampal formation were best visualized (Fig. 1). On average, two slices per animal met these criteria, and the slices were typically located in the anterior body of the hippocampus. Unlike humans (14), identifying the entorhinal cortex and the subregions of the hippocampus proper in a single slice is difficult. Therefore, measurements of the entorhinal cortex were selected in more anterior slices, typically at the level of the uncus (Fig. 1). For each slice, visualized anatomical landmarks were used together with standard atlases (28) to identify the general locale of four subregions: the entorhinal cortex, the dentate gyrus, the CA1 subfield, and the subiculum. To minimize volume-averaging artifacts, the values of the four highest pixels per subregion were averaged, reflecting gray matter CBV. A mean total CBV measurement was calculated by averaging signal from both slices, and this CBV measurement for each hippocampal subregion was used for group data analysis. To test the reliability of the anatomical criteria, region of interests were determined independently by two separate investigators, and reliability analysis revealed a high level of correspondence across all subregions, ranging from an alpha of 0.8 to 0.88.
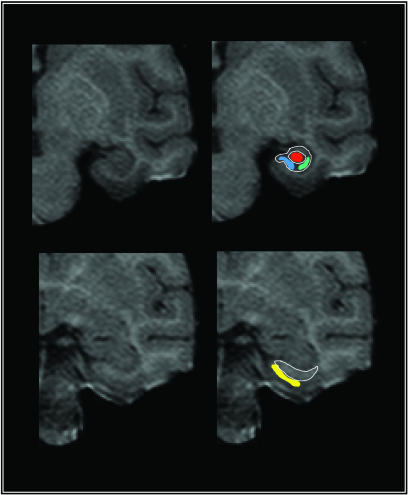
Fig. 1.
High-resolution images visualize individual hippocampal subregions. Maps of CBV were generated from T1-weighted images that possess high spatial and anatomical resolution. Images were acquired perpendicular to the hippocampal long axis, and the upper left image shows an example in which the hippocampal subregions can be visualized. The external morphology and the internal architecture of the hippocampus (white lines) are used as landmarks to segment the hippocampus into the subiculum (blue), the CA1 subfield (green), and the dentate gyrus (red). Note that border zones between the subiculum and CA1 subregion are excluded from analysis. The entorhinal cortex (yellow) is identified in more anterior images, as shown in the lower left image, relying on the underlying white matter and the collateral sulcus as anatomical landmarks.
Hippocampal-dependent memory tests were performed in a subgroup of animals. Delayed nonmatching-to-sample testing was conducted in a manual apparatus as described in detail elsewhere (29). Briefly, delayed nonmatching-to-sample trials consisted of a sample object presentation followed by a recognition test. The sample appeared over the baited central well of the test tray, and, after a response, an opaque barrier was lowered to impose a retention interval. The sample item was subsequently presented together with a novel object that covered a food reward. Objects were selected from a large pool such that a new pair was presented on each trial. During initial testing, monkeys learned the nonmatching rule with a 10-sec retention interval, to a criterion of 90% correct or better (across 100 trials, 20 trials per day, intertrial interval = 30 sec throughout testing). Demands on recognition memory were subsequently increased by imposing successively longer delays of 15, 30, 60, 120 sec (100 trials total at each delay, 20 trials per day) and 600 seconds (50 trials total, 5 trials per day).
Mapping Arc Expression in Rats. Experiments were conducted with male F344 rats (Harlan Sprague–Dawley) from three different age groups (9, 15, and 24 months old; 10 animals per age group). Animals were handled before behavioral exposure to ensure acclimatization to the experimenter. Environment A was a circular platform 0.36 m2 in area, divided into nine 0.04-m2 grids, containing 30-cm-high walls. Environment B was a square platform 0.36 m2 in area, divided into nine 0.04-m2 grids placed on a 10-cm-high platform, with no walls, and located in a different room. In both cases, the entire environment could be uniformly explored. The animals were allowed to explore the entire environment by being picked up by the handler and released in the center of one of nine different grids at random every 10 sec during the 5-min exploration. The rats were allowed to explore environment A for 5 min, were returned to their home cages for 20 min, and then allowed another 5 min exposure to either environment A or environment B for 5 min before being killed. Brains were rapidly removed after either the environment A/A or environment A/B exposure and processed for in situ hybridization for Arc gene expression.
