Expansion of GGGGCC repeats in C9orf72 causes familial amyotrophic lateral sclerosis (ALS) and frontotemporal dementia, but the underlying mechanism is unclear. Using RNA pulldown and immunohistochemistry in ALS biosamples, Cooper-Knock et al. identify proteins that bind to the repeat expansions. Disrupted RNA splicing and/or nuclear export may underlie C9orf72-ALS pathogenesis.
Keywords: amyotrophic lateral sclerosis, pathology, genetics, fluorescence imaging
Abstract
GGGGCC repeat expansions of C9orf72 represent the most common genetic variant of amyotrophic lateral sclerosis and frontotemporal degeneration, but the mechanism of pathogenesis is unclear. Recent reports have suggested that the transcribed repeat might form toxic RNA foci that sequester various RNA processing proteins. Consensus as to the identity of the binding partners is missing and whole neuronal proteome investigation is needed. Using RNA fluorescence in situ hybridization we first identified nuclear and cytoplasmic RNA foci in peripheral and central nervous system biosamples from patients with amyotrophic lateral sclerosis with a repeat expansion of C9orf72 (C9orf72+), but not from those patients without a repeat expansion of C9orf72 (C9orf72−) or control subjects. Moreover, in the cases examined, the distribution of foci-positive neurons correlated with the clinical phenotype (t-test P < 0.05). As expected, RNA foci are ablated by RNase treatment. Interestingly, we identified foci in fibroblasts from an asymptomatic C9orf72+ carrier. We next performed pulldown assays, with GGGGCC5, in conjunction with mass spectrometry analysis, to identify candidate binding partners of the GGGGCC repeat expansion. Proteins containing RNA recognition motifs and involved in splicing, messenger RNA nuclear export and/or translation were significantly enriched. Immunohistochemistry in central nervous system tissue from C9orf72+ patients with amyotrophic lateral sclerosis demonstrated co-localization of RNA foci with SRSF2, hnRNP H1/F, ALYREF and hnRNP A1 in cerebellar granule cells and with SRSF2, hnRNP H1/F and ALYREF in motor neurons, the primary target of pathology in amyotrophic lateral sclerosis. Direct binding of proteins to GGGGCC repeat RNA was confirmed in vitro by ultraviolet-crosslinking assays. Co-localization was only detected in a small proportion of RNA foci, suggesting dynamic sequestration rather than irreversible binding. Additional immunohistochemistry demonstrated that neurons with and without RNA foci were equally likely to show nuclear depletion of TDP-43 (χ2 P = 0.75) or poly-GA dipeptide repeat protein inclusions (χ2 P = 0.46). Our findings suggest two non-exclusive pathogenic mechanisms: (i) functional depletion of RNA-processing proteins resulting in disruption of messenger RNA splicing; and (ii) licensing of expanded C9orf72 pre-messenger RNA for nuclear export by inappropriate association with messenger RNA export adaptor protein(s) leading to cytoplasmic repeat associated non-ATG translation and formation of potentially toxic dipeptide repeat protein.
Introduction
Expanded GGGGCC repeats in intron 1 of the C9orf72 gene represent the most common cause of familial amyotrophic lateral sclerosis (ALS) and familial frontotemporal degeneration (DeJesus-Hernandez et al., 2011; Renton et al., 2011), though how this genetic change results in neuronal injury is not yet understood. Three potential mechanisms have been proposed: (i) haploinsufficiency through disrupted expression of the expanded allele (DeJesus-Hernandez et al., 2011); (ii) RNA mediated gain-of-function toxicity by the transcribed expanded intronic sequence; and (iii) protein mediated gain-of-function toxicity by dipeptide repeat protein aberrantly translated from the repeat sequence by repeat associated non-ATG translation (Ash et al., 2013; Mori et al., 2013b). Evidence for haploinsufficiency is mixed; several groups have reported reduced expression of the C9orf72 messenger RNA, but this finding is not consistent (Sareen et al., 2013). Furthermore no additional loss of function mutations have been found in the C9orf72 gene (Harms et al., 2013) and we and others have shown that smaller repeat lengths, which are considered pathogenic (Byrne et al., 2013; Gomez-Tortosa et al., 2013), do not reduce transcription (Cooper-Knock et al., 2013; Xi et al., 2013). More evidence is being gathered for a gain-of-function toxicity mediated either by RNA foci formed from the expanded intron or through repeat associated non-ATG translation.
Recently, a number of studies reported that molecular phenotypes correlated with the presence of RNA foci (Donnelly et al., 2013; Lagier-Tourenne et al., 2013; Lee et al., 2013; Mizielinska et al., 2013; Sareen et al., 2013). Two of these studies corrected the observed phenotype by targeted degradation of the foci using antisense oligonucleotides (Donnelly et al., 2013; Sareen et al., 2013). One study suggested that foci burden in the frontal cortex positively correlated with disease severity in eight patients with C9orf72 frontotemporal degeneration (Mizielinska et al., 2013). Two of these reports identified co-localization of RNA foci with various proteins (Donnelly et al., 2013; Sareen et al., 2013) and suggested that pathogenic sequestration might be occurring. A similar process has been observed in myotonic dystrophy type 1, another neuromuscular disease caused by an intronic expansion (Jiang et al., 2004). Previously two groups generated candidate binding partners of the GGGGCC repeat expansion, but did not include co-localization studies with RNA foci (Mori et al., 2013a; Xu et al., 2013). Further work to characterize protein binding partners of the RNA foci is required, particularly because many of the studies thus far are in disagreement as to the most important interactions.
Observations regarding toxicity of repeat associated non-ATG translation are still at an early stage: the produced dipeptide repeat protein appears to be toxic in a cell model (Zu et al., 2013), but levels of the aberrantly translated protein observed do not correlate with neurodegeneration in autopsy material (Mackenzie et al., 2013). An important question remains over the mechanism by which the transcribed repeat sequence is exported to the cytoplasm to allow repeat associated non-ATG translation. Clearly, normal control of messenger RNA nuclear export would be expected to inhibit this movement. However, several studies report cytoplasmic RNA foci in CNS tissue (Donnelly et al., 2013, Mizielinska et al., 2013).
We have used fluorescence in situ hybridization (FISH) to examine the abundance and location of RNA foci in cerebellum, where p62-positive protein inclusion pathology is characteristic of C9orf72+ disease (Cooper-Knock et al., 2012), and in motor neurons of the ventral horn. We also examined the relationship between RNA foci and characteristic neuropathology of C9orf72+ ALS: first, the loss of nuclear TDP-43 in motor neurons, which is the pathological hallmark of ALS (Neumann et al., 2006) and has been shown to correlate with neuronal loss (Brettschneider et al., 2013); and second, the presence of cytoplasmic aggregates containing dipeptide repeat protein, which are a hallmark of C9orf72+ disease (Ash et al., 2013; Mackenzie et al., 2013; Mori et al., 2013b). We have then identified protein binding partners of the RNA repeat expansion, initially in an in vitro RNA pulldown assay using both cerebellum and neuronal cell-line extracts, and then subsequently in CNS tissue from C9orf72+ patients with ALS by immunohistochemistry. Protein–RNA UV-crosslinking confirmed in vitro direct interactions with the repeat sequence. We add novel insights to this growing field and in particular, our focus on motor neurons from the ventral horn of the spinal cord has allowed us to characterize RNA foci and their interactions in the neuronal population most vulnerable to neurodegeneration in ALS.
It should be noted that other groups have observed RNA foci transcribed from the repeat sequence in an antisense direction consisting of a GGCCCC repeat (Gendron et al., 2013; Lagier-Tourenne et al., 2013; Mizielinska et al., 2013); antisense foci were not examined in this study.
