Abstract
DNA methylation plays a critical role during the development of acquired chemoresistance. The aim of this study was to identify candidate DNA methylation drivers of cisplatin (DDP) resistance in non-small cell lung cancer (NSCLC). The A549/DDP cell line was established by continuous exposure of A549 cells to increasing concentrations of DDP. Gene expression and methylation profiling were determined by high-throughput microarrays. Relationship of methylation status and DDP response was validated in primary tumor cell culture and the Cancer Genome Atlas (TCGA) samples. Cell proliferation, apoptosis, cell cycle, and response to DDP were determined in vitro and in vivo. A total of 372 genes showed hypermethylation and downregulation in A549/DDP cells, and these genes were involved in most fundamental biological processes. Ten candidate genes (S100P, GDA, WISP2, LOXL1, TIMP4, ICAM1, CLMP, HSP8, GAS1, BMP2) were selected, and exhibited varying degrees of association with DDP resistance. Low dose combination of 5-aza-2′-deoxycytidine (5-Aza-dC) and trichostatin A (TSA) reversed drug resistance of A549/DDP cells in vitro and in vivo, along with demethylation and restoration of expression of candidate genes (GAS1, TIMP4, ICAM1 and WISP2). Forced expression of GAS1 in A549/DDP cells by gene transfection contributed to increased sensitivity to DDP, proliferation inhibition, cell cycle arrest, apoptosis enhancement, and in vivo growth retardation. Together, our study demonstrated that a panel of candidate genes downregulated by DNA methylation induced DDP resistance in NSCLC, and showed that epigenetic therapy resensitized cells to DDP.
Keywords: 5-aza-2′-deoxycytidine, GAS1, cisplatin, lung cancer, methylation, trichostatin A
Introduction
Lung cancer is the leading cause of cancer-related deaths worldwide. Non-small cell lung cancer (NSCLC) accounts for approximately 85% of overall lung cancers.1 Platinum-based combination chemotherapy is currently the standard chemotherapy treatment for NSCLC, and cisplatin (DDP) is widely used.2 However, the response to chemotherapy differs among cancer patients, and the efficacy of DDP treatment is often impaired by the emergence of resistance to the drug.3 Therefore, a better understanding of the mechanisms of chemoresistance is required and strategies are needed to identify patients who are unlikely to benefit from the treatment.
DDP induces DNA interstrand and intrastrand crosslinks in tumor cells that subsequently inhibit cell replication and transcription. Chemoresistance to DDP is a complex phenotype that involves many cellular processes, including alterations in drug influx or efflux, apoptosis, cell cycle, DNA repair capacity, and other cellular pathways required for proper response to DNA damage.4 Multiple cellular events, such as DNA methylation, single nucleotide polymorphisms, non-coding RNA, and stem cells, have been reported to be involved in the processes of acquired chemoresistance.5-9
DNA methylation is a major epigenetic modification that leads to gene silencing at the transcriptional level. These epigenetic alterations are heritable and play important roles in carcinogenesis and progression. Substantial DNA methylation either alone or in combination with genetic changes during the acquisition of platinum resistance has been widely reported in various cell line models. Previous studies have reported changes in DNA methylation at promoter CpG islands and associated transcriptional gene silencing in DDP-resistant cancer cells.10-13 However, data in NSCLC has been relatively insufficient.
Unlike genetic mutations, DNA methylation is a reversible process. Therefore, although the mechanisms of demethylation and restoration of expression are not yet fully understood, epigenetic cancer therapy holds great promise for overcoming chemoresistance.14 In this study, we identified a panel of candidate DNA methylation drivers of DDP resistance in NSCLC. Furthermore, we demonstrated that a combination therapy of epigenetic agents could resensitize cancer cells to DDP.
Results
Gene expression and methylation profiling
To identify possible candidate genes involved in DPP chemoresistance, we conducted gene expression and methylation profiling of the A549/DDP cell line, which was established by continuous exposure of A549 cells to increasing concentrations of DDP, using high-throughput microarrays. We filtered the genes according to the following criteria: expression log fold change A549 vs. A549/DDP > 1.0 and methylation delta_β A549 vs. A549/DDP < –0.7. These criteria identified a total of 372 genes, which covered 861 CpG sites (Fig. S1; Table S1). These hypermethylated and downregulated genes were considered candidate genes that may play a potential role in DDP resistance.
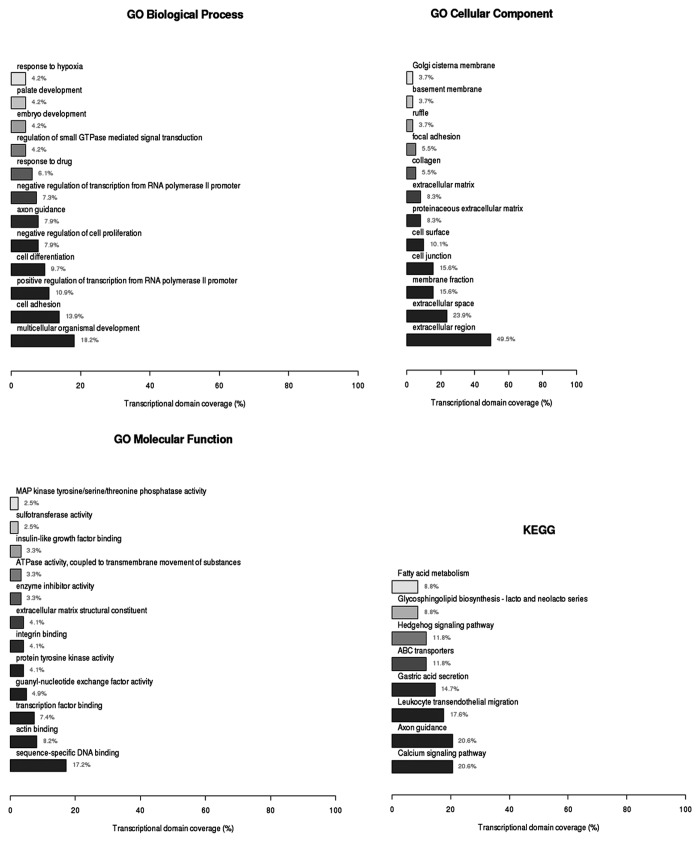
To identify possible cellular functions of these genes, we performed gene ontology (GO) analysis using three domains: biological process, which includes multicellular organismal development, cell adhesion, positive regulation of transcription from RNA polymerase II promoter, cell differentiation, negative regulation of cell proliferation, and response to drug (Table S2); cellular component, mostly involving the extracellular region (Table S3); and molecular function (Table S4), which is mainly related to sequence-specific DNA binding, actin binding, and transcription factor binding KEGG Pathway analysis included the calcium signaling pathway, ABC transporters and Hedgehog signaling pathway. Four key genes in the Hedgehog signaling pathway (encoding GAS1, BMP2, WNT5A, and WNT6) showed a significant difference between the paired cell lines (P < 0.05, Table S5; Fig. 1).
Figure 1. Function and pathway analysis of the 372 hypermethylated genes identified in the A549/DDP cell line. Genes were identified using the Illumina Infinium HumanMethylation450 BeadArray platform. Gene ontology (GO) analysis by three domains: Biological Process (A), Cellular Component (B) and Molecular Function (C). KEGG Pathway analysis (D).
Validation of the gene expression and methylation status
To verify the results of the microarray, a self-assembling quantitative PCR array was employed to evaluate the differentially expressed genes in A549 and A549/DDP cells. In general, the expression of 62 candidate coding or non-coding genes from the PCR Array was consistent with results from the Agilent microarray. Moreover, 61.3% (38/62) of the candidate genes showed a >5-fold decrease in expression in A549/DDP cells compared with A549 cells (Table S6).
