Abstract
Bradyrhizobium japonicum RegSR regulatory proteins belong to the family of two-component regulatory systems, and orthologs are present in many Proteobacteria where they globally control gene expression mostly in a redox-responsive manner. In this work, we have performed a transcriptional profiling of wild-type and regR mutant cells grown under anoxic denitrifying conditions. The comparative analyses of wild-type and regR strains revealed that almost 620 genes induced in the wild type under denitrifying conditions were regulated (directly or indirectly) by RegR, pointing out the important role of this protein as a global regulator of denitrification. Genes controlled by RegR included nor and nos structural genes encoding nitric oxide and nitrous oxide reductase, respectively, genes encoding electron transport proteins such as cycA (blr7544) or cy 2 (bll2388), and genes involved in nitric oxide detoxification (blr2806-09) and copper homeostasis (copCAB), as well as two regulatory genes (bll3466, bll4130). Purified RegR interacted with the promoters of norC (blr3214), nosR (blr0314), a fixK-like gene (bll3466), and bll4130, which encodes a LysR-type regulator. By using fluorescently labeled oligonucleotide extension (FLOE), we were able to identify two transcriptional start sites located at about 35 (P1) and 22 (P2) bp upstream of the putative translational start codon of norC. P1 matched with the previously mapped 5′end of norC mRNA which we demonstrate in this work to be under FixK2 control. P2 is a start site modulated by RegR and specific for anoxic conditions. Moreover, qRT-PCR experiments, expression studies with a norC-lacZ fusion, and heme c-staining analyses revealed that anoxia and nitrate are required for RegR-dependent induction of nor genes, and that this control is independent of the sensor protein RegS.
Introduction
The Rhizobiales order of α-Proteobacteria include Gram-negative nitrogen-fixing soil bacteria collectively named rhizobia which have the unique ability to establish N2-fixing symbioses with legume roots and stems of some aquatic legumes, leading to the formation of new plant organs called nodules. Expression of nitrogen fixation and other symbiosis-related genes requires low-oxygen conditions [1]–[2]. To cope with oxygen limitation prevailing under microoxic free-living conditions or in so-called bacteroids existing within plant cells of nodules, rhizobial species express the high-affinity cbb 3 oxidase encoded by the fixNOQP operon [3]. Moreover, some rhizobial species are able to use nitrate as final electron acceptor to support respiration under microoxic or anoxic conditions [4]–[6]. The switch from oxygen to nitrate respiration leads to a reduction in the ATP yield, yet it allows bacteria to survive and multiply under oxygen-limiting conditions [7]. Denitrification has been defined as the dissimilatory reduction of nitrate (NO3 −) or nitrite (NO2 −) to N2 via the gaseous intermediates nitric oxide (NO) and nitrous oxide (N2O) with concomitant ATP generation [8]. This process requires four separate enzymatic reactions catalyzed by nitrate-, nitrite-, nitric oxide-, and nitrous oxide reductases, encoded by nar/nap, nir, nor, and nos genes, respectively [9]–[11].
In recent years, it has emerged that many rhizobial species have denitrification genes [4]–[6]. Among them, the soybean symbiont Bradyrhizobium japonicum is considered the model organism for studying rhizobial denitrification. In this bacterium, denitrification depends on the napEDABC, nirK, norCBQD, and nosRZDYFLX genes that encode a periplasmic nitrate reductase (Nap), a copper-containing nitrite reductase (NirK), a c-type nitric oxide reductase (cNor), a nitrous oxide reductase (Nos), respectively [4]. Similar to many other denitrifiers, expression of denitrification genes in B. japonicum requires both oxygen limitation and the presence of nitrate or a derived nitrogen oxide [4]. Perception and transduction of the “low-oxygen signal” are mediated by conserved regulatory proteins that are integrated into species-specific networks in different rhizobia [1]–[2]. Two interlinked oxygen responsive regulatory cascades are present in B. japonicum, the FixLJ-FixK2 and the RegSR-NifA cascades [12]. A moderate decrease in the oxygen concentration in the gas phase (≤5%) is sufficient to activate expression of FixLJ-FixK2-dependent targets [12]. This “low-oxygen” signal is sensed by the heme-based sensory kinase FixL which auto-phosphorylates and transfers the phosphoryl group to the FixJ response regulator which then activates transcription of fixK2. In turn, FixK2 induces expression of regulatory genes such as rpoN 1, fixK 1, nnrR [13]–[15], genes associated with microoxic metabolism, e.g. fixNOQP [14]–[16] as well as genes involved in denitrification such as nap, nirK, nor, and nos [4], [13]–[14], [17]. Induction of genes controlled by the RegSR-NifA cascade requires very low oxygen concentration (≤0.5%) because of the pronounced oxygen sensitivity of NifA. The response regulator RegR of the RegSR two-component regulatory system induces expression of the fixR-nifA operon which is preceded by two overlapping promoters, P1 and P2 [18]–[20]. RegR activates transcription originating from P2 under all oxygen conditions via binding to a DNA element located around position −67 upstream of the transcription start site. Upon a switch to low-oxygen or anoxic conditions, the redox-responsive NifA protein in concert with RNA polymerase containing RpoN (σ54) enhances its own synthesis via activation of the −24/−12-type promoter P1. In B. japonicum, RpoN is encoded by the two highly similar and functionally equivalent genes (rpoN 1 and rpoN 2) [21]. Since rpoN 1 is under the control of FixK2, this gene represents the link between the two regulatory cascades. Targets of NifA include nif and fix genes, which are directly or indirectly involved in nitrogen fixation, and also genes that are not essential for this process or have an unknown function [1], [22]–[23]. Recent results from our group showed that NifA is also required for maximal expression of nap, nirK, and nor genes, suggesting a new role for the RegSR-NifA regulatory cascade in the control of the denitrification genes in B. japonicum [24]. A large number of members of the RegR regulon have been identified by transcriptome analysis of a B. japonicum regR mutant grown under oxic and microoxic free-living conditions and also in bacteroids [25]. However, no data are available about the RegR regulon in cells grown under denitrifying conditions. Moreover, we have recently demonstrated that the level of NorC is significantly lower in membranes isolated from a B. japonicum regR mutant compared to the wild type when cells were grown under denitrifying conditions [26]. However, the involvement of RegSR in norCBQD genes expression has not been investigated so far.
Here, we have performed a comparative transcriptome analysis of B. japonicum wild type and a regR mutant grown under denitrifying conditions. Among the novel RegR targets, nor genes encoding the nitric oxide reductase were identified. By different approaches we also demonstrated that RegR control of nor genes induction requires anoxia and nitrate, and that this control is independent on the sensor protein RegS.
Materials and Methods
Bacterial Strains and Growth Conditions
The wild-type strain B. japonicum 110spc4 [27] and its derivatives 2426 (ΔregR), 2409 (ΔregS), [20], and 9043 (ΔfixK 2) [15] were used in this study. Strain 2499 [13] is B. japonicum 110spc4 containing a norC–lacZ fusion. In this work, plasmid pRJ2499 containing the norC–lacZ fusion [13], was integrated by homologous recombination into the chromosome of the regR and regS mutant strains resulting in strains 2499RR and 2499RS, respectively. B. japonicum strains were grown oxically at 30°C in peptone-salts-yeast extract (PSY) medium supplemented with 0.1% L-arabinose [14], [27]. Growth under oxygen-limiting conditions was performed in Bergersen minimal medium [28] with succinate as carbon source and supplemented (BSN) or not (BS) with 10 mM KNO3. For comparison with previous experiments, yeast extract-mannitol (YEM) medium [29] supplemented with 10 mM KNO3 was used for anoxic cultures of the wild type and fixK2 mutant in some primer extension experiments. Once they were inoculated to an OD600 of about 0.2, cultures were subjected to oxygen-limiting conditions generated by two different experimental procedures. In the first set of experiments, 17 ml serum tubes or 500 ml flasks containing 5 or 200 ml medium, respectively, were sealed with rubber septa stoppers and the headspace atmosphere was replaced by a gas mixture (2% oxygen, 98% argon) before cultures were incubated. In the second set, cells were incubated in completely filled, sealed 200 ml bottles or 17 ml tubes without gas exchange. The latter conditions are referred to as anoxic conditions throughout the manuscript. Antibiotics were added to B. japonicum cultures at the following concentrations (µg ml−1): cloramphenicol 20, spectinomycin 200, streptomycin 100, tetracycline 100. Escherichia coli strains were cultured in Luria-Bertani (LB) medium [30] at 37°C. E. coli S17-1 [31] served as the donor for conjugative plasmid transfer. Tetracycline was used at 10 µg ml−1 in E. coli cultures.