Materials and methods
Human samples
The study was approved by the South Sheffield Research Ethics Committee and informed consent was obtained for all samples. Brain and spinal cord tissues were donated to the Sheffield Brain Tissue Bank for research with the consent of the next of kin. Immunohistochemistry and RNA FISH were performed on formalin fixed paraffin-embedded tissues from up to five C9orf72+ ALS cases, three C9orf72− ALS cases and three neurologically normal controls. Lymphoblastoid cells and fibroblasts from three C9orf72+ ALS cases, one C9orf72+ asymptomatic carrier, three C9orf72− ALS cases and three controls were used for RNA FISH. Lymphoblastoid cell lines were obtained from the Wellcome Trust/Motor Neurone Disease Association ALS/MND UK DNA and Lymphoblastoid cell line Bank. Fibroblasts were obtained from the Sheffield MND Biosamples Bank.
RNA fluorescence in situ hybridization
A 5’ TYE-563-labelled LNA (16-mer fluorescent)-incorporated DNA probe was used against the sense RNA hexanucleotide repeat (Exiqon, Inc., batch number 607323). Slides with tissue, lymphoblastoid cells or fibroblasts were fixed in 4% paraformaldehyde for 10 min. Before use, formalin fixed paraffin-embedded tissue sections were deparaffinized. Slides were blocked with hybridization solution [50% formamide, 2× saline sodium citrate (SSC), 100 mg/ml dextran sulphate, 50 mM sodium phosphate pH 7.0] for 3 h at 66°C and then incubated with 400 ng/ml of denatured probe in hybridization solution overnight at 66°C. After hybridization, slides were washed once in 2×SSC/0.1% Tween-20 at room temperature and three times in 0.1× SSC at 65°C. Slides were mounted with mounting medium containing DAPI (Vector Labs, Inc.). All solutions were made with DEPC-treated water.
Visualization of RNA foci
Primary visualization of foci was performed using a Leica SP5 confocal microscope system with a ×63/1.4 oil immersion objective lens. The presence of foci was assessed within a high resolution (1433 µm2 per image, 511 × 511 pixels) z-stack made up of images at 0.13-µm intervals through the entire nuclear volume of the cell under consideration.
Biotinylated RNA pulldown assays
Total extracts were prepared by homogenizing and lysing cells/tissue in RNA-pulldown (RPD) lysis buffer [25 mM Tris pH 7.4, 100 mM NaCl, 1 mM DTT, 10% (v/v) glycerol, 0.5% (v/v) Triton™ X-100]. Lysates were cleared by centrifugation and supernatants taken for experiments. Nuclear extracts from SH-SY5Y cells were prepared using the Dignam method (Dignam et al., 1983). We chose to use two methods of lysis because cell lysis has been shown to influence the composition of ribonucleoprotein complexes (Mili and Steitz, 2004).
AAAAUU5 and GGGGCC5 RNA molecules with 3’ biotin modifications were used to identify protein binding partners in pulldown assays. 60 μl aliquots of streptavidin sepharose (GE Healthcare) were blocked overnight on a spinning wheel at 4°C with RPD lysis buffer containing 2% bovine serum albumin. Total extracts were lysed in RPD lysis buffer whereas cerebellum homogenates and SH-SY5Y whole cell or nuclear extracts were mixed 1:1 with RPD lysis buffer (2×) supplemented with protease and RNase inhibitors. 1-2 mg of the appropriate total cellular or nuclear lysate was mixed with 15 µg biotin-labelled RNA, incubated at room temperature for 30 min and then on ice for 30 min. Mixtures were then transferred to a 6-cm petri dish and UV irradiated on ice at 0.3 J/cm2 in a UV crosslinker (Fisher). Mixtures were then applied to blocked streptavidin sepharose and incubated at 4°C for 2 h with agitation. Following binding, beads were washed three times with RPD lysis buffer and then twice with RPD wash buffer (25 mM Tris pH 7.4, 100 mM NaCl, 1 mM DTT). Complexes were eluted by addition of RPD elution buffer (25 mM Tris pH 7.4, 25 mM NaCl, 1 mM EDTA) and 10 µg RNase A followed by agitation at room temperature for 30 min. Eluates were analysed by SDS-PAGE and proteins identified by mass spectrometry or western immunoblotting.
Mass spectrometry
In solution tryptic digestions were performed on the eluted fractions by the addition of 100 mM final concentration ammonium bicarbonate and 0.1% ProteaseMAX™ surfactant. Trypsin was added to a mass ratio of (1:50) and incubated at 37°C overnight. Digestions were stopped with the addition of 1–2 µl glacial acetic acid and subsequently dried under vacuum. Tryptic digests were resuspended in 0.1% final concentration of trifluoroacetic acid. Five microlitres was used for liquid chromatography–mass spectrometry/mass spectrometry (LC–MS/MS) analysis. Peptides were separated using an UltiMate™ 3000 RSLC nano liquid chromatography system (Dionex), using a 150 mm × 75 µm I.D. PepMap™ reversed phase column (Dionex). Linear gradient elution was performed from 95% buffer A (0.1% formic acid) to 50 % buffer B (0.1% formic acid, 95 % acetonitrile) at a flow rate of 300 nl/min in 60 mins. MS/MS analysis was performed using a maXis UHR TOF mass spectrometer (Bruker Daltonics) using an automated acquisition approach. MS and MS/MS scans (m/z 50–2000) were acquired in positive ion mode. Lock mass calibration was performed using HP 1221.990364. Line spectra data were then processed into peak list by data analysis using the following settings. The sum peak finder algorithm was used for peak detection using a signal to noise ratio of 10, a relative to base peak intensity of 0.1% and an absolute intensity threshold of 100. Spectra were deconvoluted and the peak lists exported as Mascot Generic Files (MGF) and searched using Mascot 2.2 server (Matrix Science). The Swiss-Prot database (Swiss-Prot Release 10.5m5, 20 April 2010, 516604 sequences) was searched using the following parameters (analysis peptide tolerance = ±0.01 Da, MS/MS tolerance = ±0.01 Da and peptide charge 2+ and 3+). Search parameters were as follows: enzyme; trypsin; fixed modifications: carbamidomethyl (C); variable modifications: deamidation (NQ), oxidation (M); maximum missed cleavages: 1. Deamidation (NQ) were chosen as variable modifications. Additionally, we also used a peptide MOWSE score of <25 as a cut-off as calculated by Mascot. The false discovery rate was estimated to be 1% for peptide IDs after searching reverse databases. Protein identifications were based on a minimum of two unique peptides.
RNA-binding ultraviolet crosslinking assays
RNA-binding assays were carried out as described previously (Hautbergue et al., 2008, 2009). GGGGCC5 RNA was 5’ end labelled with γ32P-ATP using T4 polynucleotide kinase (Fermentas). Reaction mixes were made up in RNA binding buffer [15 mM HEPES pH 7.5, 150 mM NaCl, 5 mM MgCl2, 10% (v/v) glycerol, 0.05% (v/v) Tween-20] with 50 ng radiolabelled RNA and 5 µg purified recombinant protein. Mixes were incubated for 20 min at room temperature and 20 min on ice before being UV-irradiated on ice at full power. Complexes were analysed by SDS-PAGE and stained with Coomassie blue before being vacuum-dried and exposed on a phosphoimage screen.
