Abstract
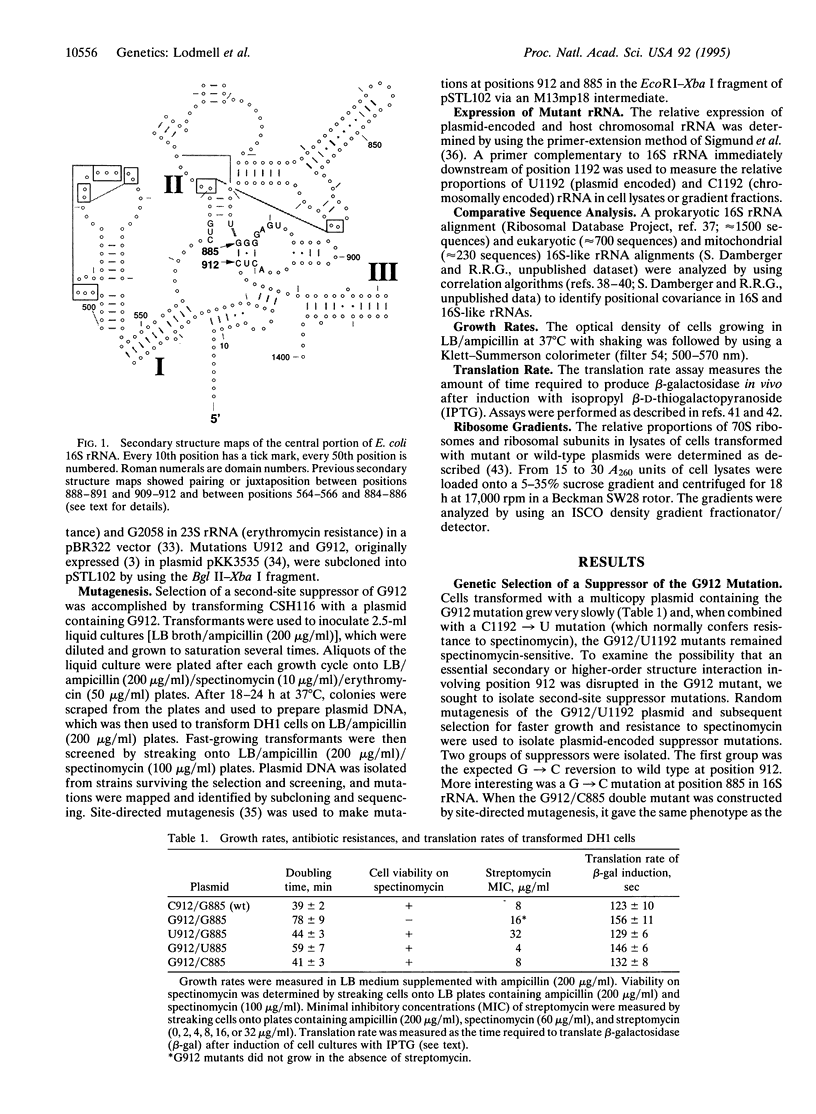
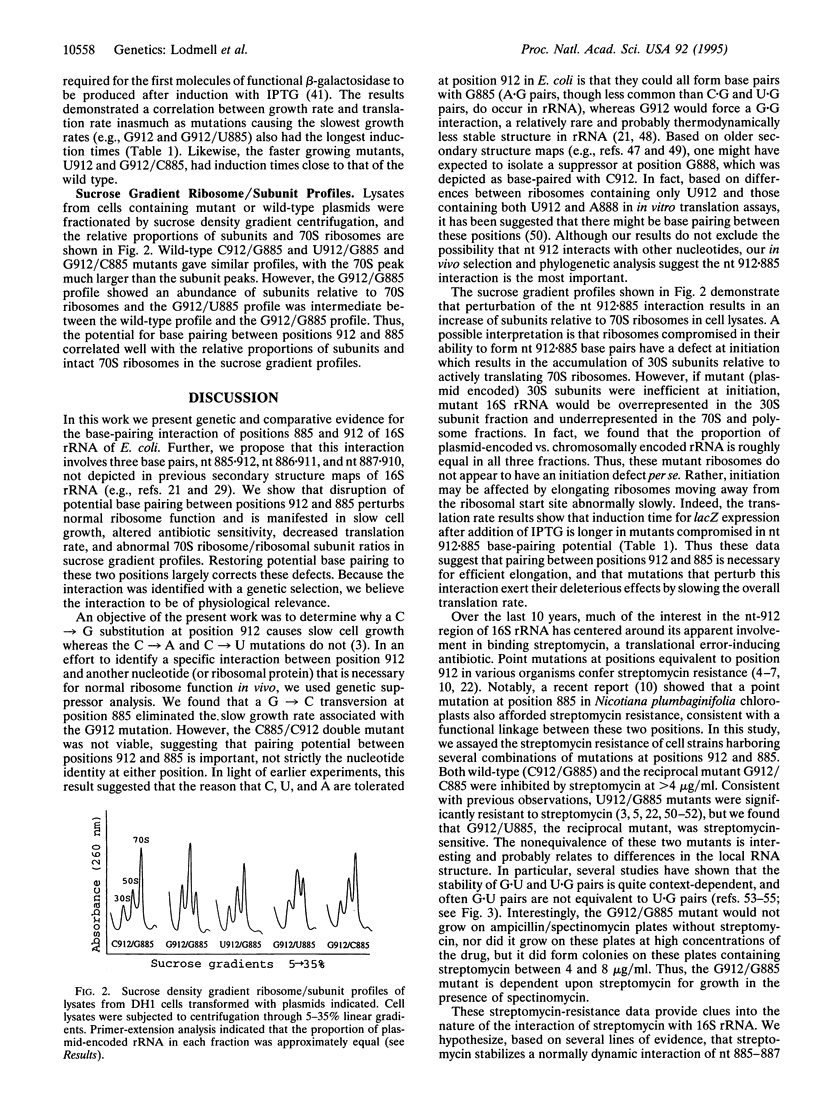
Mutations at position 912 of Escherichia coli 16S rRNA result in two notable phenotypes. The C-->U transition confers resistance to streptomycin, a translational-error-inducing antibiotic, while a C-->G transversion causes marked retardation of cell growth rate. Starting with the slow-growing G912 mutant, random mutagenesis was used to isolate a second site mutation that restored growth nearly to the wild-type rate. The second site mutation was identified as a G-->C transversion at position 885 in 16S rRNA. Cells containing the G912 mutation had an increased doubling time, abnormal sucrose gradient ribosome/subunit profile, increased sensitivity to spectinomycin, dependence upon streptomycin for growth in the presence of spectinomycin, and slower translation rate, whereas cells with the G912/C885 double mutation were similar to wild type in these assays. Comparative analysis showed there was significant covariation between positions 912 and 885. Thus the second-site suppressor analysis, the functional assays, and the comparative data suggest that the interaction between nt 912 and nt 885 is conserved and necessary for normal ribosome function. Furthermore, the comparative data suggest that the interaction extends to include G885-G886-G887 pairing with C912-U911-C910. An alternative secondary structure element for the central domain of 16S rRNA is proposed.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abad J. P., Amils R. Location of the streptomycin ribosomal binding site explains its pleiotropic effects on protein biosynthesis. J Mol Biol. 1994 Jan 28;235(4):1251–1260. doi: 10.1006/jmbi.1994.1078. [DOI] [PubMed] [Google Scholar]
- Allen P. N., Noller H. F. Mutations in ribosomal proteins S4 and S12 influence the higher order structure of 16 S ribosomal RNA. J Mol Biol. 1989 Aug 5;208(3):457–468. doi: 10.1016/0022-2836(89)90509-3. [DOI] [PubMed] [Google Scholar]
- Andersson D. I., Bohman K., Isaksson L. A., Kurland C. G. Translation rates and misreading characteristics of rpsD mutants in Escherichia coli. Mol Gen Genet. 1982;187(3):467–472. doi: 10.1007/BF00332630. [DOI] [PubMed] [Google Scholar]
- Bonny C., Montandon P. E., Marc-Martin S., Stutz E. Analysis of streptomycin-resistance of Escherichia coli mutants. Biochim Biophys Acta. 1991 Jun 13;1089(2):213–219. doi: 10.1016/0167-4781(91)90010-j. [DOI] [PubMed] [Google Scholar]