RNA Isolation, cDNA Synthesis, and Microarray Analysis
Cultures of B. japonicum wild type and regR mutant strains were grown anoxically in BSN medium to an OD600 of about 0.4. Cell harvest, isolation of total RNA, cDNA synthesis, fragmentation, labeling and conditions for microarray hybridization were done as described previously [22], [25], [32]–[33]. Details of the custom designed Affymetrix B. japonicum gene chip BJAPETHa520090 (Santa Clara, CA) have also been described previously [22]. For each strain, a minimum of four biological replicates was analyzed. Details on data processing, normalization, and further analysis are described elsewhere [33]. GeneSpring GX 7.3.1 software (Agilent Technologies, Santa Clara, CA) was used for comparative analyses. Only the probe sets that were called “present” or “marginal” in ≥75% of the replicates of each experiment were considered for further analysis. The student t-test with a P value threshold of 0.025 was applied for statistical comparisons. We considered genes passing the statistical tests as differentially expressed only if the relative change in expression (n-fold) was ≥2 or ≤−2 when different conditions or strains were compared. Operon predictions were done as described in Hauser et al. (2007) [22] and Mesa et al. (2008) [14]. An operon-like organization of genes (bicistronic or larger) was assumed if they were orientated in the same direction and separated by less than 32 bp. This distance was enlarged to 100 bp if the first three letters in the gene names were identical.
Quantitative Real-Time PCR
Expression of nosZ, nosY, norC, blr2808, napE, napA, cycA, copC, bll3466, bll4130 and bll2388 genes was also analyzed by quantitative reverse transcription-PCR (qRT-PCR) using an iQ™5 Optical System (Bio-Rad, CA). B. japonicum wild-type and regR cultures, RNA isolation and cDNA synthesis were performed as described for microarray experiments. Primers for the PCR reactions (Table S1 in supplemental material) were designed with Primer3Web v.0.4.0 (http://frodo.wi.mit.edu/primer3/input.htm) to have a melting temperature of 57°C to 62°C and generate PCR products of 50 to 100 bp. Each PCR reaction contained 9.5 µl of iQ™ SYBR Green Supermix (Bio-Rad), 2 µM (final concentration) of individual primers and appropriate dilutions of different cDNAs in a total volume of 19 µl. Reactions were run in triplicates. Melting curves were generated to verify the specificity of the amplification. Relative changes in gene expression were calculated as described elsewhere [34]. Expression of the primary sigma factor gene sigA was used as a reference for normalization (primers SigA-1069F and SigA-1155R; [25]).
Heme-Staining Analysis
Cells of B. japonicum grown oxically in 150 ml PSY medium were harvested by centrifugation at 8,000× g for 5 min, washed twice with BS or BSN, resuspended in 500 ml of the same medium, and cultured under anoxic conditions or with 2% initial O2 concentration for 48 hours (final resulting OD600 about 0.5). Cells were disrupted using a French pressure cell (SLM Aminco, Jessup, MD, USA) and membranes were isolated as described previously [35]. Membrane protein aliquots were diluted in sample buffer [124 mM Tris-HCl, pH 7.0, 20% glycerol, 4.6% sodium dodecyl sulfate (SDS) and 50 mM 2-mercaptoethanol], and incubated at room temperature for 10 min. Membrane proteins were separated at 4°C in 12%-SDS polyacrylamide gel electrophoresis (20 µg protein per lane), transferred to a nitrocellulose membrane and stained for heme-dependent peroxidase activity as described previously [36] using the chemiluminescence detection kit “SuperSignal” (Pierce, Thermo Fisher Scientific, IL, USA).
NO Consumption Activity
Cells of B. japonicum were cultured anoxically during 48 hours in BSN (OD600 of about 0.5). Cells were harvested by centrifugation at 8,000× g for 10 min at 4°C, and washed with 50 mM Tris-HCl buffer (pH 7.5). NO consumption rates were determined with a 2 mm ISONOP NO electrode APOLO 4000 (World Precision Inst., Sarasota, FL) in a 2 ml temperature-controlled, magnetically stirred reaction chamber [37]. The membrane-covered electrode was situated at the bottom of the chamber above the stirrer and reactants were injected with a Hamilton syringe through the port in the glass stopper. The chamber was filled with 760 µl of 25 mM phosphate buffer (pH 7.4), 900 µl of cell suspension (4–5 mg protein), 100 µl of an enzyme mix of Aspergillus niger glucose oxidase (40 units ml−1) and bovine liver catalase (250 units ml−1) (Sigma-Aldrich, St. Louis, MO), 90 µl 1 M sodium succinate, and 100 µl of 320 mM glucose. Once a steady base line was observed, 50 µl of a saturated NO solution (1.91 mM at 20°C) was added to the cuvette to start the reaction. Each assay was run until the NO detection had dropped to zero, i.e. when all NO was oxidized.
β-Galactosidase Assays
To measure β-galactosidase activity, strains 2499, 2499RR and 2499RS were grown oxically in PSY medium, collected by centrifugation at 8,000× g for 10 min at 4°C, washed twice with BS or BSN medium and cultured anoxically or under 2% initial O2 in the same medium for 48 h (OD600 of about 0.5). Activity was determined with permeabilized cells from at least three independently grown cultures assayed in triplicate for each strain and condition. β-Galactosidase assays were performed essentially as previously described [30]. The absorbance data for A 420, A 550, and A 600 were determined for all samples in a plate reader (SUNRISE Absorbance Reader, TECAN, Männedorf, Switzerland) using the XFluor4 software (TECAN). Data were then transferred to Microsoft Excel to calculate the specific activities in Miller units.
Fluorescently Labeled Oligonucleotide Extension (FLOE)
B. japonicum wild type, regR and fixK2 mutant strains were cultured as indicated above for microarray and qRT-PCR experiments. Cells were harvested and total RNA was isolated using the hot phenol extraction procedure described by Babst and coworkers (1996) [38]. To determine the transcription start site of norC gene, the NorC53 reverse primer was synthesized, HPLC-purified, and labeled with 6-carboxyfluorescein (6-FAM) at the 5′-end (Eurofins MWG Operon, Ebersberg, Germany). The sequence of NorC53 was 5′-GAGCCGCCGTAGAAGACGTTTC-3′ which corresponds to positions 53-31 downstream of the annotated translation start codon of norC. The primer extension assay was performed using Avian Myeloblastosis Virus Reverse Transcriptase (AMV RT; Promega, Wisconsin, USA). The reaction mixture (50 µl) contained MgCl2 (5 mM), dNTPs (2 mM each), AMV RT 1X buffer, 50 pmoles FAM-labeled primer, 7–10 µg of RNA and water. Reactions were incubated at 65°C for 15 minutes, then 2 µl of the AMV/RNasin mixture (1∶1; Promega) was added. Next, the mixtures were kept at 15°C for 10 minutes followed by incubation at 45°C and 95°C for 45 and 5 minutes, respectively. Finally, 2 µl of RNase (2 mg ml−1) was added to the reactions which were further incubated at 37°C for 90 minutes. The length of the reverse transcribed cDNA products was analyzed in an ABI Prism 3100 Genetic Analyzer capillary electrophoresis instrument (Newbiotechnic S.A., Seville, Spain). GeneScan version 3.1.2 (Applied Biosystems, Austin TX) was used to analyze the data, i.e. peak identification and determination of the length and abundance of cDNA.
Electrophoretic Mobility Shift Assays (EMSAs)
Binding of RegR to putative target promoters was tested by EMSA using radiolabeled PCR fragments obtained with the primers listed in Table S1. PCR fragments were end labeled with [γ-32P] ATP using T4 polynucleotide kinase (MBI Fermentas), and subsequently purified over Micro Bio-Spin 6 chromatography columns (Bio-Rad, Spain). His-tagged RegR was overexpressed and purified as described previously [39]. For in vitro phosphorylation, RegR protein (40 µM final concentration) was incubated with 25 mM acetyl phosphate (Sigma-Aldrich, St. Louis, MO) in DNA binding buffer [20] for 1 h at 30°C. Phosphorylated RegR (0 to 7.5 µM) was incubated with column-purified DNA fragments (0.5 to 1 µg) in DNA binding buffer in a total volume of 20 µl. After 15-min incubation at 30°C, samples were mixed with loading dye and separated on 8% non-denaturing polyacrylamide gels in Tris-borate 89 mM and EDTA 2 mM electrophoresis buffer pH 8.2 for 2 h at 70 V. Gels were dried, and radiolabeled bands were visualized with a phosphorimager (Bio-Rad, Spain).
