Abstract
Many membrane and secretory proteins that fail to pass quality control in the endoplasmic reticulum (ER) are dislocated into the cytosol and degraded by the proteasome. In applying rigid rules, however, quality control sometimes discharges proteins that, even though defective, retain their function. The unnecessary removal of such proteins represents the pathogenetic hallmark of diverse genetic diseases, in the case of ΔF508 mutant of cystic fibrosis transmembrane conductance regulator being probably the best known example. Recently, the inappropriate proteasomal degradation of skeletal muscle sarcoglycans (α, β, γ and δ) with missense mutation has been proposed to be at the bases of mild-to-severe forms of limb girdle muscular dystrophy (LGMD) known as type 2D, 2E, 2C and 2F, respectively. The quality control pathway responsible for sarcoglycan mutant disposal, however, is so far unexplored. Here we reveal key components of the degradative route of V247M α-sarcoglycan mutant, the second most frequently reported mutation in LGMD-2D. The disclosure of the pathway, which is led by the E3 ligases HRD1 and RFP2, permits to identify new potential druggable targets of a disease for which no effective therapy is at present available. Notably, we show that the pharmacological inhibition of HRD1 activity rescues the expression of V247-α-sarcoglycan both in a heterologous cell model and in myotubes derived from a LGMD-2D patient carrying the L31P/V247M mutations. This represents the first evidence that the activity of E3 ligases, the enzymes in charge of mutant fate, can be eligible for drug interventions to treat sarcoglycanopathy.
INTRODUCTION
Up to one-third of newly synthesized proteins mature into the endoplasmic reticulum (ER). Continuous monitoring of ER functional status is essential for cellular homeostasis. In fact, occasionally, proteins fail to fold properly because of transcriptional and translational errors or the imbalanced production of complex subunits. In addition, gene mutations might also lead to the formation of misfolded/unfolded polypeptides. To avoid their potentially harmful accumulation, such aberrant proteins are (i) recognized by the quality control system, (ii) transported to ER extraction sites, (iii) polyubiquitinated by ER-associated E3 ligases and (iv) retrotranslocated into the cytoplasm where (v) degradation by the 26S proteasome occurs (1,2). These sequential processes, collectively called ER-associated degradation (ERAD), are carried out by different specialized pathways depending on the position in the protein of structural defects. In fact, defects exposed in ER luminal, transmembrane or cytosolic domains, identify misfolded polypeptides as ERAD-L, ERAD-M or ERAD-C substrates (3). A number of ER luminal chaperones and lectins initially recognize and deliver ERAD substrates to membrane export sites (4,5). Here, a multi-protein complex, led by ER-associated E3 ubiquitin ligases, such as HRD1, gp78, TEB4 or RMA1, marks the misfolded protein with ubiquitin (6,7), an event also favoring the dislocation into the cytosol, where the 26S proteasome resides. The polyubiquitin chain, in fact, offers a handle to p97, an AAA-ATPase that eventually provides the driving force to eradicate ERAD substrates from the membrane (8–10).
We have recently shown that several mutants of α-sarcoglycan are eliminated by the ubiquitin–proteasome system (11). α-Sarcoglycan, together with β-, γ- and δ-sarcoglycan, forms an essential tetrameric complex of striated muscles (12). Sarcoglycans are co-translationally translocated in the ER, where they mature and assemble to be eventually targeted to the cell membrane. Their correct assembly is crucial for a stable localization into the plasma membrane, alongside the dystrophin associated protein complex (13). Mutations in any one of the sarcoglycans compromise complex stability, in fact, loss of the defective protein is accompanied by the reduction or absence of the other subunits. This leads to the development of mild-to-severe, world-wide rare, muscular dystrophies, collectively named sarcoglycanopathies and identified as LGMD-2C-F, according to the sarcoglycan involved (14). They are characterized by a progressive limb-girdle muscle weakness and wasting, with frequent respiratory insufficiency and in some cases cardiomyopathy (15). Sarcoglycanopathy clinical phenotype is very heterogeneous for age of onset, rate of progression and severity, often correlated to the level of residual sarcoglycans. Most of the LGMD-2D cases, due to defects of the α-sarcoglycan coding gene, are associated with missense mutations that could lead to a misfolded protein that becomes an ERAD substrate (11,16,17). Loss or strong reduction of α-sarcoglycan usually causes the absence or reduction of the other subunits, thus depriving muscle fibers of a critical complex (18) and identifying ERAD action as the key pathogenetic mechanism of LGMD-2D (12,16). Even though many disease-causing missense mutations do not have functional consequences, because of the quality control rigid rules, these mutants are prematurely degraded. However, treatments interfering with either ERAD or proteasome attenuated the degradation of misfolded but ‘functional’ sarcoglycans and allowed a significant fraction of the surviving protein to reach the cell surface (11,16,17).
α-Sarcoglycan is expected to be substrate of the ERAD-LM pathway (19). In fact α-sarcoglycan is a type I, membrane-tethered protein with most disease-causing mutations located in the N-terminal domain (12), i.e. the region exposed to the ER lumen during maturation. Yet, none of the ERAD components responsible for the removal of α-sarcoglycan mutants have been identified.
To explore the ERAD pathway, we studied V247M α-sarcoglycan, the second most frequently reported disease-causing mutant (Leiden Muscular Dystrophy pages at www.dmd.nl). Our results show that disposal of V247M α-sarcoglycan is operated by an ERAD-LM pathway comprising the E3 ligase HRD1, its adaptor protein SEL1L, the E2 conjugase UBE2J1, and the dislocon components Derlin-1 and p97. An additional E3 ligase, RFP2, also contributes to the disposal of V247M mutant. Notably, the study demonstrates that the pharmacological inhibition of HRD1 ligase activity is effective in rescuing the expression of α-sarcoglycan mutant both in a heterologous cell system and in myotubes derived from a LGMD-2D patient. This finding represents a very important advancement for the development of a pharmacological therapy to treat sarcoglycanopathy, as, at present, no effective cure is available.
RESULTS
This study was carried out in HEK-293 cells, where wild-type α-sarcoglycan is transported to the cell membrane even in the absence of the other sarcoglycan subunits (20,21). Importantly, in HEK-293 cells constitutively expressing the wild-type forms of β-, γ- and δ-sarcoglycan, most of transiently transfected α-sarcoglycan mutants, like, for example, the V247M protein, is intercepted and degraded by the proteasome and only traces of the sarcoglycan complex are recognizable at the plasma membrane (11), in this resembling the condition present in skeletal muscle of LGMD-2D subjects (18). Here we utilized HEK-293 cells in which only the V247M mutant of α-sarcoglycan was stably transfected (hereafter named V247M cells). All V247M cells express the mutant protein, although with slightly different efficiency (Supplementary Material, Fig. S1B). Inhibition of proteasome led to the accumulation in V247M cells of both mature and deglycosylated forms of the mutant (Supplementary Material, Fig. S1A and B). Moreover, after proteasomal block, a faint α-sarcoglycan signal became evident at the surface of several not permeabilized cells (Supplementary Material, Fig. S1B). Thus, we took advantage of this simplified and efficient cell system to investigate the ERAD pathway of sarcoglycan mutants.
The E3 ubiquitin ligase HRD1 is required for the disposal of V247M α-sarcoglycan mutant
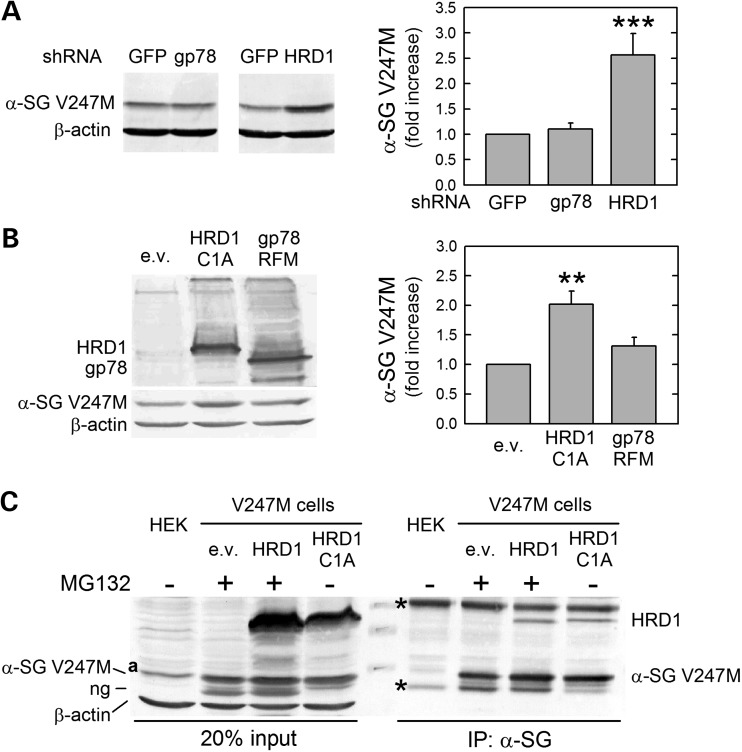
In mammals, two ERAD-LM pathways have been reported so far to process membrane-tethered proteins with luminal defects, like α-sarcoglycan. One is built around the E3 ligase HRD1 and the other to the ligase gp78 (19). Therefore, we first investigated, by RNA interference experiments, the putative involvement of these RING finger E3 ligases in controlling the disposal of V247M α-sarcoglycan mutant. In V247M cells we demonstrated that downregulation of HRD1 by a specific shRNA caused a 2.5-fold increase of V247M protein level, compared with cells transfected with the control GFP shRNA (Supplementary Material, Fig. S2A). By contrast, the silencing of gp78 (Supplementary Material, Fig. S2B and C) had no effect on α-sarcoglycan mutant protein level (Fig. 1A).
