Abstract
Tuberous sclerosis complex (TSC) is a disorder arising from mutation in the TSC1 or TSC2 gene, characterized by the development of hamartomas in various organs and neurological manifestations including epilepsy, intellectual disability and autism. TSC1/2 protein complex negatively regulates the mammalian target of rapamycin complex 1 (mTORC1) a master regulator of protein synthesis, cell growth and autophagy. Autophagy is a cellular quality-control process that sequesters cytosolic material in double membrane vesicles called autophagosomes and degrades it in autolysosomes. Previous studies in dividing cells have shown that mTORC1 blocks autophagy through inhibition of Unc-51-like-kinase1/2 (ULK1/2). Despite the fact that autophagy plays critical roles in neuronal homeostasis, little is known on the regulation of autophagy in neurons. Here we show that unlike in non-neuronal cells, Tsc2-deficient neurons have increased autolysosome accumulation and autophagic flux despite mTORC1-dependent inhibition of ULK1. Our data demonstrate that loss of Tsc2 results in autophagic activity via AMPK-dependent activation of ULK1. Thus, in Tsc2-knockdown neurons AMPK activation is the dominant regulator of autophagy. Notably, increased AMPK activity and autophagy activation are also found in the brains of Tsc1-conditional mouse models and in cortical tubers resected from TSC patients. Together, our findings indicate that neuronal Tsc1/2 complex activity is required for the coordinated regulation of autophagy by AMPK. By uncovering the autophagy dysfunction associated with Tsc2 loss in neurons, our work sheds light on a previously uncharacterized cellular mechanism that contributes to altered neuronal homeostasis in TSC disease.
INTRODUCTION
Tuberous sclerosis complex (TSC) is a multisystem disease with an incidence of ∼1:6000 worldwide (1). Neurological manifestations include epilepsy, intellectual disability and autism with pathological abnormalities such as cortical tubers (2). TSC is caused by TSC1 and TSC2 mutations that abrogate the activity of the TSC1/2 protein complex. The TSC1/2 complex plays a major role in controlling mammalian target of rapamycin (mTOR), which exists in two distinct complexes: mTORC1 and mTORC2 (3). The TSC1/2 complex functions as a checkpoint for mTORC1 (4), which acts as an inhibitor of macroautophagy (hereafter referred to as autophagy) (5).
Autophagy is a cellular quality-control and metabolic process for the removal of damaged organelles and energy retrieval whereby cytoplasmic material and damaged organelles are engulfed in double membrane vesicles called autophagosomes (6). The latter are delivered to the lysosome to form the autolysosome where the cargo is degraded by lysosomal proteases. The autophagosomal lipidated microtubule-associated protein 1 light chain 3 (LC3-II), and the autophagy substrate sequestosome p62/SQSTM1 are often used to monitor autophagy (7,8). The homeostatic function of this pathway is particularly critical for the brain where alterations of autophagy have been associated with several neurological disorders (9). Previous studies have indicated that mTORC1 and the AMP-activated protein kinase (AMPK) act respectively as an autophagy repressor and inducer through differential phosphorylation of the Atg1/Unc-51-like-kinase1 (ULK1) (10–14). Despite the involvement of the mTORC1 pathway in autophagy and its critical role in neuronal function (15), most of the current knowledge on the effect of mTORC1 activity on autophagy is based on studies performed using dividing cells (16,17).
In this study, via knockdown of the Tsc2 gene in a cell culture system, we investigated how activation of mTORC1 affects autophagy in postmitotic neurons. Here, we show that loss of Tsc1/2 complex function and subsequent mTORC1 activation in neurons results in accumulation of the autophagic marker LC3-II and of the autophagy substrate p62. Our data demonstrate that concomitant to mTORC1-mediated inhibitory phosphorylation of ULK1 at Ser757, Tsc2-knockdown neurons have increased AMPK-activation and AMPK-dependent activating phosphorylation of ULK1 at Ser555. As a result, Tsc2-knockdown neurons have AMPK-dependent autophagic flux. Importantly, similar autophagy dysfunction was observed in brain tissues of the Tsc1c/c;NestinCre and the Tsc1c/c;L7Cre conditional knockout mice and in cortical tuber specimens from TSC patients.
Our work reveals unexpected autophagy dysfunction specifically associated with loss of TSC1/2 in brain tissue and the molecular mechanism underlying these defects. In contrast to the current knowledge that mTORC1 activation by TSC1/2-loss inhibits autophagy, we show evidence that Tsc2-knockdown neurons display increased accumulation of autolysosomes and autophagic activity through AMPK-dependent activation of ULK1. The autophagy defects identified in this study may help to elucidate the mechanisms that contribute to altered neuronal function in TSC disease.