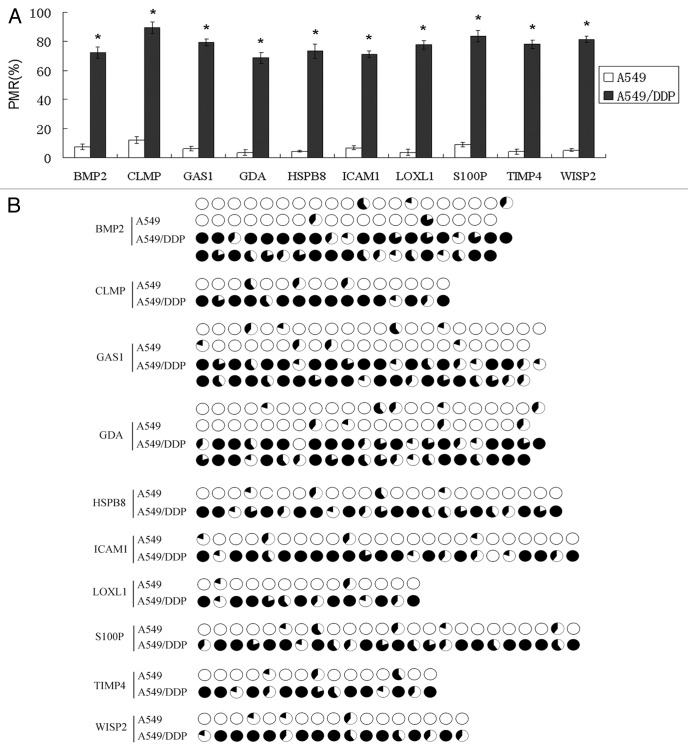
Based on the fold-change of gene expression, CpG status (low-CpG was excluded) and the literature, a total of 10 methylated genes (S100P, GDA, WISP2, LOXL1, TIMP4, ICAM1, CLMP, HSP8, GAS1, BMP2) detected in A549/DDP cells were selected for further analysis (Table 1). qMSP and BSP were used to evaluate their methylation status. All 10 candidate genes were confirmed to be hypermethylated in A549/DDP cells compared with A549 cells (Fig. 2). This data directly supports the results of the gene expression and methylation microarray.
Table 1. Candidate genes selected based on the fold change value in gene expression, CpG status and literature.
| Gene Symbol | Gene Name |
Genomic Coordinates | Log.FC.A549.vs.DDP1 | FC.A549.vs DDP2 | The main function |
|---|---|---|---|---|---|
| S100P | S100 calcium binding protein P | chr4:6698819–6698878 | 10.9517 | 11465.41 | Cell cycle progression and differentiation23,24 |
| GDA | guanine deaminase | chr9:74764241–74764903 | 10.10189 | 21100.65 | Microtubule assembly37 |
| WISP2 | WNT1 inducible signaling pathway protein 2 | chr20:43356347–43356406 | 9.287682 | 852.17 | Downstream of the WNT1 signaling pathway that is relevant to malignant transformation25,26 |
| LOXL1 | lysyl oxidase-like 1 | chr15:74241855–74241914 | 7.908342 | 411.57 | Formation of crosslinks in collagens and elastin27,28 |
| TIMP4 | Tissue inhibitor of metallopeptidase 4 | chr3:12194787–12194728 | 7.575363 | 200.16 | Remodeling of the Extracellular matrix29 |
| ICAM1 | intercellular adhesion molecule 1 | chr19:10396298–10396358 | 7.333666 | 119.84 | cell adhesion30,31 |
| CLMP | CXADR-like membrane protein | chr11:122943288–122943229 | 7.327258 | 2328.20 | A component of epithelial tight junctions38 |
| HSPB8 | heat shock 22kDa protein 8 | chr12:118100934–118101264 | 7.275802 | 1704.34 | regulation of cell proliferation, apoptosis, and carcinogenesis32 |
| GAS1 | growth arrest-specific 1 | chr9:89559953–89559894 | 6.892795 | 175.46 | Growth suppression33,34 |
| BMP2 | bone morphogenetic protein 2 | chr20:6760810–6760869 | 5.316618 | 55.52 | Cell growth, differentiation, migration and invasion35 |
1 Data from Agilent SurePrint G3 Human Gene Expression microarray; 2Data from customized PCR array; FC, fold change.
Figure 2. Validation of the methylation status by qMSP (A) and BSP (B). All 10 candidate genes were confirmed to be hypermethylated in A549/DDP cells compared with A549 cells. The amount of methylated DNA was determined by the threshold cycle number (Ct) for each sample and assessed as the percentage of methylation reference (PMR). Each pie in BSP results represented a CpG site; methylation (black) and unmethylation (white) ratios were calculated according to the five clones.
Methylation status and DDP response
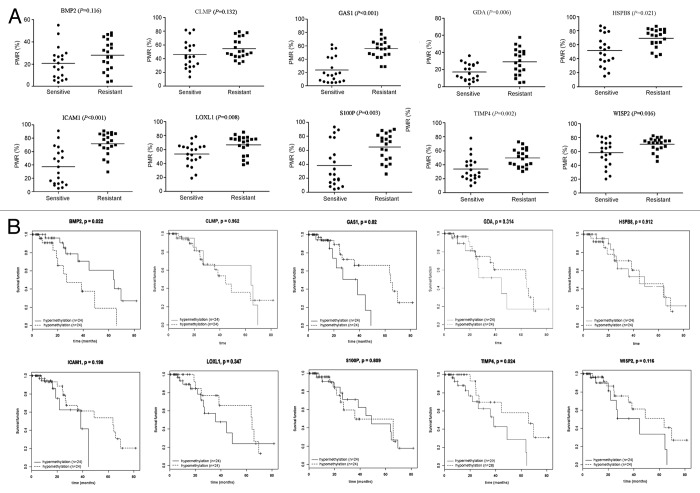
Next we evaluated the methylation status of the candidate genes in primary tumor samples. NSCLC samples were analyzed by primary tumor cell culture and drug susceptibility testing. Twenty NSCLC samples were identified as DDP sensitive (IC50 < 5 mg/L) and 20 samples were considered DDP resistant (IC50 > 10 mg/L). We tested the methylation status of the 10 candidate genes in these samples using qMSP. Eight genes (S100P, GDA, WISP2, LOXL1, TIMP4, ICAM1, HSP8, and GAS1) showed higher methylation levels in DDP-resistant samples compared with DDP-sensitive samples (P < 0.05, Fig. 3A).
Figure 3. Methylation status and DDP response. (A) By primary tumor cell culture and drug susceptibility testing, 20 NSCLC samples were considered DDP sensitive (IC50 < 5 mg/L) and 20 samples were considered DDP resistant (IC50 > 10 mg/L). Methylation status of 10 candidate genes was tested by qMSP; (B) Kaplan-Meier analysis of candidate gene methylation and overall survival in patients who received platinum-based chemotherapy using TCGA data. Hyper- or hypo-methylation was defined by the median of methylation index β-values.
We downloaded and analyzed publicly available data of lung adenocarcinoma and squamous cell carcinoma from the Cancer Genome Atlas Project (TCGA) Project (Table S7) and identified 71 patients with a history of cisplatin/carboplatin chemotherapy. Kaplan-Meier analysis showed that hypermethylation of TIMP4 or GAS1 was associated with adverse overall survival (P = 0.024 and P = 0.02, respectively), while BMP2 methylation showed a protective role in survival (P = 0.022). Although no statistical significance was found, WISP2 methylation tended to be associated with adverse overall survival (P = 0.116, Fig. 3B).