Figure 1.
E3 ubiquitin ligase HRD1 is required for the disposal of V247M α-sarcoglycan mutant. (A) Transfection of shRNAs targeting the E3 ligases HRD1 and gp78 was carried out in V247M cells. Depletion of HRD1, but not that of gp78, induced the accumulation of V247M α-sarcoglycan mutant (α-SG-V247M) as revealed by the α-sarcoglycan-specific antibody; as loading control, blots were stained for β-actin. Densitometric analysis of at least four independent experiments is reported in the graph. (B) Inactive variants of the two E3 ligases, HRD1-C1A and c-Myc-tagged gp78-RFM, or the empty vector (e.v.), were transfected with V247M cells. Blots were stained with HRD1, cMyc, α-sarcoglycan and β-actin (as loading control) specific antibodies. Inactive HRD1, but not inactive gp78, induced 2-fold V247M mutant accumulation as determined by densitometric analysis of at least four independent experiments. (C) HRD1 interacts with V247M α-sarcoglycan mutant. V247M cells were transfected with vectors encoding wild-type HRD1, inactive HRD1-C1A, or the e.v.; where indicated (+), to prevent mutant degradation proteasome was inhibited by 10 μm MG132; proteins from naive HEK-293 cells (HEK) were used as negative control. Protein lysates were immunoprecipitated with a monoclonal antibody specific for α-sarcoglycan; IgG of the same isotype were used as negative IP control (see Supplementary Material, Fig. S3); blot was probed for HRD1, α-sarcoglycan, and β-actin. In blot C: ng, non-glycosylated form of V247M α-sarcoglycan mutant (11); a, non-specific protein band recognized by the HRD1 antibody; asterisks, IgG whole molecule and IgG heavy chains; **P < 0.01; ***P < 0.001.
To validate these data, we utilized a mutated form of both HRD1, called HRD1-C1A, and gp78, called RFM-gp78. The substitution of critical RING domain residues (C291A in HRD1, C337S and C374S in gp78) exerts a dominant-negative effect on the activity of the two ligases (22,23) resulting in a reduced disposal of their ERAD substrates. As expected, transfection of V247M cells with the inactive HRD1-C1A substantially increased the level of α-sarcoglycan mutant, whereas the presence of the inactive gp78 had no effect (Fig. 1B).
Then, we examined whether HRD1 interacts with the α-sarcoglycan mutant by performing co-immunoprecipitation (IP) assays. Protein lysates from V247M cells transfected with either wild-type HRD1, or the inactive form HRD1-C1A, were incubated with the antibody-specific for α-sarcoglycan. To prolong the short-lived association between the substrate and its E3 ligase, we utilized the HRD1-C1A inactive form. Indeed, due to RING finger motif disruption, substrate ubiquitination is impaired and interaction is consequently protracted. On the other hand, in wild-type HRD1 overexpression experiments, a similar effect has been obtained by inhibiting proteasome activity. The α-sarcoglycan-specific antibody was able to immunoprecipitate the V247M mutant together with either wild-type HRD1 or inactive HRD1-C1A (Fig. 1C), confirming the close interaction of this E3 ligase with the mutant protein. All together, these results show that the V247M mutant degradative pathway is HRD1-dependent and gp78 dispensable.
The HRD1-associated proteins handling V247M mutant
In mammals, HRD1 complex is composed of several proteins that guarantee recognition, ubiquitination and retro-translocation into the cytosol of ERAD clients. A scaffolding role is played by SEL1L, an adaptor protein able to interact with E3 and E2 enzymes, the ligase substrate (either directly or by means of lectin-binding proteins), as well as with other components of the complex (5,24).
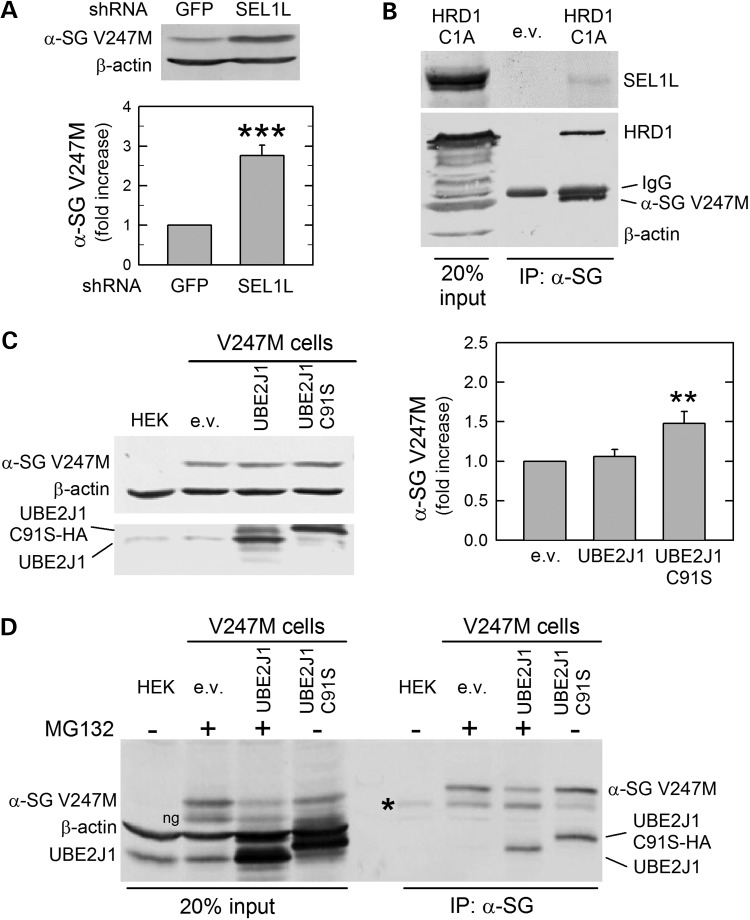
Downregulation of SEL1L by a specific shRNA (Supplementary Material, Fig. S2B) caused about a 3-fold increase of V247M mutant level (Fig. 2A), suggesting that either the altered stoichiometry of HRD1 complex might reduce the E3 ligase activity (25), or the reduction of SEL1L might compromise the nexus between the proteins recognizing the substrate, still unknown for α-sarcoglycan, and HRD1 (5). Whatever is the case, the result is the salvage of the V247M mutant. The involvement of the adaptor protein has also been confirmed by co-IP assays. In fact, as expected, from protein lysates of V247M cells transfected with HRD1-C1A, both HRD1 and SEL1L co-precipitated with the α-sarcoglycan mutant (Fig. 2B).
Figure 2.
SEL1L and UBE2J1 co-operates with HRD1 in the disposal of V247M α-sarcoglycan mutant. (A) Reduction of SEL1L, produced by the expression in V247M cells of a specific shRNA, permitted the accumulation of the α-sarcoglycan mutant; β-actin expression was probed as loading control. The graph shows the densitometric analysis of at least four independent experiments. (B) V247M mutant interacts with both SEL1L and HRD1. Proteins solubilized from V247M cells transfected with inactive HRD1-C1A or the e.v. were immunoprecipitated with a monoclonal antibody specific for α-sarcoglycan, IgG of the same isotype were used as negative IP control (see Supplementary Material, Fig. S3). Blot was probed with SEL1L, HRD1, β-actin and α-sarcoglycan-specific antibodies. (C) The inactive UBE2J1 causes the increase of V247M mutant. Wild-type UBE2J1, the inactive variant HA-tagged UBE2J1-C91S or the e.v. were transfected in V247M cells. Blots were stained with either UBE2J1, β-actin and α-sarcoglycan-specific antibodies. Densitometric analysis of at least four independent experiments is reported in the graph. (D) UBE2J1 interacts with V247M mutant. V247M cells were transfected with wild-type UBE2J1, inactive HA-tagged UBE2J1-C91S, or the e.v.; where indicated (+), to prevent mutant degradation, proteasome was inhibited by 10 μm MG132. Protein lysates were immunoprecipitated with a monoclonal antibody specific for α-sarcoglycan; IgG of the same isotype were used as negative IP control (see Supplementary Material, Fig. S3); blot was probed for β-actin, α-sarcoglycan and UBE2J1. HEK, naive HEK-293 cells, used in C and D as negative control; ng, non-glycosylated form of V247M α-sarcoglycan mutant (11); asterisk in blot D, IgG heavy chains; **P < 0.01; ***P < 0.001.
Crucial for the ubiquitination activity of the RING finger HRD1 ligase is the association with the E2 ubiquitin conjugase. Two ER membrane-associated E2 enzymes, UBE2J1 (aka UBC6e) and UBE2J2, and the soluble UBE2G2, have been identified to work in a concerted action with ER-associated E3 ligases (6). The UBE2J1 conjugase has been already demonstrated to work in conjunction with HRD1 in the disposal of misfolded MHC class I heavy chain (26), whereas the UBE2G2 has been shown to support gp78 (27). Therefore, to verify whether UBE2J1 is part of the HRD1 complex disposing V247M mutant, we transfected V247M cells with either wild-type UBE2J1 or its inactive mutant, HA-tagged UBE2J1-C91S (28). The inactive E2 conjugase, because of a critical substitution in the ubiquitin conjugating domain, was unable to attach ubiquitin to the V247M mutant resulting in a diminished degradation. On the other hand, overexpression of wild-type UBE2J1 had no effects on the mutant level (Fig. 2C).