Immunohistochemistry
The following antibodies were used for immunohistochemistry: anti-TDP-43 (Proteintech 10782-2-AP) anti-FUS (Novus NB100-2599), anti-hnRNP H1/F (Abcam ab10689), anti-hnRNP A1 (Abcam ab5832, 9H10 clone), anti-hnRNP D (Proteintech 12770-1-AP), anti-SRSF1 (phosphor, Abcam ab11826), anti-SRSF2 (Abcam ab30817), anti-ALYREF (Sigma, clone 11G5) and anti-hnRNP C1/C2 (Abcam ab10294). Poly-(Gly-Ala) dipeptide repeat protein was detected using anti-GA antibodies (mouse, clone 5F2) as previously described (Mackenzie et al., 2013). Antigen retrieval was performed by 10–30-min microwave in EDTA at pH 8.0 for all antibodies except anti-SRSF1, anti-ALYREF and anti-TDP-43 where antigen retrieval involved microwave 10–20 min in trisodium citrate at pH 6.5, and for anti-hnRNP H/F where no specific antigen retrieval was performed. After incubation with the primary antibodies, slides were washed in PBS and incubated in species specific Alexa Fluor® 488-conjugated secondary antibodies.
Results
RNA fluorescence in situ hybridization
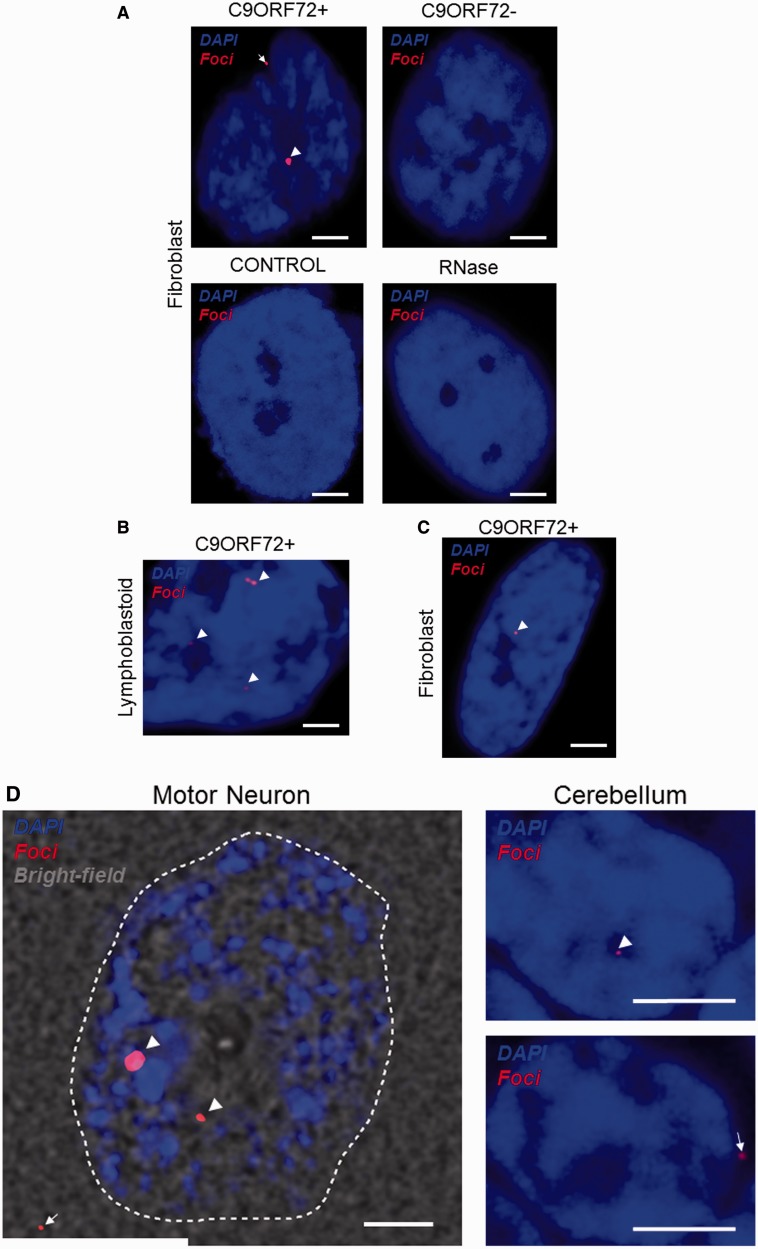
The presence of RNA foci clearly distinguished fibroblasts, lymphoblastoid cells and CNS tissue from C9orf72+ patients with ALS compared to C9orf72− patients with ALS and neurologically normal control subjects (Fig. 1A–D). To validate our RNA FISH methodology, discrete nuclear foci-like staining was quantified in a blinded study of 50 cerebellar granule neurons from each of nine cases: three C9orf72+ patients with ALS, three C9orf72− patients with ALS and three control subjects. In C9orf72+ tissue the average proportion of neurons containing nuclear RNA foci was 39% (range 21–63%); in three C9orf72− cases with ALS the average proportion of neurons containing foci-like staining was 1.6% (range 1.1–2.5%); in normal controls the average proportion of neurons containing foci-like staining was 1.4% (range 1.3–1.6%). Only seven foci-like objects were observed in 300 neurons from the six non-C9orf72+ cases and never was more than one focus-like object was observed in a single cell; in contrast the average rate in C9orf72+ tissue was two foci per cell. RNase treatment in fibroblasts ablated foci, illustrating that the labelled product is RNA and in agreement with previous studies (Fig. 1A).
Figure 1.
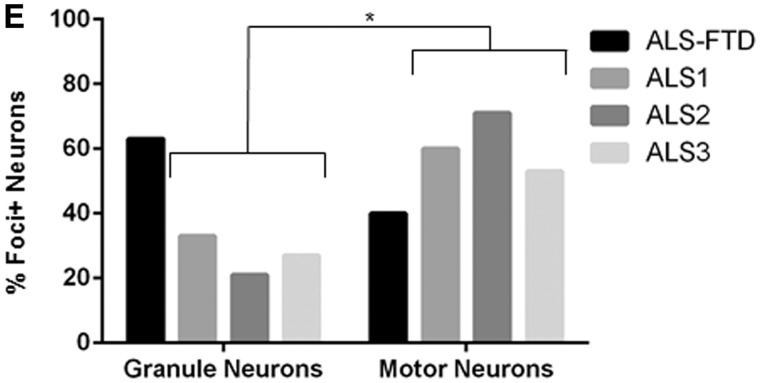
RNA FISH shows GGGGCC expanded RNA foci are found in peripheral cells and CNS tissue from C9orf72+ patients but not from C9orf72− ALS cases or controls. RNA foci (arrowheads) are present in fibroblasts (A), lymphoblastoid cells (B) and CNS tissue (D) from C9orf72+ patients with ALS, and in fibroblasts from a C9orf72+ asymptomatic carrier (C). RNA foci are ablated by RNase treatment (A). RNA foci are predominantly nuclear but cytoplasmic foci are observed in peripheral cells and CNS tissue (A and D, arrows). Abundance of foci in cerebellar granule cells and motor neurons has been quantified (E), in those cases where the initial clinical presentation was ALS the proportion of foci+ motor neurons is significantly higher (*P < 0.05). Scale bar = 3 µm. FTD = frontotemporal degeneration.
It is noteworthy that RNA foci were identified in fibroblasts derived from an asymptomatic C9orf72+ carrier (Fig. 1C). In four C9orf72+ cases the proportion of foci+ cerebellar granule neurons was quantified and compared to the proportion of foci+ motor neurons in the ventral horn (Fig. 1E). More than 35 cells of each neuron-type were examined in each case. Three of the cases presented initially with ALS (Supplementary Fig. 1); in these patients the average proportion of foci+ neurons was significantly higher in the ventral horn (61% versus 27%, t-test P < 0.05). In the fourth case, who presented with frontotemporal degeneration and later developed ALS, the pattern was reversed (40% versus 63%). Foci were primarily nuclear, however, some cytoplasmic foci were also observed in fibroblasts, cerebellar granule cells and in motor neurons (Fig. 1C).