RESULTS
Loss of Tsc1/2 in neurons results in accumulation of the autophagic marker LC3-II and autophagic organelles
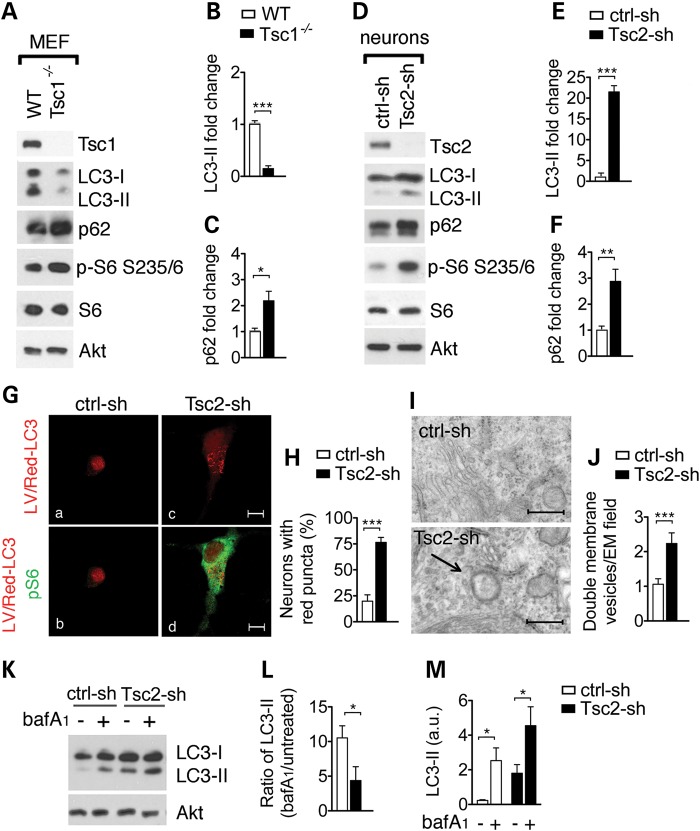
In dividing cells, mTORC1 inhibits autophagy, and accordingly, we found that mTORC1 activation in Tsc1−/− mouse embryonic fibroblasts (MEFs) reduced the level of LC3-II and increased p62 (Fig. 1A–C). Since we had previously shown that knockdown of Tsc2 results in the same morphological and biochemical changes as Tsc1 knockout in dissociated cells (18), we used shRNA interference of the Tsc2 gene to investigate the effect of neuronal Tsc1/2-loss on autophagy. We choose Akt as loading control as it was previously reported unchanged in different models of TSC-loss (19–24). As compared with controls (ctrl-sh), neurons transduced with Tsc2-shRNA (Tsc2-sh) lentivirus displayed decreased Tsc2 protein expression and increased mTORC1 activation as shown by phosphorylation of the mTORC1 downstream target S6 protein (Fig. 1D; Supplementary Material, Fig. S1). Similarly to the MEFs, the Tsc2-sh neurons had increased levels of the autophagy substrate p62 (Fig. 1D and F); however, the LC3-II accumulation (Fig. 1D–E) sharply contrasted with the LC3-II reduction seen in either the Tsc1−/− (Fig. 1A and B) or the Tsc2−/− MEFs (16). Although autophagy is mainly regulated at the posttranslational level, recent findings have shown that certain autophagy genes such as LC3 can be regulated at the transcriptional level (25–27). We confirmed that the Tsc2-sh neurons had a significant reduction in the Tsc2 mRNA expression but no difference in LC3 transcriptional regulation or Tsc1 mRNA expression (Supplementary Material, Fig. S2A–C). The LC3-II accumulation observed in the Tsc2-sh neurons was autophagy dependent as it was significantly reduced by silencing of the atg5 gene, which is essential for autophagosome formation (28) (Supplementary Material, Fig. S3A–C).
Figure 1.
Loss of Tsc1/2 in neurons results in accumulation of the autophagic marker LC3-II and autophagic organelles. (A) Representative western blot of LC3-II and p62 in wild-type (WT) and Tsc1−/− MEFs and in rat hippocampal neurons (D) transduced with control (ctrl-sh) and with Tsc2-sh lentivirus. S6 phosphorylation at Ser235/236 (p-S6) indicates mTORC1 activation. Densitometric quantification of LC3-II (MEFs in (B) n = 4; neurons in (E) n = 3) and p62 (MEFs in (C) n = 4; neurons in (F) n = 5) normalized to total Akt. Experiments performed on independent culture preparations. (G) Tsc2-sh neurons transduced with LV/Red-LC3 virus display accumulation of red puncta. Immunofluorescent analysis of LV/Red-LC3 (in red) and mTORC1 activation identified by p-S6 antibody staining (in green) in ctrl-sh and in Tsc2-sh neurons. Scale bar is 10 μm. (H) Quantification of the percentage of neurons with red puncta (n = 4; 20–60 neurons/exp.). *P < 0.05; **P < 0.01; ***P < 0.001). (I) Representative EM images from control and Tsc2-sh cultures. Scale bar is 500 nm. (J) Quantification of organelles with double membrane vesicles/EM field (50 μm2). A total of 51 control and 52 Tsc2-sh neurons were imaged and analyzed (***P < 0.001). (K) Western blot of LC3-II in ctrl-sh and Tsc2-sh neurons untreated or treated with 400 nm bafilomycin (bafA1) for 4 h. (L) Densitometry quantification of LC3-II ratio (bafA1 treated/untreated) in ctrl-sh and Tsc2-sh cultures normalized to total Akt (n = 4, *P < 0.05). (M) Densitometry quantification of LC3-II level (a.u.) in untreated and bafA1-treated neurons. (n = 6, *P < 0.05). Bars represent mean ± s.e.m.
To visualize autophagic organelles, we performed immunofluorescent (IF) analysis of neuronal cultures transduced with a lentivirus expressing the membrane associated LC3B tagged to a red fluorescent protein (LV/Red-LC3) (27). Compared with controls, Tsc2-sh cultures had a significant increase in the percentage of neurons with LV/Red-LC3-positive organelles measured as red puncta (Fig. 1G and H). Similar results were obtained when we expressed the Red-LC3 protein by transient transfection of neurons transduced with a GFP-tagged version of the Tsc2-shRNA virus. No puncta were observed when we expressed the Red alone (Supplementary Material, Fig. S4A–E). Electron microscopy (EM) is as another method often used to visualize autophagic organelles (27). Similar to the IF, EM analysis revealed that Tsc2-sh neurons had a significant accumulation of double membrane organelles with dense luminal material similar to surrounding cytosol resembling late autophagosome vesicles (Fig. 1I and J).
To examine autophagic flux, we monitored LC3-II accumulation in the presence and in the absence of the proton pump inhibitor bafilomycin A1 (bafA1) (29). BafA1 blocks the fusion between autophagosomes and autolysosomes thus an enhancement of LC3-II in the presence of bafA1 indicates presence of autophagic flux while no change indicates inhibition of autophagic degradation. In addition, the ratio of LC3-II (bafA1 treated/untreated) can be used to determine level of flux. Although the reduced ratio of LC3-II indicated that compared with controls Tsc2-sh neurons had lower autophagic flux, we still observed accumulation of LC3-II in the bafA1-treated Tsc2-sh cultures versus the untreated ones, indicating the presence of autophagic activity (Fig. 1K and M). Together, the identification of autophagic organelle accumulation and presence of autophagic activity in the Tsc2-sh neurons suggest that the mechanisms of autophagy regulation in response to reduced Tsc1/2 function differ between neurons and fibroblasts.