Then, we verified the interaction between UBE2J1 and V247M mutant, by co-IP experiments in protein lysates from V247M cells transfected with either wild-type UBE2J1 or the inactive form UBE2J1-C91S. As a consequence of inactivation, the interaction between UBE2J1-C91S and its substrate is prolonged, in that improving the co-IP assay. For the same purpose, in cells transfected with either the empty vector or the wild-type form of UBE2J1, the proteasome was inhibited by MG132. Both the E2 conjugase and the mutated α-sarcoglycan were present in the immunocomplexes recovered by the α-sarcoglycan-specific antibody (Fig. 2D). Thus, we concluded that UBE2J1 acts in concert with HRD1 ligase in targeting V247M mutant to degradation.
Dislocation from the ER of V247M mutant
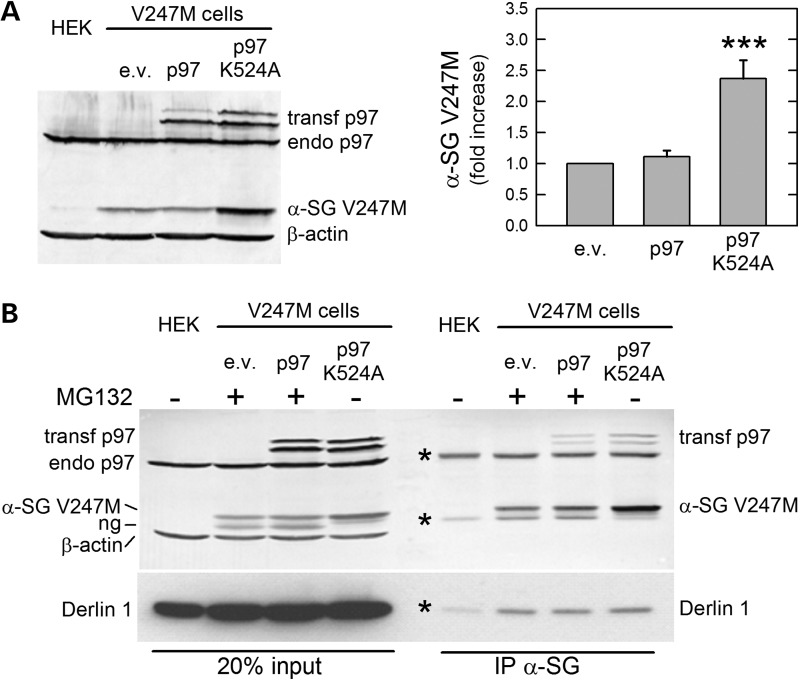
To be degraded, ERAD substrates must cross the ER membrane and reach the cytosol, where proteasome resides. Diverse ERAD factors are involved in the dislocation process (included the E3 ligase complexes), whereas the force to eradicate ubiquitinated proteins from the ER is, in most cases, provided by the cytosolic AAA-ATPase p97 (8). In this context, Derlin-1 works in concert with p97 and, because of its rhomboid architecture, by compressing and deforming the lipid bilayer, may help ERAD substrates crossing the membrane (29). Thus, we asked whether processing of α-sarcoglycan mutants also comprises these two ERAD components. Overexpression of wild-type GFP-tagged p97 (30) was without major effects on V247M α-sarcoglycan expression, suggesting that the endogenous level of p97 is sufficient to fully extract α-sarcoglycan mutant from the ER. On the contrary, the presence of the dominant-negative mutant p97-K524A, in which the K524A substitution in the Walker A domain inactivates the enzymatic activity (30), strongly reduced degradation of V247M mutant (Fig. 3A).
Figure 3.
The AAA-ATPase p97 and Derlin-1 are involved in the retro-translocation of V247M α-sarcoglycan mutant. (A) Wild-type or a dominant-negative form of p97 (p97 K524A), both GFP-tagged, or the e.v. was transfected in V247M cells. Blots were stained with p97, α-sarcoglycan and β-actin-specific antibodies. p97-specific antibody recognized both the ectopically expressed and endogenous forms of the protein. The dominant-negative form of p97 induced >2-fold V247M mutant accumulation, as determined by densitometric analysis of at least four independent experiments, whereas wild-type p97 had no effects. (B) p97 and Derlin-1 interact with V247M mutant. V247M cells were transfected with wild-type p97, inactive p97 K524A, both GFP-tagged, or the e.v.; where indicated (+), to prevent mutant degradation, proteasome was inhibited with 10 μm MG132. Protein lysates were immunoprecipitated with a monoclonal antibody-specific for α-sarcoglycan ; IgG of the same isotype were used as negative IP control (see Supplementary Material, Fig. S3); blots were probed for p97, Derlin-1, α-sarcoglycan and β-actin. Naive HEK-293 cells (HEK) were used as negative control; ng, non-glycosylated form of V247M mutant protein (11); asterisks in blot B, IgG whole molecule and IgG heavy and light chains; endo, endogenous p97; transf, transfected forms of p97; ***P < 0.001.
To verify the interaction between V247M α-sarcoglycan, p97 and, possibly, Derlin-1, we performed IP experiments by using the α-sarcoglycan-specific antibody in protein lysates from V247M cells transfected with either wild-type or inactive p97. Being the dominant-negative form of p97 unable to eradicate the protein from the ER, a large amount of α-sarcoglycan mutant co-sedimented with the AAA-ATPase even from cells where the proteasome was fully active (Fig. 3B). Inhibition of proteasome, however, by reducing mutant degradation, permitted the IP of V247M mutant also together with the wild-type form of p97, indicating a close association between the two proteins during retrotranslocation.
In Figure 3B, endogenous Derlin-1, whose molecular weight is ∼29 kDa (29), has the same electrophoretic mobility of α-sarcoglycan antibody light chains, making the interpretation of results difficult. However, it is evident that the intensity of the antibody light chain band is strongly increased in all immunoprecipitated samples, but not in the negative control (HEK), where only the antibody is present. All together these data strongly support the idea that Derlin-1 helps the extraction of the α-sarcoglycan mutant from the ER membrane, with the p97 ATPase providing the driving force for such operation.
V247M mutant interacts with the endogenous dislocon
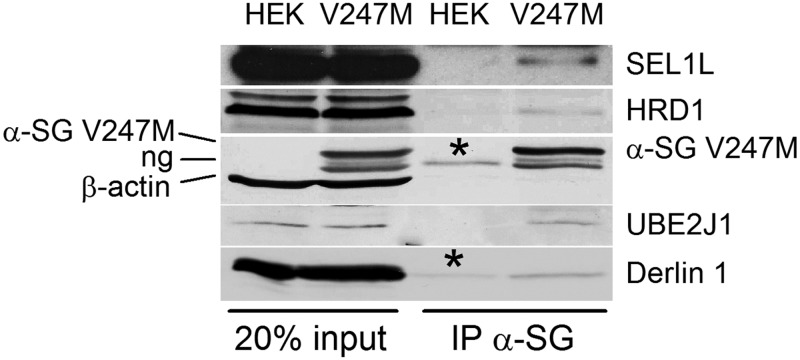
To definitely demonstrate that V247M α-sarcoglycan is the substrate of HRD1 complex, we performed an IP assay in 1% digitonin solubilized proteins from V247M cells with the antibody specific for α-sarcoglycan. To prolong the association among HRD1 complex components and α-sarcoglycan mutant, we inhibited proteasome activity with MG132. The recovered immunocomplexes contained all the endogenous ERAD partners identified with the above described experiments, i.e. SEL1L, HRD1, UBE2J1 and Derlin-1 (Fig. 4).
Figure 4.
The α-sarcoglycan V247M mutant interacts with the endogenous dislocon components. 1% Digitonin solubilized proteins from naive HEK-293 (HEK) and V247M cells, both treated for 8 h with the proteasome inhibitor MG132, were incubated overnight with the α-sarcoglycan monoclonal-specific antibody. Immunocomplexes subjected to SDS-PAGE and western blot, were probed for SEL1L, HRD1, α-sarcoglycan, β-actin, UBE2J1, and Derlin-1. ng, non-glycosylated form of V247M mutant protein (11); asterisks, IgG heavy and light chains.
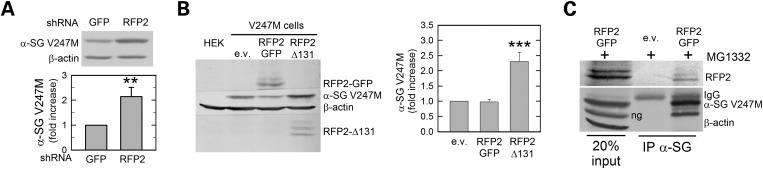
The E3 ligase RFP2 contributes to V247M mutant degradation
Besides HRD1 and gp78 (22,31), additional ER membrane-bound E3 ligases have been reported to be active in the processing of ERAD substrates, such as, for example, TEB4, RMA1, TRC8 and RFP2 (23,32–34). Interestingly, RFP2, a RING finger E3 ligase involved in ERAD of CD3-δ and the CaV 1.2 channel (34,35) is highly expressed in skeletal muscle (36). Thus, we first investigated its possible involvement in the ubiquitination of sarcoglycans by RNA interference experiments. RFP2 downregulation (Supplementary Material, Fig. S2E) caused almost the doubling of α-sarcoglycan mutant level (Fig. 5A), demonstrating that the loss of RFP2 has critical consequences on the disposal of V247M mutant. To confirm this result, we transfected V247M cells with either a mutated form of RFP2, lacking the RING domain (Δ131-RFP2) and consequently devoid of the ligase activity (35), or the wild-type GFP-tagged form of RFP2. Overexpression of wild-type RFP2, detectable in western blot as a main protein band of ∼74 kDa plus additional weak bands known to be the auto-ubiquitinated forms of the ligase (34), had no major consequences on the degradation of V247M mutant. On the other hand, the inactive Δ131-RFP2, recognizable as a low-molecular weight protein band of ∼28 kDa, remarkably prevented the degradation of V247M mutant (Fig. 5B).