Microarray Data Accession Number
The microarray data are available in the NCBI Gene Expression Omnibus database (GEO; http://www.ncbi.nlm.nih.gov/geo) under GEO Series accession number GSE56668.
Results
Transcriptional Profiling of a B. japonicum regR Mutant Grown Under Free-living Denitrifying Conditions
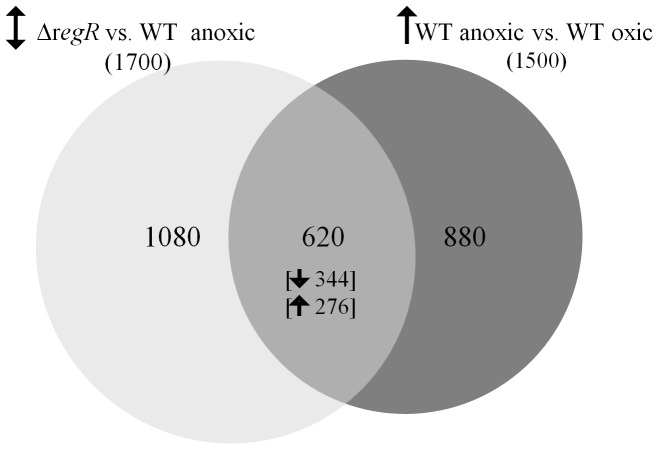
Comparative analyses of B. japonicum wild type and the regR mutant strain grown under anoxic conditions in BSN medium revealed that approximately 1,700 genes were differentially expressed in the regR mutant (Fig. 1). The main focus of this work was the identification of genes that were upregulated in the wild type under denitrifying conditions (in comparison with oxically-grown wild type) [22], [33], and at the same time regulated by RegR. The comparison of both regulons gave a total number of 620 genes (Fig. 1, Table S2 in supplemental material). Within this group, we focused on the genes positively controlled by RegR under denitrifying conditions (344 genes). Among them, we found genes involved in the denitrification process, such as nosRZDFYLX encoding nitrous oxide reductase [40], and norECBQD genes encoding nitric oxide reductase [41]. We also identified cycA which codes for cytochrome c 550 that is implicated in electron delivery to NirK, the Cu-containing nitrite reductase [42]–[43]. Of the napEDABC genes specifying periplasmic nitrate reductase [35], only napB and napC genes were identified as RegR dependently transcribed, yet the relative fold change (FC) of expression was only −2.8 and −3.4, respectively (Table S2). Surprisingly, nirK could not be identified among the RegR-controlled genes (Table S2). We also found as RegR target the cy2 gene (bll2388) that encodes the previously identified FixK2-dependent cytochrome c 2 [14], which suggests that this gene might be relevant for life under denitrifying conditions. In addition to denitrification genes, numerous other genes were identified as candidates for being RegR targets under anoxic conditions (Table S2).
Figure 1. Venn diagram based on anoxically induced genes of the B. japonicum wild type (WT; dark grey circle) and genes which, when compared with the wild type, are differentially expressed in the regR mutant both grown under anoxic denitrifying conditions (light grey circle).
The overlap shows the fraction of genes which are induced in the wild type and differentially transcribed in the regR mutant (the number of genes upregulated or downregulated in the regR mutant appears in brackets). Strains and conditions are indicated alongside the circles. Up-down arrows reflect increased and decreased gene expression in microarray analyses. Numbers in parentheses indicate the total number of differentially expressed genes.
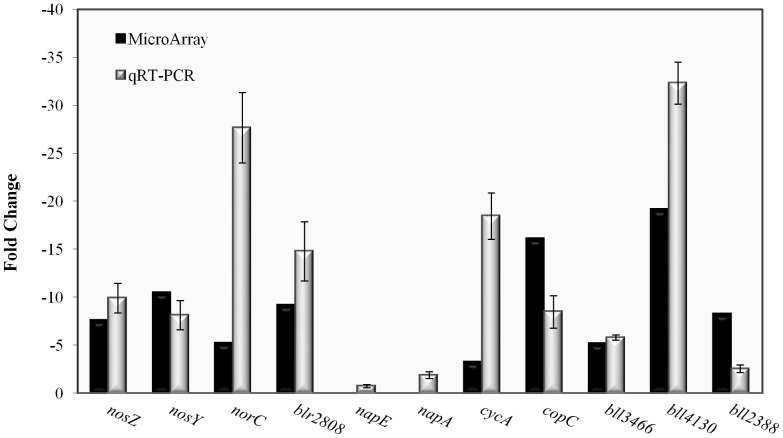
To validate the microarrays results, we performed qRT-PCR with several selected genes. Among them, we have selected genes encoding (i) denitrification enzymes (nosZ, nosY, norC, napE and napA) and electron transfer proteins (cycA, bll2388), (ii) NO detoxification proteins (blr2806-09), (iii) copper homeostasis proteins (copC), and (iv) transcription factors (bll3466 encoding a FixK-like protein and bll4130 which encodes a LysR-type regulator). As shown in Fig. 2, we confirmed by qRT-PCR RegR-dependent induction of genes for nitrous oxide reductase (nosZ, nosY), nitric oxide reductase (norC), and also cycA and bll2388. Likewise, qRT-PCR confirmed the microarray data which indicated that genes encoding the periplasmic nitrate reductase (napE and napA) are not significantly controlled by RegR. Furthermore, we could verify RegR-dependent expression of blr2808 which is part of a gene cluster (blr2806-blr2809) possibly involved in NO detoxification and nitrate assimilation [44]. Finally, qRT-PCR also confirmed RegR-dependent expression of copC, bll3466, and bll4130 (Fig. 2). Note that differences observed between FC values determined by microarray technology and qRT-PCR such as those observed for norC, cycA and bll2388 are not uncommon.
Figure 2. Comparison of expression data generated by microarray experiments (black bars) and qRT-PCR (grey bars).
RNA was isolated from the wild-type strain and the regR mutant cultured in BSN medium under anoxic conditions. qRT-PCRs were repeated in three independent experiments each including four parallel amplification reactions. Fold-change values refer to differences of expression when the regR mutant was compared with the wild type.
RegR Binding to the Promoter Region of New Target Genes
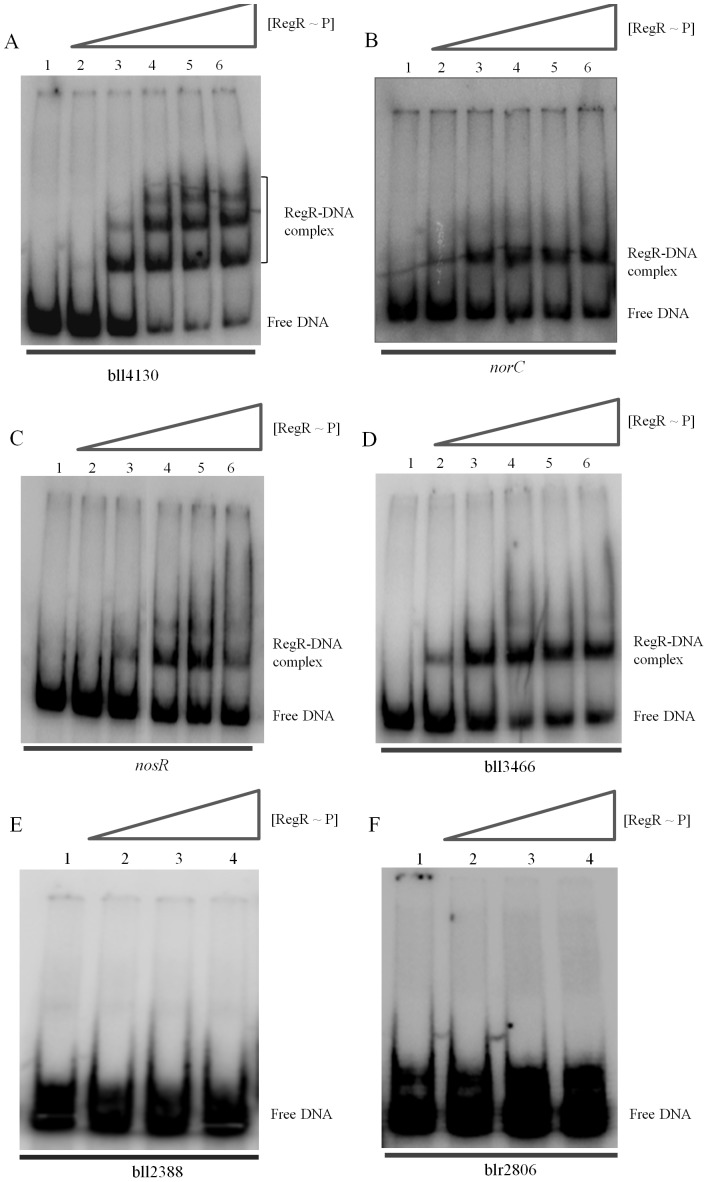
Because microarray analysis cannot discriminate directly and indirectly controlled genes, we performed DNA binding analyses by EMSA to identify direct RegR target genes. We used PCR fragments corresponding to the promoter region of six candidate genes whose expression was found to be under positive control of RegR (Table 1). 32P-labeled fragments were incubated with increasing concentrations of phosphorylated RegR (0 to 7.5 µM). As positive and negative controls, we used amplification products covering the promoter and the coding region of the bll2087 gene, respectively (Table S1) [32]. RegR displayed consistent binding to the promoter region of bll4130 (Fig. 3A). In addition to bl4130, RegR bound to the DNA probes derived from the promoter regions of norC (Fig. 3B), nosR (Fig. 3C), and bll3466 (Fig. 3D). By contrast, no binding was observed to the promoter regions of bll2388 (Fig. 3E) and blr2806 (Fig. 3F). While EMSAs with the norC, nosR and bll3466 promoter regions showed one predominat retarded band (Fig. 3B, C and D), up to 3 bands of distinct mobility were observed with the bll4130 promoter and increasing RegR concentrations (Fig. 3A).