After behavioral exploration, animals were decapitated, and the brains were quick frozen in isopentane cooled over an ethanol/dry ice bath and stored at -70°C. Twenty-micrometerthick coronal sections were cut and arranged on a slide so that all of the experimental groups were represented on each slide. Sections were thaw-mounted and dried before storage at -70°C. Riboprobes were generated from the full-length Arc cDNA (≈3 kb in length, described in ref. 30) by using a commercial RNA transcription kit (Maxiscript, Ambion, Austin, Texas) and RNA-labeling nucleotide mix containing digoxigenin-tagged UTP (Roche Molecular Biochemicals). Fluorescence in situ hybridization was performed as described (24). Briefly, riboprobes were purified on a G-50 spin column (Boehringer Ingelheim), and a small aliquot was subjected to gel electrophoresis before use for determination of yield and integrity of the riboprobes. Slides containing the sections were thawed to room temperature, fixed with freshly prepared buffered paraformaldehyde (4%), treated with 0.5% acetic anhydride/1.5% triethanolamine, incubated in methanol and acetone (1:1) for 5 min, and equilibrated in 2× saline sodium citrate (SSC). Sections were incubated with 100 μl of 1× prehybridization buffer (Sigma) for 30 min at room temperature. Approximately 100 ng of riboprobe was diluted in 1× enhanced hybridization buffer (Amersham Pharmacia), heat-denatured at 90°C, chilled on ice, and applied to each section. A coverslip was placed on each slide and incubated overnight at 56°C. Post hybridization washes started with 2× SSC, increased in stringency to 0.5× SSC at 56°C. RNase A (10 μg/ml) at 37°C was used to dissociate any single-stranded RNA. After quenching the endogenous peroxidases with 2% H2O2, slides were blocked with blocking agent (NEN Life Sciences, Boston) and incubated with an anti-digoxigenin Ab conjugated with horseradish peroxidase (Roche Molecular Biochemicals) overnight at 4°C. Slides were washed with Tris-buffered saline containing 0.05% Tween-20, and the horseradish peroxidase-antibody conjugate was detected by using NEN fluorescence in situ hybridization detection kits. Glass coverslips were applied to the slides after counterstaining with propidium iodide (Vector Laboratories) with a small amount of antifade media and sealed with nail polish.
Stained sections were imaged with a Leica TCS-4D laser scanning confocal microscope equipped with an argon-krypton laser. The entire dentate gyrus was imaged with a Nikon ×5 objective. Maximal resolution images (1024 × 1024 pixels) were acquired by using the maximum confocal pinhole setting to increase the optical section thickness so that labeled cells could be seen from a single image. After optimization of laser settings, detector gain and offsets were kept constant for each confocal session to minimize variability. The CA3c and CA1 subregions of the hippocampus were imaged with a Nikon ×40 oil objective. The entire CA3c region was included in the analysis. Two to three frames from identical anatomical regions were imaged for the CA1 subregion per slide. The total number of CA1 cells analyzed for each rat ranged from 88 to 170 cells. Areas of analysis were Z-sectioned into 1.0-μm optical sections and analyzed by an experimenter who was blind to conditions by using the metamorph image analysis software (Universal Imaging, Downington, PA). To minimize sampling errors, only cells with whole nuclei were assessed and included in the analysis. Arc RNA-positive labeling was observed in nucleus, perinucleus/cytoplasm, or both. Analysis of 164 frames in the dorsal blade of the dentate gyrus contained 222–237 cells per frame (512 × 512 pixels) with an average of 230 cells per frame for young, middle-aged, or old animals. Cells containing Arc RNA labeling were counted and normalized to the total number of cells and number of frames.
Results
Imaging Aging Monkeys. The relationship between CBV and age, as measured from each hippocampal subregion, is shown in Fig. 2. Because of hippocampal circuit properties (1), a primary site of dysfunction will influence function in other hippocampal subregion. We, therefore, first performed a multivariate stepwise regression analysis, in which CBV from each hippocampal subregion was included as the independent variable and age was included as the dependent variable. This analysis is designed to control for circuit-wide effects. This analysis revealed that the dentate gyrus was the only hippocampal subregion whose CBV significantly correlated, inversely, with age (beta = -0.77; P = 0.001). To confirm this effect in a univariate analysis, we performed a bivariate correlation between age and each individual CBV measure. Again, the dentate gyrus was the only hippocampal subregion whose CBV correlated, inversely, correlate with age (correlation coefficient = -0.77; P = 0.001). Interestingly, although a statistically insignificant decline was noted in the subiculum (correlation coefficient = -0.34; P = 0.23), age-related CBV decline was absent in the entorhinal cortex (correlation coefficient = 0.18; P = 0.53) and CA1 subregion (correlation coefficient = 0.02; P = 0.94).
