Abstract
Objective
To develop a new ImmunoFISH technique for the study of oligodendrogliomas by combining a standard immunohistochemical stain using MIB-1 antibody with a standard FISH technique using commercial 1p36 and 19q13 chromosomal probes.
Methods
Validation was performed by two observers on a series of 36 pre-selected oligodendrogliomas and compared to the results previously determined by FISH alone.
Results
The ImFISH technique is easy to perform and to analyze and is no more time-consuming than the usual FISH technique. Our results show that the inter-observer reliability of ImFISH is high (κ = 0.86 and 0.95 respectively for 1p and 19q). Compared to FISH, the ImFISH exhibits a very high sensitivity (∼100%) and specificity (∼90%) for 1p and/or 19q deleted cases. The sensitivity is high for normal cases (∼85%) and imbalanced cases (∼90%) with a specificity ranging between 50 and 85%. Finally, there were no significant differences between FISH and ImFISH results calculated on 60, 40 or 20 cells.
Conclusion
Our study demonstrates the reliability of the ImFISH technique in oligodendrogliomas and emphasizes its advantage in poorly cellular tumoral specimen.
Introduction
The study of chromosome 1p and 19q status has become an essential step in the treatment of oligodendroglial tumors in recent years [1], [2], [3], [4], [5], [6], [7], [8], [9], [10], [11], [12]. Codeletion of 1p and 19q whole arms is strongly correlated with a better response to standard treatment with radiotherapy and chemotherapy as well as a better overall survival [13], [14], [15], [16], [17], [18], [19], [20], [21].
Several molecular techniques are described in the literature to study the chromosomal status of tumor cells, including fluorescence in situ hybridization (FISH) [22], [23], [24], [25], [26], polymerase chain reaction [27], [28], quantitative microsatellite analysis [14], [29], loss of heterozygosity (LOH) by microsatellite analysis [8], [10], [13], [30], [31], [32], [33], [34], [35], comparative genomic hybridization array (CGH) [17], [36], [37], [38], [39], [40], [41] and multiplex ligation - dependent probe [42]. All these techniques have their advantages and disadvantages but the most widely used among them is FISH [26] because it can be performed by fluorescent microscopy on paraffin embedded tumor tissue sections and is thus easily accessible to most pathology laboratories.
Although some guidelines exist in the literature to harmonize the interpretation of FISH results [43], [44], [45], several authors have emphasized the difficulty that may be encountered in the interpretation of chromosomal signals, especially in polyploid cases [26], [46], [47]. The main causes of these difficulties include the thickness of the histological section, a low density of tumor cells in some specimens or conversely a high density of tumor cells with overlapping nuclear profiles, making their FISH interpretation difficult [26], [47].
To optimize the detection of chromosomal status by FISH technique, some authors have proposed to replace the standard histological section from paraffin-embedded tissue with tumor cell nuclei isolated from paraffin embedded blocks [46], frozen smears [48] or fresh tissue touch preparations [49]. Other authors have proposed to sample a larger number of tumor cells by using automatic analysis [44] or to perform a chromogenic technique using dual-color chromogenic in situ hybridization (CISH) [50].
Another technique proposed to increase the diagnostic yield of FISH involves adding an immunochemical step to permit simultaneous analysis of the genotype and the phenotype on the same tissue sample. This technique called ImmunoFISH (ImFISH) was first described in the literature in the 90 s and was originally developed for the study of hematologic malignancies [51], [52], [53]. Its use has since been extended to other neoplastic processes including those of nerve [54], breast [55], prostate [56] and the gastrointestinal tract [57].
The ImFISH technique, as originally described, combined conventional double immunofluorescence, using fluorochrome-conjugated secondary antibodies, with a standard FISH technique and its interpretation was done directly on a fluorescence microscope [51].
As part of our ongoing efforts to improve the diagnostic yield and accuracy of smaller and less cellular brain tumor samples, we decided to try the ImFISH technique on oligodendroglial tumors. We began by looking at a combination of FISH and MIB-1 immunostaining since the proliferation index, as measured with the MIB-1 antibody, is routinely reported on most brain neoplasms and the chromosome 1p and 19q status is routinely determined for all oligodendroglial tumors in our laboratory. It thus seemed appropriate to us to combine these two techniques into one.
Preliminary tests by immunofluorescence led to inconclusive results, especially in tumors with a low proliferative index, in which the identification of neoplastic cells proved difficult. Autofluorescence of erythrocyte clusters was a problem, as was distinguishing proliferating blood vessels from adjacent tumor cells, which made the analysis time consuming and difficult to reproduce. Therefore we decided to replace the immunofluorescence step with a conventional chromogenic immunohistochemical technique. Although this modification meant that the analysis could no longer be performed solely by fluorescent microscopy, we were able to use a standard digital microscopy setup to capture both fluorescent and bright-field microscopic images and superimpose the images.
The results of our preliminary assays encouraged us to extend this technique to a series of 36 oligodendroglial tumors previously analyzed in our laboratory by standard FISH technique.
To validate this technique we first studied the ImFISH reproducibility between two independent observers.
Results obtained by ImFISH technique were then compared with those obtained by standard FISH technique which served as the reference in our study.
To determine the reliability of this technique on a limited number of cells, we analyzed a decreasing number of cells (60 cells, 40 cells and 20 cells) by ImFISH and checked if the reported 1p and 19q status remained constant or not.
In our series, chromosomal data were also compared with the clinical and surgical data, and with MIB-1 labeling indices to seek any correlation existing between these different parameters.
For all our analysis, we decided to calculate chromosomal status using both the combination and ratio methods, in order to permit the widest possible comparison with the literature data.
Materials and Methods
Ethics statement
The Research Ethics Committee of the Centre Hospitalier Universitaire de Québec was consulted for this study and decided that its approval was not necessary. The committee waived the need for consent, the aim of this study being the optimization of an institutional diagnostic technique with anonymized data.
Tissue samples
Formalin fixed paraffin-embedded (FFPE) tissue from 36 brain tumor samples (biopsies or surgical resections) with previously established 1p/19q status by FISH was pre-selected for the ImFISH study. All tumors were classified and graded according to the guidelines of the World Health Organization [58] by two neuropathologists (PVG and SS). The cases included 11 WHO grade II oligodendrogliomas (OII), 22 WHO grade III anaplastic oligodendrogliomas (OIII) and 3 WHO grade IV glioblastomas with oligodendroglioma component (GBMO). FISH analysis of 1p/19q was initiated in all cases during diagnostic work-up.
FISH
FISH analysis was performed using the LSI 1p36/19q13 Dual-Color Probe kit (Abbott Molecular Inc., Abbott Park, Illinois, USA). Slides were immersed in 0.2 N HCl for 20 minutes at room temperature, rinsed with purified water and incubated in pretreatment solution (2x saline sodium citrate (SSC) for 30 minutes at 97°C. After washes with purified water and 2x SSC, pretreated slides were digested with a pepsin solution (Dako) at 37°C for 5 minutes, rinsed in 2× SSC at RT for 5 minutes, and dehydrated using graded ethanol (70%, 85%, and 100% for 1 minutes each) and finally air dried. The probe mix (5 to 15 µl) was added to each slide according to the manufacturer's instructions and the hybridization area was covered with a coverslip then sealed with rubber cement. Target DNA and probes were codenatured at 74°C for 5 minutes and incubated at 37°C overnight in a humidified hybridization chamber (ThermoBrite, Abbott Molecular Inc.). Posthybridization washes were performed in NP40 0.3%/2×SSC (pH 7) at 75°C for 2 minutes. Finally, the slides were air dried and counterstained with DAPI (4′,6-diamidino-2-phenylindole) diluted in Vectashield (Vector, Burlingame, CA, USA). Signal acquisition was performed for each slide over 10 to 15 more representative areas. These areas were automatically captured at x400 using a Metasystems station (Zeiss MetaSystems, Thornwood, NY) equipped with a Zeiss Axioplan fluorescent microscope.
