Abstract
Background
Asthma is a heterogeneous disease characterized by abnormal airway pathophysiology and susceptibility to different stimuli, as exemplified by a subset of individuals with exercise-induced bronchoconstriction (EIB). Induced sputum provides a noninvasive method to sample airway biofluids that are enriched in proteins.
Objective
We hypothesized that novel mechanisms in the pathogenesis of asthma may be revealed by studying the patterns of protein expression in induced sputum.
Methods
We used shotgun proteomics to analyze induced sputum from 5 normal individuals and 10 asthmatics, including 5 with EIB. Differential protein expression between asthmatics, asthma subphenotypes and control subjects was determined using spectral counting and computational methods.
Results
Using Gene Ontology analysis, we defined the functional landscape of induced sputum proteome and applied network analysis to construct a protein interaction map for this airway compartment. Shotgun proteomics analysis identified a number of proteins whose differential enrichment or depletion robustly distinguishedasthmatics from normal controls, and captured the effects of exercise on induced sputum proteome. Functional and network analysis identified key processes, including proteolytic activity that are known contributors to airway remodeling. Importantly, this approach highlighted previously unrecognized roles for differentially expressed proteins in pathways implicated in asthma, such as modulation of phospholipase A2 by secretoglobin, a putative role for S100A8/9 in human asthma, and selective upregulation of complement 3a in response to exercise in asthmatics.
Conclusion
Computationally-intensive analysis of induced sputum proteome is a powerful approach to understand the pathophysiology of asthma and a promising methodology to investigate other diseases of the airways.
Keywords: Asthma, exercise-induced bronchoconstriction, shotgun proteomics, induced sputum, protein network
Introduction
Noninvasive methods to measure mediators of respiratory function are important tools for the diagnosis, assessment of response to treatment, and understanding molecular mechanisms in pulmonary disorders. Induced sputum is one such validated technique that is commonly utilized for sampling airway secretions and cells1, and shown to provide diagnostic and mechanistic insights into a number of chronic lung diseases including asthma2, 3, COPD4, sarcoidosis5, and cystic fibrosis6. Induced sputum is comprised of a cellular part and a fluid phase, each of which represents constituents from various sources including airway epithelial cells, inflammatory cells, airway secretions, and even bacterial/viral components. The fluid phase of induced sputum is an abundant source of proteins, many of which have been implicated in lung disorders hallmarked by airway inflammation7.
Proteomics approaches offer great promise for the systematic analysis of complex biological samples such as induced sputum, and have the advantage of assessing the presence and abundance of gene products (i.e., proteins) that are functionally relevant to clinical phenotypes. However, very few studies have applied this approach to investigate the proteome of induced sputum in pulmonary disorders. Nicholas et al reported on using a combination of 2-dimensional (2-D) gel electrophoresis and mass spectrometry to analyze the protein content of induced sputum obtained from a single healthy individual8. They identified over 190 unique human proteins, many of which were distinct from proteins derived from saliva or bronchoalveolar lavage fluid. Despite excellent resolving power, 2-D gel proteomics has several limitations. First, it is biased in favor detecting high-abundance proteins and therefore does not provide a comprehensive proteomic profile. In addition, difficulties in reproducibility and protein identification, limit its utility for large-scale proteomics analysis across multiple samples. More recently, Gray et al applied surface-enhanced laser desorption/ionization time-of-flight (SELDI-TOF) technology to compare induced sputum proteomic signatures in asthma, COPD, cystic fibrosis, and bronchiectasis9. While SELDI-TOF has the advantage of being a high-throughput screening platform for biomarker discovery, it detects only spectral peaks as surrogates for putative proteins and formal identification of proteins must be performed using different methods such as tandem mass spectrometry or Western analysis. Indeed, the investigators identified and confirmed only eight distinct proteins—a small subset of the induced sputum proteome.
Shotgun proteomics, in which tryptic digests of complex protein mixtures are analyzed by tandem mass spectrometry, overcomes many of these limitations and is an ideal approach for large-scale protein measurements across multiple biological samples. We have previously used shotgun proteomics to study the airspace proteome as sampled by bronchoalveolar lavage in healthy subjects10 and those with cystic fibrosis11.
In the present work, we applied this approach to deeply explore the proteome of induced sputum in healthy subjects and asthmatics with and without exercise-induced bronchoconstriction (EIB). Despite being the most common chronic disease in young adults, the heterogeneous nature of asthma has hampered efforts to elucidate common pathophysiologic mechanisms. Therefore, we focused our proteomics strategy to a group of carefully phenotyped asthmatics and normal controls under baseline conditions and after an acute exercise challenge. We chose to include exercise challenge because EIB is a manifestation of indirect airway hyperresponsiveness—a hallmark feature of asthma. We tested the hypothesis that asthma and its sub types cause distinct alterations in the airway proteome, and an understanding of these perturbed proteomic signatures may provide mechanistic insights into the pathophysiology of this complex disorder.
Methods
Please see Online Repository Material for further details.
Subject recruitment and phenotyping
We utilized biological samples from two cohorts of subjects that were enrolled at the University of Washington3, 12. The University of Washington Institutional Review Board approved the study protocols, and written informed consent was obtained from all participants. Subjects 18-59 years of age were recruited who had a physician diagnosis of asthma for ≥ 1 year, and used only an inhaled β 2-agonist for asthma treatment. Participants were not enrolled if they had used an inhaled or oral corticosteroid, leukotriene modifier, long-acting antihistamine, cromone, or long-acting β2-agonist in the 30 days prior to the study. In accordance with a priori definitions, asthmatics with a methacholine PC20 ≤ 4 mg/ml were identified with ≥ 20% fall in FEV1 following exercise challenge (EIB+ group) and asthmatic controls without EIB were identified with ≤ 5% fall in FEV1 following exercise challenge (EIB− group). Comparisons were also made to non-asthmatic subjects with normal spirometry (FEV1> 80% predicted) and a negative exercise challenge test (< 5% fall in FEV1 following exercise). Inducted sputum was obtained from each subject at baseline and on a separate day 30 min after exercise challenge.
Induced sputum sample preparation
Sputum induction was performed as previously described3, 12. Briefly, subjects inhaled nebulized hypertonic 3% saline for 20 min. Sputum samples were dispersed in 6.5mM DTT and the supernatant removed after the sample was centrifuged at 1000 × g. Equal volumes and concentrations of each sputum sample were denatured, reduced and alkylated and the solution underwent proteolysis for 16 hr using a 1:20 trypsin-to-protein ratio13 followed by desalting and completely dried using a Speed-Vac (Thermo-Savant, MA).
