Abstract
The use of Assisted Reproductive Technologies (ARTs) in modern cattle breeding is an important tool for improving the production of dairy and beef cattle. A frequently employed ART in the cattle industry is in vitro production of embryos. However, bovine in vitro produced embryos differ greatly from their in vivo produced counterparts in many facets, including developmental competence. The lower developmental capacity of these embryos could be due to the stress to which the gametes and/or embryos are exposed during in vitro embryo production, specifically ovarian hormonal stimulation, follicular aspiration, oocyte in vitro maturation in hormone supplemented medium, sperm handling, gamete cryopreservation, and culture of embryos. The negative effects of some ARTs on embryo development could, at least partially, be explained by disruption of the physiological epigenetic profile of the gametes and/or embryos. Here, we review the current literature with regard to the putative link between ARTs used in bovine reproduction and epigenetic disorders and changes in the expression profile of embryonic genes. Information on the relationship between reproductive biotechnologies and epigenetic disorders and aberrant gene expression in bovine embryos is limited and novel approaches are needed to explore ways in which ARTs can be improved to avoid epigenetic disorders.
Keywords: superovulation, in vitro maturation, embryo culture, DNA methylation, genome imprinting
Introduction
The term “epigenetics” was introduced in the early 1940s by Conrad H. Waddington and describes “the events which lead to the unfolding of the genetic program.”1 Today epigenetics entails the study of changes in gene function that are mitotically or meiotically inherited, but are not based on a change in DNA sequence.2 Epigenetic changes play a crucial role in defining the temporal and tissue specific gene expression profile. While the genetic code is considered to be rather static, the epigenetic code is highly dynamic and tissue-specific in most cells of an organism during its entire life.3
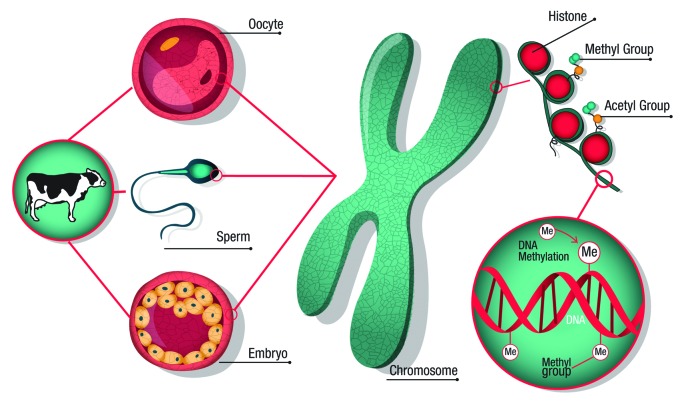
The main epigenetic changes in mammalian cells include four different mechanisms. (1) DNA methylation by addition of a methyl group to the cytosine molecule of the DNA predominantly in DNA regions known as CpG islands. With few exceptions, it is associated with gene silencing, while hypomethylation is mostly associated with gene expression.4 (2) Post-translational histone modifications: the N-termini of histone tails contain amino acid residues that can be methylated, acetylated, phosphorylated, ubiquitynated and/or sumoylated.5 (3) Chromatin remodeling: this process occurs when ATP- dependent protein complexes alter the location and/or the structure of nucleosomes.6 (4) Small noncoding RNAs: Micro RNAs (miRNA) and small interfering RNAs (siRNAs) are short RNA sequences, ~22 nucleotides in size, that are found in plants and mammals. They regulate gene expression at the post-transcriptional level7 and are involved in transcriptional changes and steps that determine cell fate and phenotype.8 A schematic representation of the epigenetic landscape is provided below (Fig. 1).
Figure 1. Epigenetic landscape during embryo development. Several epigenetic changes occurring during gamete formation and early embryo development could alter gene expression which in turn negatively affects embryo production. Histone acetylation in specific lysine residues is mostly associated with transcriptional activity, whereas methylation of other histone amino acids and DNA methylation tends to be linked with transcriptional repression.
Assisted reproductive technologies (ARTs) are well developed in the cattle industry. They include artificial insemination (AI) embryo transfer (ET), in vitro embryo production (IVP), and somatic cell nuclear transfer (SCNT). ARTs have been used to shorten the generational interval, to propagate valuable genetic stock from breeding populations, and in biomedical and reproductive research. The practical application of these technologies had a positive economic impact on beef and milk production.9,10
However, ARTs involve several steps that may exert environmental stress on gametes and early embryos. This is a reason for the growing interest in the putative link between these techniques and epigenetic modifications related to changes in gene expression profiles and imprinting disorders.11-13 Animal studies revealed a link between different ARTs and imprinting disorders, via altered DNA-methylation patterns and histone codes.
The goal of the present review is to discuss the relationship between ARTs, including ovarian stimulation, in vitro maturation, sperm manipulation, embryo culture, and freeze/thawing, and changes in gene expression and epigenetic disorders in bovine embryos. We do not include the epigenetic effects of SCNT on the embryos, because these have been extensively reviewed recently.14-16
Female Gamete Manipulation
Superovulation (SOV)
During growth and development of mammalian ovarian follicles, activation and deactivation of most genes are under control of diverse modifiers via genetic and epigenetic events.17 In the female germ line, methylation patterns are established in a gene-specific manner, predominantly during later stages of oocyte development.18-20 Most maternal imprints appear to be set by completion of meiotic metaphase II (MII). In humans, some maternal imprints may not be completed until fusion of the two pronuclei.21 Mouse studies have demonstrated that superovulation can be associated with reduced oocyte quality, delayed embryonic and fetal development,22,23 disturbances in post-zygotic genome reprogramming,24,25 and altered DNA methylation and expression patterns in oocytes, embryos, fetuses, and placentas.26-28 Similar adverse effects of superovulation may occur in humans.29-31
To increase the number of oocytes for assisted reproduction, protocols incorporate application of gonadotropins in various doses.32,33 Bovine embryos produced by superovulation may have a different gene expression profile compared with those produced by natural ovulation; this difference could be due to changes in epigenetic marks that control gene expression during oocyte maturation and ovulation.34 Recent studies reported an increased risk of imprinting disorders in children conceived via ARTs.35 Ovarian stimulation has been linked to an increased frequency of Beckwith–Wiedemann syndrome (BWS) and Angelman syndrome (AS) in ART-conceived children.29,36-38 An important factor could be ovarian stimulation with high doses of gonadotropins.
A recent study reported divergent transcriptome profiles in oocytes of stimulated vs non-stimulated cows, with over 50% of genes over-expressed in oocytes from hormonally stimulated animals.39 This could represent a response of the oocytes to the perturbation of the follicular hormonal environment. Alterations in the global DNA methylation status, in mitochondrial function and cortical granules were not detected in oocytes produced by treatment with moderate levels of gonadotropins. However, high dosages of gonadotropins induced spindle and chromosomal abnormalities in the oocytes.40 There is not yet enough information about the DNA methylation status at specific differentially methylated regions (DMRs) of imprinted genes after treatment of donor animals with different gonadotropin concentrations and/or combination of gonadotropins.
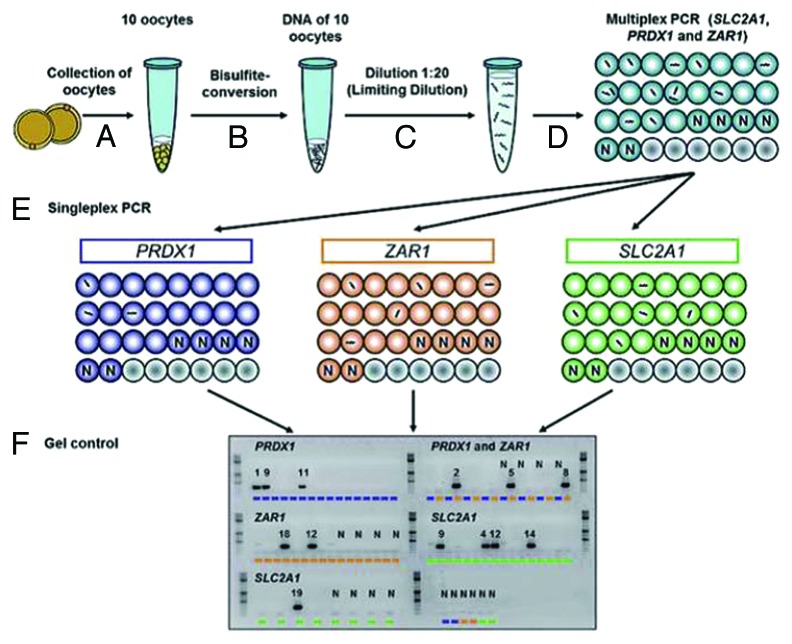
Using the Limited dilution (LD) bisulfite sequencing technique41 which allows amplification of a high number of alleles (Fig. 2), it was shown that epigenetic changes may contribute to the reduced developmental competence of oocytes from prepubertal cattle compared with that of their adult counterparts. DNA methylation patterns in three developmentally important, non-imprinted genes (SLC2A1, PRDX1, ZAR) and two satellite sequences were analyzed to determine the potential impact of age (prepubertal vs adult cattle) and hormonal treatment (FSH and IGF1) of the donor animal on oocyte quality and development. Although methylation changes were not detected in the three genes, significant changes in the satellite DNA methylation profile were observed, suggesting a role of DNA methylation in the acquisition of developmental capacity of bovine oocytes, which needs to being explored in future studies. The relative transcript abundance of selected genes was significantly different in immature and in vitro matured oocytes although only minor changes related to origin and treatment were observed.42
Figure 2. Schematic drawing of the main steps of limiting dilution bisulfite sequencing. (A) Immature and mature oocytes are collected by OPU or after IVM. Ten oocytes of a defined group are pooled. (B) DNA isolation and bisulfite conversion. (C) Dilution of the converted oocyte DNA. (D) The diluted DNA is distributed over 20 wells on a microtiter plate. Most wells contain either no or a single DNA target molecule (ideograms); few wells may contain two or more copies. In addition, six negative controls (N) are added. First-round multiplex PCR is performed with outer primers for the PRDX1, ZAR1, and SLC2A1 genes.(E) Second-round singleplex PCRs of the three studied genes in individual microtiter plates (indicated by different colors) using 1 ml multiplex PCR product as template and gene-specific inner primers. (F) Second-round PCR products are visualized on agarose gels. The color code of each lane indicates the plate (gene), numbers, and Ns of the specific well on that plate. DNA from wells containing a PCR product is analyzed by direct bisulfite sequencing.41,42,49
Although some studies have evaluated the effects of hormonal stimulation of cows on oocyte gene expression and epigenetic changes, it is still not clear whether or not changes of the gene expression after application of exogenous hormones affect the quality and competence of the produced embryos.