Figure 5.
The E3 ligase RFP2 is involved in the disposal of V247M α-sarcoglycan mutant. (A) RFP2-specific shRNA expression permitted α-sarcoglycan mutant accumulation in V247M cells compared with control (GFP); β-actin level was probed as loading control. The graph shows the densitometric analysis of at least three independent experiments. (B) Overexpression of the inactive form of RFP2 (Δ131-RFP2) delays degradation of V247M mutant, permitting a 2-fold increase of its level, whereas overexpression of wild-type GFP-tagged RFP2 (RFP2-GFP) was ineffective. Wild-type ligase is detectable as a main protein band of ∼74 kDa plus additional weak bands, probably auto-ubiquitinated forms (34); the inactive RFP2 presents a lower molecular weight due to ring deletion (35). Densitometric analysis of at least four independent experiments is reported in the graph. (C) RFP2 interacts with V247M α-sarcoglycan. V247M cells were transfected with wild-type RFP2 or the e.v. and incubated for 8h before lysis with the proteasome inhibitor MG132. Protein lysates were immunoprecipitated with a monoclonal antibody specific for α-sarcoglycan; IgG of the same isotype were used as negative IP control (see Supplementary Material, Fig. S3). Blots were probed for RFP2, α-sarcoglycan and β-actin. ng, non-glycosylated V247M α-sarcoglycan mutant (11); **P < 0.01; *** P < 0.001.
Then, we assessed the interaction of the E3 ligase RFP2 with the α-sarcoglycan mutant by co-IP assay. To prolong the putative interaction between the enzyme and its substrate, V247M cells, transfected with the GFP-tagged wild-type form of RFP2, were treated with the proteasome inhibitor MG132. Figure 5C shows that the antibody specific for α-sarcoglycan was able to sediment the mutant protein together with RFP2. These data strongly suggest that also this E3 ligase acts in the disposal of V247M mutant.
Both HRD1 and RFP2 ubiquitinate V247M mutant
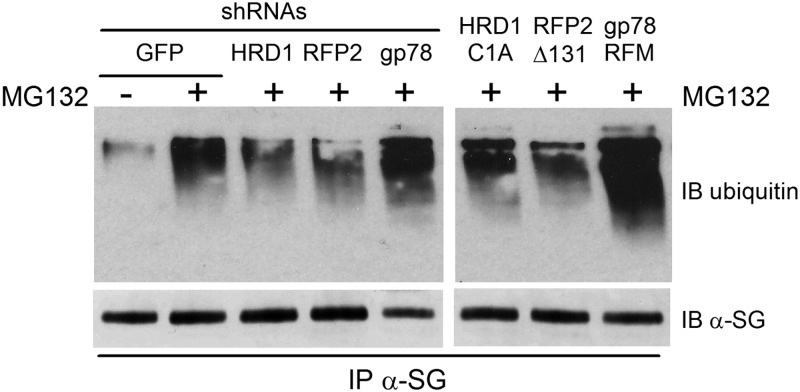
Next, we examined the functional role of HRD1 and RFP2 E3 ligases by evaluating the level of V247 mutant ubiquitination after knocking down either HRD1, RFP2 or gp78. IP of α-sarcoglycan in V247M cells transfected with the control shRNA (GFP) and treated with the proteasome inhibitor MG132 evidenced a large increase of mutant ubiquitination compared with untreated cells (Fig. 6, left). Importantly, in the same cells, V247M protein polyubiquitination was substantially reduced when HRD1 or RFP2 were depleted by the specific shRNAs, whereas, as expected, the level of ubiquitination was unchanged when the shRNA for gp78 was used. Similar results were obtained by overexpressing the inactive forms of the three ligases. In fact, the presence of either HRD1-C1A or Δ131-RFP2 substantially decreased the ubiquitination of V247M mutant, whereas gp78-RFM was ineffective (Fig. 6, right).
Figure 6.
HRD1 and RFP2 are the E3 ligases responsible for the V247M mutant ubiquitination. Left blot, V247M cells were transfected with the indicated shRNAs. Right blot, V247M cells were transfected with the indicated inactive forms of the E3 ubiquitin ligases (22,23,35). Eight hours before lysis, cells were treated with either 10 μm MG132 (+), in order to block protein degradation through the proteasome, or the vehicle DMSO (−). Two hundred micrograms of proteins solubilized in RIPA buffer were incubated overnight with the α-sarcoglycan monoclonal-specific antibody. After SDS PAGE, blots were probed with antibodies-specific to mono and polyubiquitinated proteins (IB ubiquitin) and to α-sarcoglycan (IB α-SG).
Pharmacological inhibition of HRD1 leads to V247M mutant recovery
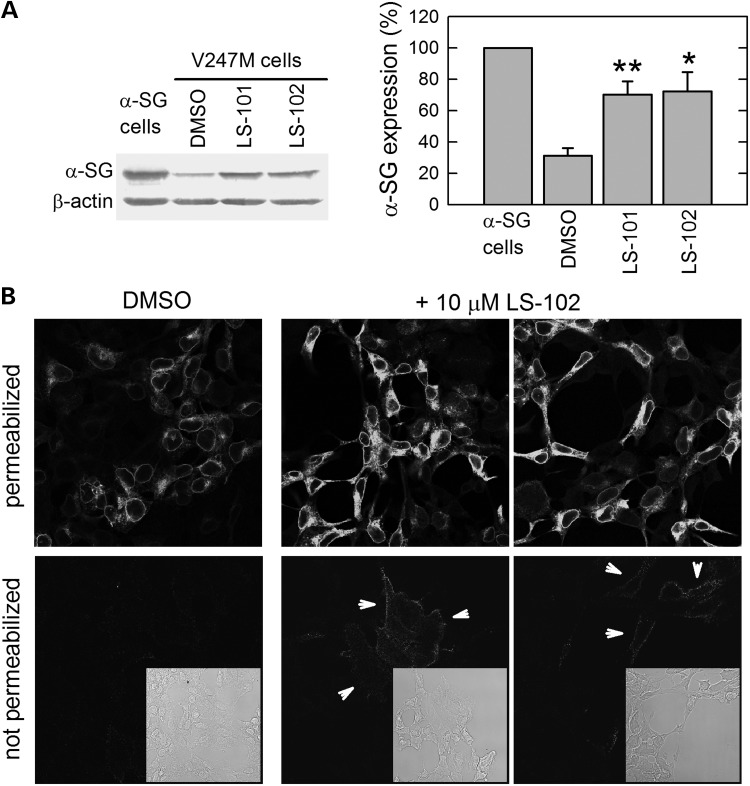
The ultimate goal of our work was the identification of new pharmacological targets for the treatment of sarcoglycanopathy. To this end, we tested two small molecules, LS-101 and LS-102, recently identified by high-throughput screening as strong inhibitors of the E3 ligase HRD1, and proposed as drug candidates for the treatment of Rheumatoid arthritis (37). Incubation of V247M cells for 8 h with either 15 μm LS-101 or 20 μm LS-102 led to a 3-fold increase of α-sarcoglycan mutant level, practically reaching that of the wild-type protein (Fig. 7A). Then, to evaluate the cytolocalization of the protein after LS-102 treatment, we performed an experiment in which V247M cells, grown on glass coverslips, were treated for 8 h with either 10 μm LS-102 or its vehicle, dimethyl sulfoxide (DMSO). In permeabilized cells (Fig. 7B), it is well evident the intracellular increase of α-sarcoglycan mutant level after incubation with LS-102, in comparison to DMSO treatment. Importantly, this short treatment induced the appearance of some of the rescued V247M mutant in the plasma membrane of several not permeabilized cells (see arrowheads in Fig. 7B).
Figure 7.
Rescue of V247M mutant in V247M cells after pharmacological inhibition of the E3 ligase HRD1. (A) V247M cells were treated for 8 h with either 15 μm LS-101 or 20 μm LS-102, specific HRD1 ligase inhibitors (37). DMSO, the vehicle of HRD1 inhibitors, was used as negative control. Cells expressing wild-type α-sarcoglycan were used as positive control. β-actin level was used to normalize protein loading. Densitometric analysis of at least three independent experiments, in which the level of wild-type α-sarcoglycan was set as 100%, is reported in the graph. *P < 0.05; **P < 0.01. (B) V247M cells were treated for 8 h with the HRD1 inhibitor LS102 (10 μm) or its vehicle DMSO (1‰). At the end of treatments, cells were either permeabilized with Triton X-100 or left intact (not permeabilized) and immunodecorated with the monoclonal α-sarcoglycan-specific antibody. Localization of α-sarcoglycan was revealed with the secondary DyLight 488-conjugated anti-mouse antibody. Insets are the transmitted light images of the same IF fields. All images, collected with a Leica SP2 confocal microscope, are at the same magnification. It is well evident that, after HRD1 inhibitor treatment, the intensity of the intracellular α-sarcoglycan signal increased, and that, in several not permeabilized cells (arrowheads) a faint membrane stain became evident.