Shotgun proteomics analysis
Equal sample concentrations were analyzed by liquid chromatography tandem mass spectrometry (LC-MS/MS) as previously described14 to a hybrid linear ion trap-orbitrap mass spectrometer (Thermo Fisher, CA). All precursor ion survey scans were performed from m/z 400 to 2000. Collision-induced dissociation (CID) was performed in the linear ion trap. Subsequently, tandem MS were acquired in the Orbitrap. Data were acquired using data-dependent ion selection where the five most intense ions were selected for CID. The following two m/z ranges were surveyed and sampled: 400-675, 676-2000.
Protein identification and quantification
Tandem mass spectral RAW (ThermoFinnigan) files were first converted to mzXML format15 and then matched to a protein sequence in the IPI Human 3.53 database using SEQUEST16. A protein was considered to be identified only if: (i) ProteinProphet probability was greater than 0.817 and (ii) if greater than one unique peptide was found for each protein. Relative protein levels were determined using spectral counting as previously described18, 19. Briefly, spectral counts were determined by the number of times a peptide that matched a given protein was selected for CID. The final protein's spectral count value was the sum of the spectral counts acquired over the specified two gas phase fractions. To assess differences in relative protein abundance between biological conditions, we applied a validated non-parametric “spectral index” metric and determined statistical significance (95% confidence level) using random permutation (n = 1000)11, 20.
Western analysis
Western blots were conducted to measure differences in the levels of SERPINA1, SCGB1A1, SMR3B, C3a, and HPX in induced sputum supernatant. Polyclonal rabbit antibodies were used to probe for SERPINA1 (GenWay Biotech, CA), SCGB1A1 (Santa Cruz, CA), and HPX (Abnova, CA), and murine monoclonal antibodies were used to probe for SMR3B (Novus Biologics, CO) and C3a (Abcam, MA). Membranes were incubated with the primary antibody overnight at 4°C and then subsequently incubated with either anti-rabbit or anti-mouse HRP-linked antibody as appropriate (Cell Signaling, MA) for 1 hour at RT. Peroxidase activity was detected using LumiGLO ECL reagents (Cell Signaling, MA).
Statistical analysis
Statistical comparison between immunoblot intensities was performed using Student's t-test with 2-tailed P-values adjusted for unequal variances. Paired t-test or Wilcoxon matched pairs testing was performed when appropriate (GraphPad Prism 5, CA).
Functional analysis
Functional annotation of all identified proteins in the induced sputum of the subjects was obtained from the Gene Ontology (GO) database21. Overrepresented functional categories in induced sputum proteins relative to the human proteome were determined using the Database for Annotation, Visualization, and Integrated Discovery (DAVID)22. Enriched functional categories were required to be significant at a false discovery rate < 0.05 using a random permutation analysis (n = 1000). The GO hierarchical relationships among the biological modules were mapped using a web-based software program23.
Unsupervised classification
Two-dimensional hierarchical clustering of the proteomics profiles between subjects was performed using the average linkage method and the Pearson correlation metric24. Principal components analysis was performed based on the covariance matrix of protein spectral counts24.
Network analysis
A protein network was generated based on experimentally verified gene product interactions derived from Ingenuity System's database25 and several publicly available protein-protein interaction resources incorporated under a web-tool interface known as Search Tool for the Retrieval of Interacting Genes/Proteins (STRING)26.
Results
Phenotyping of subjects
Induced sputum samples from fifteen subjects were utilized in this study (see Table 1 for characteristics). Ten individuals had a confirmed diagnosis of asthma based on a methacholine PC20 ≤ 4 mg/ml. As shown in Figure 1, five asthma subjects met criteria for EIB (≥ 20% fall in FEV1 after exercise, EIB+) and five subjects with asthma did not have EIB (≤ 5% fall in FEV1 after exercise, EIB−). Five additional subjects were recruited that did not have asthma based on normal spirometry and negative exercise challenge (normal controls). The baseline FEV1 values of the EIB+ and EIB− subjects were similar but significantly lower than the controls, indicating underlying airflow limitation in the asthmatic group (Figure 1). The cellular differential of leukocytes and epithelial cells from induced sputum samples did not differ significantly among the individuals at baseline and after exercise (online Supplementary Figure 1).
Table.
Comparison of the characteristics of study groups.*
| Asthma | Normal Control | P Value | ||
|---|---|---|---|---|
| EIB+ | EIB− | |||
| Age (yrs)(range) | 28.8 (24-34) | 31.4 (23-54) | 25.8 (18-40) | 0.64 |
| Gender† | 1.00 | |||
| Male (%) | 20.0% | 20.0% | 20.0% | |
| Female (%) | 80.0% | 80.0% | 80.0% | |
| Race† | 0.34 | |||
| Caucasian | 100.0% | 100.0% | 80.0% | |
| Asian | 20.0% | |||
| Lung Function | ||||
| FEV1 (% pred) | 90.0 ± 10.3 | 83.4 ± 10.2 | 106.4 ± 12.2 | 0.02 |
| FVC (% pred) | 104.6 ± 12.8 | 97.0 ± 8.7 | 111.6 ± 16.8 | 0.26 |
| FEV1/FVC | 0.7± 0.1 | 0.7 ± 0.1 | 0.8 ± 0.1 | 0.27 |
| Post Bronchodilator | ||||
| Δ FEV1 (%) | 5.9 ± 3.3 | 8.9 ± 6.8 | 3.6 ± 1.0 | 0.20 |
| Exercise-induced Bronchoconstriction | ||||
| Maximum Decrease in FEV1 | −36.4 ± 3.3 | −0.9 ± 2.0 | −0.7 ± 2.0 | < 0.001 |
| Area Under FEV1 Curve‡ | −896.6 ± 173.4 | 106.0 ± 81.9 | 47.0 ± 57.6 | <0.001 |
| Direct Bronchial Hyperresponsiveness | ||||
| PC20 Methacholine¥ | 0.2 (0.1 - 1.0) | 0.7 (0.1 - 1.0) | NA | 0.05 |
Comparisons made by ANOVA except where otherwise specified. Values reported are mean ± standard deviation unless otherwise specified
Chi-square test.
Area under the FEV1 curve over the first 30 min after exercise (% change*min)
Value represent the geometric mean (95% confidence interval). The P value is for the log-transformed value.
Figure 1. Pulmonary function profiles of subjects.