- Brimacombe R. RNA-protein interactions in the Escherichia coli ribosome. Biochimie. 1991 Jul-Aug;73(7-8):927–936. doi: 10.1016/0300-9084(91)90134-m. [DOI] [PubMed] [Google Scholar]
- Brink M. F., Verbeet M. P., de Boer H. A. Formation of the central pseudoknot in 16S rRNA is essential for initiation of translation. EMBO J. 1993 Oct;12(10):3987–3996. doi: 10.1002/j.1460-2075.1993.tb06076.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosius J., Ullrich A., Raker M. A., Gray A., Dull T. J., Gutell R. R., Noller H. F. Construction and fine mapping of recombinant plasmids containing the rrnB ribosomal RNA operon of E. coli. Plasmid. 1981 Jul;6(1):112–118. doi: 10.1016/0147-619x(81)90058-5. [DOI] [PubMed] [Google Scholar]
- Chernoff Y. O., Vincent A., Liebman S. W. Mutations in eukaryotic 18S ribosomal RNA affect translational fidelity and resistance to aminoglycoside antibiotics. EMBO J. 1994 Feb 15;13(4):906–913. doi: 10.1002/j.1460-2075.1994.tb06334.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frattali A. L., Flynn M. K., De Stasio E. A., Dahlberg A. E. Effects of mutagenesis of C912 in the streptomycin binding region of Escherichia coli 16S ribosomal RNA. Biochim Biophys Acta. 1990 Aug 27;1050(1-3):27–33. doi: 10.1016/0167-4781(90)90136-p. [DOI] [PubMed] [Google Scholar]
- Freier S. M., Kierzek R., Caruthers M. H., Neilson T., Turner D. H. Free energy contributions of G.U and other terminal mismatches to helix stability. Biochemistry. 1986 Jun 3;25(11):3209–3213. doi: 10.1021/bi00359a019. [DOI] [PubMed] [Google Scholar]
- Gautheret D., Damberger S. H., Gutell R. R. Identification of base-triples in RNA using comparative sequence analysis. J Mol Biol. 1995 Apr 21;248(1):27–43. doi: 10.1006/jmbi.1995.0200. [DOI] [PubMed] [Google Scholar]
- Gravel M., Melançon P., Brakier-Gingras L. Cross-linking of streptomycin to the 16S ribosomal RNA of Escherichia coli. Biochemistry. 1987 Sep 22;26(19):6227–6232. doi: 10.1021/bi00393a041. [DOI] [PubMed] [Google Scholar]
- Gutell R. R. Collection of small subunit (16S- and 16S-like) ribosomal RNA structures: 1994. Nucleic Acids Res. 1994 Sep;22(17):3502–3507. doi: 10.1093/nar/22.17.3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutell R. R., Larsen N., Woese C. R. Lessons from an evolving rRNA: 16S and 23S rRNA structures from a comparative perspective. Microbiol Rev. 1994 Mar;58(1):10–26. doi: 10.1128/mr.58.1.10-26.