Abstract
Purpose
The intracardiac synthesis of anthracycline alcohol metabolites (e.g., daunorubicinol) contributes to the pathogenesis of anthracycline-related cardiotoxicity. Cancer patients with Down syndrome (DS) are at increased risk for anthracycline-related cardiotoxicity. We profiled the expression of anthracycline metabolizing enzymes in hearts from donors with- and without- DS.
Methods
Cardiac expression of CBR1, CBR3, AKR1A1, AKR1C3 and AKR7A2 was examined by quantitative real time PCR, quantitative immunoblotting, and enzyme activity assays using daunorubicin. The CBR1 polymorphism rs9024 was investigated by allelic discrimination with fluorescent probes. The contribution of CBRs/AKRs proteins to daunorubicin reductase activity was examined by multiple linear regression.
Results
CBR1 was the most abundant transcript (average relative expression; DS: 81%, non-DS: 58%), and AKR7A2 was the most abundant protein (average relative expression; DS: 38%, non-DS: 35%). Positive associations between cardiac CBR1 protein levels and daunorubicin reductase activity were found for samples from donors with- and without- DS. Regression analysis suggests that sex, CBR1, AKR1A1, and AKR7A2 protein levels were significant contributors to cardiac daunorubicin reductase activity. CBR1 rs9024 genotype status impacts on cardiac CBR1 expression in non-DS hearts.
Conclusions
CBR1, AKR1A1, and AKR7A2 protein levels point to be important determinants for predicting the synthesis of cardiotoxic daunorubicinol in heart.
Keywords: Anthracyclines, anthracycline-related cardiotoxicity, Down syndrome, carbonyl reductases, aldo-keto reductases
INTRODUCTION
In humans, carbonyl reductases (CBRs) and aldo-keto reductases (AKRs) play predominant roles during the metabolism of more than 30 clinically relevant drugs including the antipsychotic agent haloperidol, the opioid receptor antagonist naltrexone, the antiemetic dolasetron, the anti-inflammatory ketoprofen, and the anticancer anthracyclines doxorubicin and daunorubicin (1–3). The use of anthracyclines for the chemotherapy of a variety of solid and hematological cancers is limited by the development of cardiotoxicity in some patients (4). For example, a study of children (n = 115) who survived leukemia revealed that 60% of those treated with anthracyclines developed preclinical abnormalities in cardiac structure and function (5). Of note, pediatric cancer patients with Down syndrome (DS, trisomy 21) constitute a population at particularly greater risk for anthracycline-related cardiotoxicity, and a safe dose of anthracyclines for cancer patients with- DS remains to be defined (6, 7). For example, children with- DS represent 15% of pediatric acute myeloid leukemia (AML) cases, and a report from the Children’s Oncology group has documented clinically symptomatic cardiomyopathy in 17.5% of DS-AML patients treated with an anthracycline-containing regimen (7, 8).
The complex pathogenesis of anthracycline-related cardiotoxicity is mediated by a combination of oxidative stress and metabolic perturbations in myocardial tissue that are induced by the C-13 anthracycline alcohol metabolites, whose formation is catalyzed by specific CBRs and AKRs (4, 9–12). A growing cumulus of evidence indicates that two monomeric CBRs, namely CBR1 and CBR3, together with the members of the AKRs superfamily AKR1A1, AKR1C3 and AKR7A2 catalyze the synthesis of cardiotoxic C-13 anthracycline alcohol metabolites (e.g., daunorubicinol and doxorubicinol) (13–16). In addition specific CBRs/AKRs genetic variants may contribute to the unpredictable pharmacological profile of anthracyclines in cancer patients (17–19). For example, a recent study from the Children’s Oncology group described the impact of functional single nucleotide polymorphisms in CBR1 and CBR3 on the risk of anthracycline-related cardiomyopathy in childhood cancer survivors (20). Thus, interindividual variability in the expression of CBRs and AKRs would impact the intracardiac formation of cardiotoxic C-13 anthracycline alcohol metabolites, and consequently the pharmacodynamics of anthracycline drugs. Furthermore, the CBR1 and CBR3 genes are located in the DS critical region of chromosome 21 (21q21-21q22.3). The altered expression of CBRs as a result of the gene dosage effect may contribute to the increased risk of anthracycline-related cardiotoxicity in cancer patients with- DS (21).
In spite of the prominent contributions of CBRs and AKRs towards the pharmacodynamics of anthracycline drugs, reports documenting gene expression levels and protein abundance in cardiac tissue are limited to the analysis of individual samples or pooled tissue samples (13, 22, 23). Thus, the main goal of this study was to document the extent of interindividual variability in the expression of CBR1, CBR3, AKR1A1, AKR1C3 and AKR7A2 in a collection of heart samples from donors with- and without- DS. The expression of CBRs and AKRs was examined by quantitative real time PCR (qRT-PCR) with specific primers, quantitative immunoblotting with specific antibodies, and enzyme activity assays using the anthracycline substrate daunorubicin. We also examined the impact of a functional polymorphism in CBR1 (rs9024), known to impact CBR1 expression and daunorubicinol synthesis in liver, on cardiac CBR1 gene expression and enzymatic activity for the substrate daunorubicin (21, 24, 25).
MATERIAL AND METHODS
Human heart samples
The Institutional Review Board of the State University of New York at Buffalo approved this research. Heart samples from donors with- (n = 9) and without- DS (n = 30) were procured from The National Disease Research Interchange (NDRI, funded by the National Center for Research Resources), The Cooperative Human Tissue Network (CHTN, funded by the National Cancer Institute), and the National Institute of Child Health and Human Development (NICHD) Brain and Tissue Bank. The postmortem to tissue recovery interval was ≤10 h. Samples (2 – 20 g, myocardium, left ventricle only) were frozen immediately after recovery and stored in liquid nitrogen until further processing. The main demographics from donors with- and without- DS are summarized in Supplemental Table I. Down syndrome status (yes/no) and relevant diagnoses (Supplemental Table II) were obtained from anonymous medical histories. Heart samples were processed following standardized procedures to isolate DNA and RNA as described (24, 26).
Array CGH analysis
Down syndrome status was confirmed by array comparative genomic hybridization (aCGH). Briefly, genomic DNA (3.0 μg) from test samples and a euploid reference DNA sample were fluorescently labeled and hybridized to high resolution Agilent 244K aCGH arrays containing +236,000 coding and non-coding human probes. Changes in DNA copy number were determined by evaluating log2 ratios across whole chromosomes. aCGH assays were performed at the Genomics core facility, Roswell Park Cancer Institute (Buffalo, NY).
Quantitative real-time PCR
Cardiac CBR1, CBR3, AKR1A1, AKR1C3, and AKR7A2 mRNA expression was analyzed by qRT-PCR with gene specific primers (Table I) following the MIQE guidelines (27). Briefly, 5 ng of total RNA was reverse transcribed and amplified using the one-step QuantiTect SYBR Green RT-PCR kit (Qiagen, Venlo, The Netherlands). Target genes and ACTB (reference gene) were amplified in parallel with the following cycling parameters: 50°C for 30 min (reverse transcription), 95°C for 10 min (Taq DNA polymerase activation), 40 cycles of 95°C for 15 s (denaturation), 56°C for 30 s (annealing) and 72°C for 30 s (extension). PCR amplification products were analyzed by electrophoresis in 1% agarose gels stained with ethidium bromide. PCR amplification products were cloned into pCR 2.1- TOPO vectors (Invitrogen, Carlsbad, CA), and their identities were verified by DNA sequencing. Cloned PCR products were diluted to generate standards for calibration curves. In all cases, the regression coefficients of the standard curves were r2 ≥ 0.95. The amplification efficiencies of the target genes (e.g., CBR1, AKR1A1) and the reference gene (ACTB) were comparable and ranged between 96 – 110%. For each gene, experimental samples and standards for calibration curves were analyzed in triplicate. For each heart sample, the copy number of the target gene and ACTB were calculated using the average Ct values and direct extrapolation from the calibration curves (Fig. 1). In all cases, cardiac mRNA levels were expressed as copy number ratios using the following expression:
Table I.