Epigenetic therapy in vitro
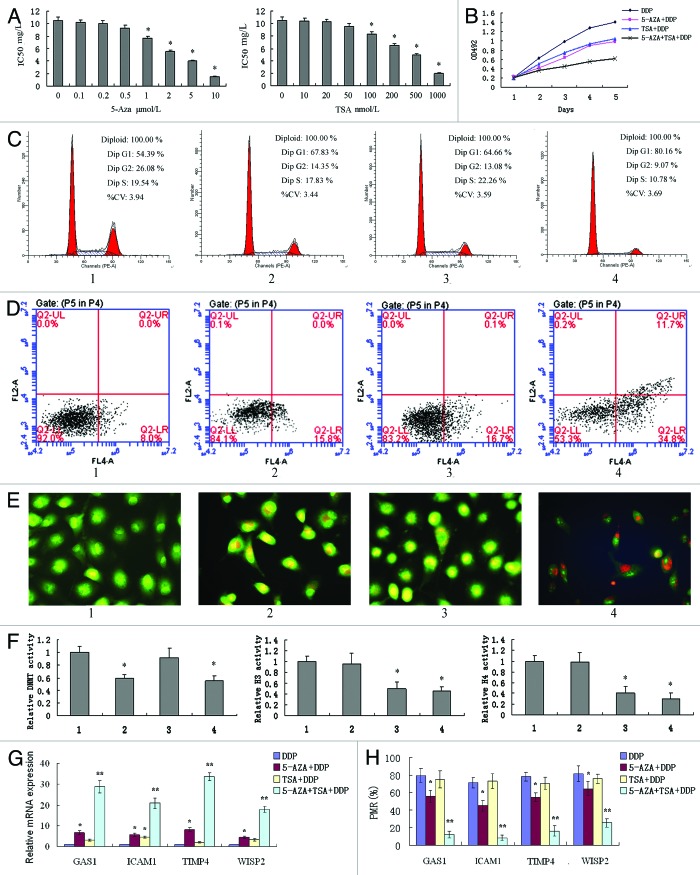
We next assessed the effects of epigenetic agents on A549/DDP cells. Epigenetic agents in experimental analysis were used at the minimum effective dose (Fig. 4A), and A549/DDP cells were cultured in RPMI-1640 medium containing 2 mg/L DDP in these experiments. Combinatorial treatments of 1 μM 5-Aza-dC and 100 nM TSA significantly inhibited cellular proliferation, induced G1 arrest and increased apoptosis of A549/DDP cells compared with either single treatment or untreated cells (Fig. 4B–E).
Figure 4. In vitro effects of the combinatorial treatments with 5-Aza-dC and TSA in the A549/DDP cell line. (A) Minimum effective dose of 5-Aza-dC and TSA determined by MTT; (B) cell proliferation determined by MTT; (C) cell cycle determined by flow cytometry; (D) apoptosis determined by flow cytometry; (E) apoptosis determined by fluorescence microscope; (F) DNMT activity and histone deacetylase H3/H4 activity; (G) mRNA expression of four candidate genes; (H) methylation status of four candidate genes. 1, A549/DDP cells; 2, A549/DDP cells treated with 1 μM 5-Aza-dC; 3, A549/DDP cells treated with 100 nM TSA; 4, A549/DDP cells treated with 1 μM 5-Aza-dC and 100 nM TSA. Cells were cultured in RPMI-1640 medium containing 2 mg/L DDP in these experiments. *P < 0.05 vs control; **P < 0.05 vs. other 3 groups.
DNA methyltransferase (DNMT) activity and histone deacetylase H3/H4 activity were inhibited by 1 μM 5-Aza-dC and 100 nM TSA, respectively, but combinatorial treatments had no synergistic inhibitory effects on DNMT and H3/H4 activity (Fig. 4F). This suggests that the synergistic anti-tumor effects of 5-Aza-dC and TSA might be due to the regulation of key gene expression. This speculation was further confirmed by the fact that hypermethylated status and downregulated expression of GAS1, TIMP4, ICAM1 and WISP2 genes in A549/DDP cells were all reversed after combinatorial treatments (Fig. 4G and H).
Epigenetic therapy in vivo
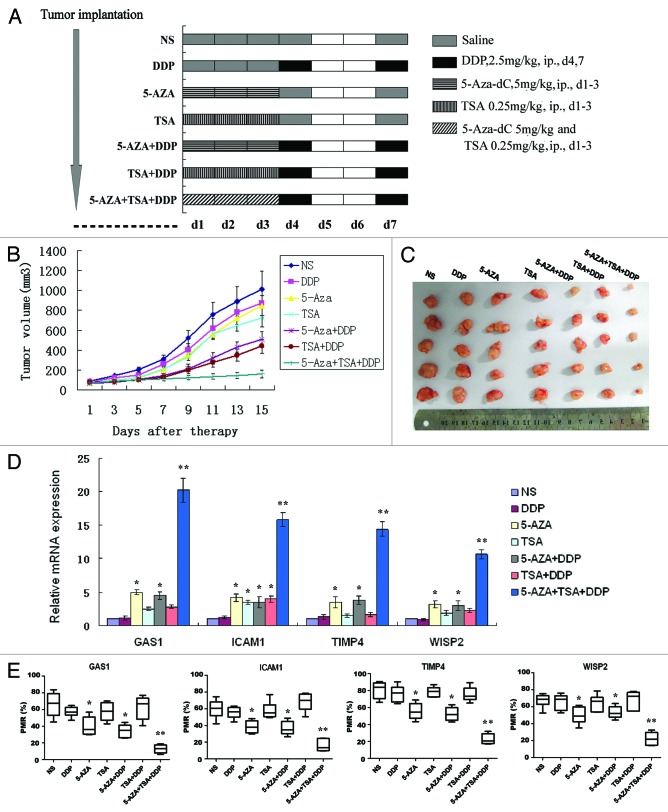
Epigenetic therapy effects were next evaluated in vivo. The doses of epigenetic agents were well tolerated by control mice without causing any serious toxicity, for instance hemorrhage, infection and death (Fig. 5A). As shown in Figure 5B and C, DDP, 5-Aza-dC or TSA alone did not result in significant suppression of tumor growth (P = 0.467, 0.316 and 0.105, respectively). However, pre-treatment with 5-Aza-dC or TSA followed by DDP caused significant tumor inhibition (>50% compared with control, P = 0.010 and 0.005, respectively), while pre-treatment with 5-Aza-dC and TSA fully inhibited tumor growth compared with other groups (P < 0.001). Together this data suggests that epigenetic pre-treatment reactivated the genes potentially related to DDP chemosensitivity.
Figure 5. Epigenetic therapy in vivo. A549/DDP cells (2 × 106/100 μL PBS) were subcutaneously inoculated into the right flank of BALB/c nu/nu mice and animals were randomly divided into 7 treatment groups as described in Methods (A). The tumor size was monitored every other day (B). Mice were sacrificed and the tumors were isolated after two weeks (C). The mRNA expression (D) and methylation (E) profiling of GAS1, TIMP4, ICAM1 and WISP2 were determined in each group. *P < 0.05 vs. NS group; **P < 0.05 vs. other 6 groups.
The methylation and expression profiles of GAS1, TIMP4, ICAM1 and WISP2 were determined in each experimental group. As expected, methylation levels were lowest and expression levels were highest in combinatorial pre-treatment groups (Fig. 5D and E).
GAS1 function in DDP chemoresistance
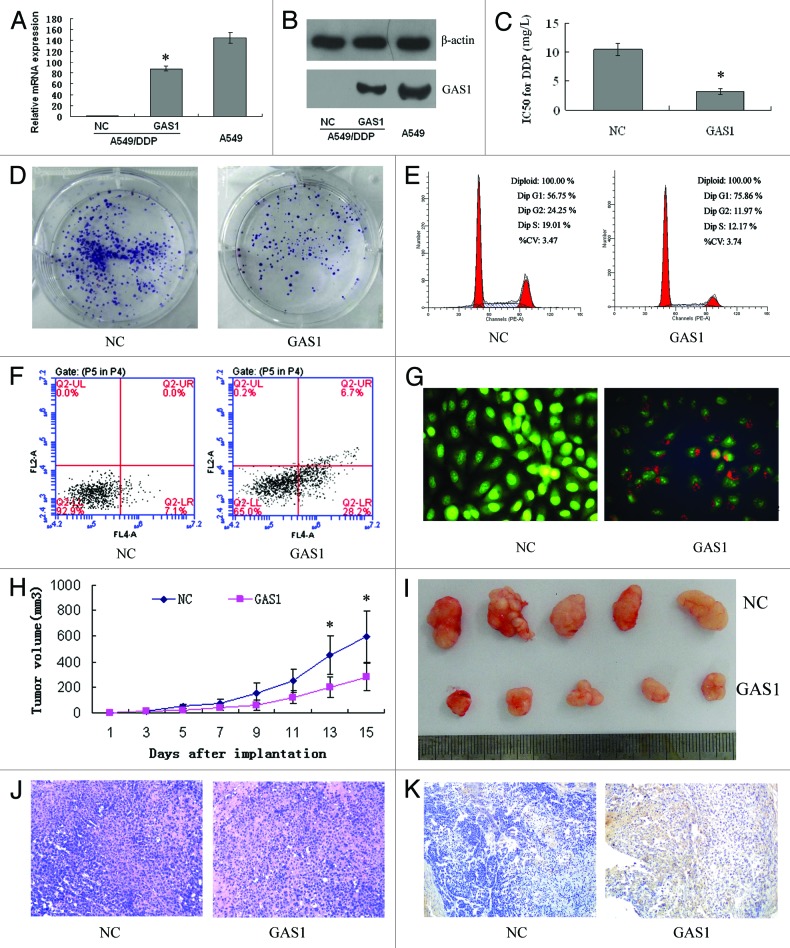
To examine the association between candidate gene expression and DDP chemosensitivity, A549/DDP cells were stably transfected with the N-eGFP-GAS1 vector. Real-time-PCR and western blot analyses confirmed that the expression of GAS1 was significantly increased in A549/DDP cells (P < 0.001), but still lower than the level in parental A549 cells (Fig. 6A and B).