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutell R. R., Power A., Hertz G. Z., Putz E. J., Stormo G. D. Identifying constraints on the higher-order structure of RNA: continued development and application of comparative sequence analysis methods. Nucleic Acids Res. 1992 Nov 11;20(21):5785–5795. doi: 10.1093/nar/20.21.5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Harris E. H., Burkhart B. D., Gillham N. W., Boynton J. E. Antibiotic resistance mutations in the chloroplast 16S and 23S rRNA genes of Chlamydomonas reinhardtii: correlation of genetic and physical maps of the chloroplast genome. Genetics. 1989 Oct;123(2):281–292. doi: 10.1093/genetics/123.2.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L., Kierzek R., SantaLucia J., Jr, Walter A. E., Turner D. H. Nearest-neighbor parameters for G.U mismatches: [formula; see text] is destabilizing in the contexts [formula; see text] and [formula; see text] but stabilizing in [formula; see text]. Biochemistry. 1991 Nov 19;30(46):11124–11132. doi: 10.1021/bi00110a015. [DOI] [PubMed] [Google Scholar]
- Honoré N., Cole S. T. Streptomycin resistance in mycobacteria. Antimicrob Agents Chemother. 1994 Feb;38(2):238–242. doi: 10.1128/aac.38.2.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclerc D., Melançon P., Brakier-Gingras L. Mutations in the 915 region of Escherichia coli 16S ribosomal RNA reduce the binding of streptomycin to the ribosome. Nucleic Acids Res. 1991 Jul 25;19(14):3973–3977. doi: 10.1093/nar/19.14.3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclerc D., Melançon P., Brakier-Gingras L. The interaction between streptomycin and ribosomal RNA. Biochimie. 1991 Dec;73(12):1431–1438. doi: 10.1016/0300-9084(91)90175-z. [DOI] [PubMed] [Google Scholar]
- Maidak B. L., Larsen N., McCaughey M. J., Overbeek R., Olsen G. J., Fogel K., Blandy J., Woese C. R. The Ribosomal Database Project. Nucleic Acids Res. 1994 Sep;22(17):3485–3487. doi: 10.1093/nar/22.17.3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier A., Kirschner P., Bange F. C., Vogel U., Böttger E. C. Genetic alterations in streptomycin-resistant Mycobacterium tuberculosis: mapping of mutations conferring resistance. Antimicrob Agents Chemother. 1994 Feb;38(2):228–233. doi: 10.1128/aac.38.2.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D., Noller H. F. Interaction of antibiotics with functional sites in 16S ribosomal RNA. Nature. 1987 Jun 4;327(6121):389–394. doi: 10.1038/327389a0. [DOI] [PubMed] [Google Scholar]
- Moazed D., Noller H. F. Transfer RNA shields specific nucleotides in 16S ribosomal RNA from attack by chemical probes. Cell. 1986 Dec 26;47(6):985–994. doi: 10.1016/0092-8674(86)90813-5. [DOI] [PubMed] [Google Scholar]