Tsc2-sh neurons have mTORC1-independent accumulation of autolysosomes
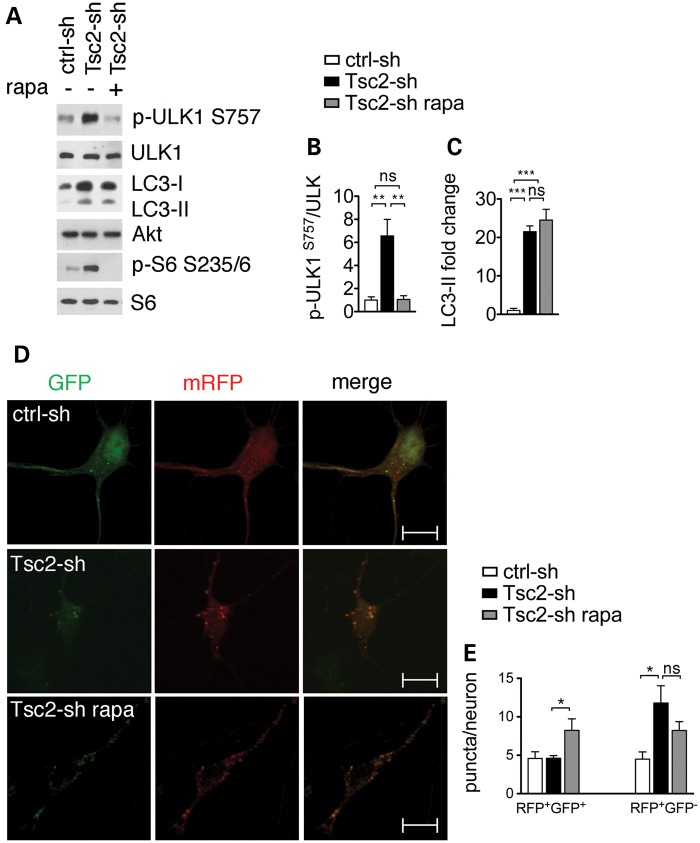
The identification of autophagic flux in the Tsc2-sh neurons was unexpected given that mTORC1 is a known repressor of the autophagy-initiating kinase ULK1 (10–13). To assess how mTORC1 activity affected autophagy in Tsc2-sh neurons, we examined regulation of ULK1 in neurons in the presence and absence of the mTORC1-inhibitor rapamycin. Tsc2-knockdown neurons showed a rapamycin-sensitive increase in ULK1 phosphorylation at the mTORC1-dependent inactivating site S757 (Fig. 2A and B). In addition, rapamycin reversed p62 accumulation as assessed by both western blot and IF staining of Tsc2-sh neurons (Supplementary Material, Fig. S5A–D). Together, these findings suggest that mTORC1 acts as a repressor of autophagy in Tsc2-sh neurons.
Figure 2.
Tsc2-sh neurons have mTORC1-dependent inhibition of ULK1 at S757 and mTORC1-independent accumulation of autolysosomes. (A) Representative western blot of ULK1 and LC3-II regulation in ctrl-sh and Tsc2-sh neurons untreated or treated with 20 nm rapamycin (rapa) for 24 h. Effective mTORC1 inhibition by rapamycin treatment is shown by reduced pS6 phosphorylation at S235/6. Densitometry quantification of p-ULK1 S757 (n = 4) normalized to total ULK1 (B) and LC3-II (n = 3) normalized to total Akt (C) (**P < 0.01; ***P < 0.001). (D) Expression of the td-tag-LC3 vector in control and in Tsc2-sh neurons untreated or treated with 20 nm rapamycin (rapa) for 24 h. Scale bar is 10 μm (E) Quantification of the number of RFP+/GFP+ puncta (autophagosomes) per neurons or RFP+/GFP− puncta (autolysosomes) per neurons (n ≥ 3, *P < 0.05). Bars represent mean ± s.e.m.
Intriguingly, rapamycin did not alter the elevated LC3-II levels in the Tsc2-knockdown neurons (Fig. 2A and C). To directly examine whether mTORC1 activity affected the LC3-II transit through the autophagy pathway, we used the tandem-tagged mCherry-EGFP-LC3B (td-tag-LC3) assay in the absence and presence of rapamycin. The td-tag-LC3 reporter construct is pH sensitive; thus, it allows the expression of LC3 exhibiting both red and green fluorescence when in the autophagosome (RFP+/GFP+ puncta) and red fluorescence only when in the autolysosome (RFP+/GFP− puncta) (27,30). The td-tag-LC3 reporter was able to track autophagic flux faithfully (Supplementary Material, Fig. S6). Compared with controls, the Tsc2-sh neurons displayed a baseline increase in the number of autolysosomes and no change in the number of autophagosomes (Fig. 2D and E). While the autolysosome number was largely insensitive to mTORC1 inhibition, induction of autophagy with rapamycin significantly increased the autophagosomes. The findings that the baseline number of autophagosomes was unchanged despite ULK1 inhibition and that accumulation of autolysosomes was rapamycin insensitive indicate that Tsc2-sh neurons have mTORC1-independent mechanism to regulate autophagy.
Tsc2-sh neurons have AMPK-dependent ULK1 activation and autolysosome accumulation
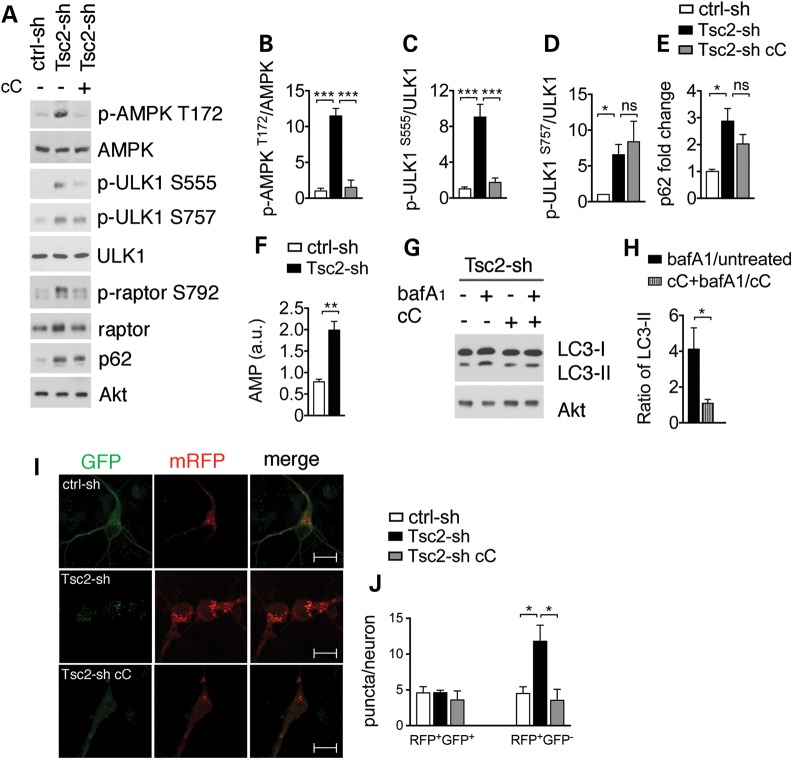