Figure 6. Verification of GAS1 function. A549/DDP cells were stably transfected with N-eGFP-NC or N-eGFP-GAS1 vector, and GAS1 expression levels were detected via real-time PCR (A) and western blot (B) 48 h post-transfection. (C) MTT assay revealed that cells with upregulated GAS1 expression were more sensitive to DDP. (D) Colony formation assay revealed suppression of cellular proliferation with upregulated GAS1 expression. (E) Flow cytometric analysis of cell cycle indicated that upregulation of GAS1 significantly induced G1 phase arrest. Flow cytometric analysis (F) and fluorescence microscope (G) indicated that upregulation of GAS1 significantly induced apoptosis. (H) Growth curve of tumors derived from GAS1-transfected A549/DDP cells and NC cells. Each point represents the mean ± SD of 5 mice. (I) Representative photographs of tumors formed 2 wk after the subcutaneous transplantation. Transplanted tumors with H&E staining (J), and immunohistochemistry of GAS1 (K).
Next, MTT assay was used to determine chemosensitivity of cells overexpressing GAS1 to DDP. Compared with mock-transfected ones, the IC50 values for DDP in GAS1-transfected A549/DDP cells was significantly reduced by 66.8% (10.45 ± 0.95 mg/L vs. 3.27 ± 0.48 mg/L, respectively) (P < 0.001, Fig. 6C). Colony formation assay results showed that upregulation of GAS1 led decreased proliferation of A549/DDP cells (P < 0.001, Fig. 6D). Flow cytometry assay showed that enforced expression of GAS1 in A549/DDP cells resulted in cell cycle arrest in G1 phase (P = 0.011, Fig. 6E) and a dramatic increase of apoptosis (P = 0.001, Fig. 6F). Morphological features of apoptotic cells and necrosis were also observed in GAS1-transfected A549/DDP cells (Fig. 6G).
A549/DDP cells transfected with GAS1-expressing or mock plasmid were injected subcutaneously into nude mice, followed by DDP treatment. As shown in Figure 6H, tumors derived from GAS1-transfected A549/DDP cells grew more slowly compared with controls. All mice were sacrificed 2 wk after the transplantation, and the average size of tumors derived from GAS1-transfected A549/DDP cells was significantly reduced by 53.3% (P = 0.041, Fig. 6I). These results suggested that GAS1 downregulation by methylation was associated with DDP chemoresistance.
Discussion
DNA methylation at CpG islands within or near promoter regions has been demonstrated as an important epigenetic regulatory mechanism of gene expression. Hypermethylation could lead to an alteration of the chromatin framework, thereby directly repressing transcription and leading to the downregulation or silencing of tumor suppressor genes, thus contributing to cancer initiation and progression.15 Previous studies have demonstrated an association between abnormal methylation and cancer diagnosis or prognosis in various cancers.16 In addition, DNA methylation plays a critical role in the development of acquired chemoresistance via transcriptional inactivation of several key genes. Clinically, colorectal cancers with hMLH1 methylation are less aggressive, but nonreactive to 5-Fu.17 The correlations between CHFR methylation and sensitivity to microtubule inhibitors,18 and MGMT gene methylation and response to alkylating agents in patients with glioblastoma19 have also been confirmed.
In this study, integrated analysis of DNA methylation and gene expression in a genome-wide profiling revealed specific methylated and silenced genes associated with DDP resistance in a lung adenocarcinoma cell line. GO analysis showed these 372 genes are involved in nearly all the fundamental biological processes, such as multicellular organismal development, cell adhesion, positive regulation of transcription from RNA polymerase II promoter, cell differentiation, negative regulation of cell proliferation, and drug response. KEGG pathway analysis revealed functions in the calcium signaling pathway, ABC transporters and Hedgehog signaling pathway. Except for a few genes (such as S100P), our results were not fully consistent with previously reports, which identified p16, hMLH1, RASSF1A, and IGFBP3 methylation, Wnt, JNK/p38 MAPK, p53 and their signaling pathways in DDP resistance.20-22 The differences may be due to different cell lines and microarray analysis approaches used. Furthermore, as chemoresistance processes involve multiple major cellular pathways, it is likely that the underlying mechanisms are broad and complex, and thus each of these studies only provides a narrow glimpse of an intricate regulatory network.
In this study, our gene microarray results were verified by a low-throughput, self-assembling quantitative PCR array. Ten genes (S100P, GDA, WISP2, LOXL1, TIMP4, ICAM1, CLMP, HSP8, GAS1, BMP2) were initially screened for analysis, based on fold-change of gene expression, CpG status and literature. These genes were involved in the regulation of cell growth, cell cycle, differentiation, apoptosis, adhesion, migration and tumor microenvironment.23-36 Except GDA37 and CLMP,38 the other genes have been proven to have important significance in tumor development, and many have critical roles in drug resistance.39-42 For example, S100P, a 95 amino acid protein, acts as an autocrine growth and survival factor. Decreased S100P expression was observed in at least five platinum-resistant cell lines, including one uterine cervix cancer, one liver cancer, one colon cancer and two bladder cancer cell lines.12 All 10 candidate genes were confirmed to be hypermethylated in A549/DDP cells compared with parental A549 cells. Relationships of methylation status and DDP response were further validated in primary tumor cells and samples from TCGA patients who underwent platinum-based chemotherapy, and four genes (GAS1, TIMP4, ICAM1, and WISP2) were selected for subsequent analysis. Our results confirm that hypermethylation of these genes involved in DDP resistance in NSCLC. Further research is needed to explore the mechanisms underlying their contribution to chemoresistance.
As DDP resistance was associated with downregulation of key genes by DNA methylation in NSCLC, we speculated whether epigenetic therapy could reverse DDP resistance. DNMT inhibitors can effectively induce DNA demethylation and phenotypic changes related to the reactivation of epigenetically silenced genes. These findings were later adapted to the targeting of epigenetic mutations in cancer and brought about the fundamental concept of epigenetic cancer therapy.43 In addition to DNMT inhibitors,44 histone deacetylase (HDAC) inhibitors have also contributed to shape epigenetic cancer therapy.45 In solid tumors, additional DNMT and HDAC inhibitors are currently being evaluated in both preclinical studies and clinical trials. However, a direct link between DNA demethylation and clinical responses has not yet been demonstrated, and currently there are no established DNA methylation biomarkers that accurately predict patient response. It is important to note that histone deacetylation has been shown to synergistically interact with DNA methylation in the epigenetic silencing of cancer genes.46 This observation has also provided the scientific rationale for various clinical trials with combinations of DNMT and HDAC inhibitors. Somewhat surprisingly, these studies have yet to reveal synergistic clinical effects, which might again be related to complexities in the drugs’ modes of action that are only beginning to become elucidated.