Identification of binding partners of the C9orf72 repeat expansion
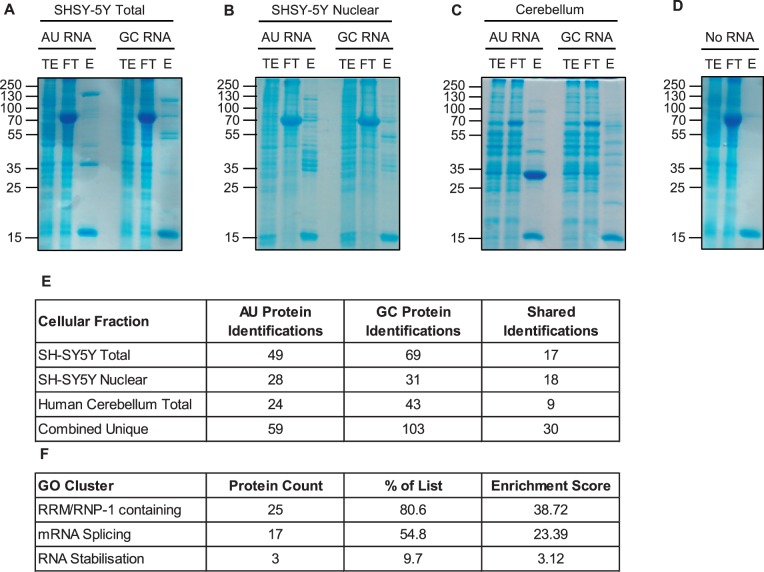
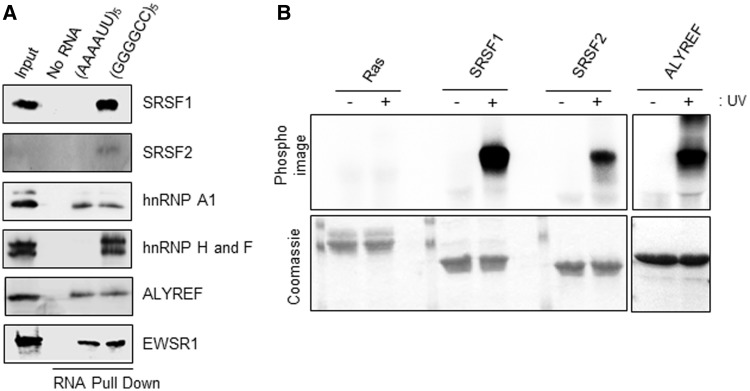
We generated 3’ biotinylated RNAs with the following sequences: 5’-[AAAAUU]5-Bio-3’ and 5’-[GGGGCC]5-Bio-3’. It has been demonstrated that the GGGGCC repeat expansion can form RNA G-quadruplexes in vitro, with the smallest repeating unit consisting of four repeats (Fratta et al., 2012; Reddy et al., 2013). To identify proteins interacting with the biotinylated RNAs, RNAs were preincubated with protein extracts and resulting complexes fixed by UV-irradiation. The RNA bait and bound proteins were captured using streptavidin sepharose and eluted after RNase A digestion. We used whole cell lysates of the human neuronal cell line SH-SY5Y, SH-SY5Y nuclear extract and dissected human cerebellum whole extract (Fig. 2A–C). Controls without RNA bait were processed in parallel (Fig. 2D). Eluted proteins were identified by mass spectrometry. In total, 103 unique proteins were identified that bind GGGGCC5, the majority of which did not bind to AAAAUU5 (Fig. 2E and Supplementary material).
Figure 2.
5’-[AAAAUU]5- and 5’-[GGGGCC]5 RNAs sequester distinct sets of proteins from human neuronal cell line fractions and dissected human cerebellar tissue. Pulldown assays using biotinylated RNAs (no RNA, 5’-[AAAAUU]5- or 5’-[GGGGCC]5) and extracts from total or nuclear fractions of SH-SY5Y cells, or human cerebellar tissue; I = input (1%); FT = flow through (1%); E = eluted (25%) (A–D). Mass spectrometry (MS) analysis of proteins co-purified with biotinylated RNAs (E). Gene ontology (GO) enrichment of SH-SY5Y nuclear hits (F).
Gene ontology (GO) enrichment analysis of each GGGGCC5-derived list of bound proteins was carried out using the Database for Annotation, Visualization and Integrated Discovery (DAVID) (Huang da et al., 2009a, b). This yielded functional categories associated with aspects of messenger RNA metabolism including splicing and stabilization, and an RNA recognition motif-containing class (Supplementary Fig. 2). This was