Abstract
Fanconi anemia (FA) is a chromosome instability syndrome characterized by increased cancer predisposition. Within the FA pathway, an upstream FA core complex mediates monoubiquitination and recruitment of the central FANCD2 protein to sites of stalled replication forks. Once recruited, FANCD2 fulfills a dual role towards replication fork recovery: (i) it cooperates with BRCA2 and RAD51 to protect forks from nucleolytic degradation and (ii) it recruits the BLM helicase to promote replication fork restart while suppressing new origin firing. Intriguingly, FANCD2 and its interaction partners are also involved in homologous recombination (HR) repair of DNA double-strand breaks, hinting that FANCD2 utilizes HR proteins to mediate replication fork recovery. One such candidate is CtIP (CtBP-interacting protein), a key HR repair factor that functions in complex with BRCA1 and MRE11, but has not been investigated as putative player in the replication stress response. Here, we identify CtIP as a novel interaction partner of FANCD2. CtIP binds and stabilizes FANCD2 in a DNA damage- and FA core complex-independent manner, suggesting that FANCD2 monoubiquitination is dispensable for its interaction with CtIP. Following cellular treatment with a replication inhibitor, aphidicolin, FANCD2 recruits CtIP to transiently stalled, as well as collapsed, replication forks on chromatin. At stalled forks, CtIP cooperates with FANCD2 to promote fork restart and the suppression of new origin firing. Both functions are dependent on BRCA1 that controls the step-wise recruitment of MRE11, FANCD2 and finally CtIP to stalled replication forks, followed by their concerted actions to promote fork recovery.
INTRODUCTION
Fanconi anemia (FA) is a recessively inherited genome instability syndrome characterized by bone marrow failure and cancer predisposition. FA patient cells are sensitive to DNA interstrand crosslinks (ICLs) and show spontaneous chromosomal aberrations that are further exacerbated upon treatment with replication-inhibiting agents such as hydroxyurea (HU) or aphidicolin (APH) (1,2). The 16 known FA proteins participate in a common pathway. Following replication fork stalling, an upstream FA core complex (8 FA proteins) monoubiquitinates the central FA pathway members FANCD2 and FANCI, followed by their recruitment to chromatin and into DNA repair foci (3,4). Monoubiquitinated FANCD2 (FANCD2Ub) functions to recruit DNA repair factors FAN1 (Fanconi-associated nuclease 1) (5–8) and SLX4 (identical to FANCP; a Holliday junction resolvase in complex with SLX1) (9–12), suggesting that chromatin-bound FANCD2Ub is a docking platform for some DNA repair nucleases. Positioned further downstream in the FA pathway are several breast cancer-associated proteins including FANCD1/BRCA2 (breast cancer-associated protein 2), which coordinates with upstream FA pathway members to promote homologous recombination (HR) repair of DNA double-stranded breaks (DNA DSBs) (13–15).
Three new studies shed light on additional FA pathway functions at sites of stalled replication forks: Schlacher et al. reported that several FA pathway members including FANCD2 and BRCA2 protect nascent DNA strands at stalled forks from nucleolytic degradation (1,16). Furthermore, we demonstrated that FANCD2 has additional roles in regulating the BLM helicase pathway (mutated in Bloom's syndrome) to restart stalled replication forks while suppressing new origin firing (17).
Intriguingly, recent reports identified several additional key HR proteins—including the MRE11 nuclease, RAD51 and XRCC3 (18,19)—as crucial factors during replication fork recovery. Moreover, all of these proteins are able to interact with FANCD2 (1,20–22), suggesting that FANCD2 promotes replication fork recovery in concert with members of the HR repair machinery.
CtIP (CtBP-interacting protein) (23) is a central HR repair factor that interacts with the MRE11/RAD50/NBS1 (MRN) complex to mediate DNA end resection, and with BRCA1 (breast cancer associated 1) to promote HR repair in a cell cycle-dependent manner (24–27). However, while CtIP's role in HR-mediated DNA DSB repair is well established, a putative function for CtIP in replication fork recovery—possibly in concert with FANCD2—has not been addressed.
In this study, we demonstrate that CtIP interacts with FANCD2 and promotes FANCD2 protein stability. Moreover, this interaction permits FANCD2 to recruit CtIP to sites of transiently stalled and collapsed replication forks. At stalled forks, CtIP cooperates with FANCD2 to promote fork restart while suppressing firing of new replication origins. Both functions are dependent on the upstream factor BRCA1 that monitors recruitment of MRE11, FANCD2 and finally CtIP to stalled replication forks, followed by their coordinated actions to promote fork recovery.
RESULTS
FANCD2 interacts with CtIP in a DNA damage- and monoubiquitination-independent manner
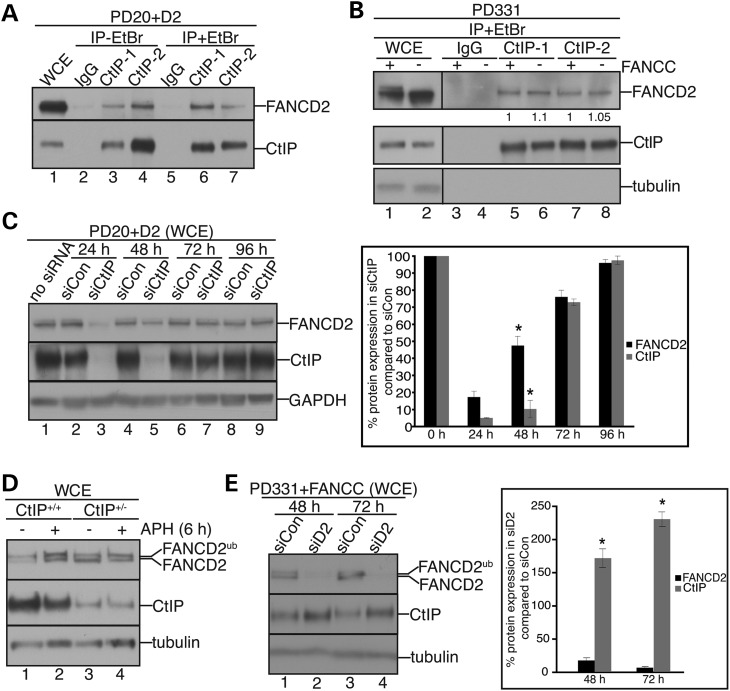
The CtIP-binding partner MRE11 interacts with FANCD2 and promotes FANCD2 protein stability (21). To test whether CtIP and FANCD2 have a comparable relationship, we immunoprecipitated CtIP from unperturbed wild-type cells (PD20+D2; a FANCD2 patient-derived cell line complemented with wild-type FANCD2), using antibodies against the N- or C-terminus of CtIP (28). Both CtIP antibodies coimmunoprecipitated FANCD2 (Fig. 1A), even in the presence of the DNA intercalator ethidium bromide (EtBr), indicating that their interaction is not mediated by DNA. Unfortunately, we were not able to analyze the FANCD2–CtIP interaction in reverse, since a FANCD2 antibody did not coimmunoprecipitate CtIP from wild-type cells, possibly due to interference with the FANCD2–CtIP interaction site. Importantly, the FANCD2–CtIP interaction was not diminished in FANCC-deficient cells (PD331) or FANCA-deficient cells (PD221) that cannot promote FANCD2Ub formation (Fig. 1B and Supplementary Material, Fig. S1A, respectively), indicating that FANCD2 monoubiquitination is dispensable for its interaction with CtIP. To further support this finding, we immunoprecipitated CtIP from wild-type cells that were either untreated or treated with APH to induce FANCD2Ub formation. We observed a decrease in CtIP-associated FANCD2 levels following FANCD2 monoubiquitination (Supplementary Material, Fig. S1B) demonstrating that FANCD2's ability to interact with CtIP is inversely correlated with its monoubiquitination status.