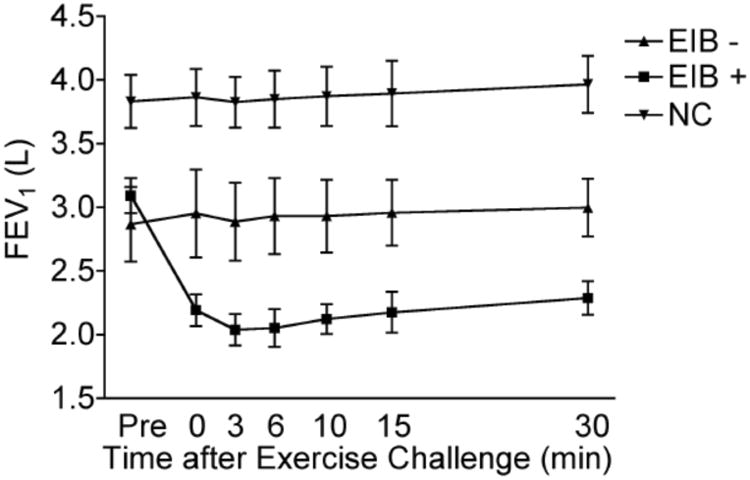
Forced expiratory volume in 1 sec (FEV1) of 10 asthmatics (5 EIB+, 5 EIB−) and 5 normal controls (NC) is shown as baseline (Pre) and temporally after exercise challenge. Note that the asthmatic group has a lower baseline FEV1, but only the EIB+ subjects develop further airflow obstruction post exercise.
Proteomic landscape of induced sputum
We identified 254 proteins in the induced sputum of all 15 subjects (5 normal, 10 asthmatics) of which 240 mapped to unique genes (online Supplementary Table 1). To better elucidate the functional role played by these proteins, we performed Gene Ontology analysis on this dataset and statistically determined biological modules that were enriched in induced sputum relative to the entire human proteome (Figure 2). The highly over-represented functional categories included those involved in defense response, protease inhibitor activity, immunity, response to inflammation and wounding, and complement activation. This finding implies that the induced sputum proteome is selectively enriched in proteins that map to specific functional roles. We did not observe significant differences in the distribution of these biological processes when we limited the proteins to those identified in normal subjects or asthmatics, suggesting that the global functional architecture of induced sputum proteome is not perturbed in asthma.
Figure 2. Functional analysis of induced sputum proteome.
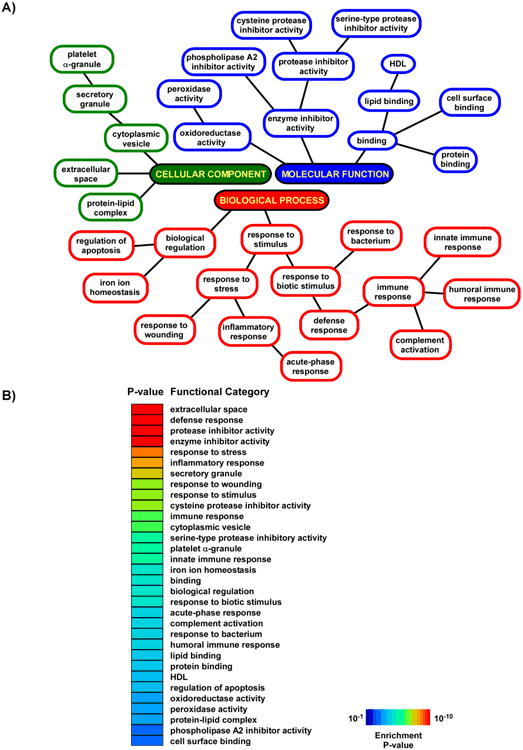
A. Enriched biological modules representing biological processes, molecular functions and cellular components are depicted based on the hierarchical structure of Gene Ontology annotation. B. The same functional categories are rank-ordered based on their enrichment P-values.
To further investigate relationships among the identified proteins, we queried gene product interaction knowledgebases and created a protein-protein network representation of the induced sputum proteome (Figure 3). This relational network or “interactome” was comprised of 80 nodes (approximately one-third of the identified proteins) and depicts a complex functional map of mediator interactions in the human airways (see also online Supplementary Table 1). The biological relevance of this putative interactome was assessed by systematically searching PubMed using PubMatrix27 for associations between network proteins and pulmonary disease in humans and animal models. Almost 90% of the nodes (70 out of 80) have been linked to lung disorders and 66% (53 out of 80) with airway disorders such as asthma.
Figure 3. Protein interaction network of induced sputum.
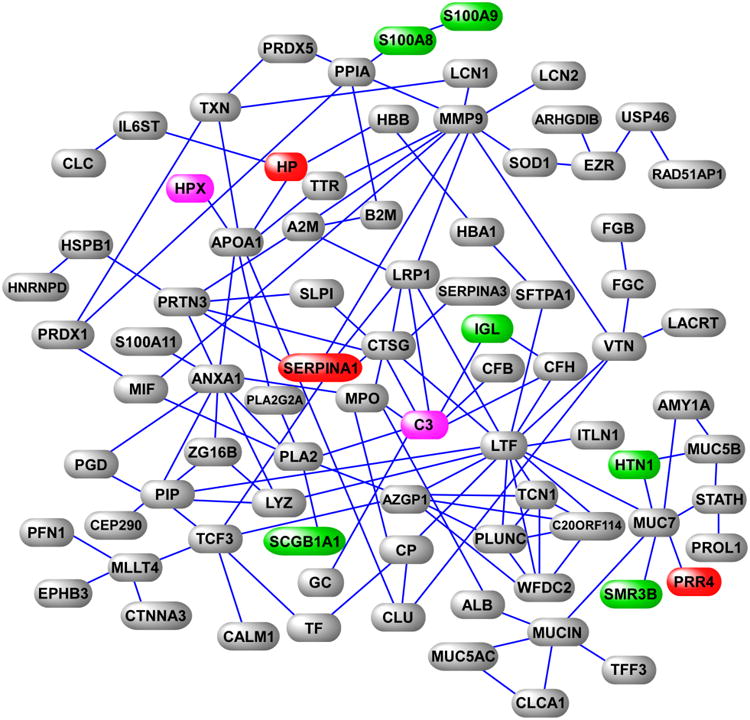
This interactome is a subset of the entire induced sputum proteome and is characterized by experimentally verified functional interaction of its member gene products. Selected proteins upregulated (red) and downregulated (green) in asthma are highlighted, as are two proteins upregulated in EIB+ subjects in response to exercise challenge (magenta). Note the interaction of the downregulated protein SCGB1A with phospholipase A2, and the high connectivity of the C3 hub (please see text for discussion).
