Abstract
Although synaptogenesis within the retina is obviously essential for vision, mechanisms responsible for the initiation and maintenance of retinal synapses are poorly understood. In addition to its scientific interest, understanding retinal synapse formation is becoming clinically relevant with ongoing efforts to develop transplantation-based approaches for the treatment of retinal degenerative disease. To extend our understanding, we have focused on the chick model system and have studied the neuroligin family of neuronal adhesion factors that has been shown to participate in synapse assembly in the brain. We identified chicken orthologs of neuroligins 1, -3, and -4, but could find no evidence of neuroligin 2. We investigated temporal and spatial patterns of mRNA and protein expression during development using standard polymerase chain reaction (RT-PCR), quantitative PCR (QPCR), laser-capture microdissection (LCM), and confocal microscopy. At the mRNA level, neuroligins were detected at the earliest period tested, embryonic day (ED)5, which precedes the period of inner retina synaptogenesis. Significant alternative splicing was observed through development. While neuroligin gene products were generally detected in the inner retina, low levels of neuroligin 1 mRNA were also detected in the photoreceptor layer. Neuroligin 3 and -4 transcripts, on the other hand, were only detected in the inner retina. At retinal synapses neuroligin 1 protein was detected in the inner plexiform layer, but its highest levels were detected in the outer plexiform layer on the tips of horizontal cell dendrites. This work lays the groundwork for future studies on the functional roles of the neuroligins within the retina.
Indexing Terms: chick, horizontal cell, neurexin, photoreceptor, development, synaptogenesis
Synaptogenesis, a key component in the development of neural circuits, involves a series of overlapping processes in which pre- and postsynaptic proteins must be transcribed, translated, and transported to specific cellular compartments in a carefully choreographed manner (Lise and El-Husseini, 2006). In the chick retina, rods and four spectral classes of cone photoreceptors make connections at the outer plexiform layer (OPL) where they form stereotypical triad structures comprised of photoreceptor nerve endings with invaginating dendrites of horizontal and bipolar cells. Although less well characterized than other retinal cell types, some 8–9 classes of morphologically distinct bipolar cells can be distinguished in the avian retina with fine dendrites projecting to the OPL and single axons to the inner plexiform layer (IPL) where they innervate with amacrine and ganglion cell dendrites (Mariani, 1987). In addition, there exist at least three classes of morphologically distinct horizontal cells that vary in the extent of their dendritic arbor surface area, the depth of their projections to different strata in the OPL, and in some cases the presence of lateral axons (Gallego, 1986; Tanabe et al., 2006; Fischer et al., 2007; Edqvist et al., 2008; Rompani and Cepko, 2008). Lastly, amacrine cells are situated in the half of the inner nuclear layer (INL) proximal to the IPL where they modulate signals moving between bipolar and ganglion cells.
Two groups of proteins whose important role in synaptogenesis have been well demonstrated outside of the eye are the neuroligin (NLGN) and neurexin (NRXN) families (Lise and El-Husseini, 2006; Chih et al., 2006; Huang and Scheiffele, 2008). These adhesion molecules are thought to participate in synapse organization by bringing into register the pre- and postsynaptic elements responsible for neurotransmitter release and reception, respectively (Ichtchenko et al., 1996; Scheiffele et al., 2000; Taniguchi et al., 2007). Studies of central synapses from the brain in vitro have demonstrated that their ectopic expression can promote excitatory or inhibitory synapse formation depending on the combination of NLGN and NRXN molecules present (Scheiffele et al., 2000; Dean et al., 2003; Chih et al., 2006; Huang and Scheiffele, 2008). For instance, interactions between NLGN2 and its respective binding partners, the α- and β-NRXNs, are thought to promote formation and/or stabilization of inhibitory synapses through recruitment of GABAergic signaling machinery, while NLGN1, -3, and -4 are believed to influence excitatory synapses through recruitment of glutamatergic signaling components (Chih et al., 2006; Craig and Kang, 2007; Huang and Scheiffele, 2008; Kang et al., 2008). The recent discovery of NLGN3 at both GABAergic (inhibitory) and glutamatergic (excitatory) synapses, however, suggests that this situation might be more complex than previously appreciated (Budreck and Scheiffele, 2007).
In humans, the three NRXN genes give rise to considerable diversity through alternative promoter use and internal splicing. An upstream promoter gives rise to the more abundant larger α-NRXNs while a downstream promoter gives rise to the smaller b-NRXNs (Ushkaryov et al., 1992; Ushkaryov and Südhof, 1993). The NLGNs are encoded by five genes in humans (NLGN1–4X, 4Y) and by four genes (NLGN1–4) in mice and several other species. Although they too undergo alternative splicing, their degree of variability is considerably less than the NRXN's (Ichtchenko et al., 1996; Philibert et al., 2000; Bolliger et al., 2008). The inclusion of short alternative exons within the NLGN1 gene at three sites (designated A1, A2, and B) account for much of the variability in their binding affinity to NRXN ligands. NLGN2 mRNA undergoes similar splicing at the A2 site, thereby generating two different isoforms. The B site is absent from NLGN2, -3, and -4. While all NLGN genes are capable of producing transcripts bearing the A2 splice site, only NLGN1 and -3 have both A1 and A2 splice inserts (Bolliger et al., 2008). Strong polar charges at the A- sites and/or inhibitory N-linked glycosylation at the B- site influence interactions between alternatively spliced α- and β-NRXNs and NLGNs. Splice variants that preferentially bind β-NRXNs promote synapse formation, while those that bind both α-and β-NRXNs promote synapse expansion (Comoletti et al., 2003; Boucard et al., 2005; Comoletti et al., 2006).
Cell replacement therapy holds promise as an approach for the treatment of blinding retinal diseases such as the retinal degenerations; however, establishing synapse formation between graft and host tissues remains a major challenge (Adler, 2008). Overcoming this obstacle requires a better understanding of the molecules involved in synaptogenesis. As a step in this direction, we have concentrated on the NLGN family, and have focused on the chick retina model system because it is well suited to both gain-of-function and loss-of-function studies. To this end, we have experimentally derived the full open reading frames for each of the chick NLGN genes (NLGN1, -3, and 4), including their numerous retinal isoforms, studied their developmental patterns of expression, and investigated their distribution throughout the retina.
Materials and Methods
Animals
All procedures were carried out in accordance with animal protocols approved by the Animal Care and Use Committee at Johns Hopkins University. White Leghorn chick embryos (Gallus domesticus; B&E Eggs, York Springs, PA) were incubated at 37°C in a forced-air incubator with periodic rotation until the desired embryonic stage.
Gene analysis
Total RNA was isolated from chicken retinas by Trizol extraction and reverse transcription was carried out with Superscript III according to the manufacturer's protocol (Invitrogen, Gaithersburg, MD). Gene-specific primers were designed to recognize splice sites in the three chicken NLGN genes (Table 1). The identity of PCR products was verified by sequencing. Real-time QPCR was performed in 96-well plates with the Bio-Rad (Hercules, CA) IQ5 Multicolor Real Time PCR system. Each 20-μL reaction contained 10 μL of 2× SYBR Green Supermix (Bio-Rad) and 300 nM of target gene primer mix. QPCR conditions included denaturation at 95°C, followed by 40 cycles of amplification (95°C for 10 seconds, 56°C for 30 seconds, and 72 for 30 seconds). Relative gene expression was calculated as described by Pfaffl (2001). The housekeeping gene GAPDH was used for normalization.
Table 1. Oligonucleotides Used for Cloning and Gene Analysis.
| Gene name | Accession #. | Forward primer | Reverse primer | Size (bp) |
|---|---|---|---|---|
| Cell markers & housekeeping | ||||
| GAPDH | NM_204305 | CCATTCCTCCACCTTTGATG | GGAACAGAACTGGCCTCTCA | 280 |
| Brn3 | NM_204759 | CCATCGCCGAGAAGTTAGAC | CAAACCCTGCCCAGTTCT | 208 |
| Lim1 | L35569 | AGTCCTCCGTTCTCCTCGATA | TCCCGAGGTGACATTGTCTT | 211 |
| Pax6 | D87837 | CACCACTTCCACAGGTCTCAT | TCGTGCCGAACCGATATAAT | 281 |
| Opsin (green) | NM_205490 | GAATTCTCCTCCCTGCTTCC | TTCTCCGTGCTGCTTCTGTT | 595 |
| Full coding sequences | ||||
| Neuroligin-1 | N/A | ATGGCCTTTCCAAGATCCA | CTACATTACCCTGGTTGTTGAGTGTGA | 2592,2565, 2532,2472 |
| Neuroligin-3 | N/A | ATGTGGCTGATACTCTGGAG | CTATACCCTGGTGGTGGAGTGTGAG | 2565,2499, 2439 |
| Neuroligin-4 | N/A | ATGTCGAAACCAATGGGACT | CTATACCCTAGTTGTTGAATGTCCA | 2541,2481, 2382 |
| Neuroligin – no splice site | ||||
| Neuroligin-1 | N/A | ACCCATGCACAAGAAGAGGA | TGGGCGTCATTAAGGGAATA | 169 |
| Neuroligin-3 | N/A | CGGTGGTGACAGCTGACCT | CTGTTGTAGGCGGGTGAGAT | 585 |
| Neuroligin-4 | N/A | CCCAGAGAAACACAACCAATG | GTATGCAAAGGCTGCATTCC | 245 |
| Splice site ‘A’ | ||||
| NL1-A site | N/A | ATCTATGTCCCAACCGAGGA | CCAGTGCCTTCCATGTAGGAC | 219,159,99 |
| NL3 –A site | N/A | CCCAACGAGGACTGTCTGTA | AGAAGGCGATGTTCTCACTG | 418,358,298 |
| NL4 –A site | N/A | ACCTGGATGAGAGGTCGTTG | AAGATCCGCCATGGATGTAG | 252, 192 |
| Splice site ‘B’ | ||||
| NL1- B site | N/A | CATGCGTCAATCTGTTGACTTT | GAGTGCTGTTCCACTTTGAGC | 113, 86 |
| NL3- B site | N/A | CATCACCGTCTTCGGTTCC | GGATGATGGCTCTCTGGAAG | 146 |
| NL4- B site | N/A | GATCTGGGGCAGGAGCTT | CGAAGGCATTCAACCAAATCAG | 202 |
| Neurexin | ||||
| NRXN1 | CGTAGTACGTTTCACGAGAAGTGG | AGACCAGAGAGCTGGCCTTG | 171, 261 | |
| NRXN3 | TGACAGTGCTCCTGGACTTG | TGATGTAGTTTCGCGTCTCG | 1511, 1268, 1208, 896, 887,704,326 |
Cloning of open reading frames
Full-length chicken orthologs of human NLGN genes were identified using genomic and expressed sequence tag (EST) information from publicly available databases at NCBI and TIGR (www.tigr.org). BLAT (Blast-like alignment tool) alignments from the University of California at Santa Cruz (UCSC) genome browser were used to identify phylogenetically conserved sequences. When significant gaps at translation start or stop sites became problematic we performed RACE PCR (see below) to obtain full-length coding sequences. Contig assembly was performed using Sequencher 4.7 software (Genecodes, Ann Arbor, MI) or Geneious Pro (Biomatters, New Zealand). Amplification of full-length open reading frames, including alternatively spliced transcripts, was carried out with Phusion Hot Start high fidelity DNA polymerase (Finnzymes, Espoo, Finland) using the included HF buffer or GC buffer supplemented with 3% DMSO for difficult to amplify sequences. HA-tags were introduced by overlap extension PCR, adjacent to the signal peptide cleavage site (Supporting Fig. S3). Blunt-end PCR products were directionally inserted into pENTR-D-TOPO (Invitrogen, Carlsbad, CA) and sub-cloned into a pCAGEN (Addgene) expression plasmid custom modified to include gateway recombination sites (CMV enhancer/chicken β-actin promoter). The identity of all clones was verified by sequencing.
Rapid amplification of cDNA ends (RACE): suppression and SMART-RACE
Suppression PCR based 3′ RACE-PCR was used to obtain 3′ prime sequence information (Innis, 1990; Ozawa et al., 2004). A random 9-mer containing a unique polylinker (5′-ACA GCA GGT CAG TCA AGC AGT AGC AGC AGT TCG ATA AGC GGC CGC CAT GGA NNNNNNNNN-3′) was used to prime reverse transcription. The SMART (Switching Mechanism At 5′ end of RNA Transcript; ClonTech, Palo Alto, CA) method was used to complete 5′ end sequences. Briefly, reverse transcription with MMLV reverse transcriptase was followed by addition of 3–5 residues (predominantly dC) to the 3′ end of the first-strand cDNA by the endogenous terminal transferase activity of MMLV. A GC-rich SMART oligo was annealed to the dC-rich tail, thus serving as a primer for second-strand formation. Using universal (sense) and gene specific (antisense) primers RACE products were PCR-amplified, subcloned into pENTR-D-TOPO.
Protein alignment and sequence analysis
Open reading frames were identified as described above and the deduced protein sequences were aligned using the ClustalW (Id matrix) plugin provided with Geneious Pro (http://www.geneious.com). Sequences from Anolis carolinensis (anole lizard), Apis mellifera (European honeybee), Acyrthosiphon pisum (pea aphid), Aedes aegypti (mosquito), Bos taurus (bovine), Danio rerio (zebrafish), Equus caballus (horse), Gallus gallus (chicken), Homo sapiens (human), Mus musculus (mouse), Pan troglodytes (chimpanzee), Rattus norvegicus (rat) obtained with accession numbers provided in Figure 1 were collected for tree building. Prior to alignment, signal peptide sequences were trimmed and gaps at internal splice sites were joined. Percent identity between human and chicken orthologs was calculated using ClustalW (http://www.ebi.ac.uk/Tools/clustalw) (Thompson et al., 1994). Identity along the NLGN sequence is graphically illustrated in Supporting Figure S1. Signal peptide cleavage sites were predicted with SignalP, a Web-based predictor of signal peptides (Bendtsen et al., 2004; Emanuelsson et al., 2007). PHYML was used to make a bootstrapped (100 iterations) maximum-likelihood phylogenetic tree using the JTT substitution model and was optimized for topology and branch length and rate parameters (Guindon and Gascuel, 2003).
Figure 1.