In vitro oocyte maturation
In cattle, IVM of oocytes is an integral part of current in vitro embryo production protocols. However, only approximately 30% of the in vitro matured oocytes produce embryos that reach the blastocyst stage. In contrast, the blastocyst rate could be raised up to 60% using in vivo matured oocytes followed by IVF,43 indicating a major role of maturation conditions for acquisition of oocyte developmental competence. Some studies have shown a significant increase in the rates of viable embryos derived from IVM oocytes by changing the follicular development to produce developmentally competent bovine oocytes,44 or by modification of the conventional maturation system. The simulated physiological oocyte maturation (SPOM) constitutes a novel in vitro maturation system that substantially improves bovine embryo development.45
This divergence in oocyte competence could at least partially be explained by significant differences in the transcriptomic profile between in vivo and in vitro matured oocytes.46 Moreover, differences in the methylation profile of embryos produced in vitro, in vivo, or by somatic cell nuclear transfer could be related to the production method.47,48 Recently, the influence of different maturation systems, (in vivo vs. in vitro) using two different media (i.e., TCM and mSOF that are commonly used in bovine IVP), on the methylation profile in DMRs of three imprinted genes (PEG3, H19, and SNRPN) was evaluated for the first time in bovine oocytes. The study did not find significant differences in epigenetic marks in IVM derived matured oocytes compared with their in vivo matured counterparts, indicating that current IVM protocols have none or only marginal effects on these critical epigenetic marks. However, the study reported different mRNA expression profiles in genes with epigenetic importance between in vivo-matured oocytes vs. their in vitro-matured counterparts (Fig. 3), suggesting an influence of regulatory mechanisms other than DNA methylation.49 The paternally imprinted genes H19 and IGF2R and the maternally imprinted gene PEG3 were significantly up-regulated in both groups of in vitro-matured oocytes (TCM and mSOF) compared with in vivo matured oocytes, while the methyltransferases DNMT1a, DNMT3a and DNMT3b were significantly up-regulated in in vitro matured oocytes, irrespective of the maturation system, compared with in vivo matured oocytes.49
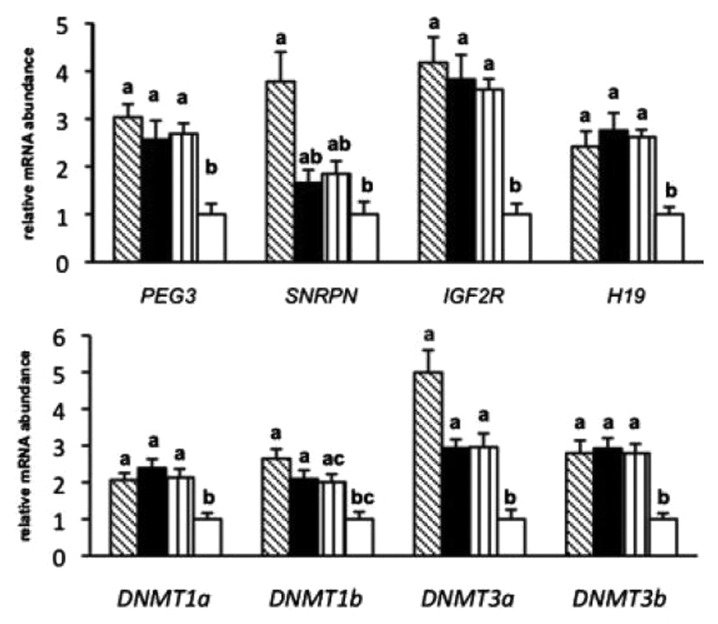
Figure 3. Relative poly(A) mRNA abundance of imprinted genes, methyltransferases, based on single cell preparations of 8–15 oocytes per group: Immature (shaded), mSOF (black), TCM (vertically lined), and in vivo (white).49
Racedo and collaborators measured the methylation status of H3K9, acetylation of H4K12 and satellite DNA methylation status at different stages during bovine oocyte maturation.50 The H3K9me2 signal was present at GV stage and remained detectable until the end of the maturation period. The H4K12ac antibody gave a stronger signal in GV and GVBD oocytes, but was markedly decreased after GVBD. The signal showing the methylation of DNA was present during the entire maturation period. G9A, SUV39H1, DNMT1, DNMT3b and ZAR1 showed a gene-specific mRNA expression profile during oocyte maturation. These results contribute to the understanding of epigenetic modifications implicated in bovine oocyte in vitro maturation and their possible relationship with the acquisition of developmental competence during follicular maturation.50
Recently, a study revealed that the methylation status in the intragenic DMR of the IGF2 locus in bovine oocytes differs with oocyte size and developmental competence.51 This may be useful as molecular marker in studies of oocyte competence, potentially contributing to improvement of in vitro embryo production.
Given that IVM of bovine oocytes is a crucial step in the in vitro production of embryos, in-depth molecular evaluation of the oocyte is required for a better understanding of developmental competence acquisition. There is evidence for differential gene expression and different methylation profiles in competent and non-competent oocytes, which could be used to improve ARTs.
Sperm Handling
The spermatozoon is a highly specialized cell that delivers the paternal haploid genome to the oocyte. Epigenetic changes or changes in gene regulatory properties and mechanisms assist in the preparation of the paternal genome to contribute to zygote formation and subsequently embryogenesis.52 Sexing of spermatozoa—separating male and female sperm according to relative DNA contents on Y and X-chromosomes—by means of flow cytometry was developed in the 1980s.53 This technology can greatly enhance breeding programs by allowing the production of animals of the desired gender; the use of sexed spermatozoa increases the rate of genetic progress, especially in combination with genomic selection of sires.54 However, the high cost, the limited number of sperm samples to be used for insemination, and the frequently reduced pregnancy rates, both in artificial insemination and embryo transfer programs,55,56 have so far limited a wider application of this technology in cattle breeding. The FAO emphasized that sperm or embryo sexing, in combination with other biotechnologies, including genomics, proteomics or phenomics, or sperm-mediated gene transfer57,58 are promising to help meeting the increasing demand for animal derived food production.54
Sex-sorted sperm are exposed to several hazardous conditions. For the sorting process, the membrane-permeable bisbenzimidazole fluorescent dye, Hoechst 33342, is used to stain the DNA and the flow cytometric system recognizes and separates living X- and Y-chromosome-bearing sperm according to the relative amount of fluorescence.53 The effects of sperm sexing by flow cytometry on the methylation patterns of the genes IGF2 and IGF2R were recently evaluated in bull sperm. Sex-sorting did not affect the DNA methylation pattern on the DMR located in the last exon of the IGF2 gene, and neither did it affect the DMR located in the second imprinting control region (ICR) of the IGF2R gene. However, the study revealed variable methylation patterns for individual bulls. Furthermore, a highly specific methylation pattern was observed in the IGF2R gene, probably due to an epigenetic characteristic of Bos indicus cattle.59
Prior to IVF, spermatozoa are subjected to a process that selects for motile spermatozoa which includes centrifugation in Percoll gradients. The Percoll volume, the duration of centrifugation, and higher centrifugation forces had no negative effect on chromatin integrity.60 Obviously, bovine sperm chromatin is resistant to X-irradiation screening, and embryos resulting from such sperm did not show an impaired development.61 Other studies on bull semen have detected alterations in sperm DNA integrity caused by the cryopreservation protocol and even the in vitro incubation period.62 The potential long-term effects of these epigenetic changes are unknown.
Spermatozoa deliver not only the paternal genome into the oocyte, but also carry remnant mRNAs from spermatogenesis.63 The sperm transcriptome harbors a complex mixture of messengers implicated in a wide array of cell functions. RNA profiling could be used for assessing sperm quality, and could determine whether the contribution of paternal RNA is associated with epigenetic changes.64 A recent study revealed an abundance of miRNAs in bovine spermatozoa, of which seven were differentially expressed (hsa-aga-3155, -8197, -6727, -11796, -14189, -6125, -13659) between males with low and high fertility. The relative abundance of miRNAs in spermatozoa and the differential expression in sperm from high vs. low fertility bulls suggests that miRNAs could possibly play an important role in regulating mechanisms of bovine spermatozoa function and in early embryo development.65
An important aspect in this context is the divergent time lines in the acquisition of paternal vs. maternal imprints that may have important implications for ARTs. Overall, isolation and treatment of male germ cells for IVP occur after male-specific methylation reprogramming. Therefore, it is plausible to assume that the aberrant methylation patterns that have been observed in IVF/ICSI sperm in human and mice66-68 may be due to impaired spermatogenesis of the donors, and not to ART itself. In contrast, IVC of oocytes, superovulation, and IVF may well interfere with the proper acquisition of maternal methylation imprints during oogenesis.69 A recent study analyzed three imprinted genes in mice produced by ICSI. These mice maintained primary epimutations in somatic tissue, whereas the epimutations were corrected in the germ line by epigenetic reprogramming and thus not propagated to subsequent generations.70
Embryo Culture
The successful in vitro culture of preimplantation embryos has contributed substantially to the success of assisted reproduction techniques.71-73 The high number of papers from laboratories around the globe reflects the intensity of research toward improving culture conditions and reducing the deficiencies that might lead to changes in gene expression and an increased frequency of epigenetic disorders. During early embryogenesis the parental genomes undergo a wave of de- and re-methylation rendering early embryos specifically vulnerable to ART- induced epigenetic defects.69 This epigenetic reprogramming of the genome after fertilization creates the methylation patterns needed for normal development by activation and silencing of specific genes.74,75 Global methylation of the bovine genome declines to a nadir at the 6–8 cell stage and increases thereafter; methylation is lower in female embryos than in male embryos at the blastocyst stage and lower in the ICM than TE.76 Using immunostaining, it was shown that in vitro culture (IVC) of bovine embryos may affect DNA methylation patterns and thus early embryo developmental capacity.77 The imprinting status of the gene encoding the small nuclear ribonucleoprotein polypeptide N (SNRPN) was evaluated in bovine embryos produced by AI, IVP or SCNT. The allelic expression profile was compared with the methylation pattern of a DMR located in the promoter region. Prolonged in vitro culture and SCNT were associated with abnormal reprogramming of several imprinted gene loci, including SNPRN, PEG3, PEG10, PEG11, IGF2, and IGF2R, suggesting that these regions are sensitive to environmental factors which in turn could lead to epigenetic disorders.48,78
Genomic imprinting is an epigenetic phenomenon in which only one allele of a specific gene is transcriptionally active, while the other allele is silenced based on the parent-of-origin.35 Approximately 200 genes are imprinted in the mammalian genome.79 More than 70 genes in mice and at least 50 genes in humans have been reported to be imprinted. Table 1 contains a summary of bovine imprinted genes (http://www.geneimprint.com, http://igc.otago.ac.nz). The imprinting status is conserved for some genes in humans, mouse, and cattle. Imprinting disorders are more prevalent in gametes and embryos after ART than in their counterparts derived from in vivo production. In the mouse model, it was shown that embryo culture media may affect gene imprinting.80-82 Anomalies in DNA methylation and disorders in gene imprinting in bovine embryos produced by SCNT have been extensively reviewed.15,16
Table 1. Imprinted genes in cattle.
