Abstract
Bromodomain- and PHD finger-containing protein 1 (BRPF1) is a unique epigenetic regulator that contains multiple structural domains for recognizing different chromatin modifications. In addition, it possesses sequence motifs for forming multiple complexes with three different histone acetyltransferases, MOZ, MORF, and HBO1. Within these complexes, BRPF1 serves as a scaffold for bridging subunit interaction, stimulating acetyltransferase activity, governing substrate specificity and stimulating gene expression. To investigate how these molecular interactions are extrapolated to biological functions of BRPF1, we utilized a mouse strain containing a knock-in reporter and analyzed the spatiotemporal expression from embryos to adults. The analysis revealed dynamic expression in the extraembryonic, embryonic, and fetal tissues, suggesting important roles of Brpf1 in prenatal development. In support of this, inactivation of the mouse Brpf1 gene causes lethality around embryonic day 9.5. After birth, high expression is present in the testis and specific regions of the brain. The 4-dimensional expression atlas of mouse Brpf1 should serve as a valuable guide for analyzing its interaction with Moz, Morf, and Hbo1 in vivo, as well as for investigating whether Brpf1 functions independently of these three enzymatic epigenetic regulators.
Keywords: epigenetic reader, histone acetyltransferase, KAT6A, KAT6B, KAT7, BRPF1
Introduction
Histone modifications are frequently recognized by “readers” that contain structural modules, including the bromodomain, the PHD finger and the PWWP domain.1-3 Composed of multiple such domains,4,5 bromodomain- and PHD finger-containing protein 1 (BRPF1) was initially identified as BR140 (bromodomain protein with an estimated molecular weight of 140 kDa) and peregrin.6 During expression screening with a polyclonal antibody against affinity-purified placental integrin,6 BRPF1 was incidentally cloned in 1994 as a zinc finger protein with a bromodomain similar to those two of the 250-kDa subunit (TAFII250, now known as TAF1) of the transcription initiation factor TFIID,6 suggesting a role in chromatin control and transcriptional regulation. Molecular cloning of leukemia-associated fusion partners on chromosomes 10 and 17 (AF10 and AF17, respectively) in the mid-1990s unraveled the interesting feature that they share a Cys/His-rich domain with BR140.7-9 The domain contains two PHD (plant homeodomain-linked) fingers joined by a C2HC zinc knuckle and was later named the PZP (PHD-zinc knuckle-PHD) module.10 In light of this striking sequence feature, new proteins with the module were sought after, thereby leading to the identification of BRL (BR140 like protein), also known as BRD1 (bromodomain protein 1) and BRPF2.11 Afterwards, BRPF3, a third human paralog, was uncovered during genome annotation. These three proteins are highly homologous to each other and form a unique family of multidomain chromatin regulators.1,3,4
The PZP module is present in other chromatin modifiers, including the leukemia-associated methyltransferase NSD1 and the demethylase GASC1,10 suggesting a role in chromatin regulation. In support of this, the PZP modules of BRPF1 and BRPF2 recognize the unmodified N-terminus of histone H312,13 and the second PHD finger of BRPF2 binds directly to DNA.14 The PZP module is located in the middle part, slightly N-terminal to the bromodomain. Located at the C-terminal end is a well-conserved PWWP domain.4 The bromodomain and PWWP domain of BRPF1 have the ability to bind specifically to acetylated and methylated forms of histone H3, respectively.15-17 BRPFs contain two more sequence motifs flanking the PZP module and displaying similarity to EPC (enhancer of polycomb) proteins.10,18 Biochemical purification revealed that, in HeLa cells, BRPFs form tetrameric complexes with the histone acetyltransferase MOZ (or the paralog MORF), ING5 and EAF6.18 The N-terminal enhancer of polycomb (EPC)-like motif may contribute to interaction with MOZ (also known as MYST3 and KAT6A) and MORF (also referred to as MYST4 and KAT6B), while the C-terminal EPC-like motif binds to ING5 (inhibitor of growth 5) and EAF6 (homolog of yeast Esa1-associated factor),18,19 indicating that BRPFs serve as scaffolds for complex formation.19 The PZP module and EPC-like motifs are conserved in JADEs (proteins encoded by genes [J] for apoptosis and differentiation in epithelia), which form similar tetrameric complexes with HBO1 (also referred to as KAT7), ING4/5, and EAF6.18,20 Recent studies have revealed that HBO1 also forms tetrameric complexes with BRPFs, ING4/5, and EAF6.13,21 Strikingly, HBO1 association with JADE1 or BRPF1 switches the substrate specificity from nucleosomal histone H4 to H3,13 indicating a dominant role of BRPF1 in determining the substrate specificity. Thus, BRPF1 and perhaps also its paralogs BRPF2 and BRPF3 promote formation of multiple acetyltransferase complexes and govern their enzymatic activity, substrate specificity, and ability to stimulate transcription. Compared with BRPF2 and BRPF3, BRPF1 possesses an extra potential zinc finger at the N-terminal end, with no known functions. This potential finger is conserved in the sole fly BRPF protein, suggesting a potentially important role.
A crucial question is how molecular interactions of BRPF1 with MOZ, MORF, and HBO1, drawn from biochemical and cell line-based studies,13,18,19 can be extrapolated to functional relevance at the organismal level. For example, one important issue is whether BRPF1 functions globally in all tissues or only in certain tissues at specific developmental stages. The MOZ and MORF genes are rearranged in hematological malignancies and the MORF gene is subject to chromosome translocation in uterine leiomyomata.4,22,23 In addition, MORF is mutated in different developmental disorders, including Noonan syndrome-like disorder,24 Ohdo syndrome,25 and Genitopatellar syndrome,26,27 so another important issue is whether BRPF1 serves a “modifier” of these diseases. Molecular and cell-based studies suggest that this may be the case.13,18,19 Related to these two important issues, very little information is available on when and where mammalian BRPF1 is expressed.4,5
To determine the physiological and pathological functions of mammalian BRPF1, it is necessary to know its spatiotemporal expression. Such information (or the 4-dimensional expression atlas) serves an important roadmap to dissect functions in different tissues through a genetic approach. With these considerations, we utilized a mouse strain containing a knock-in β-galactosidase reporter at the Brpf1 locus and determined the expression from embryos to adults. We found dynamic expression of Brpf1 in the placenta, yolk sac, limb buds, brain, spinal cord, retina, nose, bone, and brown fat at the prenatal stages. After birth, high expression was found in the testis and specific regions of the brain. Moreover, inactivation of the Brpf1 gene results in embryonic lethality, which, together with the 4D expression atlas of mouse Brpf1, suggests that mammalian BRPF1 also has roles independent of the three acetyltransferases MOZ, MORF, and HBO1.
Results
Brpf1 expression during embryonic development
To substantiate our previous molecular studies on the interaction of BRPF1 with MOZ and MORF,18,19 we sought to investigate biological relevance at the tissue and organismal levels. For this, we first utilized a mouse strain containing a promoterless LacZ cassette and two LoxP sites inserted at the Brpf1 locus (Fig. 1A and C).28-30 By use of the convenient X-gal staining to determine β-galactosidase activity, the LacZ cassette serves as a useful and efficient reporter for determination of transcriptional activity from the locus at a single-cell resolution.28-30 To establish the 4-dimensional expression atlas, we first performed whole-mount β-galactosidase staining of mouse concepti or embryos from E8.5–12.75 (Fig. 2). The choice of this time window is due to the observations that organogenesis initiates after E8.0 and the whole-mount staining method works not so well for embryos at or older than E13.5 due to X-gal penetration problems. While no staining was detected in the wild-type E8.5 conceptus (i.e., the embryo proper plus the extraembryonic tissues; Fig. 2A, right), high β-galactosidase activity was detected in the heterozygous mutant embryo and extraembryonic tissues, including the yolk sac and ectoplacental cone (Fig. 2A, left). In the embryo proper, high activity was found in the primitive neural tube and the headfold (Fig. 2A, left). At E9.75, high β-galactosidase activity was also detected in the mutant but not wild-type yolk sac and ectoplacental cone (Fig. 2B; data not shown). Ubiquitous low expression was present in the embryo proper, with enriched expression in the spinal cord, forelimb buds and pharyngeal arches (Fig. 2B).