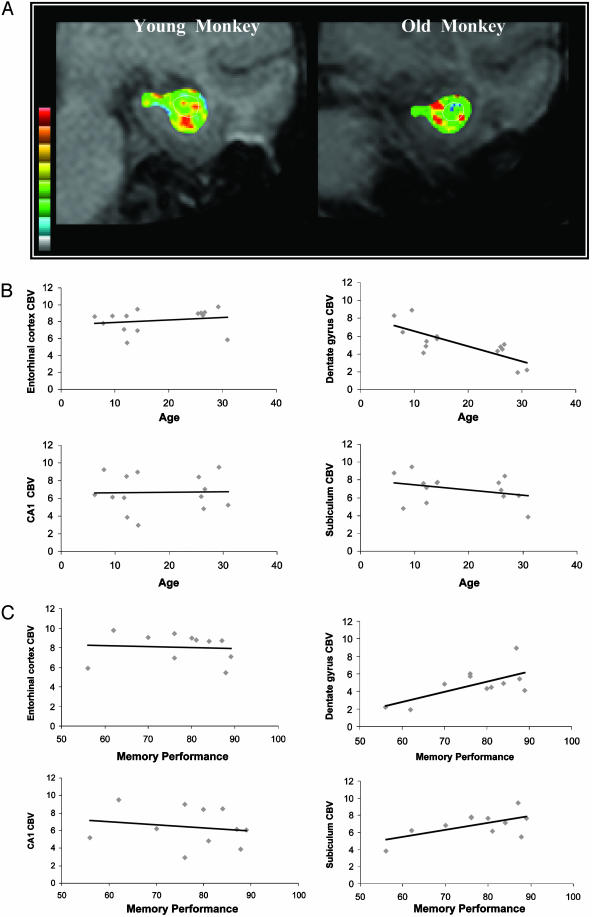
Fig. 2.
CBV measured from the dentate gyrus differentially correlates with age in rhesus monkeys. (A) Individual examples of gadolinium-induced change in MRI signal, a measure of CBV and a correlate of brain metabolism, acquired from a young and old rhesus monkey. Warmer colors indicate higher CBV. The white circle, identified in precontrast images, overlies the dentate gyrus. (B) CBV measurements are plotted against age for individual hippocampal subregions. A significant age-related decline in CBV was observed only the dentate gyrus. (C) CBV measurements are plotted against memory performance on the delayed nonmatching-to-sample test. A significant relationship between CBV and memory was observed only in the dentate gyrus.
To test whether CBV measurements are related to cognitive function, a bivariate correlation was performed between memory performance on a delayed nonmatching-to-sample task (mean percent correct recognition across delays) and each individual CBV measure (Fig. 2B). Results revealed that the dentate gyrus was the only hippocampal subregion whose CBV significantly correlated with performance on (correlation coefficient = 0.83; P = 0.026). The relationships between memory and CBV for the other hippocampal subregions were not statistically significant (entorhinal cortex correlation coefficient = -0.13; subiculum correlation coefficient = 0.48; CA1 correlation coefficient = -0.17).
Imaging Aging Rats. The numbers of cells that express Arc RNA after behavioral exploration was determined by using the cat-FISH method. Dentate gyrus granule cells and pyramidal cells in hippocampal regions CA1 and CA3 were counted in each of the three age groups (Fig. 3). An ANOVA was used to first compare the number of cells that expressed Arc mRNA across subregions. Independent of age, pyramidal cells of the CA3 subregion (t = -17.6; P < 0.001) and of the CA1 subregion (t = -43.7; P < 0.001) expressed a significantly higher number of Arc-positive cells compared with the granule cells of the dentate gyrus. This difference is consistent with prior electrophysiological studies showing that, compared with pyramidal neurons, a majority of granule cells exhibit low firing rates while rats perform a spatial task (31).