Figure 1.
FANCD2 interacts with CtIP and requires CtIP for its own protein stability. (A) FANCD2 interacts with CtIP. WCE were prepared from PD20 + D2 cells (lane 1) and subjected to IP with rabbit IgG (lanes 2 and 5; neg. control) or the anti-CtIP antibodies CtIP-1 (N-terminus, lanes 3 and 6) or CtIP-2 (C-terminus, lanes 4 and 7). IPs were performed in the absence or presence of EtBr as indicated. WCE and IP samples were analyzed for the presence of FANCD2 and CtIP by western blot. (B) The FANCD2-CtIP interaction is FA core complex independent. WCE were prepared from PD331 and PD331 + FANCC cells and subjected to IP using rabbit IgG (lanes 3 and 4, neg. control), and CtIP antibodies (lanes 5–8). WCE and equal volumes of IP samples were analyzed for the presence of CtIP and FANCD2. Immunoblot signals for FANCD2 levels interacting with CtIP were analyzed by densitometry and normalized against CtIP levels using ImageJ software. (C) CtIP promotes FANCD2 protein stability. Left panel: WCE were prepared from human PD20+D2 cells (lanes 1–9) that had been untreated (lane 1), or treated with control siRNA (siCon) or CtIP siRNA (siCtIP) for the indicated time points and analyzed for FANCD2 and CtIP. GAPDH, loading control. Right panel: Immunoblot signals shown in the left panel were analyzed by densitometry and normalized against GAPDH signals using ImageJ. The graph shows the percentage of FANCD2 and CtIP protein levels in siCtIP- compared with siCon-treated cells. Error bars, standard errors of the means (two independent experiments). *P < 0.01. (D) HCT116 CtIP+/− cells maintain wild-type-like FANCD2 protein levels. WCE were prepared from human HCT116 CtIP+/+ cells (lanes 1 and 2) and HCT116 CtIP+/− cells (lanes 3 and 4) that had been untreated (lanes 1 and 3) or treated with 30 μm APH for 6 h (lanes 2 and 4) and analyzed for the presence of FANCD2 and CtIP by western blot. Tubulin, loading control. (E) siRNA-mediated FANCD2 knockdown triggers upregulation of CtIP protein levels. Left panel: WCE were prepared from PD331+FANCC cells that had been treated with siCon (lanes 1 and 3) or siFANCD2 (lanes 2 and 4) for the indicated time points and analyzed for FANCD2 and CtIP by western blot. Tubulin, loading control. Right panel: Immunoblot signals shown in the left panel were analyzed by densitometry and normalized against GAPDH signals using ImageJ software. The graph shows the percentage of FANCD2 and CtIP protein levels in siFANCD2- compared with siCon-treated cells. Error bars, standard errors of the means (two independent experiments). *P < 0.01.
Interestingly, FANCD2 levels dropped precipitously following CtIP knockdown (siCtIP, 50 nm) at 24 h (CtIP: 5.3%; FANCD2: 13.9%) (Fig. 1C). On the other hand, while CtIP protein levels remained strongly reduced at 48 h after siCtIP treatment (15.4%), FANCD2 protein levels had returned to >50% (Fig. 1C). Moreover, treatment of cells with higher siCtIP concentrations (300 nm) delayed restoration of CtIP levels beyond 96 h, whereas FANCD2 protein levels were completely restored at 72 h (Supplementary Material, Fig. S1). Thus, while CtIP appeared to stabilize FANCD2, low amounts of CtIP were sufficient. In agreement with this, a heterozygous CtIP knockout cell line (HCT116 CtIP+/−) containing ∼20% residual CtIP protein levels nonetheless exhibited wild-type-like FANCD2 protein concentrations (Fig. 1D). Interestingly, siRNA-mediated knockdown of FANCD2 did not reduce CtIP protein levels; in fact, FANCD2 knockdown triggered a mild upregulation of cellular CtIP protein levels (Fig. 1E), hinting that cells may attempt to counteract the dwindling FANCD2 protein synthesis by stabilizing existing FANCD2 protein via CtIP.
FANCD2 recruits CtIP to transiently and terminally stalled replication forks
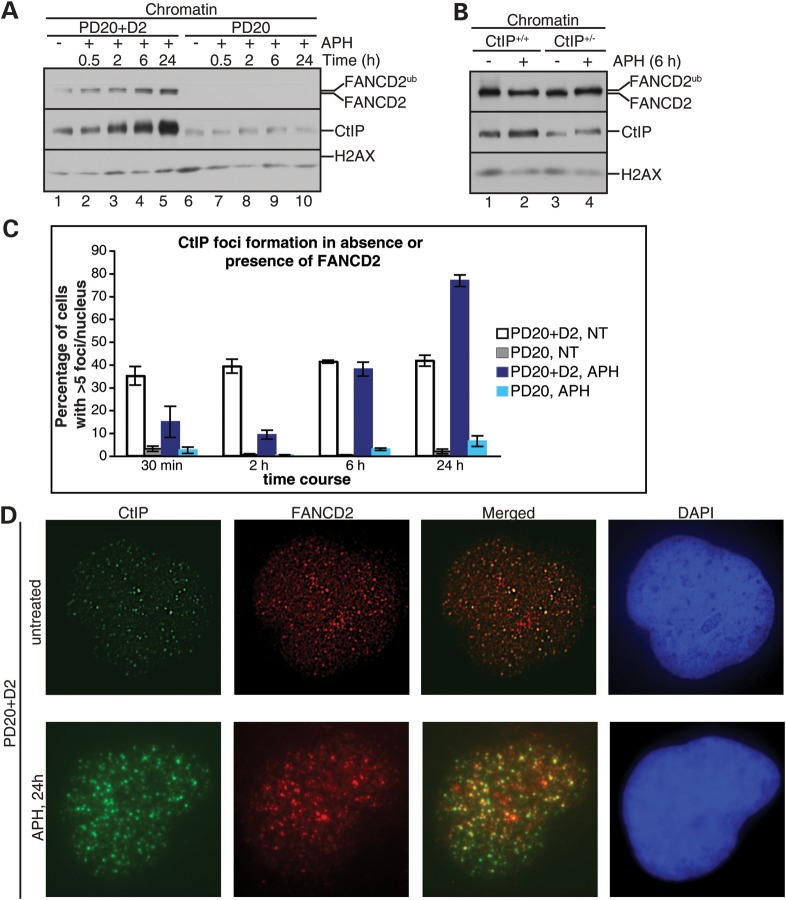
FANCD2 recruits BLM and BLM-associated DNA repair proteins to transiently stalled replication forks in order to promote fork restart (17). To test if CtIP is recruited to stalled forks in a FANCD2-dependent manner, we isolated chromatin fractions from FANCD2-proficient (PD20 + D2) and -deficient (PD20) cells that had been untreated or treated with APH for different lengths of time (30 min, 2, 6 or 24 h). Importantly in this context, APH- or HU-stalled replication forks remain stable and restart proficient for several hours (30 min–6 h time points), but do eventually collapse into DNA DSBs that cannot be restarted (24 h time point) (17,19). We observed that FANCD2-proficient cells contained chromatin-bound CtIP (CtIP-chr) even in the absence of APH. Following APH addition, CtIP-chr levels increased continuously with a strong peak at 24 h (Fig. 2A). In contrast, CtIP-chr levels were already reduced in untreated FANCD2-deficient cells and failed to increase further at any time following cellular exposure to APH. Thus, FANCD2 recruits CtIP to (i) unperturbed replicating chromatin, (ii) transiently stalled forks and (iii) fork collapse-induced DNA DSBs. In contrast, CtIP+/− cells (20% residual CtIP protein levels) did not exhibit reduced FANCD2-chr levels in the absence or presence of APH (Fig. 2B); moreover, these cells were fully capable of promoting FANCD2 monoubiquitination upon APH treatment (Fig. 1D). These results indicate that FANCD2 acts upstream of CtIP during normal replication and following APH treatment, and is crucial to recruit CtIP to transiently stalled, as well as collapsed, replication forks.