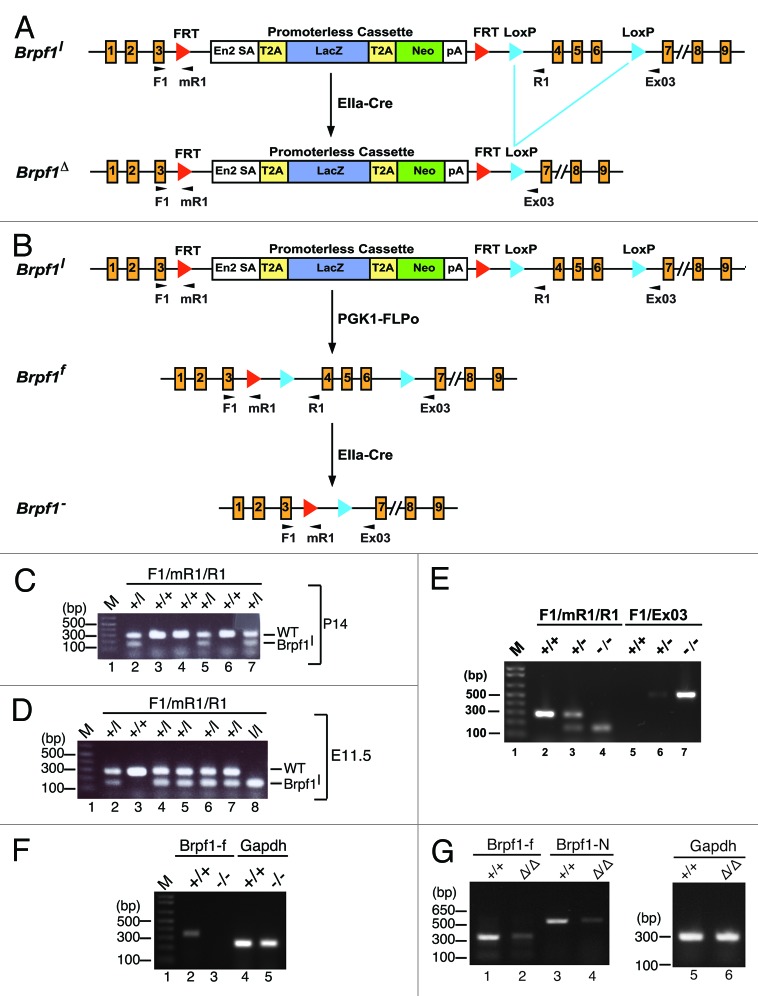
Figure 1. Analysis of the Brpf1l knock-in allele. (A) Schematic representation of the knock-in allele. Within the mutant locus, two FRT sites flank the promoterless LacZ-Neo cassette, which itself contains an En2 splicing acceptor (En2 SA) and two copies of the coding sequence for the porcine teschovirus-1 peptide 2A (T2A) ribosome-skipping signal along with a polyadenylation signal (pA). For Cre-mediated excision, LoxP sites are placed before and after exons 4–6 of the Brpf1 gene. Positions of the three primers used for genotyping are indicated with arrowheads. (B) Generation of the Brpf1-/ - mice. Brpf1l mice were crossed with PGK1-FLPo mice to remove the promoterless cassette and obtain the conditional Brpf1f allele. Through EIIa-Cre mediated recombination, the LoxP-flanked region spanning exons 4–6 was deleted to yield the Brpf1- allele. Mice heterozygous for the Brpf1- allele were mated to yield the mutant homozygotes. The genotyping primers were indicated with tiny arrowheads. (C) Representative genomic PCR analysis of a litter of six two-week old pups from the cross of a Brpf1l/+ heterozygote with a wild-type mouse. Primers Brpf1-F1, -mR1 and -R1, depicted as arrowheads in (A), were used to amplify the wild-type (227 bp) and Brpf1l (162 bp) alleles. According to the result, the litter contained 3 wild-type pups (lanes 3, 4, and 6) and 3 heterozygotes (lanes 2, 5, and 7). (D) Representative PCR genotyping of a litter of 7 embryos (E11.5) from the intercross between Brpf1l/+ male and female mice. The primers were the same as those used in (C). Based on the result, the litter contained one wild-type embryo (lane 3), 5 heterozygotes (lanes 2 and 4–7) and one homozygote (lane 8). (E) Primers Brpf1-F1, -mR1, and -R1 were used to detect the wild type (227 bp) and mutant (162 bp) alleles. Primers Brpf1-F1 and -ex03 were used to amplify the Brpf1- allele (460 bp). M, 100 bp DNA ladder. (F) RT-PCR analysis of Brpf1 mRNA from wild type and Brpf1−/− embryos at E9.5. A 339-bp fragment spanning the floxed exons was amplified to determine the efficiency of EIIa-Cre mediated excision. Gapdh was used as an internal control. (G) RT-PCR analysis of Brpf1 mRNA from wild type and Brpf1△/△embryos at E9.5. A 339-bp fragment spanning the floxed exons and a 577-bp fragment covering the coding sequence for the N-terminal portion of Brpf1 were amplified to determine the efficiency of the knockout-first strategy. Gapdh was used as the internal control.
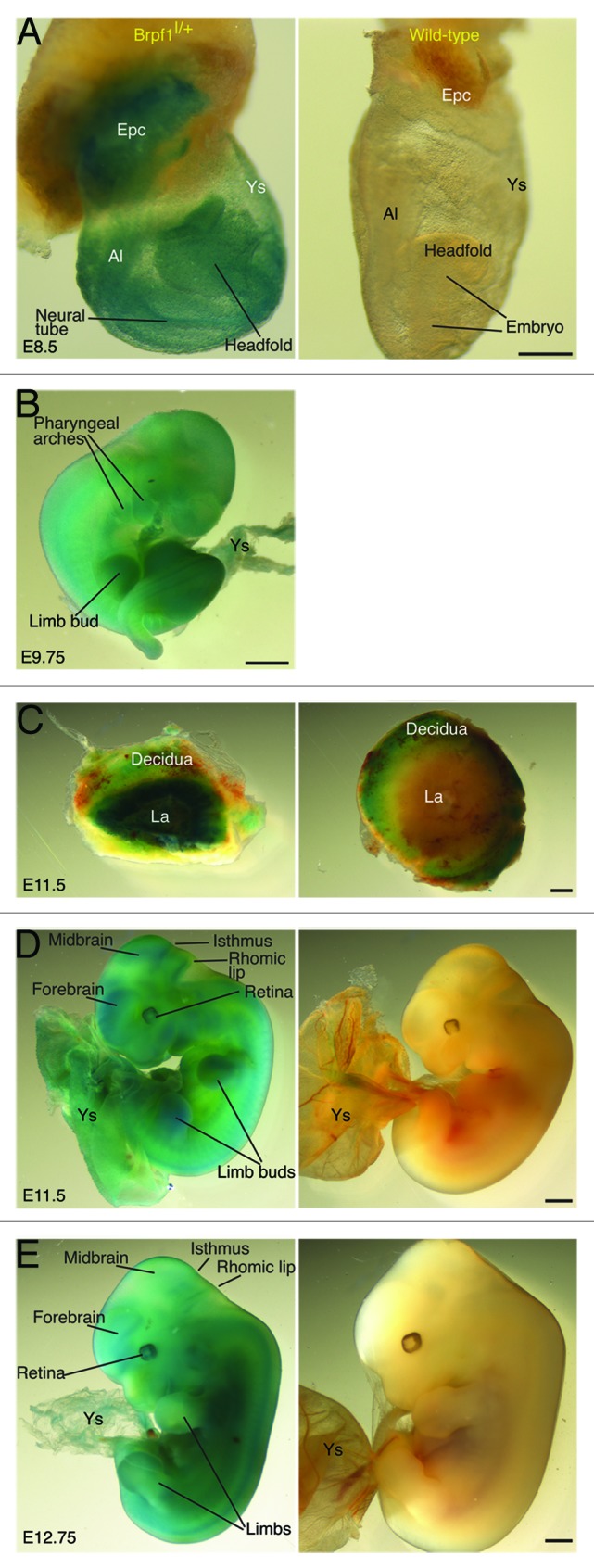
Figure 2. Brpf1 expression from E8.5 to E12.75. The expression was assessed by whole-mount β-galactosidase staining of Brpf1l/+ and wild-type E8.5 concepti (A), E9.75 embryos (B), E11.5 placentae (C) and embryos (D), and E12.75 embryos (E). For E9.75, only the Brpf1l/+ embryo is shown, and no staining was detected in the wild-type embryo, yolk sac or placenta (data not shown). At E8.5, ubiquitous staining was observed in the embryo proper, allantois, ectoplacental cone, and yolk sac (A). At E9.5, Brpf1 was widely expressed in the embryo, with the highest level in the forelimb buds (B). At E11.5, Brpf1 was highly expressed in the placental labyrinth (C, where only one half is shown for the mutant placenta and the wild-type placenta exhibited some non-specific staining at the decidua), limb buds, telencephalic and mesencephalic vesicles, isthmus (midbrain-hindbrain border), rhomic lip (cerebellar primordium), neural tube, eye, and yolk sac (D). The expression remained similar at E12.75, except for the decreased signal in the limbs (E). Abbreviations: Al, allantois; Epc, ectoplacental cone; La, labyrinth; Ys, yolk sac. Scale bars, 200 μm (A) and 500 μm (B–E).
At E11.5, specific expression was detected in the labyrinth but not decidua of the mutant placenta (Fig. 2C), as well as in the mutant yolk sac (Fig. 2D). In the mutant embryo proper, high specific expression was present in the telencephalon (part of the forebrain), mesencephalon (midbrain), isthmus (midbrain-hindbrain border), rhomic lip (cerebellar neuroepithelium), spinal cord, retina, and limb buds (Fig. 2D). About one day later, at E12.75, the expression at the extraembryonbic tissues such as the yolk sac and placenta remained high (Fig. 2E; data not shown). In the embryo itself, expression in the mesencephalon, isthmus and limb buds decreased but the level sustained in the telencephalon, spinal cord, retina, and rhomic lip (Fig. 2E). Thus, the whole-mount staining analysis revealed dynamic expression of Brpf1 in the embryonic and extraembryonic tissues from E8.5–12.75, suggesting potential roles in development of the placenta, yolk sac, brain, spinal cord, limbs, and retina during mouse embryonic development.
Brpf1 expression in embryos, fetuses, and neonates
The whole-mount analysis provided valuable information at the tissue level, but additional investigation is needed to gain insights into the expression at the cellular level. We thus utilized frozen sections for X-gal staining to determine β-galactosidase activity at the single-cell level. At E8.5, X-gal staining of frozen sections revealed strong expression in the ectoplacental cone (Fig. 3A and C), neuroepithelial cells of the neural tube (Fig. 3A and B) and blood islands of the yolk sac (Fig. 3D) from the heterozygous but not wild-type concepti. One day later, at E9.5, specific expression was found in the neuroepithelium of the neural tube (Fig. 4A and B), labyrinth of the placenta (Fig. 4A and C) and blood islands of the yolk sac (Fig. 4D) from the heterozygous but not wild-type concepti (data not shown). Strong expression also appeared in the mutant somites (Fig. 4B).