List of PCR primers for the amplification of cardiac CBRs and AKRs mRNAs
| Gene | Forward primer | Reverse primer | Size (bp) |
|---|---|---|---|
| CBR1 | 5′-TCAAGCTGAAGTGACGATGA-3′ | 5′-GGTGCACTCCCTTCTTTGTA-3′ | 239 |
| CBR3 | 5′-AACCTCATGGGAGAGTGGTG-3′ | 5′-TCCTCGATAAGACCGTGACC-3′ | 231 |
| AKR1A1 | 5′-CCTGGTCAGGTAAAAGCAGC-3′ | 5′-CCCGCTCAAAGGCATAAGG-3′ | 286 |
| AKR1C3 | 5′-CCTCCAGAGGTTCCGAGAAG-3′ | 5′-GTGGACCAAAGCTTTGAAGTG-3′ | 173 |
| AKR7A2 | 5′-CCTCCAGAGGTTCCGAGAAG-3′ | 5′-GTGGACCAAAGCTTTGAAGTG-3′ | 168 |
| ACTB | 5′-GGACTTCGAGCAAGAGATGG-3′ | 5′-AGCACTGTGTTGGCGTACAG-3′ | 234 |
Fig. 1.
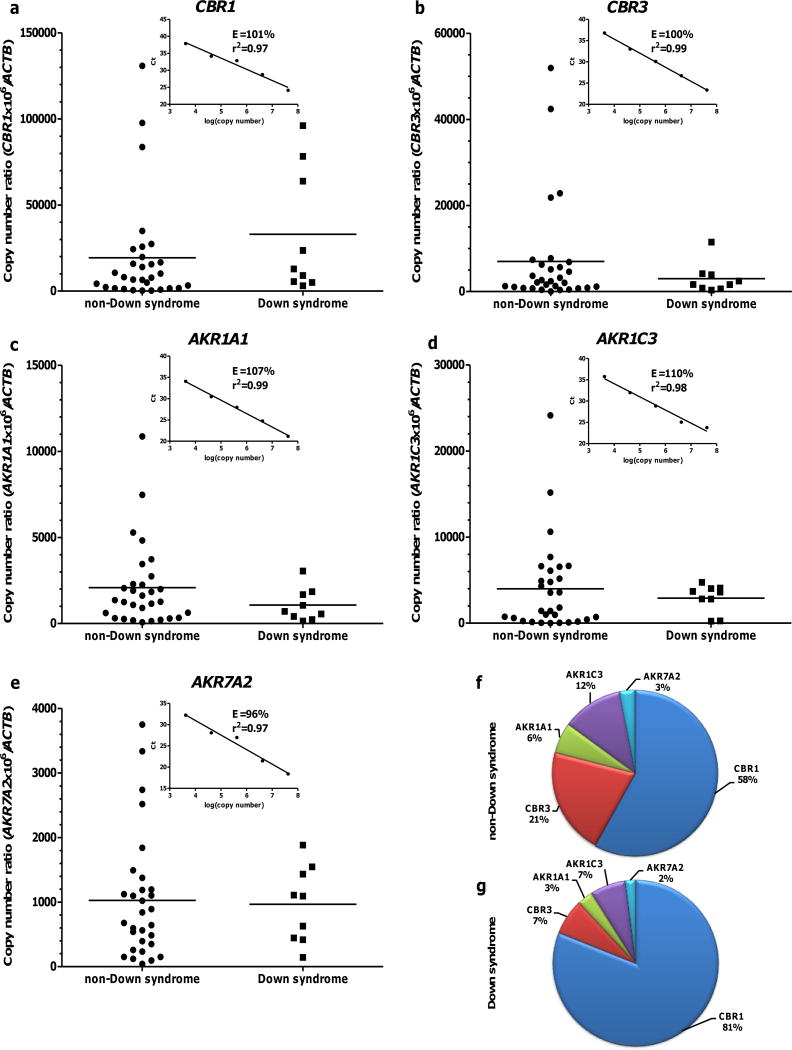
Cardiac CBR1, CBR3, AKR1A1, AKR1C3, and AKR7A2 mRNA expression in samples from donors with- (n = 9) and without- DS (n = 30) (Panels: a – e). Each symbol depicts the average of individual samples. Horizontal lines indicate group means. Insets show the corresponding calibration curves. Samples and standards for calibration curves were analyzed in triplicates. Relative abundance of cardiac CBRs and AKRs mRNAs in donors without- (f) and with- DS (g).
Quantitative immunoblotting
Heart tissue cytosols (~50 mg) were obtained using the Minute Cytoplasmic and Nuclear Extraction Kit (Invent Biotechnologies, Eden Prairie, MN) following the manufacturer instructions. Protein concentration was determined using a BCA assay (Pierce, Rockford, IL) with bovine serum albumin as a standard.
Cytosolic proteins were denatured with NuPAGE LDS sample buffer (Invitrogen) containing NuPAGE sample reducing agent (Invitrogen), and boiled at 70°C for 10 minutes prior to use. Briefly, cytosolic protein (12.5 μg for CBR1, AKR1A1, AkR1C3 and AKR7A2, and 40 μg for CBR3) and corresponding recombinant protein standards (Abcam, Cambridge, England) were separated by gel electrophoresis using NuPAGE Novex 4 – 12% Bis-Tris precast gels, and transferred onto nitrocellulose membranes using the iBlot Gel Transfer Device (Invitrogen). Membranes were blocked with 5% non-fat milk in 0.2% Tween 20-PBS for 1 hour at room temperature and then probed with specific polyclonal anti-human CBR1 (1:3000; Santa Cruz Biotechnology, Dallas, TX), CBR3 (1:500; Santa Cruz Biotechnology), AKR1A1 (1:3000; Abnova, Taipei City, Taiwan), AKR1C3 (1:3000; Abnova), and AKR7A2 (1:3000; Abnova) antibodies overnight at 4°C. Next, membranes were incubated with a secondary goat or rabbit anti-IgG antibodies conjugated with horseradish peroxidase (1:10000; Santa Cruz Biotechnology) for 1.5 hours at room temperature. To normalize for protein loading, membranes were stripped with Restore Western Blot Stripping Buffer (Thermo Scientific, Waltham, MA), and re-probed with anti-GAPDH antibody (1:1000; Santa Cruz Biotechnology). Immunoreactive bands were visualized with the ECL Plus Western blotting substrate (GE Healthcare, Little Chalfont, England) on a ChemiDoc XRS imager (Bio-Rad, Berkeley, CA). Densitometric analysis was performed using Image Lab software (Bio-Rad). Cardiac CBR1, CBR3, AKR1A1, AKR1C3, and AKR7A2 protein levels were estimated by direct extrapolation from the corresponding recombinant protein standard curves. Detection of recombinant CBR1, CBR3, AKR1A1, AKR1C3, and AKR7A2 proteins was linear (range: 0.01 – 0.1 μg; r2 > 0.99). The limits of detection (LOD) and limits of quantification (LOQ) were calculated based on the standard deviation of the band intensity and the slope of the calibration curve (Supplemental Fig. 1). Results are as follows: LODCBR1=0.34 ng, LOQCBR1=0.80 ng; LODCBR3=0.49 ng, LOQCBR3=1.49 ng; LODAKR1A1=0.58 ng, LOQAKR1A1=1.76 ng; LODAKR1C3=0.33 ng, LOQAKR1C3=0.98 ng; LODAKR7A2=0.44ng, LOQAKR7A2=1.34 ng.