Having identified autophagic flux in the Tsc2-sh neurons, we hypothesized the existence of an autophagy initiation signal counteracting mTORC1. We examined regulation of the AMPK, which induces autophagy by direct activating phosphorylation of ULK1 under conditions of energetic stress (11,14). Compared with controls, Tsc2-sh neurons had a significant increase of AMPK activation with higher phosphorylation at T172 (31); moreover, ULK1 phosphorylation was increased at the AMPK-dependent site S555 (Fig. 3A–C). Notably, Tsc2-sh neurons displayed increased AMP levels indicating energetic stress, which is a prerequisite for AMPK activation (Fig. 3F). To confirm that ULK1 phosphorylation at S555 was AMPK dependent, we treated the Tsc2-sh neurons with the pharmacological AMPK inhibitor, compound C (cC) (32). Compared with vehicle-treated neurons, Tsc2-sh neurons treated with cC had a reduction of AMPK activation as shown by decreased phosphorylation of AMPK at T172 and of its direct substrate the regulatory associated protein of mTORC1 (raptor) at S792 (33). Furthermore, cC treatment specifically reduced phosphorylation of ULK1 at S555 while it did not change the mTORC1-dependent ULK1 phosphorylation site S757 or p62 level (Fig. 3A, C and E). Interestingly, ULK1 activation by AMPK was specific to neurons, as Tsc1−/− MEFs had no AMPK activation or ULK1-S555 phosphorylation (Supplementary Material, Fig. S7). We next addressed whether AMPK inhibition affected autophagic flux. cC treatment abrogated the LC3-II accumulation seen in the Tsc2-sh neurons in the presence of bafA1 and significantly reduced the number of autolysosomes in the td-tag-LC3 assay (Fig. 3G–L). Together, these data indicate that autophagic flux and autolysosome accumulation we observe in Tsc2-knockdown neurons are AMPK dependent.
Figure 3.
Tsc2-sh neurons have AMPK-dependent ULK1 activation and autolysosome accumulation. (A) Representative western blot of ULK1 and AMPK regulation in ctrl-sh and Tsc2-sh neurons untreated or treated with 5 μm cC for 3 h. Quantification of p-AMPK (T172) (n = 3) (B), p-ULK1 S555 (n = 5) (C), p-ULK1 S757 (n = 4) (D) and p62 (n = 3) (E). (F) Quantification of AMP level by LC–MC analysis in ctrl-sh and Tsc2-sh neurons (n = 4, **P < 0.01). (G) Western blot of LC3-II in ctrl-sh and Tsc2-sh neurons untreated or treated with 400 nm bafilomycin (bafA1) for 4 h in the presence or in the absence of cC. (H) Densitometry quantification of LC3-II ratio (bafA1 treated/untreated) versus cC+ bafA1/cC. (I) Expression of the td-tag-LC3 vector in control and in Tsc2-sh neurons untreated or treated with cC. Scale bar is 10 μm. (J) Quantification of the RFP+/GFP+ puncta and the RFP+/GFP− puncta per neuron. *P < 0.05; **P < 0.01; ***P < 0.001. Bars represent mean ± s.e.m. (n ≥ 3).
LC3-II is increased in brain tissue of TSC mouse models and in cortical tubers of TSC patients
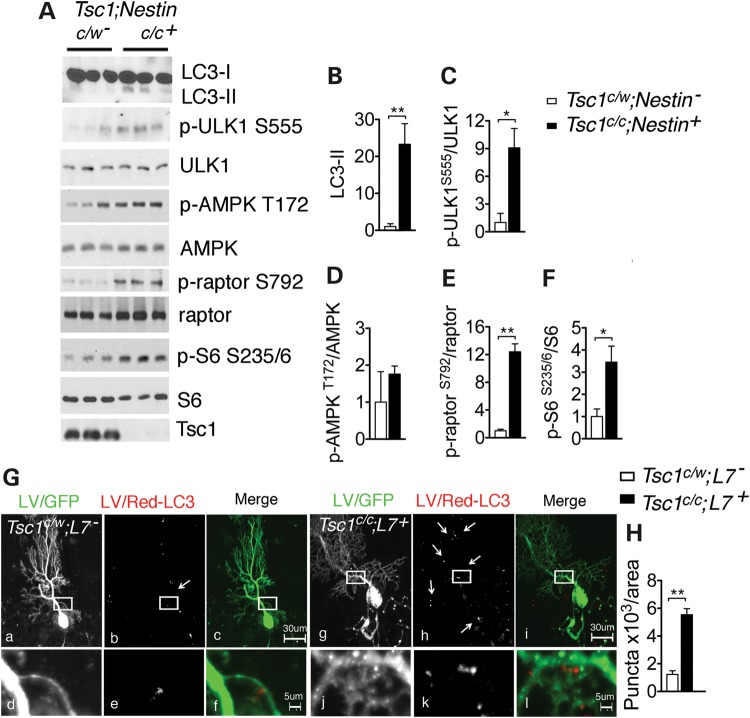
To investigate whether the autophagy defect seen in the Tsc2-sh neurons also occurs in vivo, we examined Tsc1c/c; Nestin+ and Tsc1c/c; L7+ mice, which have mTORC1 signaling activation in neuronal progenitor cells or cerebellar Purkinje cells (PC), respectively (21,34). Activation of the mTORC1 pathway by Tsc1 gene silencing was confirmed by the increased phosphorylation of S6 at S235/6 in the Tsc1c/c; Nestin+ (Fig. 4A and F). Notably, compared with controls, the brains of Tsc1c/c; Nestin+ mice showed a significant increase in LC3-II (Fig. 4A and B). In addition, the Tsc1c/c; Nestin+ brain lysates displayed significant increase in the phosphorylation of ULK1 at S555 and of raptor at S792, indicating AMPK activation similar to what we observed in Tsc2-knockdown cultured neurons (Fig. 4A, C and E).
Figure 4.