FISH interpretation for 1p and 19q status using the combination method was performed according to the guidelines of the International Society of Paediatric Oncology (E-SIOP Neuroblastoma Study Group) for studies of peripheral neuroblastic tumours [43]. For each case, two independent and blinded observers (FS and SS) assessed 100 non-overlapping nuclei for red ‘R’ (marker) and green ‘G’ (reference) signals. The frequencies of signal patterns for 1p (R) and 1q (G) on one slide, and for 19p (G) and 19q (R) on another slide, were noted. The cut-off of nuclei that had to show deletion was calculated on a series of 10 non-neoplastic brain tissue samples (from epilepsy surgery cases and autopsy brains). This cut-off was calculated using mean +3 SD and was set at 50% for both 1p and 19q. Cases above the cutoff were considered deleted and those under the cut off were considered normal or imbalanced according to the literature guidelines [43], [44].
For each case the signal ratio of red signals to green signals per cell was also established. A ratio ≤0.8 was considered to indicate a deletion whereas a ratio between 0.8 and 1.1 was considered to indicate a normal status on the chromosomal arm. A ratio over 1.1 was considered to indicate polysomy and was classified in the imbalanced status subgroup [2], [30].
ImmunoFISH
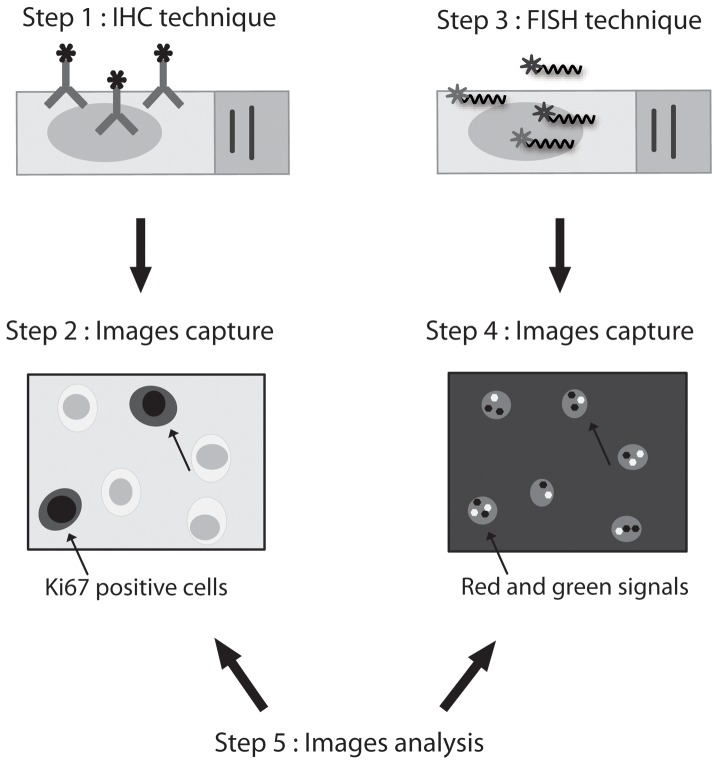
Immunohistochemistry (IHC) procedures were performed on 4-µm-thick sections obtained from FFPE brain tumor tissues. Sections were deparaffinized in 2 xylene washes for 5 min and rehydrated in 100% ethanol baths. Antigen retrieval was then carried out in a PT Link pretreatment system (Dako, Mississauga, Ontario, Canada). After rinsing the slides in purified water and in wash buffer (PBS-Tween 20x, Dako), the sections were incubated in a humid chamber at room temperature (RT) with monoclonal mouse primary antibody against Ki67 (MIB-1; Beckman Coulter, Fullerton, CA, USA) diluted 1∶100 in EnVision Flex antibody diluent (Dako) for 15 minutes. Immunolabelling was revealed with the EnVision G2/AP System with permanent red chromogen (Dako) according to the manufacturer's instructions except that all incubations were performed at room temperature for 15 min. The sections were then counterstained with hematoxylin. Tumor cells with red staining were considered positive, regardless of IHC staining intensity. MIB-1 signal acquisition was performed for each slide over 15 to 30 areas that were selected by an observer (CD). These areas were automatically captured at x400 using the same microscope and image analysis software than previously described for the FISH (Figure 1). After MIB-1 signal acquisition, slides were washed in a 100% ethanol bath to remove the red aminoethylcarbazol (AEC) chromogen staining from tissues then air dried. FISH analysis was performed on the same slide using the same procedure as described above. Fluorescent signals were automatically recorded at x400 in the same preregistered positions as for MIB-1 signal acquisitions (Figure 1).
Figure 1. Overview of ImmunoFish procedure.
In the first step, immunohistochemistry with MIB-1 antibody is performed on FFPE oligodendrogliomas. Digital images of the 10 most labelled areas are taken at high magnification (x 400). Then the slide is washed and used for a second FISH step using 1p36 or 19q13 probes. Digital images of the same 10 areas that were selected based on MIB-1 labelling are taken at the same magnification. Analysis of the two sets of images is done simultaneously on two separate screens. Only cells with MIB-1 labelling are taken into account for FISH analysis.
IHC and FISH images from the ImFISH technique were analysed simultaneously on two separate computers screens (Figure 1) by two independent and blinded observers (CD and SS).
The MIB-1 labelling index (LI) was calculated for each case on IHC images by counting 100 contiguous cells in the most positive areas.
For each probe 1p and 19q, a total of 60 MIB-1 positive nuclei were analyzed for the chromosomal status using the same procedures previously described for FISH. For the combination method, and in the absence of positive immunostaining in our non-tumor brain control series, we decided to set our cut-off at the median value of our tumor series which corresponds to a value of 65% for both 1p and 19q. For the ratio method, established values were the same as for the FISH.
Statistical analyses
All statistical analyses were carried out with the R statistical environment (http://www.R-project.org/). Inter-observer agreement of ImFISH analysis was estimated by calculating Cohen's kappa coefficient (κ) with the Kappa function of the R package vcd. We considered a κ value between 0.6 and 0.8, as good agreement and a value >0.8 as high concordance.
ImFISH reliability was studied by calculation of the sensitivity and the specificity of the method compared to FISH independently for each of the two observers' values.
Chi-square test was performed for group comparisons between FISH and ImFISH analysis for 20 cells, 40 cells and 60 cells. A significant correlation between two parameters was noted at the 95% confidence interval and a P value <0.05 was considered as a significant difference between two groups.
Mib1 labelling indices, histologic and clinical data were compared with each other according to the chromosomal status using logistic regression models. P values ≤0.05 were considered as statistically significant.
Results
Patient characteristics
Our 36 patients included 22 females and 14 males (respectively 60% and 40% of the cohort). The age of patients ranged from 26 to 82 years with a median age of 55 years, and 14 patients were younger than 50 years.
The patients underwent open surgery with gross total or partial tumor resection (n = 26; 72%) or stereotactic biopsies (n = 10; 28%).
For statistical analysis purposes tumor location was considered to be the lobe of the brain within which the largest volume of the glioma resided if more than one lobe was affected. In our series, tumor locations in order of decreasing frequency were frontal (n = 20; 55%), temporal (n = 10; 28%), parietal (n = 5; 14%) and insular lobe (n = 1; 3%).
Six cases among the 36 cases were recurrent tumors (17%).
Interpretation of ImFISH results
For a given sample area, the ImFISH technique generates two distinct images, MIB-1 IHC and 1p or 19q FISH, that are easily interpreted by the user. These images are easily stackable by image software for analysis together on the same screen or can be analyzed separately on two separate screens (Figure 2). We preferred the second solution for this study because of its simplicity, knowing that many people who examine chromosomal status by FISH do not necessarily use sophisticated image processing and image fusion software.