particularly striking in the list of GGGGCC RNA-binders isolated from nuclear extracts of the SH-SY5Y human neuronal cell line (Fig. 2F). Another strongly represented group was messenger RNA export adaptors, which promote nuclear export via remodelling of the NXF1/TAP export receptor (Hautbergue et al., 2008), including ALYREF and the shuttling splicing factors SRSF1 (SF2/ASF), SFRS3 (SRp20) and SFRS7 (9G8) (Walsh et al., 2010).
Cellular distribution of RNA foci and RNA recognition motif-containing proteins
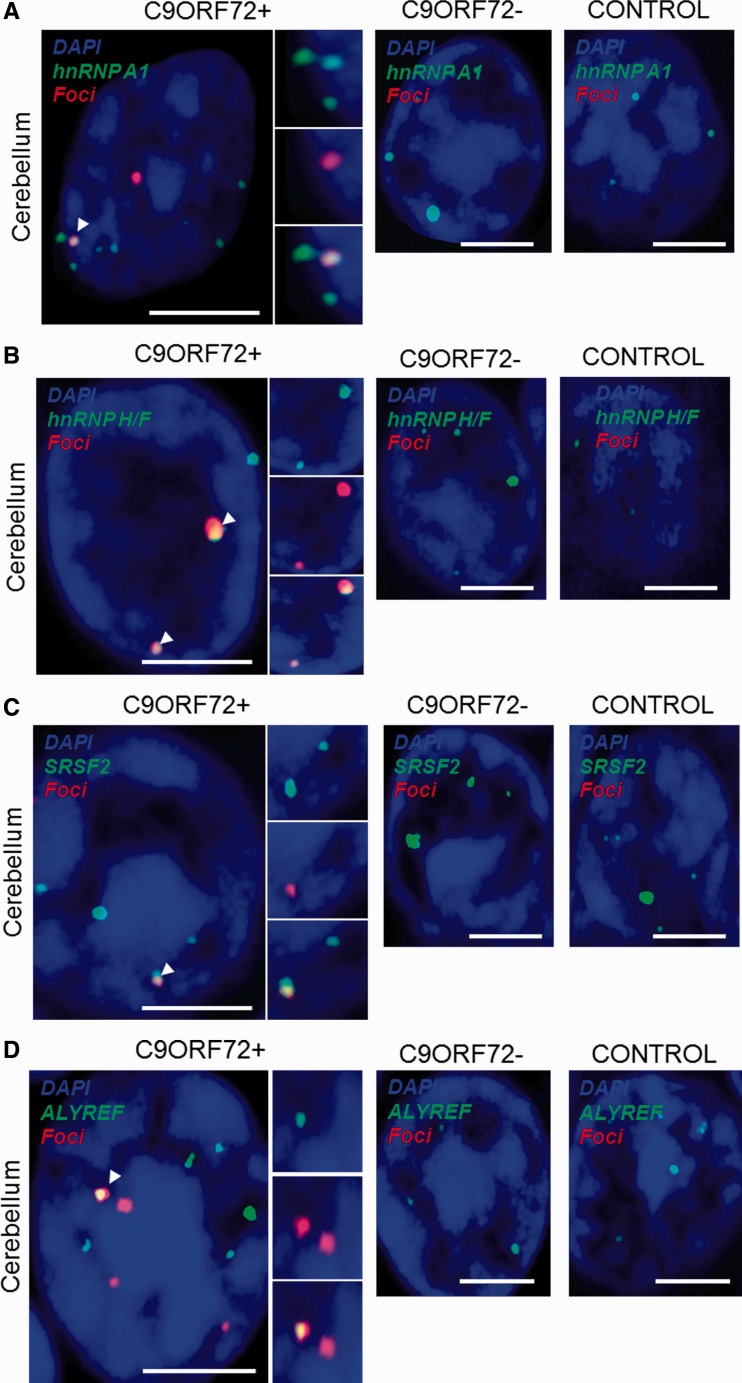
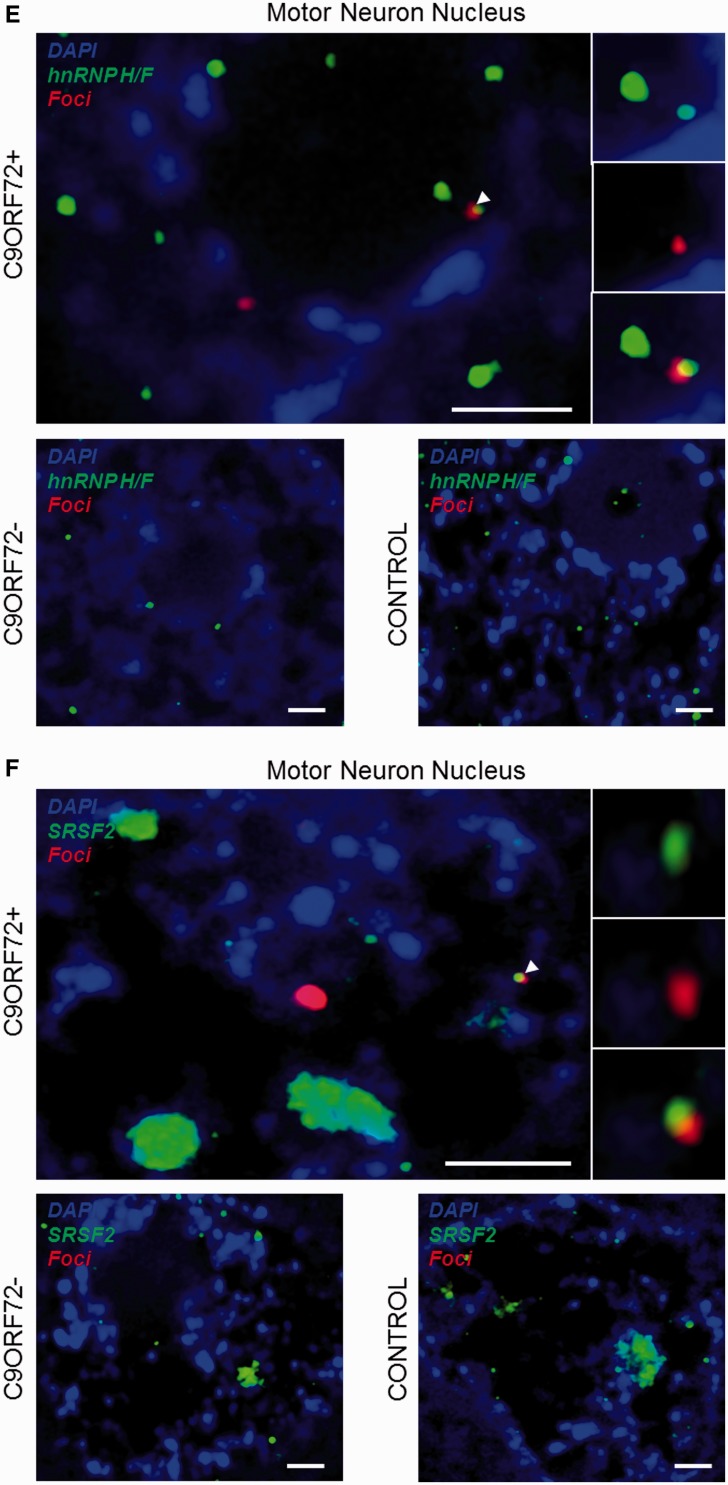
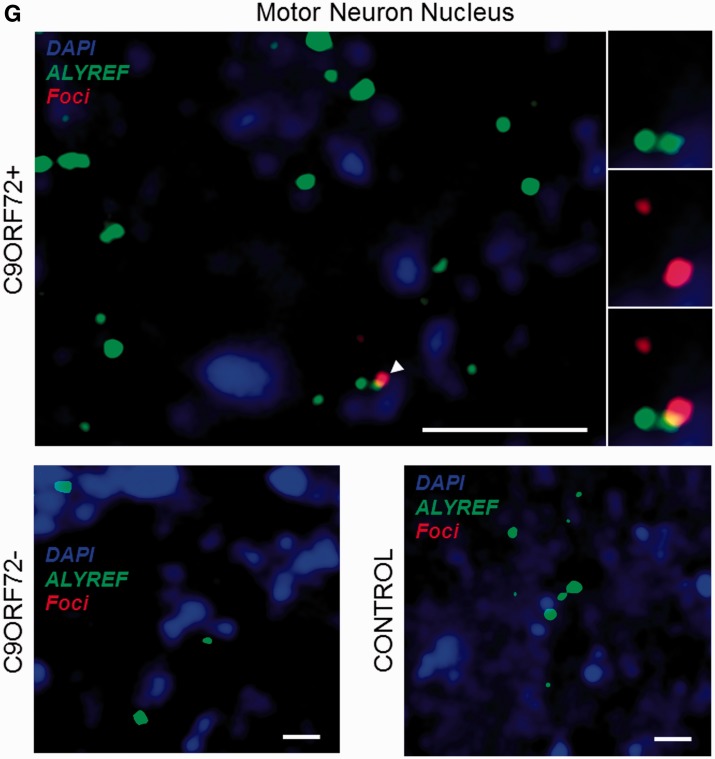
We used confocal microscopy to validate in vivo some of the hits identified by mass spectrometry. For this purpose, eight well-described RNA recognition motif-containing proteins including splicing factors and messenger RNA nuclear export adaptors were prioritized and selected depending on available and efficacious commercial antibodies. The distribution of each protein relative to RNA foci was examined in approximately 200 cerebellar granule neurons and 50 motor neurons from a minimum of three C9orf72+ cases with ALS. Simultaneous co-staining was carried out in parallel in C9orf72− cases with ALS and neurologically normal control subjects. For all tested candidates, overall cellular protein distribution was not grossly different between C9orf72+ cases, C9orf72− cases and controls except for areas where co-localization was demonstrated. In cerebellar granule cells we demonstrated co-localization of hnRNP A1, hnRNP H1/F, ALYREF and SRSF2 with 27%, 30%, 26% and 33% of RNA foci, respectively (Fig. 3A–D). In motor neurons, the cell type most vulnerable to the neurodegenerative process in ALS, we demonstrated co-localization of hnRNP H1/F, ALYREF and SRSF2 with 19%, 29% and 30% RNA foci, respectively (Fig. 3E–G). In contrast, we were unable to detect any evidence of co-localization of other identified GGGGCC-binding partners SRSF1, FUS, hnRNP C or hnRNP D with sense foci in either the cerebellar granule layer or the ventral horn (Supplementary Fig. 3).