Therefore, epigenetic cancer therapy combined with chemotherapy may be a direction worth exploring. Combining 5-Aza-dC with cytotoxic drugs has been found to increase sensitivity of neuroblastoma cells to chemotherapeutic drugs.47 In a phase II clinical trial, low-dose decitabine altered DNA methylation of genes and cancer pathways, restoring sensitivity to carboplatin in patients with heavily pretreated ovarian cancer and resulting in a high response rate and prolonged progression-free survival.48 In this study, pretreatment of low-dose 5-Aza-dC and TSA significantly increased DDP chemosensitivity of resistant cells both in vitro and in vivo. Re-expression of key genes, such as GAS1, TIMP4, ICAM1, and WISP2, was considered to account for the underlying mechanisms. However, global demethylation does not only affect cancer-specific genes but also affects some developmental genes.49 Moreover, epigenetic side effects could severely curtail the specificity of therapy.50 Dose limitation may be an effective way to reduce side effects, while a combination of low-dose epigenetic agents improves the efficacy. The combined modes, dose of epigenetic agents, and cytotoxic drugs still need further study to achieve optimal therapeutic effect.
To directly verify candidate DNA methylation drivers of DDP resistance in NSCLC, the effect of GAS1 transfection on A549/DDP cells chemosensitivity was examined. Forced expression of GAS1 in A549/DDP cells by gene transfection contributed to increased sensitivity to DDP, proliferation inhibition, cell cycle arrest, apoptosis enhancement, and in vivo growth retardation. Thus, GAS1 downregulation by methylation had a close relationship with DDP chemoresistance. GAS1, a glycosylphosphatidyl inositol-linked protein, was originally identified as a protein involved in the contact inhibition of fibroblasts.51 As a tumor suppressor gene, it is directly related to cell cycle arrest in the G0 to S phase transition.52 A previous study also indicated that GAS1 was an epirubicin resistance-related gene in gastric cancer cells because upregulation of GAS1 reversed the chemoresistance of gastric cancer. GAS1 is considered to enhance hedgehog signaling activity,53 activate p38MAPK54 and prevent the activation of Akt,55,56 all of which are crucial mediators of survival and cellular proliferation pathways.
Taken together, we identified a panel of candidate genes downregulated by DNA methylation and demonstrated that these genes induced DDP resistance in NSCLC. Epigenetic therapy resensitized cells to DDP both in vitro and in vivo, along with the demethylation and restoration of expression of candidate genes. Finally, we proved that overexpression of GAS1 directly affected the DDP chemosensitivity of resistant cells. Our study enhanced the understanding of DDP resistance mechanisms and contributed to developments for treatment for NSCLC.
Materials and Methods
Cell culture
The A549 human lung adenocarcinoma cell line was purchased from Shanghai Institutes for Biological Sciences, Chinese Academy of Cell Resource Center. The DDP-resistant A549/DDP variant cell line was established by continuous exposure to increasing concentrations of DDP for a 10-mo period following the methodology described previously (Fig. S2).13,57 A549 and A549/DDP cell lines were maintained in RPMI-1640 medium (Life Technologies) supplemented with 10% fetal calf serum (Gibco) in a humid atmosphere containing 5% CO2 at 37 °C. The A549/DDP cells medium additionally contained 2 mg/L DDP to maintain the cells’ drug-resistant phenotype. Cells in the logarithmic phase of growth were used for all experiments. For treatment with 5-aza-2′-deoxycytidine (5-Aza-dC, Sigma), a specific DNA methyltransferase inhibitor, or trichostatin A (TSA, Sigma), a specific histone deacetylase inhibitor, the cell culture medium was changed every 24 h.
Methylome analysis
DNA methylation was assessed using the Illumina Infinium HumanMethylation450 BeadArray platform, as previously described.58 DNA methylation index (MI) is reported as β-values, calculated from mean methylated (M) and unmethylated (U) signal intensities for each locus for each sample using the formula (β = M/[M + U]).
Gene expression microarrays
The cDNA from cell lines was labeled with Cy3 dye and hybridized to Agilent SurePrint G3 Human Gene Expression microarrays (8?60K) to assess gene expression according to the manufacturer’s protocol (Agilent Technologies). Fluorescence images of the hybridized arrays were generated using the Agilent DNA Microarray Scanner, and the intensities were extracted with Agilent Feature Extraction software ver.10.7.3.1. The fold change was log2 transformed.
PCR array
This study used a customized PCR array from CT Bioscience59 to compare the expression profile of a selected group of genes in A549 and A549/DDP cells. Sixty-two potential genes based on methylation and expression microarray were selected as the target mRNAs. GAPDH, β2-MG, ACTB, RPL27, HPRT1, and OAZ1 were used as housekeeping genes for normalization. After RNA isolation, 1 μg of total RNA was used for reverse transcription using an RT kit (CT Bioscience, Cat # CTB101) in a 20 μL volume. The PCR array employs SYBR Green I-based real-time PCR to quantify gene expression level. Gene specific primers are pre-deposited into wells of a 96-well PCR plate in the array.
Culture and DDP treatment of NSCLC tissues
To measure the sensitivity of the primary NSCLC samples to DDP, fresh tumors were minced, passed through a nylon mesh and enzyme disaggregated with collagenase type II and hyaluronidase in Dulbecco’s modified Eagle’s medium nutrient mixture F-12/Ham media (Sigma Aldrich) with antibiotics. Cells were resuspended in 96-well microtiter plates followed by exposure to various concentrations of DDP as described previously.13
Validation in the cancer genome atlas samples
To verify associations of methylation observed in microarray with overall survival in NSCLC patients, we downloaded and analyzed data publicly available from the Cancer Genome Atlas Project (TCGA; http://tcga-data.nci.nih.gov/). Patients who received platinum-based chemotherapy were selected. Survival curves were calculated using the Kaplan-Meier method and compared by log-rank testing. Hyper- or hypo-methylation was defined by the median of methylation index β-values.
Real-time quantitative PCR
Total RNA was isolated using TRIzol reagent (Invitrogen). Reverse transcription reaction was performed using 2 μg of total RNA with a first strand cDNA kit (Takara). PCR amplification was performed for 20 s at 95 °C, followed by 40 cycles at 95 °C for 5 s, and annealing/extension at 60 °C for 30 s in ABI 7300 Thermocycler (Applied Biosystems), using the SYBR Premix Ex Taq kit (Takara). The specific primer sequences for each gene were listed in Table S8. Data analysis was done using the 2-△△CT method for relative quantification, and all samples were normalized to GAPDH.
Detection of methylation status
After genomic DNA extraction and spectrophotometric quantization, 1 μg of genomic DNA was bisulfite-treated with the EZ-DNA methylation Gold Kit (Zymo Research). Quantitative methylation-specific PCR (qMSP) was then performed in an ABI 7300 Thermocycler, using the SYBR Premix Ex Taq kit (Takara) as described previously.60 The specific primer sequences for each gene were designed by Methyl Primer Express® Software v1.0 and are listed in Table S8. β-actin (ACTB) was used to normalize for the input DNA. The amount of methylated DNA was determined by the threshold cycle number (Ct) for each sample and assessed as the percentage of methylation reference (PMR) using the formula: [(gene/ACTB)sample: (gene/ ACTB)SssI-treated genomic DNA] × 100. Samples with ≥ 4% of PMR were regarded as hypermethylated. For bisulphite sequencing (BSP), the bisulphite DNA was amplified by PCR with BSP primers and PCR products were cloned into the pUC57 vector (Genscript). Five clones were selected and sequenced for each sample.
MTT assay for cell growth inhibition
A549/DDP cells (cultured in RPMI-1640 medium containing 2 mg/L DDP) were seeded at a density of 2 × 103 cells in each well of the 96-well plates. 5-Aza-dC (1 μM) and/or TSA (100 nM) were added to the wells for 24, 48 and 72 h. MTT (5 g/L, 20 μL/well) was added to each well and incubated at 37 °C for 4 h. DMSO was then added (150 μL/well) to each well to dissolve any crystals and the plates were agitated for 10 min. Absorbance values at 492 nm were detected by the microplate reader.
Cell viability assay
A549/DDP cells (cultured in RPMI-1640 medium without DDP) were seeded into 96-well plates (2 × 103 cells/well) 24 h after drug treatment or transfection and allowed to attach overnight. Freshly prepared DDP was then added at different final concentrations for 48 h. Cell viability was assessed via a standard MTT assay and IC50 was calculated. All assays were performed in quintuplicate and repeated at least three times.
Transfection of cells
GAS1 gene-expressing plasmid (N-eGFP-GAS1) and the mock N-eGFP-negative control (N-eGFP-NC) were purchased from GenePharma. All plasmid DNA was extracted using a Plasmid Mini Kit (Qiagen). Cells were transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol. G418 (600 mg/L, Sigma) was used to screen the stably transfected cells. GAS1 expression was confirmed by real-time PCR and western blotting.