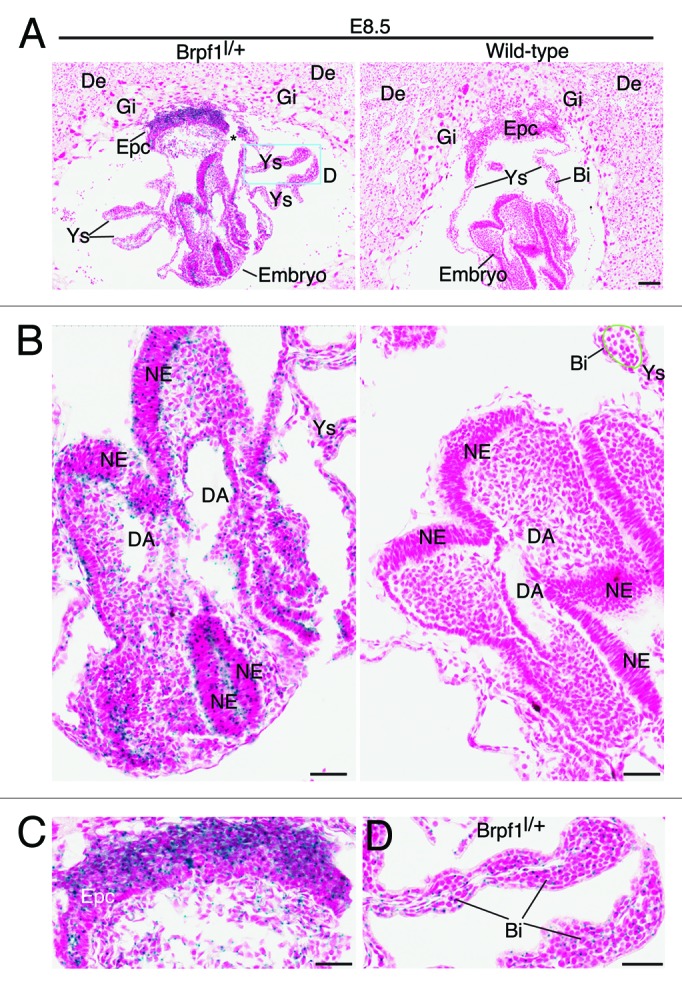
Figure 3. Analysis of Brpf1 expression in sections of E8.5 concepti. (A) β-Galactosidase staining was performed with frozen sections from E8.5 Brpf1l/+ and wild-type concepti. Representative images are shown. (B–D) The embryo proper (B), ectoplacental cone (C), and a portion of the yolk sac (D), boxed in (A), are presented at high magnification. Positive staining was detected in the embryonic neuroepithelium, ectoplacental cone and yolk sac of the heterozygous but not wild-type conceptus. Embryonic and extraembryonic structures were labeled according to published atlases.52,53 Abbreviations: Bi, blood island; DA, dorsal aorta; De, decidua; Epc, ectoplacental cone; Gi, giant trophoblasts; NE, neuroepithelium; Ys, yolk sac. Scale bars, 100 μm (A) and 50 μm (B-D).
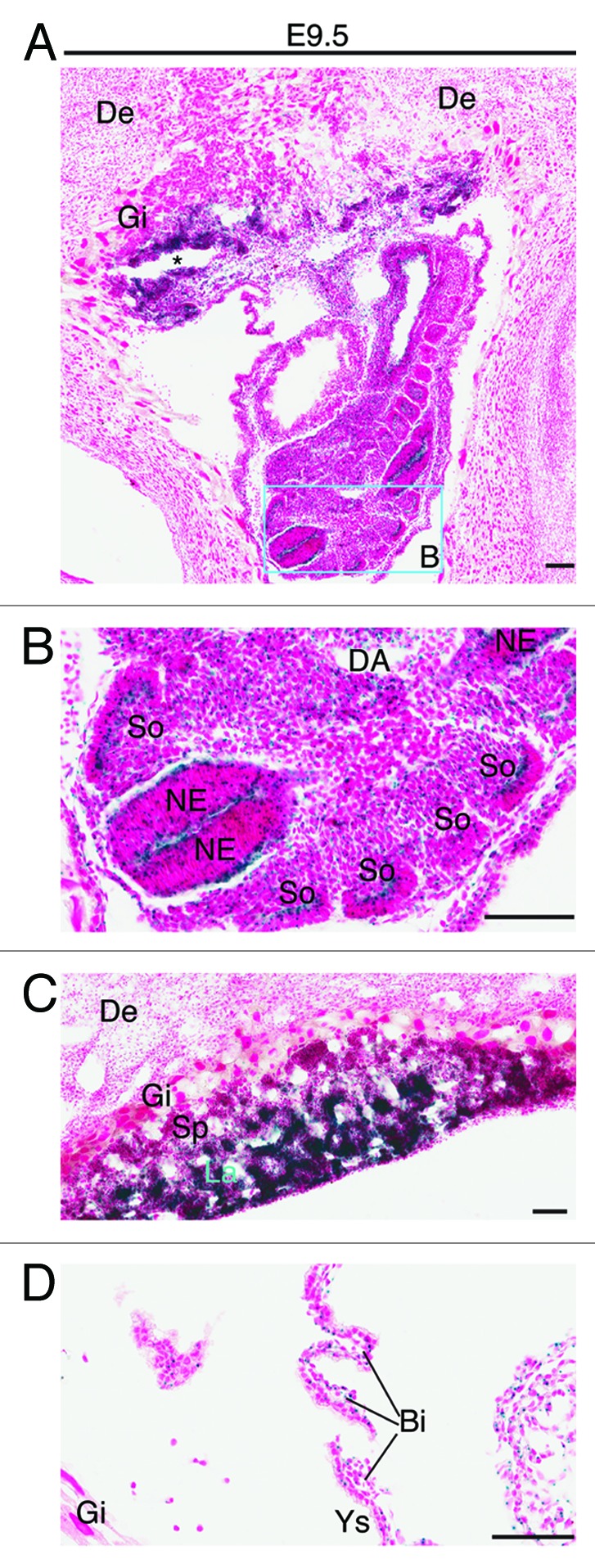
Figure 4. Analysis of Brpf1 expression in E9.5 concepti. (A) β-Galactosidase staining was performed with frozen sections from Brpf1l/+ concepti at E9.5. A representative image of mutant concepti is shown, and no staining was detected in sections from the wild-type concepti (data not shown). (B) Magnified image of the regions boxed in (A). (C) Magnified image for the placenta from a section different from that used in (A). (D) Enlarged image for the yolk sac from a section different from that used in (A). Strong staining was detected in the neuroepithelium and somites of the embryo proper (B), labyrinth of the placenta (C) and blood islands of the yolk sac (D). Abbreviations: Bi, blood island; DA, dorsal aorta; De, decidua; Gi, giant trophoblast; La, labyrinth; NE, neuroepithelium; So, somites; Sp, spongiotrophoblast; Ys, yolk sac. Scale bars, 100 μm.
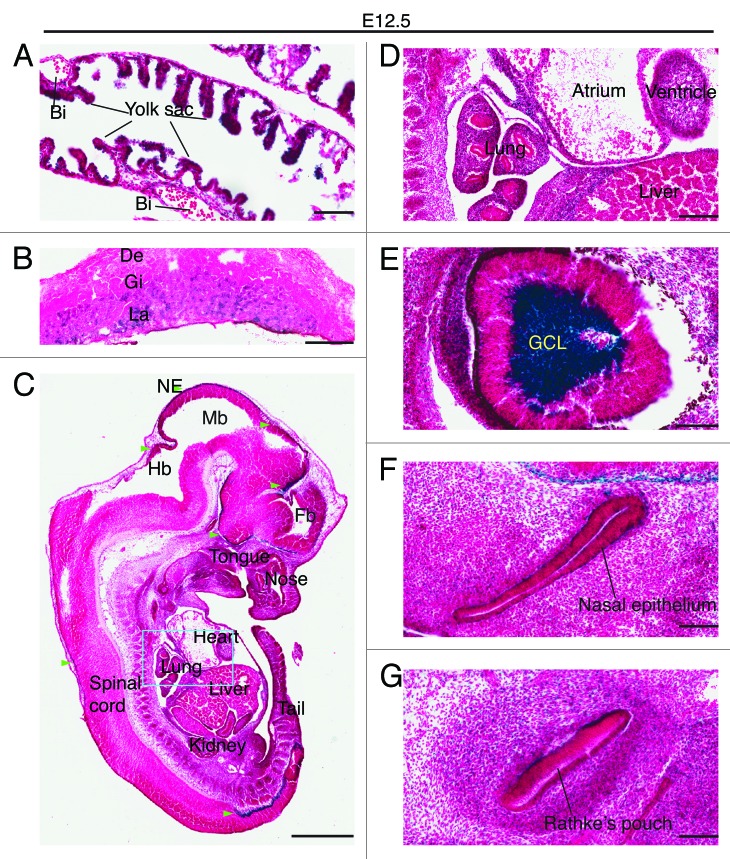
At E12.5, expression remained strong in the yolk sac endoderm but no positive staining was detected in the blood islands (Fig. 5A). High expression also sustained in the labyrinth of the placenta (Fig. 5B). In the embryo itself, high expression was still present in the neuroepithelium of the brain and spinal cord (Fig. 5C). Weak expression was detectable in the lung and heart, but not in the liver (Fig. 5D). This is in stark contrast to the well-established role of Moz in definitive hematopoiesis in the fetal liver.31,32 It should be noted, however, that careful analysis is still needed to examine the role of Brpf1 in this important developmental process. Unexpectedly, particularly robust expression was found in the ganglion cell layer of the retina (Fig. 5E), with the expression level even much higher than that in the neuroepithelium of the brain or spinal cord (Fig. 5C). In addition, specific expression was detected in the nasal epithelium (Fig. 5F) and Rathke's pouch (Fig. 5G), the pituitary primordium.
Figure 5. Brpf1 expression in E12.5 embryonic and extraembryonic sections. (A–C) β-Galactosidase staining was performed on frozen sections of the Brpf1l/+ yolk sac (A), placenta (B) and embryo (C) at E12.5. Representative images are presented here, and no positive staining was detected in sections from the wild-type counterparts (data not shown). (D) Magnified image of the region boxed in (C). (E-G) Enlarged images of the eye, nasal epithelium and Rathke's pouch taken from parasagittal sections parallel to that shown in (C). While strong expression was detected in the yolk sac endoderm, blood islands appeared negative (A). The staining became diffused in the labyrinth, which is expanding at this stage of development (B). In the embryo proper, strong activity was detected at the roof of the midbrain, superficial striatum of the thalamus and hypothalamus, and mantle region of the spinal cord in the lumbosacral zone (C). Note the particularly strong staining in the ganglion cell layer of the retina (E). Abbreviations: Bi, blood island; De, decidua; Fb, forebrain; GCL, ganglion cell layer of the retina; Gi, giant trophoblasts; Hb, hindbrain; La, labyrinth layer; Mb, midbrain; NE, neuroepithelium. Scale bars, 100 μm (A) and (E–G), 0.5 mm (B), 1 mm (C), and 200 μm (D).