Figure 2.
FANCD2 recruits CtIP to chromatin and into chromatin foci. (A) FANCD2 recruits CtIP to chromatin during normal replication and following replication stress. Chromatin fractions were isolated from FANCD2-proficient (PD20+D2) or -deficient (PD20) cells that had been untreated or treated with APH for the indicated time points, and analyzed for the presence of FANCD2 and CtIP. H2AX: loading control. (B) HCT116 CtIP+/− cells are proficient for FANCD2 chromatin recruitment. HCT116 CtIP+/+ cells (lanes 1 and 2) and HCT116 CtIP+/− (lanes 3 and 4) were untreated (lanes 1 and 3) or treated with 30 μm APH for 6 h (lanes 2 and 4). Chromatin fractions were isolated from the cells and analyzed for presence of FANCD2 and CtIP. H2AX: loading control. (C) FANCD2 mediates CtIP foci formation during normal replication and following long-term (24 h) replication stress. FANCD2-proficient (PD20+D2) and -deficient cells (PD20) were untreated or treated with 30 μm APH for the indicated times (30 min, 2, 6 and 24 h) and cellular nuclei were analyzed for the presence of CtIP foci. Nuclei with >5 foci were considered positive for CtIP foci formation. (D) FANCD2 and CtIP colocalize in nuclear foci in unperturbed cells and following APH-triggered fork collapse. FANCD2-proficient cells (PD20+D2) cells were untreated or treated with 30 μm APH, followed by immunofluorescence staining with anti-FANCD2 and anti-CtIP antibodies. Nuclei were counterstained with DAPI.
FANCD2 mediates CtIP foci formation in absence or presence of replication stress
CtIP forms nuclear foci in unperturbed cells and in response to DNA DSBs triggered by cellular treatment with ionizing radiation (IR) (26,29,30). We analyzed CtIP foci formation in FANCD2-proficient (PD20+D2) and -deficient (PD20) cells that had been untreated or treated with APH for 30 min, 2, 6 or 24 h. Approximately 40% of wild-type cells contained discrete punctuate CtIP foci in unperturbed conditions (Fig. 2C and Supplementary Material, Fig. S2A). Interestingly, APH treatment caused an initial decrease in CtIP foci-containing cells (10% cells at 2 h post-APH) followed by a significant increase at 24 h (75% cells at 24 h post-APH), suggesting that CtIP foci formation does not occur at APH-stalled replication forks prior to their collapse into DNA DSBs. Notably, FANCD2-deficient cells failed to form CtIP foci at any conditions, including at 24 h post-APH treatment, indicating that FANCD2 recruits CtIP to chromatin foci during normal replication and following DNA DSB generation at collapsed forks. In strong support of these findings, we obtained very similar results in FANCD2-proficient and -deficient cells treated with HU (Supplementary Material, Fig. S2B). Moreover, FANCD2-deficient cells failed to support CtIP localization into chromatin foci even in response to directly IR-triggered DNA DSBs (Supplementary Material, Fig. S2C). Supportive of strictly FANCD2-dependent CtIP foci formation, FANCD2 and CtIP exhibited significant foci colocalization both in untreated cells and following APH treatment for 24 h (Fig. 2D). Interestingly, both types of CtIP foci-small, punctuate foci in untreated cells and large, bright foci in 24 h APH-treated cells exhibited significant colocalization with RAD51 (Supplementary Material, Fig. S2D), a protein known to be involved in replication fork protection, fork restart and DNA DSB repair (16,18,19). Thus, these foci likely represent different types of DNA damage: endogenous, replication-encountered sites of DNA lesions (small foci) and fork collapse-induced DNA DSBs (large foci). Together, these results indicate that FANCD2 globally regulates CtIP foci formation, during normal replication at endogenously generated sites of DNA repair and following exogenously (directly or indirectly) triggered DNA DSBs.
CtIP has FANCD2-dependent and -independent roles during DNA replication
FANCD2 cooperates with BLM to restart APH-stalled replication forks while simultaneously suppressing firing of new replication origins (17). FANCD2 and BLM also have independent roles: FANCD2 protects stalled replication forks from nucleolytic degradation independently of BLM, whereas BLM supports normal replication fork velocity independently of FANCD2 (1,17,31). We tested if CtIP was involved in one or more of these replication-associated functions using two sets of CtIP-proficient and -deficient cells: Set A, PD20+D2 (“wild type”) cells treated with control siRNA (siCon) or CtIP siRNA (siCtIP) for 48 h. At this time point, CtIP protein levels are still strongly reduced (15%), whereas FANCD2 protein levels have returned to at least 50% (see Fig. 1C).
Set B, wild-type HCT116 cells (CtIP+/+) or HCT116 cells carrying one CtIP knockout allele (CtIP+/−). Since CtIP+/− cells contain low CtIP protein levels (20%) yet wild-type-like FANCD2 protein levels (Fig. 1D), any phenotypes observed in these cells would be due to the absence of CtIP, not FANCD2.
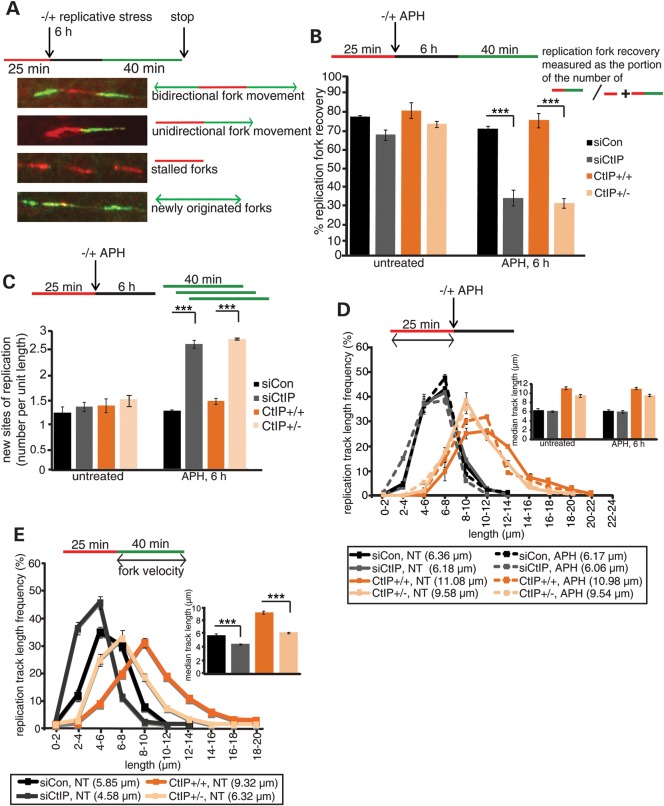
Replication events in these cells were monitored with a dual-labeling DNA fiber assay. Replication tracts were first labeled with DigU (red label) for 25 min, then untreated or treated with APH for 6 h to cause replication fork arrest, followed by second labeling with BioU (green label) for 40 min (Fig. 3A). Different from the efficient fork restart in wild-type cells, the proportion of replication forks competent for restart after APH treatment was severely reduced in both CtIP-deficient cell types (Fig. 3B, siCtIP: 32.50%; HCT116 CtIP+/−: 29.90%; P < 0.0001). Simultaneously, the number of newly originated replication tracts increased significantly in the CtIP-deficient cells (Fig. 3C, ∼2-fold; P < 0.0001), indicating that CtIP has crucial roles in replication fork restart and suppression of new origin firing. In contrast, CtIP-deficient cells did not exhibit shortening of nascent DNA strands (red label tract lengths) at APH-stalled forks compared with wild-type cells (Fig. 3D, siCtIP: 6.18 and 6.06 μm; CtIP+/−: 9.58 and 9.54 μm), suggesting that CtIP is dispensable for protection of stalled replication forks from nucleolytic degradation. Interestingly, we observed that in unperturbed conditions, CtIP-deficient cells exhibited shorter replication tracts per se (Fig. 3 E, siCon versus siCtIP: 5.85 and 4.58 μm (78% of wild type); CtIP+/+ versus CtIP+/−: 9.32 and 6.32 μm (68% of wild type; P < 0.0001) indicating an unanticipated role for CtIP in maintaining normal cellular replication fork velocity.