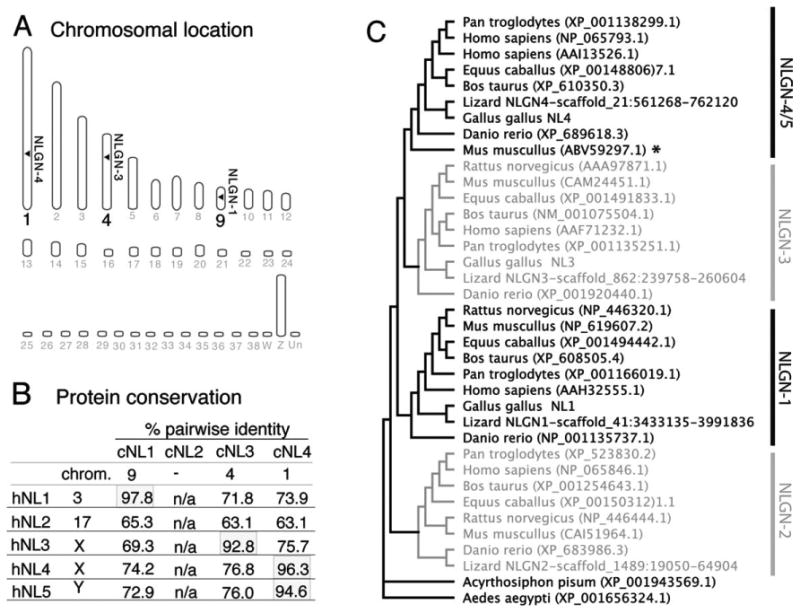
Identification and phylogenetic analysis of neuroligin family members. A: There are 38 autosomes and two sex chromosomes (W,Z) in the chicken. The identity of the chromosomes carrying possible NLGN homologs were identified with the Ensembl gene browser and are indicated by horizontal arrowheads. B: Percent identity between the coding regions of human and chicken NLGN genes calculated using a ClustalW alignment shows the degree of homology between the chicken and human orthologs. C: A rooted tree layout cladogram displaying the evolutionary relationships between vertebrate and non-vertebrate animals includes Anolis carolinensis (Lizard), Acyrthosiphon pisum (pea aphid), Aedes aegypti (yellow fever mosquito), Bos taurus (bovine), Danio rerio (zebrafish), Equus caballus (horse), Gallus gallus (chicken), Homo sapiens (human), Mus musculus (mouse), Pan troglodytes (chimpanzee), and Rattus norvegicus (rat). Accession numbers used for alignments and tree building are listed within the dendrogram.
Western blot analysis
Tissues from chicken retinas and brain or COS-7 cells transfected with HA-tagged NLGN expression plasmids were probed by Western blot. Transfections were carried out with Lipofectamine 2000 according to the manufacturer's recommendations. Tissues or cells were homogenized in ice-cold lysis buffer (150 mM NaCl, 50 mM Tris pH 8.0, 2 mM EDTA, 1% Triton X-100, plus “complete mini” protease inhibitor), incubated for 40 minutes on ice, and supernatants were sequentially spun at 4°C at 3,000 rpm for 10 minutes and 12,000g for 15 minutes. Then 30 μg of tissue lysate or 5 μg from transfected COS cells were separated on NuPAGE Novex 4-12% bis-tris gels (Invitrogen) and transferred to nitrocellulose membranes (iBlot system, Invitrogen). Membranes were blocked in 1 × casein and sequentially incubated with rat anti-HA or mouse anti-neuroligin antibodies followed by alkaline phosphatase-conjugated secondary antibodies (1:2,000; Sigma, St. Louis, MO). Protein bands were detected with NBT/BCIP in Tris buffer containing 100 mM NaCl, 100 mM Tris pH 9.5, 50 mM MgCl2.
Laser-capture microdissection
Embryonic day (ED) 20 chick embryos were euthanized by halothane overdose and samples processed as previously described (Wahlin et al., 2008). The anterior pole and vitreous from enucleated eyes were removed and resulting eyecups were incubated in 6.75, 12.5, and 25% sucrose for 10, 20, and 30 minutes, respectively, equilibrated in OCT-sucrose, and snap-frozen. Seven-μm cryosections were thaw-mounted onto PEN-foil slides (Leica Microsystems, Deerfield, IL), fixed in 70% cold ethanol for 30 seconds, briefly rinsed in DEPC-treated water, stained with hematoxylin for 10 seconds, and dehydrated in 70% ethanol for 30 seconds and 100% ethanol for 1 minute. Samples from the outer nuclear layer (ONL), inner nuclear layer (INL), and ganglion cell layer (GCL) (Fig. 4) were collected from the eye using an LMD6000 laser capture micro-dissection microscope (Leica Microsystems). RNA concentrations were determined with an Agilent (Palo Alto, CA) Bioanalyzer. Total RNA was purified using Qiagen's RNeasy microkit (Valencia, CA), primed with oligo(dT15) and cDNAs synthesized using Superscript III reverse transcriptase. Assuming an average cell diameter of 5-7 μm and 10 pg of total RNA per cell, of which 2% is mRNA, estimated sample sizes were ≈85,000 photoreceptor cells with 17 ng mRNA, 110,000 INL cells containing 22 ng mRNA, and 105,000 GCL cells with 21 ng mRNA.
Figure 4.
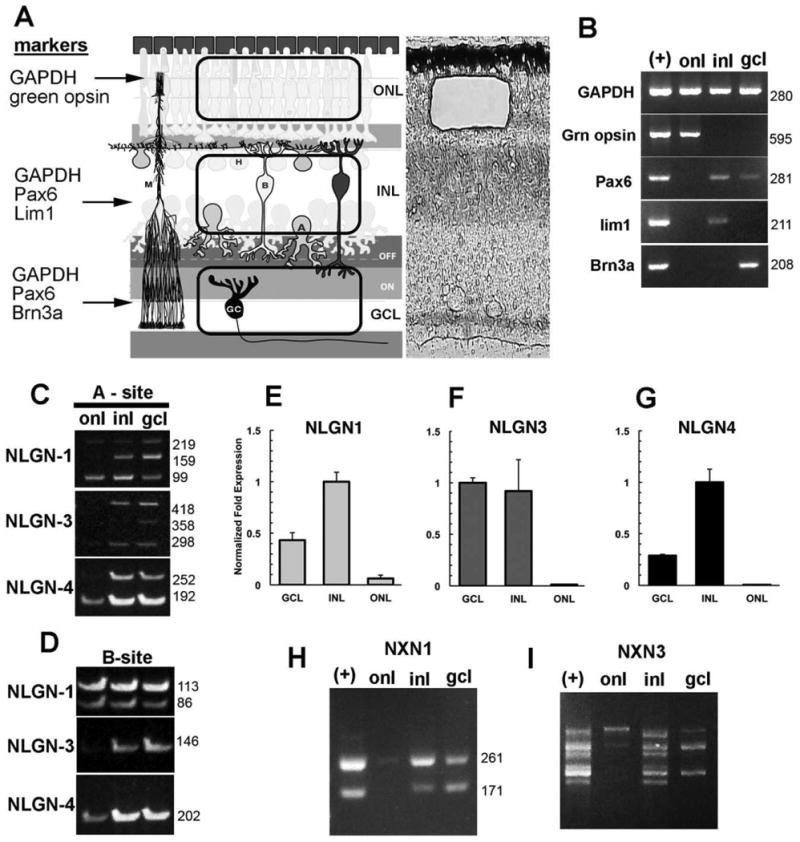
Laminar position of alternatively spliced neuroligin and neurexin transcripts in the chick retina. A: The retina is compartmentalized into outer and inner nuclear layers and the ganglion cell layer, each separated by the synaptic plexiform layers. A representative laser capture microdissection cut of the outer nuclear layer is indicated here. B: LCM samples were validated for purity and lack of contamination of RNA from other retinal layers by PCR using primers specific for photoreceptors (green opsin), inner nuclear layer horizontal cells (Lim1), inner retina (Pax6), or the ganglion cells (Brn3). Detection of NLGN1, -3, or -4 alternatively spliced variants in isolated nuclear layers is revealed by analysis with primers flanking splice sites -A (C) or -B (D). E–G: Real-time PCR was used to show relative abundance of NLGN1, -3 and -4 transcripts in relation to the housekeeping gene GAPDH. Different sized alternative spliced NRXN1 (H) and NRXN3 (I) transcripts reveal differences in splicing.
Antibody characterization
Please see Table 2 for a detailed list of all antibodies used.
Table 2. Information on Primary Antibodies Used in This Study (See Also Materials and Methods).
| Antibody | Immunogen used | Source | Dilution IHC | Dilution Western |
|---|---|---|---|---|
| Calbindin-D281 | Full-length calbindin- purified from chicken gut | Sigma, mouse monoclonal, CL3002 | 1:500 or 1:100,0001 | 1:1,000 |
| Calretinin | Full-length recombinant human protein | SWANT, rabbit polyclonal, 7699/4 | 1:1,000 | |
| CASK | Amino acids 318-415 of rat CASK/Lin2 | UC Davis NeuroMab Facility, mouse monoclonal, clone K56A/50 | 1:200 | 1:200 or 1:1,000 |
| GABA | GABA conjugated to bovine serum albumin | Sigma, rabbit polyclonal, A2052 | 1:2,500 | |
| GluR4 | Peptide sequence RQSSGLAVIASDLP of rat GluR4 | Chemicon, rabbit polyclonal, AB1508 | 1:500 | N/A |
| Hemagluttinin (HA-tag) | HA peptide sequence [YPYDVPDYA] from the human hemagglutinin protein. | Roche, rat monoclonal, 1-867-423 clone 3F10 | N/A | 1:1,000 |
| Islet-1 | Amino acids 247-349 at the C-terminus of Islet1 | DSHB, mouse monoclonal, 40.2D6 | 1:35 | N/A |
| LIM 1+2 | Amino acid residues 1-360 of rat Lim2 protein | DSHB, mouse monoclonal, 4F2 | 1:20 | N/A |
| LIM3 | Recombinant full- length murine Lim3 fused to GST | DSHB, mouse monoclonal, 67.4E12 | 1:50 | 1:50 |
| Neuroligin-11 | Intracellular C-terminal domain (aa 718-843) of rat NLGN1 | UC Davis NeuroMab Facility, mouse monoclonal, N97/A31- or N97/A31-purified | 1:10 1:500 purified or 1:5,0001 | 1:500 purified |
| Pax6 | Peptide QVPGSEPDMSQYWPRLQ from the C-terminus of mouse Pax6 | Covance, rabbit polyclonal, PRB-278P | 1:8,000 | N/A |
| Pax6 | Amino acids 1–223 of chicken Pax6 | DSHB, mouse monoclonal, Pax6-concentrate | 1:200 | N/A |
| Prox1 | C-terminal homeo- and Prospero domains (aa 546-736) of human Prox1. | Chemicon, rabbit polyclonal, AB5475 | 1:10,000 | N/A |
| PSD95 | Amino acids 77-299 of the human PSD95. | 75-028 UC Davis NeuroMab Facility, mouse monoclonal, clone K28/43 | 1:1,000 - 1,1500 | 1:2,000 |
| Ribeye (CtBP2/ribeye) | Amino acids 361-445 from the C-terminal region of mouse CtBP2. | BD-Biosciences, mouse monoclonal, 612044 | 1,000-1,2000 | N/A |
| TrkA | Extracellular domain of chicken trkA purified as a secreted protein from transfected COS cells by chroma- tography on wheat germ agglutinin, a His-Bind nickel-chelating column, and ion-exchange chromatography on Mono Q. | Dr. F. Lefcort, Montana State University, rabbit polyclonal | 1:6,000 | N/A |
| Gα t1 (K-20; transducin) | Gα t1 of human origin corresponding to amino acids 90-109. | SantaCruz, rabbit polyclonal, sc-389 | 1:1,000 | N/A |
| SV2 | From purified cholinergic synaptic vesicles from the electric organ of Discopyge ommato. | DSHB, mouse monoclonal, SV2 | 1:50-1:100 | N/A |
Mouse monoclonal, M; rat monoclonal, Mr; rabbit polyclonal, R.
Hybridoma supernatatnt; IHC, immunohistochemistry.
Tyramide signal amplification (TSA).
No longer available (but available through SWANT). DSHB, developmental studies hybridoma bank; N/A, not applicable.
Mouse anti-calbindin is recognized in a 2D gel (manufacturers observation); specificity shown by indirect immunofluorescence, immunoenzymatic labeling and radioimmunoassay, and in a comparison of retinas from wildtype and calbindin −/− mice (Wässle et al., 1998). It labels cones, subsets of bipolar and amacrine cells, and occasional cells in the GCL in the chicken retina as reported (Ellis et al., 1991).
Rabbit anti-calretinin showed a single band of ≈28–29 kDa by Western blot and labels horizontal cells by immunohistochemistry (IHC) across many species including chicken (Pasteels et al., 1990; Ellis et al., 1991; Ikenaga et al., 2006; Fischer et al., 2007).
Mouse anti-CASK recognizes bands of ≈75 kDa and 125 kDa on Western blots of chick retinas consistent with the two mammalian variants in GenBank (Wahlin et al., 2008).
GABA antiserum selectively labels amacrine and horizontal cells by IHC from multiple species, including chicken, as previously reported (Fischer et al., 1998).
GluR4 antiserum recognizes a single band of expected size ≈100–110 kDa in size on Western blots of chick retina and in horizontal cells by IHC as described (Silveira dos Santos Bredariol and Hamassaki-Britto, 2001; Wahlin et al., 2008).
Mouse anti-islet1 recognizes both Islet-1 and -2 and has a well-established pattern of cell labeling by IHC and in situ hybridization (ISH) in chicken motor and retinal neurons (Tsuchida et al., 1994; Fischer and Reh, 2002).
The specificity of anti-Lim1/2 antibodies was verified by IHC and ISH showing specific expression in only a subset of horizontal cells as previously reported (Tsuchida et al., 1994; Lui et al., 2010).
Lim3 antibodies label bipolar cells by IHC equivalent to patterns observed by ISH (Tsuchida et al., 1994; Zhadanov et al., 1995; Ericson et al., 1997; Glasgow et al., 1997; Edqvist and Hallbook, 2004; Edqvist et al., 2006). A band of expected size ≈45–50 kDa (unpublished data) is observed by Western blot.