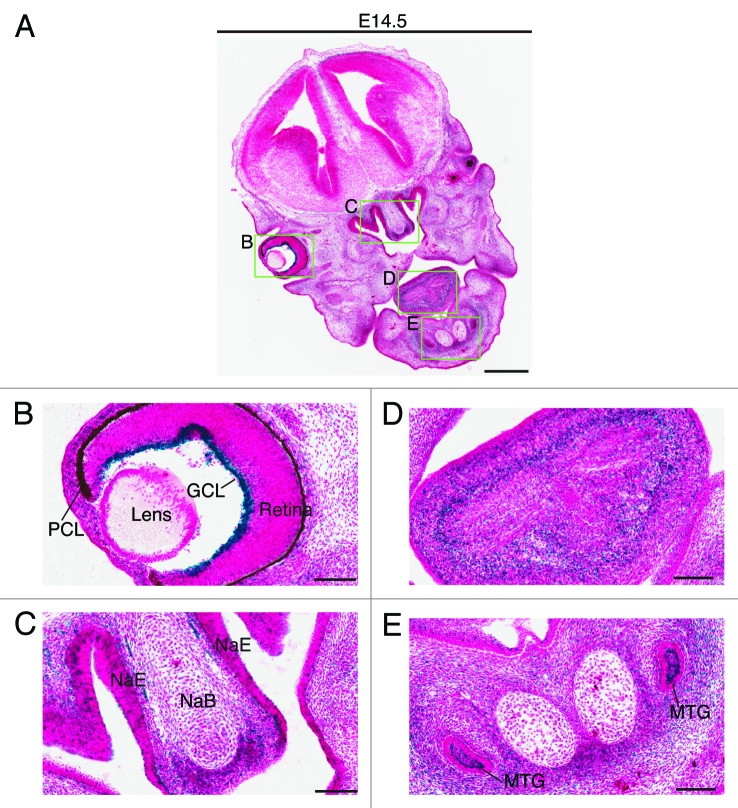
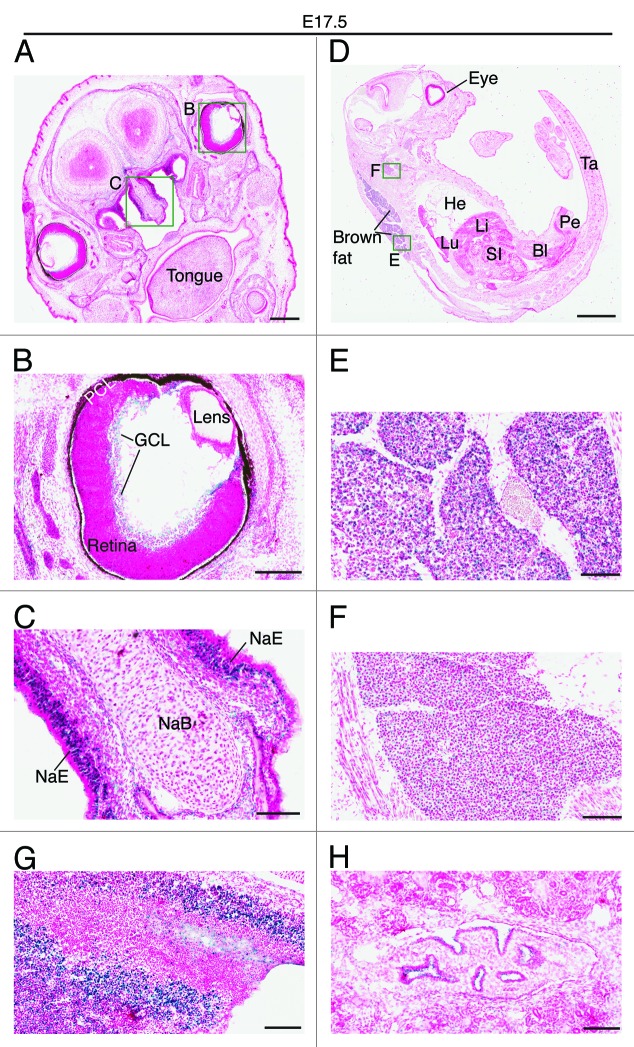
Two days later, at E14.5, very strong expression was also detected in the ganglion cell layer of the retina (Fig. 6A and B). Weaker but still strong expression was detected in the nasal epithelium and the growth plate of the nasal bone (Fig. 6A and C), as well as in the tongue (Fig. 6D) and the submandibular region (Fig. 6E). At E17.5, the expression in the ganglion cell layer decreased dramatically (Fig. 7A and B), whereas the strong expression in the nasal epithelium sustained (Fig. 7A and C). At this stage, the strongest expression was found in the brown fat (Fig. 7D and E). Positive signals were also detected in the neck area (Fig. 7D and F), the back of the nose (Fig. 7G), the pelvis of the kidney (Fig. 7H) and the trigeminal ganglion (data not shown). No or only very weak expression was detected in major organs such as the heart, lung, liver and bladder (Fig. 7D).
Figure 6. Brpf1 expression in E14.5 fetal sections. (A) β-Galactosidase staining was performed on coronal head sections from Brpf1l/+ E14.5 fetuses. A representative image is shown. (B–E) Enlarged images of four regions boxed in (A). As at E12.5 (Fig. 5), the expression in the ganglion cell layer was particularly strong (B). Strong staining was also detected in the nasal epithelium and nasal bone mesenchyme (C), in the tongue (D) and in the molar tooth germs and the mesenchyme adjacent to Meckel’s cartilages (E). Abbreviations: GCL, ganglion cell layer of the retina; MTG, molar tooth germ; NaE, nasal epithelium; NaB, nasal bone primordium; PCL, pigment cell layer of the retina. Scale bars, 500 μm (A) and 100 μm (B–E).
Figure 7. Brpf1 expression in E17.5 fetal sections. (A–C) β-Galactosidase staining was performed on coronal head sections from E17.5 Brpf1l/+ fetuses. A representative image is shown. The two areas boxed in (A) are shown in (B) and (C) at higher magnification. (D–F) β-Galactosidase staining was performed on sagittal sections of E17.5 Brpf1l/+ fetuses. The two areas boxed in (D) are enlarged in (E) and (F). (G and H) β-Galactosidase staining was performed on parasagittal sections of E17.5 Brpf1l/+ fetuses. Two images showing strong galactosidase activity in the back of the nose (an area containing olfactory nerve fibers) and in the pelvis of the kidney are presented in (G) and (H), respectively. At E17.5, expression was relatively low in the entire embryo, except the brown fat (E), nasal epithelium (C) and some other regions shown (C and F). Abbreviations: Bl, bladder; GCL, ganglion cell layer; He, heart; Li, liver; Lu, lung; NaB, nasal bone primordium; NaE, nasal epithelium; PCL, pigment cell layer; Pe, penis; SI, small intestine; Ta, tail. Scale bars, 500 μm (A), 200 μm (B), 100 μm (C), 2 mm (D), and 100 μm (E–H).
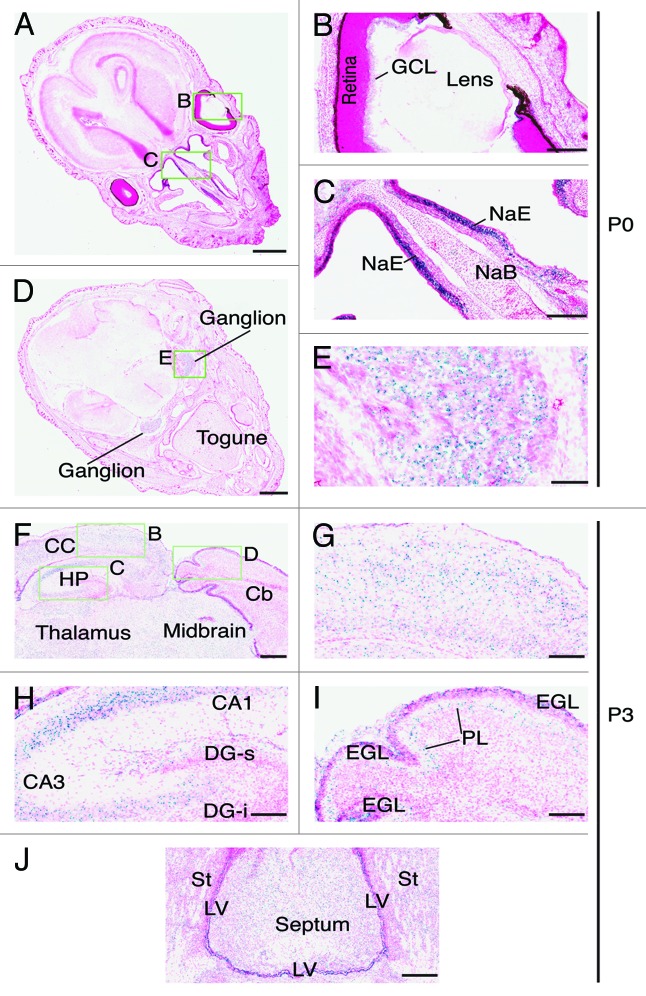
Immediately after birth, the expression in the brown fat disappeared (data not shown) and the level in the retina was also low (Fig. 8A and B). However, the expression in the nasal epithelium remained robust (Fig. 8A and C). Other regions with prominent expression were the dorsal root ganglions (Fig. 8D and E). It is important to note that the expression in major organs such as the brain, heart, lung, liver and cortex of the kidney was minimal from E14.5-P0 (Figs. 5–8), suggesting that Brpf1 may not be important in these organs.