LC3-II increase in brain tissue of TSC mouse models. (A) Western blot of LC3-II, ULK1 and AMPK activity in protein lysates from P0 brains of control Tsc1c/w;Nestin and mutant Tsc1c/c;Nestin mice. Loss of Tsc1/2 complex and high mTORC1 activity are demonstrated by reduced expression of the Tsc1 protein and increased p-S6 at S235/6 in the mutant mice. Quantification of the western blots by densitometry analysis. LC3-II (B) and p-S6 S235/6 (F) values were expressed as fold change normalized to total S6 level; p-ULK1 S555 (C), p-AMPK T172 (D) and p-raptor (S792) (E) were normalized to the respective total protein bands (n = 3–6 mice/group *P < 0.05, **P < 0.01). (G) Representative confocal images of control Tsc1c/w;L7 (a–f) and mutant Tsc1c/c;L7 (g–l) mice injected with LV/GFP and LV/Red-LC3. GFP-fluorescence was used to identify injected PC. White boxes indicate zoom area in the controls (d–f) and in the mutant mice (j–l). GFP-positive PC of mutant mice display discrete LV/Red-LC3 puncta. Scale bars are 30 μm in top row and scale bar is 5 μm in bottom row. (H) Quantification of the number of LV/Red-LC3 puncta in area unit of 103 μm2 (n = 3 mice/group, **P < 0.01).
The Tsc1c/c; L7+ mice have Tsc1 gene loss in the PC. These mice display autistic-like behaviors including abnormal social interaction, repetitive behavior and vocalization in addition to altered axonal morphology and PC excitability (34). Here, we used a viral vector approach to monitor autophagy. Cerebella of 3- to 3.5-week-old controls and mutant mice were transduced with a mixture of DsRed-LC3 (LV/Red-LC3) and GFP (LV/GFP) lentivirus. The mice were sacrificed 3 weeks after injection, and LV/Red-LC3-positive organelles were identified by examining red puncta in GFP expressing PC. Compared with controls, Tsc1c/c; L7+ mice had increased appearance of LV/Red-LC3-positive puncta indicating accumulation of autophagic organelles (Fig. 4G and H). Therefore, the evaluation of autophagy in the brains of both the Tsc1c/c; Nestin+ and the Tsc1c/c; L7+ mice supports our findings that LC3-II and autophagic organelles accumulate under neuronal TSC loss.
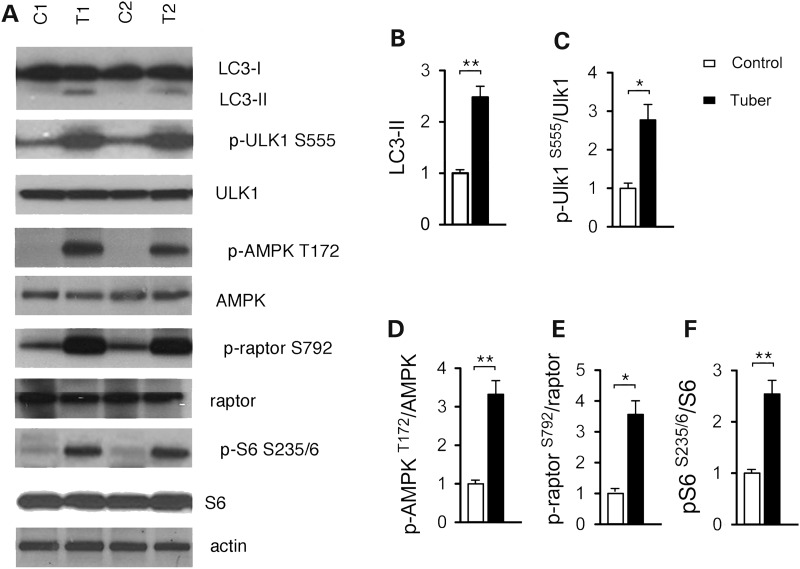
To address regulation of autophagy in TSC brain specimen, we used cortical tubers resected from TSC patients in comparison with age-matched and region-match controls. Activation of mTORC1 was confirmed by increased pS6 phosphorylation at S235/6 in the cortical tuber lysates compared with controls (Fig. 5A and F). A significant increase was observed in LC3-II relative to controls (Fig. 5A and B). In addition, the phosphorylation of ULK1 S555, AMPK T172 and raptor S792 were higher in the TSC cortical tubers compared with controls (Fig. 5A, C–E). The identification of LC3-II accumulation and increased AMPK/ULK1 activation in the cortical tubers is consistent with our findings in the Tsc2-sh cultures. Together, these data further support a role for the TSC1/2 complex in the control of neuronal autophagy through AMPK-dependent regulation of ULK1.
Figure 5.
Autophagy in cortical tubers of TSC patients. (A) Representative western blots of LC3-II, ULK1 and AMPK activation in protein lysates from control brain (C1 and C2) and cortical tubers (T1 and T2). (B–F) Quantification of the western blots by densitometry analysis. LC3-II (B) was normalized to actin; p-ULK1 S555 (C), p-AMPK T172 (D), p-raptor (S792) (E) and p-S6 S235/6 (F) were normalized to the respective total protein bands. Values were expressed as fold change relative to control levels (n = 4 samples/group; *P < 0.05, **P < 0.01). Bars represent mean ± s.e.m.
DISCUSSION
Autophagy is a tightly regulated cellular quality-control mechanism (9). Our work demonstrates that in neurons the Tsc1/2 complex represents a critical node for AMPK-dependent regulation of autophagy. It was previously reported that Tsc1/2-deficient dividing cells have reduced autophagy via mTORC1-dependent inhibition and phosphorylation of ULK1 at S757. However, here we show that Tsc2-sh neurons have autophagic activity through activation of ULK1 via AMPK-dependent phosphorylation at S555 concomitant with mTORC1 inhibition of ULK1. The overall effect of AMPK activation of autophagy is autophagic flux and accumulation of autolysosomes.
Autophagy plays an essential cytoprotective role to help prevent buildup of protein aggregates and damaged organelles particularly critical to postmitotic neurons that cannot dilute the byproducts of cellular metabolism or stress by cell division (6). Several lines of evidence have previously indicated that loss of Tsc1/2 in neurons results in increased stress detrimental for neuronal homeostasis: (i) Tsc2-knockdown in neurons leads to higher susceptibility to endoplasmic reticulum and oxidative stress (19), (ii) Tsc1/2-dependent inhibition of mTORC1 is required to regulate autophagy in response to reactive oxygen species (35) and (iii) mouse models of Tsc1/2 loss display increased stress responses (19,21,34). In this study, we also show that Tsc2-knockdown neurons have increased AMP levels thus indicating energetic stress. An open question arising from this work is whether AMPK activation in Tsc2-knockdown neurons may represent a positive feedback on autophagy to help prevent the long-term pathological effect of cellular stress as depicted in our working model (Fig. 6). Consistent with our findings, higher levels of the autophagic organelle marker LC3-II was also found in hippocampal cultures transduced with Tsc1 shRNA, and the same study reported that induction of efficient autophagic flux by Tsc1 overexpression was neuroprotective in a model of cerebral ischaemia (36). Our data show that despite the AMPK-dependent autophagic activity, Tsc2-sh neurons have p62 accumulation, which was mTORC1 dependent as it was sensitive to rapamycin. Growing evidence suggests that aside from ULK1 suppression, mTORC1 activation can affect autophagy through the regulation of lysosomal biogenesis and function (37,38). Hence, in the Tsc2-sh neurons reduced p62 degradation may result from mTORC1-dependent direct inhibition of autophagy at the termination stages.