Mouse anti-NLGN1 recognized a doublet band of expected size ≈100 kDa by Western blot of ED15 and postnatal (P)0 chicken retinas and brain tissues (see Fig. 5). Blots with lysates from COS7 cells overexpressing HA-tagged chicken NLGN1 (AAB), NLGN3 (A1A2), and NLGN4 (A2) showed no cross-reactivity to NLGN3 or -4. Knockout mice are negative by IHC and Western blot (data from M. Bolliger and T. Sudhof; NeuroMab).
The two antibodies against Pax6 recognize different epitopes yet show identical patterns of cell staining in chicken and mammalian retinas, namely, in horizontal, amacrine, and ganglion cells (Belecky-Adams et al., 1997; Ashery-Padan et al., 2000; Fischer and Reh, 2000).
Rabbit anti-Prox1 strongly labels chicken horizontal and amacrine cell subpopulations similar to previously reported studies (Edqvist and Hallbook, 2004; Fischer et al., 2007; Edqvist et al., 2008).
Mouse anti-PSD95 detects multiple, closely sized bands of expected size ≈95–110 kDa on a Western blot of chick retina and brain lysates and in the synaptic layers of the retina by IHC (Wahlin et al., 2008). No signal is present in knockout mice (Béïque et al., 2006).
In the retina, mouse anti-ribeye/CtBP2 Western blots revealed bands of ≈50 kDa and 130 kDa corresponding to the expected sizes for the short nuclear and longer synaptic ribbon specific variants, while in the brain only the short nuclear variant was detected with an IHC pattern of synaptic labeling similar to other species (Schmitz et al., 2000; Wahlin et al., 2008).
Rabbit anti-rod α-transducin Gαt1 (K-20), which is an affinity-purified rabbit polyclonal antibody raised against a peptide corresponding to amino acids 90–109 of human rod α-transducin, has been shown to react with α-transducin of mouse, rat, and human origin (Boughter et al., 1997). Its specificity was verified in chicken retinas using fluorescent promoter viruses that selectively label chicken rod photoreceptors in vivo (Semple-Rowland et al., 2007).
Mouse anti-SV2 recognizes a band of ≈75–110 kD in immunoblots of brain synaptosomes from rat, chick, and frog (Buckley and Kelly, 1985; Feany et al., 1993). In the chicken retina, SV2 labels synaptic terminals in the outer and inner plexiform layers along with PSD95, CASK, and ribeye (Wahlin and Adler, 2007a; Wahlin et al., 2008).
Rabbit anti-trkA was raised to the entire extracellular domain of chicken trkA which was expressed in COS7 cells, purified, and used to produce antibodies in rabbits (Oakley et al., 1997). To demonstrate specificity human kidney 293 cells were transfected with trkA, trkB, or trkC and reacted with antibodies to trkA. Its role as a retinal horizontal cell marker has been extensively studied by immunohistochemistry, showing a similar pattern to ISH studies with expression in a subset of horizontal cells and a few types of amacrine cells (Lefcort et al., 1996; Gallo et al., 1997; Karlsson et al., 2001; Fischer et al., 2007).
Figure 5.
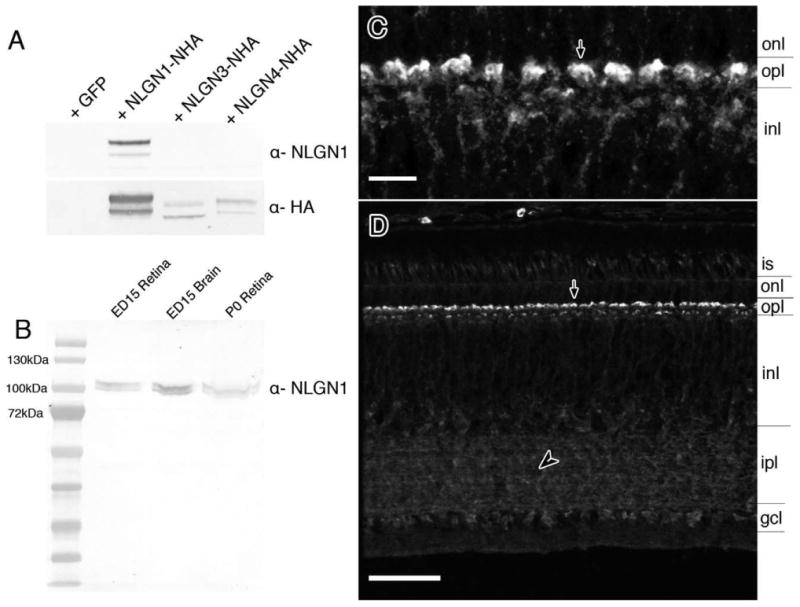
Detection of NLGN1 protein in the chick retina and antibody specificity. A: Western blots with HA-tagged recombinant chicken NLGN1, -3, or -4 proteins were probed with NLGN1 antibodies to demonstrate a lack of cross-reactivity to other NLGN family members. Samples run in parallel were probed with anti-rat HA antibodies as a loading control. B: Native protein from ED15 and P0 chick retinas and ED15 chick forebrain were separated by SDS-PAGE gel electrophoresis and probed with antibodies against NLGN1. C,D: Tissue sections from ED20 retinas were probed with NLGN1 antibodies and detected by fluorescence immunohistochemistry. Panel C is a magnified image of the outer plexiform layer with NLGN1 positive dendrites projecting from the outer inner nuclear layer into the outer plexiform layer (arrow). is, photoreceptor inner segment; onl, outer nuclear layer; opl, outer plexiform; inl, inner nuclear layer; ipl, inner plexiform layer; gcl, ganglion cell layer. Scale bars = 5 μm in C; 25 μm in D.
Tissue processing for IHC
Tissue processing was performed as described (Wahlin et al., 2008). Enucleated eyes were opened along the ora serrata, vitreous removed, and eyecups fixed for 20 minutes in cold 4% paraformaldehyde in 0.1 M phosphate buffer, pH 7.4, and subsequently immersed in 6.75, 12.5, and 25% sucrose in phosphate buffer for several hours each, followed by overnight incubation in 25% sucrose and 1 hour in a 2:1 ratio of 25% sucrose-0.1M phosphate buffer and OCT Tissue-Tek (Ted Pella, Redding, CA), and by snap-freezing on dry ice / isopentane. Cryostat sections were cut at 8–10 μm, thaw-mounted onto Super-Frost Plus glass slides (Fisher, Pittsburgh, PA). In general, sections were blocked and permeabilized in 10% goat serum in phosphate-buffered saline (PBS) containing 0.1% Triton X-100 and incubated overnight in primary antibody in 2% goat serum containing 0.02% Triton X-100. Incubation of NLGN1, GluR4, LIM 1+2, and LIM3 antibodies was done overnight at 4°C in 1% goat serum with 0.1% Triton X-100 with or without previous blocking in 1%goat serum. Primary antibody sources and dilutions are listed in Table 2. Alexa647 conjugated peanut lectin agglutinin (PNA) (Invitrogen) was used at 1:1,000. Secondary antibodies were goat antimouse and goat antirabbit IgGs coupled to Alexafluor-488, -546, -594, or -647 (Invitrogen; 1:1,000). Ten μg/mL Hoechst 3342 (Molecular Probes, Eugene, OR) was used to visualize cell nuclei. Double labeling with primary antibodies from the same species was performed using tyramide signal amplification (TSA) with TSA plus reagents (Perkin Elmer, Boston, MA) according to the manufacturer's recommendations. Following primary antibody incubation, endogenous peroxidases were quenched with 1% hydrogen peroxide in phosphate buffer for 30 minutes at room temperature and sequentially incubated with biotinylated goat antimouse IgG (1:1,000; Vector Labs, Burlingame, CA), horseradish peroxidase conjugated streptavidin (1:1,000; Vector Labs), and fluorescein-tyramide (1:500 dilution). A second mouse monoclonal antibody was then applied at standard working concentrations (see Table 2) and visualized with Alexafluor-647 conjugated goat antimouse. Control reactions using ultralow antibody concentrations were detectable with tyramide amplification but were undetectable by standard fluorescent microscopy. Serial sections processed similarly but without primary antibody were used as negative controls to rule out endogenous peroxidase or nonspecific background.
Microscopy and image processing
Confocal mages were acquired with a Zeiss LSM510 laser scanning microscope and images were adjusted for brightness and contrast using ImageJ freeware (NIH; http://rsb.info.nih.gov/ij/) or Adobe Photoshop (San Jose, CA). Similar settings were used for laser power, photomultiplier gain and offset, and a pinhole diameter of one Airey unit. Brightest point projections of z-stacks (5–10 optical sections, 0.5–1.0 μm thickness, 0.5–1.0 μm step size) were performed to highlight 3D elements of synaptic terminals such as the alignment of pre- and postsynaptic elements. Very thin optical sections (≈0.5 μm) were used to study subcellular localization or colocalization.
Results
Cloning and gene structure of the chicken neuroligins
Full-length open reading frames for chicken NLGN1, -3, and -4 were identified by analysis of genomic DNA, ESTs, and mRNA sequences from publicly available databases and experimentally derived RACE PCR products. These were subsequently cloned and sequence verified. ENSEMBL alignments localized them to chromosomes 9, 4, and 1, respectively (Fig. 1A); however, the exact gene lengths are unknown due to intronic sequence gaps that exist even in the most recent revision of the publicly available chicken genome (June 17, 2006 v11draft assembly). With the exception of NLGN1, for which there was ample sequence data, we had to rely on several complementary molecular approaches (e.g., RACE PCR and comparative genomic alignments) to identify the remaining chicken NLGN full-length coding sequences. Among the three NLGN genes, a total of 11 isoforms were amplified from chick retina or brain cDNA (GenBank accession numbers: NLGN1 isoforms, A1A2B (GU574580), A1A2 (GU080337), A1B (GU080338), A2B (GU080339), B (GU080340); NLGN3 isoforms, A1A2 (GU574581), A2 (GU080341), (−) (GU080342); and NLGN4 isoforms, A2 (GU080343), (−) (GU080344), x3 (GU080345). One of the NLGN1 variants is equivalent (with minor differences) to a previously identified isoform isolated from ED15 chick ciliary ganglion (Conroy et al., 2007). Primers spanning the translation initiation and termination sites of NLGN1 allowed isolation from the retina of five distinct transcripts that vary at two positions within the extracellular carboxyesterase domain, which are designated splice sites A and B. These sites are similar to those already identified in mammals (Fig. 2). These transcripts produce precleaved proteins of 874 (NLGN1 A1A2B), 865 (NLGN1 A1A2), 843 (NLGN1 A1B), 843 (NLGN1 A2B), and 823 (NLGN1 B) amino acids with predicted molecular weights of ≈96.6, 95.5, 94, 94, and 92 kDa, respectively. Splice variation for NLGN4 was observed at the A splice site, but not the B-site, yielding precleaved proteins of 847 (NLGN4 A2), 827 (NLGN4 -), and 776 (NLGN4 -x3) amino acids in length. The NLGN4 -x3 variant was unique in that it lacked both site A2 and the adjacent exon 3. We are not aware of a mammalian equivalent. For NLGN3, we needed to perform suppression PCR-RACE and SMART RACE (ClonTech) to obtain full sequence data. A BLAT alignment of NLGN1 and -3 contig sequences revealed seven coding exons while NLGN4 had only six (Fig. 2B–D). Despite repeated attempts, an avian equivalent for NLGN2 was not identified using degenerate PCR or through extensive online database searches of the chicken genome.
Figure 2.
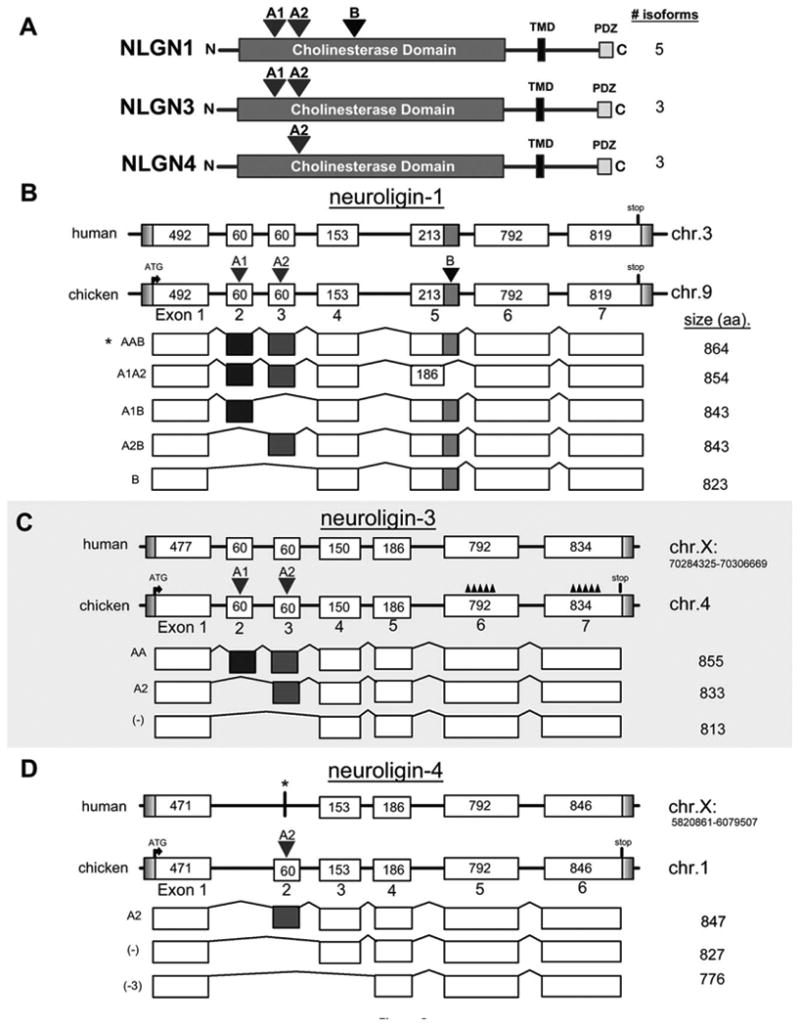
Genomic structure of alternatively spliced isoforms identified in the chick nervous system. A: Cartoon representation of NLGN1, -3, and -4 determined experimentally by sequencing of chicken transcripts from either retina and/or brain. Peptide sequences were aligned to the chicken genome using BLAT. The intron–exon structure of the coding regions for NLGN1 (B), NLGN3 (C), and NLGN4 (D) depict the two major splice sites indicated by inverted red and blue triangles above the A and B splice sites, respectively. The length in basepairs of each exon is indicated within each box and this diagram is not drawn to scale. The amino acid length of each isoform is indicated on the right. The chromosomal positions were included for human NLGN3 and -4 to distinguish between the two genes that are both located on the X-chromosome. The accession numbers used for constructing the human NLGN3 and NLGN4 exon maps were AAF71232.1 and NP_065793.1, respectively. The human NLGN3 did not contain A1 and A2 sites so they were added prior to doing a BLAT alignment. A similar approach was taken for NLGN4. The asterisk above human NLGN4 indicates that NLGN4 is represented in the human genome. The jagged peaks represented by “∧∧∧∧∧∧∧” above coding exons 5 and 6 for chicken NLGN3 signify probable gaps in the current draft of the chicken genome.