| Name | Gene Symbol | Expressed allele | Chromosome location |
Reference |
|---|---|---|---|---|
| Paternally expressed 10 | PEG10 | Paternal | 4 | 83,84 |
| Mesoderm specific transcript homolog (mouse) | MEST, PEG1 | Paternal | 4 | 84 |
| Nucleosome assembly protein 1-like 5 | NAP1L5 | Paternal | 6 | 85 |
| Insulin-like growth factor 2 receptor | IGF2R | Maternal | 9 | 84,86 |
| Pleiomorphic adenoma gene-like 1 | PLAGL1 | Paternal | 9 | 84 |
| GNAS complex locus | GNAS, NESP55 | Maternal | 13 | 87 |
| Neuronatin | NNAT | Paternal | 13 | 85,88 |
| MER1 repeat containing imprinted transcript 1 (non-protein coding) | MIMT1 ITUP1, USP29) |
Paternal | 18 | 89 |
| Paternally expressed 3 | PEG3 | Paternal | 18 | 89,90 |
| Maternally expressed 3 (non-protein coding) | MEG3, GLT2 | Maternal | 21 | 91 |
| Small nuclear ribonucleoprotein polypeptide N | SNRPN | Paternal | 21 | 78,84 |
| Retrotransposon-like 1 | RTL1, PEG11 | Paternal | 21 | 83 |
| Maternally expressed gene 8 | MEG8 | Maternal | 21 | 92 |
| MAGE-like 2 | MAGEL2 | Paternal | 21 | 83 |
| Tumor suppressing subtransferable candidate 4 | TSSC4 | Maternal | 29 | 83 |
| H19, imprinted maternally expressed transcript (non-protein coding) | H19 | Maternal | 29 | 85,93 |
| Insulin-like growth factor 2 | IGF2 | Paternal | 29 | 91 |
| Pleckstrin homology-like domain, family A, member 2 | PHLDA2 | Maternal | 29 | 94 |
| X (inactive)-specific transcript | XIST | Paternal | X | 91,95 |
Epigenetic alterations and changes in chromatin configuration may occur during extended in vitro culture periods.96 The development of effective chromatin immunoprecipitation (ChIP) protocols has enabled studies of protein-DNA interactions and mapping of histone modifications to the DNA.97,98 ChIP assays have recently been refined to allow analysis of small cell samples.99 The feasibility of histone modification analysis on individual gene promoters in bovine blastocysts was demonstrated recently for the first time.100 The gene expression patterns in the ICM and TE of bovine blastocysts were consistent with the histone modification patterns on the promoter of selected genes, including POU5F1 (OCT4), NANOG, INFT, GAPDH, SLC2A3, and IGF1.100 Only few studies reported effects of IVC on chromatin configuration changes in bovine embryos, and alterations in histone modifications in in vitro produced embryos101 and in parthenotes102 have been described. A recent study suggested that cloned bovine embryos were reprogrammed with histone modifications similar to that of IVF embryos, both IVF-derived and cloned embryos showed a homogeneous distribution of histone modifications in morulae and blastocysts.103
Bovine embryos are increasingly accepted as valuable model for studies of epigenetic alterations because bovine embryos are a better model for early human embryonic development than the laboratory mouse.104 Studies on the effects of embryo culture condition on the development of bovine embryos usually require in vivo counterparts as “physiological controls” for all stages of preimplantation development. Advanced ultrasound guided follicular aspiration and laparoscopical techniques are used to isolate oocytes and oviductal embryonic stages with minimal invasiveness from female cattle.105,106
The differences in gene expression in IVC vs. in vivo derived bovine embryos have been proposed as strategy to identify molecular mechanisms and pathways susceptible to culture conditions and could thus provide clues to enhance in vivo development of blastocysts.37,105,107 Altered phenotypes from in vitro produced and cloned bovine embryos may be the result of an aberrant expression profile of imprinted and/or non-imprinted genes caused by the failure to properly establish or maintain DNA methylation and histone modifications.108,109 The aberrant expression of IGF2R was correlated with the incidence of the Large Offspring Syndrome (LOS) in sheep110 and aberrant expression of imprinted and non-imprinted genes has been observed in fetuses, placentas and offspring derived from IVP.111-113 Expression levels of both IFN-tau and IGF2R depended on embryo density when the embryos were maintained in droplet culture.114 Up-regulated IFN-tau expression and down-regulated IGF2R expression were observed when embryos were cultured in groups of 25 embryos, while no differences were found in the well-of-the-well (WOW) system culture.114 Increased embryo density appears to enhance the accumulation of toxic by-products of embryo metabolism such as ammonium.115 Ammonium induced aberrant expression of the imprinting gene H19 in mice blastocysts, but did not affect the rate of blastocyst formation.115
Differences in growth rates and metabolism between male and female mammalian embryos have been widely documented. These differences appear already prior to sexual differentiation of the gonads and, could not be explained by sex-related hormonal differences.116 Differences in growth rate, metabolism, gene expression and epigenetic programming during preimplantation development indicate that male and female embryos may respond differently to environmental conditions and suggest that early perturbations may have sex-specific effects, not only during preimplantation development, but also in fetal and postnatal development.117,118 The methylation pattern of a DNA sequence adjacent to a variable number of tandem repeats (VNTR) was higher in males (39.8%) than in females (23.7%). In addition, differences with regard to gene expression between sexes were observed for genes related to cytosine methylation and histone methylation, including DNMT3a, DNMT3b, HMT1, and ILF3.118
One of the main differences between male and female embryos during preimplantation development is the relative abundance of X-linked transcripts. The expression of X-linked genes was higher in IVP derived embryos compared with their in vivo produced counterparts,119,120 suggesting that X-linked expression in IVP blastocysts is aberrant and may lead to higher XIST expression than in their in vivo counterparts. A recent study showed that HDAC inhibition using a low trichostatin (TSA) concentration had no effect on cell cycle progression. Increased histone acetylation levels and XIST expression in female bovine embryos were related to HDAC and HDAC inhibition decreased XIST mRNA levels.121
Effects of Storage of Oocytes and Embryos
Storage of oocytes and embryos is routine procedure in ARTs. The success rates after transfer of cryopreserved or vitrified bovine embryos have been increased significantly over the past years.122 Few studies addressed the safety of oocytes and embryo cryopreservation at the DNA level, and most of these focused on apoptosis.123,124 and gene expression in various signaling and metabolic pathways;125,126 very few studies investigated epigenetics.127 Vitrification caused aberrant methylation at H19 ICRs in murine embryos, with compensation of the disordered H19/IGF2 expression in IVF embryos, but did not affect H19 or Igf2 expression in placentas.127 Vitrification did not significantly alter the methylation patterns of CpG islands in the promoter region of DNMT1o, HAT1, or HDAC1, but decreased expression of DNMT1o in mouse MII oocytes.128 In slowly frozen bovine embryos, expression of developmentally important genes was evaluated and significant differences compared with non-frozen controls were detected for DNMT3A129 which could be linked with epigenetic aberrations. Global DNA methylation levels were significantly lower after slow freezing and vitrification of bovine oocytes.130 Vitrification significantly increased the methylation level at ICR of H19 in 2-cell embryos.131 These preliminary findings suggest that even well-established cryoprotection protocols could be associated with epigenetic deviations. To what extent these may affect viability of the oocytes/embryos remains to be determined.
What is Happening with the Organs?
In vitro embryo production has emerged as a useful tool to multiply superior genotypes and is an alternative to conventional embryo transfer, and thus being increasingly used commercially in many countries around the globe.9 However, phenotypic alterations have been reported in fetuses and offspring derived from in vitro produced embryos, including aberrant placental development, extended gestation length, sudden perinatal death, breathing difficulties, a skewed sex ratio with more male calves, and large size at birth.108,132,133 These alterations in phenotype were called LOS, with the predominant feature of increased birth weights134; LOS has been observed in cattle, sheep,135,136 and mice137-139 produced by ART. However, a better understanding of the necessary culture conditions led to the development of semi-defined media, with embryos incubated in the absence of feeder cells with little or no serum added, which in turn significantly decreased the incidence of LOS.140 Numerous studies have been undertaken to improve the efficiency of embryo production and eventually the synthetic oviductal fluid (SOF)-BSA medium, originally based on the biochemical composition of sheep uterine tubal fluid,141 as well as Charles Rosenkrans medium142 became popular bovine embryo culture media. Most systems used serum and co-culture; however, these constituents were associated with the incidence of LOS.134,143 This problem could be eliminated by replacing serum/co-culture with SOF, not only in cattle, but also in sheep.144,145
Fetuses resulting from the transfer of IVP embryos were reported to display disproportionate organ development in some studies,144,146 but not in others.133,147 In addition, alterations in the histological development of fetal muscle148,149 and placental tissue150 have been reported in pregnancies from embryos produced in vitro. Recently, in vitro embryo production was found to be associated with subtle changes in fetal development as well as altered expression of both imprinted and non-imprinted genes.151 Fetuses at Day 70 of gestation derived from embryos produced in vitro had decreased crown-rump length and increased paired kidney weights. Fetuses from in vitro produced embryos also had a decreased expression level of mRNAs for IGF1 in liver and IGF2R in both liver and skeletal muscle, compared with fetuses from in vivo produced embryos.151 The insulin-like growth factor type 2 receptor (IGF2R) is an imprinted gene that regulates fetal and placental development in cattle and other species.152,153 The primary function of the IGF2 receptor is to bind IGF2, it is imprinted in cattle, acts as a powerful mitogen, and serves as target for lysosomal degradation.154 The level of bovine AIRN ncRNA, which is required for regular imprinted expression of IGF2R in fetuses during the post-implantation period, was altered relative to the production method of pre-implantation embryos; the mRNA expression was significantly reduced in livers of Day 70 bovine fetuses from IVP embryos compared with that of in vivo produced embryos.155
Accumulating evidence suggests that epigenetic mechanisms are disturbed in gametes and embryos by extracorporal handling and/or culture conditions in various species.156-158 The effects of two in vitro fertilization protocols (IVF1 and IVF2) on fetal phenotype and genomic cytosine methylation levels were assessed in bovine fetal liver, skeletal muscle, and brain.159 One IVF protocol employed 0.01 U/ml FSH and LH in oocyte maturation medium and 5% estrous cow serum (ECS) in embryo culture medium, whereas the second IVF protocol employed 0.2 U/ml FSH and no LH for oocyte maturation and 10% ECS for embryo culture. Fetuses derived from the second IVF protocol displayed an overgrowth phenotype and were significantly heavier (19.9%) and longer (4.7%), and showed increased heart (25.2%) and liver (27.9%) weights. DNA hypomethylation was found in liver and muscle of fetuses derived from the first IVF and significant hypermethylation was determined in liver of fetuses from the second IVP protocol. The 5mC level of cerebral DNA was not affected by IVF protocol. These data indicate that bovine IVF procedures can affect fetal genomic 5mC levels in a protocol- and tissue-specific manner and show that hepatic hypermethylation may be associated with fetal overgrowth and its correlated endocrine changes.159
The bicistronic gene SNURF-SNRPN, referred here as SNRPN, has been extensively studied in mice and humans due to the correlation between abnormal DMR methylation and the incidence of neurodevelopmental disorders, known as Prader-Willi or Angelman syndrome. Interestingly, decreased levels of DNA methylation of the maternal allele in the SNRPN DMR have been observed in children conceived by ART, suggesting that the SNRPN methylation pattern is directly affected by in vitro culture systems.19,160 The SNRPN gene is maternally imprinted in preimplantation bovine embryos.161 Bi-allelic SNRPN gene expression was found in in vitro cultured preimplantation embryos; loss of methylation was also found in embryonic and extra-embryonic tissues of pregnancies derived from IVF embryos cultured in vitro.78 This may be a good model to study the etiology of the Prader-Willi and Angelman syndromes in human patients.