Kinetic analysis
Anthracycline reductase activity was measured in heart cytosols using the substrate daunorubicin as described previously (21). Validation experiments with pooled heart cytosols showed that 400 μM daunorubicin [S] ensured conditions of Vmax (zero-order kinetics). Incubation mixtures (0.5 mL) contained potassium phosphate buffer (0.1 M; pH 7.4), cytosolic protein (400 μg) and daunorubicin (400 μM; Sigma-Aldrich, St. Louis, MO). Mixtures were equilibrated for 5 minutes at 37°C. Reactions were initiated by the addition of NADPH (200 μM; Sigma). Reaction mixtures were flash frozen with liquid nitrogen after 2 minutes, and stored at −80°C until daunorubicinol quantification. Daunorubicinol was quantified using a high performance liquid chromatography (UPLC) fluorescent detection method (28). The metabolite was extracted, and then quantified using a daunorubicinol standard (Toronto Research Chemicals, Toronto, ON) as previously described (29).
CBR1 Genotyping
The CBR1 polymorphism (rs9024) was investigated by allelic discrimination with fluorescent probes and real-time PCR (Assays-by-Design; Applied Biosystems, Foster City, CA) as described previously (24, 30). Rs9024 genotype status in DS samples (i.e., trisomy 21) was determined with a validated allelic discrimination assay as described (21).
Data analysis
Statistical comparisons were performed using Excel 2007 (Microsoft Office; Microsoft, Redmond, WA) and GraphPad Prism version 4.03 (GraphPad Software Inc., La Jolla, CA). The Kolmogorov–Smirnov test was used to analyze the normality of data sets. The Mann-Whitney U test or Student’s t test were used to compare group means. Pearson’s coefficient of correlation (rp) was used to analyze data sets with normal distributions. Comparison tests were considered significant at the P < 0.05.
Multiple linear regression models were fit using the R programming language (http://www.r-project.org/). Multiple linear regression analysis was restricted to non-DS hearts due to sample size limitations. For the regression modeling, the observations that have a missing value in at least one of the protein expression measurements were eliminated from the data set. CBR3 was not considered for the regression modeling due to the number of missing values. In the union of missing data for the remaining proteins: CBR1, AKR1A1, AKR1C3, and AKR7A2, there were 12 observations with missing values for at least one of the measured proteins. These observations were eliminated for model fitting, and the remaining subset of 18 observations (9 males and 9 females), was used for the regression. The full model was represented as:
where βj for j = 0, 1, …, 5 represent the regression coefficients for the predictors in the model, β0 is the intercept, and ε ~ N(0,1), is an error term. CBR1, AKR1A1, AKR1C3, and AKR7A2 represent the normalized protein concentration (nmol/gram cytosolic protein) in heart. The term Sex was represented as a binary factor.
RESULTS
CBR1, CBR3, AKR1A1, AKR1C3 and AKR7A2 mRNA expression in hearts from donors with- and without- DS
In all cases, DS status from medical records (i.e., trisomy 21) was confirmed by aCGH. Cardiac CBR1, CBR3, AKR1A1, AKR1C3 and AKR7A2 mRNA levels in donors with- and without- DS are shown in Fig. 1 (panels: a – e). Analysis of the average relative expression levels showed that CBR1 mRNA is the most abundant transcript in heart samples from donors with- (81%) and without- DS (58%. Fig. 1f and 1g). On average, the remaining transcripts showed similar trends in their relative expression levels in samples from donors with- (CBR3≈AKR1C3>AKR1A1>AKR7A2. Fig. 1g) and without- DS (CBR3>AKR1C3>AKR1A1>AKR7A2. Fig. 1f). The cardiac expression of CBR1, CBR3, AKR1A1, AKR1C3 and AKR7A2 mRNA displayed considerable interindividual variability in samples from donors with- and without- DS (Table II). The cardiac expression of CBRs and AKRs mRNAs did not significantly differ between samples from donors with- and without- DS (Table II).
Table II.
Cardiac CBR1, CBR3, AKR1A1, AKR1C3, and AKR7A2 mRNA expression in samples from donors without- and with- DS
| Non-DS | DS | ||||
|---|---|---|---|---|---|
|
| |||||
| Transcript | Copy number ratio | Copy number ratio | P value | ||
| Mean ± SD | Range | Mean ± SD | Range | ||