Conservation and sequence analysis of the chicken neuroligin proteins
Alignment of the deduced protein-coding regions when compared with those of other vertebrates, including human, showed a high degree of interspecies conservation (Supporting Fig. S1) which is consistent with a recently published report including partial chicken NLGN sequences (Bolliger et al., 2008). Analysis of the mature cleaved protein showed a 97.5% identity between human and mouse NLGN1, 92.8% identity for NLGN3, and 96.4 or 94.6 for NLGN4X and -4Y, respectively (Fig. 1B). Since the N-terminal signal peptide sequence is cleaved from the mature protein (Supporting Fig. S3) and is less conserved we did not include that in the phylogenetic analysis. Bolliger et al. (2008) previously uncovered partial evidence of three neuroligin orthologs (NLGN1, -3, and -4) in the chicken and no evidence of NLGN2. Such identification was based on limited EST sequences and/or computationally predicted sequences and did not reflect experimentally derived open reading frame sequences. Our own phylogenetic analysis, shown in Figure 1C, builds on this by including direct experimentally verified open reading frame sequences that fills in significant sequence gaps at 5′ and 3′ ends of the NLGN3 and -4 coding sequence. Figure 1C illustrates the evolutionary relationship among a wide range of species from insects to primates with orthologs from the class Insecta forming the root of this tree. Throughout the vertebrate clade the root lays with NLGN2; however, this gene appears to be absent from chicken. As expected, primate NLGNs are clustered together, while those from avian and teleost species are generally offset at adjacent nodes, as would be expected of their evolutionary distance. A noteworthy outlier is mouse NLGN4 that is situated far away from other mammals. This evolutionary difference has been attributed to rapid mutations in the mouse genome (Bolliger et al., 2008). While the sequencing of lizard genomes are far from complete, we identified sequences from Anolis carolinensis that are homologous to NLGN2 (Supporting Fig. S1). Furthermore, we noted that some syntenic regions adjacent to the human NLGN2 gene are scattered throughout the chicken genome, while others are not (Supporting Fig. S4). For instance, genes immediately upstream to human NLGN2 were not found and genes downstream mapped remotely to chicken Chr18. It is noteworthy that a striking number of genes flanking the NLGN2 gene (e.g., SPEM1, FGF11, etc.) were found in human but not in chick. The absence of NLGN2 in the chick warrants further exploration of animals closer to their evolutionary lineage.
The degree of sequence conservation between the chicken NLGN family members and their human homologs tended to be within the extracellular esterase domain, with less conservation evident in the linker before the transmembrane domain (green) (Supporting Fig. S2). For each NLGN an N-terminal signal peptide sequence, an esterase domain, a linker with O-glycosylation sites, a transmembrane domain, putative N-glycosylation sites, and a cytoplasmic tail containing a PDZ interacting domain were identified. Although the signal peptide region was relatively poorly conserved at intra- and interspecies levels, the predicted cleavage sites appeared similar: at amino acids 45–46 for NLGN1 (human and chick), 37–38 (human), and 39–40 (chick) for NLGN3, and 33–34 for NLGN4 (human and chick). (Complete signal peptide sequences are highlighted in Supporting Fig. S3C.)
Temporal aspects of neuroligin gene expression
To assess the developmental regulation of NLGN in the retina we performed RT-PCR (Fig. 3). In general, the NLGNs were detected in the retina considerably before the period when synaptogenesis is thought to occur. Using standard PCR conditions, NLGN1, -3, and -4 mRNAs were each detected in the developing retina at the earliest time evaluated (ED5; not shown), rising only slightly between ED6 and ED20 (Fig. 3B). A more careful examination using quantitative real-time PCR with oligonucleotides flanking the B splice site, however, uncovered levels of gene expression for NLGN1, -3, and -4 that progressively increased during development, peaking between ED12 and -14 (Fig. 3C). NLGN1 transcripts remained high while transcripts for NLGN3 and -4 decreased after the initial peak.
Figure 3.
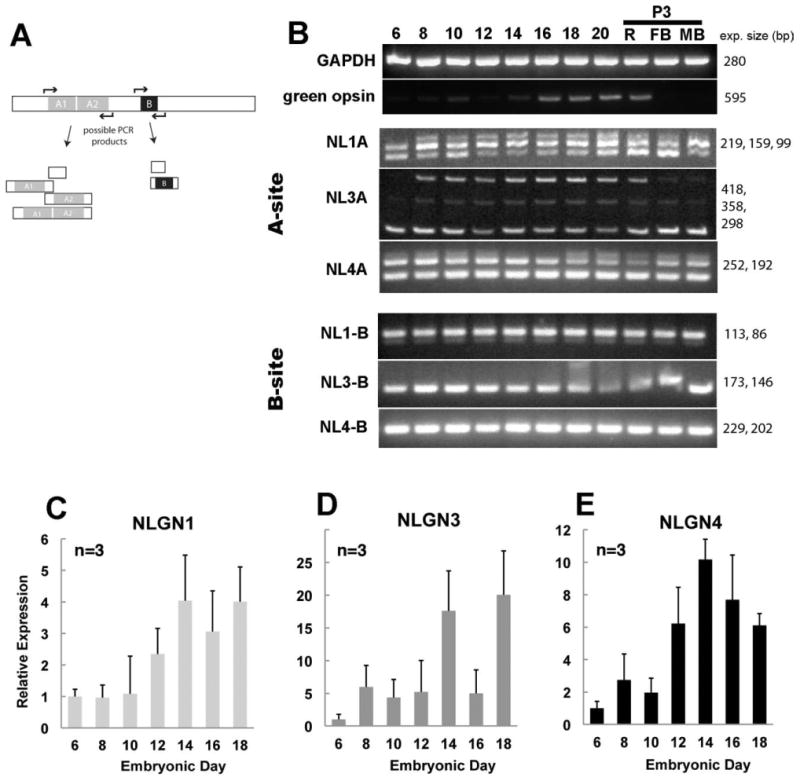
Alternatively spliced neuroligin family members expressed during retinal development and relative abundance of gene expression. A: PCR primers flanking each of the conserved splice sites (sites A and B) for NLGN were used to detect NLGN isoform variation and give rise to several combinations of transcript variants. B: PCR amplification of cDNAs from a developmental time course of retinal tissues was separated by gel electrophoresis. C–E: Quantitative real-time PCR shows the relative abundance of NLGN1 (C), NLGN3 (D), and NLGN4 (E) transcripts in relation to the GAPDH housekeeping control gene. Error bars represent standard deviation (n = 3).
The temporal pattern of alternative splicing was evaluated by PCR. NLGN1, -3, and -4 mRNA spliced transcripts were monitored with gene-specific oligonucleotides flanking the conserved A sites such that PCR products yielded 1, 2, or 3 bands, depending on the degree of splicing (Fig. 3B). Although there are actually four possible transcripts at the A site, only three can be resolved by electrophoresis, with a small 99 bp band for transcripts lacking at the A site, an intermediate band of 159 bp for those containing the A1 or A2 splice sites, and a higher band 219 bp band for those containing both the A1 and A2 sites. At ED6, NLGN1 amplification gave both short and intermediate sized bands, representing none or only one “A” splice site, and by ED8 a combination of all three transcript sizes were present. Control samples from forebrain (FB) or midbrain brain (MB) also had evidence of alternative splicing. NLGN1 splice site B showed two DNA bands of unequal intensity; one intense band represents a single splice insert while the other much less intense band indicates the absence of this site. NLGN3 and -4 had splicing at site “A” but showed no variation at site B. NLGN3 transcripts had 1 short band at ED6 and three bands at ED8. Although they were generally similar, they had slightly different profiles than NLGN1. The high and low molecular weight NLGN3 bands were several-fold more abundant than the less intensely stained intermediate band, while with NLGN1 the intermediate band was more prevalent. Analysis of NLGN4 transcripts revealed two prominent bands throughout development, indicating the presence of splice site A2 for the larger molecular weight band. Interestingly, beginning at ED18 we saw an additional larger molecular weight DNA band that, although not verified by sequencing, suggests that additional splice variants may exist.
Laminar position of alternatively spliced isoforms
In order to analyze the laminar pattern of expression of the various NLGN genes and their spliced isoforms within the retina, we performed RT-PCR on pooled laser-capture microdissected ONL, INL, and GCL samples (Fig. 4). Demonstrating specificity of the LCM samples, green opsin was detected only in the photoreceptor layer (ONL); Brn3 was only detected in the GCL samples; Lim1, a marker specific for a subset of horizontal cells, was detected only in the INL samples; and Pax6 was detected in the INL and GCL but not the ONL (Fig. 4B).
Although NLGN1, -3, and -4 were each detected in all cell layers, the signal for NLGN3 in the ONL was so low that it was barely above background (Fig. 4C,D). To determine the transcript abundance in retinal cell layers, we used quantitative real-time PCR (Fig. 4E–G). Based on normalization to GAPDH expression, NLGN1 was most abundant in the INL and least abundant in the ONL, with intermediate levels of expression in the GCL. Similar levels of NLGN3 were seen among the INL and GCL samples, while NLGN4 had ≈3-fold higher levels in the INL than in the GCL. Both NLGN3 and -4 showed low levels of expression in photoreceptors that were barely detectable. Analysis of NLGN1 transcripts in the ONL by standard PCR and gel electrophoresis revealed two DNA bands of different length; one small molecular weight signal presumably belonging to transcripts lacking the A site and another barely detectable band with a higher molecular weight that was suggestive of having both A1 and A2 splice inserts. Both the INL and GCL samples had three bands representing all combinations of alternative splicing at splice site A. Qualitatively, we observed that the smallest molecular weight band was most intense in the INL, while the intermediate band was more prominent in the GCL. NLGN3 transcripts were also more abundant in the inner retina, with the INL expressing both small and a large molecular weight transcripts and the GCL exhibiting all three bands, the largest molecular sizes band being the most prominent. For NLGN4 we only observed two bands, suggesting that only one splice site is present, which is consistent with both experimental cloning and genomic analysis and is similar to that observed in mammals that have no splice insert at the A1 splice site. It should be noted that we did not see the third upper band that was occasionally noted in cDNA preparations from late-stage retinas. At the alternatively spliced B site we only observed variation for NLGN1, with NLGN3 and -4 being consistently negative.
Alternative splicing of NRXN1 and -3 were studied in a similar fashion. While NRXN1 (Fig. 4H) was restricted primarily to the INL and GCL layers, NRXN3 (Fig. 4I) was more variable, expressing both large molecular weight transcripts in the photoreceptor samples and lower molecular weight transcripts in the INL and GCL. In order to detect a broader range of NRXN1 and -3 transcripts the oligonucleotides used in this study do not discriminate between α- and β-neurexin isoforms. Many more neurexin isoforms do exist and are currently being investigated separately. To date, we have uncovered the open reading frames from the retina of at least seven β-NRXN3 transcripts (unpubl. data), and this preliminary analysis did not include the α-NRXNs for which many more are likely to exist.
Localization of neuroligin 1 to cells in the OPL and IPL
The specificity of the antibody against NLGN1 was confirmed by Western blot and immunohistochemistry (Fig. 5A,B). Expression patterns in vivo showed that many cells of the inner retina, including those in the INL and GCL, showed moderate to strong NLGN1 signals (Fig. 5C,D). The strongest NLGN1 signal was evident in the two plexiform layers, particularly the OPL, where rod and cone synapses are known to exist. An especially strong signal was apparent in the uppermost S1 sublayer of the OPL. Other regions where NLGN1 was abundantly expressed were those occupied by amacrine and ganglion cell synapses.
Developmental patterns of expression of neuroligin 1 in the chick retina
As noted above, NLGN mRNA transcripts can be detected at early stages of retinal development, significantly earlier than synapses are thought to exist (Fig. 3). To determine whether protein expression follows suite, we used immunohistochemistry to analyze expression from ED5 to ED20 (Fig. 6). As early as ED5 (Fig. 6A), faint expression was seen throughout the neuroepithelium, with slightly higher deposits along the vitreal border. At ED7, expression increased in the ganglion cell layer and in cell bodies and processes likely to belong to amacrine cells (Fig. 6B). At ED8, staining increased in those cells and the range of cells expressing NLGN1 expanded to include putative horizontal cells (arrow) as determined by their proximity to the nascent OPL (Fig. 6C). One day later, at ED9, after horizontal cells have completed their outward migration to the outer aspect of the inner nuclear layer, expression was readily detected in their cell soma (Fig. 6D). In the IPL, expression was seen as early as ED7 and peaked at ≈ED14 (Fig. 6F). Although its distribution became more diffuse and less intense over time, it still persisted even after the retina was fully mature. In the OPL, expression was observed as early as ED8 and peaked at around ED17 (Fig. 6G). By ED20 (Fig. 6H) it became more discrete in appearance throughout the OPL, decorating what appeared to be the apical tips of horizontal cell dendrites.
Figure 6.