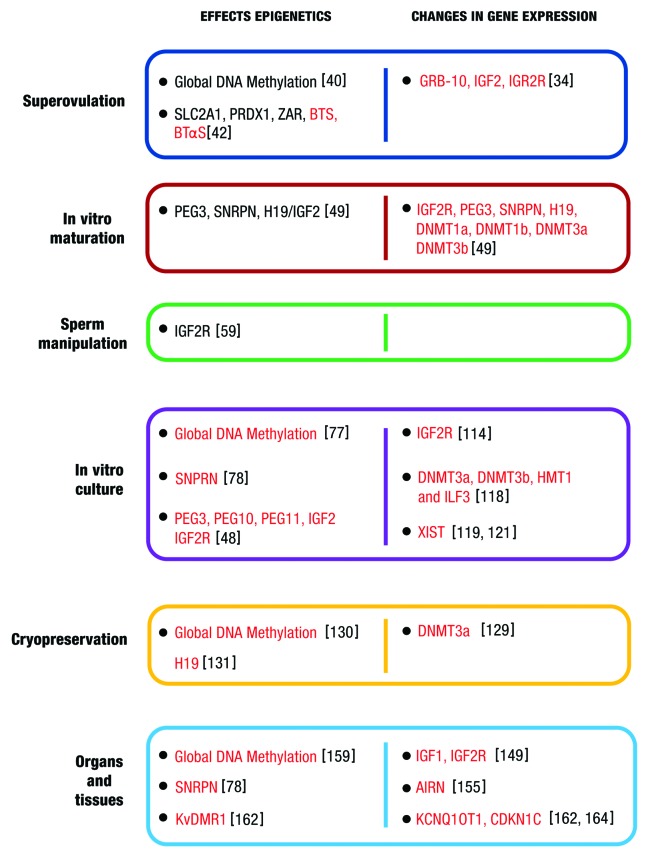
Imprinted gene expression of KCNQ1OT1, CDKN1C, H19, and PLAGL1 and the methylation patterns at the KvDMR1 and H19/IGF2 ICRs are conserved between humans and cattle.162-164 Phenotypic and epigenetic similarities between LOS and BWS were observed, and it was proposed that LOS in animals is promising to investigate the etiology of BWS.164 Hori et al. described for first time the abnormal hypomethylation of the KvDMR1 domain and subsequent changes in the gene expression profile of KCNQ1OT1 and CDKN1C in organs of calves produced by IVP or SCNT.162 Another study showed that KCNQ1OT1 which is the most-often dysregulated imprinted gene in BWS, was bi-allelically expressed in various organs in two out of seven oversized conceptuses from the IVC group, but showed mono-allelic expression in all tissues of AI conceptuses. Furthermore, bi-allelic expression of KCNQ1OT1 was associated with a loss of methylation at the KvDMR1 on the maternal allele and with down-regulation of the maternally expressed allele.164 Figure 4 shows a summary of the effects of superovulation, in vitro maturation, sperm manipulation, in vitro culture, cryopreservation on oocytes, sperm, embryos, organs and tissues domestic cattle.
Figure 4. Effects of SOV, IVM, sperm manipulation, IVC, and cryopreservation on epigenetic marks and changes in expression of genes in oocytes, sperm, embryos, organs and tissues. Genes with aberrant pattern are marked in red; genes with normal pattern are marked black.
Few studies have evaluated potential effects of IVP on gestation length and birth weight by comparing offspring produced by IVP with their counterparts produced by artificial insemination or natural breeding. An average increase of 8% in birth weight of Holstein calves from IVP embryos was found compared with artificial insemination (AI), with 34% of IVP offspring > 50 kg.165 Overweight calves from IVP embryos have also been reported for other cattle breeds, incl. Angus,133 Japanese black166 and Hanwoo.167 Gestation length can also be affected by in vitro embryo production.145,167 Recently, it was shown in a large cohort of IVP calves that in vitro embryo production with serum and co-culture can alter phenotypic characteristics of Gyr calves by increasing the birth weight at calving but with little effects on gestation length.168
Concluding Remarks
This review clearly shows that, although ARTs are useful tools for improving reproduction in the cattle industry, some of the procedures involved could potentially affect gametes and embryos by causing epigenetic disorders which in turn may lead to aberrant gene expression (Fig. 5). The differences between embryos produced in vivo with respect to those produced in vitro, can be linked to molecular differences, including epigenetic patterns, which could explain differences in metabolism, cell number, ultrastructure and cryotolerance. Despite the widespread application of ARTs under commercial conditions, the exact mechanisms leading to epigenetic disorders and aberrant gene expression are not yet fully understood not only in the bovine species, but also in the mouse model and in humans.
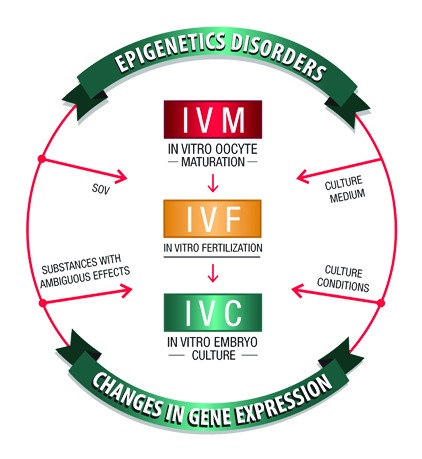
Figure 5. Factors inducing epigenetic disorders and changes in gene expression in the in vitro embryo production. Protocols for superovulation, substances with ambiguous effects such as fetal calf serum (FCS), culture conditions including changes in pH, osmolality, temperature and various basic culture media may affect the normal epigenetic phenotype during early development, and thereby decrease the quality of the embryos.
To improve the results of ARTs, further studies are necessary to understand how epigenetic regulation is affected by ART in gametes, early embryos and post-implantation. A battery of diagnostic tests to identify, prevent and/or reduce epigenetic disorders and changes in gene expression after use of bovine assisted reproductive technologies could be beneficial in this respect.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
R.U. is in the PhD program of the Animal Science, Antioquia University, Colombia and is supported by a Doctoral Scholarship from COLCIENCIAS. We would like to acknowledge the sustainability strategy (2013-2014) from CODI (Universidad de Antioquia). We also thank Jose Tamayo for assistance in the Figures 1, 4, and 5.
References
- 1.Waddington CH. The epigenotype. 1942. Int J Epidemiol. 2012;41:10–3. doi: 10.1093/ije/dyr184. [DOI] [PubMed] [Google Scholar]
- 2.Wu Ct, Morris JR. Genes, genetics, and epigenetics: a correspondence. Science. 2001;293:1103–5. doi: 10.1126/science.293.5532.1103. [DOI] [PubMed] [Google Scholar]
- 3.Li ZX, Ma X, Wang ZH. A differentially methylated region of the DAZ1 gene in spermatic and somatic cells. Asian J Androl. 2006;8:61–7. doi: 10.1111/j.1745-7262.2006.00069.x. [DOI] [PubMed] [Google Scholar]
- 4.Biermann K, Steger K. Epigenetics in male germ cells. J Androl. 2007;28:466–80. doi: 10.2164/jandrol.106.002048. [DOI] [PubMed] [Google Scholar]
- 5.Rajender S, Avery K, Agarwal A. Epigenetics, spermatogenesis and male infertility. Mutat Res. 2011;727:62–71. doi: 10.1016/j.mrrev.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 6.Narlikar GJ, Fan HY, Kingston RE. Cooperation between complexes that regulate chromatin structure and transcription. Cell. 2002;108:475–87. doi: 10.1016/S0092-8674(02)00654-2. [DOI] [PubMed] [Google Scholar]
- 7.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brait M, Sidransky D. Cancer epigenetics: above and beyond. Toxicol Mech Methods. 2011;21:275–88. doi: 10.3109/15376516.2011.562671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rodriguez-Martinez H. Assisted reproductive techniques for cattle breeding in developing countries: a critical appraisal of their value and limitations. Reprod Domest Anim. 2012;47(Suppl 1):21–6. doi: 10.1111/j.1439-0531.2011.01961.x. [DOI] [PubMed] [Google Scholar]
- 10.Madan ML. Animal biotechnology: applications and economic implications in developing countries. Rev Sci Tech. 2005;24:127–39. [PubMed] [Google Scholar]
- 11.Niemann H, Wrenzycki C. Alterations of expression of developmentally important genes in preimplantation bovine embryos by in vitro culture conditions: implications for subsequent development. Theriogenology. 2000;53:21–34. doi: 10.1016/S0093-691X(99)00237-X. [DOI] [PubMed] [Google Scholar]
- 12.Iliadou AN, Janson PCJ, Cnattingius S. Epigenetics and assisted reproductive technology. J Intern Med. 2011;270:414–20. doi: 10.1111/j.1365-2796.2011.02445.x. [DOI] [PubMed] [Google Scholar]
- 13.Denomme MM, Mann MRW. Genomic imprints as a model for the analysis of epigenetic stability during assisted reproductive technologies. Reproduction. 2012;144:393–409. doi: 10.1530/REP-12-0237. [DOI] [PubMed] [Google Scholar]
- 14.Niemann H, Tian XC, King WA, Lee RSF. Epigenetic reprogramming in embryonic and foetal development upon somatic cell nuclear transfer cloning. Reproduction. 2008;135:151–63. doi: 10.1530/REP-07-0397. [DOI] [PubMed] [Google Scholar]
- 15.Smith LC, Suzuki J, Jr., Goff AK, Filion F, Therrien J, Murphy BD, Kohan-Ghadr HR, Lefebvre R, Brisville AC, Buczinski S, et al. Developmental and epigenetic anomalies in cloned cattle. Reprod Domest Anim. 2012;47(Suppl 4):107–14. doi: 10.1111/j.1439-0531.2012.02063.x. [DOI] [PubMed] [Google Scholar]
- 16.Rodriguez-Osorio N, Urrego R, Cibelli JB, Eilertsen K, Memili E. Reprogramming mammalian somatic cells. Theriogenology. 2012;78:1869–86. doi: 10.1016/j.theriogenology.2012.05.030. [DOI] [PubMed] [Google Scholar]
- 17.Pan Z, Zhang J, Li Q, Li Y, Shi F, Xie Z, Liu H. Current advances in epigenetic modification and alteration during mammalian ovarian folliculogenesis. J Genet Genomics. 2012;39:111–23. doi: 10.1016/j.jgg.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 18.Kerjean A, Couvert P, Heams T, Chalas C, Poirier K, Chelly J, Jouannet P, Paldi A, Poirot C. In vitro follicular growth affects oocyte imprinting establishment in mice. Eur J Hum Genet. 2003;11:493–6. doi: 10.1038/sj.ejhg.5200990. [DOI] [PubMed] [Google Scholar]
- 19.Lucifero D, Chaillet JR, Trasler JM. Potential significance of genomic imprinting defects for reproduction and assisted reproductive technology. Hum Reprod Update. 2004;10:3–18. doi: 10.1093/humupd/dmh002. [DOI] [PubMed] [Google Scholar]
- 20.Ciccone DN, Su H, Hevi S, Gay F, Lei H, Bajko J, Xu G, Li E, Chen T. KDM1B is a histone H3K4 demethylase required to establish maternal genomic imprints. Nature. 2009;461:415–8. doi: 10.1038/nature08315. [DOI] [PubMed] [Google Scholar]
- 21.El-Maarri O, Buiting K, Peery EG, Kroisel PM, Balaban B, Wagner K, Urman B, Heyd J, Lich C, Brannan CI, et al. Maternal methylation imprints on human chromosome 15 are established during or after fertilization. Nat Genet. 2001;27:341–4. doi: 10.1038/85927. [DOI] [PubMed] [Google Scholar]
- 22.Ertzeid G, Storeng R. The impact of ovarian stimulation on implantation and fetal development in mice. Hum Reprod. 2001;16:221–5. doi: 10.1093/humrep/16.2.221. [DOI] [PubMed] [Google Scholar]
- 23.Van der Auwera I, D’Hooghe T. Superovulation of female mice delays embryonic and fetal development. Hum Reprod. 2001;16:1237–43. doi: 10.1093/humrep/16.6.1237. [DOI] [PubMed] [Google Scholar]
- 24.Shi W, Haaf T. Aberrant methylation patterns at the two-cell stage as an indicator of early developmental failure. Mol Reprod Dev. 2002;63:329–34. doi: 10.1002/mrd.90016. [DOI] [PubMed] [Google Scholar]