Figure 8. Brpf1 expression in neonate head sections. (A–E) β-Galactosidase staining was performed on transverse sections of neonate heads at P0. Two representative images are presented in (A) and (D). The three areas boxed in (A) and (D) are shown at higher magnification in (B), (C), and (E) as indicated. At P0, Brpf1 was highly expressed in the nasal epithelium (C) and ganglion (E). (F) β-Galactosidase staining was performed on frozen transverse head sections from P3 pups. Only a portion of a representative image is shown. (G–I) Enlarged images of the three areas boxed in (F) as indicated. (J) Image of the expression in the septum and the lateral ventricular zone. At P3, Brpf1 was widely expressed in the brain, including the neocortex (G), hippocampus (H), cerebellum (I), and mitral cell layer in the olfactory bulb (data not shown). Structures of the mouse brain were labeled according to published atlases.54-56 Abbreviations: CA1, cornu ammonis 1; CA3, cornu ammonis 3; DG-s, suprapyramidal blade of the dentate gyrus; DG-i, infrapyramidal blade of the dentate gyrus; GCL, ganglion cell layer of the retina; EGL, external germinal layer; HP, hippocampus; LV, lateral ventricle; NaE, nasal epithelium; NaB, nasal bone PL, Purkinje neuron layer; St, striatum. Scale bars, 1 mm (A) and (D), 200 μm (B and C), 100 μm (E and G–J) and 300 μm (F).
The above expression data from frozen sections indicate that Brpf1 is specifically expressed in the yolk sac, labyrinth of the placenta, neuroepithelium of the neural tube, limb buds, ganglion cell layer of the retina, nasal epithelium, and tongue muscle, suggesting a potential role in these specific tissues during pre- and perinatal development.
Brpf1 expression during postnatal development
The general low levels of Brpf1 expression in neonates (Fig. 8) were interesting, so we asked whether expression increases at later stages. To address this, we performed whole-mount staining of major organs from adult male and female mice. As shown in Table S1, expression remained low in major organs such as the heart, lung, liver and kidney, but the level was high in the brain and significant expression was also detected in the testis, suggesting potential roles of Brpf1 in these two sites, but not in those major organs with low or undetectable signals, during postnatal development.
The high expression in the brain was intriguing, so we analyzed sagittal and parasagittal slices by whole-mount staining. The analysis revealed a striking expression pattern (Fig. S2). Strong expression was detected in at least four specific structures: neocortex, hippocampus, cerebellum and olfactory bulb. While it was rather uniform in the neocortex, the expression was very strong in the pyramidal layer of the CA regions and the granular layer of the dentate gyrus within the hippocampus (Fig. S2). The expression was also high in the subiculum and postsubiculum, two accessory structures of the hippocampus (Fig. S2C). The hippocampus and accessory structures are important for learning and memory,33 implying that Brpf1 may play a role in these important mental processes.
Within the cerebellum, the expression was very strong in the granular layer and Purkinje neurons (Fig. S2B and D; data not shown), suggesting potential roles in regulating cerebellar development and functions. This is consistent with the observation that at the embryonic stage, Brpf1 expression was detected in the rhomic lip (Fig. 2D and E), a region from which the cerebellum develops. The cerebellum is a key structure for motor coordination and precision,34 so it will be interesting to investigate whether Brpf1 plays a role in related processes. Expression was also detected in the olfactory bulb, especially the mitral layer, and the accessory olfactory bulb (Fig. S2B; data not shown), suggesting a potential role in olfaction.
The strong expression in the adult brain is in stark contrast to no or low expression in E17.5 and P0 brain sections (Figs. 7 and 8), suggesting that Brpf1 expression dramatically increases in the brain after birth. To substantiate this, we analyzed transverse brain sections from P3 pups. Strong expression was detected in different regions of the brain, including the neocortex (Fig. 8F and G), hippocampus (Fig. 8H), cerebellum (Fig. 8I), and septum (Fig. 8J). In the hippocampus, strong expression was present in the pyramidal layer of the CA region but not in granular layer of the dentate gyrus (Fig. 8H), indicating that Brpf1 expression in the dentate gyrus appears after P3. In the cerebellum, strong signals were present in the external germinal layer and Purkinje neurons (Fig. 8I). The external germinal layer contains progenitors that will migrate inwards to the internal granular layer and terminally differentiate into granular neurons there.35 Consistent with the result from this section analysis, Brpf1 was expressed in the internal granular layers and Purkinje neurons of the adult brain (Fig. S2B and D). Together, these results support dynamic expression of Brpf1 during postnatal brain development.
Brpf1 inactivation leads to embryonic lethality at E9.5
The dynamic expression of Brpf1 during embryonic, fetal and postnatal development suggests that it may play an important role in these developmental processes. To gain insights into this important issue, we investigated the impact of Brpf1 inactivation. The Brpf1l allele was designed based on the knockout-first strategy (Fig. 1A).28-30 Because Brpf1+/l mice were rather indistinguishable from the wild-type, we performed intercross between male and female heterozygous Brpf1+/l mice to assess survival of homozygous embryos and neonates. In the first litter that we obtained from the mating, there were 8 pups and all of them appeared normal. PCR genotyping identified 4 wild-type pups and 4 heterozygotes. The lack of homozygotes in the litter led us to ask whether Brpf1 disruption causes prenatal lethality, so we retrieved fetuses from two pregnant mice at E17.5, which yielded 9 normal fetuses and 4 implantation sites that contained necrotic fetal tissues or no fetuses at all. No genotypes could be obtained for these 4 implantation sites due to the lack of good-quality tissues for DNA analysis. Genotyping of the 9 normal fetuses identified 4 wild-type fetuses and 5 heterozygotes. Based on these results, we wondered whether potential lethality occurs much earlier than E17.5. To substantiate this, we analyzed three pregnant mice at E11.5, which yielded 19 normal embryos and 4 dead ones, as well as 4 implantation sites that contained only necrotic embryonic tissues or no embryos at all. PCR genotyping results were really striking: among the 19 normal embryos, 5 were wild-type and 14 were heterozygous, whereas the 4 dead embryos were all homozygous (Fig. 1D; data not shown). No genotypes could be obtained for the 4 implantation sites containing only necrotic embryonic tissues or no embryos. Together, these analyses indicate that Brpf1 inactivation causes embryonic lethality prior to E11.5.
To investigate this further, we generated the mutant allele Brpf1Δ (Fig. 1A). The Brpf1l allele was deigned based on the knockout-first strategy,28-30 but we found in some other strains that this strategy may or may not lead to inactivation (data not shown). As such, we mated Brpf1+/l mice with the EIIa-Cre strain, which expresses the Cre recombinase at the pre-implantation stage,36 to generate global excision at the LoxP sites. This mating strategy resulted in Brpf1l/+;EIIa-Cre (or Brpf1+/Δ) mice (Fig. 1A), and further intercross between the heterozygous male and female mice was expected to generate homozygous Brpf1Δ/Δ mice. Surprisingly, no viable pups were born (Table S2), suggesting that the homozygous mutation might have caused lethality before birth. We thus analyzed the viability of embryos or fetuses at different days from E8.75–17.5. As shown in Table S2, at E8.75, embryos of three expected genotypes were recovered at the normal Mendelian ratio. However, most homozygous embryos displayed severe abnormalities at E9.5. Thereafter, development of the embryos did not progress properly, and they increasingly deteriorated. No viable ones could be obtained at or after E11.5. RT-PCR confirmed the inactivation of the Brpf1 gene (Fig. S1F and G). Thus, Brpf1 loss causes embryonic lethality around E9.5.
As the mutant allele Brpf1Δ still possesses the promoterless LacZ cassette (Fig. 1A). To avoid potential complication from this cassette, we removed it by crossing Brpf1+/l mice with the PGK1-FLP0 strain (Fig. 1B). This strain displays ubiquitous expression of the FLPo recombinase and induces excision at the FRT sites in all lineages, including the germ lines.37 The cross generated the floxed allele Brpf1f (Fig. 1B). Further mating with the EIIa-Cre strain36 resulted in Brpf1+/f;EIIa-Cre (or Brpf1+/-) mice, and intercross between the resulting heterozygous male and female mice was expected to generate homozygous Brpf1-/- mice (Fig. 1E and F). As observed with intercross of Brpf1+/Δ mice (Table S2), almost no viable pups were born and no normal embryos were recovered after E9.5 (Table S3), further supporting that Brpf1 inactivation leads to embryonic lethality around E9.5. This phenotype is unexpected as it is much more severe than inactivation of the Moz gene, which results in lethality at E14.5.31,32 The severe phenotype of Brpf1 inactivation suggests potential roles independent of Moz.
Discussion
Molecular and cell-based analyses have firmly established that BRPF1 forms multiple acetyltransferase complexes with three members of the MYST family, MOZ, MORF, and HBO1, to govern their enzymatic activity, substrate specificity, and ability to stimulate transcription.13,18,19 As the first step to extrapolate these molecular interactions to the biological functions at the tissue and organismal levels, we have analyzed the spatiotemporal expression of mouse Brpf1. This analysis has revealed that Brpf1 is dynamically expressed in extraembryonic, embryonic, fetal and adult tissues (Figs. 2–8; Table S1). At the adult stage, it was highly expressed in the testis and specific structures of the brain (Table S1; Fig. S2). Together, the spatiotemporal expression information forms a 4-dimensional expression atlas, which will serve as a valuable guide for further genetic analysis of Brpf1 and dissection of its functional interaction with Moz, Morf, and Hbo1 in individual tissues at specific developmental stages. In support of the dynamic expression in extraembryonic and embryonic tissues, inactivation of the mouse Brpf1 gene led to embryonic lethality at E9.5. Moreover, the mutant Brpf1 allele contains sequence elements for excision by FLP and Cre recombinases (Fig. 1A)—therefore, mating the mutant mice with tissue-specific Cre-expressing strains will yield insights into biological functions of Brpf1 in the tissues where it is expressed, according to the 4D expression atlas established herein.