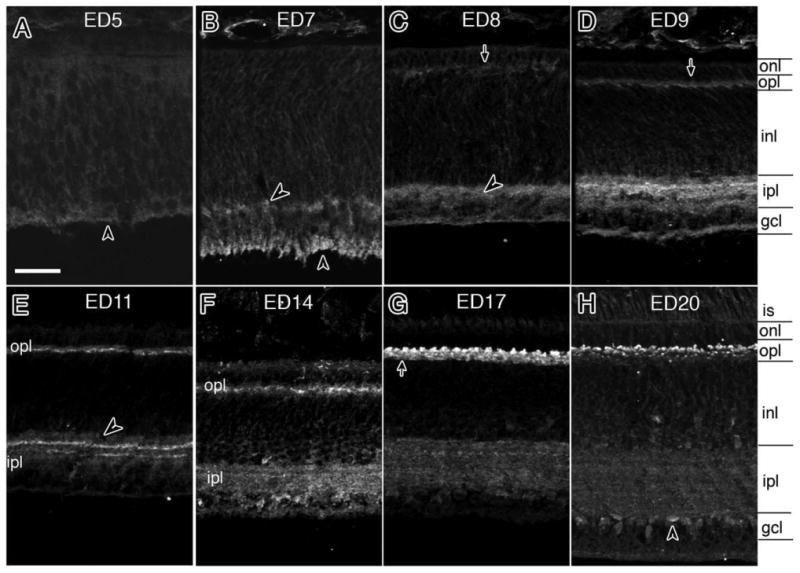
Developmental patterns of NLGN1 expression during development. A–H: A developmental series of NLGN1 immunoreactivity in the synaptic plexiform layers of the chick retina was carried out from between embryonic day 5 (ED5) and ED20. Arrows indicate preferential detection in the OPL while arrowheads are used to indicate inner plexiform layer features. is, inner segments; onl, outer nuclear layer; opl, outer plexiform layer; sublamina of the outer plexiform layer are indicated as sublayers 1–3 (sublayer1 is located proximal to the onl, sublayer 3 is positioned distal and sublayer 2 is located in between); see text for details. Scale bar = 20 μm.
Neuroligin 1 protein is present adjacent to but not within photoreceptor terminals
NLGN1 protein was observed in “mushroom cap”-shaped structures in the OPL in a fashion reminiscent of horizontal cell dendrite labeling. To localize NLGN1 more precisely to individual cell types we used double label immunohistochemistry with a panel of synaptic markers that selectively label photoreceptor terminals (Fig. 7). Labeling of posthatch (P3) chick retinas was analyzed with antibodies against NLGN1 and ribeye (Fig. 7A–D), PSD95 (Fig. 7D–G), and SV2 (Fig. 7H–K). NLGN1 colocalized just beneath ribeye positive terminals in all three sublayers of the OPL, indicating that most, if not all, photoreceptors retain NLGN1 near their nerve terminals (Fig. 7D; arrows). Similar to the situation in the pigeon, the trilaminar structure of the chick OPL is separated into three sublayers with the strata most proximal to photoreceptors composed of rod and double cone terminals, a medial strata composed of red and green cone terminals, and a lower distal strata closest to the INL composed of terminals for blue and violet sensitive photoreceptors. Thus, the staining pattern at all three sublayers of the OPL reflects synaptic terminals of all major classes of photoreceptors. Similar results were observed for PSD95 and SV2 that also labeled all OPL sublayers. Identification of NLGN1 at postsynaptic terminals is most conspicuous near the base of double cone terminals in the uppermost strata (S1), as these are easily identified by their position and their comparatively larger synaptic terminal diameters as compared to terminals in the medial (S2) and distal (S3) aspects of the OPL, which are considerably smaller. Top-down orthogonal slice views of the OPL were constructed from z-stacks in which ribeye and PSD95 (Fig. 7B1,B2,F1,F2) were respectively used to visualize NLGN1-positive structures in the OPL. Positive signals in each sublayer could be distinguished by size since S1 terminals have a significantly larger average diameter (≈5 μm) as compared with the smaller terminals in S2 that are only 2 μm on average. Since rods are the only other cell type to project to S1 we performed double labeling with NLGN1 and rod transducin (Fig. 8). Although the separation between the photoreceptor terminal and horizontal cell dendrites was less obvious with this type of synaptic connection than with cones, there was slightly more NLGN1 signal just beneath the transducin positive rod terminal rather than strictly colocalized with it. Thus, both cones and rods are likely to retain NLGN1 as part of their synaptic complex.
Figure 7.
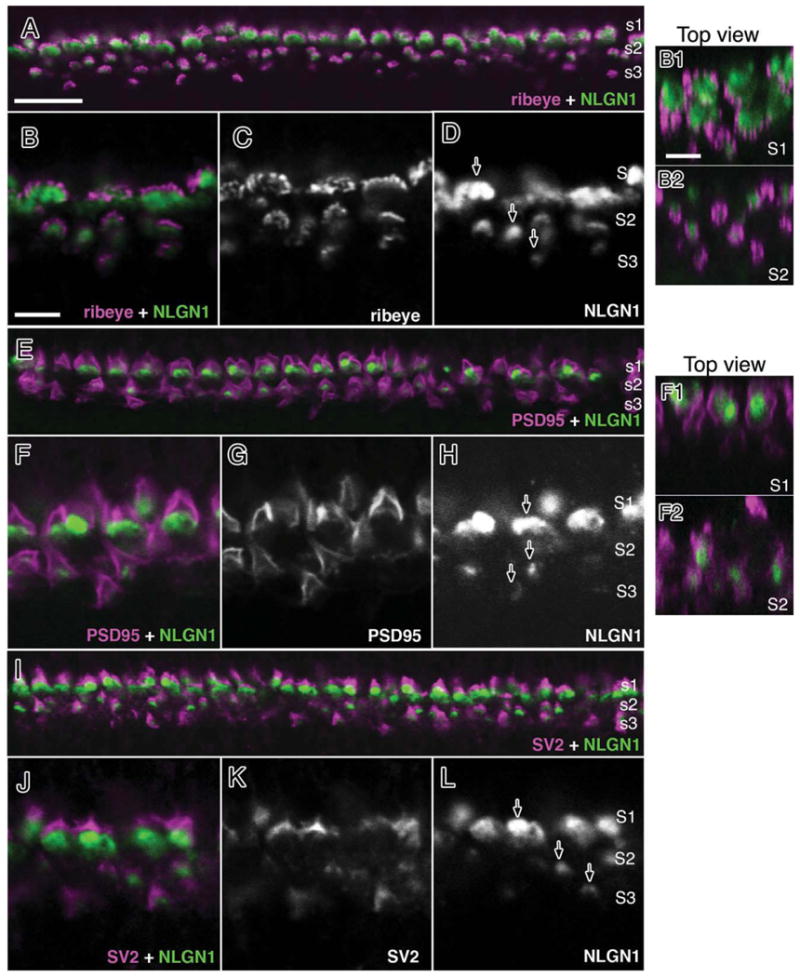
NLGN1 expression adjacent to photoreceptor terminals in the mature chick retina. Double-labeled immunohistochemistry for NLGN1 and ribeye (A–D), PSD95 (E–H) or SV2 (I–L) is shown in a 21-day-old (ED21) chick retina at the level of the outer plexiform layer. Accumulation of NLGN1 and presynaptic proteins in three distinct strata (S1–3) showed separate labeling patterns between NLGN1 immunoreactive bodies and the boundaries of photoreceptor terminals. Single arrows indicate the ribbed appearance of synaptic ribbon-like structures (D), the peripheral staining of PSD95 (E), or the SV2 positive vesicle pool (I) of double cone terminals. Panels to the right (B1,B2,F1,F2) are top-down orthogonal views showing size differences between synaptic terminals in S1 and S2. Scale bars = 15 μm in A; 5 μm in B,B1.
Figure 8.
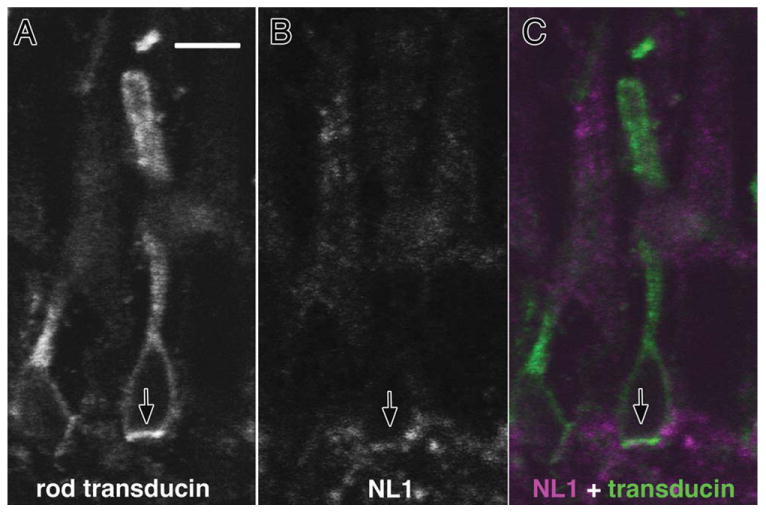
Neuroligin expression in rod photoreceptors. Double labeling with antibodies against NLGN1 and rod transducin were carried on cryosections from 21-day-old chick retinas. Thin optical sections showing (A) transducin-positive rod photoreceptors were superimposed with (B) NLGN1-positive signals to reveal (C) positive signals at the base of rod photoreceptor terminals. Arrows indicate identical regions of the tissue section across different fluorescent filter sets. Scale bar = 10 μm.
Since strong neuroligin signals were present in the outer aspects of the OPL (S1), where double cone terminals reside, we sought to further verify that these signals were not present within double cones but were merely projecting to them. For this purpose we performed double staining with antibodies against calbindin (Fig. 9), an established marker of the principal component of double cones (Fischer et al., 2007; Wahlin et al., 2008). Staining showed intense NLGN1 staining in S1 at the base of calbindin-positive terminals with little actual staining within the photoreceptor itself (not shown). Peanut lectin agglutinin (PNA) has also been extensively used as a cone marker, where it has a high affinity for inner and outer segments of all cones and subset of their synaptic terminals (Adler et al., 1984; Blanks and Johnson, 1984). In addition, it has also been employed as a marker for double cone synaptic terminals (Tanabe et al., 2006). We therefore used PNA to add additional positional information to NLGN1 expression (Fig. 9B). Surprisingly, we observed that while NLGN1-positive terminals were distributed throughout the OPL, PNA-positive terminals were only present in the medial strata (S2) and not in the outer strata where the double cone terminals exist (Fig. 9B–G). Furthermore, when PNA was incubated with calbindin antibodies there was a clear separation between the calbindin (+) principal cones in S1 and PNA in S2 (Fig. 9C). Since ribeye labels all three OPL sublamina (S1–3), its association with PNA in S2 provides the clearest bit of evidence that PNA does not label double cone terminals (Fig. 9E) as previously reported (Tanabe et al., 2006). It is also helpful that TrkA antibodies label only a subset of horizontal cells projecting to double cone terminals (Fischer et al., 2007). When TrkA antibodies and PNA were incubated together, trkA (+) terminals predictably projected to S1, while PNA was restricted to a region distinctly below that (Fig. 9D). In the report that used PNA as a marker for double cones, retinal tissues studied were from ED16 chick retinas. To investigate whether this discrepancy might be related to the age of the retina, we compared PNA staining in retinas harvested from ED16 and ED21 chick eyes. In this fashion, we demonstrated that at ED16 the OPL is only comprised of one tight sublayer as opposed to the three loose sublayers present at maturity (Fig. 9F,F1,G,G1). Since red and green cones reside in the medial S2 sublamina of the OPL, our data suggest that, rather than being a marker for double cones, PNA actually labels red and/or green principal cone synapses. Although we currently do know whether green or red cones are being labeled, this observation should help us study this synaptic sublayer in greater detail.
Figure 9.
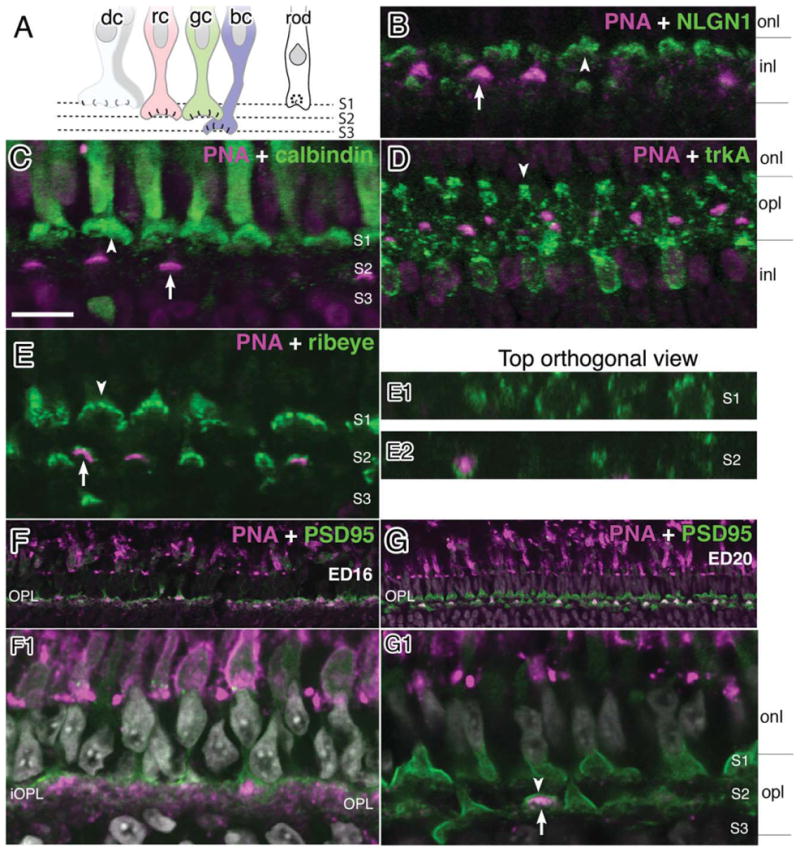
Neuroligin expression in specific sublayers of the OPL. Double labeling immunohistochemistry was performed with antibodies against NLGN1 and markers known to localize to different OPL strata in 21-day-old (ED21) chick retinas. A: A cartoon diagram of photoreceptor projections illustrates the projections of different photoreceptor types that project to different OPL strata. B: AlexaFluor conjugated PNA labeling (magenta) in the medial OPL is indicated with an inverted arrow while NLGN1 signal is in multiple strata (green). C: Calbindin (+) double cone terminals are located in the outer aspect of the OPL (arrowhead) while PNA is located in the medial aspect of the OPL (arrow). D: TrkA positive dendrites extending to the outer OPL terminate near the base of double cones (arrowhead). E: Ribeye signals (arrow) are stratified in all three sublamina with PNA reactivity (arrowhead) restricted to the medial position. Panels to the right (E1,E2) are top-down orthogonal views showing size differences between synaptic terminals in S1 and S2, respectively, and show PNA-negative terminals in S1 and positive PNA terminals in S2 in a number of terminals. A single row of synaptic terminals (arrowhead) is present at ED16 (F,F1). At ED21, three sublaminae (arrowheads) are present (G,G1). dc, double cones; rc, red cones; gc, green cones; bc, blue or violet cones. Scale bar = 10 μm.