Figure 3.
CtIP has crucial functions during normal replication and in response to replication stress. (A) Images of DNA fibers with a schematic of defining sites of replication. Red tracks: DigU; green tracks: BioU. (B) CtIP is required for stalled fork restart. The efficiency of replication restart in wild type (siCon and HCT116 CtIP+/+) and respectively matching, CtIP-deficient cells (siCtIP and HCT116 CtIP+/−) was measured as the number of restarted replication forks after APH-mediated fork stalling (DigU → BioU tracts), compared with the total number of DigU-labeled tracts (DigU plus DigU → BioU); ***P < 0.0001. (C) CtIP acts to suppress new origin firing during replication blockade. The fraction of new sites of replication originating during the 40 min recovery period after APH treatment was compared between wild-type (siCon and HCT116 CtIP+/+) and respectively matching, CtIP-deficient cells (siCtIP and HCT116 CtIP+/−). Fractions were measured as the number of green-only (BioU) tracts compared with the total number of spreading replication tracts (BioU plus DigU→BioU); ***P < 0.0001. (D) CtIP does not protect replication forks from degradation. Lengths of nascent replication fork tracts indicating fork stability (labeled with DigU only) were measured before and after 6 h of APH treatment. Preformed DigU track lengths did not shorten during 6 h APH-treatment in siCtIP- versus siCon-treated wild-type cells, or in HCT116 CtIP+/− versus HCT116 CtIP+/+ cells. Median track lengths are indicated below each panel. Inset: plotted median track lengths. (E) CtIP supports normal replication fork velocity. BioU track length distributions were determined on DigU → BioU double-labeled DNA fibers isolated from untreated (NT) wild-type cells (siCon and HCT116 CtIP+/+) or respectively matching, CtIP-deficient cells (siCtIP and HCT116 CtIP+/−). Median track lengths are indicated below each panel. Inset: plotted median track length; ***P < 0.0001. [NB: Wild-type HCT116 cells exhibit a higher average fork velocity than the (complemented or non-complemented) PD20 cells. This might be due to different genetic backgrounds of the two cell types: PD20 cells are FA patient-derived fibroblasts, whereas HCT116 is a non-FA, colorectal cancer-derived cell line.]
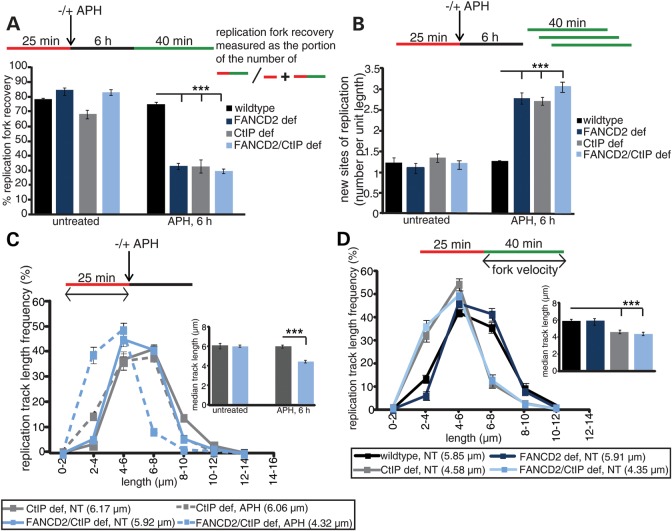
Together, these results suggested partial cooperation between CtIP and FANCD2 during replication fork recovery. To test this, we treated PD20+D2 or PD20 cells with control siRNA or CtIP siRNA to generate wild-type, FANCD2-, CtIP- or FANCD2/CtIP double-deficient cells, followed by dual-label DNA fiber analysis. As expected, FANCD2- and CtIP-deficient cells exhibited similarly reduced replication fork restart events (32.80 and 32.75%, P < 0.0001); moreover, this defect was not further exacerbated in FANCD2/CtIP double-depleted cells (29.55%, P < 0.0001), demonstrating that FANCD2 and CtIP act in the same pathway to restart replication forks (Fig. 4A). In parallel, the proportion of newly originated replication tracts increased significantly and equally in FANCD2-, CtIP- and FANCD2/CtIP double-deficient cells compared with wild-type cells (Fig. 4B, ∼2-fold; P < 0.0001). In contrast, only the absence of FANCD2 but not CtIP caused significant degradation of nascent DNA strands at APH-stalled replication forks (Fig. 4C, 4.32 versus 6.06 μm; P < 0.0001), confirming that FANCD2 protects stalled replication forks even in absence of CtIP. In addition, CtIP knockdown caused shortening of replication tract lengths in unperturbed FANCD2-proficient cells (4.58 μm, 78% of wild type, P < 0.0001) or FANCD2-deficient cells (4.35 μm, 74% of FANCD2 deficient, P < 0.0001; Fig. 4D), demonstrating that CtIP acts independently of FANCD2 to promote replication fork velocity.
Figure 4.
CtIP acts in concert with FANCD2 to mediate replication fork restart and suppression of new origin firing. (A) FANCD2 and CtIP cooperate to mediate replication fork restart. The efficiency of replication restart in wild-type, FANCD2-, CtIP- and CtIP/FANCD2 double-deficient cells was measured as the number of restarted replication forks after APH-mediated fork stalling (DigU → BioU tracts), compared with the total number of DigU-labeled tracts (DigU plus DigU → BioU); ***P < 0.0001. (B) FANCD2 and CtIP act in concert to suppress new origin firing during replication blockade. The fraction of new sites of replication originating during the 40 min recovery period after APH treatment was compared between wild-type, FANCD2-, CtIP- and CtIP/FANCD2 double-deficient cells. Fractions were measured as the number of green-only (BioU) tracts compared with the total number of spreading replication tracts (BioU plus DigU→BioU); ***P < 0.0001. (C) FANCD2 protects replication forks independently of CtIP. Lengths of nascent replication fork tracts indicating fork stability (labeled with DigU only) were measured before and after 6 h of APH treatment. Preformed DigU tract lengths do not shorten during APH treatment in CtIP-deficient cells compared with CtIP/FANCD2 double-deficient cells. Median track lengths are indicated below each panel. Insets: plotted median track lengths. (D) CtIP maintains replication fork velocity independently of FANCD2. BioU track length distributions were determined on DigU → BioU double-labeled DNA fibers isolated from untreated (NT) wild-type, FANCD2-, CtIP- and CtIP/FANCD2 double-deficient cells. Median track lengths are indicated below each panel. Insets: plotted median track lengths.