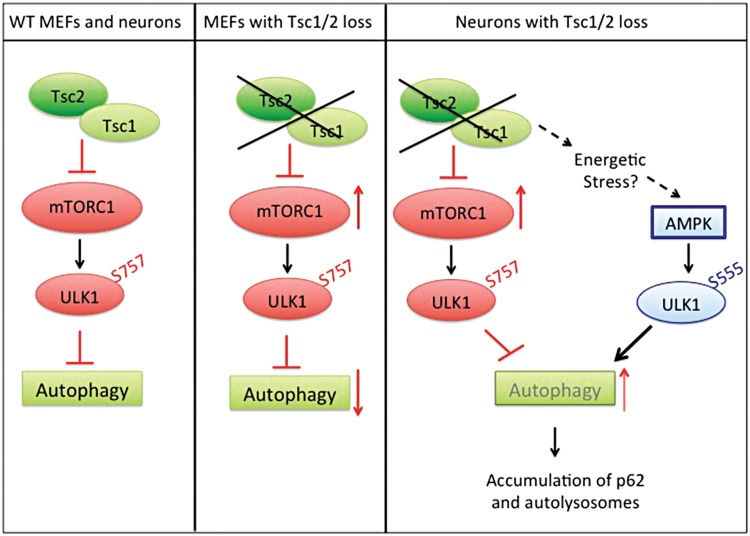
Figure 6.
Model for regulation of autophagy in the Tsc2-sh neurons. In wild-type cells, Tsc1/2 complex keeps mTORC1 under control and allows autophagy. In fibroblasts, when Tsc1/2 complex activity is lost, mTORC1 activation inhibits autophagy via ULK1 phosphorylation at S757. Neuronal Tsc1/2 complex allows autophagy by acting as a checkpoint on mTORC1. Prolonged Tsc1/2 complex loss causes a buildup of neuronal stress providing a positive feedback on autophagy via AMPK-dependent activation of ULK1 by phosphorylation at S555. Dysfunctional autophagy leads to accumulation of the autophagic substrate p62 and of autolysosomes.
Defective autophagy is a hallmark of several neurodegenerative diseases (39). Although TSC has never been classified as a neurodegenerative disease, the increased stress response in the Tsc1c/c; L7+ mice (34) together with the accumulation of autophagic organelles identified by this study (Fig. 4G and H) raise the possibility that uncleared protein aggregates and cellular debris may contribute to the altered cerebellar axonal morphology and PC loss in the mutant mice. Furthermore, the accumulation of autolysosomes we identified in the Tsc2-sh neurons may explain the presence of dysfunctional organelles observed in the enlarged giant cells of the Tsc1/Nestin-rtTA knockout mice at late ages and of vacuole accumulation in the giant cells seen in human tubers (22).
In the brain, Tsc1/2 complex-mediated regulation of mTORC1 is crucial for growth cone dynamics and proper axonal connectivity (20). Remarkably, ULK1 has been implicated in synaptic vesicles transport in Drosophila (40), in axonal elongation in Caenorhabditis elegans (41,42) and in neurite extension in mouse cerebellar granule cells (43). Although it is currently unclear whether the role of ULK1 in autophagy is connected to its other roles in neuronal function, our findings raise the possibility that alteration in ULK1 regulation might contribute to both the defects in autophagy and in neuronal connectivity in TSC.
Depending on the disease status, autophagy has been found beneficial or detrimental. Recently, significant advances have been made in the discovery of autophagy inducers since several studies have shown the beneficial effect of autophagy induction in the clearance of protein aggregates in neuronal tissues affected by defective autophagy (44–46). The finding that increased AMPK-dependent activation of ULK1 counteracts its inhibition by mTORC1 in Tsc2-knockdown neurons suggests that the use of autophagy enhancers may be a potential therapeutic strategy for the neuronal pathology of TSC.
In conclusion, by uncovering that neuronal Tsc1/2 complex controls autophagy through AMPK-dependent regulation of ULK1, our study provides fundamental insights for the mechanisms that may underlie the neurological manifestations in TSC or TSC-related proteinopathies with defective autophagy.
MATERIALS AND METHODS
Rat hippocampal neuronal cultures were prepared as previously published (19). HEK293T cells (ATCC) were used for these studies. Autophagy was monitored by LC3-I to LC3-II conversion, by quantification of Red fluorescent puncta in neuronal cultures transduced with Red-LC3 lentivirus or transiently transfected with the Red-LC3 construct, by the td-tag-LC3 report construct, by EM, by autophagic flux, by LV/Red-LC3 injection in cerebellar vermis and by p62/SQSTM1 accumulation. The Tsc1ccNestin+ and the Tsc1c/c; L7+ mice used in this study were previously described (21,34). Experimental procedures were performed in compliance with animal protocols approved by the Animal Research Committee of Boston Children's Hospital. Human TSC specimens were collected from patients who underwent surgery for medically intractable epilepsy at New York University Langone Medical Center (NYULMC), New York, NY, USA. Postmortem control samples were obtained from the Brain and Tissue Bank for Developmental Disorders at the University of Maryland, Baltimore, MD, USA. The study was approved by the NYULMC Institutional Review Board. Detailed descriptions are provided in Supplementary Material, Methods.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by Nancy Lurie Marks Family Foundation, Autism Speaks and Boston Children's Hospital Translational Research Program (to M.S.), National Institute of Health [P01 NS024279 (to D.J.K.) and K08 NS083733 (to P.T.T.)], the Tuberous Sclerosis Alliance (to A.D.N.), FACES [Finding a Cure for Epilepsy and Seizures (to D.M.T.)]. Control human tissue was obtained from the National Institute of Child Health and Human Development- Brain and Tissue Bank for Developmental Disorders at the University of Maryland, Baltimore, MD, USA, contract HHSN275200900011C, Ref. No. N01-HD-9-0011.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank members of the Sahin lab for critical reading of the manuscript, Elizabeth Petri Henske, Hilaire C. Lam and David Clapham for helpful discussions, Anthony Hill and Boston Children's Hospital Intellectual and Developmental Disabilities Research Center for technical assistance (supported by P30HD18655).