Table 1. Summary of RegR binding studies with promoter regions of anoxically induced, RegR-controlled B. japonicum genes.
| Gene no.a | Gene nameb | Descriptionc | Genomic regiond | Shifte | Putative RegR-box positionf | Sequence of putative RegR-boxg |
| blr0314 | nosR | Nitrous oxide reductase expression regulator | −195 to +46 | + | −73 | TGCGTCAACGGCGA |
| −39 | CGCGGCCCGGTCGG | |||||
| bll2388 | cy2 | Cytochrome c2 | −143 to +31 | − | None found | |
| blr2806 | Nitrite extrusion protein | −214 to +34 | − | −64 | CGCGCCTCCGTGGCCG | |
| −49 | GGAGGCAGAGCCTG | |||||
| blr3214 | norC | Nitric oxide reductase subunit C | −149 to +53 | + | −64 | CGCGCGAAGCGGC |
| −123 | CGTGTCGGCCGTCGT | |||||
| bll3466 | fixK | Transcriptional regulator FixK-type | −160 to +58 | + | −78 | TGCGACATCGGCGGC |
| −88 | CGAGCCGGAGTGCGAC | |||||
| bll4130 | Transcriptional regulatory protein LysR-family | −114 to +61 | + | −51 | TGCGGCTTTCGTGCC | |
| −98 | TGCGGCAAAGGAGCC |
All listed genes are differentially expressed (FC<−2) in a comparison of the wild type with the ΔregR mutant both grown under the anoxic, denitrifying conditions as described in this work.
Gene name as indicated in the EMBL-EBI database.
Protein description according to Kaneko and coworkers, 2002 [67].
Genomic region included in the PCR fragment used for EMSAs. Coordinates refer to the first nucleotide position relative to the annotated translation start site of the genes listed in column 1.
Indicates qualitatively whether (+) or not (−) RegR binding was observed to the DNA fragments specified in column 4.
Position of the 5′-end nucleotide of the putative RegR box (column 7) relative to the annotated translational start site of the associated gene.
Sequence of the putative RegR binding sites. Conserved nucleotides are highlighted.
Figure 3. Analysis of RegR binding to the promoter region of several putative target genes by EMSA.
Increasing amounts of purified RegR∼P were incubated with constant amounts (0.5 to 1 µg) of double-stranded 32P-labeled PCR amplified products from the promoter region of B. japonicum bll4130, norC, nosR, bll3466, bll2388 and blr2806 genes. In panels A to D, RegR concentrations were 1.5 µM (lanes 2), 3 µM (lanes 3), 4.5 µM (lanes 4), 6 µM (lanes 5) and 7.5 µM (lanes 6). In panels E and F, RegR concentrations were 0.8 µM (lanes 2), 1.6 µM (lanes 3) and 3.2 µM (lanes 4). No RegR protein was added to the control reactions loaded in lane 1 of all panels. Samples were run on 6% non-denaturing polyacrylamide gels and visualized with a phosphorimager.
Inspection of DNA sequences included in the EMSA assays by using the previously proposed RegR box as query [25] allowed us to identify two putative RegR binding sites in all promoter regions except for bll2388 (Table 1). As described previously [25], half sites of the proposed RegR boxes are differently spaced and not perfectly conserved (Table 1).
Mapping of norC Transcripts
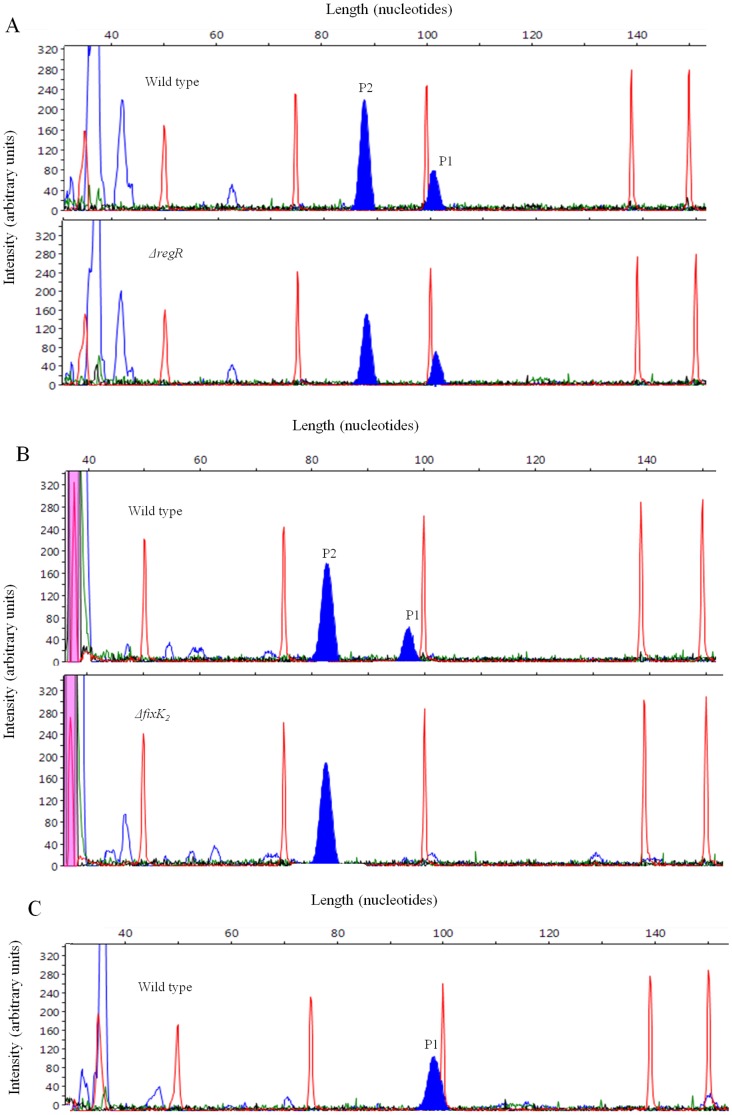
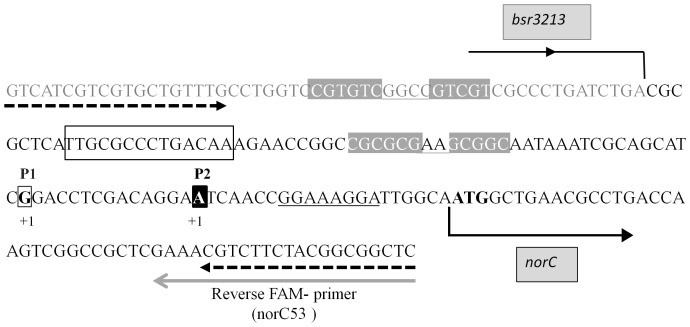
In order to substantiate RegR control of nor genes, we mapped the transcriptional start site of norC in wild-type and regR cells grown under dentrifying conditions. Using FLOE technique we identified two signals with cDNA derived from wild-type RNA (Fig. 4, panel A, upper electropherogram) suggesting the presence of two transcriptional start sites. The cDNA signal marked as P1 corresponds to the previously proposed start site of norC [41] which maps to a guanosine 35 bp upstream of the translational start codon of NorC (Fig. 5). The cDNA signal designed as P2 corresponds to a second start site which maps to an adenosine located 14 bp downstream of P1 and 21 bp upstream of the norC start codon (Fig. 5). As the peak area is directly proportional to the number of cDNA molecules [45], the FLOE technique allows quantification of intensity of each signal in arbitrary units. The average area of P2 from 6 different replicates indicated that the abundance of P2 cDNA obtained with RNA from the regR mutant (Fig. 4A, lower electropherogram) was reduced by approximately 40% compared to wild type-derived cDNA (Fig. 4A, upper electropherogram). However, the area of the P1 cDNA peak did not significantly change in the regR mutant relative to the wild type (Fig. 4A). This suggests that RegR is needed for efficient synthesis of the P2 transcript under anoxic conditions. The transcriptional start site P1 is located 45 bp downstream of a FixK2 box (Fig. 5), which supports our previous notion that norC transcription depends on the FixK2 transcription factor [41]. To confirm this hypothesis, we also have performed primer extension experiments with RNA isolated from a B. japonicum fixK2 mutant. As shown in Fig. 4B (lower electropherogram), the primer extension signal corresponding to P1 was absent in cDNA derived from the fixK 2 mutant but present in cDNA obtained with wild-type RNA (Fig. 4B, upper electropherogram).