GluR4 and neuroligin 1 are present in the majority of horizontal cells
NLGN1 signal at the nascent OPL and adjacent cell soma at ED9 (Fig. 6D) pointed to a horizontal cell identity, so we performed additional IHC analysis with markers previously described to label horizontal cells at maturity. AMPA-type GluR4 receptors in the OPL appear to be present in many (if not all) horizontal cells (Wahlin et al., 2008). While NLGN1 (+) signals were most readily observed in discrete “mushroom cap-like” structures located at the distal tips of OPL dendrites, GluR4 was additionally present along the entire cell surface, thus providing a useful silhouette of each horizontal cell with which to correlate NLGN1 expression. GluR4 provided a better silhouette at ED21 than in the posthatch setting, so this period was used for further analysis (not shown). Thus, by using double-labeling IHC we observed that virtually all GluR4 (+) cells in the OPL were also positive for NLGN1 (Fig. 10A–O). The synaptic terminals of photoreceptors were essentially negative for both NLGN1 and GluR4 (Fig. 10A–C); however, the presence of a very faint signal in the inner segment region prevents ruling out the possibility of low expression in these cells (Fig. 5D).
Figure 10.
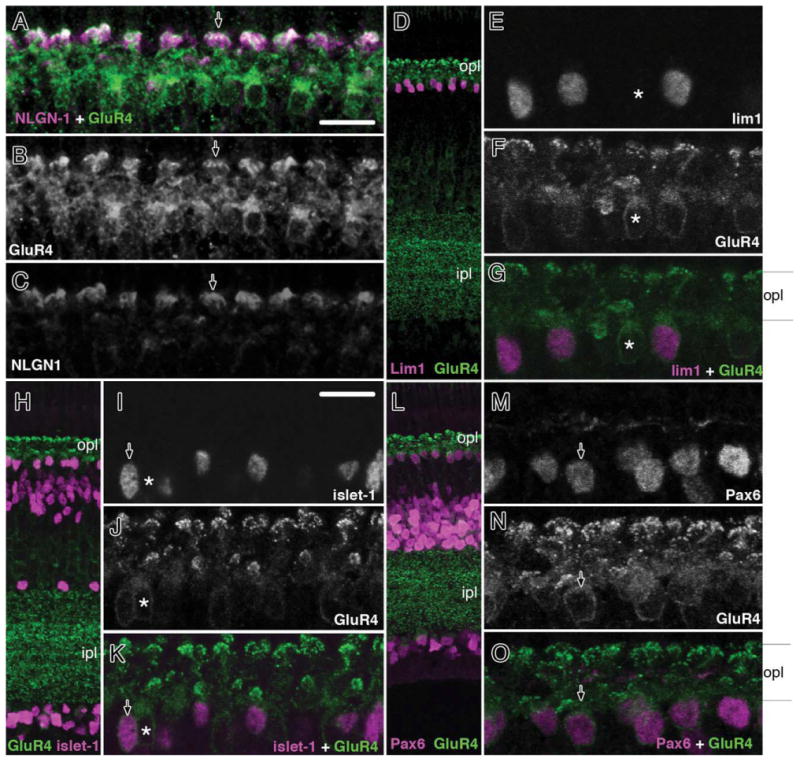
NLGN1 and GluR4 expression in dendrites of retinal horizontal cell subpopulations. Double labeling of embryonic day 21 chick retinas (ED21) with NLGN and GluR4 (A–C) or GluR4 and lim1 (D–G), islet1 (H–K), and Pax6 (L–O) showed strong NLGN labeling in different retinal horizontal cell subgroups. Arrowhead (A–C) indicates dendritic tips of double-labeled NLGN1 and GluR4-positive cells. Arrows (G,K,O) indicate GluR4/lim1, GluR4/islet1, and GluR4/Pax6-positive horizontal cells. A single asterisk indicates a horizontal cell that is positive for GluR4 but negative for the above transcription factors. onl, outer nuclear layer; opl, outer plexiform; inl, inner nuclear layer; ipl, inner plexiform layer; gcl, ganglion cell layer. Scale bars = 10 μm.
Since there are at least three horizontal cell subgroups based on morphology, and possibly four or more if one accounts for differences in dendritic field arborization and protein expression, one cannot be certain that all horizontal cells express NLGN1 simply by looking at NLGN1 and GluR4 colocalization. We therefore used an indirect approach to determine that NLGN1 is present in all horizontal cell types (Mariani, 1987; Fischer et al., 2007). Several homeodomain transcription factors have been previously established as markers for subpopulations of HCs: lim1 labels type I axon-bearing horizontal cells with “brush-shaped” dendrites, islet1 labels nuclei of axon-less types II and III HCs, and Prox1 labels all horizontal cells (Fischer et al., 2007; Edqvist et al., 2008). In addition, at the age studied (P0) Pax6 also labels all horizontal cells, as reported elsewhere (Fig. 10L–O) (Edqvist et al., 2008). While these markers are all useful for identifying nuclear bodies of these cell types, the separation between NLGN1 signals and the cell nuclei did not provide enough overlap to ensure that transcription factor (+) signals and NLGN1 belonged to the same cells. Fortunately, double labeling with GluR4 and transcription factors can be extrapolated with the strong overlap between GluR4 and NLGN1 to indirectly explore whether or not NLGN1 is expressed in all horizontal cells or simply subsets. In this fashion, we observed that GluR4 was present in all cells that expressed Lim1 (Fig. 10D–G), Islet1 (Fig. 10H–K), Pax6 (Fig. 10L–O), and Prox1 (not shown). Since this spectrum of markers covers all horizontal cell types and there was a very strong association between GluR4 and NLGN1, it is likely that at the age tested all horizontal cells express both GluR4 and NLGN1.
Neuroligin 1 localizes to dendrites of all major horizontal cell subgroups
While it is likely that the anti-NLGN1 antibody was labeling dendrites of all horizontal cell types, the data above was only indirect. Direct evidence was accomplished by using horizontal cell marker antibodies that preferentially label specific horizontal cell subgroups (Fig. 11). TrkA (+) cells have a smaller somata and bulbous dendritic endings, which project to the S1 synaptic sublamina. Double labeling with trkA and NLGN1 showed positive signals at the thin distal tips of trkA (+) cells often crossing one another in a chiasm-like formation (Fig. 11B–D; arrowhead). Extensive labeling of NLGN1 in the surrounding areas was also seen in dendrites from trkA (−) cells. The morphology and cell type distribution of GABA and calretinin has been shown to be similar and is primarily in trkA (−) cells (Fischer et al., 2007). While thickly branched GABA (+) dendrites were less refined than those seen with calretinin labeling, this approach allowed us to further visualize GABA (+) shafts which extended right up to NLGN1-positive structures (Fig. 11E–H). Calretinin-positive cells, on the other hand, showed very fine “brushed-like” structures with calretinin-immunoreactive features overlapped by NLGN1 (+) signals (Fig. 11I–L). Together, these findings suggest that NLGN1 is expressed in most, if not all, horizontal cells in a fashion that links them directly to photoreceptor synaptic terminals.
Figure 11.
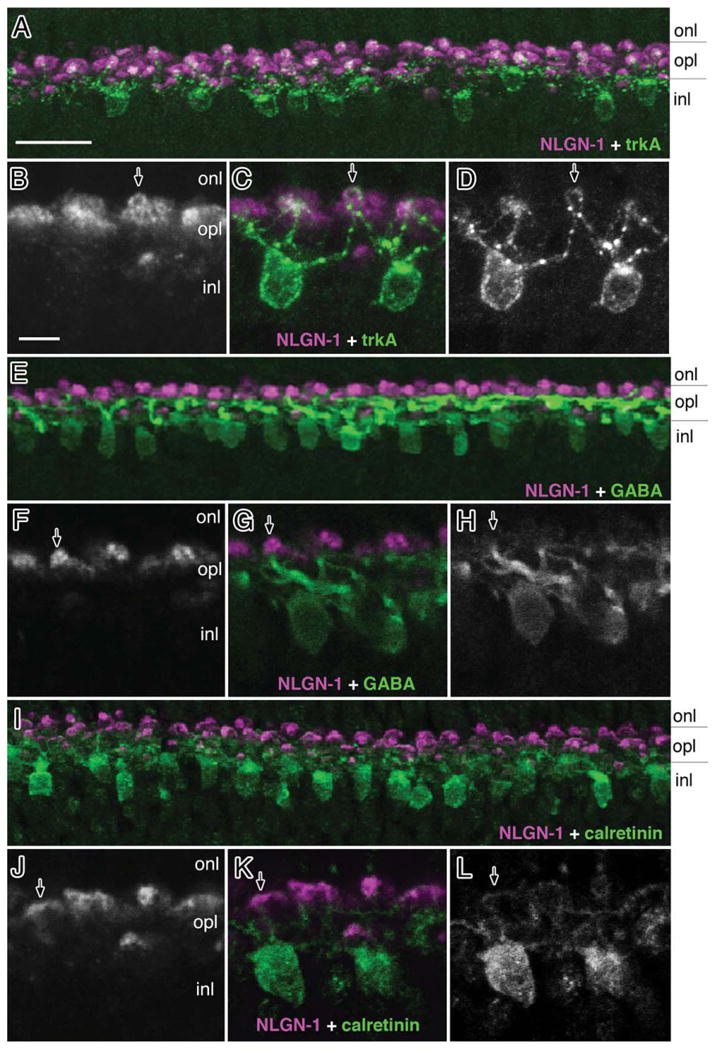
Neuroligin 1 expression in horizontal cell subsets. Double labeling of embryonic day 21 chick retinas (ED21) with NLGN1 and trkA (A–D), GABA (E–H) or calretinin (I–L) showed colocalization with markers for different retinal horizontal cell subgroups. Arrows indicate NLGN1-positive dendritic tips of horizontal cells that are also positive for trkA(C), GABA(G), and calretinin(K). onl, outer nuclear layer; opl, outer plexiform; inl, inner nuclear layer; ipl, inner plexiform layer; gcl, ganglion cell layer. Scale bars = 15 μm in A; 5 μm in B.
While all horizontal cell types were immunoreactive for NLGN1, it was often difficult to determine whether all dendrites, or just a significant portion, of a particular cell type were positive. For example, the broad “brush-shaped” dendritic branches of calretinin (+) HC1 cells were clearly positive for NLGN1, even though we could not trace all dendrites due to their fine interwoven structure. Similarly, GABA-positive HC2 cells (medium-to-wide field or “stellate” cells) were strongly positive for GABA along their thick dendritic trunks, while at their distal tips GABA reactivity was considerably less. In contrast to the difficulty associated with identifying HC1 and HC2 cells, NLGN1-positive HC3 (“candelabrum” shaped) cells were much easier to identify since trkA reactivity was continuous throughout the cell, including the distal dendritic tips where NLGN1 resides. The fine projections of bipolar cells, on the other hand, were much more difficult to study since in addition to there being up to nine morphologically distinct subclasses they have comparatively fewer immunocytochemical markers to identify them. Considering these technical hurdles, NLGN1 was not easily observed in bipolar cell dendrites, so we cannot rule out the possibility that NLGN1 plays a role there too. Future work should address these technical issues.
Discussion
Through a detailed exploration of the NLGNs expressed in the chick retina, we have found that: 1) chicken NLGN1, -3 and -4 are well conserved with their mammalian counterparts, but an avian homolog of NLGN2 appears to be absent. 2) Alternative splicing generates considerable diversity of the NLGN1, -3, and -4 family of proteins, through alternative splicing occurring at conserved “A” and “B” splice sites, similar to that observed in mammals. In total, five unique NLGN1 transcripts, three NLGN3 transcripts, and three NLGN4 transcripts were identified. 3) NLGN expression at the mRNA level begins relatively early in development, by ED5, which represents the earliest age tested. This is potentially interesting since it is considerably earlier than when synaptogenesis is thought to occur in either the inner or outer retina. Although alternative splicing exists at ED6, by ED8 (the period of inner retina synaptogenesis) elevated levels of alternatively spliced variants were evident. In general, levels of neuroligin mRNA increase over time, peaking at ED14 for NLGN1 and NLGN4. NLGN3, on the other hand, continues to rise throughout development. 4) Although RT-PCR of LCM-derived samples allowed detection of NLGN1 and -4 transcripts in all three nuclear layers, more careful quantitative analysis revealed that NLGN1 expression is markedly higher in the inner retina than in the outer retina, and NLGN3 and -4 are only expressed in the inner retina. Alternative splicing was most complex in the ganglion cell layer and less variable in the inner nuclear layer. 5) At the protein level, NLGN1 is present in both synaptic plexiform layers. 6) NLGN1 protein was first detected in the radial processes of the neuroepithelium by ED5, in ganglion cells and presumptive amacrine cells at ED7, and in the soma of horizontal cells by ED8. At ED9 both the outer and inner plexiform layers were clearly positive. 7) Photoreceptor synaptic terminal active zone markers such as ribeye, PSD95, and SV2 failed to colocalize with NLGN1, while horizontal cell markers such as GluR4, trkA, calretinin, and GABA did, indicating a postsynaptic distribution in the dendrites of horizontal cells.
With the exception of the NLGN2 gene, which appears to be absent in the chick, the NLGN gene family is well conserved between chicken and other vertebrate species. Humans have five NLGN genes. Although it was previously thought that rodents only have three, a fourth gene with a dramatically divergent sequence was recently discovered in mouse (Bolliger et al., 2008). The absence of NLGN2 from chicken EST databases and from the available chicken genome does not appear to be due to limitations in the state of the chicken genome project, since in its most current state (June 17, 2006 draft assembly) the chicken genome has a 6.6-fold coverage with ≈95% of sequences anchored to chromosomes and the remainder concatenated into a virtual chromosome (“chrUn_random”); however, its possible presence in a so-called “difficult to clone” region cannot be excluded. It is noteworthy that fish, which are presumed to be more ancient than birds or mammals, have all four NLGN genes, as does the lizard Anolis carolinensis that branched off from the evolutionary tree more recently after birds and snakes diverged. A genomic contraction after this evolutionary split could offer a possible explanation for this difference (Veyrunes et al., 2008). In addition, there are numerous genes flanking the human NLGN2 gene for which there are no regions of shared synteny with the chicken genome, further supporting the notion that portions of the chicken genome may have been lost through evolution.