Figure 3.
Combined RNA FISH and immunohistochemistry demonstrate co-localization of nuclear speckle components with RNA foci in CNS tissue. hnRNP A1 (A), hnRNP H1/F (B), SRSF2 (C) and ALYREF (D) are observed to co-localize with RNA foci (arrowheads) in cerebellar granule cells from C9orf72+ patients with ALS. hnRNP H1/F (E), SRSF2 (F) and ALYREF (G) are observed to co-localize with RNA foci (arrowheads) in nuclei of motor neurons from C9orf72+ patients with ALS. Co-localization events are enlarged and unmerged protein and RNA foci are shown for comparison. The normal staining pattern of the two proteins in C9orf72− cases with ALS and control subjects is included for comparison. Scale bar = 3 µm.
For six of the proteins identified in the mass spectrometry analysis, including those proteins observed to co-localize with RNA foci in vivo, specificity of interaction with the (GGGGCC)5 RNA was assessed using RNA pull down assays from whole neuronal SH-SY5Y cell extract and western immunoblotting (Fig. 4A). Direct binding of some of these proteins to (GGGGCC)5 RNA repeat was also confirmed in a UV-cross linking assay using radiolabelled RNA and recombinant proteins which were expressed and purified from E. coli (Fig. 4B).
Figure 4.
Identified RNA-binding candidates interact specifically and directly with GGGGCC5 RNA. (A) Neuronal SH-SY5Y whole cell extract was incubated with either no RNA, AU-rich or GC-rich biotinylated RNA coated onto streptavidin beads before UV-cross linking. Bound proteins were eluted using RNase A and further identified using SDS-PAGE and western immunoblotting with the indicated antibodies. It is noted that the weak signal for SRSF2 is due to difficulty finding an antibody that is efficacious in western immunoblotting. The anti-hnRNP H1/F antibody recognizes both proteins, which are similar (Garneau et al., 2005). (B) Hexa-histidine-tagged recombinant SRSF1 11-196, GB1-tagged SRSF2 9-101 and ALYREF full length were expressed in E. coli and purified using metal ion affinity chromatography in 1 M NaCl containing buffers to remove potentially bound RNA from E. coli (bottom). GGGGCC5 RNA was separately end-labelled with poly nucleotide kinase using [γ-32P]-ATP, before incubation with purified proteins. RNA was covalently bound (+) or not (−) after UV irradiation. Absence of radioactive signal (top; PhosphoImage) in absence of UV irradiation demonstrates specificity of direct binding observed after UV treatment. All gels shown in the different panels were exposed simultaneously for the same amount of time (5 h).
We also examined the co-incidence of RNA foci with depletion of TDP-43 from the nuclei of motor neurons of C9orf72+ patients with ALS. Mislocalization of TDP-43 is the pathophysiological hallmark of ALS (Neumann et al., 2006). All surviving motor neurons were examined in formalin fixed paraffin-embedded sections from three C9orf72+ ALS cases. The majority of cells with nuclear depletion of TDP-43 contained nuclear RNA foci, but this was not significantly different to the proportion of cells with nuclear TDP-43 expression that contained RNA foci (66% versus 60%, χ2 P = 0.75) (Supplementary Fig. 4).
In view of our prediction that the repeat sequence might sequester several proteins important for messenger RNA export, we wanted to explore the relationship between repeat associated non-ATG translated protein and RNA foci in specific neuronal populations. As expression of dipeptide repeat proteins is reported to be rare in the ventral horn of C9orf72+ patients with ALS (Mackenzie et al., 2013), we chose to focus on cerebellar granule cells. Fifty per cent of the neurons which stained for poly-GA, the most abundant dipeptide repeat protein, contained nuclear RNA foci; this was not significantly different to the proportion of neurons with nuclear RNA foci which did not stain for poly-GA (50% versus 40%, χ2 P = 0.46) (Supplementary Fig. 5).
Discussion
There is an urgent need to understand the mechanisms of neuronal injury in C9orf72+ disease. This genetic variant is the most common identified cause of ALS and frontotemporal degeneration. We and others (DeJesus-Hernandez et al., 2011; Donnelly et al., 2013; Lagier-Tourenne et al., 2013; Lee et al., 2013; Mizielinska et al., 2013; Sareen et al., 2013) have identified RNA foci formed from the intronic GGGGCC repeat sequence in peripheral cells and CNS tissue from C9orf72+ patients. We have particularly focused on characterizing RNA foci within spinal motor neurons, which are the primary target of pathology in ALS. Indeed we have shown that RNA foci are present in a higher proportion of motor neurons of the ventral horn compared to cerebellar granule cells in patients where the initial clinical presentation was ALS; in a single patient where the initial clinical presentation was with extra-motor disease the opposite was true. This is consistent with toxicity initiated by RNA foci. However, this finding will require validation in a larger number of cases.
We have identified a number of putative binding partners of the RNA repeat expansion which are consistent with previous observations (Lee et al., 2013; Mori et al., 2013a; Sareen et al., 2013; Xu et al., 2013). Of the RNA recognition motif-containing proteins we found to be co-localized with RNA foci in C9orf72+ tissue, hnRNP A1 (Sareen et al., 2013), hnRNP H1/F and SRSF2 (Lee et al., 2013) have been similarly observed by others. Interestingly, our study provides the first evidence for co-localization of RNA foci with the general messenger RNA nuclear export adaptor ALYREF (Stutz et al., 2000). Observed co-localization with RNA recognition motif-containing proteins was present in a relatively low percentage of RNA foci. We suggest that this is consistent with a process of dynamic sequestration. Indeed, irreversible binding of these candidates, many of which are key regulators of essential processes such as pre-messenger RNA splicing, is unlikely to be consistent with the relatively late age of disease onset seen in C9orf72+ patients. The key pathogenic step may be downstream from protein sequestration by the expansion, such as export of the repeat expansion to enable repeat associated non-ATG translation or an accumulation of aberrant splicing events. Importantly we have confirmed a direct interaction in vitro between our protein candidates and the GGGGCC repeat RNA by UV-crosslinking.
SRSF2 is a well-known marker for nuclear speckles, nuclear domains implicated in the storage and supply of splicing factors to active transcription sites (Spector and Lamond , 2011). All of the proteins we have shown to co-localize with RNA foci, many of the binding partners identified in our RNA pulldown, and a number of the proteins implicated in genetic variants of ALS including TARDBP, EWSR1, FUS, HNRNPA1 and HNRNPA2B1, have been localized to nuclear speckles (Zhou et al., 2000; Saitoh et al., 2004; Casafont et al., 2009). Other neuromuscular diseases have been associated with depletion of normal components of nuclear speckles including myotonic dystrophy type 1 (Smith et al., 2007; Bengoechea et al., 2012). It is possible that disruption of the normal function of nuclear speckles, either by a direct mutation of one of the key protein components, or via RNA foci-mediated dynamic depletion of essential protein constituents, is a key pathogenic mechanism in ALS. Analysis of the transcriptome of pathologically affected neurons will be key to elucidating whether the interactions we have identified have a toxic effect through disruption of messenger RNA splicing.