Figure 4. Transcription start site mapping of B. japonicum norC by FLOE.
Panel A corresponds to FLOE electropherograms obtained by using RNA from the wild type (upper panel) and regR mutant (bottom panel) both cultured anoxically in BSN medium. In panel B, RNA was isolated from the wild type (upper panel) and fixK 2 mutant (botton panel) both cultured anoxically in YEM medium containing 10 mM KNO3. In panel C, RNA was isolated from the wild type cultured in BSN medium under 2% initial O2. The red peaks are GeneScan-500 ROXTM internal size markers. Filled blue peaks in each panel correspond to primer extension products P1 and P2.
Figure 5. Structure of the B. japonicum norC promoter.
Nucleotides corresponding to the start site of transcripts P1 and P2 are shown in bold, marked +1 below and highlighted with an open black box and a solid black box, respectively. The putative FixK2 and RegR binding sites are indicated with an open black box and two pairs of solid grey boxes, respectively. The norC translation start codon (ATG) as annotated in the B. japonicum genome database (http://kazusa.or.jp/rhizobase) is shown in bold face letters. A potential Shine-Dalgarno sequence of norC is underlined. Forward and reverse primers for amplification of the DNA region used in EMSAs are indicated with dashed arrows. The continuous grey arrow indicates the FAM-labeled primer used in FLOE analyses. Continuous black arrows indicate the 3′ end of bsr3213 and the 5′ end of norC.
In other bacteria, RegSR orthologous two-component regulatory systems (e.g. RegBA, PrrBA, or ActSR) respond to different oxygen levels to adapt their respiration accordingly [46]. Therefore, we were interested to determine whether the P2 transcript proposed to be RegR-modulated under anoxic conditions is also synthesized under another oxygen-limiting condition. To do so, we performed primer extension experiments using RNA from cells grown in BSN medium and flushed with 2% oxygen at the beginning of the incubation. As shown in Fig. 4C, no P2 cDNA was detected when RNA was isolated from wild-type cells grown under these conditions. By contrast, P1 cDNA corresponding to the FixK2-dependent transcript was present when wild-type cells were cultured under both anoxic or 2% initial oxygen conditions (Fig. 4A, C). Similar results were obtained when we used RNA from regR mutant cells grown under the same conditions (data not shown). These results suggest that, in contrast to P1, P2 transcript is specific for anoxic conditions.
RegR Control of nor Genes Requires Anoxia and Nitrate and is RegS Independent
In order to investigate whether or not anoxic conditions are required for RegR-dependent induction of nor genes, we analyzed expression of nor genes in wild-type and regR mutant cells cultured anoxically or under 2% initial O2 in the absence or the presence of nitrate (Table 2). The presence of nitrate induced expression of a norC-lacZ transcriptional fusion in wild-type cells grown under anoxic conditions or 2% initial O2 about 14-fold and 9-fold, respectively. Interestingly, β-galactosidase activity derived from the norC-lacZ transcriptional fusion was about 38-fold lower in the regR mutant compared to wild-type levels in cells grown anoxically in BSN medium. However, regR mutation did not decrease norC expression compared to wild-type levels when cells were cultured in the same medium under 2% initial O2. By contrast, under the latter conditions levels of β-galactosidase activity were even higher in the regR mutant compared to the wild type, regardless of the presence or absence of nitrate. These results suggest that both nitrate and anoxic conditions are required for RegR-dependent induction of nor genes.
Table 2. β-Galactosidase activity derived from a norC–lacZ fusion present in B. japonicum wild type (2499), regR (2499RR) or regS (2499RS) mutant cells.
| Strain | Relevant genotype | Miller units | |||
| Anoxia | 2% O2 | ||||
| − nitrate | + nitrate | − nitrate | + nitrate | ||
| 2499 | wild type | 76.2 (6.8) | 1079 (70) | 39.8 (11.3) | 347.3 (52) |
| 2499RR | regR | 144 (41) | 28.6 (3.9) | 169.6 (25.7) | 499.5 (22.6) |
| 2499RS | regS | nd | 1266 (90) | nd | nd |
Cells were cultured under anoxic conditions or under 2% initial O2 in Bergersen minimal medium without (BS) or with nitrate (BSN). Data are means with standard error (in parenthesis) from at least three independent cultures, assayed in triplicate. nd, not determined.
Next, we studied the potential involvement of the sensor protein RegS on RegR-dependent induction of nor genes. As shown in Table 2, expression of nor genes in the regS mutant grown anoxically in BSN medium was very similar to that observed in wild-type cells, suggesting that RegR control of norC induction is independent on RegS. Likewise, only a weak influence of RegS on norC expression was observed in qRT-PCR analyses. While norC expression was 27-fold lower in the regR mutant compared to the wild-type strain, when cells were grown anoxically in BSN medium, only an about 3-fold decrease of norC expression relative to the wild type was observed in the regS mutant (data not shown).
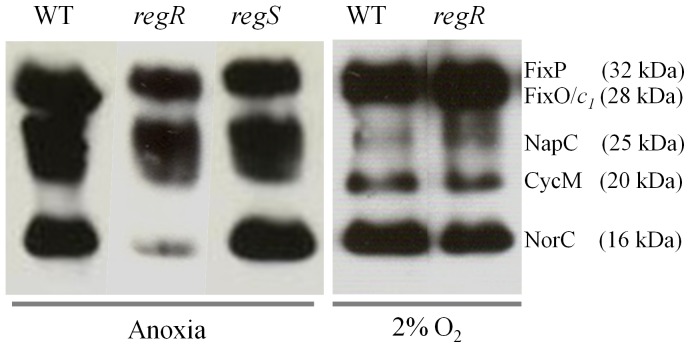
We also analyzed synthesis of NorC by heme c staining. In cells grown anoxically in BSN medium, NorC protein levels decreased in the regR mutant relative to the wild type (Fig. 6; lanes 1, 2). However, no difference of NorC expression was observed in regS mutant cells compared to wild-type cells (Fig. 6; lanes 1, 3). Furthermore, when cells were incubated under 2% initial O2 concentration in BSN medium, NorC expression did not decrease in the regR mutant (Fig. 6; lanes 4, 5).
Figure 6. Heme-stained membrane proteins from cells of B. japonicum wild type (WT; lanes 1 and 4), regR mutant strain 2426 (lanes 2 and 5), and regS mutant strain 2409 (lane 3) cultured in BSN medium under anoxic conditions or 2% initial O2.
About 20 µg membrane proteins were loaded per lane. Heme-stained c-type cytochromes identified previously are specified at the right margin along with their predicted mass. Note that all the samples were run in parallel on the same gel.
Finally, we also tested the involvement of RegR in nor expression at the level of Nor activity by measuring NO consumption of wild-type and regR mutant cells cultured anoxically in BSN medium. As expected, NO consumption by regR mutant cells was drastically decreased (54±7 nmoles NO mg protein−1 min−1) compared to wild-type cells (832±7 nmoles NO mg protein−1 min−1).