BRCA1 acts upstream of FANCD2 and CtIP during replication fork recovery
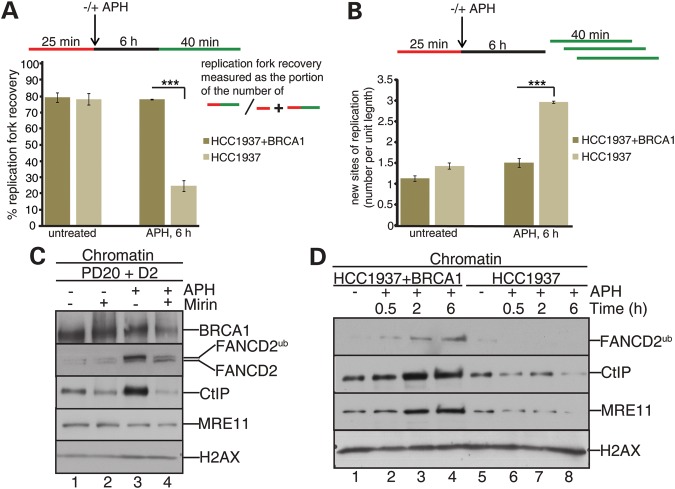
BRCA1 interacts with CtIP and recruits CtIP to chromatin and into nuclear DNA repair foci following IR-induced DNA DSB formation (32,33). Similarly, BRCA1 colocalizes with FANCD2 and promotes FANCD2 localization into nuclear foci after cellular IR treatment (3). Moreover, BRCA1-like FANCD2-protects stalled replication forks from nucleolytic degradation (1). Thus, we speculated that BRCA1 might cooperate with FANCD2 and CtIP to promote replication fork restart and suppression of new origin firing. To address this hypothesis, we utilized the BRCA1-deficient cell line HCC1937 and its complemented counterpart, HCC1937+BRCA1. Following APH treatment for 6 h, BRCA1-deficient cells showed extreme defects in replication fork restart (Fig. 5A, 24.7%; P < 0.0001; Fig. 5A), and a simultaneous increase in new origin firing compared with wild-type cells (∼2-fold; P < 0.0001; Fig. 5B), comparable with the phenotypes observed in FANCD2- and/or CtIP-deficient cells.
Figure 5.
BRCA1 regulates recruitment of FANCD2, MRE11 and CtIP to stalled replication forks and promotes fork restart. (A) BRCA1 mediates stalled fork restart. The efficiency of replication restart in wild-type (HCC1937+BRCA) and BRCA1-deficient (HCC1927) cells was measured as the number of restarted replication forks after APH-mediated fork stalling (DigU → BioU tracts), compared with the total number of DigU-labeled tracts (DigU plus DigU → BioU); ***P < 0.0001. (B) BRCA1 acts to suppress new origin firing during replication blockade. The fraction of new sites of replication originating during the 40 min recovery period after APH treatment was compared between wild-type and BRCA1-deficient cells. Fractions were measured as the number of green-only (BioU) tracts compared with the total number of spreading replication tracts (BioU plus DigU→BioU); ***P < 0.0001. (C) MRE11 nuclease activity promotes recruitment of BRCA1, FANCD2 and CtIP to stalled replication forks. Chromatin fractions were isolated from wild-type (PD20+D2) cells that had been untreated or exposed to APH for 6 h, in the presence or absence of mirin. Chromatin fractions were analyzed for the presence of BRCA1, FANCD2, CtIP and MRE11. H2AX: loading control. (D) BRCA1 promotes recruitment of MRE11, FANCD2 and CtIP to stalled replication forks. Chromatin fractions were isolated from wild-type (HCC1937+BRCA1) or BRCA1-deficient (HCC1937) cells that had been untreated or APH treated for the indicated time points. Chromatin fractions were analyzed for the presence of FANCD2, CtIP and MRE11. H2AX: loading control.
Previous studies showed that DNA DSB-triggered foci formation of BRCA1 and FANCD2 occurred in an MRE11-dependent manner (21,34), whereas CtIP and MRE11 localized to DNA DSBs independently of one another (29). Thus, we set out to test if the same hierarchy was maintained during these proteins' recruitment to transiently APH-stalled replication forks. Cellular treatment with the MRE11 nuclease inhibitor mirin significantly reduced APH-triggered recruitment of BRCA1, FANCD2 and CtIP to chromatin (Fig. 5C). Moreover, BRCA1-deficient cells exhibited reduced chromatin-bound levels of MRE11, FANCD2 and CtIP following APH treatment for 0.5–6 h (Fig. 5D). These results indicate that BRCA1 and MRE11 stabilize each other at sites of stalled replication forks, and then recruit FANCD2, followed by CtIP, to promote fork recovery. Supportive of this idea, knockdown of MRE11 did not further increase the replication fork restart defect observed in CtIP-deficient cells (Supplementary Material, Fig. S3). Considering this hierarchy, we reasoned that the S/G2-phase-dependent BRCA1-CtIP-MRN complex formation (29) might occur in a FANCD2-dependent manner. However, antibodies against CtIP coimmunoprecipitated BRCA1 and MRE11 from FANCD2-proficient and -deficient cells (Supplementary Material, Fig. S4). Thus, interactions of CtIP with its upstream regulators BRCA1 and MRE11 remain stable in FANCD2-deficient cells but are not sufficient to stably recruit CtIP to stalled replication forks.
DISCUSSION
In this study, we identify CtIP as novel protein interactor of FANCD2 and demonstrate FANCD2-dependent and -independent roles of CtIP during DNA replication and the replication stress response.
Our findings add CtIP to a growing list of FANCD2-interacting proteins such as MRE11 and BLM that promote replication fork recovery as well as HR-mediated DNA DSB repair, strongly suggesting that fork restart requires HR-related mechanisms (18,21,31,35,36). The interaction of FANCD2 with CtIP-akin to its interactions with MRE11 or BLM (17,21)-occurs in a DNA damage and FANCD2Ub-independent manner, providing additional evidence that FANCD2 monoubiquitination is dispensable for some of its cellular functions.
The fact that the absence of CtIP destabilizes FANCD2, combined with previous findings that both FANCD2 and CtIP are degraded in the absence of MRE11 (21,37) suggests that CtIP is the true stabilizer of FANCD2, likely via a direct physical interaction. Interestingly, dependency of one protein's stability on the presence of another does not necessarily reflect the functional hierarchy between the two: FANCD2 is recruited to stalled replication fork downstream of MRE11 but upstream of CtIP (Figs 5C and 2A), yet both MRE11 and CtIP regulate (indirectly or directly) FANCD2 protein stability.
FANCD2-dependent chromatin recruitment of CtIP occurs on several levels: (i) during unperturbed replication, (ii) at stalled forks and (iii) at DNA DSBs generated by fork collapse or IR treatment. These results suggest that FANCD2 controls CtIP functions not only during replication fork restart but also during HR-mediated DNA DSB repair. In support of this idea, independent studies by Nakanishi et al. (14) and Chen et al. (29) showed that FANCD2- and CtIP-deficient cells exhibit comparable deficiencies in the HR-mediated repair of DNA DSBs. Future studies should reveal if and how FANCD2 regulates CtIP-dependent functions during HR repair, perhaps via supporting end resection at DNA DSBs.