We provide evidence for cytoplasmic RNA foci, not only in peripheral cells and in the cerebellar granule layer, but also in motor neurons from the ventral horn of the spinal cord. Cytoplasmic localization of RNA foci formed from an intronic repeat sequence in peripheral cells might be consistent with extrusion during mitosis. However, in non-dividing neurons of the cerebellum and ventral horn this is not a possibility. The alternative scenario relates to nuclear export of the transcribed GGGGCC repeat expansion. Our RNA pulldown screen for binding of the repeat expansion identified multiple messenger RNA export adapters including ALYREF (Stutz et al., 2000), SRSF1, SFRS3 and SFRS7 (Huang et al., 2003; Hargous et al., 2006; Tintaru et al., 2007). In the case of ALYREF we have also demonstrated co-localization with RNA foci by immunohistochemistry, and a direct interaction with the expansion by protein-RNA UV-crosslinking. An interesting possibility is that local enrichment of messenger RNA export adaptors onto C9orf72 GGGGCC repeat pre-messenger RNA molecules overrides the normal nuclear retention of pre-messenger RNA, for example through an inappropriate interaction of ALYREF with the TAP/NXF1 nuclear export receptor. It seems unlikely that the RNA foci are exported intact, particularly because of their size and activity of DEAD box RNA helicases such as Dbp5/DDX19, on the cytoplasmic side of the nuclear pore which would be expected to unwind G-quadruplex structures (Linder, 2008). However, it is conceivable that aberrantly expanded C9orf72 pre-messenger RNA molecules are exported from the nucleus and reform into foci within the cytoplasm.
Nuclear export of GGGGCC repeat RNA is likely to be a key step leading to repeat associated non-ATG translation in the cytoplasm. If dipeptide repeat proteins formed in this manner are eventually identified as the key mediator of pathogenicity in C9orf72+ disease then blocking this export represents an attractive therapeutic target. One report has suggested that the production of repeat associated non-ATG translated protein is mutually exclusive to the presence of RNA foci (Donnelly et al., 2013). In contrast, we found an equal proportion of poly-GA staining in neurons that did or did not contain RNA foci.
We did not observe a significant correlation between nuclear loss of TDP-43 and the presence of RNA foci. This does not mean that RNA foci are not instrumental in the disease pathogenesis, but may reflect the fact that they occur significantly upstream of TDP-43 mislocalization. In this regard it is important to note that we and others (Lagier-Tourenne et al., 2013) have identified RNA foci in fibroblasts derived from asymptomatic C9orf72+ carriers.
We await confirmation of our findings by other groups. We have suggested two ways in which the interactions identified may be pathogenic: (i) through disruption of the normal function of factors involved in nuclear speckles and thus messenger RNA splicing; and (ii) through inappropriate licensing of the transcribed C9orf72 expansion for nuclear export thereby facilitating repeat associated non-ATG translation. Either or both may be important, but it should be noted that if inappropriate licensing of RNA foci for export is a key pathogenic step, then overexpression of the sequestered protein will not be of therapeutic benefit and may even have an adverse effect. On the contrary if loss of the normal function of these proteins is most important, then increasing the nuclear expression of proteins sequestered by the expansion may be of value as a neuroprotective strategy.
Supplementary Material
Acknowledgements
We are grateful to all of the patients with ALS and their family members who donated biosamples for research.
Glossary
Abbreviations
- ALS
amyotrophic lateral sclerosis
- FISH
fluorescence in situ hybridization
Funding
We acknowledge grants from the EU Framework 7 (Euromotor No259867) and the SOPHIA project (funded by JPND and MRC) to P.J.S. and J.K. P.J.S. is an NIHR Senior Investigator. J.C.K. and J.R.H. are supported by MND Association/MRC Lady Edith Wolfson Fellowship awards ([MR/K003771/1] and [G0 800380] respectively). Biosample collection was supported by the MND Association and the Wellcome Trust (P.J.S.). M.J.D. acknowledges support from the Engineering and Physical Sciences Research Council (UK) and the Biotechnology and Biological Sciences Research Council (UK). D.E. was supported by the Helmholtz Young Investigator program HZ-NG-607.
Supplementary material
Supplementary material is available at Brain online.
References
- Ash PE, Bieniek KF, Gendron TF, Caulfield T, Lin WL, Dejesus-Hernandez M, et al. Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron. 2013;77:639–46. doi: 10.1016/j.neuron.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengoechea R, Tapia O, Casafont I, Berciano J, Lafarga M, Berciano MT. Nuclear speckles are involved in nuclear aggregation of PABPN1 and in the pathophysiology of oculopharyngeal muscular dystrophy. Neurobiol Dis. 2012;46:118–29. doi: 10.1016/j.nbd.2011.12.052. [DOI] [PubMed] [Google Scholar]
- Brettschneider J, Del Tredici K, Toledo JB, Robinson JL, Irwin DJ, Grossman M, et al. Stages of pTDP-43 pathology in amyotrophic lateral sclerosis. Ann Neurol. 2013;74:20–38. doi: 10.1002/ana.23937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne S, Heverin M, Elamin M, Walsh C, Hardiman O. Intermediate repeat expansion length in C9orf72 may be pathological in amyotrophic lateral sclerosis. Amyotroph Lateral Scler Frontotemporal Degener. 2013;15:148–50. doi: 10.3109/21678421.2013.838586. [DOI] [PubMed] [Google Scholar]
- Casafont I, Bengoechea R, Tapia O, Berciano MT, Lafarga M. TDP-43 localizes in mRNA transcription and processing sites in mammalian neurons. J Struct Biol. 2009;167:235–41. doi: 10.1016/j.jsb.2009.06.006. [DOI] [PubMed] [Google Scholar]
- Cooper-Knock J, Hewitt C, Highley JR, Brockington A, Milano A, Man S, et al. Clinico-pathological features in amyotrophic lateral sclerosis with expansions in C9ORF72. Brain. 2012;135:751–64. doi: 10.1093/brain/awr365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper-Knock J, Higginbottom A, Connor-Robson N, Bayatti N, Bury JJ, Kirby J, et al. C9ORF72 transcription in a frontotemporal dementia case with two expanded alleles. Neurology. 2013;81:1719–21. doi: 10.1212/01.wnl.0000435295.41974.2e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeJesus-Hernandez M, Mackenzie I, Boeve B, Boxer A, Baker M, Rutherford N, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-Linked FTD and ALS. Neuron. 2011;72:245–56. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–89. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly CJ, Zhang PW, Pham JT, Heusler AR, Mistry NA, Vidensky S, et al. RNA Toxicity from the ALS/FTD C9ORF72 expansion is mitigated by antisense intervention. Neuron. 2013;80:415–28. doi: 10.1016/j.neuron.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fratta P, Mizielinska S, Nicoll AJ, Zloh M, Fisher EM, Parkinson G, et al. C9orf72 hexanucleotide repeat associated with amyotrophic lateral sclerosis and frontotemporal dementia forms RNA G-quadruplexes. Sci Rep. 2012;2:1016. doi: 10.1038/srep01016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau D, Revil T, Fisette JF, Chabot B. Heterogeneous nuclear ribonucleoprotein F/H proteins modulate the alternative splicing of the apoptotic mediator Bcl-x. J Biol Chem. 2005;280:22641–50. doi: 10.1074/jbc.M501070200. [DOI] [PubMed] [Google Scholar]