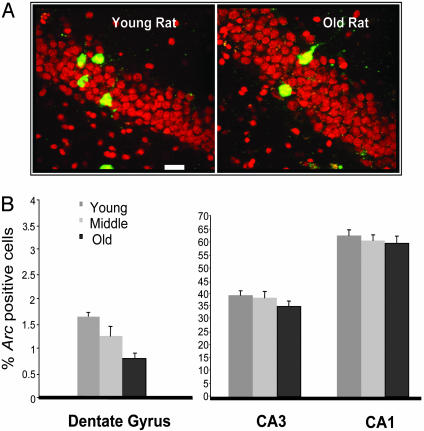
Fig. 3.
Arc expression in the granule cells of the dentate gyrus differentially correlates with age in rats. (A) Individual examples of behaviorally induced Arc RNA staining, a correlate of spike activity, in hippocampal granule cells of a young and an old rat. Red is the nuclear counterstain propidium iodide, and green is fluorescent-tagged (CY3) Arc RNA. (Scale bar, 20 μm.) (B) The average percent of Arc-positive neurons measured from the three different age groups (9, 15, and 24 months) are shown for individual hippocampal subregions. Note that there are significantly fewer Arc-positive granule cells in old rats compared with stability in the numbers of Arc-positive pyramidal cells. On average, older rats express Arc RNA in fewer cells than did young rats, and this significant age-related decline in Arc expression was observed only in the dentate gyrus.
As in the monkeys, a circuit analysis was first performed by including Arc levels from each hippocampal subregion into a multivariate regression model. The granule cells of the dentate gyrus were the only neurons with a significant inverse relationship with age group (beta = -0.51; P = 0.005). This finding was confirmed in a univariate ANOVA model, where a significant effect was found only for the dentate gyrus, in which an age-related decline in Arc-containing granule cells was observed [P < 0.002, ANOVA with Fisher's PLSD (protected least significant difference) post hoc test]. Importantly, as in the imaging studies, pyramidal-cell subregions of the hippocampus showed equivalent numbers of Arc RNA-expressing cells, suggesting that these cell types were unaffected by advancing age (Fig. 3).
Discussion
The functional organization of the hippocampal formation presents an obstacle when attempting to isolate the primary site of memory dysfunction (6). The hippocampal subregions are interconnected, and, therefore, a lesion in an individual subregion leads to secondary alterations in downstream subregions by means of transsynaptic mechanisms (1). Therefore, techniques that assess the function of the hippocampus globally and techniques that assess the function of an individual subregion in isolation are not ideally suited to pinpoint the primary source of dysfunction. By relying on approaches that map the functional integrity of the hippocampal subregions in living brains, we have isolated subregions differentially sensitive and resistant to the aging process.
Although age-related hippocampal dysfunction is observed in all mammalian species, rats are the most popular animal model of normal aging (32), and monkeys most closely resemble the human aging process (33) and human hippocampal formation (34). Both species were investigated in this study by using different but complimentary imaging techniques. The hippocampus of aging rhesus monkeys was investigated with an MRI measure of CBV. CBV was the first hemodynamic variable to be successfully measured with MRI technology (35), and a wide range of studies has established its intimate relationship to blood flow and brain metabolism (for examples, see refs. 15 and 16). Most relevant to this study, MRI measures of CBV have been found to linearly correlate with basal glucose metabolism (18), and CBV has successfully detected the effects of aging (36) and hippocampal dysfunction in Alzheimer's and other diseases (20, 21). Here we used an approach that generates CBV images with high spatial resolution (17), allowing us to visualize and analyze individual hippocampal subregions. Our first imaging study found that the dentate gyrus is the hippocampal subregion most sensitive to the effects of advancing age in rhesus monkeys.
Despite the ability to visualize individual hippocampal subregions in living brains, MRI does not possess cellular resolution. Furthermore, because it is inherently an indirect measure of brain metabolism, CBV, like any hemodynamic variable, is prone to misinterpretations (22). For example, an age-related change in vascular biophysics may lead to hemodynamic changes independent of underlying brain metabolism (37, 38). On both counts, directly imaging behaviorally induced Arc expression in hippocampal neurons compliments MRI. Arc expression effectively reports on spike activity (24, 25), and, therefore, its expression is a direct correlate of neuronal physiology. Furthermore, Arc expression is associated with long-term potentiation (23), a mechanism of plasticity linked to memory and impaired in age-related memory decline (1). Our second imaging study found that the dentate gyrus is the hippocampal subregion most sensitive to the effects of advancing age in rats.