Unexpectedly, the acute cellular response to APH- or HU-mediated replication fork stalling includes a significant reduction in CtIP foci formation, only to be followed by a subsequent increase in CtIP foci numbers that peak at 24 h, when forks have collapsed into DNA breaks (17,19). We observed that nucleotide incorporation was completely blocked at 2 h post-APH exposure (data not shown), indicating that replication forks are fully stalled at this time. Moreover, the decrease in APH-triggered CtIP foci formation at 30 min and 2 h was accompanied by a simultaneous increase in CtIP chromatin binding. Thus, while CtIP foci formation does not necessarily mirror its chromatin binding, APH-mediated replication fork stalling does actively block CtIP foci formation. Interestingly, a similar phenomenon was recently shown for RAD51, a protein that-like CtIP-promotes both replication fork restart and HR-mediated repair: Petermann et al. demonstrated that following HU exposure, the number of RAD51 foci-positive cells decreased at 1 and 2 h, but increased significantly at 24 h (19). The fact that we observed strong colocalization of CtIP and RAD51 foci in untreated conditions (small foci) as well as after 24 h of APH treatment (large foci), indicates that these two proteins cooperate during the repair of endogenously and exogenously triggered DNA lesions. While these findings require further investigation, we speculate that DNA repair proteins may not form foci at replication forks stalled in absence of a DNA lesion. The fact that CtIP and RAD51 foci reappear in great numbers once forks have collapsed into DNA DSBs supports a model where foci formation of these proteins is restricted to sites of DNA breaks. Alternatively, the rather small and punctuate CtIP and RAD51 foci observed in unperturbed cells may represent other types of DNA damage recognized by the cell through collision with the replication machinery. A recent study showed that CtIP foci in untreated cells colocalize with PCNA and BrdU, possibly representing sites of DNA lesion repair at unbroken, but currently halted forks (30).
CtIP's novel role in maintaining replication fork speed provides a mechanistic basis for previous observations that mouse and chicken (DT40) CtIP knockout cells show prolonged S-phase stages and slower overall proliferation profiles (38). The notion that CtIP performs this role independently of FANCD2 is further strengthened by a recent study showing that FANCD2 is present exclusively at stalled, not moving replication forks (39). Since FANCD2-deficient cells exhibit normal replication fork speed despite reduced chromatin-bound CtIP levels (Fig. 2A), low concentrations of chromatin-bound CtIP must be sufficient to maintain replication velocity.
Another novel role for CtIP is its FANCD2-dependent function during replication fork restart, which it shares with other FANCD2-interacting proteins such as BLM and XRCC3. However, unlike FANCD2, we find that CtIP is dispensable for replication fork protection against nucleolytic degradation. This was somewhat unexpected considering that CtIP is crucial for the recruitment of RAD51, at least to sites of DNA DSBs (38), and RAD51 is indispensable for the protection of stalled forks (19). Hence, RAD51's recruitment to stalled forks must occur independently of CtIP, predicting different mutual regulation rules among DNA repair proteins at stalled replication forks versus DNA DSBs. In this regard, we also observed that BRCA1-the other newly identified replication fork restart factor in this study-and MRE11 appear to communicate differently during replication fork restart compared with DNA DSB repair: MRE11 acts strictly upstream of BRCA1 (34) during DNA DSB repair and does not rely on BCRA1 for its own access to DNA breaks (40–42). In contrast, our results indicate mutual stabilization of BRCA1 and MRE11 at sites of stalled replication forks. This mutual regulation between DNA repair proteins appears to be an emerging theme. For example, the FA core complex member FANCM recruits FANCD2 to replicating chromatin, but also relies on FANCD2 for its own chromatin binding (43). Similarly, the MRN complex recruits ATM to DNA DSBs, but then requires ATM for its own activation (44). These mutual stabilization events at stalled replication forks or DNA lesions likely empower cells with additional flexibility, by enabling proteins that act at downstream steps of fork recovery or DNA repair to provide regulatory feedback to their upstream mediators.
In summary, our results show that the central FA pathway member FANCD2 is a major regulator of CtIP functions during replication fork recovery in cooperation with BRCA1 and MRE11 (Supplementary Material, Fig. S5). These findings support accumulating evidence that FANCD2 coordinates key HR proteins to mediate replication fork protection and allow successful replication fork restart.
MATERIALS AND METHODS
Cell culture
PD20 (FANCD2 deficient), PD20+D2 (retrovirally complemented with wild-type human FANCD2), PD221 (FANCA deficient) and PD221+A (retrovirally complemented with wild-type human FANCA) patient cells were obtained from the FA Cell Repository at the Oregon Health & Science University. PD331 (FANCC-deficient FA patient cells) and PD331+C (retrovirally complemented with wild-type human FANCC) were a kind gift from Dr Fillipo Roselli. HCC1937 (BRCA1-deficient, tumor-derived cells) and HCC1937+BRCA1 (retrovirally complemented with wild-type human BRCA1) cell lines were a kind gift from Dr Robert M. Brosh Jr PD20 and PD331 cell derivatives were cultured in DMEM (GIBCO). HCC1937 cell derivatives were grown in RPMI 1640 (GIBCO). HCT116 cell derivatives were cultured in McCoy A medium (GIBCO).
Construction of a human CtIP+/− heterozygous somatic cell line (see also Supplementary Material, Figure S6)
Expression of CtIP protein in HCT116 CtIP+/− compared with HCT116 CtIP+/+ cells was analyzed by western blot of whole cell extracts (WCE) (Fig. 1D).
Antibodies
Antibodies against CtIP have been described (28). Commercial antibodies were used against human FANCD2 (Santa Cruz, sc-20022 and Abcam, ab2187), GAPDH (Genetex, GTX627408), BRCA1 (Santa Cruz, sc-642), MRE11 (Santa Cruz, sc-5859), tubulin (Abcam, ab7291) and H2AX (Bethyl, A300-083A).
Immunoblotting
Protein samples were separated on gradient gels and transferred to Immobilon P membranes (Millipore). After blocking in 5% milk, membranes were incubated with the following primary antibodies: anti-FANCD2 (1:2000), anti-BRCA1 (1:500), anti-CtIP (mouse monoclonal, 1:40), anti-H2AX (1:10 000), anti-GAPDH (1:1000) and anti-tubulin (1:10 000). Horseradish peroxidase-conjugated rabbit secondary antibodies (Jackson Labs) or mouse secondary antibodies (Bio-Rad) were used at dilutions of 1:10 000 and 1:3000, respectively. Protein bands were visualized using an ECL Plus system (Amersham).
Preparation of WCE and chromatin fractions from human cells
Human WCE and chromatin fractions were prepared as previously described (17,45).
Immunoprecipitation from human cells
Cells were lysed in IP buffer as previously described (17). Lysates were precleared with rabbit or mouse IgG and subjected to immunoprecipitation with anti-FANCD2, anti-CtIP, anti-BRCA1 or IgG antibody in absence or presence of EtBr (10 μg/ml) at 4°C overnight. One hundred microliters of Sepharose 4B beads (50% slurry) were added and rotated for 30 min at 4°C. The beads were pelleted from solution, washed in cell lysis buffer, boiled in 1× NuPAGE buffer (Invitrogen) and analyzed by SDS–PAGE and western blotting.
siRNA experiments
siRNA duplexes were purchased from Dharmacon research (Thermo Scientific, MA, USA). The sequence of CtIP siRNA is GCUAAAACAGGAACGAAUCUU (46). The sequence of MRE11 siRNA is GCUAAUGACUCUGAUGAUAUU. siGENOME non-targeting siRNA was used as a control. Transfections were performed using DharmaFECT1 transfection reagent according to the manufacturer's protocol.
Immunofluorescence
Indirect immunofluorescence was carried out essentially as described (47). Primary antibodies used: FANCD2 (Abcam, ab2187, 1:4000), CtIP (mouse monoclonal, 1:400). Secondary antibodies used: alexa Fluor 594-conjugated goat anti-rabbit (1:1000) and Alexa Fluor 488-conjugated goat anti-mouse (1:1000; Molecular Probes). For statistical analysis of nuclear foci formation, images were taken using a Leica DM LB2 microscope with a Hamamatsu Orca-ER camera. Quantification of CtIP foci was carried out using ImageJ. To analyze foci colocalization of two proteins, images were captured using a Deltavision microscope (Applied Precision) and analyzed using Deltavision softWoRx 5.5 software.