Conflict of Interest statement. None declared.
REFERENCES
- 1.Han J.M., Sahin M. TSC1/TSC2 signaling in the CNS. FEBS Lett. 2011;585:973–980. doi: 10.1016/j.febslet.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tsai P., Sahin M. Mechanisms of neurocognitive dysfunction and therapeutic considerations in tuberous sclerosis complex. Curr. Opin. Neurol. 2011;24:106–113. doi: 10.1097/WCO.0b013e32834451c4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang J., Manning B.D. A complex interplay between Akt, TSC2 and the two mTOR complexes. Biochem. Soc. Trans. 2009;37:217–222. doi: 10.1042/BST0370217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Inoki K., Li Y., Zhu T., Wu J., Guan K.L. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat. Cell Biol. 2002;4:648–657. doi: 10.1038/ncb839. [DOI] [PubMed] [Google Scholar]
- 5.Arsham A.M., Neufeld T.P. Thinking globally and acting locally with TOR. Curr. Opin. Cell Biol. 2006;18:589–597. doi: 10.1016/j.ceb.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 6.Mizushima N., Levine B., Cuervo A.M., Klionsky D.J. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pankiv S., Clausen T.H., Lamark T., Brech A., Bruun J.A., Outzen H., Overvatn A., Bjorkoy G., Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 9.Murrow L., Debnath J. Autophagy as a stress-response and quality-control mechanism: implications for cell injury and human disease. Annu. Rev. Pathol. 2013;8:105–137. doi: 10.1146/annurev-pathol-020712-163918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang Y.Y., Neufeld T.P. An Atg1/Atg13 complex with multiple roles in TOR-mediated autophagy regulation. Mol. Biol. Cell. 2009;20:2004–2014. doi: 10.1091/mbc.E08-12-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim J., Kundu M., Viollet B., Guan K.L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jung C.H., Jun C.B., Ro S.H., Kim Y.M., Otto N.M., Cao J., Kundu M., Kim D.H. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol. Biol. Cell. 2009;20:1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ganley I.G., Lam du H., Wang J., Ding X., Chen S., Jiang X. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J. Biol. Chem. 2009;284:12297–12305. doi: 10.1074/jbc.M900573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Egan D.F., Shackelford D.B., Mihaylova M.M., Gelino S., Kohnz R.A., Mair W., Vasquez D.S., Joshi A., Gwinn D.M., Taylor R., et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331:456–461. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garelick M.G., Kennedy B.K. TOR on the brain. Exp. Gerontol. 2011;46:155–163. doi: 10.1016/j.exger.2010.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parkhitko A., Myachina F., Morrison T.A., Hindi K.M., Auricchio N., Karbowniczek M., Wu J.J., Finkel T., Kwiatkowski D.J., Yu J.J., et al. Tumorigenesis in tuberous sclerosis complex is autophagy and p62/sequestosome 1 (SQSTM1)-dependent. Proc. Natl. Acad. Sci. USA. 2011;108:12455–12460. doi: 10.1073/pnas.1104361108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qin L., Wang Z., Tao L., Wang Y. ER stress negatively regulates AKT/TSC/mTOR pathway to enhance autophagy. Autophagy. 2010;6:239–247. doi: 10.4161/auto.6.2.11062. [DOI] [PubMed] [Google Scholar]
- 18.Choi Y.J., Di Nardo A., Kramvis I., Meikle L., Kwiatkowski D.J., Sahin M., He X. Tuberous sclerosis complex proteins control axon formation. Genes Dev. 2008;22:2485–2495. doi: 10.1101/gad.1685008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Di Nardo A., Kramvis I., Cho N., Sadowski A., Meikle L., Kwiatkowski D.J., Sahin M. Tuberous sclerosis complex activity is required to control neuronal stress responses in an mTOR-dependent manner. J. Neurosci. 2009;29:5926–5937. doi: 10.1523/JNEUROSCI.0778-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nie D., Di Nardo A., Han J.M., Baharanyi H., Kramvis I., Huynh T., Dabora S., Codeluppi S., Pandolfi P.P., Pasquale E.B., et al. Tsc2-Rheb signaling regulates EphA-mediated axon guidance. Nat. Neurosci. 2010;13:163–172. doi: 10.1038/nn.2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anderl S., Freeland M., Kwiatkowski D.J., Goto J. Therapeutic value of prenatal rapamycin treatment in a mouse brain model of tuberous sclerosis complex. Hum. Mol. Genet. 2011;20:4597–4604. doi: 10.1093/hmg/ddr393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goto J., Talos D.M., Klein P., Qin W., Chekaluk Y.I., Anderl S., Malinowska I.A., Di Nardo A., Bronson R.T., Chan J.A., et al. Regulable neural progenitor-specific Tsc1 loss yields giant cells with organellar dysfunction in a model of tuberous sclerosis complex. Proc. Natl. Acad. Sci. USA. 2011;108:E1070–E1079. doi: 10.1073/pnas.1106454108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee C.H., Hong C.H., Yu H.S., Chen G.S., Yang K.C. Transforming growth factor-beta enhances matrix metalloproteinase-2 expression and activity through AKT in fibroblasts derived from angiofibromas in patients with tuberous sclerosis complex. Br. J. Dermatol. 2010;163:1238–1244. doi: 10.1111/j.1365-2133.2010.09971.x. [DOI] [PubMed] [Google Scholar]
- 24.Sato A., Kasai S., Kobayashi T., Takamatsu Y., Hino O., Ikeda K., Mizuguchi M. Rapamycin reverses impaired social interaction in mouse models of tuberous sclerosis complex. Nat. Commun. 2012;3:1292. doi: 10.1038/ncomms2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rouschop K.M., van den Beucken T., Dubois L., Niessen H., Bussink J., Savelkouls K., Keulers T., Mujcic H., Landuyt W., Voncken J.W., et al. The unfolded protein response protects human tumor cells during hypoxia through regulation of the autophagy genes MAP1LC3B and ATG5. J. Clin. Invest. 2010;120:127–141. doi: 10.1172/JCI40027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moussay E., Kaoma T., Baginska J., Muller A., Van Moer K., Nicot N., Nazarov P.V., Vallar L., Chouaib S., Berchem G., et al. The acquisition of resistance to TNFalpha in breast cancer cells is associated with constitutive activation of autophagy as revealed by a transcriptome analysis using a custom microarray. Autophagy. 2011;7:760–770. doi: 10.4161/auto.7.7.15454. [DOI] [PubMed] [Google Scholar]