Flow cytometric analysis
Cells were harvested directly or 48 h after drug treatment or transfection and washed with ice-cold phosphate-buffered saline (PBS). The PI/RNase staining kits (Multisciences) and annexin V-fluorescein isothiocyanate (FITC) apoptosis detection kits (KeyGEN Biotech) were used to detect cell cycle and apoptosis in a FACScan instrument (Becton Dickinson), respectively.
Fluorescence microscope
For drug treatment, A549/DDP cells (cultured in RPMI-1640 medium containing 2 mg/L DDP) were seeded at 5 × 104 in each well of 24-well plates, and 5-Aza-dC (1 μM) and/or TSA (100 nM) were added to the wells for 48 h. For functional analysis, A549/DDP cells transfected with GAS1 gene-expressing or mock plasmid were seeded in 24-well plates and cultured in RPMI 1640 media without DDP for 48 h. Then cells were harvested and suspended in PBS containing fluorescence dye AO/EB (both AO and EB were at the concentration of 100 mg/L in PBS), following by preparation for slides. The morphology of the cells was observed under a fluorescence microscope (IX71, Olympus) and photographed.
Colony formation assay
A total of 1000 cells were seeded into six-well plates with 2 mL culture medium. After culturing in RPMI 1640 media supplemented with 10% FBS at 37 °C and 5% CO2 for 14 d, cells were washed twice with PBS, fixed with methanol and stained with 0.1% crystal violet. Visible colonies were manually counted. The cloning efficiency (%) = (the number of clones/the number of seed cells) × 100%.
In vivo experiments
The animal study protocol was approved by the Animal Experimentation Ethics Committee of the Jinling Hospital. For epigenetic therapy, A549/DDP cells (2 × 106/100 μL PBS) were subcutaneously inoculated into the right flank of BALB/c nu/nu mice. When the average tumor size reached approximately 50 mm3, animals were randomly divided into 7 groups, with five in each group, and subjected to the corresponding treatment. The epigenetic therapy used in this study was 5-Aza-dC 5 mg/kg and/or TSA 0.25 mg/kg, ip, d1–3. DDP chemotherapy regimen was 2.5 mg/kg, ip, twice per week. Combinatorial treatments were performed by pre-treatment with 5-Aza-dC and/or TSA for 3 d and then DDP was added on the 4th and 7th day. The experimental groups were as follows: (1) normal saline (NS); (2) DDP alone; (3) 5-Aza-dC alone; (4) TSA alone; (5) pre-treatment with 5-Aza-dC followed by DDP; (6) pre-treatment with TSA followed by DDP; (6) pre-treatment with 5-Aza-dC and TSA followed by DDP. Tumor volume was estimated every other day by the formula: 0.5 × length × width2.
For GAS1 function, A549/DDP cells (2 × 106/100 μL PBS) transfected with GAS1 gene-expressing or mock plasmid were injected subcutaneously into nude mice on the right flank (n = 5 mice per group). When the average tumor size reached approximately 50 mm3, DDP was given via intraperitoneal injection at a dose of 2.5 mg/kg, two times a week. All mice were sacrificed after 2 wk. Tumor tissues were excised, paraffin-embedded, and formalin-fixed, and H&E staining was performed. Immunostaining analysis for GAS1 protein was performed according to a previous report.36
Statistical analysis
The SPSS 16.0 software system (SPSS) was used for statistical analysis. Data are expressed as the mean ± standard error. The differences between groups were analyzed using a Student t test when only two groups were compared or one-way analysis of variance when more than two groups were compared. All tests performed were two-sided. P < 0.05 was taken as statistical significance.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This study was supported by the Natural Science Foundation of China (No. 81101650). The valuable help of Prof Haoyan Chen, Shanghai Jiao Tong University is greatly appreciated.
Glossary
Abbreviations:
- 5-Aza-dC
5-aza-2′-deoxycytidine
- BSP
bisulphite sequencing
- DNMT
DNA methyltransferase
- GAS1
growth arrest-specific gene 1
- HDAC
histone deacetylase
- NSCLC
non-small cell lung cancer
- PMR
percentage of methylation reference
- TSA
trichostatin A
References
- 1.Siegel R, Ward E, Brawley O, Jemal A. Cancer statistics, 2011: the impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J Clin. 2011;61:212–36. doi: 10.3322/caac.20121. [DOI] [PubMed] [Google Scholar]
- 2.Schiller JH, Harrington D, Belani CP, Langer C, Sandler A, Krook J, Zhu J, Johnson DH, Eastern Cooperative Oncology Group Comparison of four chemotherapy regimens for advanced non-small-cell lung cancer. N Engl J Med. 2002;346:92–8. doi: 10.1056/NEJMoa011954. [DOI] [PubMed] [Google Scholar]
- 3.Stewart DJ. Mechanisms of resistance to cisplatin and carboplatin. Crit Rev Oncol Hematol. 2007;63:12–31. doi: 10.1016/j.critrevonc.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 4.Marsh S, McLeod H, Dolan E, Shukla SJ, Rabik CA, Gong L, Hernandez-Boussard T, Lou XJ, Klein TE, Altman RB. Platinum pathway. Pharmacogenet Genomics. 2009;19:563–4. doi: 10.1097/FPC.0b013e32832e0ed7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tan XL, Moyer AM, Fridley BL, Schaid DJ, Niu N, Batzler AJ, Jenkins GD, Abo RP, Li L, Cunningham JM, et al. Genetic variation predicting cisplatin cytotoxicity associated with overall survival in lung cancer patients receiving platinum-based chemotherapy. Clin Cancer Res. 2011;17:5801–11. doi: 10.1158/1078-0432.CCR-11-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bian HB, Pan X, Yang JS, Wang ZX, De W. Upregulation of microRNA-451 increases cisplatin sensitivity of non-small cell lung cancer cell line (A549) J Exp Clin Cancer Res. 2011;30:20. doi: 10.1186/1756-9966-30-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang Y, Li H, Hou S, Hu B, Liu J, Wang J. The noncoding RNA expression profile and the effect of lncRNA AK126698 on cisplatin resistance in non-small-cell lung cancer cell. PLoS One. 2013;8:e65309. doi: 10.1371/journal.pone.0065309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barr MP, Gray SG, Hoffmann AC, Hilger RA, Thomale J, O’Flaherty JD, Fennell DA, Richard D, O’Leary JJ, O’Byrne KJ. Generation and characterisation of cisplatin-resistant non-small cell lung cancer cell lines displaying a stem-like signature. PLoS One. 2013;8:e54193. doi: 10.1371/journal.pone.0054193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo R, Wu G, Li H, Qian P, Han J, Pan F, Li W, Li J, Ji F. Promoter methylation profiles between human lung adenocarcinoma multidrug resistant A549/cisplatin (A549/DDP) cells and its progenitor A549 cells. Biol Pharm Bull. 2013;36:1310–6. doi: 10.1248/bpb.b13-00153. [DOI] [PubMed] [Google Scholar]
- 10.Yu W, Jin C, Lou X, Han X, Li L, He Y, Zhang H, Ma K, Zhu J, Cheng L, et al. Global analysis of DNA methylation by Methyl-Capture sequencing reveals epigenetic control of cisplatin resistance in ovarian cancer cell. PLoS One. 2011;6:e29450. doi: 10.1371/journal.pone.0029450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zeller C, Dai W, Steele NL, Siddiq A, Walley AJ, Wilhelm-Benartzi CS, Rizzo S, van der Zee A, Plumb JA, Brown R. Candidate DNA methylation drivers of acquired cisplatin resistance in ovarian cancer identified by methylome and expression profiling. Oncogene. 2012;31:4567–76. doi: 10.1038/onc.2011.611. [DOI] [PubMed] [Google Scholar]