Figure 2. ImFISH interpretation.
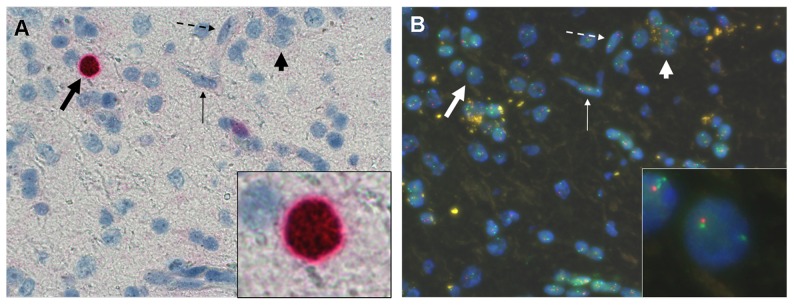
ImFISH technique allows a simultaneous analysis of nuclear staining with MIB-1 antibody by conventional immunohistochemistry (A) and in situ hybridization with chromosomal 1p and 19q probes (B) on two separate screens. The light haematoxylin counterstaining of the immunohistochemistry step allows an easy identification of the majority of the cells analyzed: oligodendrocytes (thick arrows), astrocytes (thin arrows), neurons (short arrows) and endothelial cells (dotted arrows). Only MIB-1 labeled nuclei with an oligodendroglial morphology are analysed by FISH (framed nuclei on A and B).
A phosphatase alkaline system using an AEC chromogen proved superior to the widely-used peroxidase system with its brown diaminobenzidine (DAB) chromogen. DAB chromogen remained adherent to the target despite successive washes causing non specific fluorescence and difficulties of interpretation during the FISH analysis, whereas AEC chromogen was very easy to remove with an ethanol bath.
After a short time to adapt to the two screen method used, the user can easily identify the positive nuclei on immunohistochemical labeling images and their counterparts on FISH images. An average of 15 minutes per case is necessary to analyze 60 cells, which is not much more longer than the time required in routine practice to count 100 to 200 non-overlapping cells by standard FISH technique in our experience at our institution.
The immunohistochemical counter-staining by hematoxylin in the MIB-1 IHC allows a better histological quality control of selected area than those disclosed by DAPI on FISH images. This allows a more detailed morphological analysis of the cell nuclei, and an easier identification of oligodendrocytes with their classical rounded nuclei (Figure 2, thick arrow); astrocytes displaying usually an elongated nuclei (Figure 2, thin arrow) and neurons displaying a prominent nucleoli (Figure 2, short arrow).
In anaplastic oligodendrogliomas and GBMO, nuclear MIB-1 labeling can be observed in neoplastic cells but also in some histiocytes associated with necrotic areas and some proliferating endothelial cells. To avoid these false positives in our study, we decided to exclude peri-necrotic areas from our analysis to prevent inadvertent inclusion of macrophages. Proliferating endothelial cells, for their part, were easily identified on counterstain and have been excluded from MIB-1 and FISH analysis (Figure 2, dotted arrow).
The sharpness of the nuclear staining with MIB-1 antibody facilitated nuclear identification and delineation even in the case of overlapping nuclear profiles, allowing the assessment of chromosome status for these cells which are usually excluded from analysis with standard FISH.
Inter-observer reliability of ImFISH technique
The inter-observer reliability of ImFISH was calculated by the Kappa score.
A good concordance was observed between the two readers by the ratio method for 1p (κ = 0.71) and 19q (κ = 0.72). These results are very similar to our internal institutional control values for the FISH technique. A higher concordance was observed by the combination method for both 1p (κ = 0.86) and 19q (κ = 0.95).
Comparison between FISH and ImFISH techniques
FISH yielded interpretable results in 100% of cases for 1p and 19q (Table 1). Chromosome 1p/19q alterations included both deletions (1p: n = 22 and 19q: n = 25 respectively 60% and 69%) and imbalances (1p: n = 9 and 19q: n = 8 respectively 25% and 22%). Combined 1p/19q deletion was detected in 22 cases (60%). Solitary 1p imbalance was noted in 2 cases (6%) and solitary 19q loss was noted in 3 cases (8%). In our series, no case showed 1p deletion alone. One case was imbalanced for both 1p and 19q. A minority of cases showed no abnormality (1p: n = 5 and19q: n = 3 respectively 14% and 8%).
Table 1. Histological grade, MIB1 labelling Index (LI) and 1p/19q ImmunoFISH and FISH status, by ratio and combination methods.
| Patient # | WHO Grade | MIB1 LI (%) | Ratio | Combination % | ||||||||||||||||||||||
| 1p | Status | 19q | Status | 1p | Status | 19q | Status | |||||||||||||||||||
| Obs. #1 | Obs. #2 | ImFISH | FISH | Obs. #1 | Obs. #2 | ImFISH | FISH | obs. #1 | obs. #2 | obs. #1 | obs. #2 | |||||||||||||||
| D | N | I | D | N | I | ImFISH | FISH | D | N | I | D | N | I | ImFISH | FISH | |||||||||||
| 13 | II | 3 | 1.02 | 1.14 | N/I | N | 1.02 | 1.02 | N | N | 20 | 55 | 25 | 15 | 40 | 45 | N | N | 15 | 70 | 15 | 15 | 50 | 35 | N | I * |
| 33 | II | 5 | 0.44 | 0.47 | D | D | 0.42 | 0.49 | D | D | 92 | 3 | 5 | 88 | 5 | 7 | D | D | 98 | 2 | 0 | 88 | 7 | 5 | D | D |
| 16 | II | 6 | 0.38 | 0.68 | D | D | 0.48 | 0.54 | D | D | 75 | 15 | 10 | 62 | 18 | 20 | D | D | 80 | 15 | 5 | 70 | 17 | 13 | D | D |
| 23 | II | 6 | 1.13 | 1.16 | I | N * | 0.81 | 0.95 | N | D * | 12 | 57 | 31 | 22 | 35 | 43 | N/I | N | 37 | 52 | 11 | 28 | 47 | 25 | N | D * |
| 24 | II | 6 | 0.44 | 0.60 | D | D | 0.46 | 0.51 | D | D | 90 | 8 | 2 | 82 | 7 | 12 | D | D | 82 | 10 | 8 | 90 | 5 | 5 | D | D |
| 31 | II | 6 | 0.43 | 0.55 | D | D | 0.5 | 0.67 | D | D | 87 | 12 | 1 | 80 | 10 | 10 | D | D | 80 | 13 | 7 | 67 | 12 | 22 | D | D |