Inhibition of HRD1 rescues sarcoglycan complex in primary myotubes from a LGMD-2D patient
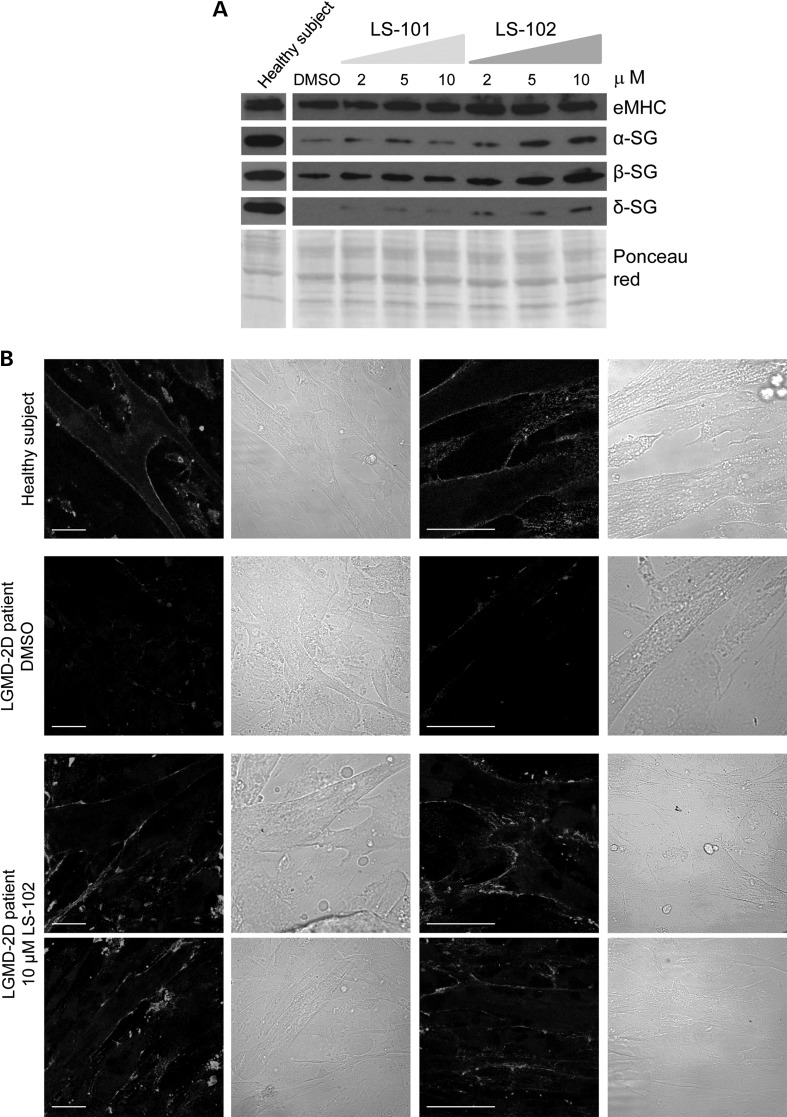
We finally tested the efficacy of the small molecules LS-101 and LS-102 in rescuing α-sarcoglycan mutant in skeletal muscle cells derived from a heterozygous LGMD-2D patient carrying the L31P and V247M substitutions on the α-sarcoglycan alleles. A small bioptic fragment from the LGMD-2D patient, received by the Telethon Genetic Biobank, was utilized to induce satellite cell sprouting, growth and differentiation into myotubes. At the fifth day of differentiation, increasing concentration of either LS101 or LS102, or the vehicle DMSO, were added to the myotubes. Forty-eight hours later, cells were lysed and the expression of different sarcoglycans was analyzed by western blot (Fig. 8A). For comparison, the total cell lysate from 7-day differentiated control myotubes, derived from immortalized satellite cells of a healthy subject, was probed for α-, β- and δ-sarcoglycan and embryonic myosin heavy chain expression. The latter marker was used to check the differentiation homogeneity of the different samples. It is well evident that, by inhibiting the HRD1 E3 ligase activity in the LGMD-2D patient myotubes, the amount of α-, β- and δ-sarcoglycan substantially increased. The effect is more pronounced for α- and δ-sarcoglycan as their basal level, in untreated cell, is very low or almost absent. It seems also clear that LS102, the more specific HRD1 inhibitor (37), produced a more effective, dose-dependent recovery of sarcoglycans. In parallel experiments, myoblasts from the LGMD-2D patient, were grown and differentiated on glass coverslips and treated with either 10 μm LS-102 or 1‰ DMSO for 48 h. Thereafter, not permeabilized myotybes were probed with an antibody specific for an extracellular epitope of α-sarcoglycan. As positive control, myotubes derived from the healthy subject were also immunodecorated. Figure 8B shows that α-sarcoglycan is homogeneously expressed at the surface of all myotubes from the healthy subject, whereas only traces of α-sarcoglycan staining are evident in untreated myotubes from the LGMD-2D patient. Instead, inhibition of HRD1 activity with LS-102 induced a marked, although not homogeneous localization of α-sarcoglycan mutant at the cell membrane of almost all myotubes derived from the LGMD-2D patient (Fig. 8B).
Figure 8.
Inhibition of the E3 ubiquitin ligase HRD1induced sarcoglycan rescue in myotubes from a LGMD-2D patient. (A) Five-day differentiated myotubes of a LGMD-2D patient, carrying the L31P and V247M mutations on the α-sarcoglycan alleles, were incubated for 48 h with either DMSO, used as negative control, or increasing concentration of LS-101 and LS-102, as indicated. At the end of incubation, myotubes were lysed and the expression of α-sarcoglycan (α-SG), β-sarcoglycan (β-SG) and δ-sarcoglycan (δ-SG) was analyzed by western blotting. To verify the differentiation homogeneity of the different samples the expression of embryonic myosin heavy chain (eMHC) (38) was measured. By comparison, the expression of the same proteins was evaluated in 7-day old myotubes derived from a healthy subject (39). As loading control, the Ponceau red staining of the membranes is reported. (B) Myoblasts form the LGMD-2D patient were grown on glass coverslips and differentiated for 7 days. At the fifth day of differentiation, either DMSO (negative control) or 10 μm LS102 were added. Two days later, not permeabilized myotubes were immunodecorated with the rabbit polyclonal antibody specific for an extracellular epitope of α-sarcoglycan. As positive control 7-day differentiated, not permeabilized myotubes derived from a healthy subject, were immunolabeled with the same antibody. The primary antibody was visualized by a DyLight 488-conjugated anti-rabbit secondary antibody. On the right of each fluorescence image, the transmitted light image of the same field is reported; in each images bars indicate 30 μm. Images were recorded with a Leica SP2 laser scanning confocal microscope at the same setting conditions.
DISCUSSION
Maturation and assembly of the skeletal muscle membrane sarcoglycan complex occur in the ER (13). However, little is known about the ER processing of the four sarcoglycans (α, β, γ and δ), how the hetero-tetrameric complex assembles, and, more importantly, which ER quality control mechanisms control the fate of misfolded or orphan subunits. Our lack of knowledge, particularly about the latter point, is a serious limit to develop suitable therapeutic intervention when the inappropriate disposal of sarcoglycan mutants causes sarcoglycanopathy. Most of sarcoglycanopathy cases are in fact associated with missense mutations that can lead to a misfolded protein. However, many disease-causing missense mutations are without functional consequences (12,17), nonetheless, the mutant is intercepted by the quality control system and delivered to the proteasome. The premature degradation precludes the possibility for the mutated, but probably functional protein, to assemble with its partners, exit the ER, and move to the plasma membrane. The result is, de facto, a loss of function responsible for sarcoglycanopathy.
Importantly, recent evidence indicates that pharmacological treatments interfering with either the initial maturation steps (17) or the ubiquitin–proteasome system (11) might prevent the removal of some of the α-sarcoglycan mutants and promote their functional recovery. As at present, no effective cure is available to treat sarcoglycanopathy, the final goal of our work was the identification of new molecular targets potentially eligible for the pharmacological treatment of the disease. To this intent, we decided to decipher the ERAD pathway controlling the disposal of V247M α-sarcoglycan. The results demonstrated that the V247M α-sarcoglycan mutant, a typical type I membrane protein with the defect located in the luminal domain, follows the ERAD-LM pathway, and that two E3 ligases, HRD1 and RFP2, are responsible for its ubiquitination and delivery to proteasome.
First, by using different and selective molecular approaches, we demonstrated that the naturally occurring V247M mutant of α-sarcoglycan is handled by the HRD1 complex. In mammals, basic components of this complex are the E3 ligase HRD1, the adaptor protein SEL1L, that exerts a fundamental role in presenting substrates to the ligase (19,24), and one E2 conjugase, that working in concert with the RING finger E3 ligase, links ubiquitin to the substrate (6). Our data demonstrate that HRD1, SEL1L and the E2 conjugase UBE2J1 interact with V247M mutant as they are co-IP by the α-sarcoglycan-specific antibody. These ERAD components play a key role in the disposal of V247M mutant, as either their knock down or the overexpression of specific inactive forms of such proteins led to α-sarcoglycan mutant accumulation (Figs 1 and 2). The activity of the HRD1 complex is usually assisted by the cytosolic AAA-ATPase, p97, that provides the force to eradicate polyubiquitinated proteins form the ER membrane (9,40), and by Derlin-1, that facilitates the extraction of substrates from the membrane (29). Accordingly, IP assays and experiments with dominant-negative forms, show that p97 and Derlin-1 are two fundamental ERAD players in the disposal of V247M mutant (Fig. 3).