- 12.Chang X, Monitto CL, Demokan S, Kim MS, Chang SS, Zhong X, Califano JA, Sidransky D. Identification of hypermethylated genes associated with cisplatin resistance in human cancers. Cancer Res. 2010;70:2870–9. doi: 10.1158/0008-5472.CAN-09-3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ibanez de Caceres I, Cortes-Sempere M, Moratilla C, Machado-Pinilla R, Rodriguez-Fanjul V, Manguán-García C, Cejas P, López-Ríos F, Paz-Ares L, de CastroCarpeño J, et al. IGFBP-3 hypermethylation-derived deficiency mediates cisplatin resistance in non-small-cell lung cancer. Oncogene. 2010;29:1681–90. doi: 10.1038/onc.2009.454. [DOI] [PubMed] [Google Scholar]
- 14.Balch C, Nephew KP. Epigenetic targeting therapies to overcome chemotherapy resistance. Adv Exp Med Biol. 2013;754:285–311. doi: 10.1007/978-1-4419-9967-2_14. [DOI] [PubMed] [Google Scholar]
- 15.Razin A, Kantor B. DNA methylation in epigenetic control of gene expression. Prog Mol Subcell Biol. 2005;38:151–67. doi: 10.1007/3-540-27310-7_6. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Y, Wang R, Song H, Huang G, Yi J, Zheng Y, Wang J, Chen L. Methylation of multiple genes as a candidate biomarker in non-small cell lung cancer. Cancer Lett. 2011;303:21–8. doi: 10.1016/j.canlet.2010.12.011. [DOI] [PubMed] [Google Scholar]
- 17.Carethers JM, Chauhan DP, Fink D, Nebel S, Bresalier RS, Howell SB, Boland CR. Mismatch repair proficiency and in vitro response to 5-fluorouracil. Gastroenterology. 1999;117:123–31. doi: 10.1016/S0016-5085(99)70558-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Satoh A, Toyota M, Itoh F, Sasaki Y, Suzuki H, Ogi K, Kikuchi T, Mita H, Yamashita T, Kojima T, et al. Epigenetic inactivation of CHFR and sensitivity to microtubule inhibitors in gastric cancer. Cancer Res. 2003;63:8606–13. [PubMed] [Google Scholar]
- 19.Hegi ME, Diserens AC, Godard S, Dietrich PY, Regli L, Ostermann S, Otten P, Van Melle G, de Tribolet N, Stupp R. Clinical trial substantiates the predictive value of O-6-methylguanine-DNA methyltransferase promoter methylation in glioblastoma patients treated with temozolomide. Clin Cancer Res. 2004;10:1871–4. doi: 10.1158/1078-0432.CCR-03-0384. [DOI] [PubMed] [Google Scholar]
- 20.Su HY, Lai HC, Lin YW, Liu CY, Chen CK, Chou YC, Lin SP, Lin WC, Lee HY, Yu MH. Epigenetic silencing of SFRP5 is related to malignant phenotype and chemoresistance of ovarian cancer through Wnt signaling pathway. Int J Cancer. 2010;127:555–67. doi: 10.1002/ijc.25083. [DOI] [PubMed] [Google Scholar]
- 21.Mansouri A, Ridgway LD, Korapati AL, Zhang Q, Tian L, Wang Y, Siddik ZH, Mills GB, Claret FX. Sustained activation of JNK/p38 MAPK pathways in response to cisplatin leads to Fas ligand induction and cell death in ovarian carcinoma cells. J Biol Chem. 2003;278:19245–56. doi: 10.1074/jbc.M208134200. [DOI] [PubMed] [Google Scholar]
- 22.Green JA, Berns EM, Coens C, van Luijk I, Thompson-Hehir J, van Diest P, Verheijen RH, van de Vijver M, van Dam P, Kenter GG, et al. EORTC Gynaecological Cancer Group Alterations in the p53 pathway and prognosis in advanced ovarian cancer: a multi-factorial analysis of the EORTC Gynaecological Cancer group (study 55865) Eur J Cancer. 2006;42:2539–48. doi: 10.1016/j.ejca.2006.06.015. [DOI] [PubMed] [Google Scholar]
- 23.Bartling B, Rehbein G, Simm A, Silber RE, Hofmann HS. Porcupine expression is associated with the expression of S100P and other cancer-related molecules in non-small cell lung carcinoma. Int J Oncol. 2010;36:1015–21. doi: 10.3892/ijo_00000582. [DOI] [PubMed] [Google Scholar]
- 24.Rehbein G, Simm A, Hofmann HS, Silber RE, Bartling B. Molecular regulation of S100P in human lung adenocarcinomas. Int J Mol Med. 2008;22:69–77. [PubMed] [Google Scholar]
- 25.Ji J, Jia S, Ji K, Jiang WG. Wnt1 inducible signalling pathway protein-2 (WISP‑2/CCN5): roles and regulation in human cancers (review) Oncol Rep. 2014;31:533–9. doi: 10.3892/or.2013.2909. [DOI] [PubMed] [Google Scholar]
- 26.Banerjee S, Dhar G, Haque I, Kambhampati S, Mehta S, Sengupta K, Tawfik O, Phillips TA, Banerjee SK. CCN5/WISP-2 expression in breast adenocarcinoma is associated with less frequent progression of the disease and suppresses the invasive phenotypes of tumor cells. Cancer Res. 2008;68:7606–12. doi: 10.1158/0008-5472.CAN-08-1461. [DOI] [PubMed] [Google Scholar]
- 27.Wu G, Guo Z, Chang X, Kim MS, Nagpal JK, Liu J, Maki JM, Kivirikko KI, Ethier SP, Trink B, et al. LOXL1 and LOXL4 are epigenetically silenced and can inhibit ras/extracellular signal-regulated kinase signaling pathway in human bladder cancer. Cancer Res. 2007;67:4123–9. doi: 10.1158/0008-5472.CAN-07-0012. [DOI] [PubMed] [Google Scholar]
- 28.Lee GH, Kim DS, Chung MJ, Chae SW, Kim HR, Chae HJ. Lysyl oxidase-like-1 enhances lung metastasis when lactate accumulation and monocarboxylate transporter expression are involved. Oncol Lett. 2011;2:831–8. doi: 10.3892/ol.2011.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dammann R, Strunnikova M, Schagdarsurengin U, Rastetter M, Papritz M, Hattenhorst UE, Hofmann HS, Silber RE, Burdach S, Hansen G. CpG island methylation and expression of tumour-associated genes in lung carcinoma. Eur J Cancer. 2005;41:1223–36. doi: 10.1016/j.ejca.2005.02.020. [DOI] [PubMed] [Google Scholar]
- 30.Arnold JM, Cummings M, Purdie D, Chenevix-Trench G. Reduced expression of intercellular adhesion molecule-1 in ovarian adenocarcinomas. Br J Cancer. 2001;85:1351–8. doi: 10.1054/bjoc.2001.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang HW, Zhang L, Chen JH, Du JJ. [The clinical significance of vascular endothelial growth factor and intercellular adhesion molecule-1 expression in non-small cell lung cancer] Zhonghua Wai Ke Za Zhi. 2005;43:354–7. [PubMed] [Google Scholar]
- 32.Cui XY, Wang N, Yang BX, Gao WF, Lin YM, Yao XR, Ma XT. HSPB8 is methylated in hematopoietic malignancies and overexpression of HSPB8 exhibits antileukemia effect. Exp Hematol. 2012;40:14–21. doi: 10.1016/j.exphem.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 33.Wang H, Zhou X, Zhang Y, Zhu H, Zhao L, Fan L, Wang Y, Gang Y, Wu K, Liu Z, et al. Growth arrest-specific gene 1 is downregulated and inhibits tumor growth in gastric cancer. FEBS J. 2012;279:3652–64. doi: 10.1111/j.1742-4658.2012.08726.x. [DOI] [PubMed] [Google Scholar]
- 34.Jiang Z, Xu Y, Cai S. Down-regulated GAS1 expression correlates with recurrence in stage II and III colorectal cancer. Hum Pathol. 2011;42:361–8. doi: 10.1016/j.humpath.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 35.Langenfeld EM, Bojnowski J, Perone J, Langenfeld J. Expression of bone morphogenetic proteins in human lung carcinomas. Ann Thorac Surg. 2005;80:1028–32. doi: 10.1016/j.athoracsur.2005.03.094. [DOI] [PubMed] [Google Scholar]