By demonstrating a consistent pattern of age-related decline in the dentate gyrus, in two species that do not develop Alzheimer's disease, these results also act to confirm a prior human imaging study (14) and prior findings in aging rats (39). Thus, despite a variety of functional imaging techniques applied to a range of species, results converge on the same finding: The dentate gyrus is the hippocampal subregion most vulnerable to advancing age, whereas the pyramidal cell subregions are relatively spared, in rats, monkeys, and humans.
By identifying the neuronal population most sensitive to aging, these results set the stage for isolating the biochemical factors within granule cells that underlie the physiologic defect. Prior studies have observed, for example, a reduction in glutamate receptors in the granule cells of aging rhesus monkeys, a finding that can account for our findings (40). Nevertheless, by explicitly comparing molecular profiles of granule and pyramidal hippocampal neurons across the lifespan, future studies can begin to identify molecules most relevant to the aging process.
Our results, furthermore, clarify a distinction between normal aging and Alzheimer's disease, a pathological process that also targets the aging hippocampal formation. In contrast to aging, Alzheimer's disease causes clear histological changes, and the pattern of cell loss in Alzheimer's disease and other markers of disease have been used to identify vulnerable subregions of the hippocampal formation. The vast majority of studies have documented that the pyramidal cells of the hippocampal formation are most vulnerable, whereas the granule cells are relatively resistant, to Alzheimer's disease pathology (41–48). When changes have been observed in the dentate gyrus (49), they are postulated to occur secondarily to upstream lesions in the entorhinal cortex. Thus, by establishing differential patterns of hippocampal dysfunction, these studies confirm that aging and Alzheimer's disease are indeed distinct entities. Furthermore, imaging the functional integrity of individual hippocampal subregions is an effective method to detect the earliest stage of Alzheimer's disease, when it behaviorally overlaps with normal aging.
Acknowledgments
We thank David Amaral, Cynthia Erickson, Sania Fong, Arun Nava, Mary Roberts, Jeffrey Roberts, John Ryan, and the staff at the California National Primate Research Center for their work. This work was supported in part by National Institutes of Health Grants AG08702, AG00949, AG003376, AG09219, RR000169, and AG10606; the Beeson Faculty Scholar Award from the American Federation of Aging; the Institute for the Study on Aging; and the McKnight Endowment Fund for Neuroscience.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviation: CBV, cerebral blood volume.
References
- 1.Barnes, C. A. (1994) Trends Neurosci. 17, 13-18. [DOI] [PubMed] [Google Scholar]
- 2.Gallagher, M. & Rapp, P. R. (1997) Annu. Rev. Psychol. 48, 339-370. [DOI] [PubMed] [Google Scholar]
- 3.Small, S. A., Stern, Y., Tang, M. & Mayeux, R. (1999) Neurology 52, 1392-1396. [DOI] [PubMed] [Google Scholar]
- 4.Amaral, D. G. & Witter, M. P. (1989) Neuroscience 31, 571-591. [DOI] [PubMed] [Google Scholar]
- 5.Zhao, X., Lein, E. S., He, A., Smith, S. C., Aston, C. & Gage, F. H. (2001) J. Comp. Neurol. 441, 187-196. [DOI] [PubMed] [Google Scholar]