| CBR1 | 19283.8 ±30824.3 | 245.5–130786.4 | 33029.3 ± 36187.0 | 3103.1–96061.0 | 0.23 |
| CBR3 | 6975.3 ± 12271.4 | 0.4–51914.3 | 2992.8 ± 3465.6 | 334.5–11487.5 | 0.46 |
| AKR1A1 | 2078.4 ± 2396.1 | 66.0–10865.2 | 1073.6 ± 955.5 | 144.2–3048.4 | 0.42 |
| AKR1C3 | 3986.8 ± 5264.9 | 14.3–24131.8 | 2913.3 ± 1631.6 | 227.0–4732.8 | 0.69 |
| AKR7A2 | 1026.1 ± 955.7 | 42.3–3747.9 | 965.8 ± 590.6 | 143.9–1883.0 | 0.98 |
CBR1, CBR3, AKR1A1, AKR1C3 and AKR7A2 protein expression in hearts from donors with- and without- DS
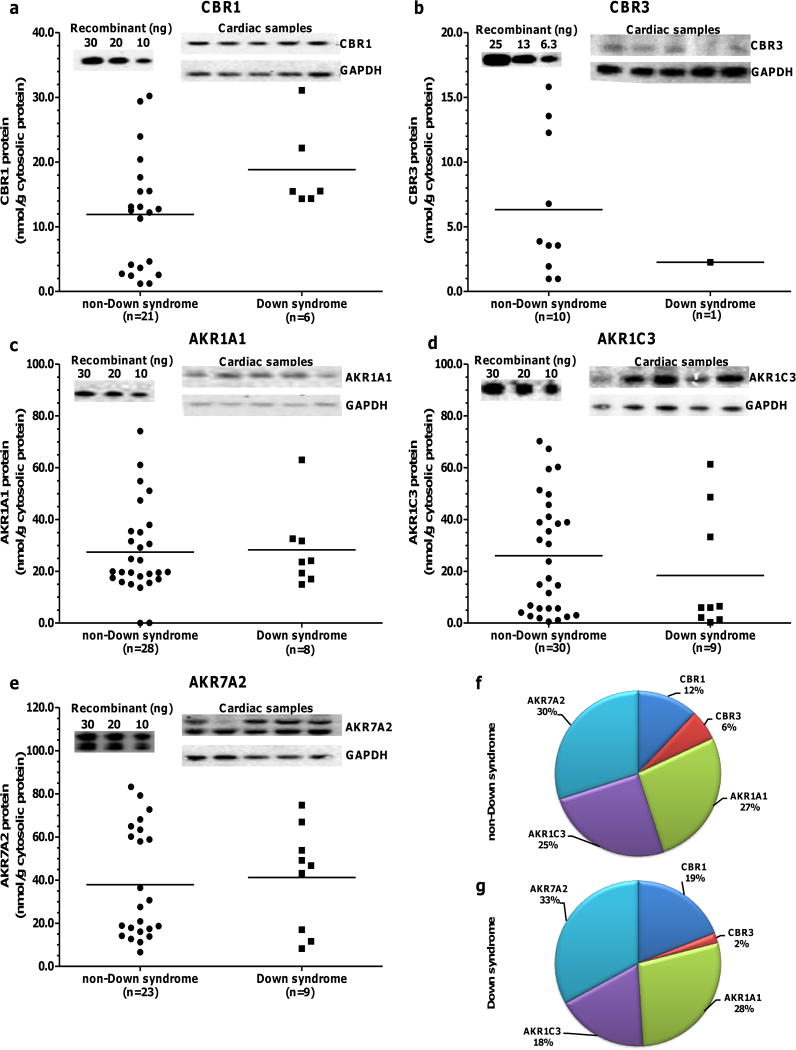
Cardiac CBR1, CBR3, AKR1A1, AKR1C3 and AKR7A2 protein expression levels in samples from donors with- and without- DS are shown in Fig. 2 (panels: a – e). Eight out of 9 heart samples from donors with- DS exhibited CBR3 protein levels below the LOQ (Supplemental Fig. 2). AKR7A2 showed the highest average expression levels in hearts from both groups, accounting for 33% of total CBRs-AKRs in donors with- DS and 30% of total CBRs-AKRs in donors without- DS. On average, the pattern of relative cardiac expression was similar in samples from donors with- (AKR7A2>AKR1A1>AKR1C3>CBR1>CBR3. Fig. 2g) and without- DS (AKR7A2>AKR1A1≈AKR1C3>CBR1>CBR3. Fig. 2f). Table III shows cardiac CBRs and AKRs protein levels in samples from donors with- and without- DS. The average cardiac expression of CBR1 was 57% higher in samples from donors with- DS than in samples from donors without- DS (Fig. 2 and Table III). The expression levels of AKR1A1, AKR1C3 and AKR7A2 did not differ between samples from donors with- and without- DS (Table III).
Fig. 2.
Cardiac CBR1, CBR3, AKR1A1, AKR1C3, and AKR7A2 protein expression in samples from donors with- and without- DS. Each symbol depicts the average of individual samples. Horizontal lines indicate group means. Samples and standards for calibration curves were analyzed in duplicates for CBR3 and in triplicates for CBR1, AKR1A1, AKR1C3, AKR7A2. Samples exhibiting protein levels below the LOQs were excluded (Supplemental Fig. 1). Insets show representative immunoblots for recombinant standards (left) and cytosolic CBRs-AKRs plus GADPH (right). Relative abundance of cardiac CBRs and AKRs proteins in donors without- (f) and with- DS (g).
Table III.
Cardiac CBR1, CBR3, AKR1A1, AKR1C3, and AKR7A2 protein expression in samples from donors without- and with- DS
| Non-DS | DS | ||||
|---|---|---|---|---|---|
|
| |||||
| Transcript | nmol/g cytosolic protein | nmol/g cytosolic protein | P value | ||
| Mean ± SD | Range | Mean ± SD | Range | ||
| CBR1 | 11.9 ± 8.9 | 1.2 – 30.2 | 18.8 ± 6.7 | 14.3 – 31.1 | 0.04 |
| CBR3 | 6.3 ± 5.5 | 1.0 – 15.8 | 2.3 | N/A | N/A |
| AKR1A1 | 27.4 ± 17.3 | 0.0 – 74.1 | 28.3 ± 15.4 | 15.0 – 63.0 | 0.86 |
| AKR1C3 | 26.0 ± 22.4 | 0.5 – 70.3 | 18.4 ± 23.2 | 0.3 – 61.4 | 0.32 |
| AKR7A2 | 31.0 ± 21.0 | 5.4 – 68.1 | 33.8 ± 19.6 | 6.8 – 61.2 | 0.97 |
N/A., not applicable (n = 1)
Maximal daunorubicin reductase activity in hearts from donors with- and without- DS
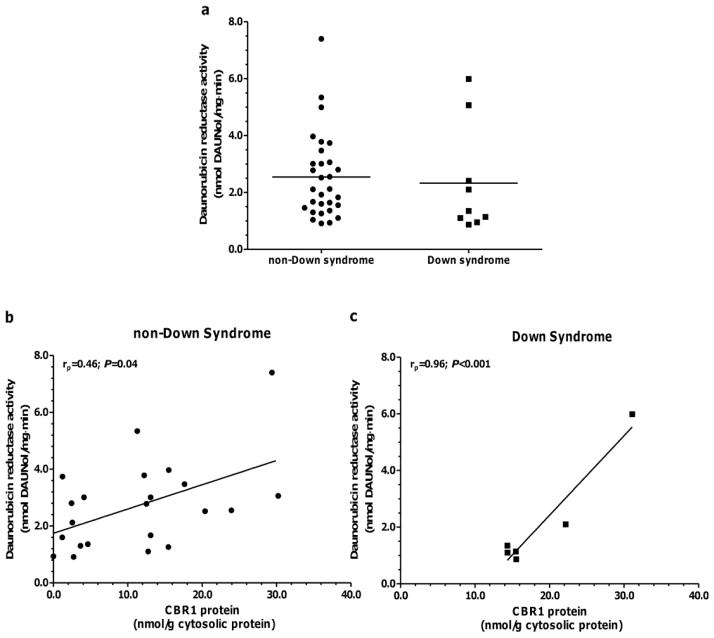
Fig. 3A shows maximal daunorubicin reductase activities in hearts from donors with- and without- DS. Variable levels of daunorubicin reductase activity were observed in cardiac samples from donors with- DS (range: 0.9 – 6.0 nmol/mg·min; 7-fold) and without- DS (range: 0.9 – 7.4 nmol/mg·min; 8-fold. Fig. 3a). Statistical comparisons of the means demonstrated that cardiac maximal daunorubicin reductase activities were similar between both groups (Activitynon-DS=2.5 ± 1.5 vs. ActivityDS = 2.3 ± 1.9, P = 0.28. Fig. 3a). Linear regression analysis was used to test for associations between CBRs-AKRs protein levels and cardiac maximal daunorubicin reductase activities (Table IV). There existed a significant positive association between CBR1 protein levels and daunorubicin reductase activity in samples from donors with- and without- DS (Table IV and Fig. 3b and 3c).
Fig. 3.
Daunorubicin reductase activity in donors with- (n=9) and without- DS (n=30) (a). Each symbol depicts the average of individual heart samples. Each sample was analyzed in duplicates. Group means are indicated by horizontal lines. Linear regression analysis of maximal daunorubicin reductase activity versus cardiac CBR1 protein expression in samples from donors without- and with- DS (panels b and c).