Differential expression of proteins in induced sputum of asthmatics
Initially, we asked whether the entire proteomic profile of induced sputum could be used to discriminate subjects with and without asthma at baseline. Using the spectral counts of all proteins identified in the 15 subjects, we performed principal components analysis to group the subjects based on variability in protein expression, and did not observe distinct clusters corresponding to clinical phenotypes (online Supplementary Figure 2). Therefore, the presence of asthma does not appear to perturb the overall proteomic variability of induced sputum.
Next, we investigated whether specific subsets of proteins were differentially expressed between asthmatics and normal controls. We assessed the relative abundance of protein expression in the induced sputum using a non-parametric test we have developed called the spectral index20. We applied this metric to the spectral counts of the proteins identified at baseline in all 15 subjects and used a permutation-based method to determine differential protein expression between the two groups at a confidence level of 95%. Seventeen proteins were significantly different between healthy subjects and those with asthma, among them calcium binding proteins S100A9 and S100A8, serpin peptidase inhibitor (SERPINA1), submaxillary gland androgen regulated protein 3B (SMR3B) and secretoglobin (SCGB1A1, also known as Clara cell 10-kD protein, CC10)(Figure 4A). This subset of differentially expressed proteins robustly classified normal individuals from asthmatics based on unsupervised hierarchical cluster analysis. Functional analysis of these proteins revealed that they mapped to processes involved in defense response, extracellular space, inflammatory response and response to stress (false discovery rate < 5%). We biochemically confirmed the differential expression of three of these proteins (SERPINA1, SMR3B, SCGB1A1) between controls and asthmatics (Figure 4B-D). We did not observe a difference in the levels of these three proteins between the asthma subgroups (EIB+ vs. EIB−) (online Supplementary Figure 3).
Figure 4. Proteomic signature of induced sputum in asthma.
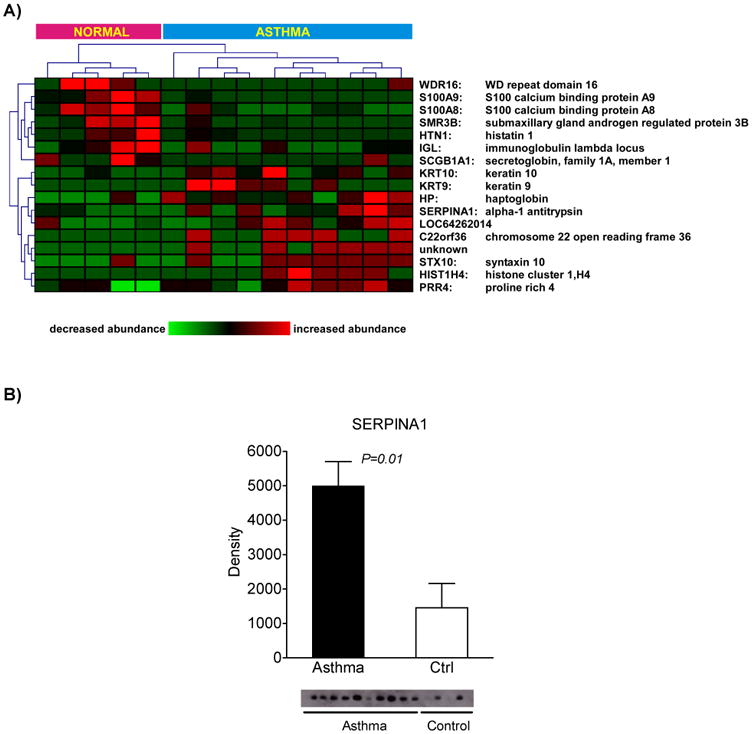
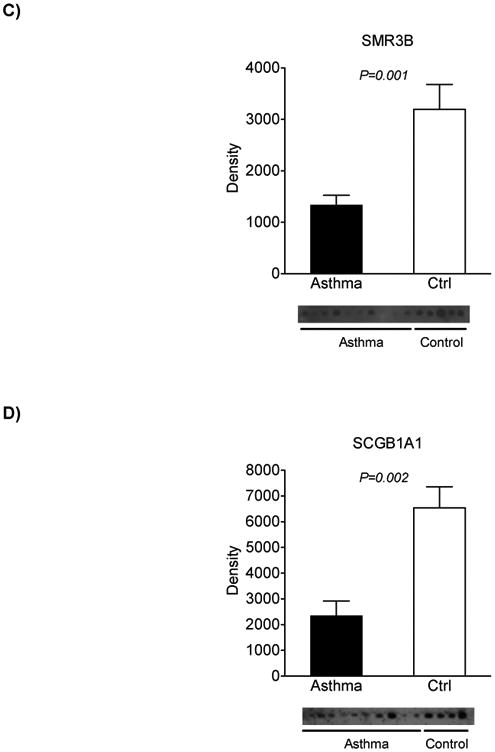
A. Hierarchical clustering of differentially expressed proteins based on spectral counting is represented as a heatmap and demonstrates robust discrimination between asthmatics and control subjects. B-D. Confirmation of proteomics findings for select proteins using Western analysis.
Proteomic signatures between asthma phenotypes and in response to exercise
We applied the spectral index metric to determine whether EIB+ and EIB− subjects could be distinguished based on their baseline induced sputum proteome. At a confidence level of 95%, we identified 5 proteins with significant differences in abundance between the two groups (online Supplementary Figure 4).
Since EIB+ individuals are uniquely characterized by their susceptibility to exercise-induced bronchoconstriction, we then assessed the effects of exercise challenge on the induced sputum of the 5 EIB+ subjects (pre vs. post, Figure 4A). Nine proteins met our 95% significance threshold, including increased expression of complement component 3 (C3) and hemopexin (HPX). We confirmed these differences by Western analysis, and demonstrated that there was a significant increase in the active complement component 3 (C3a) (Figure 5B and C).
Figure 5. Exercise-induced alterations of induced sputum proteome in asthmatics with EIB.