| 6 | II | 7 | 0.8 | 0.79 | N | N | 0.93 | 0.87 | N | N | 38 | 23 | 39 | 43 | 13 | 43 | I | I | 13 | 60 | 27 | 27 | 48 | 25 | N | I * |
| 14 | II | 9 | 0.81 | 0.84 | N | N | 0.88 | 0.87 | N | N | 46 | 28 | 26 | 43 | 22 | 35 | I | I | 23 | 37 | 40 | 28 | 17 | 55 | I | I |
| 30 | II | 9 | 0.64 | 0.93 | D/N | D | 0.82 | 0.74 | D | D | 28 | 49 | 23 | 30 | 40 | 30 | N | D * | 32 | 51 | 17 | 32 | 47 | 22 | N | D * |
| 28 | II | 10 | 0.54 | 0.63 | D | D | 0.54 | 0.58 | D | D | 73 | 10 | 17 | 62 | 12 | 27 | D | D | 67 | 18 | 15 | 65 | 12 | 23 | D | D |
| 20 | II | 11 | 0.55 | 0.53 | D | D | 0.53 | 0.51 | D | D | 83 | 10 | 7 | 83 | 7 | 10 | D | D | 83 | 12 | 5 | 88 | 7 | 5 | D | D |
| 9 | III | 5 | 1 | 0.94 | N | N | 0.98 | 1.11 | N | N | 13 | 58 | 29 | 29 | 38 | 32 | N | I * | 23 | 59 | 18 | 20 | 43 | 37 | N | I * |
| 27 | III | 5 | 0.9 | 1.20 | N/I | D | 0.95 | 0.93 | N | D | 30 | 60 | 10 | 22 | 39 | 39 | N | D * | 10 | 76 | 14 | 20 | 57 | 23 | N | D * |
| 18 | III | 6 | 0.99 | 1.09 | N | I * | 1.02 | 1.11 | N/I | I | 22 | 57 | 21 | 18 | 43 | 38 | N | I * | 20 | 59 | 21 | 13 | 63 | 23 | N | I * |
| 25 | III | 8 | 0.48 | 0.58 | D | D | 0.44 | 0.50 | D | D | 80 | 10 | 10 | 75 | 8 | 17 | D | D | 75 | 8 | 17 | 80 | 8 | 12 | D | D |
| 1 | III | 10 | 0.55 | 0.62 | D | D | 0.58 | 0.53 | D | D | 67 | 13 | 20 | 60 | 3 | 37 | D | D | 68 | 15 | 17 | 65 | 3 | 32 | D | D |
| 12 | III | 11 | 0.99 | 1.01 | N | D * | 0.96 | 0.97 | N | N | 8 | 69 | 23 | 20 | 47 | 33 | N | I * | 12 | 48 | 40 | 7 | 77 | 17 | N | N |
| 4 | III | 12 | 1.01 | 1.05 | N | N | 0.43 | 0.43 | D | D | 8 | 65 | 27 | 10 | 47 | 43 | N | N | 85 | 12 | 3 | 85 | 12 | 3 | D | D |
| 11 | III | 12 | 1.19 | 1.26 | I | N | 0.95 | 1.01 | N | N | 8 | 50 | 42 | 10 | 37 | 53 | I | I | 12 | 57 | 31 | 17 | 40 | 43 | N | N |
| 26 | III | 13 | 0.5 | 0.54 | D | D | 0.43 | 0.55 | D | D | 88 | 5 | 7 | 92 | 2 | 7 | D | D | 87 | 7 | 6 | 80 | 8 | 12 | D | D |
| 34 | III | 13 | 0.49 | 0.46 | D | D | 0.49 | 0.59 | D | D | 90 | 3 | 7 | 92 | 3 | 5 | D | D | 88 | 7 | 5 | 77 | 15 | 8 | D | D |
| 21 | III | 16 | 0.49 | 0.56 | D | D | 0.5 | 0.55 | D | D | 87 | 8 | 5 | 78 | 15 | 7 | D | D | 87 | 6 | 7 | 75 | 13 | 12 | D | D |
| 32 | III | 20 | 0.518 | 0.61 | D | D | 0.53 | 0.53 | D | D | 67 | 5 | 28 | 52 | 10 | 38 | D | D | 83 | 13 | 4 | 80 | 12 | 8 | D | D |
| 2 | III | 21 | 0.52 | 0.50 | D | D | 0.52 | 0.49 | D | D | 83 | 7 | 10 | 83 | 3 | 13 | D | D | 82 | 12 | 6 | 87 | 8 | 5 | D | D |
| 17 | III | 21 | 0.5 | 0.49 | D | D | 0.58 | 0.54 | D | D | 75 | 7 | 18 | 65 | 5 | 30 | D | D | 53 | 15 | 32 | 58 | 7 | 35 | D | D |
| 5 | III | 22 | 0.57 | 0.49 | D | D | 0.52 | 0.57 | D | D | 73 | 10 | 17 | 75 | 12 | 13 | D | D | 68 | 10 | 22 | 65 | 13 | 22 | D | D |
| 8 | III | 22 | 1.1 | 1.03 | N | N | 1.03 | 1.17 | N/I | N | 22 | 52 | 26 | 15 | 25 | 60 | N/I | N | 17 | 53 | 30 | 13 | 40 | 47 | N | N |
| 36 | III | 23 | 0.52 | 0.54 | D | D | 0.45 | 0.57 | D | D | 65 | 15 | 20 | 59 | 13 | 28 | D | D | 78 | 7 | 15 | 60 | 17 | 23 | D | D |
| 7 | III | 26 | 1.23 | 1.47 | I | N * | 1.05 | 1.01 | N | N | 8 | 32 | 60 | 17 | 28 | 55 | I | I | 1 | 47 | 52 | 15 | 22 | 63 | I | I |
| 35 | III | 29 | 0.33 | 0.38 | D | D | 0.38 | 0.42 | D | D | 94 | 3 | 3 | 93 | 3 | 3 | D | D | 83 | 7 | 10 | 87 | 2 | 12 | D | D |
| 15 | III | 34 | 0.84 | 0.89 | N | N | 0.65 | 0.70 | D | D | 37 | 37 | 26 | 28 | 22 | 50 | I | I | 57 | 28 | 15 | 42 | 15 | 43 | I | I |
| 22 | III | 34 | 0.57 | 0.51 | D | D | 0.45 | 0.60 | D | D | 74 | 13 | 13 | 75 | 3 | 22 | D | D | 78 | 5 | 17 | 53 | 15 | 32 | D | D |
| 29 | III | 34 | 0.52 | 0.53 | D | D | 0.45 | 0.59 | D | D | 65 | 10 | 25 | 60 | 17 | 23 | D | D | 77 | 5 | 18 | 65 | 10 | 25 | D | D |
| 10 | IV | 14 | 1.37 | 1.27 | I | N * | 1.03 | 1.09 | N | N | 8 | 43 | 49 | 17 | 37 | 47 | I | I | 13 | 54 | 33 | 12 | 33 | 55 | N/I | I |
| 19 | IV | 14 | 0.51 | 0.62 | D | D | 0.55 | 0.51 | D | D | 70 | 22 | 8 | 73 | 20 | 7 | D | D | 67 | 20 | 13 | 72 | 15 | 13 | D | D |
| 3 | IV | 19 | 1.05 | 1.16 | N/I | N | 0.54 | 0.49 | D | D | 18 | 47 | 35 | 13 | 37 | 50 | N | N | 83 | 10 | 7 | 92 | 3 | 5 | D | D |
D: deletion, N: no deletion and no imbalance, I: imbalance
*ImFISH/FISH discordance
ImFISH was calculated on 60 nuclei and FISH on 100.
ImFISH also yielded interpretable results in 100% of cases for 1p and 19q (Table 1) however in 4 out of 72 tests (36 for 1p and 36 for 19q) the technique failed to identify 60 MIB-1 positive cells as initially planned (mean∶31 nuclei; min∶20 – max∶47). Chromosome 1p/19q alterations included both deletions (1p: n = 20 and 19q: n = 22 respectively 55% and 60%) and imbalances (1p: n = 6 and 19q: n = 3 respectively 17% and 8%). Combined 1p/19q deletion was detected in 20 cases (55%). Solitary 1p imbalance was noted in 3 cases (8%) and solitary 19q loss was noted in 2 cases (6%). No case showed 1p deletion alone and one case was imbalanced for both 1p and 19q. A minority of cases showed no abnormality (1p: n = 10 and 19q: n = 11 respectively 28% and 31%).
Compared to FISH, the ImFISH exhibits a very high sensitivity (100%) and a high specificity (91%) for 1p and/or 19q deleted cases. Combination and ratio methods give nearly identical results (Table 2).