- Gendron TF, Bieniek KF, Zhang YJ, Jansen-West K, Ash PE, Caulfield T, et al. Antisense transcripts of the expanded C9ORF72 hexanucleotide repeat form nuclear RNA foci and undergo repeat-associated non-ATG translation in c9FTD/ALS. Acta Neuropathol. 2013;126:829–44. doi: 10.1007/s00401-013-1192-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Tortosa E, Gallego J, Guerrero-Lopez R, Marcos A, Gil-Neciga E, Sainz MJ, et al. C9ORF72 hexanucleotide expansions of 20–22 repeats are associated with frontotemporal deterioration. Neurology. 2013;80:366–70. doi: 10.1212/WNL.0b013e31827f08ea. [DOI] [PubMed] [Google Scholar]
- Hargous Y, Hautbergue GM, Tintaru AM, Skrisovska L, Golovanov AP, Stevenin J, et al. Molecular basis of RNA recognition and TAP binding by the SR proteins SRp20 and 9G8. EMBO J. 2006;25:5126–37. doi: 10.1038/sj.emboj.7601385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms MB, Cady J, Zaidman C, Cooper P, Bali T, Allred P, et al. Lack of C9ORF72 coding mutations supports a gain of function for repeat expansions in amyotrophic lateral sclerosis. Neurobiol Aging. 2013;34:2234.e2213–39. doi: 10.1016/j.neurobiolaging.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hautbergue GM, Hung ML, Golovanov AP, Lian LY, Wilson SA. Mutually exclusive interactions drive handover of mRNA from export adaptors to TAP. Proc Natl Acad Sci USA. 2008;105:5154–9. doi: 10.1073/pnas.0709167105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hautbergue GM, Hung ML, Walsh MJ, Snijders AP, Chang CT, Jones R, et al. UIF, a New mRNA export adaptor that works together with REF/ALY, requires FACT for recruitment to mRNA. Curr Biol. 2009;19:1918–24. doi: 10.1016/j.cub.2009.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009a;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009b;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Huang Y, Gattoni R, Stévenin J, Steitz JA. SR splicing factors serve as adapter proteins for TAP-dependent mRNA export. Mol Cell. 2003;11:837–43. doi: 10.1016/s1097-2765(03)00089-3. [DOI] [PubMed] [Google Scholar]
- Jiang H, Mankodi A, Swanson MS, Moxley RT, Thornton CA. Myotonic dystrophy type 1 is associated with nuclear foci of mutant RNA, sequestration of muscleblind proteins and deregulated alternative splicing in neurons. Hum Mol Genet. 2004;13:3079–88. doi: 10.1093/hmg/ddh327. [DOI] [PubMed] [Google Scholar]
- Lagier-Tourenne C, Baughn M, Rigo F, Sun S, Liu P, Li HR, et al. Targeted degradation of sense and antisense C9orf72 RNA foci as therapy for ALS and frontotemporal degeneration. Proc Natl Acad Sci USA. 2013;110:E4530–9. doi: 10.1073/pnas.1318835110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YB, Chen HJ, Peres JN, Gomez-Deza J, Attig J, Stalekar M, et al. Hexanucleotide repeats in ALS/FTD form length-dependent RNA foci, sequester RNA binding proteins, and are neurotoxic. Cell Rep. 2013;5:1178–86. doi: 10.1016/j.celrep.2013.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder P. mRNA export: RNP remodeling by DEAD-box proteins. Curr Biol. 2008;18:R297–9. doi: 10.1016/j.cub.2008.02.027. [DOI] [PubMed] [Google Scholar]
- Mackenzie IR, Arzberger T, Kremmer E, Troost D, Lorenzl S, Mori K, et al. Dipeptide repeat protein pathology in C9ORF72 mutation cases: clinico-pathological correlations. Acta Neuropathol. 2013;126:859–79. doi: 10.1007/s00401-013-1181-y. [DOI] [PubMed] [Google Scholar]
- Mili S, Steitz JA. Evidence for reassociation of RNA-binding proteins after cell lysis: implications for the interpretation of immunoprecipitation analyses. RNA. 2004;10:1692–4. doi: 10.1261/rna.7151404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizielinska S, Lashley T, Norona FE, Clayton EL, Ridler CE, Fratta P, et al. C9orf72 frontotemporal lobar degeneration is characterised by frequent neuronal sense and antisense RNA foci. Acta Neuropathol. 2013;126:845–57. doi: 10.1007/s00401-013-1200-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K, Lammich S, Mackenzie IR, Forne I, Zilow S, Kretzschmar H, et al. hnRNP A3 binds to GGGGCC repeats and is a constituent of p62-positive/TDP43-negative inclusions in the hippocampus of patients with C9orf72 mutations. Acta Neuropathol. 2013a;125:413–23. doi: 10.1007/s00401-013-1088-7. [DOI] [PubMed] [Google Scholar]
- Mori K, Weng SM, Arzberger T, May S, Rentzsch K, Kremmer E, et al. The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science. 2013b;339:1335–8. doi: 10.1126/science.1232927. [DOI] [PubMed] [Google Scholar]
- Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–3. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- Reddy K, Zamiri B, Stanley SY, Macgregor RB, Jr, Pearson CE. The disease-associated r(GGGGCC)n repeat from the C9orf72 gene forms tract length-dependent uni- and multimolecular RNA G-quadruplex structures. J Biol Chem. 2013;288:9860–6. doi: 10.1074/jbc.C113.452532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renton AE, Majounie E, Waite A, Simón-Sánchez J, Rollinson S, Gibbs JR, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–68. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh N, Spahr CS, Patterson SD, Bubulya P, Neuwald AF, Spector DL. Proteomic analysis of interchromatin granule clusters. Mol Biol Cell. 2004;15:3876–90. doi: 10.1091/mbc.E04-03-0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sareen D, O'Rourke JG, Meera P, Muhammad AK, Grant S, Simpkinson M, et al. Targeting RNA foci in iPSC-derived motor neurons from ALS patients with a C9ORF72 repeat expansion. Sci Transl Med. 2013;5:208ra149. doi: 10.1126/scitranslmed.3007529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KP, Byron M, Johnson C, Xing Y, Lawrence JB. Defining early steps in mRNA transport: mutant mRNA in myotonic dystrophy type I is blocked at entry into SC-35 domains. J Cell Biol. 2007;178:951–64. doi: 10.1083/jcb.200706048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector DL, Lamond AI. Nuclear speckles. Cold Spring Harb Perspecti Biol. 2011;3:pii: a000646. doi: 10.1101/cshperspect.a000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stutz F, Bachi A, Doerks T, Braun IC, Seraphin B, Wilm M, et al. REF, an evolutionary conserved family of hnRNP-like proteins, interacts with TAP/Mex67p and participates in mRNA nuclear export. RNA. 2000;6:638–50. doi: 10.1017/s1355838200000078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tintaru AM, Hautbergue GM, Hounslow AM, Hung ML, Lian LY, Craven CJ, et al. Structural and functional analysis of RNA and TAP binding to SF2/ASF. EMBO Rep. 2007;8:756–62. doi: 10.1038/sj.embor.7401031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh MJ, Hautbergue GM, Wilson SA. Structure and function of mRNA export adaptors. Biochem Soc Trans. 2010;38:232–6. doi: 10.1042/BST0380232. [DOI] [PubMed] [Google Scholar]
- Xi Z, Zinman L, Moreno D, Schymick J, Liang Y, Sato C, et al. Hypermethylation of the CpG island near the GC repeat in ALS with a C9orf72 expansion. Am J Hum Genet. 2013;92:981–9. doi: 10.1016/j.ajhg.2013.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Poidevin M, Li X, Li Y, Shu L, Nelson DL, et al. Expanded GGGGCC repeat RNA associated with amyotrophic lateral sclerosis and frontotemporal dementia causes neurodegeneration. Proc Natl Acad Sci USA. 2013;110:7778–83. doi: 10.1073/pnas.1219643110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Luo MJ, Straesser K, Katahira J, Hurt E, Reed R. The protein Aly links pre-messenger-RNA splicing to nuclear export in metazoans. Nature. 2000;407:401–5. doi: 10.1038/35030160. [DOI] [PubMed] [Google Scholar]
- Zu T, Liu Y, Banez-Coronel M, Reid T, Pletnikova O, Lewis J, et al. RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proc Natl Acad Sci USA. 2013;110:E4968–77. doi: 10.1073/pnas.1315438110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.