- 25.de Waal E, Yamazaki Y, Ingale P, Bartolomei MS, Yanagimachi R, McCarrey JR. Gonadotropin stimulation contributes to an increased incidence of epimutations in ICSI-derived mice. Hum Mol Genet. 2012;21:4460–72. doi: 10.1093/hmg/dds287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fauque P, Jouannet P, Lesaffre C, Ripoche MA,, Dandolo L, Vaiman D, Jammes H. Assisted Reproductive Technology affects developmental kinetics, H19 Imprinting Control Region methylation and H19 gene expression in individual mouse embryos. BMC Dev Biol. 2007;7:116. doi: 10.1186/1471-213X-7-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fortier AL, Lopes FL, Darricarrère N, Martel J, Trasler JM. Superovulation alters the expression of imprinted genes in the midgestation mouse placenta. Hum Mol Genet. 2008;17:1653–65. doi: 10.1093/hmg/ddn055. [DOI] [PubMed] [Google Scholar]
- 28.Market-Velker BA, Zhang L, Magri LS, Bonvissuto AC, Mann MRW. Dual effects of superovulation: loss of maternal and paternal imprinted methylation in a dose-dependent manner. Hum Mol Genet. 2010;19:36–51. doi: 10.1093/hmg/ddp465. [DOI] [PubMed] [Google Scholar]
- 29.Ludwig M, Katalinic A, Gross S, Sutcliffe A, Varon R, Horsthemke B. Increased prevalence of imprinting defects in patients with Angelman syndrome born to subfertile couples. J Med Genet. 2005;42:289–91. doi: 10.1136/jmg.2004.026930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baart EB, Martini E, Eijkemans MJ, Van Opstal D, Beckers NGM, Verhoeff A, Macklon NS, Fauser BCJM. Milder ovarian stimulation for in-vitro fertilization reduces aneuploidy in the human preimplantation embryo: a randomized controlled trial. Hum Reprod. 2007;22:980–8. doi: 10.1093/humrep/del484. [DOI] [PubMed] [Google Scholar]
- 31.Khoueiry R, Ibala-Rhomdane S, Méry L,, Blachère T, Guérin JF, Lornage J, Lefèvre A. Dynamic CpG methylation of the KCNQ1OT1 gene during maturation of human oocytes. J Med Genet . 2008;45:583–588. doi: 10.1136/jmg.2008.057943. [DOI] [PubMed] [Google Scholar]
- 32.Bó GA, Guerrero DC, Tríbulo A, Tríbulo H, Tríbulo R, Rogan D, Mapletoft RJ. New approaches to superovulation in the cow. Reprod Fertil Dev. 2010;22:106–12. doi: 10.1071/RD09226. [DOI] [PubMed] [Google Scholar]
- 33.Mapletoft RJ, Bó GA. The evolution of improved and simplified superovulation protocols in cattle. Reprod Fertil Dev. 2011;24:278–83. doi: 10.1071/RD11919. [DOI] [PubMed] [Google Scholar]
- 34.Mundim TCD, Ramos AF, Sartori R, Dode MA, Melo EO, Gomes LF, Rumpf R, Franco MM. Changes in gene expression profiles of bovine embryos produced in vitro, by natural ovulation, or hormonal superstimulation. Genet Mol Res. 2009;8:1398–407. doi: 10.4238/vol8-4gmr646. [DOI] [PubMed] [Google Scholar]
- 35.Manipalviratn S, DeCherney A, Segars J. Imprinting disorders and assisted reproductive technology. Fertil Steril. 2009;91:305–15. doi: 10.1016/j.fertnstert.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chang AS, Moley KH, Wangler M, Feinberg AP, Debaun MR. Association between Beckwith-Wiedemann syndrome and assisted reproductive technology: a case series of 19 patients. Fertil Steril. 2005;83:349–54. doi: 10.1016/j.fertnstert.2004.07.964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wrenzycki C, Herrmann D, Lucas-Hahn A, Lemme E, Korsawe K, Niemann H. Gene expression patterns in in vitro-produced and somatic nuclear transfer-derived preimplantation bovine embryos: relationship to the large offspring syndrome? Anim Reprod Sci. 2004;82-83:593–603. doi: 10.1016/j.anireprosci.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 38.Sutcliffe AG, Peters CJ, Bowdin S, Temple K, Reardon W, Wilson L, Clayton-Smith J, Brueton LA, Bannister W, Maher ER. Assisted reproductive therapies and imprinting disorders--a preliminary British survey. Hum Reprod. 2006;21:1009–11. doi: 10.1093/humrep/dei405. [DOI] [PubMed] [Google Scholar]
- 39.Chu T, Dufort I, Sirard MA. Effect of ovarian stimulation on oocyte gene expression in cattle. Theriogenology. 2012;77:1928–38. doi: 10.1016/j.theriogenology.2012.01.015. [DOI] [PubMed] [Google Scholar]
- 40.Liu S, Feng HL, Marchesi D, Chen ZJ, Hershlag A. Effect of gonadotropins on dynamic events and global deoxyribonucleic acid methylation during in vitro maturation of oocytes: an animal model. Fertil Steril. 2011;95:1503–, e1-3. doi: 10.1016/j.fertnstert.2010.09.049. [DOI] [PubMed] [Google Scholar]
- 41.El Hajj N, Trapphoff T, Linke M, May A, Hansmann T, Kuhtz J, Reifenberg K, Heinzmann J, Niemann H, Daser A, et al. Limiting dilution bisulfite (pyro)sequencing reveals parent-specific methylation patterns in single early mouse embryos and bovine oocytes. Epigenetics. 2011;6:1176–88. doi: 10.4161/epi.6.10.17202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Diederich M, Hansmann T, Heinzmann J, Barg-Kues B, Herrmann D, Aldag P, Baulain U, Reinhard R, Kues W, Weissgerber C, et al. DNA methylation and mRNA expression profiles in bovine oocytes derived from prepubertal and adult donors. Reproduction. 2012;144:319–30. doi: 10.1530/REP-12-0134. [DOI] [PubMed] [Google Scholar]
- 43.Bermejo-Alvarez P, Rizos D, Rath D, Lonergan P, Gutierrez-Adan A. Epigenetic differences between male and female bovine blastocysts produced in vitro. Physiol Genomics. 2008;32:264–72. doi: 10.1152/physiolgenomics.00234.2007. [DOI] [PubMed] [Google Scholar]
- 44.Blondin P, Bousquet D, Twagiramungu H, Barnes F, Sirard MA. Manipulation of follicular development to produce developmentally competent bovine oocytes. Biol Reprod. 2002;66:38–43. doi: 10.1095/biolreprod66.1.38. [DOI] [PubMed] [Google Scholar]
- 45.Albuz FK, Sasseville M, Lane M, Armstrong DT, Thompson JG, Gilchrist RB. Simulated physiological oocyte maturation (SPOM): a novel in vitro maturation system that substantially improves embryo yield and pregnancy outcomes. Hum Reprod. 2010;25:2999–3011. doi: 10.1093/humrep/deq246. [DOI] [PubMed] [Google Scholar]
- 46.Wrenzycki C, Herrmann D, Niemann H. Messenger RNA in oocytes and embryos in relation to embryo viability. Theriogenology. 2007;68(Suppl 1):S77–83. doi: 10.1016/j.theriogenology.2007.04.028. [DOI] [PubMed] [Google Scholar]
- 47.Curchoe CL, Zhang S, Yang L, Page R, Tian XC. Hypomethylation trends in the intergenic region of the imprinted IGF2 and H19 genes in cloned cattle. Anim Reprod Sci. 2009;116:213–25. doi: 10.1016/j.anireprosci.2009.02.008. [DOI] [PubMed] [Google Scholar]
- 48.Niemann H, Carnwath JW, Herrmann D, Wieczorek G, Lemme E, Lucas-Hahn A, Olek S. DNA methylation patterns reflect epigenetic reprogramming in bovine embryos. Cell Reprogram. 2010;12:33–42. doi: 10.1089/cell.2009.0063. [DOI] [PubMed] [Google Scholar]
- 49.Heinzmann J, Hansmann T, Herrmann D, Wrenzycki C, Zechner U, Haaf T, Niemann H. Epigenetic profile of developmentally important genes in bovine oocytes. Mol Reprod Dev. 2011;78:188–201. doi: 10.1002/mrd.21281. [DOI] [PubMed] [Google Scholar]
- 50.Racedo SE, Wrenzycki C, Lepikhov K, Salamone D, Walter J, Niemann H. Epigenetic modifications and related mRNA expression during bovine oocyte in vitro maturation. Reprod Fertil Dev. 2009;21:738–48. doi: 10.1071/RD09039. [DOI] [PubMed] [Google Scholar]
- 51.Fagundes NS, Michalczechen-Lacerda VA, Caixeta ES, Machado GM, Rodrigues FC, Melo EO, Dode MA, Franco MM. Methylation status in the intragenic differentially methylated region of the IGF2 locus in Bos taurus indicus oocytes with different developmental competencies. Mol Hum Reprod. 2011;17:85–91. doi: 10.1093/molehr/gaq075. [DOI] [PubMed] [Google Scholar]
- 52.Emery BR, Carrell DT. The effect of epigenetic sperm abnormalities on early embryogenesis. Asian J Androl. 2006;8:131–42. doi: 10.1111/j.1745-7262.2006.00127.x. [DOI] [PubMed] [Google Scholar]
- 53.Johnson LA. Sexing mammalian sperm for production of offspring: the state-of-the-art. Anim Reprod Sci. 2000;60-61:93–107. doi: 10.1016/S0378-4320(00)00088-9. [DOI] [PubMed] [Google Scholar]
- 54.Rath D, Barcikowski S, de Graaf S, Garrels W, Grossfeld R, Klein S, Knabe W, Knorr C, Kues W, Meyer H, et al. Sex selection of sperm in farm animals: status report and developmental prospects. Reproduction. 2013;145:R15–30. doi: 10.1530/REP-12-0151. [DOI] [PubMed] [Google Scholar]
- 55.Seidel GE, Schenk JL, Herickhoff LA, Doyle SP, Brink Z, Green RD, Cran DG. Insemination of heifers with sexed sperm. Theriogenology. 1999;52:1407–20. doi: 10.1016/s0093-691x(99)00226-5. [DOI] [PubMed] [Google Scholar]
- 56.Bermejo-Alvarez P, Lonergan P, Rath D, Gutiérrez-Adan A, Rizos D. Developmental kinetics and gene expression in male and female bovine embryos produced in vitro with sex-sorted spermatozoa. Reprod Fertil Dev. 2010;22:426–36. doi: 10.1071/RD09142. [DOI] [PubMed] [Google Scholar]
- 57.De Cecco M, Spinaci M, Zannoni A, Bernardini C, Seren E, Forni M, Bacci ML. Coupling sperm mediated gene transfer and sperm sorting techniques: a new perspective for swine transgenesis. Theriogenology. 2010;74:856–62. doi: 10.1016/j.theriogenology.2010.04.010. [DOI] [PubMed] [Google Scholar]
- 58.Niemann H, Kuhla B, Flachowsky G. Perspectives for feed-efficient animal production. J Anim Sci. 2011;89:4344–63. doi: 10.2527/jas.2011-4235. [DOI] [PubMed] [Google Scholar]
- 59.Carvalho JO, Michalczechen-Lacerda VA, Sartori R, Rodrigues FC, Bravim O, Franco MM, Dode MAN. The methylation patterns of the IGF2 and IGF2R genes in bovine spermatozoa are not affected by flow-cytometric sex sorting. Mol Reprod Dev. 2012;79:77–84. doi: 10.1002/mrd.21410. [DOI] [PubMed] [Google Scholar]