Table 2. Sensitivity and specificity of ImFISH according to the FISH.
| Combination | Ratio | |||||
| D | N | I | D | N | I | |
| Sensitivity 1p | 1 | 0.84 | 0.94 | 0.96 | 0.87 | 0.82 |
| Specificity 1p | 0.91 | 0.7 | 0.67 | 0.89 | 0.5 | 0 |
| Sensitivity 19q | 1 | 0.77 | 1 | 1 | 0.89 | 0.95 |
| Specificity 19q | 0.88 | 1 | 0.44 | 0.9 | 0.83 | 0.5 |
Sensitivity and specificity was calculated for each of the 2 observers and for each of the chromosomal analysis method (combination and ratio) on 36 cases. The sum of both results is reported here (total n = 72).
The sensitivity of the technique decreases slightly for normal cases (Combination∶84%–Ratio∶87%) for both 1p and 19q. The specificity also decreases slightly for 19q normal status (Combination∶100%–Ratio∶83%) but much more for 1p normal status (Combination∶70%–Ratio∶50%).
For imbalanced cases the sensitivity remains high for both 1p (∼90%) and 19q (∼100%) imbalances regardless of the method of calculation used. Nevertheless the specificity decreases strongly to roughly 50% for both 1p and 19q imbalances (Table 2).
ImFISH results according to the number of cells analyzed
Study of the minimal number of cells needed for ImFISH analysis was made on three decreasing series of cells (60 cells, 40 cells and 20 cells). The final result of each of these series was compared to the results initially obtained by FISH (Table 3). This study was performed on 34 of our 36 cases; in two cases ImFISH failed to label at least 60 cells. Results were divided into subgroups of deleted, normal and imbalanced status and did not show a statistical difference by the Chi-square test between the cohorts of the different subgroups obtained by FISH and ImFISH technique. Likewise there were no significant difference between FISH and ImFISH results calculated on 60, 40 or 20 cells and this regardless of the method of calculation used (combination or ratio) or the type of chromosomal status studied (Chi-square test not significant). The results from both observers were very similar; hence we have shown the results of only one observer in Table 3.
Table 3. Comparison between FISH and ImFISH results.
| Combination | Ratio | |||||||
| FISH | ImFISH 20 | ImFISH 40 | ImFISH 60 | FISH | ImFISH 20 | ImFISH 40 | ImFISH 60 | |
| Codeletion | 21 | 17 | 18 | 20 | 21 | 22 | 21 | 20 |
| Deletion 1p | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Deletion 19q | 3 | 4 | 3 | 2 | 4 | 3 | 3 | 4 |
| Normal 1p | 5 | 7 | 8 | 6 | 11 | 5 | 7 | 8 |
| Normal 19q | 3 | 8 | 8 | 8 | 8 | 9 | 9 | 9 |
| Imbalancement 1p | 8 | 9 | 7 | 7 | 1 | 6 | 5 | 5 |
| Imbalancement 19q | 7 | 5 | 5 | 4 | 1 | 0 | 1 | 1 |
| Chi-square test | NS | NS | NS | NS | NS | NS | ||
NS = non significant.
Number of final diagnoses obtained by FISH and ImFISH on 20, 40 and 60 cells respectively and for each combination and ratio chromosomal analysis method. Chi-square test was applied between FISH and each of three ImFISH trials.
It should be noted that our results show that the calculation method by combination or ratio does not always produce the same result, especially for normal and imbalanced chromosome status (Table 1 and 3).
Comparison of chromosomal status with clinical, surgical and immunohistochemical data
Loss of 1p and 19q was observed in 6 (of 11) (55%) of OII, 13 (of 22) (59%) of OIII and 1 (of 3) (33%) of GBMO.
This codeletion was observed in 16 (of 30) (53%) of primary tumors and 4 (of 6) (66%) of recurrent tumors.
The frequency of 1p/19q alterations was not significantly different in WHO Grade II, Grade III or Grade IV tumors or in primary and recurrent tumors.
No correlation was observed between the sex or the age of the population and the frequency of the 1p/19q status.
Loss of 1p and 19q was identified in 15 (of 20) (75%) of frontal, 2 (of 10) (20%) of temporal, 2 (of 5) (40%) of parietal and 1 (of 1) (100%) of insular located tumors. This repartition was statistically significant (p<0.05). When analyzed separately only 1p deletion remains significantly associated with the cerebral location: 75% of the 1p deleted tumors are located in the frontal lobe (p = 0.01) and 50% of the 1p normal status tumors are located in the temporal lobe (p = 0.01).
In our series MIB-1 immunolabelling appeared as nuclear staining (Figure 1). MIB-1 mean indices ranged from 7% for OII (min: 3 – max: 11), 18% for OIII (min: 5 – max: 34) and 16% for GBMO (min: 14 – max: 19). These proliferative index values was statistically significant between the low grade and the high grade tumors (p<0.01).
Logistic regression analysis observed no significant correlation between the MIB-1 labelling index (LI) and the 1p and/or the 19q deletion.
Discussion
Our modified ImFISH technique is easy to perform and to analyse, requiring a short time to learn. In the absence of clearly established guidelines for ImFISH use in the literature, we favoured an unbiased approach performing a double independent observer analysis for each step in our study, and using both commonly used methods of FISH chromosomal status reporting∶i.e. combination and ratio.
The Kappa test shows a good concordance between the results for both observers. This concordance is higher for the combination method than the ratio method. These results may seem surprising given that our cut off for the combination method was established theoretically. However, our results highlight the fact that the two common methods for calculating the chromosomal status, although convergent in most cases are not necessarily superimposable. The combination method is more adapted to detect small groups of cells with a different chromosomal profile while the ratio method gives an overall average of all examined cells and appears less appropriate to highlight a particular chromosomal profile from a small group of cells. In the literature, the use of one method or the other to determine the chromosomal status of 1p and 19q is a matter of institutional practice and there is no clear evidence that it is better to use of one method or another.
ImFISH inter-observer results are equivalent or superior to those accepted for the FISH in our internal institutional practice. This emphasizes the benefits of the ImFISH when focusing the cellular analysis on a limited group of pre-selected cells (here MIB-1 immunoreactive cells) which reduces the field of analysis for the observer and increases greatly the probability that two independent observers will analyze the same cells.
In this series, the sensitivity of ImFISH compared to FISH is high regardless of the method of calculation used (combination or ratio) (Table 2). This confirms the value of this technique in identifying true positive cases while avoiding false negatives. Sensitivity appears satisfactory for deleted and normal cases but inadequate for imbalanced cases. Some cases are imbalanced by FISH and normal by ImFISH and vice versa, regardless of the method of calculation used (Table 1). This discrepancy is important to study further given the growing awareness of importance of the 1p/19q imbalanced population in the prognosis of oligodendroglial tumours. Recent studies show a strong correlation between the presence of an imbalanced tumor cell population and a shorter overall survival [12], [21]. Analysis of imbalanced cases by FISH is known to pose problems of interpretation due to chromosomal polyploidy and multiple probe signals [23]. In our series, changing the cut-off values used for ImFISH or extending the analysis to 100 MIB-1 labelled tumor cells instead of 60 did not eliminate the discrepancy (data not shown), suggesting a real difference in cell analysis by these two techniques, since only the MIB-1 immunoreactive cells are analyzed by ImFISH whereas in FISH cell selection is independent of cell type or cell cycle status. This difference may explain the selection by ImFISH of many more potentially false positive cells than by FISH, thereby decreasing the value of its specificity without jeopardizing its sensitivity. We feel that these potentially false positive cells are most likely genetically unstable true-positive cells not taken into account by the standard FISH technique. The present study contains few imbalanced cases, so analysis of a larger cohort would be necessary to verify these results. Other molecular analysis techniques such as LOH analysis or CGH may also be of interest to characterize the discordant cases between FISH and ImFISH. These techniques are efficient in highly cellular tumor cases since at least 70% of the cells need to be part of the tumor for a satisfactory interpretation, but they are not appropriate for tumors of low cellularity for which FISH and ImFISH are more suitable [35], [37]. For tumors of low cellularity, a correlation with overall survival data would be necessary to determine which imbalanced cases by FISH or ImFISH are technical true positives but biological false negatives.