Discussion
The ability of B. japonicum to denitrify is well documented [4]. However, so far no information has been available about target genes controlled by RegR under denitrifying conditions. Our transcriptome analyses showed that expression of approximately 1,700 genes was altered in a regR mutant compared to the wild type when cells were grown anoxically in BSN medium. If only those genes are considered which showed a fold-change value of expression ≤−5 and ≥5 (343 genes; Table S3), the large majority of them (83%) are subject to positive control by RegR. This documents that this protein predominantly acts as an activator under the applied growth conditions, which extends the results obtained previously with regR mutant cells grown under oxic or microoxic free-living conditions, or as bacteroids in root nodules [25]. Notably, among this group of 343 genes we found the previously described RegR targets such as fixR, nifA, the blr1515-blr1516 operon that encodes a predicted multidrug efflux system [25], [47], and bll4130 encoding a LysR-type regulator, which validates the approach used in this work. Among the 620 genes anoxically induced whose expression differed in the regR mutant, a group of 344 genes were positively regulated (Figure 1). Based on this observation we propose a relevant role for RegR in B. japonicum denitrification. For example, we found among RegR targets structural denitrification genes (nor, nos) and genes participating in electron transport through the denitrification pathway (cycA, cy 2), and the respective microarray data was validated by qRT-PCR. In this context, the involvement of R. sphaeroides PrrBA [48] and Brucella suis RegBA [49] in nirK regulation is worth to be mentioned. Likewise, insertional inactivation of the response regulatory Agrobacterium tumefaciens actR gene significantly reduced nirK expression as well as expression of paz encoding the electron-transport protein pseudoazurin [50]. Recently, it has been reported that the redox-responsive NtrYX and PrrBA two-component systems of Brucella abortus co-ordinately regulate the expression of denitrification in response to oxygen-limited conditions [51]–[53]. Results from our work indicate that, in contrast to nor and nos genes, nirK and nap denitrification genes were not under the control of RegR, which documents that B. japonicum denitrification genes differ with regard to their dependence on RegR. Notably, disparate regulation of nap, nirK and nor genes by FixK2 was found previously during transcription profiling studies with a B. japonicum fixK 2 mutant strain grown under micro-oxic conditions [14]. Specifically, nap, nirK, and nnrR, but not nor or nos genes were among the targets of FixK2.
We have also identified and validated as RegR targets, copCAB genes encoding proteins involved in the assembly of periplasmic and secreted cuproproteins [54] which might be involved in maturation of the Cu-containing NirK or Nos enzymes. Furthermore, genes involved in NO detoxification and nitrate assimilation (blr2806-09) [44] as well as genes encoding transcriptional regulators were found to be controlled by RegR. Among the latter group was bll3466 encoding a FixK-like protein proposed to be involved in the negative feed-back of fixK 2 expression [55] and bll4130 encoding a LysR-type regulator. Finally, phyR (bll7795) and the associated ECF σ-factor gene ecfG (blr7797) which contribute to stress tolerance and symbiotic proficiency of B. japonicum [56], were also identified as targets of RegR in this work.
Electrophoretic mobility shift experiments revealed RegR binding to the promoter regions of norC, nosR, bll3466, and bll4130. RegR binding to the latter, a LysR-type regulator gene, was also previously observed by Lindemann and co-workers (2007) [25]. Interestingly, we observed several bands of different mobilities in the case of the bll4130 promoter. It might be possible that at low concentration, RegR starts binding to one of the RegR-boxes found within this promoter region, and that at higher concentration it also binds to the secondary binding site of probably lower affinity. A hierarchical binding was also described for PhoB, the response regulator of the PhoRB two-component system which activates the transcription of several genes involved in phosphate uptake and assimilation [57]. By EMSA assays, these authors demonstrated that two PhoB dimers bind to two consecutive pho boxes in a hierarchical and cooperative manner. Using the FLOE technique we identified two transcriptional start sites in the norC promoter region (P1 and P2). P1 is the previously proposed FixK2-dependent start site [41], and P2 is a start site whose abundance was modulated by RegR under our experimental denitrifying conditions. Contrary to this work, previous primer extension analyses of norC, performed by using [γ-32P]ATP and the subsequent detection of extension products in denaturing polyacrylamide gels, only revealed the presence of the FixK2-dependent start site P1 but not the RegR-dependent start site P2 [41]. This apparent discrepancy could be due to the different growth conditions and methodological approaches used by Mesa and colleagues (2002) [41] and in this work. Anoxia and nitrate are required for RegR-dependent induction of nor genes. This is in line with microarray experiments performed previously to characterize the B. japonicum RegR regulon where nor genes were not among RegR-regulated genes in cells grown in PSY medium under microoxic conditions (max. 0.5% oxygen throughout cell cultivation) [25]. Those authors identified RegR-dependent genes in either free-living oxic or microoxic and symbiotic conditions and concluded that this protein contributes to redox regulation in B. japonicum. As shown here, RegR has also a regulatory role under anoxic conditions, and the presence of nitrate or a nitrogen oxide generated from nitrate reduction was crucial for RegR control of nor genes. Analogous to our findings, the presence of nitric oxide (NO) is required for ResDE-dependent anaerobic induction of Bacillus subtillis nasDE and hmp genes which encode a nitrite reductase and a NO-detoxifying flavohemoglobin, respectively. In this bacterium, NO inactivates the NO-sensitive NsrR transcriptional repressor of nasDE and hmp [58] leading to anaerobic induction of nasDE and hmp by ResDE. B. japonicum genome lacks genes coding for an obvious NsrR homolog [59], which, however, does not exclude that in addition to RegR other regulators contribute to the control of B. japonicum nor genes under denitrifying conditions.
RegSR two-component regulatory system comprises the membrane associated RegS histidine protein kinase and its cognate RegR response regulator [39]. The regulatory mechanism has been well-studied in the orthologous RegBA system of R. capsulatus [46], [60]. In this bacterium, the membrane-localized ubiquinone pool and the redox-active cysteine (Cys265) function as redox sensors that regulate RegB kinase activity which auto-phosphorylates and transfers the phosphoryl group to the RegA response regulator [61]–[64]. In this work, we made the intriguing observation that regulation of norC differed in regS and regR mutants. While deletion of regR abolished activation of norC under anoxic conditions in BSN medium, mutation of regS resulted in wild type-like expression norC. This is in line with previously described phenotypic differences between regR and regS mutants of B. japonicum [20], regB and regA mutants of R. capsulatus [65], or roxS and roxR mutants of Pseudomonas aeruginosa [66]. It might be possible that in the B. japonicum regS mutant, RegR is phosphorylated via cross-talk by an alternative sensor kinase. In fact, the two-component regulatory system encoded by B. japonicum genes blr1154 and blr1155 shows pronounced similaritiy to RegSR (H.M. Fischer, unpublished data), and thus is a candidate for the postulated cross-talk.
Supporting Information
List of primers used for qRT-PCR experiments and EMSA assays.
(DOCX)
Anoxically induced genes (as compared to oxic conditions) whose expression differed in the Δ regR strain relative to the wild type.
(DOCX)
Differentially expressed genes by a factor of ≤−5 or ≥5 in the Δ regR strain grown anoxically and their putative operon members.
(DOCX)
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. The microarray data are available in the NCBI Gene Expression Omnibus database (GEO; http://www.ncbi.nlm.nih.gov/geo) under GEO Series accession number GSE56668.
Funding Statement
Support was provided by Fondo Europeo de Desarrollo Regional (FEDER)-cofinanced grants AGL2010-18607, AGL2011-23383 and BIO2011-22833 from Ministerio de Economía y Competitividad (Spain) (to MJD CV SM); Spanish National Network on Extremophilic Microorganisms (BIO2011-12879-E) (to CV); Junta de Andalucía to Group BIO-275 and CVI-7293 (to EJB CV); Consejo Superior de Investigaciones Cientificas I3P Programme (to MJT); Grants from ETH Zurich and Functional Genomics Centre of ETH Zurich and University of Zurich (FGCZ) (to H-MF). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Fischer HM (1994) Genetic regulation of nitrogen fixation in rhizobia. Microbiol Rev 58: 352–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dixon R, Kahn D (2004) Genetic regulation of biological nitrogen fixation. Nat Rev Microbiol 2: 621–631. [DOI] [PubMed] [Google Scholar]
- 3. Delgado MJ, Bedmar EJ, Downie JA (1998) Genes involved in the formation and assembly of rhizobial cytochromes and their role in symbiotic nitrogen fixation. Adv Microb Physiol 40: 191–231. [DOI] [PubMed] [Google Scholar]
- 4. Bedmar EJ, Robles EF, Delgado MJ (2005) The complete denitrification pathway of the symbiotic, nitrogen-fixing bacterium Bradyrhizobium japonicum . Biochem Soc Trans 33: 141–144. [DOI] [PubMed] [Google Scholar]
- 5.Delgado MJ, Casella S, Bedmar EJ (2007) Denitrification in rhizobia-legume symbiosis. In: H Bothe, S. J Ferguson and W. E Newton, editors. Biology of the Nitrogen Cycle. Amsterdam: Elservier Science. pp. 57–66.