- 60.Machado GM, Carvalho JO, Filho ES, Caixeta ES, Franco MM, Rumpf R, Dode MA. Effect of Percoll volume, duration and force of centrifugation, on in vitro production and sex ratio of bovine embryos. Theriogenology. 2009;71:1289–97. doi: 10.1016/j.theriogenology.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 61.Hendricks KEM, Penfold LM, Evenson DP, Kaproth MT, Hansen PJ. Effects of airport screening X-irradiation on bovine sperm chromatin integrity and embryo development. Theriogenology. 2010;73:267–72. doi: 10.1016/j.theriogenology.2009.09.009. [DOI] [PubMed] [Google Scholar]
- 62.Waterhouse KE, Gjeldnes A, Tverdal A, De Angelis PM, Farstad W, Håård M, Kommisrud E. Alterations of sperm DNA integrity during cryopreservation procedure and in vitro incubation of bull semen. Anim Reprod Sci. 2010;117:34–42. doi: 10.1016/j.anireprosci.2009.04.011. [DOI] [PubMed] [Google Scholar]
- 63.Feugang JM, Rodriguez-Osorio N, Kaya A, Wang H, Page G, Ostermeier GC, Topper EK, Memili E. Transcriptome analysis of bull spermatozoa: implications for male fertility. Reprod Biomed Online. 2010;21:312–24. doi: 10.1016/j.rbmo.2010.06.022. [DOI] [PubMed] [Google Scholar]
- 64.Gilbert I, Bissonnette N, Boissonneault G, Vallée M, Robert C. A molecular analysis of the population of mRNA in bovine spermatozoa. Reproduction. 2007;133:1073–86. doi: 10.1530/REP-06-0292. [DOI] [PubMed] [Google Scholar]
- 65.Govindaraju A, Uzun A, Robertson L, Atli MO, Kaya A, Topper E, Crate EA, Padbury J, Perkins A, Memili E. Dynamics of microRNAs in bull spermatozoa. Reprod Biol Endocrinol. 2012;10:82. doi: 10.1186/1477-7827-10-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kobayashi H, Hiura H, John RM, Sato A, Otsu E, Kobayashi N, Suzuki R, Suzuki F, Hayashi C, Utsunomiya T, et al. DNA methylation errors at imprinted loci after assisted conception originate in the parental sperm. Eur J Hum Genet. 2009;17:1582–91. doi: 10.1038/ejhg.2009.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hammoud SS, Purwar J, Pflueger C, Cairns BR, Carrell DT. Alterations in sperm DNA methylation patterns at imprinted loci in two classes of infertility. Fertil Steril. 2010;94:1728–33. doi: 10.1016/j.fertnstert.2009.09.010. [DOI] [PubMed] [Google Scholar]
- 68.El Hajj N, Zechner U, Schneider E, Tresch A, Gromoll J, Hahn T, Schorsch M, Haaf T. Methylation status of imprinted genes and repetitive elements in sperm DNA from infertile males. Sex Dev. 2011;5:60–9. doi: 10.1159/000323806. [DOI] [PubMed] [Google Scholar]
- 69.El Hajj N, Haaf T. Epigenetic disturbances in in vitro cultured gametes and embryos: implications for human assisted reproduction. Fertil Steril. 2013;99:632–41. doi: 10.1016/j.fertnstert.2012.12.044. [DOI] [PubMed] [Google Scholar]
- 70.de Waal E, Yamazaki Y, Ingale P, Bartolomei M, Yanagimachi R, McCarrey JR. Primary epimutations introduced during intracytoplasmic sperm injection (ICSI) are corrected by germline-specific epigenetic reprogramming. Proc Natl Acad Sci U S A. 2012;109:4163–8. doi: 10.1073/pnas.1201990109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vajta G, Rienzi L, Cobo A, Yovich J. Embryo culture: can we perform better than nature? Reprod Biomed Online. 2010;20:453–69. doi: 10.1016/j.rbmo.2009.12.018. [DOI] [PubMed] [Google Scholar]
- 72.Lane M, Gardner DK. Embryo culture medium: which is the best? Best Pract Res Clin Obstet Gynaecol. 2007;21:83–100. doi: 10.1016/j.bpobgyn.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 73.Rienzi L, Vajta G, Ubaldi F. New culture devices in ART. Placenta. 2011;32(Suppl 3):S248–51. doi: 10.1016/j.placenta.2011.06.018. [DOI] [PubMed] [Google Scholar]
- 74.Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293:1089–93. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- 75.Haaf T. Methylation dynamics in the early mammalian embryo: implications of genome reprogramming defects for development. Curr Top Microbiol Immunol. 2006;310:13–22. doi: 10.1007/3-540-31181-5_2. [DOI] [PubMed] [Google Scholar]
- 76.Dobbs KB, Rodriguez M, Sudano MJ, Ortega MS, Hansen PJ. Dynamics of DNA methylation during early development of the preimplantation bovine embryo. PLoS One. 2013;8:e66230. doi: 10.1371/journal.pone.0066230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hou J, Liu L, Lei T, Cui X, An X, Chen Y. Genomic DNA methylation patterns in bovine preimplantation embryos derived from in vitro fertilization. Sci China C Life Sci. 2007;50:56–61. doi: 10.1007/s11427-007-0003-7. [DOI] [PubMed] [Google Scholar]
- 78.Suzuki J, Jr., Therrien J, Filion F, Lefebvre R, Goff AK, Smith LC. In vitro culture and somatic cell nuclear transfer affect imprinting of SNRPN gene in pre- and post-implantation stages of development in cattle. BMC Dev Biol. 2009;9:9. doi: 10.1186/1471-213X-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Luedi PP, Dietrich FS, Weidman JR, Bosko JM, Jirtle RL, Hartemink AJ. Computational and experimental identification of novel human imprinted genes. Genome Res. 2007;17:1723–30. doi: 10.1101/gr.6584707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zaitseva I, Zaitsev S, Alenina N, Bader M, Krivokharchenko A. Dynamics of DNA-demethylation in early mouse and rat embryos developed in vivo and in vitro. Mol Reprod Dev. 2007;74:1255–61. doi: 10.1002/mrd.20704. [DOI] [PubMed] [Google Scholar]
- 81.Doherty AS, Mann MR, Tremblay KD, Bartolomei MS, Schultz RM. Differential effects of culture on imprinted H19 expression in the preimplantation mouse embryo. Biol Reprod. 2000;62:1526–35. doi: 10.1095/biolreprod62.6.1526. [DOI] [PubMed] [Google Scholar]
- 82.Khosla S, Dean W, Brown D, Reik W, Feil R. Culture of preimplantation mouse embryos affects fetal development and the expression of imprinted genes. Biol Reprod. 2001;64:918–26. doi: 10.1095/biolreprod64.3.918. [DOI] [PubMed] [Google Scholar]
- 83.Khatib H, Zaitoun I, Kim ES. Comparative analysis of sequence characteristics of imprinted genes in human, mouse, and cattle. Mamm Genome. 2007;18:538–47. doi: 10.1007/s00335-007-9039-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.O’Doherty AM, O’Shea LC, Fair T. Bovine DNA methylation imprints are established in an oocyte size-specific manner, which are coordinated with the expression of the DNMT3 family proteins. Biol Reprod. 2012;86:67. doi: 10.1095/biolreprod.111.094946. [DOI] [PubMed] [Google Scholar]
- 85.Zaitoun I, Khatib H. Assessment of genomic imprinting of SLC38A4, NNAT, NAP1L5, and H19 in cattle. BMC Genet. 2006;7:49. doi: 10.1186/1471-2156-7-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Killian JK, Nolan CM, Wylie AA, Li T, Vu TH, Hoffman AR, Jirtle RL. Divergent evolution in M6P/IGF2R imprinting from the Jurassic to the Quaternary. Hum Mol Genet. 2001;10:1721–8. doi: 10.1093/hmg/10.17.1721. [DOI] [PubMed] [Google Scholar]
- 87.Khatib H. Imprinting of Nesp55 gene in cattle. Mamm Genome. 2004;15:663–7. doi: 10.1007/s00335-004-2331-2. [DOI] [PubMed] [Google Scholar]
- 88.Ruddock NT, Wilson KJ, Cooney MA, Korfiatis NA, Tecirlioglu RT, French AJ. Analysis of imprinted messenger RNA expression during bovine preimplantation development. Biol Reprod. 2004;70:1131–5. doi: 10.1095/biolreprod.103.022236. [DOI] [PubMed] [Google Scholar]
- 89.Kim J, Bergmann A, Choo JH, Stubbs L. Genomic organization and imprinting of the Peg3 domain in bovine. Genomics. 2007;90:85–92. doi: 10.1016/j.ygeno.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 90.Kim J, Bergmann A, Lucas S, Stone R, Stubbs L. Lineage-specific imprinting and evolution of the zinc-finger gene ZIM2. Genomics. 2004;84:47–58. doi: 10.1016/j.ygeno.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 91.Dindot SV, Farin PW, Farin CE, Romano J, Walker S, Long C, Piedrahita JA. Epigenetic and genomic imprinting analysis in nuclear transfer derived Bos gaurus/Bos taurus hybrid fetuses. Biol Reprod. 2004;71:470–8. doi: 10.1095/biolreprod.103.025775. [DOI] [PubMed] [Google Scholar]
- 92.Hou XH, Li DJ, Su H, Hu JQ, Li N, Li SJ. Molecular cloning, expression, and imprinting status of maternally expressed gene 8 (Meg8) in dairy cattle. Genetika. 2011;47:1120–5. [PubMed] [Google Scholar]
- 93.Zhang S, Kubota C, Yang L, Zhang Y, Page R, O’Neill M, Yang X, Tian XC. Genomic imprinting of H19 in naturally reproduced and cloned cattle. Biol Reprod. 2004;71:1540–4. doi: 10.1095/biolreprod.104.031807. [DOI] [PubMed] [Google Scholar]
- 94.Sikora KM, Magee DA, Berkowicz EW, Lonergan P, Evans AC, Carter F, Comte A, Waters SM, MacHugh DE, Spillane C. PHLDA2 is an imprinted gene in cattle. Anim Genet. 2012;43:587–90. doi: 10.1111/j.1365-2052.2011.02292.x. [DOI] [PubMed] [Google Scholar]
- 95.Xue F, Tian XC, Du F, Kubota C, Taneja M, Dinnyes A, Dai Y, Levine H, Pereira LV, Yang X. Aberrant patterns of X chromosome inactivation in bovine clones. Nat Genet. 2002;31:216–20. doi: 10.1038/ng900. [DOI] [PubMed] [Google Scholar]
- 96.Enright BP, Jeong BS, Yang X, Tian XC. Epigenetic characteristics of bovine donor cells for nuclear transfer: levels of histone acetylation. Biol Reprod. 2003;69:1525–30. doi: 10.1095/biolreprod.103.019950. [DOI] [PubMed] [Google Scholar]
- 97.Dahl JA, Collas P. Q2ChIP, a quick and quantitative chromatin immunoprecipitation assay, unravels epigenetic dynamics of developmentally regulated genes in human carcinoma cells. Stem Cells. 2007;25:1037–46. doi: 10.1634/stemcells.2006-0430. [DOI] [PubMed] [Google Scholar]