- Montandon P. E., Nicolas P., Schürmann P., Stutz E. Streptomycin-resistance of Euglena gracilis chloroplasts: identification of a point mutation in the 16S rRNA gene in an invariant position. Nucleic Acids Res. 1985 Jun 25;13(12):4299–4310. doi: 10.1093/nar/13.12.4299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montandon P. E., Wagner R., Stutz E. E. coli ribosomes with a C912 to U base change in the 16S rRNA are streptomycin resistant. EMBO J. 1986 Dec 20;5(13):3705–3708. doi: 10.1002/j.1460-2075.1986.tb04703.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor M., Göringer H. U., Dahlberg A. E. A ribosomal ambiguity mutation in the 530 loop of E. coli 16S rRNA. Nucleic Acids Res. 1992 Aug 25;20(16):4221–4227. doi: 10.1093/nar/20.16.4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osswald M., Greuer B., Brimacombe R., Stöffler G., Bäumert H., Fasold H. RNA-protein cross-linking in Escherichia coli 30S ribosomal subunits; determination of sites on 16S RNA that are cross-linked to proteins S3, S4, S5, S7, S8, S9, S11, S13, S19 and S21 by treatment with methyl p-azidophenyl acetimidate. Nucleic Acids Res. 1987 Apr 24;15(8):3221–3240. doi: 10.1093/nar/15.8.3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinard R., Côté M., Payant C., Brakier-Gingras L. Positions 13 and 914 in Escherichia coli 16S ribosomal RNA are involved in the control of translational accuracy. Nucleic Acids Res. 1994 Feb 25;22(4):619–624. doi: 10.1093/nar/22.4.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinard R., Payant C., Melançon P., Brakier-Gingras L. The 5' proximal helix of 16S rRNA is involved in the binding of streptomycin to the ribosome. FASEB J. 1993 Jan;7(1):173–176. doi: 10.1096/fasebj.7.1.7678560. [DOI] [PubMed] [Google Scholar]
- Powers T., Noller H. F. A functional pseudoknot in 16S ribosomal RNA. EMBO J. 1991 Aug;10(8):2203–2214. doi: 10.1002/j.1460-2075.1991.tb07756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuermann R., Tam S., Burgers P. M., Lu C., Echols H. Identification of the epsilon-subunit of Escherichia coli DNA polymerase III holoenzyme as the dnaQ gene product: a fidelity subunit for DNA replication. Proc Natl Acad Sci U S A. 1983 Dec;80(23):7085–7089. doi: 10.1073/pnas.80.23.7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra M. J., Axenson T. J., Turner D. H. A model for the stabilities of RNA hairpins based on a study of the sequence dependence of stability for hairpins of six nucleotides. Biochemistry. 1994 Nov 29;33(47):14289–14296. doi: 10.1021/bi00251a042. [DOI] [PubMed] [Google Scholar]