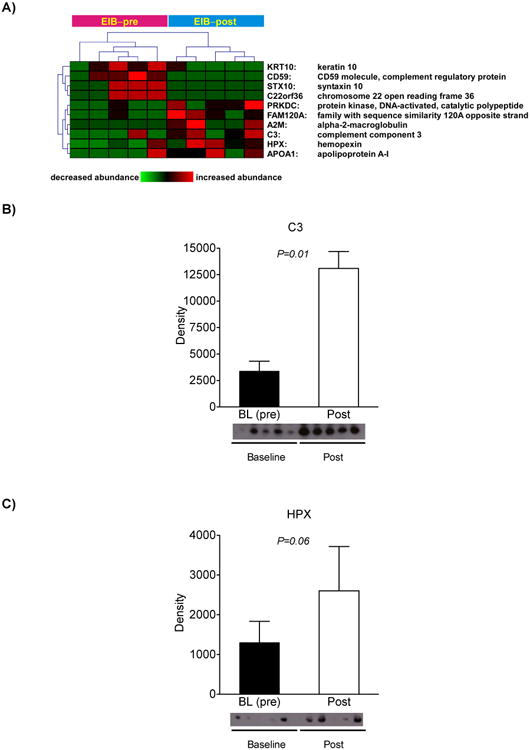
A. Heatmap depiction of differentially expressed proteins in EIB+ subjects upon exercise challenge. B-C. Immunoblot analysis confirms the shotgun proteomics results for the upregulation of C3a in response to exercise and trends towards significance for HPX.
Discussion
Induced sputum—a complex mixture of mucins, lipids, and proteins derived from tracheobronchial secretions, provides a non-invasive window into the physiologic state of the airways in health and disease28. In this work, we initially defined the proteomic landscape of induced sputum in 15 subjects and subsequently investigated its perturbation in asthma and asthma sub-phenotypes. The current study represents the most comprehensive shotgun proteomics analysis of induced sputum to date and highlights the utility of this approach to investigate the pathophysiology of airway disorders.
Our results indicate that the induced sputum proteome is highly enriched in select number of processes, most prominently those involved in defense response, immunity, protease inhibitory activity, and inflammation. We previously reported that many of these biological modules are also significantly over-represented in the proteome of bronchoalveolar lavage fluid (BALF)13, implying a substantial overlap in the functional role of biofluids sampled from the airways and those obtained from the airspaces, even though the identified proteins differ significantly between the two compartments. Using curated gene product interaction databases, we demonstrated that many of the proteins identified in induced sputum are functionally linked with each other. This protein-protein interactome provided a roadmap to understand the functional consequences of airway diseases, such as asthma, as discussed below.
An advantage of shotgun proteomics over gel-based methods is its amenability to label-free quantitative assessment of protein abundance between samples18, 29. We therefore employed a validated statistical approach known as the spectral index11, 20 to identify differentially expressed proteins in induced sputum of asthmatics relative to healthy controls. Ten proteins were significantly upregulated in asthma, including SERPINA1 (α1-antitrypsin), which we confirmed by Western analysis. Airway inflammation and remodeling in asthma involves degradation of the extracellular matrix, most prominently elastin, and is characterized by an imbalance between elastase and its primary inhibitor SERPINA1, both of which have been reported to be elevated in the induced sputum of asthmatics30. Our findings confirm these previous observations and suggest the presence of a proinflammatory and proteolytic milieu in the airways of subjects with asthma, promoting the active remodeling of airways. Interestingly, Serpina1 gene and protein expression were recently reported to be highly upregulated in the developing lung of Fisher rats—a strain known for its innate susceptibility to airway hyperresponsiveness31.
We identified seven proteins significantly downregulated in induced sputum of asthmatics, including the founding member of the secretoglobin superfamily, SCGB1A1. Allergen-challenged Scgb1a1-null mice manifest an exaggerated allergic response withprofound pulmonary eosinophilia, elevation of Th2 cytokines, and development of eosinophilic chronic rhinosinusitis32-34. In humans, the number of SCGB1A1-positive airway epithelial cells are significantly reduced in asthmatics35 and the serum levels of this protein are decreased36. Our Gene Ontology analysis mapped SCGB1A1 to the “phospholipase A2 inhibitor activity” module (Figure 2A), a functional role that has been experimentally verified37, and implicates this proteinas an inhibitor of phospholipase A2-dependent biosynthesis of eicosanoids in asthma (Figure 3). We recently reported that the phospholipase A2 pathway is over-expressed in asthmatics12, 38, and our current finding suggests a novel mechanism whereby this pathway can be activated by downregulation of SCGB1A1, leading to bronchial hyperresponsiveness.
A notable strength of our study is the meticulous phenotyping of the asthmatic group to identify subjects susceptible to exercise-induced bronchoconstriction (EIB+). Proteomics analysis of induced sputum in the EIB+ subjects identified several proteins whose abundance changed in response to exercise challenge, including significant increases in the levels of complement component3 (C3) and hemopexin (HPX) (Figure 4). C3, a member of the phylogenetically-conserved complement system, is a key component of the innate immunity and central to the activation of both the classical and alternative complement pathways. Activated C3 (C3a) is a potent chemoattractant for recruitment and activation of proinflammatory leukocytes, and can initiate a cascade of events leading to the release of allergic mediators such as leukotrienes, histamine, interleukin-1, interleukin-6 and tumor necrosis factor39. Activation of C3 can also promote airway mucus secretion, smooth muscle contraction and induce vascular permeability40. A number of studies have demonstrated that C3 levels are elevated in the airspaces of asthmatics upon allergen challenge41, 42, and in response to multiple environmental exposures including diesel exhaust43, airborne particulate matter44, ozone45, and tobacco smoke46, 47. However, to our knowledge, increased abundance of C3a in the induced sputum of EIB+ subjects following exercise challenge has not been previously recognized, but is consistent with our previous findings in support of an inflammatory basis for EIB48. C3 is a densely connected node in the induced sputum protein interaction network (Figure 3)—an observation that supports its central role in the functional stability of the interactome49-52, and suggests that it represents a rational therapeutic target for reversing the acute airflow obstruction associated withEIB53, 54. Hemopexin (HPX), an acute phase heme-binding protein, was also upregulated post exercise in the EIB+ subjects. Unlike C3, a role for HPX in airway hyperresponsiveness has not been elucidated although recent proteomic analyses on BALF have revealed elevated HPX levels in asthmatics55 and in a murine model of asthma56.