- 98.Dahl JA, Collas P. MicroChIP--a rapid micro chromatin immunoprecipitation assay for small cell samples and biopsies. Nucleic Acids Res. 2008;36:e15. doi: 10.1093/nar/gkm1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Dahl JA, Reiner AH, Klungland A, Wakayama T, Collas P. Histone H3 lysine 27 methylation asymmetry on developmentally-regulated promoters distinguish the first two lineages in mouse preimplantation embryos. PLoS One. 2010;5:e9150. doi: 10.1371/journal.pone.0009150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Herrmann D, Dahl JA, Lucas-Hahn A, Collas P, Niemann H. Histone modifications and mRNA expression in the inner cell mass and trophectoderm of bovine blastocysts. Epigenetics. 2013;8:281–9. doi: 10.4161/epi.23899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Monteiro FM, Oliveira CS, Oliveira LZ, Saraiva NZ, Mercadante ME, Lopes FL, Arnold DR, Garcia JM. Chromatin modifying agents in the in vitro production of bovine embryos. Vet Med Int. 2010;2011 doi: 10.4061/2011/694817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Maalouf WE, Alberio R, Campbell KHS. Differential acetylation of histone H4 lysine during development of in vitro fertilized, cloned and parthenogenetically activated bovine embryos. Epigenetics. 2008;3:199–209. doi: 10.4161/epi.3.4.6497. [DOI] [PubMed] [Google Scholar]
- 103.Wu X, Li Y, Xue L, Wang L, Yue Y, Li K, Bou S, Li GP, Yu H. Multiple histone site epigenetic modifications in nuclear transfer and in vitro fertilized bovine embryos. Zygote. 2011;19:31–45. doi: 10.1017/S0967199410000328. [DOI] [PubMed] [Google Scholar]
- 104.Rodriguez-Osorio N, Wang H, Rupinski J, Bridges SM, Memili E. Comparative functional genomics of mammalian DNA methyltransferases. Reprod Biomed Online. 2010;20:243–55. doi: 10.1016/j.rbmo.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 105.Kues WA, Sudheer S, Herrmann D, Carnwath JW, Havlice V, Besenfelder U, Lehrach H, Adjaye J, Niemann H. Genome-wide expression profiling reveals distinct clusters of transcriptional regulation during bovine preimplantation development in vivo. Proc Natl Acad Sci U S A. 2008;105:19768–73. doi: 10.1073/pnas.0805616105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Velazquez MA, Parrilla I, Van Soom A, Verberckmoes S, Kues W, Niemann H. Sampling techniques for oviductal and uterine luminal fluid in cattle. Theriogenology. 2010;73:758–67. doi: 10.1016/j.theriogenology.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 107.Gad A, Schellander K, Hoelker M, Tesfaye D. Transcriptome profile of early mammalian embryos in response to culture environment. Anim Reprod Sci. 2012;134:76–83. doi: 10.1016/j.anireprosci.2012.08.014. [DOI] [PubMed] [Google Scholar]
- 108.Farin PW, Piedrahita JA, Farin CE. Errors in development of fetuses and placentas from in vitro-produced bovine embryos. Theriogenology. 2006;65:178–91. doi: 10.1016/j.theriogenology.2005.09.022. [DOI] [PubMed] [Google Scholar]
- 109.Nowak-Imialek M, Wrenzycki C, Herrmann D, Lucas-Hahn A, Lagutina I, Lemme E, Lazzari G, Galli C, Niemann H. Messenger RNA expression patterns of histone-associated genes in bovine preimplantation embryos derived from different origins. Mol Reprod Dev. 2008;75:731–43. doi: 10.1002/mrd.20816. [DOI] [PubMed] [Google Scholar]
- 110.Young LE, Fernandes K, McEvoy TG, Butterwith SC, Gutierrez CG, Carolan C, Broadbent PJ, Robinson JJ, Wilmut I, Sinclair KD. Epigenetic change in IGF2R is associated with fetal overgrowth after sheep embryo culture. Nat Genet. 2001;27:153–4. doi: 10.1038/84769. [DOI] [PubMed] [Google Scholar]
- 111.Wrenzycki C, Herrmann D, Lucas-Hahn A, Korsawe K, Lemme E, Niemann H. Messenger RNA expression patterns in bovine embryos derived from in vitro procedures and their implications for development. Reprod Fertil Dev. 2005;17:23–35. doi: 10.1071/RD04109. [DOI] [PubMed] [Google Scholar]
- 112.Long JE, Cai X, He LQ. Gene profiling of cattle blastocysts derived from nuclear transfer, in vitro fertilization and in vivo development based on cDNA library. Anim Reprod Sci. 2007;100:243–56. doi: 10.1016/j.anireprosci.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 113.Perecin F, Méo SC, Yamazaki W, Ferreira CR, Merighe GK, Meirelles FV, Garcia JM. Imprinted gene expression in in vivo- and in vitro-produced bovine embryos and chorio-allantoic membranes. Genet Mol Res. 2009;8:76–85. doi: 10.4238/vol8-1gmr541. [DOI] [PubMed] [Google Scholar]
- 114.Sugimura S, Akai T, Hashiyada Y, Aikawa Y, Ohtake M, Matsuda H, Kobayashi S, Kobayashi E, Konishi K, Imai K. Effect of embryo density on in vitro development and gene expression in bovine in vitro-fertilized embryos cultured in a microwell system. J Reprod Dev. 2013;59:115–22. doi: 10.1262/jrd.2012-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lane M, Gardner DK. Ammonium induces aberrant blastocyst differentiation, metabolism, pH regulation, gene expression and subsequently alters fetal development in the mouse. Biol Reprod. 2003;69:1109–17. doi: 10.1095/biolreprod.103.018093. [DOI] [PubMed] [Google Scholar]
- 116.Burgoyne PS, Thornhill AR, Boudrean SK, Darling SM, Bishop CE, Evans EP. The genetic basis of XX-XY differences present before gonadal sex differentiation in the mouse. Philos Trans R Soc Lond B Biol Sci. 1995;350:253–60, discussion 260-1. doi: 10.1098/rstb.1995.0159. [DOI] [PubMed] [Google Scholar]
- 117.Gutiérrez-Adán A, Perez-Crespo M, Fernandez-Gonzalez R, Ramirez MA, Moreira P, Pintado B, Lonergan P, Rizos D. Developmental consequences of sexual dimorphism during pre-implantation embryonic development. Reprod Domest Anim. 2006;41(Suppl 2):54–62. doi: 10.1111/j.1439-0531.2006.00769.x. [DOI] [PubMed] [Google Scholar]
- 118.Bermejo-Alvarez P, Rizos D, Rath D, Lonergan P, Gutierrez-Adan A. Epigenetic differences between male and female bovine blastocysts produced in vitro. Physiol Genomics. 2008;32:264–72. doi: 10.1152/physiolgenomics.00234.2007. [DOI] [PubMed] [Google Scholar]
- 119.Wrenzycki C, Lucas-Hahn A, Herrmann D, Lemme E, Korsawe K, Niemann H. In vitro production and nuclear transfer affect dosage compensation of the X-linked gene transcripts G6PD, PGK, and Xist in preimplantation bovine embryos. Biol Reprod. 2002;66:127–34. doi: 10.1095/biolreprod66.1.127. [DOI] [PubMed] [Google Scholar]
- 120.Saraiva NZ, Oliveira CS, Tetzner TAD, de Lima MR, de Melo DS, Niciura SC, Garcia JM. Chemically assisted enucleation results in higher G6PD expression in early bovine female embryos obtained by somatic cell nuclear transfer. Cell Reprogram. 2012;14:425–35. doi: 10.1089/cell.2011.0077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Oliveira CS, Saraiva NZ, Cruz MHC, Mazeti B, Oliveira LZ, Lopes FL, Garcia JM. HDAC inhibition decreases XIST expression on female IVP bovine blastocysts. Reproduction. 2013;145:9–17. doi: 10.1530/REP-11-0343. [DOI] [PubMed] [Google Scholar]
- 122.Stroud B, Callesen H. IETS statement on worldwide ET statistics for 2010. Anim Reprod 2012: 210–216. [Google Scholar]
- 123.Park SY, Kim EY, Cui XS, Tae JC, Lee WD, Kim NH, Park SP, Lim JH. Increase in DNA fragmentation and apoptosis-related gene expression in frozen-thawed bovine blastocysts. Zygote. 2006;14:125–31. doi: 10.1017/S0967199406003649. [DOI] [PubMed] [Google Scholar]
- 124.Camargo LSA, Boite MC, Wohlres-Viana S, Mota GB, Serapiao RV, Sa WF, Viana JH, Nogueira LA. Osmotic challenge and expression of aquaporin 3 and Na/K ATPase genes in bovine embryos produced in vitro. Cryobiology. 2011;63:256–62. doi: 10.1016/j.cryobiol.2011.09.135. [DOI] [PubMed] [Google Scholar]
- 125.Räty M, Ketoja E, Pitkänen T, Ahola V, Kananen K, Peippo J. In vitro maturation supplements affect developmental competence of bovine cumulus-oocyte complexes and embryo quality after vitrification. Cryobiology. 2011;63:245–55. doi: 10.1016/j.cryobiol.2011.09.134. [DOI] [PubMed] [Google Scholar]
- 126.Aksu DA, Agca C, Aksu S, Bagis H, Akkoc T, Caputcu AT, Arat S, Taskin AC, Kizil SH, Karasahin T, et al. Gene expression profiles of vitrified in vitro- and in vivo-derived bovine blastocysts. Mol Reprod Dev. 2012;79:613–25. doi: 10.1002/mrd.22068. [DOI] [PubMed] [Google Scholar]
- 127.Wang Z, Xu L, He F. Embryo vitrification affects the methylation of the H19/Igf2 differentially methylated domain and the expression of H19 and Igf2. Fertil Steril. 2010;93:2729–33. doi: 10.1016/j.fertnstert.2010.03.025. [DOI] [PubMed] [Google Scholar]
- 128.Zhao XM, Ren JJ, Du WH, Hao HS, Wang D, Qin T, Liu Y, Zhu HB. Effect of vitrification on promoter CpG island methylation patterns and expression levels of DNA methyltransferase 1o, histone acetyltransferase 1, and deacetylase 1 in metaphase II mouse oocytes. Fertil Steril. 2013;100:256–61. doi: 10.1016/j.fertnstert.2013.03.009. [DOI] [PubMed] [Google Scholar]
- 129.Stinshoff H, Wilkening S, Hanstedt A, Brüning K, Wrenzycki C. Cryopreservation affects the quality of in vitro produced bovine embryos at the molecular level. Theriogenology. 2011;76:1433–41. doi: 10.1016/j.theriogenology.2011.06.013. [DOI] [PubMed] [Google Scholar]
- 130.Hu W, Marchesi D, Qiao J, Feng HL. Effect of slow freeze versus vitrification on the oocyte: an animal model. Fertil Steril. 2012;98:752–60, e3. doi: 10.1016/j.fertnstert.2012.05.037. [DOI] [PubMed] [Google Scholar]
- 131.Zhao XM, Ren JJ, Du WH, Hao HS, Wang D, Liu Y, Qin T, Zhu HB. Effect of 5-aza-2′-deoxycytidine on methylation of the putative imprinted control region of H19 during the in vitro development of vitrified bovine two-cell embryos. Fertil Steril. 2012;98:222–7. doi: 10.1016/j.fertnstert.2012.04.014. [DOI] [PubMed] [Google Scholar]
- 132.Kruip TA, Bevers MM, Kemp B. Environment of oocyte and embryo determines health of IVP offspring. Theriogenology. 2000;53:611–8. doi: 10.1016/S0093-691X(99)00261-7. [DOI] [PubMed] [Google Scholar]