- 36.Wen XZ, Akiyama Y, Baylin SB, Yuasa Y. Frequent epigenetic silencing of the bone morphogenetic protein 2 gene through methylation in gastric carcinomas. Oncogene. 2006;25:2666–73. doi: 10.1038/sj.onc.1209297. [DOI] [PubMed] [Google Scholar]
- 37.Akum BF, Chen M, Gunderson SI, Riefler GM, Scerri-Hansen MM, Firestein BL. Cypin regulates dendrite patterning in hippocampal neurons by promoting microtubule assembly. Nat Neurosci. 2004;7:145–52. doi: 10.1038/nn1179. [DOI] [PubMed] [Google Scholar]
- 38.Raschperger E, Engstrom U, Pettersson RF, Fuxe J. CLMP, a novel member of the CTX family and a new component of epithelial tight junctions. J Biol Chem. 2004;279:796–804. doi: 10.1074/jbc.M308249200. [DOI] [PubMed] [Google Scholar]
- 39.Persano L, Pistollato F, Rampazzo E, Della Puppa A, Abbadi S, Frasson C, Volpin F, Indraccolo S, Scienza R, Basso G. BMP2 sensitizes glioblastoma stem-like cells to Temozolomide by affecting HIF-1α stability and MGMT expression. Cell Death Dis. 2012;3:e412. doi: 10.1038/cddis.2012.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gonzalez-Malerva L, Park J, Zou L, Hu Y, Moradpour Z, Pearlberg J, Sawyer J, Stevens H, Harlow E, LaBaer J. High-throughput ectopic expression screen for tamoxifen resistance identifies an atypical kinase that blocks autophagy. Proc Natl Acad Sci U S A. 2011;108:2058–63. doi: 10.1073/pnas.1018157108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zheng Y, Yang J, Qian J, Qiu P, Hanabuchi S, Lu Y, Wang Z, Liu Z, Li H, He J, et al. PSGL-1/selectin and ICAM-1/CD18 interactions are involved in macrophage-induced drug resistance in myeloma. Leukemia. 2013;27:702–10. doi: 10.1038/leu.2012.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao L, Pan Y, Gang Y, Wang H, Jin H, Tie J, Xia L, Zhang Y, He L, Yao L, et al. Identification of GAS1 as an epirubicin resistance-related gene in human gastric cancer cells with a partially randomized small interfering RNA library. J Biol Chem. 2009;284:26273–85. doi: 10.1074/jbc.M109.028068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rius M, Lyko F. Epigenetic cancer therapy: rationales, targets and drugs. Oncogene. 2012;31:4257–65. doi: 10.1038/onc.2011.601. [DOI] [PubMed] [Google Scholar]
- 44.Kantarjian H, Issa JP, Rosenfeld CS, Bennett JM, Albitar M, DiPersio J, Klimek V, Slack J, de Castro C, Ravandi F, et al. Decitabine improves patient outcomes in myelodysplastic syndromes: results of a phase III randomized study. Cancer. 2006;106:1794–803. doi: 10.1002/cncr.21792. [DOI] [PubMed] [Google Scholar]
- 45.Piekarz RL, Frye R, Turner M, Wright JJ, Allen SL, Kirschbaum MH, Zain J, Prince HM, Leonard JP, Geskin LJ, et al. Phase II multi-institutional trial of the histone deacetylase inhibitor romidepsin as monotherapy for patients with cutaneous T-cell lymphoma. J Clin Oncol. 2009;27:5410–7. doi: 10.1200/JCO.2008.21.6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cameron EE, Bachman KE, Myöhänen S, Herman JG, Baylin SB. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat Genet. 1999;21:103–7. doi: 10.1038/5047. [DOI] [PubMed] [Google Scholar]
- 47.Charlet J, Schnekenburger M, Brown KW, Diederich M. DNA demethylation increases sensitivity of neuroblastoma cells to chemotherapeutic drugs. Biochem Pharmacol. 2012;83:858–65. doi: 10.1016/j.bcp.2012.01.009. [DOI] [PubMed] [Google Scholar]
- 48.Matei D, Fang F, Shen C, Schilder J, Arnold A, Zeng Y, Berry WA, Huang T, Nephew KP. Epigenetic resensitization to platinum in ovarian cancer. Cancer Res. 2012;72:2197–205. doi: 10.1158/0008-5472.CAN-11-3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Howard G, Eiges R, Gaudet F, Jaenisch R, Eden A. Activation and transposition of endogenous retroviral elements in hypomethylation induced tumors in mice. Oncogene. 2008;27:404–8. doi: 10.1038/sj.onc.1210631. [DOI] [PubMed] [Google Scholar]
- 50.Eden A, Gaudet F, Waghmare A, Jaenisch R. Chromosomal instability and tumors promoted by DNA hypomethylation. Science. 2003;300:455. doi: 10.1126/science.1083557. [DOI] [PubMed] [Google Scholar]
- 51.Evdokiou A, Cowled PA. Growth-regulatory activity of the growth arrest-specific gene, GAS1, in NIH3T3 fibroblasts. Exp Cell Res. 1998;240:359–67. doi: 10.1006/excr.1998.4011. [DOI] [PubMed] [Google Scholar]
- 52.Martinelli DC, Fan CM. The role of Gas1 in embryonic development and its implications for human disease. Cell Cycle. 2007;6:2650–5. doi: 10.4161/cc.6.21.4877. [DOI] [PubMed] [Google Scholar]
- 53.Martinelli DC, Fan CM. Gas1 extends the range of Hedgehog action by facilitating its signaling. Genes Dev. 2007;21:1231–43. doi: 10.1101/gad.1546307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Leem YE, Han JW, Lee HJ, Ha HL, Kwon YL, Ho SM, Kim BG, Tran P, Bae GU, Kang JS. Gas1 cooperates with Cdo and promotes myogenic differentiation via activation of p38MAPK. Cell Signal. 2011;23:2021–9. doi: 10.1016/j.cellsig.2011.07.016. [DOI] [PubMed] [Google Scholar]
- 55.Domínguez-Monzón G, Benítez JA, Vergara P, Lorenzana R, Segovia J. Gas1 inhibits cell proliferation and induces apoptosis of human primary gliomas in the absence of Shh. Int J Dev Neurosci. 2009;27:305–13. doi: 10.1016/j.ijdevneu.2009.03.009. [DOI] [PubMed] [Google Scholar]
- 56.Ma Y, Qin H, Cui Y. MiR-34a targets GAS1 to promote cell proliferation and inhibit apoptosis in papillary thyroid carcinoma via PI3K/Akt/Bad pathway. Biochem Biophys Res Commun. 2013;441:958–63. doi: 10.1016/j.bbrc.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 57.Plasencia C, Martínez-Balibrea E, Martinez-Cardús A, Quinn DI, Abad A, Neamati N. Expression analysis of genes involved in oxaliplatin response and development of oxaliplatin-resistant HT29 colon cancer cells. Int J Oncol. 2006;29:225–35. doi: 10.3892/ijo.29.1.225. [DOI] [PubMed] [Google Scholar]
- 58.Zeilinger S, Kühnel B, Klopp N, Baurecht H, Kleinschmidt A, Gieger C, Weidinger S, Lattka E, Adamski J, Peters A, et al. Tobacco smoking leads to extensive genome-wide changes in DNA methylation. PLoS One. 2013;8:e63812. doi: 10.1371/journal.pone.0063812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zheng Y, Wang R, Song HZ, Pan BZ, Zhang YW, Chen LB. Epigenetic downregulation of RUNX3 by DNA methylation induces docetaxel chemoresistance in human lung adenocarcinoma cells by activation of the AKT pathway. Int J Biochem Cell Biol. 2013;45:2369–78. doi: 10.1016/j.biocel.2013.07.013. [DOI] [PubMed] [Google Scholar]
- 60.Ortiz AP, Pérez J, Otero-Domínguez Y, García-Rodríguez O, Garced-Tirado S, Escalera-Maldonado F, Gaud-Quintana S, Santiago-Rodríguez E, Svensson K, Vergara-Arroyo JL, et al. Promoter methylation of CDKN2A and lack of p16 expression characterize patients with hepatocellular carcinoma. BMC Cancer. 2010;10:31. doi: 10.1186/1471-2407-10-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.