Table IV.
Linear regression analysis of cardiac daunorubicin reductase activities versus CBRs/AKRs protein levels in samples from donors without- and with- DS
| Non-DS | DS | |||
|---|---|---|---|---|
|
| ||||
| Protein | Pearson correlation coefficient | P value | Pearson correlation coefficient | P value |
| CBR1 | 0.46 | 0.04 | 0.96 | 0.002 |
| CBR3 | 0.20 | 0.58 | N/A | N/A |
| AKR1A1 | 0.00 | 0.99 | −0.03 | 0.95 |
| AKR1C3 | −0.16 | 0.41 | 0.32 | 0.41 |
| AKR7A2 | 0.28 | 0.19 | −0.22 | 0.58 |
N/A., not applicable (n = 1).
Our collection included three samples from pediatric donors with- DS (Supplemental Table III). Age-related changes in the cardiac expression of CBRs/AKRs during the developmental continuum encompassing birth and adolescence may impact the metabolism of anthracycline substrates in pediatric patients. Exploratory analyses revealed no differences in the cardiac expression of CBRs/AKRs (mRNA, protein and daunorubicin reductase activity) in samples from pediatric donors in comparison to samples from non-pediatric donors (Supplemental Table III).
Multiple linear regression analysis was used to examine the impact of cardiac CBRs/AKRs protein levels on daunorubicin reductase activities in samples from donors without- DS. The results (Supplemental Table IV) indicate Sex is the most significant factor (P < 0.001), followed by AKR7A2, CBR1, and AKR1A1 (P < 0.05). AKR1C3 was not significant and removed from the model in an effort to identify the most parsimonious model of daunorubicin reductase activity (Table V). As expected, the removal of this term did not change the overall model, and the terms that were significant in the full model remained significant in the reduced model (Table V).
Table V.
Results of the reduced multiple regression model fit.
| Term | β estimate | Std. Error | t-stat | P value | Sig. Codes |
|---|---|---|---|---|---|
| Intercept | −2.527 | 1.023 | −2.471 | 0.028 | *** |
| Sex | 2.560 | 0.571 | 4.481 | 0.001 | **** |
| CBR1 | 2.380 | 1.001 | 2.377 | 0.033 | ** |
| AKR1A1 | 1.739 | 0.592 | 2.939 | 0.012 | ** |
| AKR7A2 | 1.242 | 0.357 | 3.479 | 0.004 | *** |
The significant codes are as follows:
for P< 0.001,
for P< 0.01,
P< 0.05,
P< 0.1. The r2 is 0.73
CBR1 rs9024 genotype-phenotype associations in heart samples from donors with- and without- DS
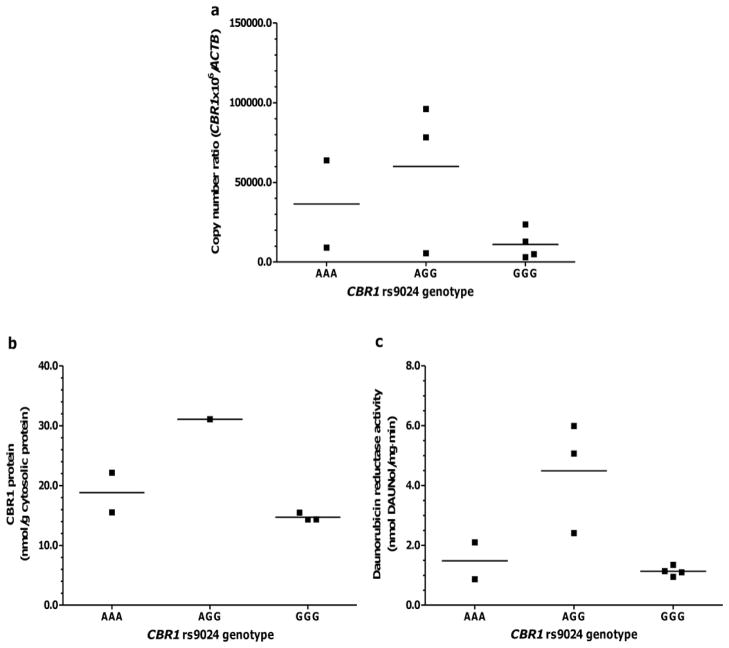
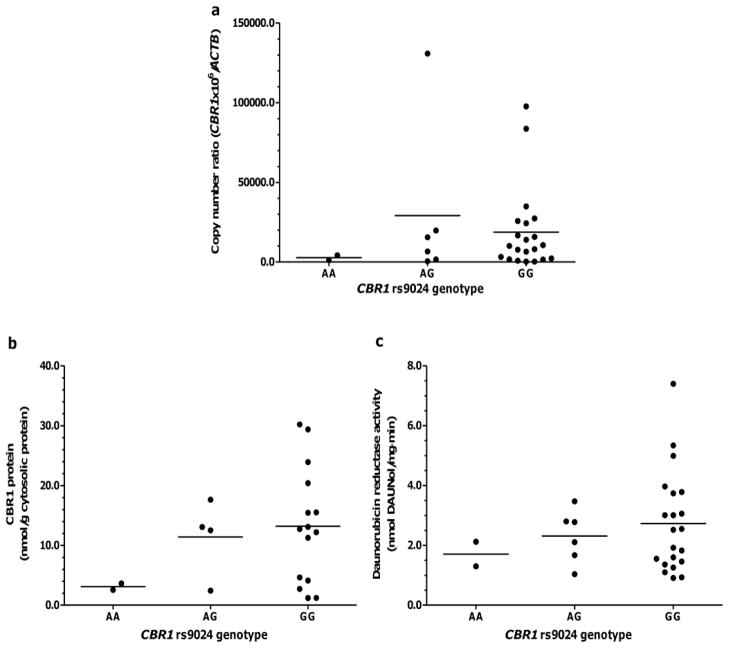
Tables VI and VII show cardiac CBR1 expression in samples from donors with- and without- DS stratified by CBR1 rs9024 genotype status. No statistical comparisons are shown for donors with- DS due to sample size limitations. Visual inspection of the data shown in Fig. 4 suggests the absence of an evident association between rs9024 genotype status and cardiac CBR1 mRNA and protein expression in samples from donors with- DS (Fig. 4a and 4b). Similarly, visual inspection does not suggest any association between CBR1 rs9024 genotype status and maximal cardiac daunorubicin reductase activity in samples from donors with- DS (Fig. 4c). The analysis of donors without- DS showed that, on average, relative CBR1 mRNA levels tend to be higher in samples with CBR1 rs9024 homozygous G/G genotype compared to samples with homozygous A/A genotype; the difference between genotype groups did not reach statistical significance (Table VII and Fig. 5a). Further analysis showed a trend towards higher CBR1 protein levels (≈4-fold) in samples from donors with CBR1 rs9024 homozygous G/G genotype in comparison to samples with homozygous A/A genotype (Table VII and Fig. 5b). In line, heart cytosols from donors with homozygous CBR1 rs9024 G/G genotypes displayed higher maximal rates of daunorubicinol synthesis (≈1.6-fold) than cytosols from donors with homozygous A/A genotype (Table VII and Fig. 5c).
Table VI.