- Sigmund C. D., Ettayebi M., Borden A., Morgan E. A. Antibiotic resistance mutations in ribosomal RNA genes of Escherichia coli. Methods Enzymol. 1988;164:673–690. doi: 10.1016/s0076-6879(88)64077-8. [DOI] [PubMed] [Google Scholar]
- Stern S., Powers T., Changchien L. M., Noller H. F. RNA-protein interactions in 30S ribosomal subunits: folding and function of 16S rRNA. Science. 1989 May 19;244(4906):783–790. doi: 10.1126/science.2658053. [DOI] [PubMed] [Google Scholar]
- Tapprich W. E., Dahlberg A. E. A single base mutation at position 2661 in E. coli 23S ribosomal RNA affects the binding of ternary complex to the ribosome. EMBO J. 1990 Aug;9(8):2649–2655. doi: 10.1002/j.1460-2075.1990.tb07447.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triman K., Becker E., Dammel C., Katz J., Mori H., Douthwaite S., Yapijakis C., Yoast S., Noller H. F. Isolation of temperature-sensitive mutants of 16 S rRNA in Escherichia coli. J Mol Biol. 1989 Oct 20;209(4):645–653. doi: 10.1016/0022-2836(89)92000-7. [DOI] [PubMed] [Google Scholar]
- Vila A., Viril-Farley J., Tapprich W. E. Pseudoknot in the central domain of small subunit ribosomal RNA is essential for translation. Proc Natl Acad Sci U S A. 1994 Nov 8;91(23):11148–11152. doi: 10.1073/pnas.91.23.11148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R., Fox G. E., Zablen L., Uchida T., Bonen L., Pechman K., Lewis B. J., Stahl D. Conservation of primary structure in 16S ribosomal RNA. Nature. 1975 Mar 6;254(5495):83–86. doi: 10.1038/254083a0. [DOI] [PubMed] [Google Scholar]
- Woese C. R., Gutell R., Gupta R., Noller H. F. Detailed analysis of the higher-order structure of 16S-like ribosomal ribonucleic acids. Microbiol Rev. 1983 Dec;47(4):621–669. doi: 10.1128/mr.47.4.621-669.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R., Magrum L. J., Gupta R., Siegel R. B., Stahl D. A., Kop J., Crawford N., Brosius J., Gutell R., Hogan J. J. Secondary structure model for bacterial 16S ribosomal RNA: phylogenetic, enzymatic and chemical evidence. Nucleic Acids Res. 1980 May 24;8(10):2275–2293. doi: 10.1093/nar/8.10.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh K. C., To K. Y., Sun S. W., Wu M. C., Lin T. Y., Chen C. C. Point mutations in the chloroplast 16s rRNA gene confer streptomycin resistance in Nicotiana plumbaginifolia. Curr Genet. 1994 Aug;26(2):132–135. doi: 10.1007/BF00313800. [DOI] [PubMed] [Google Scholar]