Our results show that another E3 ligase, RFP2, cooperates with HRD1 in the processing of V247M mutant. This is not surprising, as emerging evidence demonstrates that two, or even more, E3 ligases might assist the processing of certain ERAD substrates. For example, HRD1 cooperates with gp78 in the degradation of mutant neuroserpin (41), whereas gp78 together with Trc8 operates the sterol-mediated degradation of 3-hydroxy-3-methylglutaryl coenzyme A reductase (42), and the orphan CD3-δ subunit is client of both gp78 and RFP2 E3 ligases (34). The latter ligase also mediates the degradation of CaV 1.2 channels (35) and regulates the expression of the p53 tumor suppressor by degrading MDM2 (43). Considering that RFP2 is highly expressed in skeletal muscle, as well as in HEK-293 cells (36), we thought RFP2 as an additional E3 ligase handling sarcoglycan. Knocking down experiments or inactive RFP2 overexpression confirmed this hypothesis (Figs. 5 and 6). Interestingly, RFP2 ligase activity has been linked to the UBE2J1 conjugase (34), Derlin-1 and p97 (35), ERAD elements also working with the HRD1 ligase, suggesting that α-sarcoglycan mutants follow a common ERAD pathway converging on alternative E3 ligases. In this light, it will be a stimulating challenge to understand the functional meaning of the cross-talk between ubiquitin ligases. As RFP2 is known to remove orphan subunits of multimeric complexes (34,35), our data are the first demonstration that RFP2 also handles misfolded mutated proteins. However, as α-sarcoglycan is member of a tetramer, and our experiments have been carried out in cells expressing solely this protein, RFP2 role in V247M α-sarcoglycan disposal might be linked to the propensity of this ligase to take care of unassembled subunits. Additional experiments are needed to test this possibility.
In conclusion, by deciphering the ERAD pathway responsible for the processing of V247M mutant we have focused the interest on the two key elements of the pathway, HRD1 and RFP2. The E3 ligases, for their fundamental role in driving substrate disposal, are emerging as significant druggable targets (44) for the treatment of different diseases involving the ubiquitin–proteasome system (45). In this respect, our work, for the first time, addressed the activity of an E3 ligase involved in the disposal of an α-sarcoglycan mutant causing LGMD-2D. Two small molecules, screened as inhibitors of HRD1 (37) were actually able to recover the expression of the V247M mutant both in a cellular model (V247M cells) and, remarkably, in primary skeletal muscle cells derived from a LGMD-2D patient. In the cellular model, a short treatment (8 h) with both LS101 and LS102, produced a recovery of the mutant that almost reached the level of wild-type α-sarcoglycan expressed in the HEK293 cells (Fig. 7A) and permitted the appearance of the V247M-α-sarcoglycan on the cell surface. Moreover, inhibition of the ligase activity seems to block also the dislocation from the ER, as the recovered mutant is fully glycosylated, differently to what happens when the proteasome is blocked, a treatment that produces the accumulation of both the mature and cytosolic deglycosylated form of α-sarcoglycan (11). The availability of primary cells from a LGMD-2D patient, permitted us to test the efficacy of the two small molecules not only in the myogenic cell background but, more importantly, directly on cells of a human subject affected by the disease. The genetic analysis from this LGMD-2D patient indicated the presence of a T>C substitution at position 98 of one allele and a G>A substitution at position 739 of the other allele of the SGCA gene leading to L31P and V247M missense mutations on α-sarcoglycan. The residual α-sarcoglycan expression in the skeletal muscle tissue, as determined by western blotting analysis, was ∼5%, whereas no information was available for β-, γ- and δ-sarcoglycan (data from Telethon Genetic Biobank). At 7 days of differentiation, in comparison with sarcoglycan expression in myotubes of a healthy subject, patient myotubes show a very low level of both α- and β-sarcoglycan protein, whereas δ-sarcoglycan is almost undetectable. Both LS101 and LS102 seems to be effective in rescuing α-sarcoglycan, with the second molecule causing a much more pronounced recovery of the protein, in a dose-dependent fashion. This is true not only for the mutated α-sarcoglycan, but also for the wild-type partners, δ-sarcoglycan and β-sarcoglycan, suggesting that the beneficial effect of the treatments is extended to the entire sarcoglycan complex. Even though our data cannot establish if both, or just one, of the two allelic forms of α-sarcoglycan has been recovered, the result is very promising. In fact, the central issue is that the amount of saved α-sarcoglycan should reach a level sufficient to assemble with the other sarcoglycans which in turn, being no more orphan subunits, are preserved from degradation. It is important to underline that the severity of the disease phenotype is strictly associated with the residual level of sarcoglycans present in muscle fibers (12,18), therefore, even a small overall sarcoglycans rescue is expected to ameliorate patient condition.
Despite still preliminary, our work has identified novel potential druggable targets providing new hints for the search and/or optimization of potent and selective inhibitors of the ERAD pathway specifically involved in α-sarcoglycan mutant disposal. Moreover, the successful pharmacological intervention in both a cellular model and in a LGMD-2D patient specimen can be considered a proof of principle study on the development of a future sarcoglycanopathy therapy, specifically targeting an E3 ubiquitin ligase.
MATERIALS AND METHODS
Cell culture, transfection, and treatments
HEK-293 and V247M cells, were grown in Dulbecco's modified Eagle's medium (Sigma) supplemented with 10% fetal bovine serum (FBS) (Gibco) and maintained in a humidified atmosphere containing 5% CO2 at 37 °C. V247M cells were generated by HEK-293 cell transfection with the mutated α-sarcoglycan cDNA cloned in pcDNA3 and subsequent G418 selection.
Immortalized human myoblasts (39), from the ‘Human cell culture platform’ of the Myology Institute in Paris, were grown in Skeletal Muscle Cell Growth Medium (Promocell) supplemented with 15% FBS (Gibco), named skeletal growth medium (SGM). To start differentiation, myoblasts grown at confluence were incubated with DMEM supplemented with 2% horse serum (Euroclone), 10 µg/ml human recombinant insulin (Sigma), 100 µg/ml human Apotransferrin (Sigma), named skeletal differentiating medium (SDM). Differentiation was carried out for seven days.
For overexpression and RNAi experiments, V247M cells were seeded at ∼50 000 cells/cm2 and transiently transfected the day after seeding with Transit293 (MirusBio) according to manufacturer's instruction. Cells were lysed either 48 or 72 h (RNAi assays) after transfection and subjected to direct western blotting analysis or immunoprecipitation assay. MG132 (Sigma) or HRD1 inhibitors, kindly provided by Toshihiro Nakajima (Tokyo Medical University) (37), were added 8 h before V247M cell lysis or 8–48 h before immunofluorescence staining.
Culture and differentiation of primary cells derived from a human muscular biopsy
Thanks to the Telethon Genetic Bio-Bank facility we obtained a small fragment of a frozen biopsy from a LGMD-2D patient carrying two different missense mutations (V247M/L31P) on the α-sarcoglycan alleles. The biopsy, washed twice, was incubated with SGM containing 25% human plasma from donors, in order to form a clot. A week later, cells started to spread off and the biopsy was cut into small pieces, transferred on gelatin-coated plastic wells and incubated in SGM. Myoblasts, spreading from the biopsy fragments, were expanded and subsequently used to carry out experiments. When at confluence, myoblasts were differentiated by changing SGM with SDM. Differentiation was carried out for 7 days. E3 ligase inhibitors were added 48 h before myotubes lysis.
Plasmids
V247M-α-sarcoglycan construct was generated as previously described (11). p97-GFP and p97 K524A-GFP mutant were kindly provided by Akira Kakizuka (Kyoto University); HRD1 and HRD1-C1A mutant were obtained from Emanuel Wiertz (Leiden University); murine gp78 C337/374S RFM mutant was donated by Kazuhiro Nagata (Kyoto University); human gp78-HA and the plasmid expressing gp78-specific shRNA were kindly provided by Petek Ballar (Ege University); UBE2J1 and UBE2J1-C91S were from Hidde L. Ploegh (WIBR, Cambridge); RFP2 and Δ131-RFP2 mutant were obtained from Gerald W. Zamponi (University of Calgary). RFP2, originally cloned in pUC57 vector, was subcloned in the pEGFPN1 vector, in frame with GFP.
shRNA producing vectors
The target sequences for shRNA-mediated knock down are: GAGACAGTTTCAGATGATT for HRD1, CAGATAAAGTGAAGGAATT for RFP2, GGCTATACTGTGGCTAGAA for SEL1L, GAAGTCGTGCTGCTTCATG for GFP, GACTACACAAATCAGCGATTT for LacZ, the last two used as controls. Complementary oligonucleotides comprising the sense and antisense sequences separated by a standard TTCAAGAGA loop structure were purchased from MWG, annealed and cloned into the pSUPER vector. All constructs were checked by sequencing.
Antibodies
The antibodies utilized were: mouse monoclonal α-sarcoglycan from DBA and home-made rabbit polyclonal α-sarcoglycan antibody (raised against the peptide ‘CYDTLAPHFRVDW’ mapping in the extracellular domain of the protein, see Supplementary Material, Fig. S4 for the characterization); HRD1, Derlin1, p97, GFP and UBE2J1 from Abcam; β-actin and SEL1L from Sigma Aldrich; HA and Myc from Millipore; RFP2, gp78 and mouse IgG from Santa Cruz Biotechnology; mono and polyubiquitinated conjugates from Enzo Life Science; embryonic myosin heavy chain was a kind gift of Stefano Schiaffino (VIMM, Padova) (38); DyLight 488-conjugated anti-mouse and anti-rabbit antibodies from Jackson.