In a recent study, Gray et al applied a different proteomics approach (using SELDI-TOF) to investigate alterations in induced sputum proteome of subjects with suppurative respiratory diseases, including asthma9. They identified eight unique proteins and validated changes in abundance for five of them: S100A8, S100A9, SCGB1A1, SMR3B, and S100A12. We also report that four of these proteins (S100A8, S100A9, SCGB1A1, SMR3B) are differentially expressed in the induced sputum of asthmatics. Although our results confirm those of Gray et al regarding the downregulation of SCGB1A1 and SMR3B in asthma, our findings demonstrated that S100A8 and S100A9 were less abundant in asthmatics, which disagree with their observation of modest increases of these proteins (with significant variability within their asthma cohort). Since inflammatory cells are a source of S100A8/A9, one reason for discrepancy between the two studies may be that a subset of Gray's asthmatics may have harbored increased number of inflammatory cells in their airways, whereas we did not observe any differences in airway cell counts between our asthmatic cohort and the normal controls (online Supplementary Figure 1). S100A8/A9, also known as calgranulin A and B, form heterodimeric calcium-binding complexes that mediate diverse cellular effects including inflammation, immune response, cell growth, response to infection and oxidative defense. Although their role in asthma is not clear, recent reports demonstrate that S100A8 is a key protective molecule that, when given exogenously, modulates mast cell function and suppresses eosinophilic infiltration in OVA-challenged mice57. These findings support our results (Figure 3) and suggest that reduced abundance of S100A8/9 in the airways of asthmatics may make the environmental milieu more hospitable to mast cell activation and bronchial hyperresponsiveness.
Our study has a number of limitations. Given the complexity of shotgun proteomics, we have analyzed induced sputum samples from relatively few asthmatics and healthy controls. Although the subjects were carefully phenotyped and the current study is the most comprehensive of its kind, our findings need to be confirmed in larger cohorts. Despite possessing a wide dynamic range that is ideally suited for large-scale proteomics mapping, our shotgun approach is still biased toward the detection of larger and more abundant proteins. Therefore, smaller molecules such as cytokines are less likely to be captured by this technique. Furthermore, our statistical approaches for determining differential protein expression based on spectral counting, while powerful and validated, still represent a “semi-quantitative” assessment and require functional confirmation. Finally, it is important to note that differential abundance in a protein may simply be a marker of disease and does not necessarily imply that it plays a critical role in its pathogenesis.
In this work, we integrated shotgun proteomics with computational data analysis to define the proteomic landscape of induced sputum and then utilized this information to identify candidate proteins and putative mechanisms differentially activated in asthma and its subtypes. Our comprehensive and unbiased proteomics results were mapped into a protein network that highlighted several previously under-recognized mechanisms in asthma, including the interaction of SCGB1A1 with the phospholipase A2 pathway, significant reduction in the abundance of S100A8/A9 in airways of asthmatics, and a role for activated complement component 3 in exercise-induced bronchoconstriction. Since induced sputum can be collected noninvasively and routinely in the clinical setting, our integrated proteomics methodology represents a promising approach to study asthma and other pulmonary disorders affecting the airways.
Supplementary Material
Key Messages.
Shotgun proteomics of induced sputum is a useful method to comprehensively identify airway-derived proteins.
Asthma and its subtypes such as exercise-induced bronchoconstriction are associated with distinct proteomic signatures.
Pathway-centric computational analysis of induced sputum proteome highlights previously unrecognized roles and mechanisms in asthma for several candidate proteins.
Acknowledgments
Funding Sources: Institute of Translational Health Sciences grant (SAG), NIH R01HL89215 (TSH).
Abbreviations
- COPD
chronic obstructive pulmonary disease
- SELDI-TOF
surface-enhanced laser desorption/ionization time-of-flight
- EIB
exercise-induced bronchoconstriction
- FEV1
forced expired volume in 1 sec
- MS
mass spectrometry
- CID
collision-induced dissociation
- m/z
mass to charge ratio
- BALF
bronchoalveolar lavage fluid
References
- 1.Nicholas B, Djukanovic R. Induced sputum: a window to lung pathology. Biochem Soc Trans. 2009;37:868–72. doi: 10.1042/BST0370868. [DOI] [PubMed] [Google Scholar]
- 2.Green RH, Brightling CE, McKenna S, Hargadon B, Parker D, Bradding P, et al. Asthma exacerbations and sputum eosinophil counts: a randomised controlled trial. Lancet. 2002;360:1715–21. doi: 10.1016/S0140-6736(02)11679-5. [DOI] [PubMed] [Google Scholar]
- 3.Hallstrand TS, Wurfel MM, Lai Y, Ni Z, Gelb MH, Altemeier WA, et al. Transglutaminase 2, a novel regulator of eicosanoid production in asthma revealed by genome-wide expression profiling of distinct asthma phenotypes. PLoS One. 2010;5:e8583. doi: 10.1371/journal.pone.0008583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stanescu D, Sanna A, Veriter C, Kostianev S, Calcagni PG, Fabbri LM, et al. Airways obstruction, chronic expectoration, and rapid decline of FEV1 in smokers are associated with increased levels of sputum neutrophils. Thorax. 1996;51:267–71. doi: 10.1136/thx.51.3.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fireman E, Topilsky I, Greif J, Lerman Y, Schwarz Y, Man A, et al. Induced sputum compared to bronchoalveolar lavage for evaluating patients with sarcoidosis and non-granulomatous interstitial lung disease. Respir Med. 1999;93:827–34. doi: 10.1016/s0954-6111(99)90269-x. [DOI] [PubMed] [Google Scholar]