Molecular and cell-based studies13,18,19 suggest that human BRPF1 may function as a “disease modifier.” Related to this, the MORF gene is mutated in three developmental disorders, Noonan syndrome-like disorder,24 Ohdo syndrome,25 and Genitopatellar syndrome,26,27 with the common characteristic of intellectual disability. Specific expression in the neocortex, hippocamus, and cerebullum of mouse Brpf1 (Table S1; Fig. S2) supports that human BRPF1 may affect the severity of these disorders. In addition, the MOZ and MORF genes are rearranged in leukemia.4,23 About this, mouse Brpf1 was expressed in the yolk sac at E8.5 and E9.5 (Figs. 3 and 4), indicating a potential role in primitive hematopoiesis. No expression was detected in the fetal liver or bone marrow (Figs. 5–7). This is in stark contrast to the important role of mouse Moz in hematopoietic stem cells during embryonic and fetal development.23,31,32,38 However, these stem cells are rare and Brpf1 expression may have escaped detection by the method that we used here, so careful analysis is still needed to examine the role of Brpf1 in hematopoietic stem cells at the embryonic, fetal and postnatal stages.
The Brpf family of proteins is conserved from C. elegans to humans, with one member in the worm or fly but three in a vertebrate. It should be noted, however, that Drosophila Brpf is highly similar to Brpf1, whereas the C. elegans member Lin-49 is relatively distant and lacks the PWWP module.4 While little is known about the function of the fly protein, Lin-49 regulates Hox gene expression, neuron asymmetry, hindgut development and fecundity.39-41 Lin-49 genetically interacts with Lsy-12, which is only distantly related to MOZ/MORF.41 Inactivation of zebrafish Brpf1 affects anterior Hox gene expression and alters pharyngeal segmental identity,42 while disruption of medaka Brpf1 alters craniofacial and caudal skeletons by reducing expression of anterior and posterior Hox genes, respectively.43 Thus, fish Brpf1 regulates Hox gene expression during skeletal development. Related to this, the expression atlas of mouse Brpf1 (Figs. 6–8) suggests a potential function in the bone. Importantly, the atlas also points out potential functions in many other tissues, including the neural tube, limb buds, retina, nose and brown fat at the prenatal stages, as well as the brain and testis after birth (Figs. 2–8; Table S1). As little activity was detected in the heart, lung, stomach, intestine/colon and liver (Figs. 2–8; Table S1), Brpf1 may only play minor roles in these major organs. BRPF1 is paralogous to BRPF2 and BRPF3.18,19 Inactivation of mouse Brpf2 led to embryonic lethality at E15.5, with growth retardation, neural tube defects, abnormal eye development and faulty erythropoiesis.21 No knockout studies have been reported for mouse Brpf3. The mouse Brpf1 expression atlas established herein is also valuable for determining how the division-of-labor of these three proteins is achieved to regulate MOZ, MORF and HBO1 in a spatiotemporal manner.
In addition to Brpf1, we have analyzed the expression of Moz in the adult brain (Fig. S1 and 3). Like Brpf1 (Table S1), Moz was highly expressed in specific structures of the brain, including the neocortex, hippocampus, cerebellum, and olfactory bulb (Fig. S3). This pattern is similar but not identical to that of Brpf1 (Fig. S2), suggesting their potential interaction in the brain. It should be noted, however, that further analysis is needed to determine whether this is indeed the case. The role of Moz in brain development remains unknown even though the function of mouse and fish Moz in hematopoiesis and skeletal development has been investigated in several studies.31,32,44-47 Notably, there are also differences between the expression patterns of Moz and Brpf1. For example, Moz, but not Brpf1, was highly expressed in the striatum (Figs. S2 and 3). In this case, Brpf2 and Brpf3 may interact with Moz. In Purkinje neurons of the cerebellum, Brpf1, but not Moz, was expressed (Fig. 8; Figs. S2 and 3; data not shown). In these neurons, Morf and Hbo1 may replace Moz as the partner of Brpf1. Further analysis of Morf and Hbo1 may shed light on this possibility. Related to this, Morf-deficient mice containing 10% residual transcripts die at the weaning stage and display cerebral defects,48 but no abnormalities were observed in the cerebellum. Total loss of mouse Hbo1 led to embryonic lethality at E10.5,49 but the role in fetal and postnatal development remains unclear. Immunofluorescence microscopy revealed low ubiquitous expression of Hbo1 in the entire mouse brain (data not shown), suggesting that Hbo1 may not be a major partner of Brpf1 in Purkinje neurons. These observations imply that mouse Brpf1 may even have roles independent of Moz, Morf, and Hbo1. Related to this, Brpf1 is distantly related to Lin-49 in C. elegans (within the PZP module and EPC-like motifs), but only one worm MYST acetyltransferase (Lsy-12) is more homologous to mouse Moz, Morf, and Hbo1 than to the other two members of the MYST family (i.e., Tip60 and hMof).39-41
In summary, we have established the 4D expression atlas for mouse Brpf1, a unique multidomain chromatin regulator able to activate three different acetyltransferases, and compared its expression pattern with that of Moz in the adult brain. We have found dynamic expression of Brpf1 in the placenta, yolk sac, limb buds, brain, spinal cord, retina, nose, bone, and brown fat at the prenatal stages. After birth, high expression was detected in the testis and some specific regions of the brain. Related to its dynamic expression during embryonic development, Brpf1 is essential for embryogenesis prior to E9.5. The 4D expression atlas will serve as an important roadmap for determining the physiological and pathological functions of mammalian BRPF1, identifying the molecular mechanisms whereby its dynamic expression is controlled, analyzing its functional interaction with MOZ, MORF, and HBO1 in vivo, and investigating whether BRPF1 has any roles independent of these three important acetyltransferases.
Materials and Methods
Ethics statement of animal research
Mouse strains were maintained in a newly established animal facility at McGill University and all procedures involved in the use of mice were performed according to guidelines and protocols approved by the McGill University Animal Use Committee.
Wild-type and mutant mice
Brpf1l/+ mice were obtained from European Conditional Mouse Mutagenesis Program (EUCOMM, project 40402; http://www.knockoutmouse.org/genedetails/MGI:1926033).28-30 A promoterless LacZ cassette is placed between two FRT sites, while two LoxP sites flank exons 4–6 of Brpf1 (Fig. 1A). For genotyping, genomic PCR with primers Brpf1-F1 and -R1 generated a 227-bp band for wild-type, whereas genomic PCR with primers Brpf1-F1 and -mR1 produced a 162-bp fragment for the knock-in allele (Fig. 1A and C). The lines were maintained on the C57BL/6J background. Intercross between the male and female heterozygotes was employed to assess the mortality of homozygous embryos, fetuses and neonates.
For the Moz line, we utilized ES clones from EUCOMM to generate conditional knockouts with LoxP sites flanking the second coding exon of Moz. This exon has 109 bp and its deletion causes a reading-frame shift in the resulting transcript, triggering non-sense mRNA decay and inactivating the allele (http://www.knockoutmouse.org/martsearch/project/29690).28-30 The ES cells were injected into CD1 blastocysts to obtain male chimera mice, performed at McGill Transgenic Core, for germline transmission by further crosses with C57BL6 female mice. Crossing the resulting line, Mozl/+, with EIIa-Cre mice (Jackson Laboratory) yielded Mozl/+; EIIa-Cre mice and further intercross generated Moz−/− embryos. The fetuses displayed lethality at E14.75 (Fig. S1), indicating that the conditional allele functioned as expected.
Mouse genotyping
Mice were genotyped by PCR with genomic DNA extracted from yolk sac, tail cut or ear punch samples. Genomic DNA was isolated according to the protocol from Jackson Laboratory (http://jaxmice.jax.org/support/genotyping/dna-isolation-protocols.html). The primers Brpf1-F1 (TGTGCCCTGT AGAGTGTTGC) and Brpf1-R1 (GCCTTGAGTG GCACAACATA) were used to amplify a 227-bp band for the wild-type allele, whereas the primers Brpf1-F1 and Brpf1-mR1 (TTGGTGATAT CGTGGTATCG TT) yielded a 162-bp fragment for the mutant allele. Brpf1-F1 and Brpf1-ex03 (CAGCTTTAAT GAGAAAAAAA ATCA) were used to detect a 460-bp fragment for FLP0-mediated and Cre-mediated recombination of the mutant Brpf1 allele. Cre01 (GCATTACCGG TCGATGCAAC GAGTG) and Cre02 (GAACGCTAGA GCCTGTTTTG CACGTTC) were used for detection of a 374-bp fragment of the Cre transgene. Primers M5 (AGAGAGCCCT TCCTCTCAGT ACCG), LAR3 (CAACGGGTTC TTCTGTTAGT CC) and M3 (ACAGATGGGG ATTGGTTCAG CAACC) were used to amplify fragments from the wild-type (531 bp) and Mozl (284 bp) alleles. Primers LoxF (AAATGAATGC AATTGTTGTT GT), LoxR1 (GGCACTAAGA GAGAAACTCC CA) and LoxR (TGAACTGATG GCGAGCTCAG ACC) were used to detect a 382-bp fragment from the unrecombined Mozl allele and a 287-bp fragment from the recombined Moz- allele. A 10 µl PCR reaction was set up with 5 µl of the 2x GoTaq Green Master Mix (Promega, PRM5122), 1 µl genomic DNA, 0.5 µl of each primer (10 pmol/µl) and 2.5–3 µl of sterile nuclease-free water. PCR cycling conditions were as follows: 95 °C × 5 min, 30 PCR cycles (95 °C × 15 s, 50 °C × 15 s and 72 °C × 45 s) and 72 °C × 7 min, except that 60 °C was used as the annealing temperature for the Moz primers. The amplified products were kept at 4 °C or -20 °C until further analysis by agarose gel electrophoresis.
RT-PCR
Total RNA from embryos at E9.5 was extracted with the TRIZOL reagent (Invitrogen, 15596018). Embryos at E8.75 were polled for each genotype before RNA extraction with the miRNeasy Mini kit (GIAGEN, 217004). cDNA from 1 µg of total RNA was synthesized with the QuantiTect Reverse Transcription kit (QIAGEN, 205311). The synthesized cDNA was used as the template for PCR with two sets of primers (CAGTAAGATC ACCAACCGCC vs GAGGAAAGGG GTCAGCTGCA and CAGCCCCTCT GAAGTCTCAC vs CTAGTGCATT GGGGTCACCT for amplifications of the coding sequences for the Floxed and N-terminal regions of Brpf1, respectively) with the cycling conditions—95 °C for 5 min, 28 amplification cycles (95 °C for 15 s, 50 °C for 15 s, and 72 °C for 45 s) and 72 °C for 7 min.