The discovery that NLGN1 is expressed in developing and mature mouse and chick retinas raises the possibility that neuroligin/neurexin family members may also play a role in retinal synapse formation or function (von Kriegstein and Schmitz, 2003; Paraoanu et al., 2006). In the mouse retina, NLGN1 has been observed at synaptic plexiform layers, but the identity of the cells expressing it was not clear (von Kriegstein and Schmitz, 2003). Until now, the identity of cells expressing NLGN1 in the chick was even less clear since the only published report involved mRNAs extracted from whole retina (Paraoanu et al., 2006). Although we now know that NLGN1 protein is expressed in chick horizontal, amacrine, and ganglion cells, we have been unable to identify NLGN1 at photoreceptor terminals even though we detected low levels of mRNA from LCM-prepared photoreceptor samples. One possibility is that the protein is expressed but simply at a level below our threshold for detection. Alternatively, there are an increasing number of examples where post-transcriptional regulation of protein synthesis limits the translation of mRNA (Decembrini et al., 2006; Filipowicz et al., 2008). As for NLGN2, it is expressed in both the outer and inner nuclear layers of the mouse retina, where, in addition to being present at inhibitory sites alongside components of the dystroglycan complex in the inner plexiform layer, it is found at presynaptic terminals of photoreceptor ribbon synapses (Lui et al., 2010). Since inhibitory GABA receptors are also expressed by photoreceptors, this finding is not altogether surprising except for the fact that thus far we have been unable to identify NLGN2 in the chicken, let alone their photoreceptors. NLGN3 is reportedly expressed in mice by olfactory ensheathing glia and astrocytes of the retina, where it has been labeled as a retinal “gliotactin” (Gilbert et al., 2001). Given the reported observation that NLGN3 is “strikingly absent” from the chick retina, we were surprised to detect abundant mRNA levels throughout retinal development (Paraoanu et al., 2006). Further examination of NLGN3 and -4 protein expression patterns in the chicken, which so far has been limited by the availability of suitable antibodies, would clearly be of great interest.
Although it is not known precisely which neuroligin–protein interactions occur within the retina, a fair amount is already known about neuroligin-interacting molecules in other central nervous system (CNS) regions that could be applicable here. First, there are many proteins recruited as part of PDZ containing scaffolds at pre- and postsynaptic sites; one interaction involves NRXN and CASK proteins associated together at presynaptic sites, while another involves NLGN and PSD95 coupled NMDA receptors at postsynaptic sites (Hata et al., 1996; Missler et al., 1998; Bolliger et al., 2001). The neurexin1 and -3 transcripts we identified in the outer retina gives some degree of evidence that neurexins could interact with neuroligins at photoreceptor synapses. In addition, we previously reported that its binding partner CASK is present on all presynaptic photoreceptor terminals in the chick retina (Koulen et al., 1998; Wahlin and Adler, 2007b; Wahlin et al., 2008; Fischer et al., 2008). Similar to other vertebrates, PSD95 is expressed on chick photo-receptor terminals rather than at more typical postsynaptic sites, such as the horizontal or bipolar cell dendrites, where abundant levels of glutamate receptors exist (Koulen et al., 1998; Wahlin and Adler, 2007b; Wahlin et al., 2008). The association of PSD95 with plasma membrane Ca2+ pumps at photoreceptor synapses raises the possibility that it could have an altogether different role at photoreceptor synapses (Aartsen et al., 2009). In hippocampal and cortical cultures, NLGN1 and the AMPA-type glutamate receptor GluR4 can cluster together at sites of contact in artificial and neuronal synapses (Sia et al., 2007). It is conceivable that this specific interaction might also occur in the eye, since a variety of glutamate receptor subunits are known to be present in the outer plexiform layer. For example, GluR4 is robustly expressed in the outer plexiform layer where it decorates horizontal cell postsynaptic elements. In these cells the extensive overlap between NLGN1 and GluR4 (+) dendrites suggests that NLGN1 could participate in the function, assembly, or maintenance of photoreceptor-to-horizontal cell synapses; however, functional studies are needed to draw any further conclusions.
All vertebrate species studied thus far have distinct horizontal cell subtypes. In the chicken, there are three or more types of horizontal cells that are classified by morphology (dendritic field size and presence or absence of an axon) and/or expression of selective cell markers (Mariani, 1987; Karlsson et al., 1998; Tanabe et al., 2006; Fischer et al., 2007; Rompani and Cepko, 2008; Edqvist et al., 2008). Although it is currently unknown how such connections are made, each horizontal cell type has a unique series of projections from specific cone and rod subpopulations in the OPL; double cones and rod terminals project to the proximal or “upper” strata (S1), red and green cones to the medial strata (S2), and blue and violet sensitive cones to the distal or “lower” strata (S3). By colabeling NLGN with horizontal cell markers we demonstrated that essentially all horizontal cell types in the chick retina express NLGN1 at their dendritic tips, making it unlikely that NLGN is responsible for the unique targets that form between photoreceptors and horizontal cells, although it is possible that alternatively spliced variants could account for some degree of specificity (Mariani, 1987; Tanabe et al., 2006; Fischer et al., 2007; Rompani and Cepko, 2008; Edqvist et al., 2008).
It is worth mentioning that dynamic changes in synaptic protein expression, cell markers, or morphology can occur even over short periods during development and this could influence how cell connections are described. For example, peanut lectin agglutinin was reported in ED16 chick retinas as a synaptic marker for double cones, where it was used to monitor dendritic targeting of horizontal cells by cadherin (Tanabe et al., 2006). Contrary to this, we showed that double cone terminals are, in fact, negative for PNA; the actual target lies within the medial stratum, not the proximal stratum, of the OPL where red and green cone terminals reside. While it is possible that dynamic changes in expression of the ligand for PNA account for this difference, a more plausible explanation is that only upon maturity do horizontal cell dendrites stratify into multiple readily discernible sublaminae (Gallego, 1986). Furthermore, the immature OPL at ED16 has only limited amounts of synaptic ribbon-associated protein (e.g., RIBEYE) and lacks the characteristic trilaminar structure of the mature retina evident days later (Wahlin et al., 2008). Such findings underscore the need for a more complete understanding of timing for OPL morphogenesis. Developmental changes in gene expression should also be considered when classifying horizontals cells or the nature of their connections, particularly when this is done on the basis of cytochemical markers. For example, varying levels of GABA are seen in posthatch chicks from between P0 and P7, with high expression levels in all HC subsets at the time of hatching, but a week later these high levels only persist in HC2 cells (Edqvist et al., 2008). Differences in Pax6 levels in horizontal cells of the posthatch chick retina have also been observed (Fischer et al., 2007; Edqvist et al., 2008).
Understanding how the immature retina acquires its final state comprised of a fine network of highly organized connections is critical for understanding the molecular basis of eye development and for improving cell transplantation-based approaches for restoring vision in people with debilitating retinal degenerations. The present study contributes to this effort by offering a comprehensive analysis in the eye of NLGN gene family products that appear to play a role in synapse assembly in other parts of the nervous system. Future attention to the role of alternative splicing in the diversity of cellular connections during normal development and disease will be important areas of research to address. Functional studies are currently under way to dissect the role of NLGN1 alternative splicing in vivo and in vitro.
Supplementary Material
Acknowledgments
The authors thank Drs. Alex Kolodkin, Nick Marsh-Armstrong, and Richard Huganir (Johns Hopkins Department of Neuroscience) for guidance and advice. We also thank Dr. Valeria Canto-Soler, Minda McNally, David O'Brien, and David Kays for advice and technical assistance; Dr. F. Lefcort of Montana State University for generously providing the trkA antibody; and Peter Schieffele at Columbia University for providing neuroligin expressing plasmids and antibodies.
Grant sponsor: National Institutes of Health (NIH); Grant numbers: EY04859, T32-07143, EY1765, EY009769; Grant sponsor: Foundation Fighting Blindness, an unrestricted departmental grant from Research to Prevent Blindness (New York, NY); Grant sponsor: Michael B. Panitch Macular Degeneration Laboratory at the Wilmer Eye Institute; Grant sponsor: generous gift from Mr. Matthew Kovner.
Footnotes
Additional supporting information may be found in the online version of this article.
Literature Cited
- Aartsen WM, Arsanto JP, Chauvin JP, Vos RM, Versteeg I, Cardozo BN, Bivic AL, Wijnholds J. PSD95beta regulates plasma membrane Ca2+ pump localization at the photoreceptor synapse. Mol Cell Neurosci. 2009;41:156–165. doi: 10.1016/j.mcn.2009.02.003. [DOI] [PubMed] [Google Scholar]
- Adler R. Curing blindness with stem cells: hope, reality, and challenges. Adv Exp Med Biol. 2008;613:3–20. doi: 10.1007/978-0-387-74904-4_1. [DOI] [PubMed] [Google Scholar]
- Adler R, Lindsey JD, Elsner CL. Expression of cone-like properties by chick embryo neural retina cells in glial-free monolayer cultures. J Cell Biol. 1984;99:1173–1178. doi: 10.1083/jcb.99.3.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashery-Padan R, Marquardt T, Zhou X, Gruss P. Pax6 activity in the lens primordium is required for lens formation and for correct placement of a single retina in the eye. Genes Dev. 2000;14:2701–2711. doi: 10.1101/gad.184000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Béïque J, Lin D, Kang M, Aizawa H, Takamiya K, Huganir R. Synapse-specific regulation of AMPA receptor function by PSD-95. Proc Natl Acad Sci U S A. 2006;103:19535–19540. doi: 10.1073/pnas.0608492103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belecky-Adams T, Tomarev S, Li HS, Ploder L, McInnes RR, Sundin O, Adler R. Pax-6, Prox 1, and Chx10 homeobox gene expression correlates with phenotypic fate of retinal precursor cells. Invest Ophthalmol Vis Sci. 1997;38:1293–1303. [PubMed] [Google Scholar]
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- Blanks JC, Johnson LV. Specific binding of peanut lectin to a class of retinal photoreceptor cells. A species comparison. Invest Ophthalmol Vis Sci. 1984;25:546–557. [PubMed] [Google Scholar]
- Bolliger MF, Frei K, Winterhalter KH, Gloor SM. Identification of a novel neuroligin in humans which binds to PSD-95 and has a widespread expression. Biochem J. 2001;356(Pt 2):581–588. doi: 10.1042/0264-6021:3560581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolliger MF, Pei J, Maxeiner S, Boucard AA, Grishin NV, Sudhof TC. Unusually rapid evolution of Neuroligin-4 in mice. Proc Natl Acad Sci U S A. 2008;105:6421–6426. doi: 10.1073/pnas.0801383105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucard AA, Chubykin AA, Comoletti D, Taylor P, Sudhof TC. A splice code for trans-synaptic cell adhesion mediated by binding of neuroligin 1 to alpha- and beta-neurexins. Neuron. 2005;48:229–236. doi: 10.1016/j.neuron.2005.08.026. [DOI] [PubMed] [Google Scholar]
- Boughter JD, Jr, Pumplin DW, Yu C, Christy RC, Smith DV. Differential expression of alpha-gustducin in taste bud populations of the rat and hamster. J Neurosci. 1997;17:2852–2858. doi: 10.1523/JNEUROSCI.17-08-02852.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley K, Kelly RB. Identification of a transmembrane glycoprotein specific for secretory vesicles of neural and endocrine cells. J Cell Biol. 1985;100:1284–1294. doi: 10.1083/jcb.100.4.1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budreck EC, Scheiffele P. Neuroligin-3 is a neuronal adhesion protein at GABAergic and glutamatergic synapses. Eur J Neurosci. 2007;26:1738–1748. doi: 10.1111/j.1460-9568.2007.05842.x. [DOI] [PubMed] [Google Scholar]
- Chih B, Gollan L, Scheiffele P. Alternative splicing controls selective trans-synaptic interactions of the neuroligin-neurexin complex. Neuron. 2006;51:171–178. doi: 10.1016/j.neuron.2006.06.005. [DOI] [PubMed] [Google Scholar]
- Comoletti D, Flynn R, Jennings LL, Chubykin A, Matsumura T, Hasegawa H, Sudhof TC, Taylor P. Characterization of the interaction of a recombinant soluble neuroligin-1 with neurexin-1beta. J Biol Chem. 2003;278:50497–50505. doi: 10.1074/jbc.M306803200. [DOI] [PubMed] [Google Scholar]
- Comoletti D, Flynn RE, Boucard AA, Demeler B, Schirf V, Shi J, Jennings LL, Newlin HR, Südhof TC, Taylor P. Gene selection, alternative splicing, and post-translational processing regulate neuroligin selectivity for beta-neurexins. Biochemistry. 2006;45:12816–12827. doi: 10.1021/bi0614131. [DOI] [PubMed] [Google Scholar]
- Conroy WG, Nai Q, Ross B, Naughton G, Berg DK. Post-synaptic neuroligin enhances presynaptic inputs at neuronal nicotinic synapses. Dev Biol. 2007;307:79–91. doi: 10.1016/j.ydbio.2007.04.017. [DOI] [PubMed] [Google Scholar]