- 6.Small, S. A. (2001) Arch. Neurol. (Chicago) 58, 360-364. [DOI] [PubMed] [Google Scholar]
- 7.Pakkenberg, B., Pelvig, D., Marner, L., Bundgaard, M. J., Gundersen, H. J., Nyengaard, J. R. & Regeur, L. (2003) Exp. Gerontol. 38, 95-99. [DOI] [PubMed] [Google Scholar]
- 8.West, M. J. (1993) Neurobiol. Aging 14, 287-293. [DOI] [PubMed] [Google Scholar]
- 9.Gallagher, M., Landfield, P. W., McEwen, B., Meaney, M. J., Rapp, P. R., Sapolsky, R. & West, M. J. (1996) Science 274, 484-485. [DOI] [PubMed] [Google Scholar]
- 10.Terry, R. D. & Katzman, R. (2001) Neurobiol. Aging 22, 347-348; discussion 353-354. [DOI] [PubMed] [Google Scholar]
- 11.Price, J. L., McKeel, D. W., Jr. & Morris, J. C. (2001) Neurobiol. Aging 22, 351-352. [DOI] [PubMed] [Google Scholar]
- 12.Morrison, J. H. (2001) Neurobiol. Aging 22, 349-350. [DOI] [PubMed] [Google Scholar]
- 13.Small, S., Wu, E., Bartsch, D., Lacefield, C., DeLaPaz, R., Mayeux, R., Stern, Y. & Kandel, E. (2000) Neuron 653-664. [DOI] [PubMed]
- 14.Small, S. A., Tsai, W. Y., DeLaPaz, R., Mayeux, R. & Stern, Y. (2002) Ann. Neurol. 51, 290-295. [DOI] [PubMed] [Google Scholar]
- 15.van Zijl, P. C., Eleff, S. M., Ulatowski, J. A., Oja, J. M., Ulug, A. M., Traystman, R. J. & Kauppinen, R. A. (1998) Nat. Med. 4, 159-167. [DOI] [PubMed] [Google Scholar]
- 16.Hyder, F., Kida, I., Behar, K. L., Kennan, R. P., Maciejewski, P. K. & Rothman, D. L. (2001) NMR Biomed. 14, 413-431. [DOI] [PubMed] [Google Scholar]
- 17.Lin, W., Celik, A. & Paczynski, R. P. (1999) J. Magn. Reson. Imaging 9, 44-52. [DOI] [PubMed] [Google Scholar]
- 18.Gonzalez, R. G., Fischman, A. J., Guimaraes, A. R., Carr, C. A., Stern, C. E., Halpern, E. F., Growdon, J. H. & Rosen, B. R. (1995) Am. J. Neuroradiol. 16, 1763-1770. [PMC free article] [PubMed] [Google Scholar]
- 19.Harris, G. J., Lewis, R. F., Satlin, A., English, C. D., Scott, T. M., Yurgelun-Todd, D. A. & Renshaw, P. F. (1998) Am. J. Neuroradiol. 19, 1727-1732. [PMC free article] [PubMed] [Google Scholar]
- 20.Bozzao, A., Floris, R., Baviera, M. E., Apruzzese, A. & Simonetti, G. (2001) Am. J. Neuroradiol. 22, 1030-1036. [PMC free article] [PubMed] [Google Scholar]
- 21.Wu, R. H., Bruening, R., Noachtar, S., Arnold, S., Berchtenbreiter, C., Bartenstein, P., Drzezga, A., Tatsch, K. & Reiser, M. (1999) J. Magn. Reson. Imaging 9, 435-440. [DOI] [PubMed] [Google Scholar]
- 22.Mueggler, T., Sturchler-Pierrat, C., Baumann, D., Rausch, M., Staufenbiel, M. & Rudin, M. (2002) J. Neurosci. 22, 7218-7224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guzowski, J. F., Lyford, G. L., Stevenson, G. D., Houston, F. P., McGaugh, J. L., Worley, P. F. & Barnes, C. A. (2000) J. Neurosci. 20, 3993-4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guzowski, J. F., McNaughton, B. L., Barnes, C. A. & Worley, P. F. (1999) Nat. Neurosci. 2, 1120-1124. [DOI] [PubMed] [Google Scholar]
- 25.Guzowski, J. F., Setlow, B., Wagner, E. K. & McGaugh, J. L. (2001) J. Neurosci. 21, 5089-5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laakso, M. P., Juottonen, K., Partanen, K., Vainio, P. & Soininen, H. (1997) Magn. Reson. Imaging 15, 263-265. [DOI] [PubMed] [Google Scholar]
- 27.Kuppusamy, K., Lin, W., Cizek, G. R. & Haacke, E. M. (1996) Radiology 201, 106-112. [DOI] [PubMed] [Google Scholar]
- 28.Amaral, D. G. & Insausti, R. (1990) in The Human Nervous System., ed. Paxinos, R. (Academic, San Diego).