Cardiac CBR1 mRNA expression, CBR1 protein expression and daunorubicin reductase activity in hearts from donors with- DS stratified by CBR1 rs9024 genotype status
| CBR1 rs9024 genotype (n) |
CBR1 mRNA copy number ratio Mean ± SD |
CBR1 protein (nmol/g cytosolic protein) Mean ± SD |
Daunorubicin reductase activity (nmol daunol/mg. min) Mean ± SD |
|---|---|---|---|
| AA (2) | 36423.3 ± 38775.3 | 18.8 ± 4.7 | 1.5 ± 0.9 |
| AG (6) | 59954.5 ± 9322.3 | 31.1 | 4.5 ± 1.9 |
| GG (21) | 11138.5 ± 47942.6 | 14.7 ± 0.7 | 1.1 ± 0.2 |
Table VII.
Cardiac CBR1 mRNA expression, CBR1 protein expression and daunorubicin reductase activity in hearts from donors without- DS stratified by CBR1 rs9024 genotype status
| CBR1 rs9024 genotype (n) |
CBR1 mRNA copy number ratio Mean ± SD |
CBR1 protein (nmol/g cytosolic protein) Mean ± SD |
Daunorubicin reductase activity (nmol daunol/mg. min) Mean ± SD |
|---|---|---|---|
| AA (2) | 2719.1 ± 2206.0 | 3.1 ± 0.8 | 1.7 ± 0.6 |
| AG (6) | 29148.5 ± 50373.5 | 11.4 ± 6.4 | 2.3 ± 0.9 |
| GG (21) | 18726.8 ± 25996.4 | 13.2 ± 9.6 | 2.7 ± 1.7 |
Fig. 4.
Impact of CBR1 rs9024 genotype status on: cardiac CBR1 expression (a), cardiac CBR1 protein expression (b), and daunorubicin reductase activity (c) in donors with- DS. Each symbol represents individual heart samples. Horizontal lines indicate group means.
Fig. 5.
Impact of CBR1 rs9024 genotype status on: cardiac CBR1 expression (a), cardiac CBR1 protein expression (b), and daunorubicin reductase activity (c) in donors without- DS. Each symbol represents individual heart samples. Horizontal lines indicate group means.
DISCUSSION
There is a paucity of reports describing the expression of prominent drug metabolizing enzymes in tissues from subjects with- DS. Within the context of multi-agent chemotherapy that includes anthracyclines for the treatment of pediatric leukemia, there is still considerable uncertainty about the optimal dose requirements for patients with- DS (31). We propose that variability in the cardiac expression of specific CBRs and AKRs may contribute to the unpredictable pharmacodynamics of anthracycline drugs. The goal of this study was to document the expression of CBR1, CBR3, AKR1A1, AKR1C3 and AKR7A2 in a collection of heart samples from donors with- and without- DS.
CBR1, CBR3, AKR1A1, AKR1C3 and AKR7A2 mRNAs are expressed at variable levels in heart samples from donors with- and without- DS (Fig. 1 and Table II). Of all the transcripts, CBR3 mRNA exhibited the widest range of expression in samples from donors with- (34-fold) and without- DS (>1.3 x 105 fold). The cardiac expression of CBR1 and CBR3 mRNAs are of particular interest due to the location of the CBR1 and CBR3 genes in chromosome 21, and the expected ≈1.5-fold increase in gene expression due to “gene dosage effect” in tissue samples with trisomy 21. Studies on transcriptional profiles in the DS setting suggest that the gene-dosage effect for most of the chromosome 21 transcripts (≈70%) is compensated by natural variation in gene expression (32). The compensation effect appears to be dependent upon the cellular context. For example, the expression of CBR1 and CBR3 mRNAs is increased in trisomic fibroblasts (≈1.5-fold) but not in trisomic lymphoblastoid cell lines (33). In this study, the expression of CBR3 mRNA was similar in samples from donors with- and without- DS. Thus, it appears that natural interindividual variability in CBR3 mRNA expression compensates the expected gene dosage effect in trisomic heart tissues. In contrast, CBR1 mRNA expression tended to be higher (1.7-fold) in samples from donors with- DS than in samples from donors without- DS (Fig. 1 and Table II). Trisomic heart samples also showed a concomitant ≈1.6-fold increase in CBR1 protein expression (Fig. 2 and Table III). These findings are in agreement with our previous observation documenting increased cardiac CBR1 mRNA and CBR1 protein expression in a subset of 4 samples from donors with- DS (21). It is assumed that the overexpression of “dosage-sensitive” genes from chromosome 21 is involved in specific DS phenotypes (e.g., cognitive impairments, facial dysmorphisms, and congenital heart disease (34). In this context, our data suggest that CBR1 overexpression in the cardiac tissue from donors with- DS may contribute to phenotypic abnormalities in trisomic hearts.
At the protein level, AKR7A2 was the most abundant anthracycline reductase in hearts from donors with- and without- DS, representing ≈30% of the total reductase content (Fig. 2). In non-DS hearts, linear regression analysis of protein content versus maximal enzymatic activity for the substrate daunorubicin suggests that AKR7A2 content accounts for ≈8% of the total variance in cardiac daunorubicin reductase activity (Table IV). A similar analysis indicates that cardiac CBR1 protein content accounts for ≈21% of the variance in daunorubicin reductase activity in samples from donors without- DS. The contribution of CBR1 expression to daunorubicin reductase activity in hearts from donors with- DS appears to be substantial. In this case, the regression analysis suggests that CBR1 content accounts for ≈90% of the variance in cardiac daunorubicin reductase activity in hearts from donors with- DS (Fig. 3 and Table IV). These findings suggest that CBR1 protein expression is an important predictor of daunorubicinol synthesis in human heart.
CBR1 rs9024 genotype status has been associated with the hepatic expression of CBR1 and anthracycline reductase activity for the substrate doxorubicin. In this study, the impact of CBR1 rs9024 genotype status on cardiac CBR1 expression was apparent in the group of heart samples from donors without- DS (Fig. 4 and Table VII). Samples with homozygous G/G genotype exhibited higher daunorubicin reductase activity (≈60% increase) than samples with homozygosis for the A allele. At first glance, the impact of CBR1 rs9024 on cardiac daunorubicin reductase activity may appear relatively modest. However, certain therapeutic regimens such as combination chemotherapies for pediatric acute myeloid leukemia require the administration of several doses of daunorubicin (n > 5). In this “chronic” context, the effect size of CBR1 rs9024 genotype status may become more relevant in terms of favoring the intracardiac build-up of cardiotoxic daunorubicinol. It appears that CBR1 rs9024 genotype status does not impact the cardiac expression of CBR1 in heart samples from donors with- DS. A recent study described regulation of CBR1 expression by microRNAs (hsa-miR-574-5p and hsa-miR-921) dependent on the allele status of the CBR1 rs9024 (25). New evidence suggests that the expression of specific microRNAs is dysregulated in lymphocytes from subjects with- DS (35). Thus, it is plausible that alterations in the cardiac expression of hsa-miR-574-5p and/or hsa-miR-921 in the DS setting would in turn disrupt the regulation of CBR1 dependent upon rs9024 genotype status. Our laboratory is exploring this intriguing possibility by examining the expression of hsa-miR-574-5p and hsa-miR-921 in hearts and cell lines from donors with- and without- DS.