In our study, ImFISH technique gives substantially similar results to FISH even having analyzed 60, 40 or just 20 cells. These results were expected for deleted cases because these latter cases are usually very straightforward and easily analyzable even on a small number of cells [26], [45]. To our surprise, the results of normal and imbalanced cases, although less similar, remain statistically significant and underline the interest of the ImFISH. Unfortunately, for normal and imbalanced cases, the FISH literature does not offer clear quantitative data allowing us to more detailed comparisons between these two techniques.
Nevertheless, our results highlight the benefits of practicing ImFISH on tumor samples of low cellularity (diffuse low grade gliomas, periphery of the tumor or biopsy material) since the analysis of 20 cells still allows to provide a satisfactory result on 1p and 19q chromosomal status (Table 3).
Despite the preselected character of our series, the majority of our cases were codeleted as typically described in the literature [13], [17], [22], [30], [59] and there was no chromosomal abnormalities differences between the primary and the recurrent tumoral population underlining the genetic stability of the majority of recurrent oligodendrogliomas [23].
In our series, there was a significant association between frontal lobe location and the allelic loss of chromosomes 1p and 19q and between temporal lobe location and maintenance of 1p/19q status as described in many studies of the literature [1], [18], [60], [61]. These findings suggest that molecular subsets of oligodendroglial tumors might arise from site-specific precursor cells and arise preferentially in certain lobes, with tumors having deletion on 1p and 19q occurring most frequently in the nontemporal lobes [34], [60], [62].
Finally MIB-1 LI appears useful for the discrimination between low and high grade oligodendroglioma but did not show a significant association with 1p19q deletion status in concordance with the literature [34], [58], [63], [64]. These results suggest that the neoplastic protective effect observed in 1p19q deleted oligodendrogliomas does not involve the cell cycle mechanism.
Conclusion
The ImFISH technique has never been applied before in the study of gliomas to our knowledge. The technique described in this study is easy to perform and analyze and does not require additional technical equipment or interpretation expertise to be achieved. It is reproducible and generates similar results to the classic FISH technique. It appears best suited to low density tumoral areas or poorly cellular tumoral samples (low grade gliomas, peritumoral area or biopsy material).
The present study lacks a correlation between molecular markers and survival times because the length of follow-up for our patient cohort is too short for meaningful statistical analysis. Future studies are planned to broaden the application of this technique which offers many advantages for the study of pure or mixed oligodendrogliomas and other gliomas. Among the possibilities we note the interest to combine immunohistochemical analysis of other known prognostic factors in gliomas such as TP53, MGMT, EGFR and IDH1 expression with the chromosome 1p and 19q status within a given sub-population of tumor cells [9], [19], [34], [64], [65], [66]. Other oligodendroglial chromosomal abnormalities such as gain or loss of chromosome 10 or 7 could also been studied by this technique [64], [67], [68].
Finally ImFISH analysis can be easily automated, which may lead to easier validation of automated image analysis software for FISH within each institution. By focusing analysis on a limited number of pre-selected cells, ImFISH removes a potential source of inter-observer variability whether a human or computer examines the cells.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All data are included within the manuscript.
Funding Statement
This study was supported by the Fondation des hôpitaux Enfant-Jésus – Saint Sacrement (Quebec city - Quebec): grant number 682/8317. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Reifenberger G, Louis DN (2003) Oligodendroglioma: Toward molecular definitions in diagnostic neuro-oncology. J Neuropathol Exp Neurol 62: 111–126. [DOI] [PubMed] [Google Scholar]
- 2. Van den Bent MJ, Looijenga LHJ, Langenberg K, Dinjens W, Graveland W, et al. (2003) Chromosomal anomalies in oligodendroglial tumors are correlated with clinical features. Cancer 97: 1276–1284. [DOI] [PubMed] [Google Scholar]
- 3. Fuller CE, Perry A (2005) Molecular diagnostics in central nervous system tumors. Adv Anat Pathol 12: 180–194. [DOI] [PubMed] [Google Scholar]
- 4. Hartmann C, Mueller W, Lass U, Kamel-Reid S, von Deimling A (2005) Molecular genetic analysis of oligodendroglial tumors. J Neuropathol Exp Neurol 64: 10–14. [DOI] [PubMed] [Google Scholar]
- 5. Kanner AA, Staugaitis SM, Castilla EA, Chernova O, Prayson RA, et al. (2006) The impact of genotype on outcome in oligodendroglioma: validation of the loss of chromosome arm 1p as an important factor in clinical decision making. J Neurosurg 104: 542–550. [DOI] [PubMed] [Google Scholar]
- 6. Aldape K, Burger PC, Perry A (2007) Clinicopathologic aspects of 1p/19q loss and the diagnosis of oligodendroglioma. Arch Pathol Lab Med 131: 242–251. [DOI] [PubMed] [Google Scholar]
- 7. Cairncross G, Jenkins R (2008) Gliomas with 1p/19q codeletion: a.k.a. oligodendroglioma. Cancer J 14: 352–357. [DOI] [PubMed] [Google Scholar]
- 8. Gadji M, Fortin D, Tsanaclis AM, Drouin R (2009) Is the 1p/19q deletion a diagnostic marker of oligodendrogliomas? Cancer Genet Cytogenet 194: 12–22. [DOI] [PubMed] [Google Scholar]
- 9. Hirose T, Ishizawa K, Shimada S (2010) Utility of in situ demonstration of 1p loss and p53 overexpression in pathologic diagnosis of oligodendroglial tumors. Neuropathology 30: 586–596. [DOI] [PubMed] [Google Scholar]
- 10. Ramirez C, Bowman C, Maurage CA, Dubois F, Blond S, et al. (2010) Loss of 1p, 19q, and 10q heterozygosity prospectively predicts prognosis of oligodendroglial tumors—towards individualized tumor treatment? Neuro Oncol 12: 490–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hartmann C, Hentschel B, Tatagiba M, Schramm J, Schnell O, et al. (2011) Molecular markers in low-grade gliomas: predictive or prognostic? Clin Cancer Res 17: 4588–4599. [DOI] [PubMed] [Google Scholar]
- 12. Wiens AL, Cheng L, Bertsch EC, Johnson KA, Zhang S, et al. (2012) Polysomy of chromosomes 1 and/or 19 is common and associated with less favorable clinical outcome in oligodendrogliomas: fluorescent in situ hybridization analysis of 84 consecutive cases. J Neuropathol Exp Neurol 71: 618–624. [DOI] [PubMed] [Google Scholar]
- 13. Smith JS, Alderete B, Minn Y, Borell TJ, Perry A, et al. (1999) Localization of common deletion regions on 1p and 19q in human gliomas and their association with histological subtype. Oncogene 18: 4144–4152. [DOI] [PubMed] [Google Scholar]
- 14. McDonald JM, See SJ, Tremont IW, Colman H, Gilbert MR, et al. (2005) The prognostic impact of histology and 1p/19q status in anaplastic oligodendroglial tumors. Cancer 104: 1468–1477. [DOI] [PubMed] [Google Scholar]