- 6.Sagel SD, Sontag MK, Wagener JS, Kapsner RK, Osberg I, Accurso FJ. Induced sputum inflammatory measures correlate with lung function in children with cystic fibrosis. J Pediatr. 2002;141:811–7. doi: 10.1067/mpd.2002.129847. [DOI] [PubMed] [Google Scholar]
- 7.Dragonieri S, Tongoussouva O, Zanini A, Imperatori A, Spanevello A. Markers of airway inflammation in pulmonary diseases assessed by induced sputum. Monaldi Arch Chest Dis. 2009;71:119–26. doi: 10.4081/monaldi.2009.357. [DOI] [PubMed] [Google Scholar]
- 8.Nicholas B, Skipp P, Mould R, Rennard S, Davies DE, O'Connor CD, et al. Shotgun proteomic analysis of human-induced sputum. Proteomics. 2006;6:4390–401. doi: 10.1002/pmic.200600011. [DOI] [PubMed] [Google Scholar]
- 9.Gray RD, MacGregor G, Noble D, Imrie M, Dewar M, Boyd AC, et al. Sputum proteomics in inflammatory and suppurative respiratory diseases. Am J Respir Crit Care Med. 2008;178:444–52. doi: 10.1164/rccm.200703-409OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen J, Ryu S, Gharib SA, Goodlett DR, Schnapp LM. Exploration of the normal human bronchoalveolar lavage fluid proteome. Proteomics Clinical Applications. 2008;2:585–95. doi: 10.1002/prca.200780006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gharib SA, Vaisar T, Aitken ML, Park DR, Heinecke JW, Fu X. Mapping the lung proteome in cystic fibrosis. J Proteome Res. 2009;8:3020–8. doi: 10.1021/pr900093j. [DOI] [PubMed] [Google Scholar]
- 12.Hallstrand TS, Chi EY, Singer AG, Gelb MH, Henderson WR., Jr Secreted phospholipase A2 group X overexpression in asthma and bronchial hyperresponsiveness. Am J Respir Crit Care Med. 2007;176:1072–8. doi: 10.1164/rccm.200707-1088OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen J, Ryu S, Gharib SA, Goodlett DR, Schnapp LM. Exploration of the normal human bronchoalveolar lavage fluid proteome. Proteomics Clin Appl. 2008;2:585–95. doi: 10.1002/prca.200780006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scherl A, Shaffer SA, Taylor GK, Kulasekara HD, Miller SI, Goodlett DR. Genome-specific gas-phase fractionation strategy for improved shotgun proteomic profiling of proteotypic peptides. Anal Chem. 2008;80:1182–91. doi: 10.1021/ac701680f. [DOI] [PubMed] [Google Scholar]
- 15.Pedrioli PG, Eng JK, Hubley R, Vogelzang M, Deutsch EW, Raught B, et al. A common open representation of mass spectrometry data and its application to proteomics research. Nat Biotechnol. 2004;22:1459–66. doi: 10.1038/nbt1031. [DOI] [PubMed] [Google Scholar]
- 16.Kersey PJ, Duarte J, Williams A, Karavidopoulou Y, Birney E, Apweiler R. The International Protein Index: an integrated database for proteomics experiments. Proteomics. 2004;4:1985–8. doi: 10.1002/pmic.200300721. [DOI] [PubMed] [Google Scholar]
- 17.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Analytical Chemistry. 2003;75:4646–58. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 18.Liu H, Sadygov RG, Yates JR., 3rd A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem. 2004;76:4193–201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 19.Wang W, Zhou H, Lin H, Roy S, Shaler TA, Hill LR, et al. Quantification of proteins and metabolites by mass spectrometry without isotopic labeling or spiked standards. Anal Chem. 2003;75:4818–26. doi: 10.1021/ac026468x. [DOI] [PubMed] [Google Scholar]
- 20.Fu X, Gharib SA, Green PS, Aitken ML, Frazer DA, Park DR, et al. Spectral index for assessment of differential protein expression in shotgun proteomics. J Proteome Res. 2008;7:845–54. doi: 10.1021/pr070271+. [DOI] [PubMed] [Google Scholar]
- 21.Gene Ontology Consortium. Creating the gene ontology resource: design and implementation. Genome Res. 2001;11:1425–33. doi: 10.1101/gr.180801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 23.Zhang B, Kirov S, Snoddy J. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005;33:W741–8. doi: 10.1093/nar/gki475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–8. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 25.Calvano SE, Xiao W, Richards DR, Felciano RM, Baker HV, Cho RJ, et al. A network-based analysis of systemic inflammation in humans. Nature. 2005;437:1032–7. doi: 10.1038/nature03985. [DOI] [PubMed] [Google Scholar]
- 26.Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, et al. STRING 8--a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37:D412–6. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Becker KG, Hosack DA, Dennis G, Jr, Lempicki RA, Bright TJ, Cheadle C, et al. PubMatrix: a tool for multiplex literature mining. BMC Bioinformatics. 2003;4:61. doi: 10.1186/1471-2105-4-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brightling CE. Clinical applications of induced sputum. Chest. 2006;129:1344–8. doi: 10.1378/chest.129.5.1344. [DOI] [PubMed] [Google Scholar]
- 29.Zhang B, VerBerkmoes NC, Langston MA, Uberbacher E, Hettich RL, Samatova NF. Detecting differential and correlated protein expression in label-free shotgun proteomics. J Proteome Res. 2006;5:2909–18. doi: 10.1021/pr0600273. [DOI] [PubMed] [Google Scholar]
- 30.Vignola AM, Bonanno A, Mirabella A, Riccobono L, Mirabella F, Profita M, et al. Increased levels of elastase and alpha1-antitrypsin in sputum of asthmatic patients. Am J Respir Crit Care Med. 1998;157:505–11. doi: 10.1164/ajrccm.157.2.9703070. [DOI] [PubMed] [Google Scholar]
- 31.Carpe N, Mandeville I, Ribeiro L, Ponton A, Martin JG, Kho AT, et al. Genetic influences on asthma susceptibility in the developing lung. Am J Respir Cell Mol Biol. 2010;43:720–30. doi: 10.1165/rcmb.2009-0412OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen LC, Zhang Z, Myers AC, Huang SK. Cutting edge: altered pulmonary eosinophilic inflammation in mice deficient for Clara cell secretory 10-kDa protein. J Immunol. 2001;167:3025–8. doi: 10.4049/jimmunol.167.6.3025. [DOI] [PubMed] [Google Scholar]
- 33.Hung CH, Chen LC, Zhang Z, Chowdhury B, Lee WL, Plunkett B, et al. Regulation of TH2 responses by the pulmonary Clara cell secretory 10-kd protein. J Allergy Clin Immunol. 2004;114:664–70. doi: 10.1016/j.jaci.2004.05.042. [DOI] [PubMed] [Google Scholar]