Immunoprecipitation and immunoblotting
For western blotting analysis, total protein homogenate was obtained by lysing cells with 5% sodium deoxycholate solution supplemented with complete protease inhibitor (Roche). For co-immunoprecipitation assays, cells were lysed with 50 mm TRIS pH 7.4, 150 mm NaCl, 5 mm EDTA pH 7.4, 1% Triton X-100 supplemented with protease inhibitors and 2 mm N-ethylmaleimide (NEM) (Sigma). Analyses were performed on the Triton X-100 soluble fraction. Where indicated, proteins were solubilized in 1% digitonin or radioimmunoprecipitation assay (RIPA) buffer supplemented with protease inhibitors and 2 mm NEM (Sigma). Proteins were quantified by the bicinchoninic acid protein assay kit (Thermo Scientific) according to manufacturer's instructions. For immunoprecipitation assays, 200 μg of solubilized proteins were incubated overnight at 4°C with either the mouse monoclonal α-sarcoglycan-specific antibody or with mouse IgG of the same isotype, as the negative control. The following day, Protein G-magnetic beads (Millipore) were added and the mixture incubated 1 h in the same condition. Beads were recovered and extensively washed with lysis buffer and finally aspirated to dryness. After Laemmli buffer addition, samples were analyzed by sodium dodecyl sulfate (SDS-PAGE) and western blotting using specific antibodies.
Immunofluorescence
To visualize membrane-resident α-sarcoglycan, immunofluorescence experiments were performed on intact cells. V247M cells, seeded and grown on poly-lysine-treated glass coverslips, or human myoblasts seeded, grown and differentiated on gelatin/fibronectin-treated glass coverslips, at the end of specific drug treatments, were chilled at 4°C for 30 min, washed with cold PBS supplemented with Ca2+ and Mg2+. Cells were subsequently incubated at 4°C with either the mouse monoclonal or the rabbit polyclonal antibody specific for α-sarcoglycan, recognizing an extracellular epitope. After washing, cells were incubated with the specific DyLight 488-conjugated secondary antibody (Jackson). At the end of incubation, cells were washed and fixed with 4% PFA.
To visualize intracellular α-sarcoglycan, immunofluorescence experiments were performed on permeabilized cells. V247M cells cultivated as above described, were first fixed with 4% PFA and subsequently permeabilized with 0.5% Triton X-100 for 15 min. Incubation with primary and secondary antibodies was performed as described above. Cells were then examined with a Leica SP2 confocal microscope.
Statistical analysis
Data from at least three independent experiments are expressed as mean ± SEM. Statistical differences were determined by paired two-tailed Student's t-test or ANOVA (using GraphPad Prism 5).
SUPPLEMENTARY MATERIAL
AUTHORS’ CONTRIBUTIONS
E.B., R.B. and D.S. conceived and designed the experiments; M.F. and K.M. provided human specimens; E.B. and D.S. performed the experiments; E.B., R.B. and D.S. analyzed the data; E.B., R.B. and D.S. wrote the paper.
FUNDING
This work was supported by Association Francaise contre les Myopathies (grant number 14999, 15969 to R.B.), Italian Telethon (grant number GEP12058 to D.S.) and institutional funds from the Consiglio Nazionale delle Ricerche to R.B. and University of Padova to D.S. Funding to pay the Open Access publication charges for this article was provided by Italian Telethon.
Supplementary Material
ACKNOWLEDGEMENTS
Toshihiro Nakajima, Tokyo Medical University, is gratefully thanked for providing the HRD1 inhibitors. Vincent Mouly and the ‘Human cell culture platform’ of the Myology Institute Université Pierre et Marie Curie, Paris 6 are gratefully thanked for providing immortalized human myoblasts derived from a healthy subject. Corrado Angelini and the Telethon Genetic Bio-Bank facility, University of Padova are gratefully thanked for providing the skeletal muscle biopsy of a LGMD-2D patient. Stefano Schiaffino is gratefully thanked for providing the embryonic myosin heavy chain mouse monoclonal antibody.
Conflict of Interest statement. The authors declare no conflict of interest.
REFERENCES
- 1.Smith M.H., Ploegh H.L., Weissman J.S. Road to ruin: targeting proteins for degradation in the endoplasmic reticulum. Science. 2011;334:1086–1090. doi: 10.1126/science.1209235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bagola K., Mehnert M., Jarosch E., Sommer T. Protein dislocation from the ER. Biochim. Biophys. Acta. 2011;1808:925–936. doi: 10.1016/j.bbamem.2010.06.025. [DOI] [PubMed] [Google Scholar]
- 3.Carvalho P., Goder V., Rapoport T.A. Distinct ubiquitin–ligase complexes define convergent pathways for the degradation of ER proteins. Cell. 2006;126:361–373. doi: 10.1016/j.cell.2006.05.043. [DOI] [PubMed] [Google Scholar]
- 4.Hosokawa N., Wada I., Nagasawa K., Moriyama T., Okawa K., Nagata K. Human XTP3-B forms an endoplasmic reticulum quality control scaffold with the HRD1-SEL1L ubiquitin ligase complex and BiP. J. Biol. Chem. 2008;283:20914–20924. doi: 10.1074/jbc.M709336200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Christianson J.C., Shaler T.A., Tyler R.E., Kopito R.R. OS-9 and GRP94 deliver mutant alpha1-antitrypsin to the Hrd1-SEL1L ubiquitin ligase complex for ERAD. Nat. Cell. Biol. 2008;10:272–282. doi: 10.1038/ncb1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hirsch C., Gauss R., Horn S.C., Neuber O., Sommer T. The ubiquitylation machinery of the endoplasmic reticulum. Nature. 2009;458:453–460. doi: 10.1038/nature07962. [DOI] [PubMed] [Google Scholar]
- 7.Kostova Z., Tsai Y.C., Weissman A.M. Ubiquitin ligases, critical mediators of endoplasmic reticulum-associated degradation. Semin. Cell. Dev. Biol. 2007;18:770–779. doi: 10.1016/j.semcdb.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ye Y., Meyer H.H., Rapoport T.A. The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature. 2001;414:652–656. doi: 10.1038/414652a. [DOI] [PubMed] [Google Scholar]
- 9.Flierman D., Ye Y., Dai M., Chau V., Rapoport T.A. Polyubiquitin serves as a recognition signal, rather than a ratcheting molecule, during retrotranslocation of proteins across the endoplasmic reticulum membrane. J. Biol. Chem. 2003;278:34774–34782. doi: 10.1074/jbc.M303360200. [DOI] [PubMed] [Google Scholar]
- 10.Hampton R.Y., Sommer T. Finding the will and the way of ERAD substrate retrotranslocation. Curr. Opin. Cell Biol. 2012;24:460–466. doi: 10.1016/j.ceb.2012.05.010. [DOI] [PubMed] [Google Scholar]
- 11.Gastaldello S., D'Angelo S., Franzoso S., Fanin M., Angelini C., Betto R., Sandonà D. Inhibition of proteasome activity promotes the correct localization of disease-causing alpha-sarcoglycan mutants in HEK-293 cells constitutively expressing beta-, gamma-, and delta-sarcoglycan. Am. J. Pathol. 2008;173:170–181. doi: 10.2353/ajpath.2008.071146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sandonà D., Betto R. Sarcoglycanopathies: molecular pathogenesis and therapeutic prospects. Expert Rev. Mol. Med. 2009;11:e28. doi: 10.1017/S1462399409001203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Noguchi S., Wakabayashi E., Imamura M., Yoshida M., Ozawa E. Formation of sarcoglycan complex with differentiation in cultured myocytes. Eur. J. Biochem. 2000;267:640–648. doi: 10.1046/j.1432-1327.2000.00998.x. [DOI] [PubMed] [Google Scholar]
- 14.Kirschner J., Lochmüller H. Sarcoglycanopathies. Handb. Clin. Neurol. 2011;101:41–46. doi: 10.1016/B978-0-08-045031-5.00003-7. [DOI] [PubMed] [Google Scholar]
- 15.Politano L., Nigro V., Passamano L., Petretta V., Comi L.I., Papparella S., Nigro G., Rambaldi P.F., Raia P., Pini A., et al. Evaluation of cardiac and respiratory involvement in sarcoglycanopathies. Neuromuscul. Disord. 2001;11:178–185. doi: 10.1016/s0960-8966(00)00174-7. [DOI] [PubMed] [Google Scholar]
- 16.Bartoli M., Gicquel E., Barrault L., Soheili T., Malissen M., Malissen B., Vincent-Lacaze N., Perez N., Udd B., Danos O., Richard I. Mannosidase I inhibition rescues the human alpha-sarcoglycan R77C recurrent mutation. Hum. Mol. Genet. 2008;17:1214–1221. doi: 10.1093/hmg/ddn029. [DOI] [PubMed] [Google Scholar]
- 17.Soheili T., Gicquel E., Poupiot J., N'Guyen L., Le Roy F., Bartoli M., Richard I. Rescue of sarcoglycan mutations by inhibition of endoplasmic reticulum quality control is associated with minimal structural modifications. Hum. Mutat. 2012;33:429–439. doi: 10.1002/humu.21659. [DOI] [PubMed] [Google Scholar]