- 15. Jenkins RB, Blair H, Ballman KV, Giannini C, Arusell RM, et al. (2006) A t(1;19)(q10;p10) mediates the combined deletions of 1p and 19q and predicts a better prognosis of patients with oligodendroglioma. Cancer Res 66: 9852–9861. [DOI] [PubMed] [Google Scholar]
- 16. Kouwenhoven MCM, Kros JM, French PJ, Biemond-ter Stege EM, Graveland WJ, et al. (2006) 1p/19q loss within oligodendroglioma is predictive for response to first line temozolomide but not to salvage treatment. Europ J Cancer 42: 2499–2503. [DOI] [PubMed] [Google Scholar]
- 17. Kros JM, GorliaT, Kouwenhoven MC, Zheng PP, Collins VP, et al. (2007) Panel review of anaplastic oligodendroglioma from European Organization for Research and Treatment of Cancer trial 26951: assessment of consensus in diagnosis, influence of 1p/19q loss, and correlations with outcome. J Neuropathol Exp Neurol 66: 545–551. [DOI] [PubMed] [Google Scholar]
- 18. Giannini C, Burger PC, Berkey BA, Cairncross JG, Jenkins RB, et al. (2008) Anaplastic oligodendroglial tumors: refining the correlation among histopathology, 1p 19q deletion and clinical outcome in intergroup radiation therapy oncology group trial 9402. Brain Pathol 18: 360–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kanamori M, Kumabe T, Sonoda Y, Nishino Y, Watanabe M, et al. (2009) Predictive factors for overall and progression-free survival, and dissemination in oligodendroglial tumors. J Neurooncol 93: 219–228. [DOI] [PubMed] [Google Scholar]
- 20. Scheie D, Meling TR, Cvancarova M, Skullerud K, Mørk S, et al. (2011) Prognostic variables in oligodendroglial tumors: a single-institution study of 95 cases. Neuro Oncol 13: 1225–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Senetta R, Verdun di Cantogno L, Chiusa L, Castellano I, Gugliotta P, et al. (2013) A “weighted” fluorescence in situ hybridization strengthens the favorable prognostic value of 1p/19q codeletion in pure and mixed oligodendroglial tumors. J Neuropathol Exp Neurol 72: 432–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fuller CE, Perry A (2002) Fluorescence In Situ Hybridization (FISH) in diagnostic and investigative neuropathology. Brain Pathol 12: 67–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fallon KB, Palmer CA, Roth KA, Nabors LB, Wang W, et al. (2004) Prognostic value of 1p, 19q, 9p, 10q, and EGFR-FISH analyses in recurrent oligodendrogliomas. J Neuropathol Exp Neurol 63: 314–322. [DOI] [PubMed] [Google Scholar]
- 24. Gonzales M, Dale S, Susman M, Mills J (2006) Quantitation of chromosome 1p and 19q deletions in glial tumours by interphase FISH on formalin-fixed paraffin-embedded tissue. J Clin Neurosci 13: 96–101. [DOI] [PubMed] [Google Scholar]
- 25. Jeon YK, Park K, Park CK, Paek SH, Jung HW, et al. (2007) Chromosome 1p and 19q status and p53 and p16 expression patterns as prognostic indicators of oligodendroglial tumors: a clinicopathological study using fluorescence in situ hybridization. Neuropathology 27: 10–20. [DOI] [PubMed] [Google Scholar]
- 26. Reddy KS (2008) Assessment of 1p/19q deletions by fluorescence in situ hybridization in gliomas. Cancer Genet Cytogenet 184: 77–86. [DOI] [PubMed] [Google Scholar]
- 27. Burger PC, Minn AY, Smith JS, Borell TJ, Jedlicka AE, et al. (2001) Losses of Chromosomal Arms 1p and 19q in the Diagnosis of Oligodendroglioma. A Study of Paraffin-Embedded Sections. Mod Pathol 14: 842–853. [DOI] [PubMed] [Google Scholar]
- 28. Broholm H, Born PW, Guterbaum D, Dyrbye H, Laursen H (2008) Detecting chromosomal alterations at 1p and 19q by FISH and DNA fragment analysis – a comparative study in human gliomas. Clin Neuropathol 27: 378–387. [DOI] [PubMed] [Google Scholar]
- 29. Nigro JM, Takahashi MA, Ginzinger DG, Law M, Passe S, et al. (2001) Detection of 1p and 19q loss in oligodendroglioma by quantitative microsatellite analysis, a real-time quantitative polymerase chain reaction assay. Am J Pathol 158: 1253–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Smith JS, Perry A, Borell TJ, Lee HK, O'Fallon J, et al. (2000) Alterations of chromosome arms 1p and 19q as predictors of survival in oligodendrogliomas, astrocytomas, and mixed oligoastrocytomas. J Clin Oncol 18: 636–645. [DOI] [PubMed] [Google Scholar]
- 31. Sasaki H, Zlatescu MC, Betensky RA, Johnk LB, Cutone AN, et al. (2002) Histopathological-molecular genetic correlations in referral pathologist-diagnosed low-grade “oligodendroglioma”. J Neuropathol Exp Neurol 61: 58–63. [DOI] [PubMed] [Google Scholar]
- 32. Law ME, Templeton KL, Kitange G, Smith J, Misra A, et al. (2005) Molecular cytogenetic analysis of chromosomes 1 and 19 in glioma cell lines. Cancer Genet Cytogenet 160: 1–14. [DOI] [PubMed] [Google Scholar]
- 33. Mohapatra G, Betensky RA, Miller ER, Carey B, Gaumont LD, et al. (2006) Glioma test array for use with formalin-fixed, paraffin-embedded tissue array comparative genomic hybridization correlates with loss of heterozygosity and fluorescence in situ hybridization. J Mol Diagn 8: 268–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Huang L, Jiang T, Yuan F, Li GL, Liu EZ, et al. (2009) Correlation of chromosomes 1p and 19q status and expressions of O6-methylguanine DNA methyltransferase (MGMT), p53 and Ki-67 in diffuse gliomas of World Health Organization (WHO) grades II and III: a clinicopathological study. Neuropathol Appl Neurobiol 35: 367–379. [DOI] [PubMed] [Google Scholar]
- 35. Jha P, Sarkar C, Pathak P, Sharma MC, Kale SS, et al. (2011) Detection of allelic status of 1p and 19q by microsatellite-based PCR versus FISH limitations and advantages in application to patient management. Diagn Mol Pathol 20: 40–47. [DOI] [PubMed] [Google Scholar]
- 36. Bigner SH, Matthews MR, Rasheed BKA, Wiltshire RN, Friedman HS, et al. (1999) Molecular genetic aspects of oligodendrogliomas including analysis by comparative genomic hybridization. Am J Pathol 155: 375–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Koschny R, Koschny T, Froster UG, Krupp W, Zuber MA (2002) Comparative genomic hybridization in glioma: a meta-analysis of 509 cases. Cancer Genet Cytogenet 135: 147–159. [DOI] [PubMed] [Google Scholar]
- 38. Kitange G, Misra A, Law M, Passe S, Kollmeyer TM, et al. (2005) Chromosomal imbalances detected by array comparative genomic hybridization in human oligodendrogliomas and mixed oligoastrocytomas. Genes Chromosomes Cancer 42: 68–77. [DOI] [PubMed] [Google Scholar]
- 39. Koschny R, Holland H, Koschny T, Vitzthum HE (2006) Comparative genomic hybridization pattern of non-anaplastic and anaplastic oligodendrogliomas – A meta-analysis. Pathol Res Pract 202: 23–30. [DOI] [PubMed] [Google Scholar]
- 40. Trost D, Ehrler M, Fimmers R, Felsberg J, Sabel MC, et al. (2007) Identification of genomic aberrations associated with shorter overall survival in patients with oligodendroglial tumors. Int J Cancer 120: 2368–2376. [DOI] [PubMed] [Google Scholar]
- 41. Ducray F, Idbah A, de Reyniès A, Bièche I, Thillet J, et al. (2008) Anaplastic oligodendrogliomas with 1p19q codeletion have a proneural gene expression profile. Mol Cancer 7: 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Franco-Hernandez C, Martinez-Glez V, de Campos JM, Isla A, Vaquero J, et al. (2009) Allelic status of 1p and 19q in oligodendrogliomas and glioblastomas: multiplex ligation-dependent probe amplification versus loss of heterozygosity. Cancer Genet Cytogenet 190: 93–96. [DOI] [PubMed] [Google Scholar]