- 6.Sanchez C, Bedmar EJ, Delgado MJ (2011) Denitrification in Legume-associated endosymbiotic Bacteria. In: J.W.B Moir, editor. Nitrogen Cycling in Bacteria. Norfolk, UK. Caister Academic Press. pp. 197–210.
- 7. Simon J, van Spanning RJ, Richardson DJ (2008) The organisation of proton motive and non-proton motive redox loops in prokaryotic respiratory systems. Biochim Biophys Acta 1777: 1480–1490. [DOI] [PubMed] [Google Scholar]
- 8. Zumft WG (1997) Cell biology and molecular basis of denitrification. Microbiol Mol Biol Rev 61: 533–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kraft B, Strous M, Tegetmeyer HE (2011) Microbial nitrate respiration-genes, enzymes and environmental distribution. J Biotechnol 155: 104–117. [DOI] [PubMed] [Google Scholar]
- 10.Richardson DJ (2011) Redox complexes of the nitrogen cycle. In: J. W. B Moir, editor. Nitrogen Cycling in Bacteria. Norkfolk, UK: Caister Academic Press. pp. 23–39.
- 11.van Spanning RJ, Richardson DJ, Ferguson SJ (2007) Introduction to the biochemistry and molecular biology of denitrification. In: Bothe H, Ferguson SJ, Newton WE, editors. Biology of the Nitrogen Cycle. Amsterdam: Elsevier Science
- 12. Sciotti MA, Chanfon A, Hennecke H, Fischer HM (2003) Disparate oxygen responsiveness of two regulatory cascades that control expression of symbiotic genes in Bradyrhizobium japonicum . J Bacteriol 185: 5639–5642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mesa S, Bedmar EJ, Chanfon A, Hennecke H, Fischer HM (2003) Bradyrhizobium japonicum NnrR, a denitrification regulator, expands the FixLJ-FixK2 regulatory cascade. J Bacteriol 185: 3978–3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mesa S, Hauser F, Friberg M, Malaguti E, Fischer HM, et al. (2008) Comprehensive assessment of the regulons controlled by the FixLJ-FixK2-FixK1 cascade in Bradyrhizobium japonicum . J Bacteriol 190: 6568–6579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nellen-Anthamatten D, Rossi P, Preisig O, Kullik I, Babst M, et al. (1998) Bradyrhizobium japonicum FixK2, a crucial distributor in the FixLJ-dependent regulatory cascade for control of genes inducible by low oxygen levels. J Bacteriol 180: 5251–5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mesa S, Ucurum Z, Hennecke H, Fischer HM (2005) Transcription activation in vitro by the Bradyrhizobium japonicum regulatory protein FixK2 . J Bacteriol 187: 3329–3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Robles EF, Sanchez C, Bonnard N, Delgado MJ, Bedmar EJ (2006) The Bradyrhizobium japonicum napEDABC genes are controlled by the FixLJ-FixK2-NnrR regulatory cascade. Biochem Soc Trans 34: 108–110. [DOI] [PubMed] [Google Scholar]
- 18. Barrios H, Fischer HM, Hennecke H, Morett E (1995) Overlapping promoters for two different RNA polymerase holoenzymes control Bradyrhizobium japonicum nifA expression. J Bacteriol 177: 1760–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Barrios H, Grande R, Olvera L, Morett E (1998) In vivo genomic footprinting analysis reveals that the complex Bradyrhizobium japonicum fixRnifA promoter region is differently occupied by two distinct RNA polymerase holoenzymes. Proc Natl Acad Sci U S A 95: 1014–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bauer E, Kaspar T, Fischer HM, Hennecke H (1998) Expression of the fixR-nifA operon in Bradyrhizobium japonicum depends on a new response regulator, RegR. J Bacteriol 180: 3853–3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kullik I, Fritsche S, Knobel H, Sanjuan J, Hennecke H, et al. (1991) Bradyrhizobium japonicum has two differentially regulated, functional homologs of the σ54 gene (rpoN). J Bacteriol 173: 1125–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hauser F, Pessi G, Friberg M, Weber C, Rusca N, et al. (2007) Dissection of the Bradyrhizobium japonicum NifA+σ54 regulon, and identification of a ferredoxin gene (fdxN) for symbiotic nitrogen fixation. Mol Genet Genomics 278: 255–271. [DOI] [PubMed] [Google Scholar]
- 23. Nienaber A, Huber A, Göttfert M, Hennecke H, Fischer HM (2000) Three new NifA-regulated genes in the Bradyrhizobium japonicum symbiotic gene region discovered by competitive DNA-RNA hybridization. J Bacteriol 182: 1472–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bueno E, Mesa S, Sanchez C, Bedmar EJ, Delgado MJ (2010) NifA is required for maximal expression of denitrification genes in Bradyrhizobium japonicum . Environ Microbiol 12: 393–400. [DOI] [PubMed] [Google Scholar]
- 25. Lindemann A, Moser A, Pessi G, Hauser F, Friberg M, et al. (2007) New target genes controlled by the Bradyrhizobium japonicum two-component regulatory system RegSR. J Bacteriol 189: 8928–8943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Torres MJ, Bueno E, Mesa S, Bedmar EJ, Delgado MJ (2011) Emerging complexity in the denitrification regulatory network of Bradyrhizobium japonicum . Biochem Soc Trans 39: 284–288. [DOI] [PubMed] [Google Scholar]
- 27. Regensburger B, Hennecke H (1983) RNA polymerase from Rhizobium japonicum . Arch Microbiol 135: 103–109. [DOI] [PubMed] [Google Scholar]
- 28.Bergersen FJ (1977) A Treatise on Dinitrogen Fixation. In: R. W Hardy and W Silver, editors. Biology: Section III. New York, USA: Wiley. pp. 519–556.
- 29. Daniel RM, Appleby CA (1972) Anaerobic-nitrate, symbiotic and aerobic growth of Rhizobium japonicum: effects on cytochrome P450, other Hemeproteins, nitrate and nitrite reductases. Biochim Biophys Acta 275: 347–354. [DOI] [PubMed] [Google Scholar]
- 30.Miller JH (1972) Experiments in Molecular Genetics. Cold Spring Harbor, NY.: Cold Spring Harbor Laboratory Press.
- 31.Simon R, Priefer U, Pühler A (1983) Vector plasmids for in vivo and in vitro manipulation of gram-negative bacteria. In: A Pühler, editor editors. Molecular genetics of the bacteria-plant interaction. Heidelberg, Germany: Springer-Verlag. pp. 98–106.