- Craig AM, Kang Y. Neurexin-neuroligin signaling in synapse development. Curr Opin Neurobiol. 2007;17:43–52. doi: 10.1016/j.conb.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean C, Scholl FG, Choih J, DeMaria S, Berger J, Isacoff E, Scheiffele P. Neurexin mediates the assembly of presynaptic terminals. Nat Neurosci. 2003;6:708–716. doi: 10.1038/nn1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decembrini S, Andreazzoli M, Vignali R, Barsacchi G, Cremisi F. Timing the generation of distinct retinal cells by homeobox proteins. PLoS Biol. 2006;4:e272. doi: 10.1371/journal.pbio.0040272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edqvist PH, Hallbook F. Newborn horizontal cells migrate bi-directionally across the neuroepithelium during retinal development. Development. 2004;131:1343–1351. doi: 10.1242/dev.01018. [DOI] [PubMed] [Google Scholar]
- Edqvist PH, Myers SM, Hallbook F. Early identification of retinal subtypes in the developing, pre-laminated chick retina using the transcription factors Prox1, Lim1, Ap2alpha, Pax6, Isl1, Isl2, Lim3 and Chx10. Eur J Histochem. 2006;50:147–154. [PubMed] [Google Scholar]
- Edqvist PH, Lek M, Boije H, Lindback SM, Hallbook F. Axon-bearing and axon-less horizontal cell subtypes are generated consecutively during chick retinal development from progenitors that are sensitive to follistatin. BMC Dev Biol. 2008;8:46. doi: 10.1186/1471-213X-8-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis JH, Richards DE, Rogers JH. Calretinin and calbindin in the retina of the developing chick. Cell Tissue Res. 1991;264:197–208. doi: 10.1007/BF00313956. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc. 2007;2:953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- Ericson J, Rashbass P, Schedl A, Brenner-Morton S, Kawakami A, van Heyningen V, Jessell TM, Briscoe J. Pax6 controls progenitor cell identity and neuronal fate in response to graded Shh signaling. Cell. 1997;90:169–180. doi: 10.1016/s0092-8674(00)80323-2. [DOI] [PubMed] [Google Scholar]
- Feany MB, Yee AG, Delvy ML, Buckley KM. The synaptic vesicle proteins SV2, synaptotagmin and synaptophysin are sorted to separate cellular compartments in CHO fibroblasts. J Cell Biol. 1993;123:575–584. doi: 10.1083/jcb.123.3.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Fischer AJ, Reh TA. Identification of a proliferating marginal zone of retinal progenitors in postnatal chickens. Dev Biol. 2000;220:197–210. doi: 10.1006/dbio.2000.9640. [DOI] [PubMed] [Google Scholar]
- Fischer AJ, Reh TA. Exogenous growth factors stimulate the regeneration of ganglion cells in the chicken retina. Dev Biol. 2002;251:367–379. doi: 10.1006/dbio.2002.0813. [DOI] [PubMed] [Google Scholar]
- Fischer AJ, Seltner RL, Poon J, Stell WK. Immunocytochemical characterization of quisqualic acid- and N-methyl-D-aspartate-induced excitotoxicity in the retina of chicks. J Comp Neurol. 1998;393:1–15. [PubMed] [Google Scholar]
- Fischer AJ, Stanke JJ, Aloisio G, Hoy H, Stell WK. Heterogeneity of horizontal cells in the chicken retina. J Comp Neurol. 2007;500:1154–1171. doi: 10.1002/cne.21236. [DOI] [PubMed] [Google Scholar]
- Fischer AJ, Foster S, Scott MA, Sherwood P. Transient expression of LIM-domain transcription factors is coincident with delayed maturation of photoreceptors in the chicken retina. J Comp Neurol. 2008;506:584–603. doi: 10.1002/cne.21578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego A. Chapter 7 Comparative studies on horizontal cells and a note on microglial cells. Prog Retin Res. 1986;5:165–206. [Google Scholar]
- Gallo G, Lefcort FB, Letourneau PC. The trkA receptor mediates growth cone turning toward a localized source of nerve growth factor. J Neurosci. 1997;17:5445–5454. doi: 10.1523/JNEUROSCI.17-14-05445.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert M, Smith J, Roskams A, Auld V. Neuroligin 3 is a vertebrate gliotactin expressed in the olfactory ensheathing glia, a growth-promoting class of macroglia. Glia. 2001;34:151–164. doi: 10.1002/glia.1050. [DOI] [PubMed] [Google Scholar]
- Glasgow E, Karavanov A, Dawid I. Neuronal and neuroendocrine expression of lim3, a LIM class homeobox gene, is altered in mutant zebrafish with axial signaling defects. Dev Biol. 1997;192:405–419. doi: 10.1006/dbio.1997.8761. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Hata Y, Butz S, Sudhof TC. CASK: a novel dlg/PSD95 homolog with an N-terminal calmodulin-dependent protein kinase domain identified by interaction with neurexins. J Neurosci. 1996;16:2488–2494. doi: 10.1523/JNEUROSCI.16-08-02488.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang ZJ, Scheiffele P. GABA and neuroligin signaling: linking synaptic activity and adhesion in inhibitory synapse development. Curr Opin Neurobiol. 2008;18:77–83. doi: 10.1016/j.conb.2008.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichtchenko K, Nguyen T, Sudhof TC. Structures, alternative splicing, and neurexin binding of multiple neuroligins. J Biol Chem. 1996;271:2676–2682. doi: 10.1074/jbc.271.5.2676. [DOI] [PubMed] [Google Scholar]
- Ikenaga T, Huesa G, Finger TE. Co-occurrence of calcium-binding proteins and calcium-permeable glutamate receptors in the primary gustatory nucleus of goldfish. J Comp Neurol. 2006;499:90–105. doi: 10.1002/cne.21079. [DOI] [PubMed] [Google Scholar]
- Innis MA. PCR protocols: a guide to methods and applications. San Diego: Academic Press; 1990. [Google Scholar]
- Kang Y, Zhang X, Dobie F, Wu H, Craig AM. Induction of GABAergic postsynaptic differentiation by alpha-neurexins. J Biol Chem. 2008;283:2323–2334. doi: 10.1074/jbc.M703957200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson M, Clary DO, Lefcort FB, Reichardt LF, Karten HJ, Hallbook F. Nerve growth factor receptor TrkA is expressed by horizontal and amacrine cells during chicken retinal development. J Comp Neurol. 1998;400:408–416. [PMC free article] [PubMed] [Google Scholar]
- Karlsson M, Mayordomo R, Reichardt LF, Catsicas S, Karten H, Hallbook F. Nerve growth factor is expressed by postmitotic avian retinal horizontal cells and supports their survival during development in an autocrine mode of action. Development. 2001;128:471–479. doi: 10.1242/dev.128.4.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koulen P, Fletcher EL, Craven SE, Bredt DS, Wassle H. Immunocytochemical localization of the postsynaptic density protein PSD-95 in the mammalian retina. J Neurosci. 1998;18:10136–10149. doi: 10.1523/JNEUROSCI.18-23-10136.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefcort F, Clary DO, Rusoff AC, Reichardt LF. Inhibition of the NT-3 receptor TrkC, early in chick embryogenesis, results in severe reductions in multiple neuronal subpopulations in the dorsal root ganglia. J Neurosci. 1996;16:3704–3713. doi: 10.1523/JNEUROSCI.16-11-03704.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lise MF, El-Husseini A. The neuroligin and neurexin families: from structure to function at the synapse. Cell Mol Life Sci. 2006;63:1833–1849. doi: 10.1007/s00018-006-6061-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui L, Levinson JN, Noel G, Handrigan GR, Richman JM, El-Husseini A, Moukhles H. Synaptic localization of neuroligin 2 in the rodent retina: comparative study with the dystroglycan-containing complex. J Neurosci Res. 2010;88:837–849. doi: 10.1002/jnr.22258. [DOI] [PubMed] [Google Scholar]
- Mariani AP. Neuronal and synaptic organization of the outer plexiform layer of the pigeon retina. Am J Anat. 1987;179:25–39. doi: 10.1002/aja.1001790105. [DOI] [PubMed] [Google Scholar]
- Missler M, Fernandez-Chacon R, Sudhof TC. The making of neurexins. J Neurochem. 1998;71:1339–1347. doi: 10.1046/j.1471-4159.1998.71041339.x. [DOI] [PubMed] [Google Scholar]
- Oakley RA, Lefcort FB, Clary DO, Reichardt LF, Prevette D, Oppenheim RW, Frank E. Neurotrophin-3 promotes the differentiation of muscle spindle afferents in the absence of peripheral targets. J Neurosci. 1997;17:4262–4274. doi: 10.1523/JNEUROSCI.17-11-04262.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa T, Kondo M, Isobe M. 3′ rapid amplification of cDNA ends (RACE) walking for rapid structural analysis of large transcripts. J Hum Genet. 2004;49:102–105. doi: 10.1007/s10038-003-0109-0. [DOI] [PubMed] [Google Scholar]
- Paraoanu LE, Becker-Roeck M, Christ E, Layer PG. Expression patterns of neurexin-1 and neuroligins in brain and retina of the chick embryo: Neuroligin-3 is absent in retina. Neurosci Lett. 2006;395:114–117. doi: 10.1016/j.neulet.2005.10.076. [DOI] [PubMed] [Google Scholar]
- Pasteels B, Rogers J, Blachier F, Pochet R. Calbindin and calretinin localization in retina from different species. Vis Neurosci. 1990;5:1–16. doi: 10.1017/s0952523800000031. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philibert RA, Winfield SL, Sandhu HK, Martin BM, Ginns EI. The structure and expression of the human neuroligin-3 gene. Gene. 2000;246:303–310. doi: 10.1016/s0378-1119(00)00049-4. [DOI] [PubMed] [Google Scholar]
- Rompani SB, Cepko CL. Retinal progenitor cells can produce restricted subsets of horizontal cells. Proc Natl Acad Sci U S A. 2008;105:192–197. doi: 10.1073/pnas.0709979104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheiffele P, Fan J, Choih J, Fetter R, Serafini T. Neuroligin expressed in nonneuronal cells triggers presynaptic development in contacting axons. Cell. 2000;101:657–669. doi: 10.1016/s0092-8674(00)80877-6. [DOI] [PubMed] [Google Scholar]
- Schmitz F, Konigstorfer A, Sudhof TC. RIBEYE, a component of synaptic ribbons: a protein's journey through evolution provides insight into synaptic ribbon function. Neuron. 2000;28:857–872. doi: 10.1016/s0896-6273(00)00159-8. [DOI] [PubMed] [Google Scholar]
- Semple-Rowland SL, Eccles KS, Humberstone EJ. Targeted expression of two proteins in neural retina using self-inactivating, insulated lentiviral vectors carrying two internal independent promoters. Mol Vis. 2007;13:2001–2011. [PubMed] [Google Scholar]
- Sia GM, Beique JC, Rumbaugh G, Cho R, Worley PF, Huganir RL. Interaction of the N-terminal domain of the AMPA receptor GluR4 subunit with the neuronal pentraxin NP1 mediates GluR4 synaptic recruitment. Neuron. 2007;55:87–102. doi: 10.1016/j.neuron.2007.06.020. [DOI] [PubMed] [Google Scholar]
- Silveira dos Santos Bredariol A, Hamassaki-Britto DE. Ionotropic glutamate receptors during the development of the chick retina. J Comp Neurol. 2001;441:58–70. doi: 10.1002/cne.1397. [DOI] [PubMed] [Google Scholar]
- Tanabe K, Takahashi Y, Sato Y, Kawakami K, Takeichi M, Nakagawa S. Cadherin is required for dendritic morphogenesis and synaptic terminal organization of retinal horizontal cells. Development. 2006;133:4085–4096. doi: 10.1242/dev.02566. [DOI] [PubMed] [Google Scholar]
- Taniguchi H, Gollan L, Scholl FG, Mahadomrongkul V, Dobler E, Limthong N, Peck M, Aoki C, Scheiffele P. Silencing of neuroligin function by postsynaptic neurexins. J Neurosci. 2007;27:2815–2824. doi: 10.1523/JNEUROSCI.0032-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchida T, Ensini M, Morton SB, Baldassare M, Edlund T, Jessell TM, Pfaff SL. Topographic organization of embryonic motor neurons defined by expression of LIM homeobox genes. Cell. 1994;79:957–970. doi: 10.1016/0092-8674(94)90027-2. [DOI] [PubMed] [Google Scholar]
- Ushkaryov YA, Südhof TC. Neurexin III alpha: extensive alternative splicing generates membrane-bound and soluble forms. Proc Natl Acad Sci U S A. 1993;90:6410–6414. doi: 10.1073/pnas.90.14.6410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushkaryov YA, Petrenko AG, Geppert M, Südhof TC. Neurexins: synaptic cell surface proteins related to the alpha-latrotoxin receptor and laminin. Science. 1992;257:50–56. doi: 10.1126/science.1621094. [DOI] [PubMed] [Google Scholar]
- Veyrunes F, Waters PD, Miethke P, Rens W, McMillan D, Alsop AE, Grutzner F, Deakin JE, Whittington CM, Schatzkamer K, Kremitzki CL, Graves T, Ferguson-Smith MA, Warren W, Marshall Graves JA. Bird-like sex chromosomes of platypus imply recent origin of mammal sex chromosomes. Genome Res. 2008;18:965–973. doi: 10.1101/gr.7101908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Kriegstein K, Schmitz F. The expression pattern and assembly profile of synaptic membrane proteins in ribbon synapses of the developing mouse retina. Cell Tissue Res. 2003;311:159–173. doi: 10.1007/s00441-002-0674-0. [DOI] [PubMed] [Google Scholar]
- Wahlin KJ, Adler R. (4467/B54) Experimental analysis of photoreceptor synaptogenesis: cloning of chicken neuroligins, and development of bioassays for their functional investigation. ARVO. 2007a abstract. [Google Scholar]
- Wahlin KJ, Adler R. ARVO. Fort Lauderdale, FL: IOVS; 2007b. Experimental analysis of photoreceptor synaptogenesis: cloning of chicken neuroligins, and development of bioassays for their functional investigation; p. 4467. [Google Scholar]
- Wahlin KJ, Moreira EF, Huang H, Yu N, Adler R. Molecular dynamics of photoreceptor synapse formation in the developing chick retina. J Comp Neurol. 2008;506:822–837. doi: 10.1002/cne.21582. [DOI] [PubMed] [Google Scholar]
- Wässle H, Peichl L, Airaksinen M, Meyer M. Calcium-binding proteins in the retina of a calbindin-null mutant mouse. Cell Tissue Res. 1998;292:211–218. doi: 10.1007/s004410051052. [DOI] [PubMed] [Google Scholar]
- Zhadanov A, Bertuzzi S, Taira M, Dawid I, Westphal H. Expression pattern of the murine LIM class homeobox gene Lhx3 in subsets of neural and neuroendocrine tissues. Dev Dyn. 1995;202:354–364. doi: 10.1002/aja.1002020405. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