- 43. Ambros PF, Ambros IM (2001) Pathology and biology guidelines for resectable and unresectable neuroblastic tumors and bone marrow examination guidelines. Med Pediatr Oncol 37: 492–504. [DOI] [PubMed] [Google Scholar]
- 44. Belaud-Rotureau MA, Meunier N, Eimer S, Vital A, Loiseau H, et al. (2006) Automatized assessment of 1p36-19q13 status in gliomas by interphase FISH assay on touch imprints of frozen tumours. Acta Neuropathol 111: 255–263. [DOI] [PubMed] [Google Scholar]
- 45. Woehrer A, Sander P, Haberler C, Kern S, Maier H, et al. (2011) FISH-based detection of 1p 19q codeletion in oligodendroglial tumors: procedures and protocols for neuropathological practice – a publication under the auspices of the research committee of the European Confederation of Neuropathological Societies (Euro-CNS). Clin Neuropathol 30: 47–55. [DOI] [PubMed] [Google Scholar]
- 46. Gelpi E, Ambros IM, Birner P, Luegmayr A, Drlicek M, et al. (2003) Fluorescent In Situ Hybridization on isolated tumor cell nuclei: a sensitive method for 1p and 19q deletion analysis in paraffin-embedded oligodendroglial tumor specimens. Mod Pathol 16: 708–715. [DOI] [PubMed] [Google Scholar]
- 47. Horbinski C, Hamilton RL, Nikiforov Y, Pollack IF (2010) Association of molecular alterations, including BRAF, with biology and outcome in pilocytic astrocytomas. Acta Neuropathol 119: 641–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bouvier C, Roll P, Quilichini B, Metellus P, Calisti A, et al. (2004) Deletions of chromosomes 1p and 19q are detectable on frozen smears of gliomas by FISH: usefulness for stereotactic biopsies. J Neurooncol 68: 141–149. [DOI] [PubMed] [Google Scholar]
- 49. Scheie D, Andresen PA, Cvancarova M, Bø AS, Helseth E, et al. (2006) Fluorescence in situ hybridization (FISH) on touch preparations: a reliable method for detecting loss of heterozygosity at 1p and 19q in oligodendroglial tumors. Am J Surg Pathol 30: 828–837. [DOI] [PubMed] [Google Scholar]
- 50. Lass U, Hartmann C, Capper D, Herold-Mende C, von Deimling A, et al. (2013) Chromogenic in situ hybridization is a reliable alternative to fluorescence in situ hybridization for diagnostic testing of 1p and 19q loss in paraffin-embedded gliomas. Brain Pathol 23: 311–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Weber-Matthiesen K, Winkemann M, Müller-Hermelink A, Schlegelberger B, Grote W (1992) Simultaneous fluorescence immunophenotyping and interphase cytogenetics: a contribution to the characterization of tumor cells. J Histochem Cytochem 40: 171–175. [DOI] [PubMed] [Google Scholar]
- 52. Martinez-Ramirez A, Cigudosa JC, Maestre L, Rodriguez-Perales S, Haralambieva E, et al. (2004) Simultaneous detection of the immunophenotypic markers and genetic aberrations on routinely processed paraffin sections of lymphoma samples by means of the FICTION technique. Leukemia 18: 348–353. [DOI] [PubMed] [Google Scholar]
- 53. Calattini S, Sereti I, Scheinberg P, Kimura H, Childs RW, et al. (2010) Detection of EBV genomes in plasmablasts/plasma cells and non-B cells in the blood of most patients with EBV lymphoproliferative disorders by using Immuno-FISH. Blood 116: 4546–4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Perry A, Roth KA, Banerjee R, Fuller CE, Gutmann DH (2001) NF1 deletions in S-100 protein-positive and negative cells of sporadic and neurofibromatosis 1 (NF1)- associated plexiform neurofibromas and malignant peripheral nerve sheath tumors. Am J Pathol 159: 57–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Gatta LB, Incardona P, Cadei M, Grigolato P, Simoncelli S, et al. (2012) Simultaneous fluorescence immunophenotyping and Her-2/neu genotyping (FICTION) in breast carcinoma candidates to target therapy. Appl Immunohistochem Mol Morphol 20: 413–420. [DOI] [PubMed] [Google Scholar]
- 56. Trudel D, Zafarana G, Sykes J, Have CL, Bristow RG, et al. (2013) 4FISH-IF, a four-color dual-gene FISH combined with p63 immunofluorescence to evaluate NKX3.1 and MYC status in prostate cancer. J Histochem Cytochem 61: 500–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Campos M, Luque R, Jiménez J, Martinez R, Warleta F, et al. (2013) Simultaneous phenotypic and genetic characterization of single circulating tumor cells from colon cancer patients. Histol Histopathol 28: 1439–1450. [DOI] [PubMed] [Google Scholar]
- 58.Louis DN, Ohgaki H, Wiestler OD, Cavenee KC (2007) WHO Classification of Tumours of the Central Nervous System. International Agency for Research on Cancer (IARC): Lyon.
- 59. Bigner SH, Rasheed BKA, Wiltshire R, McLendon RE (1999) Morphologic and molecular genetic aspects of oligodendroglial neoplasms1. Neuro Oncol 1: 52–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zlatescu MC, Tehraniyazdi A, Sasaki H, Megyesi JF, Betensky RA, et al. (2001) Tumor location and growth pattern correlate with genetic signature in oligodendroglial neoplasms. Cancer Res 61: 6713–6715. [PubMed] [Google Scholar]
- 61. Snuderl M, Eichler AF, Ligon KL, Vu Qu, Silver M, et al. (2009) Polysomy for chromosomes 1 and 19 predicts earlier recurrence in anaplastic oligodendrogliomas with concurrent 1p/19q loss. Clin Cancer Res 15: 6430–6437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Mueller W, Hartmann C, Hoffmann A, Lanksch W, Kiwit J, et al. (2002) Genetic signature of oligoastrocytomas correlates with tumor location and denotes distinct molecular subsets. Am J Pathol 161: 313–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Coons SW, Johnson PC, Perl DK (1997) The prognostic significance of Ki-67 labeling indices for oligodendroglioma. Neurosurgery 41: 878–885. [DOI] [PubMed] [Google Scholar]
- 64. Preusser M, Hoeftberger R, Woehrer A, Gelpi E, Kouwenhoven M, et al. (2012) Prognostic value of Ki67 index in anaplastic oligodendroglial tumours – a translational study of the European Organization for Research and Treatment of Cancer Brain Tumor Group. Histopathology 60: 885–894. [DOI] [PubMed] [Google Scholar]
- 65. Mahajan D, Prayson RA (2009) Repeated molecular testing in gliomas. A retrospective study of 53 Cases. Am J Clin Pathol 132: 118–124. [DOI] [PubMed] [Google Scholar]
- 66. Shukla B, Agarwal S, Suri V, Pathak P, Sharma MC, et al. (2009) Assessment of 1p/19q status by fluorescence in situ hybridization assay: a comparative study in oligodendroglial, mixed oligoastrocytic and astrocytic tumors. Neurol India 57: 559–566. [DOI] [PubMed] [Google Scholar]
- 67. Horbinski C, Miller CR, Perry A (2011) Gone FISHing: clinical lessons learned in brain tumor molecular diagnostics over the last decade. Brain Pathol 21: 57–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Horbinski C, Nikiforova MN, Hobbs J, Bortoluzzi S, Cieply K, et al. (2012) The importance of 10q status in an outcomes-based comparison between 1p/19q fluorescence in situ hybridization and polymerase chain reaction-based microsatellite loss of heterozygosity analysis of oligodendrogliomas. J Neuropathol Exp Neurol 71: 73–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All data are included within the manuscript.