Tissue preparation
All mice were anesthetized with isoflurane through inhalation before tissue or embryo collection. For the mice used for whole-mount tissue staining, animals were perfused transcardially with cold PBS, followed by 4% paraformaldehyde (PFA) in PBS, prior to tissue collection.
X-gal staining
β-Galactosidase staining was performed as described.50,51 For whole-mount staining, embryos and extraembryonic tissues were dissected out fresh, fixed in the fixative solution (0.1 M phosphate buffer [pH 7.3], 5 mM EGTA, 0.2% glutaraldehyde and 2 mM MgCl2) for 10–30 min depending on the size of the embryo or tissue, rinsed in the detergent rinse (0.1 M phosphate buffer [pH7.3], 2 mM MgCl2, and 0.02% NP-40) three times for 15 min each at room temperature, and incubated in the staining solution (0.1 M phosphate buffer [pH7.3], 20 mM TRIS-HCl pH7.3, 2 mM MgCl2, 0.02% NP-40, 5 mM potassium ferricyanide, 5 mM potassium ferrocyanide, 0.01% sodium deoxycholate, and 1 mg/ml X-gal) at 37 °C for 4–16 h in the dark until a desired intensity of blue color was developed. After the staining, tissues were post-fixed in 4% PFA and rinsed in PBS before imaging under a dissecting microscope (SteREO Lumar.V12, Zeiss), linked to an Axiocam HRc color camera and controlled by Axiovision Re 4.8. Images were exported to Adobe Photoshop for processing and then imported to Adobe Illustrator for final presentation. For staining of the postnatal brain or other organs, mice were perfused first with cold PBS and then with 4% cold PFA in PBS, with additional 2 h post-fixation in 4% PFA at 4 °C. The brain was cut sagittally in the middle and then several parasagittal slices were prepared manually with a blade. The staining procedure was performed as just described for embryos.
For staining of frozen sections, embryos or the whole concepti were fixed in 1% PFA at 4 °C for 1 h, rinsed in PBS, cryoprotected in 30% sucrose in PBS at 4 °C overnight until the samples sank to the bottom, embedded in the Tissue-Tek O.C.T. compound (Sakura Finetek, 4583) on dry ice and stored in -80 °C. Sections were cut at 15 µm on a Cryotome (Thermo Electron, 77200187). Slides were dried and stored at -80 °C or directly used for staining. The slides were postfixed in the fixative solution (0.1 M PIPES pH 6.9, 5 mM EGTA, 2 mM MgCl2, and 2% PFA) for 10 min on ice, rinsed in PBS containing 2 mM MgCl2, and immersed in the same solution for 10 min on ice. The slides were then immersed in the detergent rinse for 10 min on ice and stained in the staining solution for 4–16 h at 37 °C as described above. After staining, the slides were washed twice in PBS containing 2 mM MgCl2 (5 min each) at room temperature, rinsed in distilled H2O and counterstained in 0.1% nuclear fast red (RICCA, R5463200) for 5 min. Afterwards, the slides were rinsed in distilled H2O, dehydrated in a gradient of ethanol, cleared in xylene and mounted with Permount. The slides were either examined under a regular light microscope or digitized with a Scanscope (Aperio, ScanScope XT) for further analysis.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Maxime Bouchard, Katie Stewart, and Yojiro Yamanaka for help on analysis of the embryos and mice. Grants were from Canadian Institutes of Health Research, Canadian Cancer Society, Canada Foundation for Innovation, and Ministère du Développement Économique, Innovation et Exportation du Québec (to X.J.Y.).
References
- 1.Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct Mol Biol. 2007;14:1025–40. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Latham JA, Dent SY. Cross-regulation of histone modifications. Nat Struct Mol Biol. 2007;14:1017–24. doi: 10.1038/nsmb1307. [DOI] [PubMed] [Google Scholar]
- 3.Musselman CA, Lalonde ME, Côté J, Kutateladze TG. Perceiving the epigenetic landscape through histone readers. Nat Struct Mol Biol. 2012;19:1218–27. doi: 10.1038/nsmb.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang XJ, Ullah M. MOZ and MORF, two large MYSTic HATs in normal and cancer stem cells. Oncogene. 2007;26:5408–19. doi: 10.1038/sj.onc.1210609. [DOI] [PubMed] [Google Scholar]
- 5.Klein BJ, Lalonde ME, Côté J, Yang XJ, Kutateladze TG. Crosstalk between epigenetic readers regulates the MOZ/MORF HAT complexes. Epigenetics. 2014;9:186–93. doi: 10.4161/epi.26792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thompson KA, Wang B, Argraves WS, Giancotti FG, Schranck DP, Ruoslahti E. BR140, a novel zinc-finger protein with homology to the TAF250 subunit of TFIID. Biochem Biophys Res Commun. 1994;198:1143–52. doi: 10.1006/bbrc.1994.1162. [DOI] [PubMed] [Google Scholar]
- 7.Chaplin T, Bernard O, Beverloo HB, Saha V, Hagemeijer A, Berger R, Young BD. The t(10;11) translocation in acute myeloid leukemia (M5) consistently fuses the leucine zipper motif of AF10 onto the HRX gene. Blood. 1995;86:2073–6. [PubMed] [Google Scholar]
- 8.Prasad R, Leshkowitz D, Gu Y, Alder H, Nakamura T, Saito H, Huebner K, Berger R, Croce CM, Canaani E. Leucine-zipper dimerization motif encoded by the AF17 gene fused to ALL-1 (MLL) in acute leukemia. Proc Natl Acad Sci U S A. 1994;91:8107–11. doi: 10.1073/pnas.91.17.8107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saha V, Chaplin T, Gregorini A, Ayton P, Young BD. The leukemia-associated-protein (LAP) domain, a cysteine-rich motif, is present in a wide range of proteins, including MLL, AF10, and MLLT6 proteins. Proc Natl Acad Sci U S A. 1995;92:9737–41. doi: 10.1073/pnas.92.21.9737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perry J. The Epc-N domain: a predicted protein-protein interaction domain found in select chromatin associated proteins. BMC Genomics. 2006;7:6. doi: 10.1186/1471-2164-7-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McCullagh P, Chaplin T, Meerabux J, Grenzelias D, Lillington D, Poulsom R, Gregorini A, Saha V, Young BD. The cloning, mapping and expression of a novel gene, BRL, related to the AF10 leukaemia gene. Oncogene. 1999;18:7442–52. doi: 10.1038/sj.onc.1203117. [DOI] [PubMed] [Google Scholar]
- 12.Qin S, Jin L, Zhang J, Liu L, Ji P, Wu M, Wu J, Shi Y. Recognition of unmodified histone H3 by the first PHD finger of bromodomain-PHD finger protein 2 provides insights into the regulation of histone acetyltransferases monocytic leukemic zinc-finger protein (MOZ) and MOZ-related factor (MORF) J Biol Chem. 2011;286:36944–55. doi: 10.1074/jbc.M111.244400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lalonde ME, Avvakumov N, Glass KC, Joncas FH, Saksouk N, Holliday M, Paquet E, Yan K, Tong Q, Klein BJ, et al. Exchange of associated factors directs a switch in HBO1 acetyltransferase histone tail specificity. Genes Dev. 2013;27:2009–24. doi: 10.1101/gad.223396.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu L, Qin S, Zhang J, Ji P, Shi Y, Wu J. Solution structure of an atypical PHD finger in BRPF2 and its interaction with DNA. J Struct Biol. 2012;180:165–73. doi: 10.1016/j.jsb.2012.06.014. [DOI] [PubMed] [Google Scholar]
- 15.Poplawski A, Hu K, Lee W, Natesan S, Peng D, Carlson S, Shi X, Balaz S, Markley JL, Glass KC. Molecular Insights into the Recognition of N-Terminal Histone Modifications by the BRPF1 Bromodomain. J Mol Biol. 2014(forthcoming doi: 10.1016/j.jmb.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vezzoli A, Bonadies N, Allen MD, Freund SM, Santiveri CM, Kvinlaug BT, Huntly BJ, Göttgens B, Bycroft M. Molecular basis of histone H3K36me3 recognition by the PWWP domain of Brpf1. Nat Struct Mol Biol. 2010;17:617–9. doi: 10.1038/nsmb.1797. [DOI] [PubMed] [Google Scholar]
- 17.Wu H, Zeng H, Lam R, Tempel W, Amaya MF, Xu C, Dombrovski L, Qiu W, Wang Y, Min J. Structural and histone binding ability characterizations of human PWWP domains. PLoS One. 2011;6:e18919. doi: 10.1371/journal.pone.0018919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doyon Y, Cayrou C, Ullah M, Landry AJ, Côté V, Selleck W, Lane WS, Tan S, Yang XJ, Côté J. ING tumor suppressor proteins are critical regulators of chromatin acetylation required for genome expression and perpetuation. Mol Cell. 2006;21:51–64. doi: 10.1016/j.molcel.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 19.Ullah M, Pelletier N, Xiao L, Zhao SP, Wang K, Degerny C, Tahmasebi S, Cayrou C, Doyon Y, Goh SL, et al. Molecular architecture of quartet MOZ/MORF histone acetyltransferase complexes. Mol Cell Biol. 2008;28:6828–43. doi: 10.1128/MCB.01297-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saksouk N, Avvakumov N, Champagne KS, Hung T, Doyon Y, Cayrou C, Paquet E, Ullah M, Landry AJ, Côté V, et al. HBO1 HAT complexes target chromatin throughout gene coding regions via multiple PHD finger interactions with histone H3 tail. Mol Cell. 2009;33:257–65. doi: 10.1016/j.molcel.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mishima Y, Miyagi S, Saraya A, Negishi M, Endoh M, Endo TA, Toyoda T, Shinga J, Katsumoto T, Chiba T, et al. The Hbo1-Brd1/Brpf2 complex is responsible for global acetylation of H3K14 and required for fetal liver erythropoiesis. Blood. 2011;118:2443–53. doi: 10.1182/blood-2011-01-331892. [DOI] [PubMed] [Google Scholar]