- 18.Barresi R., Confalonieri V., Lanfossi M., Di Blasi C., Torchiana E., Mantegazza R., Jarre L., Nardocci N., Boffi P., Tezzon F., et al. Concomitant deficiency of beta- and gamma-sarcoglycans in 20 alpha-sarcoglycan (adhalin)-deficient patients: immunohistochemical analysis and clinical aspects. Acta Neuropathol. 1997;94:28–35. doi: 10.1007/s004010050668. [DOI] [PubMed] [Google Scholar]
- 19.Bernasconi R., Galli C., Calanca V., Nakajima T., Molinari M. Stringent requirement for HRD1, SEL1L, and OS-9/XTP3-B for disposal of ERAD-LS substrates. J. Cell Biol. 2010;188:223–235. doi: 10.1083/jcb.200910042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sandonà D., Gastaldello S., Martinello T., Betto R. Characterization of the ATP-hydrolysing activity of alpha-sarcoglycan. Biochem. J. 2004;381:105–112. doi: 10.1042/BJ20031644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Draviam R.A., Wang B., Shand S.H., Xiao X., Watkins S.C. Alpha-sarcoglycan is recycled from the plasma membrane in the absence of sarcoglycan complex assembly. Traffic. 2006;7:793–810. doi: 10.1111/j.1600-0854.2006.00433.x. [DOI] [PubMed] [Google Scholar]
- 22.Kikkert M., Doolman R., Dai M., Avner R., Hassink G., van Voorden S., Thanedar S., Roitelman J., Chau V., Wiertz E. Human HRD1 is an E3 ubiquitin ligase involved in degradation of proteins from the endoplasmic reticulum. J. Biol. Chem. 2004;279:3525–3534. doi: 10.1074/jbc.M307453200. [DOI] [PubMed] [Google Scholar]
- 23.Morito D., Hirao K., Oda Y., Hosokawa N., Tokunaga F., Cyr D.M., Tanaka K., Iwai K., Nagata K. Gp78 cooperates with RMA1 in endoplasmic reticulum-associated degradation of CFTRDeltaF508. Mol. Biol. Cell. 2008;19:1328–1336. doi: 10.1091/mbc.E07-06-0601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mueller B., Klemm E.J., Spooner E., Claessen J.H., Ploegh H.L. SEL1L nucleates a protein complex required for dislocation of misfolded glycoproteins. Proc. Natl. Acad. Sci. USA. 2008;105:12325–12330. doi: 10.1073/pnas.0805371105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iida Y., Fujimori T., Okawa K., Nagata K., Wada I., Hosokawa N. SEL1L protein critically determines the stability of the HRD1-SEL1L endoplasmic reticulum-associated degradation (ERAD) complex to optimize the degradation kinetics of ERAD substrates. J. Biol. Chem. 2011;286:16929–16939. doi: 10.1074/jbc.M110.215871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burr M.L., Cano F., Svobodova S., Boyle L.H., Boname J.M., Lehner P.J. HRD1 and UBE2J1 target misfolded MHC class I heavy chains for endoplasmic reticulum-associated degradation. Proc. Natl. Acad. Sci. USA. 2011;108:2034–2039. doi: 10.1073/pnas.1016229108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen B., Mariano J., Tsai Y.C., Chan A.H., Cohen M., Weissman A.M. The activity of a human endoplasmic reticulum-associated degradation E3, gp78, requires its Cue domain, RING finger, and an E2-binding site. Proc. Natl. Acad. Sci. USA. 2006;103:341–346. doi: 10.1073/pnas.0506618103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Claessen J.H., Mueller B., Spooner E., Pivorunas V.L., Ploegh H.L. The transmembrane segment of a tail-anchored protein determines its degradative fate through dislocation from the endoplasmic reticulum. J. Biol. Chem. 2010;285:20732–20739. doi: 10.1074/jbc.M110.120766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Greenblatt E.J., Olzmann J.A., Kopito R.R. Derlin-1 is a rhomboid pseudoprotease required for the dislocation of mutant alpha-1 antitrypsin from the endoplasmic reticulum. Nat. Struct. Mol. Biol. 2011;18:1147–1152. doi: 10.1038/nsmb.2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kobayashi T., Tanaka K., Inoue K., Kakizuka A. Functional ATPase activity of p97/Valosin-containing protein (VCP) is required for the quality control of endoplasmic reticulum in neuronally differentiated mammalian PC12 cells. J. Biol. Chem. 2002;277:47358–47635. doi: 10.1074/jbc.M207783200. [DOI] [PubMed] [Google Scholar]
- 31.Fang S., Ferrone M., Yang C., Jensen J.P., Tiwari S., Weissman A.M. The tumor autocrine motility factor receptor, gp78, is a ubiquitin protein ligase implicated in degradation from the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA. 2001;98:14422–14427. doi: 10.1073/pnas.251401598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hassink G., Kikkert M., van Voorden S., Lee S.J., Spaapen R., van Laar T., Coleman C.S., Bartee E., Früh K., Chau V., Wiertz E. TEB4 is a C4HC3 RING finger-containing ubiquitin ligase of the endoplasmic reticulum. Biochem. J. 2005;388:647–655. doi: 10.1042/BJ20041241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee J.P., Brauweiler A., Rudolph M., Hooper J.E., Drabkin H.A., Gemmill R.M. The TRC8 ubiquitin ligase is sterol regulated and interacts with lipid and protein biosynthetic pathways. Mol. Cancer Res. 2010;8:93–106. doi: 10.1158/1541-7786.MCR-08-0491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lerner M., Corcoran M., Cepeda D., Nielsen M.L., Zubarev R., Pontén F., Uhlén M., Hober S., Grandér D., Sangfelt O. The RBCC gene RFP2 (Leu5) encodes a novel transmembrane E3 ubiquitin ligase involved in ERAD. Mol. Biol. Cell. 2007;18:1670–1682. doi: 10.1091/mbc.E06-03-0248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Altier C., Garcia-Caballero A., Simms B., You H., Chen L., Walcher J., Tedford H.W., Hermosilla T., Zamponi G.W. The Cavbeta subunit prevents RFP2-mediated ubiquitination and proteasomal degradation of L-type channels. Nat. Neurosci. 2011;14:173–180. doi: 10.1038/nn.2712. [DOI] [PubMed] [Google Scholar]
- 36.Baranova A., Hammarsund M., Ivanov D., Skoblov M., Sangfelt O., Corcoran M., Borodina T., Makeeva N., Pestova A., Tyazhelova T., et al. Distinct organization of the candidate tumor suppressor gene RFP2 in human and mouse: multiple mRNA isoforms in both species- and human-specific antisense transcript RFP2OS. Gene. 2003;321:103–112. doi: 10.1016/j.gene.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 37.Yagishita N., Aratani S., Leach C., Amano T., Yamano Y., Nakatani K., Nishioka K., Nakajima T. RING-finger type E3 ubiquitin ligase inhibitors as novel candidates for the treatment of rheumatoid arthritis. Int. J. Mol. Med. 2012;30:1281–1286. doi: 10.3892/ijmm.2012.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schiaffino S., Gorza L., Sartore S., Saggin L., Carli M. Embryonic myosin heavy chain as a differentiation marker of developing human skeletal muscle and rhabdomyosarcoma. A monoclonal antibody study. Exp. Cell Res. 1986;163:211–220. doi: 10.1016/0014-4827(86)90574-4. [DOI] [PubMed] [Google Scholar]
- 39.Zhu C.H., Mouly V., Cooper R.N., Mamchaoui K., Bigot A., Shay J.W., Di Santo J.P., Butler-Browne G.S., Wright W.E. Cellular senescence in human myoblasts is overcome by human telomerase reverse transcriptase and cyclin-dependent kinase 4: consequences in aging muscle and therapeutic strategies for muscular dystrophies. Aging Cell. 2007;6:515–523. doi: 10.1111/j.1474-9726.2007.00306.x. [DOI] [PubMed] [Google Scholar]
- 40.Jentsch S., Rumpf S. Cdc48 (p97): a “molecular gearbox” in the ubiquitin pathway? Trends Biochem. Sci. 2007;32:6–11. doi: 10.1016/j.tibs.2006.11.005. [DOI] [PubMed] [Google Scholar]
- 41.Ying Z., Wang H., Fan H., Wang G. The endoplasmic reticulum (ER)-associated degradation system regulates aggregation and degradation of mutant neuroserpin. J. Biol. Chem. 2011;286:20835–20844. doi: 10.1074/jbc.M110.200808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jo Y., Lee P.C., Sguigna P.V., DeBose-Boyd R.A. Sterol-induced degradation of HMG CoA reductase depends on interplay of two Insigs and two ubiquitin ligases, gp78 and Trc8. Proc. Natl. Acad. Sci. USA. 2011;108:20503–20508. doi: 10.1073/pnas.1112831108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Joo H.M., Kim J.Y., Jeong J.B., Seong K.M., Nam S.Y., Yang K.H., Kim C.S., Kim H.S., Jeong M., An S., Jin Y.W. Ret finger protein 2 enhances ionizing radiation-induced apoptosis via degradation of AKT and MDM2. Eur. J. Cell Biol. 2011;90:420–431. doi: 10.1016/j.ejcb.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 44.Goldenberg S.J., Marblestone J.G., Mattern M.R., Nicholson B. Strategies for the identification of ubiquitin ligase inhibitors. Biochem. Soc. Trans. 2010;38:132–136. doi: 10.1042/BST0380132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guerriero C.J., Brodsky J.L. The delicate balance between secreted protein folding and endoplasmic reticulum-associated degradation in human physiology. Physiol. Rev. 2012;92:537–576. doi: 10.1152/physrev.00027.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.