There are some limitations in this study, the main one being the relatively small number of cardiac samples from donors with- DS. Our procurement rates for samples from donors with- DS is low (≈ 1 sample every 9 – 12 months), even after working with national cooperative resources such as NDRI, CHTN, and the NICHD Brain and Tissue Bank. The scarcity of representative tissue samples from donors with- DS continues to impair the execution of large-scale quantitative studies to tackle fundamental topics related to the metabolism and disposition of commonly used drugs in this group of subjects (36, 37). Second, studies based on the use of tissue samples from cadaveric donors are prone to many issues including, but not limited to sample degradation, incomplete/null medical histories and/or demographics, use of concomitant medications/drugs/smoking status. Nevertheless, the collection of samples that is the object of this study may still represent an informative window to assess the extent of variability in the cardiac expression of anthracycline reductases in subjects with- and without- DS.
Weiss highlighted the need for performing comprehensive cardiac drug metabolism studies to characterize the system response at the organ level during pharmacotherapy with common drugs (e.g., anthracyclines) (38). In this study, multiple regression analysis suggests that sex together with CBR1, AKR1A1, and AKR7A2 protein expression account for ≈70% of the total variance in cardiac daunorubicin reductase activity. Thus, the expression of specific CBRs and AKRs appears to drive the cardiac synthesis of daunorubicinol. Future modeling efforts should be directed towards the identification of distinct cardiac CBR/AKR expression profiles that dictate low and/or high rates of synthesis of cardiotoxic C-13 anthracycline alcohol metabolites. The present study represents a necessary step for the creation of novel quantitative methods to predict the variable pharmacodynamics of anthracyclines in human heart by integrating genetic and phenotypic data (e.g., CBR/AKR protein abundance, rs9024, miRNA levels, DS status, etc) for the specific CBRs and AKRs involved in the metabolism of these clinically useful drugs.
Supplementary Material
Acknowledgments
This work was supported by the National Institute of General Medical Sciences [GM073646].
ABBREVIATIONS
- aCGH
Array comparative genomic hybridization
- ACTB
Actin B
- AKR1A1
aldo-keto reductase family 1, member A1
- AKR1C3
aldo-keto reductase family 1, member C3
- AKR7A2
aldo-keto reductase family 7, member A2
- AKRs
aldo-keto reductases
- CBR1
carbonyl reductase 1
- CBR3
carbonyl reductase 3
- CBRs
carbonyl reductases
- DS
Down syndrome
- LOD
limit of detection
- LOQ
limit of quantification
References
- 1.Penningand TM, Drury JE. Human aldo-keto reductases: Function, gene regulation, and single nucleotide polymorphisms. Arch Biochem Biophys. 2007;464:241–250. doi: 10.1016/j.abb.2007.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoffmannand F, Maser E. Carbonyl reductases and pluripotent hydroxysteroid dehydrogenases of the short-chain dehydrogenase/reductase superfamily. Drug Metab Rev. 2007;39:87–144. doi: 10.1080/03602530600969440. [DOI] [PubMed] [Google Scholar]
- 3.Rosemondand MJ, Walsh JS. Human carbonyl reduction pathways and a strategy for their study in vitro. Drug Metab Rev. 2004;36:335–361. doi: 10.1081/dmr-120034154. [DOI] [PubMed] [Google Scholar]
- 4.Menna P, Paz OG, Chello M, Covino E, Salvatorelli E, Minotti G. Anthracycline cardiotoxicity. Expert Opin Drug Saf. 2012;11(Suppl 1):S21–36. doi: 10.1517/14740338.2011.589834. [DOI] [PubMed] [Google Scholar]
- 5.Wouters KA, Kremer LCM, Miller TL, Herman EH, Lipshultz SE. Protecting against anthracycline-induced myocardial damage: a review of the most promising strategies. British Journal of Haematology. 2005;131:561–578. doi: 10.1111/j.1365-2141.2005.05759.x. [DOI] [PubMed] [Google Scholar]
- 6.Krischer JP, Epstein S, Cuthbertson DD, Goorin AM, Epstein ML, Lipshultz SE. Clinical cardiotoxicity following anthracycline treatment for childhood cancer: the Pediatric Oncology Group experience. J Clin Oncol. 1997;15:1544–1552. doi: 10.1200/JCO.1997.15.4.1544. [DOI] [PubMed] [Google Scholar]
- 7.O’Brien MM, Taub JW, Chang MN, Massey GV, Stine KC, Raimondi SC, Becton D, Ravindranath Y, Dahl GV. Cardiomyopathy in children with Down syndrome treated for acute myeloid leukemia: a report from the Children’s Oncology Group Study POG 9421. J Clin Oncol. 2008;26:414–420. doi: 10.1200/JCO.2007.13.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tauband JW, Ravindranath Y. What’s up with down syndrome and leukemia-A lot! Pediatr Blood Cancer. 2011;57:1–3. doi: 10.1002/pbc.23033. [DOI] [PubMed] [Google Scholar]
- 9.Menna P, Salvatorelli E, Minotti G. Cardiotoxicity of Antitumor Drugs. Chem Res Toxicol. 2008;21:978–989. doi: 10.1021/tx800002r. [DOI] [PubMed] [Google Scholar]
- 10.Mushlin PS, Cusack BJ, Boucek RJ, Jr, Andrejuk T, Li X, Olson RD. Time-related increases in cardiac concentrations of doxorubicinol could interact with doxorubicin to depress myocardial contractile function. Br J Pharmacol. 1993;110:975–982. doi: 10.1111/j.1476-5381.1993.tb13909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Olson RD, Mushlin PS, Brenner DE, Fleischer S, Cusack BJ, Chang BK, Boucek RJ., Jr Doxorubicin cardiotoxicity may be caused by its metabolite, doxorubicinol. Proc Natl Acad Sci U S A. 1988;85:3585–3589. doi: 10.1073/pnas.85.10.3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stewart DJ, Grewaal D, Green RM, Mikhael N, Goel R, Montpetit VA, Redmond MD. Concentrations of doxorubicin and its metabolites in human autopsy heart and other tissues. Anticancer Res. 1993;13:1945–1952. [PubMed] [Google Scholar]
- 13.Bains OS, Grigliatti TA, Reid RE, Riggs KW. Naturally occurring variants of human aldo-keto reductases with reduced in vitro metabolism of daunorubicin and doxorubicin. J Pharmacol Exp Ther. 2010;335:533–545. doi: 10.1124/jpet.110.173179. [DOI] [PubMed] [Google Scholar]
- 14.Bains OS, Takahashi RH, Pfeifer TA, Grigliatti TA, Reid RE, Riggs KW. Two Allelic Variants of Aldo-Keto Reductase 1A1 Exhibit Reduced in Vitro Metabolism of Daunorubicin. Drug Metab Dispos. 2008;36:904–910. doi: 10.1124/dmd.107.018895. [DOI] [PubMed] [Google Scholar]
- 15.Lakhman SS, Ghosh D, Blanco JG. Functional significance of a natural allelic variant of human carbonyl reductase 3 (CBR3) Drug Metab Dispos. 2005;33:254–257. doi: 10.1124/dmd.104.002006. [DOI] [PubMed] [Google Scholar]