- 22.Lafon A, Chang CS, Scott EM, Jacobson SJ, Pillus L. MYST opportunities for growth control: yeast genes illuminate human cancer gene functions. Oncogene. 2007;26:5373–84. doi: 10.1038/sj.onc.1210606. [DOI] [PubMed] [Google Scholar]
- 23.Butler JS, Dent SY. The role of chromatin modifiers in normal and malignant hematopoiesis. Blood. 2013;121:3076–84. doi: 10.1182/blood-2012-10-451237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kraft M, Cirstea IC, Voss AK, Thomas T, Goehring I, Sheikh BN, Gordon L, Scott H, Smyth GK, Ahmadian MR, et al. Disruption of the histone acetyltransferase MYST4 leads to a Noonan syndrome-like phenotype and hyperactivated MAPK signaling in humans and mice. J Clin Invest. 2011;121:3479–91. doi: 10.1172/JCI43428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clayton-Smith J, O’Sullivan J, Daly S, Bhaskar S, Day R, Anderson B, Voss AK, Thomas T, Biesecker LG, Smith P, et al. Whole-exome-sequencing identifies mutations in histone acetyltransferase gene KAT6B in individuals with the Say-Barber-Biesecker variant of Ohdo syndrome. Am J Hum Genet. 2011;89:675–81. doi: 10.1016/j.ajhg.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simpson MA, Deshpande C, Dafou D, Vissers LE, Woollard WJ, Holder SE, Gillessen-Kaesbach G, Derks R, White SM, Cohen-Snuijf R, et al. De novo mutations of the gene encoding the histone acetyltransferase KAT6B cause Genitopatellar syndrome. Am J Hum Genet. 2012;90:290–4. doi: 10.1016/j.ajhg.2011.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Campeau PM, Kim JC, Lu JT, Schwartzentruber JA, Abdul-Rahman OA, Schlaubitz S, Murdock DM, Jiang MM, Lammer EJ, Enns GM, et al. Mutations in KAT6B, encoding a histone acetyltransferase, cause Genitopatellar syndrome. Am J Hum Genet. 2012;90:282–9. doi: 10.1016/j.ajhg.2011.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Skarnes WC, Rosen B, West AP, Koutsourakis M, Bushell W, Iyer V, Mujica AO, Thomas M, Harrow J, Cox T, et al. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474:337–42. doi: 10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Testa G, Schaft J, van der Hoeven F, Glaser S, Anastassiadis K, Zhang Y, Hermann T, Stremmel W, Stewart AF. A reliable lacZ expression reporter cassette for multipurpose, knockout-first alleles. Genesis. 2004;38:151–8. doi: 10.1002/gene.20012. [DOI] [PubMed] [Google Scholar]
- 30.Pettitt SJ, Liang Q, Rairdan XY, Moran JL, Prosser HM, Beier DR, Lloyd KC, Bradley A, Skarnes WC. Agouti C57BL/6N embryonic stem cells for mouse genetic resources. Nat Methods. 2009;6:493–5. doi: 10.1038/nmeth.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Katsumoto T, Aikawa Y, Iwama A, Ueda S, Ichikawa H, Ochiya T, Kitabayashi I. MOZ is essential for maintenance of hematopoietic stem cells. Genes Dev. 2006;20:1321–30. doi: 10.1101/gad.1393106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thomas T, Corcoran LM, Gugasyan R, Dixon MP, Brodnicki T, Nutt SL, Metcalf D, Voss AK. Monocytic leukemia zinc finger protein is essential for the development of long-term reconstituting hematopoietic stem cells. Genes Dev. 2006;20:1175–86. doi: 10.1101/gad.1382606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deng W, Aimone JB, Gage FH. New neurons and new memories: how does adult hippocampal neurogenesis affect learning and memory? Nat Rev Neurosci. 2010;11:339–50. doi: 10.1038/nrn2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Apps R, Garwicz M. Anatomical and physiological foundations of cerebellar information processing. Nat Rev Neurosci. 2005;6:297–311. doi: 10.1038/nrn1646. [DOI] [PubMed] [Google Scholar]
- 35.Sillitoe RV, Joyner AL. Morphology, molecular codes, and circuitry produce the three-dimensional complexity of the cerebellum. Annu Rev Cell Dev Biol. 2007;23:549–77. doi: 10.1146/annurev.cellbio.23.090506.123237. [DOI] [PubMed] [Google Scholar]
- 36.Lakso M, Pichel JG, Gorman JR, Sauer B, Okamoto Y, Lee E, Alt FW, Westphal H. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc Natl Acad Sci U S A. 1996;93:5860–5. doi: 10.1073/pnas.93.12.5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kranz A, Fu J, Duerschke K, Weidlich S, Naumann R, Stewart AF, Anastassiadis K. An improved Flp deleter mouse in C57Bl/6 based on Flpo recombinase. Genesis. 2010;48:512–20. doi: 10.1002/dvg.20641. [DOI] [PubMed] [Google Scholar]
- 38.Perez-Campo FM, Borrow J, Kouskoff V, Lacaud G. The histone acetyl transferase activity of monocytic leukemia zinc finger is critical for the proliferation of hematopoietic precursors. Blood. 2009;113:4866–74. doi: 10.1182/blood-2008-04-152017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chamberlin HM, Thomas JH. The bromodomain protein LIN-49 and trithorax-related protein LIN-59 affect development and gene expression in Caenorhabditis elegans. Development. 2000;127:713–23. doi: 10.1242/dev.127.4.713. [DOI] [PubMed] [Google Scholar]
- 40.Chang S, Johnston RJ, Jr., Hobert O. A transcriptional regulatory cascade that controls left/right asymmetry in chemosensory neurons of C. elegans. Genes Dev. 2003;17:2123–37. doi: 10.1101/gad.1117903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O’Meara MM, Zhang F, Hobert O. Maintenance of neuronal laterality in Caenorhabditis elegans through MYST histone acetyltransferase complex components LSY-12, LSY-13 and LIN-49. Genetics. 2010;186:1497–502. doi: 10.1534/genetics.110.123661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Laue K, Daujat S, Crump JG, Plaster N, Roehl HH, Kimmel CB, Schneider R, Hammerschmidt M, Tübingen 2000 Screen Consortium The multidomain protein Brpf1 binds histones and is required for Hox gene expression and segmental identity. Development. 2008;135:1935–46. doi: 10.1242/dev.017160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hibiya K, Katsumoto T, Kondo T, Kitabayashi I, Kudo A. Brpf1, a subunit of the MOZ histone acetyl transferase complex, maintains expression of anterior and posterior Hox genes for proper patterning of craniofacial and caudal skeletons. Dev Biol. 2009;329:176–90. doi: 10.1016/j.ydbio.2009.02.021. [DOI] [PubMed] [Google Scholar]
- 44.Deguchi K, Ayton PM, Carapeti M, Kutok JL, Snyder CS, Williams IR, Cross NC, Glass CK, Cleary ML, Gilliland DG. MOZ-TIF2-induced acute myeloid leukemia requires the MOZ nucleosome binding motif and TIF2-mediated recruitment of CBP. Cancer Cell. 2003;3:259–71. doi: 10.1016/S1535-6108(03)00051-5. [DOI] [PubMed] [Google Scholar]
- 45.Voss AK, Vanyai HK, Collin C, Dixon MP, McLennan TJ, Sheikh BN, Scambler P, Thomas T. MOZ regulates the Tbx1 locus, and Moz mutation partially phenocopies DiGeorge syndrome. Dev Cell. 2012;23:652–63. doi: 10.1016/j.devcel.2012.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miller CT, Maves L, Kimmel CB. moz regulates Hox expression and pharyngeal segmental identity in zebrafish. Development. 2004;131:2443–61. doi: 10.1242/dev.01134. [DOI] [PubMed] [Google Scholar]
- 47.Crump JG, Swartz ME, Eberhart JK, Kimmel CB. Moz-dependent Hox expression controls segment-specific fate maps of skeletal precursors in the face. Development. 2006;133:2661–9. doi: 10.1242/dev.02435. [DOI] [PubMed] [Google Scholar]
- 48.Thomas T, Voss AK, Chowdhury K, Gruss P. Querkopf, a MYST family histone acetyltransferase, is required for normal cerebral cortex development. Development. 2000;127:2537–48. doi: 10.1242/dev.127.12.2537. [DOI] [PubMed] [Google Scholar]
- 49.Kueh AJ, Dixon MP, Voss AK, Thomas T. HBO1 is required for H3K14 acetylation and normal transcriptional activity during embryonic development. Mol Cell Biol. 2011;31:845–60. doi: 10.1128/MCB.00159-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nagy A, Gertsenstein M, Vintersten K, Behringer R. Staining Whole Mouse Embryos for beta-Galactosidase (lacZ) Activity. CSH Protoc. 2007;2007:t4725. doi: 10.1101/pdb.prot4725. [DOI] [PubMed] [Google Scholar]
- 51.Kim GW, Li L, Gorbani M, You L, Yang XJ. Mice lacking α-tubulin acetyltransferase 1 are viable but display α-tubulin acetylation deficiency and dentate gyrus distortion. J Biol Chem. 2013;288:20334–50. doi: 10.1074/jbc.M113.464792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Theiler K. (1989) The House Mouse–Atlas of Embryonic Development, Springer-Verlag, New York. [Google Scholar]
- 53.Kaufmann MH. (1994) The atlas of mouse development, Revised 1994 ed., Elsevier, Acadmic Press, Amsterdam, Boston, Heidelberg, London, Singapore, Tokyo. [Google Scholar]
- 54.Dong HW. (2008) Allen Reference Altas, a digital brain atlas of the C57BL/6J male mouse, John Wiley & Sons, Inc, Hoboken, New Jersey. [Google Scholar]
- 55.Paxinos G, Franklin KBJ. (2008) The Mouse Brain in Stereotaxic Coordinates, 3rd ed., Academic Press. [Google Scholar]
- 56.Paxinos G, Halliday GM, Watson C, Koutcherov Y, Wang H. (2006) Atlas of the Developing Mouse Brain at E17.5, P0 and P6, 1st Edition, Elsevier, Academic Press, Amsteram, Boston, Heidelberg, London, New York. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.