DNA fiber assay
We used a DNA fiber protocol as previously described (17). All shown DNA fiber results are the means of two or three independent experiments (300 DNA fibers/experiment). Error bars represent the standard error of the mean and significance was determined by t-test and Mann–Whitney tests (Supplementary Material, Table S1). Statistical significance at P < 0.01, P < 0.001 and P < 0.0001 is indicated as *, ** and ***, respectively.
SUPPLEMENTARY MATERIAL
FUNDING
A.S. was supported by the National Science Foundation (award 1121023) and the American Cancer Society (RSG-13-039-01-DMC). E.A.H. was supported by the National Institutes of Health (GM088351) and the National Cancer Institute (CA154461).
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to R. Baer for very generously sharing several human CtIP antibodies with us. We are grateful to F. Roselli and R. M. Brosh Jr for sharing cell lines with us. We thank A.K. Bielinsky and N. Shima for helpful discussions regarding this manuscript.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Schlacher K., Wu H., Jasin M. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell. 2012;22:106–116. doi: 10.1016/j.ccr.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kee Y., D'Andrea A.D. Expanded roles of the Fanconi anemia pathway in preserving genomic stability. Genes Dev. 2010;24:1680–1694. doi: 10.1101/gad.1955310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garcia-Higuera I., Taniguchi T., Ganesan S., Meyn M.S., Timmers C., Hejna J., Grompe M., D'Andrea A.D. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol. Cell. 2001;7:249–262. doi: 10.1016/s1097-2765(01)00173-3. [DOI] [PubMed] [Google Scholar]
- 4.Smogorzewska A., Matsuoka S., Vinciguerra P., McDonald E.R., III, Hurov K.E., Luo J., Ballif B.A., Gygi S.P., Hofmann K., D'Andrea A.D., et al. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell. 2007;129:289–301. doi: 10.1016/j.cell.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kratz K., Schopf B., Kaden S., Sendoel A., Eberhard R., Lademann C., Cannavo E., Sartori A.A., Hengartner M.O., Jiricny J. Deficiency of FANCD2-associated nuclease KIAA1018/FAN1 sensitizes cells to interstrand crosslinking agents. Cell. 2010;142:77–88. doi: 10.1016/j.cell.2010.06.022. [DOI] [PubMed] [Google Scholar]
- 6.Liu T., Ghosal G., Yuan J., Chen J., Huang J. FAN1 acts with FANCI-FANCD2 to promote DNA interstrand cross-link repair. Science. 2010;329:693–696. doi: 10.1126/science.1192656. [DOI] [PubMed] [Google Scholar]
- 7.MacKay C., Declais A.C., Lundin C., Agostinho A., Deans A.J., MacArtney T.J., Hofmann K., Gartner A., West S.C., Helleday T., et al. Identification of KIAA1018/FAN1, a DNA repair nuclease recruited to DNA damage by monoubiquitinated FANCD2. Cell. 2010;142:65–76. doi: 10.1016/j.cell.2010.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smogorzewska A., Desetty R., Saito T.T., Schlabach M., Lach F.P., Sowa M.E., Clark A.B., Kunkel T.A., Harper J.W., Colaiacovo M.P., et al. A genetic screen identifies FAN1, a Fanconi anemia-associated nuclease necessary for DNA interstrand crosslink repair. Mol. Cell. 2010;39:36–47. doi: 10.1016/j.molcel.2010.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crossan G.P., van der Weyden L., Rosado I.V., Langevin F., Gaillard P.H., McIntyre R.E., Gallagher F., Kettunen M.I., Lewis D.Y., Brindle K., et al. Disruption of mouse Slx4, a regulator of structure-specific nucleases, phenocopies Fanconi anemia. Nat. Genet. 2011;43:147–152. doi: 10.1038/ng.752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim Y., Lach F.P., Desetty R., Hanenberg H., Auerbach A.D., Smogorzewska A. Mutations of the SLX4 gene in Fanconi anemia. Nat. Genet. 2011;43:142–146. doi: 10.1038/ng.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stoepker C., Hain K., Schuster B., Hilhorst-Hofstee Y., Rooimans M.A., Steltenpool J., Oostra A.B., Eirich K., Korthof E.T., Nieuwint A.W., et al. SLX4, a coordinator of structure-specific endonucleases, is mutated in a new Fanconi anemia subtype. Nat. Genet. 2011;43:138–141. doi: 10.1038/ng.751. [DOI] [PubMed] [Google Scholar]
- 12.Yamamoto K.N., Kobayashi S., Tsuda M., Kurumizaka H., Takata M., Kono K., Jiricny J., Takeda S., Hirota K. Involvement of SLX4 in interstrand cross-link repair is regulated by the Fanconi anemia pathway. Proc. Natl. Acad. Sci. USA. 2011;108:6492–6496. doi: 10.1073/pnas.1018487108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.D'Andrea A.D. Susceptibility pathways in Fanconi's anemia and breast cancer. N. Engl. J. Med. 2010;362:1909–1919. doi: 10.1056/NEJMra0809889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakanishi K., Yang Y.G., Pierce A.J., Taniguchi T., Digweed M., D'Andrea A.D., Wang Z.Q., Jasin M. Human Fanconi anemia monoubiquitination pathway promotes homologous DNA repair. Proc. Natl. Acad. Sci. USA. 2005;102:1110–1115. doi: 10.1073/pnas.0407796102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang W. Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins. Nat. Rev. Genet. 2007;8:735–748. doi: 10.1038/nrg2159. [DOI] [PubMed] [Google Scholar]
- 16.Schlacher K., Christ N., Siaud N., Egashira A., Wu H., Jasin M. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell. 2011;145:529–542. doi: 10.1016/j.cell.2011.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chaudhury I., Sareen A., Raghunandan M., Sobeck A. FANCD2 regulates BLM complex functions independently of FANCI to promote replication fork recovery. Nucleic Acids Res. 2013;41:6444–6459. doi: 10.1093/nar/gkt348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bryant H.E., Petermann E., Schultz N., Jemth A.S., Loseva O., Issaeva N., Johansson F., Fernandez S., McGlynn P., Helleday T. PARP is activated at stalled forks to mediate Mre11-dependent replication restart and recombination. EMBO J. 2009;28:2601–2615. doi: 10.1038/emboj.2009.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Petermann E., Orta M.L., Issaeva N., Schultz N., Helleday T. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol. Cell. 2010;37:492–502. doi: 10.1016/j.molcel.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taniguchi T., Garcia-Higuera I., Andreassen P.R., Gregory R.C., Grompe M., D'Andrea A.D. S-phase-specific interaction of the Fanconi anemia protein, FANCD2, with BRCA1 and RAD51. Blood. 2002;100:2414–2420. doi: 10.1182/blood-2002-01-0278. [DOI] [PubMed] [Google Scholar]
- 21.Roques C., Coulombe Y., Delannoy M., Vignard J., Grossi S., Brodeur I., Rodrigue A., Gautier J., Stasiak A.Z., Stasiak A., et al. MRE11-RAD50-NBS1 is a critical regulator of FANCD2 stability and function during DNA double-strand break repair. EMBO J. 2009;28:2400–2413. doi: 10.1038/emboj.2009.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilson J.B., Yamamoto K., Marriott A.S., Hussain S., Sung P., Hoatlin M.E., Mathew C.G., Takata M., Thompson L.H., Kupfer G.M., et al. FANCG promotes formation of a newly identified protein complex containing BRCA2, FANCD2 and XRCC3. Oncogene. 2008;27:3641–3652. doi: 10.1038/sj.onc.1211034. [DOI] [PubMed] [Google Scholar]