- 27.Klionsky D.J., Abdalla F.C., Abeliovich H., Abraham R.T., Acevedo-Arozena A., Adeli K., Agholme L., Agnello M., Agostinis P., Aguirre-Ghiso J.A., et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hara T., Nakamura K., Matsui M., Yamamoto A., Nakahara Y., Suzuki-Migishima R., Yokoyama M., Mishima K., Saito I., Okano H., et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 29.Sarkar S., Perlstein E.O., Imarisio S., Pineau S., Cordenier A., Maglathlin R.L., Webster J.A., Lewis T.A., O'Kane C.J., Schreiber S.L., et al. Small molecules enhance autophagy and reduce toxicity in Huntington's disease models. Nat. Chem. Biol. 2007;3:331–338. doi: 10.1038/nchembio883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Renna M., Schaffner C., Winslow A.R., Menzies F.M., Peden A.A., Floto R.A., Rubinsztein D.C. Autophagic substrate clearance requires activity of the syntaxin-5 SNARE complex. J. Cell Sci. 2011;124:469–482. doi: 10.1242/jcs.076489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alers S., Loffler A.S., Wesselborg S., Stork B. Role of AMPK-mTOR-Ulk1/2 in the regulation of autophagy: cross talk, shortcuts, and feedbacks. Mol. Cell. Biol. 2012;32:2–11. doi: 10.1128/MCB.06159-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meley D., Bauvy C., Houben-Weerts J.H., Dubbelhuis P.F., Helmond M.T., Codogno P., Meijer A.J. AMP-activated protein kinase and the regulation of autophagic proteolysis. J. Biol. Chem. 2006;281:34870–34879. doi: 10.1074/jbc.M605488200. [DOI] [PubMed] [Google Scholar]
- 33.Gwinn D.M., Shackelford D.B., Egan D.F., Mihaylova M.M., Mery A., Vasquez D.S., Turk B.E., Shaw R.J. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol. Cell. 2008;30:214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsai P.T., Hull C., Chu Y., Greene-Colozzi E., Sadowski A.R., Leech J.M., Steinberg J., Crawley J.N., Regehr W.G., Sahin M. Autistic-like behaviour and cerebellar dysfunction in Purkinje cell Tsc1 mutant mice. Nature. 2012;488:647–651. doi: 10.1038/nature11310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang J., Kim J., Alexander A., Cai S., Tripathi D.N., Dere R., Tee A.R., Tait-Mulder J., Di Nardo A., Han J.M., et al. A tuberous sclerosis complex signalling node at the peroxisome regulates mTORC1 and autophagy in response to ROS. Nat. Cell Biol. 2013;15:1186–1196. doi: 10.1038/ncb2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Papadakis M., Hadley G., Xilouri M., Hoyte L.C., Nagel S., McMenamin M.M., Tsaknakis G., Watt S.M., Drakesmith C.W., Chen R., et al. Tsc1 (hamartin) confers neuroprotection against ischemia by inducing autophagy. Nat. Med. 2013;19:351–357. doi: 10.1038/nm.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Settembre C., Di Malta C., Polito V.A., Garcia Arencibia M., Vetrini F., Erdin S., Erdin S.U., Huynh T., Medina D., Colella P., et al. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332:1429–1433. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Korolchuk V.I., Rubinsztein D.C. Regulation of autophagy by lysosomal positioning. Autophagy. 2011;7:927–928. doi: 10.4161/auto.7.8.15862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Son J.H., Shim J.H., Kim K.H., Ha J.Y., Han J.Y. Neuronal autophagy and neurodegenerative diseases. Exp. Mol. Med. 2012;44:89–98. doi: 10.3858/emm.2012.44.2.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Toda H., Mochizuki H., Flores R., 3rd, Josowitz R., Krasieva T.B., Lamorte V.J., Suzuki E., Gindhart J.G., Furukubo-Tokunaga K., Tomoda T. UNC-51/ATG1 kinase regulates axonal transport by mediating motor-cargo assembly. Genes Dev. 2008;22:3292–3307. doi: 10.1101/gad.1734608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ogura K., Goshima Y. The autophagy-related kinase UNC-51 and its binding partner UNC-14 regulate the subcellular localization of the Netrin receptor UNC-5 in Caenorhabditis elegans. Development. 2006;133:3441–3450. doi: 10.1242/dev.02503. [DOI] [PubMed] [Google Scholar]
- 42.Ogura K., Wicky C., Magnenat L., Tobler H., Mori I., Muller F., Ohshima Y. Caenorhabditis elegans unc-51 gene required for axonal elongation encodes a novel serine/threonine kinase. Genes Dev. 1994;8:2389–2400. doi: 10.1101/gad.8.20.2389. [DOI] [PubMed] [Google Scholar]
- 43.Tomoda T., Bhatt R.S., Kuroyanagi H., Shirasawa T., Hatten M.E. A mouse serine/threonine kinase homologous to C. elegans UNC51 functions in parallel fiber formation of cerebellar granule neurons. Neuron. 1999;24:833–846. doi: 10.1016/s0896-6273(00)81031-4. [DOI] [PubMed] [Google Scholar]
- 44.Williams A., Sarkar S., Cuddon P., Ttofi E.K., Saiki S., Siddiqi F.H., Jahreiss L., Fleming A., Pask D., Goldsmith P., et al. Novel targets for Huntington's disease in an mTOR-independent autophagy pathway. Nat. Chem. Biol. 2008;4:295–305. doi: 10.1038/nchembio.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tsvetkov A.S., Miller J., Arrasate M., Wong J.S., Pleiss M.A., Finkbeiner S. A small-molecule scaffold induces autophagy in primary neurons and protects against toxicity in a Huntington disease model. Proc. Natl. Acad. Sci. USA. 2010;107:16982–16987. doi: 10.1073/pnas.1004498107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sarkar S. Regulation of autophagy by mTOR-dependent and mTOR-independent pathways: autophagy dysfunction in neurodegenerative diseases and therapeutic application of autophagy enhancers. Biochem. Soc. Trans. 2013;41:1103–1130. doi: 10.1042/BST20130134. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.