- 16.Miura T, Nishinaka T, Terada T. Different functions between human monomeric carbonyl reductase 3 and carbonyl reductase 1. Mol Cell Biochem. 2008;315:113–121. doi: 10.1007/s11010-008-9794-5. [DOI] [PubMed] [Google Scholar]
- 17.Blanco JG, Leisenring WM, Gonzalez-Covarrubias VM, Kawashima TI, Davies SM, Relling MV, Robison LL, Sklar CA, Stovall M, Bhatia S. Genetic polymorphisms in the carbonyl reductase 3 gene CBR3 and the NAD(P)H:quinone oxidoreductase 1 gene NQO1 in patients who developed anthracycline-related congestive heart failure after childhood cancer. Cancer. 2008;112:2789–2795. doi: 10.1002/cncr.23534. [DOI] [PubMed] [Google Scholar]
- 18.Lal S, Sandanaraj E, Wong ZW, Ang PC, Wong NS, Lee EJ, Chowbay B. CBR1 and CBR3 pharmacogenetics and their influence on doxorubicin disposition in Asian breast cancer patients. Cancer Sci. 2008;99:2045–2054. doi: 10.1111/j.1349-7006.2008.00903.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lal S, Mahajan A, Chen WN, Chowbay B. Pharmacogenetics of target genes across doxorubicin disposition pathway: a review. Curr Drug Metab. 2010;11:115–128. doi: 10.2174/138920010791110890. [DOI] [PubMed] [Google Scholar]
- 20.Blanco JG, Sun CL, Landier W, Chen L, Esparza-Duran D, Leisenring W, Mays A, Friedman DL, Ginsberg JP, Hudson MM, Neglia JP, Oeffinger KC, Ritchey AK, Villaluna D, Relling MV, Bhatia S. Anthracycline-Related Cardiomyopathy After Childhood Cancer: Role of Polymorphisms in Carbonyl Reductase Genes--A Report From the Children’s Oncology Group. J Clin Oncol. 2012;30:1415–1421. doi: 10.1200/JCO.2011.34.8987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kalabus JL, Sanborn CC, Jamil RG, Cheng Q, Blanco JG. Expression of the anthracycline-metabolizing enzyme carbonyl reductase 1 in hearts from donors with Down syndrome. Drug Metab Dispos. 2010;38:2096–2099. doi: 10.1124/dmd.110.035550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kassner N, Huse K, Martin HJ, Godtel-Armbrust U, Metzger A, Meineke I, Brockmoller J, Klein K, Zanger UM, Maser E, Wojnowski L. Carbonyl reductase 1 is a predominant doxorubicin reductase in the human liver. Drug Metab Dispos. 2008;36:2113–2120. doi: 10.1124/dmd.108.022251. [DOI] [PubMed] [Google Scholar]
- 23.O’Connor T, Ireland LS, Harrison DJ, Hayes JD. Major differences exist in the function and tissue-specific expression of human aflatoxin B1 aldehyde reductase and the principal human aldo-keto reductase AKR1 family members. Biochem J. 1999;343(Pt 2):487–504. [PMC free article] [PubMed] [Google Scholar]
- 24.Gonzalez-Covarrubias V, Zhang J, Kalabus JL, Relling MV, Blanco JG. Pharmacogenetics of human carbonyl reductase 1 (CBR1) in livers from black and white donors. Drug Metab Dispos. 2009;37:400–407. doi: 10.1124/dmd.108.024547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kalabus JL, Cheng Q, Blanco JG. MicroRNAs Differentially Regulate Carbonyl Reductase 1 (CBR1) Gene Expression Dependent on the Allele Status of the Common Polymorphic Variant rs9024. PLoS One. 2012;7:e48622. doi: 10.1371/journal.pone.0048622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sultan M, Piccini I, Balzereit D, Herwig R, Saran NG, Lehrach H, Reeves RH, Yaspo ML. Gene expression variation in Down’s syndrome mice allows prioritization of candidate genes. Genome Biol. 2007;8:R91. doi: 10.1186/gb-2007-8-5-r91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 28.de Jong J, Guerand WS, Schoofs PR, Bast A, van der Vijgh WJ. Simple and sensitive quantification of anthracyclines in mouse atrial tissue using high-performance liquid chromatography and fluorescence detection. J Chromatogr. 1991;570:209–216. doi: 10.1016/0378-4347(91)80218-2. [DOI] [PubMed] [Google Scholar]
- 29.Fetterly GJ, Aras U, Lal D, Murphy M, Meholick PD, Wang ES. Development of a preclinical PK/PD model to assess antitumor response of a sequential aflibercept and doxorubicin-dosing strategy in acute myeloid leukemia. The AAPS journal. 2013;15:662–673. doi: 10.1208/s12248-013-9480-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheng Q, Yang W, Raimondi SC, Pui CH, Relling MV, Evans WE. Karyotypic abnormalities create discordance of germline genotype and cancer cell phenotypes. Nat Genet. 2005;37:878–882. doi: 10.1038/ng1612. [DOI] [PubMed] [Google Scholar]
- 31.Seewald L, Taub JW, Maloney KW, McCabe ER. Acute leukemias in children with Down syndrome. Mol Genet Metab. 2012;107:25–30. doi: 10.1016/j.ymgme.2012.07.011. [DOI] [PubMed] [Google Scholar]
- 32.Ait Yahya-Graison E, Aubert J, Dauphinot L, Rivals I, Prieur M, Golfier G, Rossier J, Personnaz L, Creau N, Blehaut H, Robin S, Delabar JM, Potier MC. Classification of human chromosome 21 gene-expression variations in Down syndrome: impact on disease phenotypes. American journal of human genetics. 2007;81:475–491. doi: 10.1086/520000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prandini P, Deutsch S, Lyle R, Gagnebin M, Delucinge Vivier C, Delorenzi M, Gehrig C, Descombes P, Sherman S, Dagna Bricarelli F, Baldo C, Novelli A, Dallapiccola B, Antonarakis SE. Natural gene-expression variation in Down syndrome modulates the outcome of gene-dosage imbalance. American journal of human genetics. 2007;81:252–263. doi: 10.1086/519248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Patterson D. Molecular genetic analysis of Down syndrome. Human genetics. 2009;126:195–214. doi: 10.1007/s00439-009-0696-8. [DOI] [PubMed] [Google Scholar]
- 35.Xu Y, Li W, Liu X, Chen H, Tan K, Chen Y, Tu Z, Dai Y. Identification of dysregulated microRNAs in lymphocytes from children with Down syndrome. Gene. 2013 doi: 10.1016/j.gene.2013.07.055. [DOI] [PubMed] [Google Scholar]
- 36.McCabeand LL, McCabe ER. Down syndrome: issues to consider in a national registry, research database and biobank. Mol Genet Metab. 2011;104:10–12. doi: 10.1016/j.ymgme.2011.03.018. [DOI] [PubMed] [Google Scholar]
- 37.Oster-Granite ML, Parisi MA, Abbeduto L, Berlin DS, Bodine C, Bynum D, Capone G, Collier E, Hall D, Kaeser L, Kaufmann P, Krischer J, Livingston M, McCabe LL, Pace J, Pfenninger K, Rasmussen SA, Reeves RH, Rubinstein Y, Sherman S, Terry SF, Whitten MS, Williams S, McCabe ER, Maddox YT. Down syndrome: national conference on patient registries, research databases, and biobanks. Mol Genet Metab. 2011;104:13–22. doi: 10.1016/j.ymgme.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weiss M. Functional characterization of drug uptake and metabolism in the heart. Expert Opin Drug Metab Toxicol. 2011;7:1295–1306. doi: 10.1517/17425255.2011.614233. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.