- 29.Rapp, P. R., Morrison, J. H. & Roberts, J. A. (2003) J. Neurosci. 23, 5708-5714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lyford, G. L., Yamagata, K., Kaufmann, W. E., Barnes, C. A., Sanders, L. K., Copeland, N. G., Gilbert, D. J., Jenkins, N. A., Lanahan, A. A. & Worley, P. F. (1995) Neuron 14, 433-445. [DOI] [PubMed] [Google Scholar]
- 31.Jung, M. W. & McNaughton, B. L. (1993) Hippocampus 3, 165-182. [DOI] [PubMed] [Google Scholar]
- 32.Gallagher, M. (1997) Philos. Trans. R. Soc. London B 352, 1711-1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lane, M. A. (2000) Exp. Gerontol. 35, 533-541. [DOI] [PubMed] [Google Scholar]
- 34.Seress, L. (1992) Epilepsy Res. 7, Suppl., 3-28. [PubMed] [Google Scholar]
- 35.Belliveau, J. W., Rosen, B. R., Kantor, H. L., Rzedzian, R. R., Kennedy, D. N., McKinstry, R. C., Vevea, J. M., Cohen, M. S., Pykett, I. L. & Brady, T. J. (1990) Magn. Reson. Med. 14, 538-546. [DOI] [PubMed] [Google Scholar]
- 36.Wenz, F., Rempp, K., Brix, G., Knopp, M. V., Guckel, F., Hess, T. & van Kaick, G. (1996) Magn. Reson. Imaging 14, 157-162. [DOI] [PubMed] [Google Scholar]
- 37.Buckner, R. L., Snyder, A. Z., Sanders, A. L., Raichle, M. E. & Morris, J. C. (2000) J. Cogn. Neurosci. 12, Suppl. 2, 24-34. [DOI] [PubMed] [Google Scholar]
- 38.D'Esposito, M., Zarahn, E., Aguirre, G. K. & Rypma, B. (1999) NeuroImage 10, 6-14. [DOI] [PubMed] [Google Scholar]
- 39.Smith, C. B., Goochee, C., Rapoport, S. I. & Sokoloff, L. (1980) Brain 103, 351-365. [DOI] [PubMed] [Google Scholar]
- 40.Gazzaley, A. H., Siegel, S. J., Kordower, J. H., Mufson, E. J. & Morrison, J. H. (1996) Proc. Natl. Acad. Sci. USA 93, 3121-3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Braak, H. & Braak, E. (1996) Acta Neurol. Scand. Suppl. 165, 3-12. [DOI] [PubMed] [Google Scholar]
- 42.Fukutani, Y., Sasaki, K., Mukai, M., Matsubara, R., Isaki, K. & Cairns, N. J. (1997) Psychiatry Clin. Neurosci. 51, 227-231. [DOI] [PubMed] [Google Scholar]
- 43.Giannakopoulos, P., Hof, P. R., Kovari, E., Vallet, P. G., Herrmann, F. R. & Bouras, C. (1996) J. Neuropathol. Exp. Neurol. 55, 1210-1220. [DOI] [PubMed] [Google Scholar]
- 44.Hof, P. R. (1997) Eur. Neurol. 37, 71-81. [DOI] [PubMed] [Google Scholar]
- 45.Price, J. L., Ko, A. I., Wade, M. J., Tsou, S. K., McKeel, D. W. & Morris, J. C. (2001) Arch. Neurol. 58, 1395-1402. [DOI] [PubMed] [Google Scholar]
- 46.Rossler, M., Zarski, R., Bohl, J. & Ohm, T. G. (2002) Acta Neuropathol. 103, 363-369. [DOI] [PubMed] [Google Scholar]
- 47.West, M. J., Coleman, P. D., Flood, D. G. & Troncoso, J. C. (1994) Lancet 344, 769-772. [DOI] [PubMed] [Google Scholar]
- 48.West, M. J., Kawas, C. H., Martin, L. J. & Troncoso, J. C. (2000) Ann. N.Y. Acad. Sci. 908, 255-259. [DOI] [PubMed] [Google Scholar]
- 49.Redwine, J. M., Kosofsky, B., Jacobs, R. E., Games, D., Reilly, J. F., Morrison, J. H., Young, W. G. & Bloom, F. E. (2003) Proc. Natl. Acad. Sci. USA 100, 1381-1386. [DOI] [PMC free article] [PubMed] [Google Scholar]