- 34.Wang H, Long XB, Cao PP, Wang N, Liu Y, Cui YH, et al. Clara cell 10-kD protein suppresses chitinase 3-like 1 expression associated with eosinophilic chronic rhinosinusitis. Am J Respir Crit Care Med. 2010;181:908–16. doi: 10.1164/rccm.200904-0597OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shijubo N, Itoh Y, Yamaguchi T, Imada A, Hirasawa M, Yamada T, et al. Clara cell protein-positive epithelial cells are reduced in small airways of asthmatics. Am J Respir Crit Care Med. 1999;160:930–3. doi: 10.1164/ajrccm.160.3.9803113. [DOI] [PubMed] [Google Scholar]
- 36.Shijubo N, Itoh Y, Yamaguchi T, Sugaya F, Hirasawa M, Yamada T, et al. Serum levels of Clara cell 10-kDa protein are decreased in patients with asthma. Lung. 1999;177:45–52. doi: 10.1007/pl00007626. [DOI] [PubMed] [Google Scholar]
- 37.Levin SW, Butler JD, Schumacher UK, Wightman PD, Mukherjee AB. Uteroglobin inhibits phospholipase A2 activity. Life Sci. 1986;38:1813–9. doi: 10.1016/0024-3205(86)90135-9. [DOI] [PubMed] [Google Scholar]
- 38.Hallstrand TS, Lai Y, Ni Z, Oslund RC, Henderson WR, Jr, Gelb MH, et al. Relationship between levels of secreted phospholipase A(2) groups IIA and X in the airways and asthma severity. Clin Exp Allergy. doi: 10.1111/j.1365-2222.2010.03676.x. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang X, Kohl J. A complex role for complement in allergic asthma. Expert Rev Clin Immunol. 2010;6:269–77. doi: 10.1586/eci.09.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wills-Karp M. Complement activation pathways: a bridge between innate and adaptive immune responses in asthma. Proc Am Thorac Soc. 2007;4:247–51. doi: 10.1513/pats.200704-046AW. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Humbles AA, Lu B, Nilsson CA, Lilly C, Israel E, Fujiwara Y, et al. A role for the C3a anaphylatoxin receptor in the effector phase of asthma. Nature. 2000;406:998–1001. doi: 10.1038/35023175. [DOI] [PubMed] [Google Scholar]
- 42.Krug N, Tschernig T, Erpenbeck VJ, Hohlfeld JM, Kohl J. Complement factors C3a and C5a are increased in bronchoalveolar lavage fluid after segmental allergen provocation in subjects with asthma. Am J Respir Crit Care Med. 2001;164:1841–3. doi: 10.1164/ajrccm.164.10.2010096. [DOI] [PubMed] [Google Scholar]
- 43.Kanemitsu H, Nagasawa S, Sagai M, Mori Y. Complement activation by diesel exhaust particles (DEP) Biol Pharm Bull. 1998;21:129–32. doi: 10.1248/bpb.21.129. [DOI] [PubMed] [Google Scholar]
- 44.Walters DM, Breysse PN, Schofield B, Wills-Karp M. Complement factor 3 mediates particulate matter-induced airway hyperresponsiveness. Am J Respir Cell Mol Biol. 2002;27:413–8. doi: 10.1165/rcmb.4844. [DOI] [PubMed] [Google Scholar]
- 45.Park JW, Taube C, Joetham A, Takeda K, Kodama T, Dakhama A, et al. Complement activation is critical to airway hyperresponsiveness after acute ozone exposure. Am J Respir Crit Care Med. 2004;169:726–32. doi: 10.1164/rccm.200307-1042OC. [DOI] [PubMed] [Google Scholar]
- 46.Shima M, Adachi M. Effects of environmental tobacco smoke on serum levels of acute phase proteins in schoolchildren. Prev Med. 1996;25:617–24. doi: 10.1006/pmed.1996.0097. [DOI] [PubMed] [Google Scholar]
- 47.Robbins RA, Nelson KJ, Gossman GL, Koyama S, Rennard SI. Complement activation by cigarette smoke. Am J Physiol. 1991;260:L254–9. doi: 10.1152/ajplung.1991.260.4.L254. [DOI] [PubMed] [Google Scholar]
- 48.Hallstrand TS, Moody MW, Wurfel MM, Schwartz LB, Henderson WR, Jr, Aitken ML. Inflammatory basis of exercise-induced bronchoconstriction. Am J Respir Crit Care Med. 2005;172:679–86. doi: 10.1164/rccm.200412-1667OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Luscombe NM, Babu MM, Yu H, Snyder M, Teichmann SA, Gerstein M. Genomic analysis of regulatory network dynamics reveals large topological changes. Nature. 2004;431:308–12. doi: 10.1038/nature02782. [DOI] [PubMed] [Google Scholar]
- 50.Gharib SA, Liles WC, Klaff LS, Altemeier WA. Noninjurious mechanical ventilation activates a proinflammatory transcriptional program in the lung. Physiol Genomics. 2009;37:239–48. doi: 10.1152/physiolgenomics.00027.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lovegrove FE, Gharib SA, Patel SN, Hawkes CA, Kain KC, Liles WC. Expression microarray analysis implicates apoptosis and interferon-responsive mechanisms in susceptibility to experimental cerebral malaria. Am J Pathol. 2007;171:1894–903. doi: 10.2353/ajpath.2007.070630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Becker L, Gharib SA, Irwin AD, Wijsman E, Vaisar T, Oram JF, et al. A macrophage sterol-responsive network linked to atherogenesis. Cell Metab. 2010;11:125–35. doi: 10.1016/j.cmet.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ames RS, Lee D, Foley JJ, Jurewicz AJ, Tornetta MA, Bautsch W, et al. Identification of a selective nonpeptide antagonist of the anaphylatoxin C3a receptor that demonstrates antiinflammatory activity in animal models. J Immunol. 2001;166:6341–8. doi: 10.4049/jimmunol.166.10.6341. [DOI] [PubMed] [Google Scholar]
- 54.Mizutani N, Nabe T, Yoshino S. Complement C3a regulates late asthmatic response and airway hyperresponsiveness in mice. J Immunol. 2009;183:4039–46. doi: 10.4049/jimmunol.0901468. [DOI] [PubMed] [Google Scholar]
- 55.Lee SH, Kim KH, Kim JM, Yoon SH, Kim TH, Park SW, et al. Relationship between Group-specific Component Protein and the Development of Asthma. Am J Respir Crit Care Med. doi: 10.1164/rccm.201006-0951OC. in press. [DOI] [PubMed] [Google Scholar]
- 56.Haenen S, Vanoirbeek JA, De Vooght V, Maes E, Schoofs L, Nemery B, et al. Proteome analysis of multiple compartments in a mouse model of chemical-induced asthma. J Proteome Res. 2010;9:5868–76. doi: 10.1021/pr100638m. [DOI] [PubMed] [Google Scholar]
- 57.Zhao J, Endoh I, Hsu K, Tedla N, Endoh Y, Geczy CL. S100A8 Modulates Mast Cell Function and Suppresses Eosinophil Migration in Acute Asthma. Antioxid Redox Signal. doi: 10.1089/ars.2010.3583. in press. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