- 32. Hauser F, Lindemann A, Vuilleumier S, Patrignani A, Schlapbach R, et al. (2006) Design and validation of a partial-genome microarray for transcriptional profiling of the Bradyrhizobium japonicum symbiotic gene region. Mol Genet Genomics 275: 55–67. [DOI] [PubMed] [Google Scholar]
- 33. Pessi G, Ahrens CH, Rehrauer H, Lindemann A, Hauser F, et al. (2007) Genome-wide transcript analysis of Bradyrhizobium japonicum bacteroids in soybean root nodules. Mol Plant Microbe Interact 20: 1353–1363. [DOI] [PubMed] [Google Scholar]
- 34. Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Delgado MJ, Bonnard N, Tresierra-Ayala A, Bedmar EJ, Müller P (2003) The Bradyrhizobium japonicum napEDABC genes encoding the periplasmic nitrate reductase are essential for nitrate respiration. Microbiology 149: 3395–3403. [DOI] [PubMed] [Google Scholar]
- 36. Vargas C, McEwan AG, Downie JA (1993) Detection of c-type cytochromes using enhanced chemiluminescence. Anal Biochem 209: 323–326. [DOI] [PubMed] [Google Scholar]
- 37. Zhang X, Broderick M (2000) Amperometric detection of nitric oxide. Mod Asp Immunobiol 1: 160–165. [Google Scholar]
- 38. Babst M, Hennecke H, Fischer HM (1996) Two different mechanisms are involved in the heat-shock regulation of chaperonin gene expression in Bradyrhizobium japonicum . Mol Microbiol 19: 827–839. [DOI] [PubMed] [Google Scholar]
- 39. Emmerich R, Panglungtshang K, Strehler P, Hennecke H, Fischer HM (1999) Phosphorylation, dephosphorylation and DNA-binding of the Bradyrhizobium japonicum RegSR two-component regulatory proteins. Eur J Biochem 263: 455–463. [DOI] [PubMed] [Google Scholar]
- 40. Velasco L, Mesa S, Xu CA, Delgado MJ, Bedmar EJ (2004) Molecular characterization of nosRZDFYLX genes coding for denitrifying nitrous oxide reductase of Bradyrhizobium japonicum . Antonie Van Leeuwenhoek 85: 229–235. [DOI] [PubMed] [Google Scholar]
- 41. Mesa S, Velasco L, Manzanera ME, Delgado MJ, Bedmar EJ (2002) Characterization of the norCBQD genes, encoding nitric oxide reductase, in the nitrogen fixing bacterium Bradyrhizobium japonicum . Microbiology 148: 3553–3560. [DOI] [PubMed] [Google Scholar]
- 42. Bueno E, Bedmar EJ, Richardson DJ, Delgado MJ (2008) Role of Bradyrhizobium japonicum cytochrome c 550 in nitrite and nitrate respiration. FEMS Microbiol Lett 279: 188–194. [DOI] [PubMed] [Google Scholar]
- 43. Velasco L, Mesa S, Delgado MJ, Bedmar EJ (2001) Characterization of the nirK gene encoding the respiratory, Cu-containing nitrite reductase of Bradyrhizobium japonicum . Biochim Biophys Acta 1521: 130–134. [DOI] [PubMed] [Google Scholar]
- 44. Cabrera JJ, Sanchez C, Gates AJ, Bedmar EJ, Mesa S, et al. (2011) The nitric oxide response in plant-associated endosymbiotic bacteria. Biochem Soc Trans 39: 1880–1885. [DOI] [PubMed] [Google Scholar]
- 45. Fekete RA, Miller MJ, Chattoraj DK (2003) Fluorescently labeled oligonucleotide extension: a rapid and quantitative protocol for primer extension. Biotechniques 35: 90–94, 97–98. [DOI] [PubMed] [Google Scholar]
- 46. Bueno E, Mesa S, Bedmar EJ, Richardson DJ, Delgado MJ (2012) Bacterial adaptation of respiration from oxic to microoxic and anoxic conditions: redox control. Antioxid Redox Signal 16: 819–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lindemann A, Koch M, Pessi G, Muller AJ, Balsiger S, et al. (2010) Host-specific symbiotic requirement of BdeAB, a RegR-controlled RND-type efflux system in Bradyrhizobium japonicum . FEMS Microbiol Lett 312: 184–191. [DOI] [PubMed] [Google Scholar]
- 48. Laratta WP, Choi PS, Tosques IE, Shapleigh JP (2002) Involvement of the PrrB/PrrA two-component system in nitrite respiration in Rhodobacter sphaeroides 2.4.3: evidence for transcriptional regulation. J Bacteriol 184: 3521–3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Abdou E, Deredjian A, Jimenez de Bagues MP, Kohler S, Jubier-Maurin V (2013) RegA, the regulator of the two-component system RegB/RegA of Brucella suis, is a controller of both oxidative respiration and denitrification required for chronic infection in mice. Infect Immun 81: 2053–2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Baek SH, Hartsock A, Shapleigh JP (2008) Agrobacterium tumefaciens C58 uses ActR and FnrN to control nirK and nor expression. J Bacteriol 190: 78–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Carrica MC, Fernandez I, Sieira R, Paris G, Goldbaum FA (2013) The two-component systems PrrBA and NtrYX co-ordinately regulate the adaptation of Brucella abortus to an oxygen-limited environment. Mol Microbiol 88: 222–233. [DOI] [PubMed] [Google Scholar]
- 52. Carrica MC, Fernandez I, Marti MA, Paris G, Goldbaum FA (2012) The NtrY/X two-component system of Brucella spp. acts as a redox sensor and regulates the expression of nitrogen respiration enzymes. Mol Microbiol 85: 39–50. [DOI] [PubMed] [Google Scholar]
- 53. Roop RM Jr, Caswell CC (2012) Redox-responsive regulation of denitrification genes in Brucella . Mol Microbiol 85: 5–7. [DOI] [PubMed] [Google Scholar]
- 54. Hernandez-Montes G, Arguello JM, Valderrama B (2012) Evolution and diversity of periplasmic proteins involved in copper homeostasis in gamma proteobacteria. BMC Microbiol 12: 249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Reutimann L, Mesa S, Hennecke H (2010) Autoregulation of fixK2 gene expression in Bradyrhizobium japonicum . Mol Genet Genomics 284: 25–32. [DOI] [PubMed] [Google Scholar]
- 56. Gourion B, Sulser S, Frunzke J, Francez-Charlot A, Stiefel P, et al. (2009) The PhyR-σEcfG signalling cascade is involved in stress response and symbiotic efficiency in Bradyrhizobium japonicum . Mol Microbiol 73: 291–305. [DOI] [PubMed] [Google Scholar]
- 57. Blanco AG, Canals A, Coll M (2012) PhoB transcriptional activator binds hierarchically to pho box promoters. Biol Chem 393: 1165–1171. [DOI] [PubMed] [Google Scholar]
- 58. Kommineni S, Yukl E, Hayashi T, Delepine J, Geng H, et al. (2010) Nitric oxide-sensitive and -insensitive interaction of Bacillus subtilis NsrR with a ResDE-controlled promoter. Mol Microbiol 78: 1280–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Rodionov DA, Dubchak IL, Arkin AP, Alm EJ, Gelfand MS (2005) Dissimilatory metabolism of nitrogen oxides in bacteria: comparative reconstruction of transcriptional networks. PLoS Comput Biol 1: e55 doi:10.1371/journal.pcbi.0010055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wu J, Bauer CE (2008) RegB/RegA, a global redox-responding two-component system. Adv Exp Med Biol 631: 131–148. [DOI] [PubMed] [Google Scholar]
- 61. Swem LR, Gong X, Yu CA, Bauer CE (2006) Identification of a ubiquinone-binding site that affects autophosphorylation of the sensor kinase RegB. J Biol Chem 281: 6768–6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Swem LR, Kraft BJ, Swem DL, Setterdahl AT, Masuda S, et al. (2003) Signal transduction by the global regulator RegB is mediated by a redox-active cysteine. EMBO J 22: 4699–4708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wu J, Bauer CE (2010) RegB kinase activity is controlled in part by monitoring the ratio of oxidized to reduced ubiquinones in the ubiquinone pool. MBio 1 (5) pii: e00272-10 doi:10.1128/mBio.00272-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wu J, Cheng Z, Reddie K, Carroll K, Hammad LA, et al. (2013) RegB kinase activity is repressed by oxidative formation of cysteine sulfenic acid. J Biol Chem 288: 4755–4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Mosley CS, Suzuki JY, Bauer CE (1994) Identification and molecular genetic characterization of a sensor kinase responsible for coordinately regulating light harvesting and reaction center gene expression in response to anaerobiosis. J Bacteriol 176: 7566–7573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Comolli JC, Donohue TJ (2002) Pseudomonas aeruginosa RoxR, a response regulator related to Rhodobacter sphaeroides PrrA, activates expression of the cyanide-insensitive terminal oxidase. Mol Microbiol 45: 755–768. [DOI] [PubMed] [Google Scholar]
- 67. Kaneko T, Nakamura Y, Sato S, Minamisawa K, Uchiumi T, et al. (2002) Complete genomic sequence of nitrogen-fixing symbiotic bacterium Bradyrhizobium japonicum USDA110. DNA Res 9: 189–197. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of primers used for qRT-PCR experiments and EMSA assays.
(DOCX)
Anoxically induced genes (as compared to oxic conditions) whose expression differed in the Δ regR strain relative to the wild type.
(DOCX)
Differentially expressed genes by a factor of ≤−5 or ≥5 in the Δ regR strain grown anoxically and their putative operon members.
(DOCX)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. The microarray data are available in the NCBI Gene Expression Omnibus database (GEO; http://www.ncbi.nlm.nih.gov/geo) under GEO Series accession number GSE56668.