- 133.Bertolini M, Mason JB, Beam SW, Carneiro GF, Sween ML, Kominek DJ, Moyer AL, Famula TR, Sainz RD, Anderson GB. Morphology and morphometry of in vivo- and in vitro-produced bovine concepti from early pregnancy to term and association with high birth weights. Theriogenology. 2002;58:973–94. doi: 10.1016/S0093-691X(02)00935-4. [DOI] [PubMed] [Google Scholar]
- 134.Young LE, Sinclair KD, Wilmut I. Large offspring syndrome in cattle and sheep. Rev Reprod. 1998;3:155–63. doi: 10.1530/ror.0.0030155. [DOI] [PubMed] [Google Scholar]
- 135.Wilmut I, Schnieke AE, McWhir J, Kind AJ, Campbell KH. Viable offspring derived from fetal and adult mammalian cells. Nature. 1997;385:810–3. doi: 10.1038/385810a0. [DOI] [PubMed] [Google Scholar]
- 136.Wilmut I, Beaujean N, de Sousa PA, Dinnyes A, King TJ, Paterson LA, Wells DN, Young LE. Somatic cell nuclear transfer. Nature. 2002;419:583–6. doi: 10.1038/nature01079. [DOI] [PubMed] [Google Scholar]
- 137.Wakayama T, Perry ACF, Zuccotti M, Johnson KR, Yanagimachi R. Full-term development of mice from enucleated oocytes injected with cumulus cell nuclei. Nature. 1998;394:369–74. doi: 10.1038/28615. [DOI] [PubMed] [Google Scholar]
- 138.Eggan K, Akutsu H, Loring J, Jackson-Grusby L, Klemm M, Rideout WM, 3rd, Yanagimachi R, Jaenisch R. Hybrid vigor, fetal overgrowth, and viability of mice derived by nuclear cloning and tetraploid embryo complementation. Proc Natl Acad Sci U S A. 2001;98:6209–14. doi: 10.1073/pnas.101118898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Fernández-Gonzalez R, Moreira P, Bilbao A, Jiménez A, Pérez-Crespo M, Ramírez MA, Rodríguez De Fonseca F, Pintado B, Gutiérrez-Adán A. Long-term effect of in vitro culture of mouse embryos with serum on mRNA expression of imprinting genes, development, and behavior. Proc Natl Acad Sci U S A. 2004;101:5880–5. doi: 10.1073/pnas.0308560101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Machaty Z, Peippo J, Peter A. Production and manipulation of bovine embryos: techniques and terminology. Theriogenology. 2012;78:937–50. doi: 10.1016/j.theriogenology.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 141.Tervit HR, Whittingham DG, Rowson LE. Successful culture in vitro of sheep and cattle ova. J Reprod Fertil. 1972;30:493–7. doi: 10.1530/jrf.0.0300493. [DOI] [PubMed] [Google Scholar]
- 142.Rosenkrans CF, Jr., Zeng GQ, MCNamara GT, Schoff PK, First NL. Development of bovine embryos in vitro as affected by energy substrates. Biol Reprod. 1993;49:459–62. doi: 10.1095/biolreprod49.3.459. [DOI] [PubMed] [Google Scholar]
- 143.Walker SK, Hartwich KM, Seamark RF. The production of unusually large offspring following embryo manipulation: Concepts and challenges. Theriogenology. 1996;45:111–20. doi: 10.1016/0093-691X(95)00360-K. [DOI] [Google Scholar]
- 144.Sinclair KD, McEvoy TG, Maxfield EK, Maltin CA, Young LE, Wilmut I, Broadbent PJ, Robinson JJ. Aberrant fetal growth and development after in vitro culture of sheep zygotes. J Reprod Fertil. 1999;116:177–86. doi: 10.1530/jrf.0.1160177. [DOI] [PubMed] [Google Scholar]
- 145.van Wagtendonk-de Leeuw AM, Mullaart E, de Roos AP, Merton JS, den Daas JH, Kemp B, de Ruigh L. Effects of different reproduction techniques: AI MOET or IVP, on health and welfare of bovine offspring. Theriogenology. 2000;53:575–97. doi: 10.1016/S0093-691X(99)00259-9. [DOI] [PubMed] [Google Scholar]
- 146.Farin PW, Farin CE. Transfer of bovine embryos produced in vivo or in vitro: survival and fetal development. Biol Reprod. 1995;52:676–82. doi: 10.1095/biolreprod52.3.676. [DOI] [PubMed] [Google Scholar]
- 147.Sangild PT, Schmidt M, Jacobsen H, Fowden AL, Forhead A, Avery B, Greve T. Blood chemistry, nutrient metabolism, and organ weights in fetal and newborn calves derived from in vitro-produced bovine embryos. Biol Reprod. 2000;62:1495–504. doi: 10.1095/biolreprod62.6.1495. [DOI] [PubMed] [Google Scholar]
- 148.Maxfield EK, Sinclair KD, Dolman DF, Staines ME, Maltin CA. In vitro culture of sheep embryos increases weight, primary fiber size and secondary to primary fiber ratio in fetal muscle at day 61 of gestation. Theriogenology. 1997;47 doi: 10.1016/S0093-691X(97)82503-4. [DOI] [Google Scholar]
- 149.Crosier AE, Farin CE, Rodriguez KF, Blondin P, Alexander JE, Farin PW. Development of skeletal muscle and expression of candidate genes in bovine fetuses from embryos produced in vivo or in vitro. Biol Reprod. 2002;67:401–8. doi: 10.1095/biolreprod67.2.401. [DOI] [PubMed] [Google Scholar]
- 150.Farin PW, Crosier AE, Farin CE. Influence of in vitro systems on embryo survival and fetal development in cattle. Theriogenology. 2001;55:151–70. doi: 10.1016/S0093-691X(00)00452-0. [DOI] [PubMed] [Google Scholar]
- 151.Farin CE, Alexander JE, Farin PW. Expression of messenger RNAs for insulin-like growth factors and their receptors in bovine fetuses at early gestation from embryos produced in vivo or in vitro. Theriogenology. 2010;74:1288–95. doi: 10.1016/j.theriogenology.2010.05.035. [DOI] [PubMed] [Google Scholar]
- 152.Baker J, Liu JP, Robertson EJ, Efstratiadis A. Role of insulin-like growth factors in embryonic and postnatal growth. Cell. 1993;75:73–82. doi: 10.1016/0092-8674(93)90680-O. [DOI] [PubMed] [Google Scholar]
- 153.Constância M, Hemberger M, Hughes J, Dean W, Ferguson-Smith A, Fundele R, Stewart F, Kelsey G, Fowden A, Sibley C, et al. Placental-specific IGF-II is a major modulator of placental and fetal growth. Nature. 2002;417:945–8. doi: 10.1038/nature00819. [DOI] [PubMed] [Google Scholar]
- 154.Wang ZQ, Fung MR, Barlow DP, Wagner EF. Regulation of embryonic growth and lysosomal targeting by the imprinted Igf2/Mpr gene. Nature. 1994;372:464–7. doi: 10.1038/372464a0. [DOI] [PubMed] [Google Scholar]
- 155.Farmer WT, Farin PW, Piedrahita JA, Bischoff SR, Farin CE. Expression of antisense of insulin-like growth factor-2 receptor RNA non-coding (AIRN) during early gestation in cattle. Anim Reprod Sci. 2013;138:64–73. doi: 10.1016/j.anireprosci.2013.01.009. [DOI] [PubMed] [Google Scholar]
- 156.Walker SK, Hartwich KM, Robinson JS. Long-term effects on offspring of exposure of oocytes and embryos to chemical and physical agents. Hum Reprod Update. 2000;6:564–77. doi: 10.1093/humupd/6.6.564. [DOI] [PubMed] [Google Scholar]
- 157.Farin CE, Farin PW, Piedrahita JA. Development of fetuses from in vitro-produced and cloned bovine embryos. J Anim Sci. 2004;82 E-Suppl(E-Suppl):E53–62. doi: 10.2527/2004.8213_supplE53x. [DOI] [PubMed] [Google Scholar]
- 158.Wrenzycki C, Niemann H. Epigenetic reprogramming in early embryonic development: effects of in-vitro production and somatic nuclear transfer. Reprod Biomed Online. 2003;7:649–56. doi: 10.1016/S1472-6483(10)62087-1. [DOI] [PubMed] [Google Scholar]
- 159.Hiendleder S, Wirtz M, Mund C, Klempt M, Reichenbach HD, Stojkovic M, Weppert M, Wenigerkind H, Elmlinger M, Lyko F, et al. Tissue-specific effects of in vitro fertilization procedures on genomic cytosine methylation levels in overgrown and normal sized bovine fetuses. Biol Reprod. 2006;75:17–23. doi: 10.1095/biolreprod.105.043919. [DOI] [PubMed] [Google Scholar]
- 160.Gosden R, Trasler J, Lucifero D, Faddy M. Rare congenital disorders, imprinted genes, and assisted reproductive technology. Lancet. 2003;361:1975–7. doi: 10.1016/S0140-6736(03)13592-1. [DOI] [PubMed] [Google Scholar]
- 161.Lucifero D, Suzuki J, Bordignon V, Martel J, Vigneault C, Therrien J, Filion F, Smith LC, Trasler JM. Bovine SNRPN methylation imprint in oocytes and day 17 in vitro-produced and somatic cell nuclear transfer embryos. Biol Reprod. 2006;75:531–8. doi: 10.1095/biolreprod.106.051722. [DOI] [PubMed] [Google Scholar]
- 162.Hori N, Nagai M, Hirayama M, Hirai T, Matsuda K, Hayashi M, Tanaka T, Ozawa T, Horike S. Aberrant CpG methylation of the imprinting control region KvDMR1 detected in assisted reproductive technology-produced calves and pathogenesis of large offspring syndrome. Anim Reprod Sci. 2010;122:303–12. doi: 10.1016/j.anireprosci.2010.09.008. [DOI] [PubMed] [Google Scholar]
- 163.Robbins KM, Chen Z, Wells KD, Rivera RM. Expression of KCNQ1OT1, CDKN1C, H19, and PLAGL1 and the methylation patterns at the KvDMR1 and H19/IGF2 imprinting control regions is conserved between human and bovine. J Biomed Sci. 2012;19:95. doi: 10.1186/1423-0127-19-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Chen Z, Robbins KM, Wells KD, Rivera RM. Large offspring syndrome: a bovine model for the human loss-of-imprinting overgrowth syndrome Beckwith-Wiedemann. Epigenetics. 2013;8:591–601. doi: 10.4161/epi.24655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Kruip TAM, den Daas JHG. In vitro produced and cloned embryos: Effects on pregnancy, parturition and offspring. Theriogenology. 1997;47:43–52. doi: 10.1016/S0093-691X(96)00338-X. [DOI] [Google Scholar]
- 166.Numabe T, Oikawa T, Kikuchi T, Horiuchi T. Birth weight and gestation length of Japanese black calves following transfer of embryos produced in vitro with or without co-culture. J Vet Med Sci. 2001;63:515–9. doi: 10.1292/jvms.63.515. [DOI] [PubMed] [Google Scholar]
- 167.Yang BS, Im GS, Park SJ. Characteristics of Korean native, Hanwoo, calves produced by transfer of in vitro produced embryos. Anim Reprod Sci. 2001;67:153–8. doi: 10.1016/S0378-4320(01)00125-7. [DOI] [PubMed] [Google Scholar]
- 168.Camargo LSA, Freitas C, de Sa WF, de Moraes Ferreira A, Serapiao RV, Viana JHM. Gestation length, birth weight and offspring gender ratio of in vitro-produced Gyr (Bos indicus) cattle embryos. Anim Reprod Sci. 2010;120:10–5. doi: 10.1016/j.anireprosci.2010.02.013. [DOI] [PubMed] [Google Scholar]