- 23.Schaeper U., Subramanian T., Lim L., Boyd J.M., Chinnadurai G. Interaction between a cellular protein that binds to the C-terminal region of adenovirus E1A (CtBP) and a novel cellular protein is disrupted by E1A through a conserved PLDLS motif. J. Biol. Chem. 1998;273:8549–8552. doi: 10.1074/jbc.273.15.8549. [DOI] [PubMed] [Google Scholar]
- 24.Escribano-Diaz C., Orthwein A., Fradet-Turcotte A., Xing M., Young J.T., Tkac J., Cook M.A., Rosebrock A.P., Munro M., Canny M.D., et al. A cell cycle-dependent regulatory circuit composed of 53BP1-RIF1 and BRCA1-CtIP controls DNA repair pathway choice. Mol. Cell. 2013;49:872–883. doi: 10.1016/j.molcel.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 25.Moynahan M.E., Cui T.Y., Jasin M. Homology-directed DNA repair, mitomycin-c resistance, and chromosome stability is restored with correction of a Brca1 mutation. Cancer Res. 2001;61:4842–4850. [PubMed] [Google Scholar]
- 26.Sartori A.A., Lukas C., Coates J., Mistrik M., Fu S., Bartek J., Baer R., Lukas J., Jackson S.P. Human CtIP promotes DNA end resection. Nature. 2007;450:509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scully R., Chen J., Plug A., Xiao Y., Weaver D., Feunteun J., Ashley T., Livingston D.M. Association of BRCA1 with Rad51 in mitotic and meiotic cells. Cell. 1997;88:265–275. doi: 10.1016/s0092-8674(00)81847-4. [DOI] [PubMed] [Google Scholar]
- 28.Yu X., Baer R. Nuclear localization and cell cycle-specific expression of CtIP, a protein that associates with the BRCA1 tumor suppressor. J. Biol. Chem. 2000;275:18541–18549. doi: 10.1074/jbc.M909494199. [DOI] [PubMed] [Google Scholar]
- 29.Chen L., Nievera C.J., Lee A.Y., Wu X. Cell cycle-dependent complex formation of BRCA1.CtIP.MRN is important for DNA double-strand break repair. J. Biol. Chem. 2008;283:7713–7720. doi: 10.1074/jbc.M710245200. [DOI] [PubMed] [Google Scholar]
- 30.Gu B., Chen P.L. Expression of PCNA-binding domain of CtIP, a motif required for CtIP localization at DNA replication foci, causes DNA damage and activation of DNA damage checkpoint. Cell Cycle. 2009;8:1409–1420. doi: 10.4161/cc.8.9.8322. [DOI] [PubMed] [Google Scholar]
- 31.Davies S.L., North P.S., Hickson I.D. Role for BLM in replication-fork restart and suppression of origin firing after replicative stress. Nat. Struct. Mol. Biol. 2007;14:677–679. doi: 10.1038/nsmb1267. [DOI] [PubMed] [Google Scholar]
- 32.Yu X., Fu S., Lai M., Baer R., Chen J. BRCA1 ubiquitinates its phosphorylation-dependent binding partner CtIP. Genes Dev. 2006;20:1721–1726. doi: 10.1101/gad.1431006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu X., Wu L.C., Bowcock A.M., Aronheim A., Baer R. The C-terminal (BRCT) domains of BRCA1 interact in vivo with CtIP, a protein implicated in the CtBP pathway of transcriptional repression. J. Biol. Chem. 1998;273:25388–25392. doi: 10.1074/jbc.273.39.25388. [DOI] [PubMed] [Google Scholar]
- 34.Yuan J., Chen J. MRE11-RAD50-NBS1 complex dictates DNA repair independent of H2AX. J. Biol. Chem. 2010;285:1097–1104. doi: 10.1074/jbc.M109.078436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chu W.K., Hanada K., Kanaar R., Hickson I.D. BLM has early and late functions in homologous recombination repair in mouse embryonic stem cells. Oncogene. 2010;29:4705–4714. doi: 10.1038/onc.2010.214. [DOI] [PubMed] [Google Scholar]
- 36.Kikuchi K., Abdel-Aziz H.I., Taniguchi Y., Yamazoe M., Takeda S., Hirota K. Bloom DNA helicase facilitates homologous recombination between diverged homologous sequences. J. Biol. Chem. 2009;284:26360–26367. doi: 10.1074/jbc.M109.029348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Buis J., Stoneham T., Spehalski E., Ferguson D.O. Mre11 regulates CtIP-dependent double-strand break repair by interaction with CDK2. Nat. Struct. Mol. Biol. 2012;19:246–252. doi: 10.1038/nsmb.2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakamura K., Kogame T., Oshiumi H., Shinohara A., Sumitomo Y., Agama K., Pommier Y., Tsutsui K.M., Tsutsui K., Hartsuiker E., et al. Collaborative action of Brca1 and CtIP in elimination of covalent modifications from double-strand breaks to facilitate subsequent break repair. PLoS Genet. 2010;6:e1000828. doi: 10.1371/journal.pgen.1000828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lossaint G., Larroque M., Ribeyre C., Bec N., Larroque C., Decaillet C., Gari K., Constantinou A. FANCD2 binds MCM proteins and controls replisome function upon activation of s phase checkpoint signaling. Mol. Cell. 2013;51:678–690. doi: 10.1016/j.molcel.2013.07.023. [DOI] [PubMed] [Google Scholar]
- 40.Wang Y., Cortez D., Yazdi P., Neff N., Elledge S.J., Qin J. BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev. 2000;14:927–939. [PMC free article] [PubMed] [Google Scholar]
- 41.Wu X., Petrini J.H., Heine W.F., Weaver D.T., Livingston D.M., Chen J. Independence of R/M/N focus formation and the presence of intact BRCA1. Science. 2000;289:11. doi: 10.1126/science.289.5476.11a. [DOI] [PubMed] [Google Scholar]
- 42.Zhang J., Willers H., Feng Z., Ghosh J.C., Kim S., Weaver D.T., Chung J.H., Powell S.N., Xia F. Chk2 phosphorylation of BRCA1 regulates DNA double-strand break repair. Mol. Cell Biol. 2004;24:708–718. doi: 10.1128/MCB.24.2.708-718.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sobeck A., Stone S., Landais I., de Graaf B., Hoatlin M.E. The Fanconi anemia protein FANCM is controlled by FANCD2 and the ATR/ATM pathways. J. Biol. Chem. 2009;284:25560–25568. doi: 10.1074/jbc.M109.007690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lavin M.F. ATM and the Mre11 complex combine to recognize and signal DNA double-strand breaks. Oncogene. 2007;26:7749–7758. doi: 10.1038/sj.onc.1210880. [DOI] [PubMed] [Google Scholar]
- 45.Ge X.Q., Jackson D.A., Blow J.J. Dormant origins licensed by excess Mcm2–7 are required for human cells to survive replicative stress. Genes Dev. 2007;21:3331–3341. doi: 10.1101/gad.457807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu X., Chen J. DNA damage-induced cell cycle checkpoint control requires CtIP, a phosphorylation-dependent binding partner of BRCA1 C-terminal domains. Mol. Cell Biol. 2004;24:9478–9486. doi: 10.1128/MCB.24.21.9478-9486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu D., Guo R., Sobeck A., Bachrati C.Z., Yang J., Enomoto T., Brown G.W., Hoatlin M.E., Hickson I.D., Wang W. RMI, a new OB-fold complex essential for Bloom syndrome protein to maintain genome stability. Genes Dev. 2008;22:2843–2855. doi: 10.1101/gad.1708608. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.