Abstract
In Caenorhabditis elegans, the dopamine transporter DAT-1 regulates synaptic dopamine (DA) signaling by controlling extracellular DA levels. In dat-1(ok157) animals, DA is not taken back up presynaptically but instead reaches extrasynpatic sites, where it activates the dopamine receptor DOP-3 on choligeneric motor neurons and causes animals to become paralyzed in water. This phenotype is called swimming-induced paralysis (SWIP) and is dependent on dat-1 and dop-3. Upstream regulators of dat-1 and dop-3 have yet to be described in C. elegans. In our previous studies, we defined a role for HLH-17 during dopamine response through its regulation of the dopamine receptors. Here we continue our characterization of the effects of HLH-17 on dopamine signaling. Our results suggest that HLH-17 acts downstream of dopamine synthesis to regulate the expression of dop-3 and dat-1. First, we show that hlh-17 animals display a SWIP phenotype that is consistent with its regulation of dop-3 and dat-1. Second, we show that this behavior is enhanced by treatment with the dopamine reuptake inhibitor, bupropion, in both hlh-17 and dat-1 animals, a result suggesting that SWIP behavior is regulated via a mechanism that is both dependent on and independent of DAT-1. Third, and finally, we show that although the SWIP phenotype of hlh-17 animals is unresponsive to the dopamine agonist, reserpine, and to the antidepressant, fluoxetine, hlh-17 animals are not defective in acetylcholine signaling. Taken together, our work suggests that HLH-17 is required to maintain normal levels of dopamine in the synaptic cleft through its regulation of dop-3 and dat-1.
Keywords: reserpine, bupropion, fluoxetine, dopamine receptor, acetylcholine signaling
In Caenorhabditis elegans and other multicellular organisms, basic helix-loop-helix (bHLH) proteins coordinate a number of developmental events, including myogenesis (Chen et al. 1994), organ morphogenesis (Tamai and Nishiwaki 2007), and mesodermal development (Harfe et al. 1998). These proteins also have vital functions during neurogenesis (Hallam et al. 2000; Krause et al. 1997). For example, the proneural protein HLH-14 is required to generate multiple neurons stemming from a variety cell lineage types, while HLH-3 is needed for the differentiation of hermaphrodite-specific motor neurons (Doonan et al. 2008; Frank et al. 2003; Poole et al. 2011). HLH-17 is the C. elegans homolog of the mammalian proneural family Olig (Ligon et al. 2006; Zhou and Anderson 2002) but does not appear to play a role in neuronal specification during embryogenesis (Yoshimura et al. 2008). Our previous studies instead demonstrated that HLH-17 is required for normal behavioral responses to dopamine signaling (Felton and Johnson, 2011).
In vertebrates and invertebrates, dopamine signaling is associated with motivation, recognition and reward, memory and adaptation, hormonal regulation, and motor control. In humans, imbalances in dopamine signaling are associated with many neurological diseases, including Parkinson disease, Alzheimer disease, ADHD, and substance abuse (Choi and Tarazi 2010; Middleton et al. 2007; Xie et al. 2010). Dopamine signaling in C. elegans involves many of the same molecules as in mammals (Chase and Koelle 2007). For example, dopamine is synthesized by the tyrosine hydroxylase enzyme CAT-2. On synthesis, dopamine is sequestered in presynaptic storage vesicles by the vesicular monoamine transporter CAT-1, where it remains until being released into the presynaptic cleft in response to a stimulus. Once in the synapse, dopamine binds to and activates D1-like (DOP-1) and D-2 like receptors (DOP-2 and DOP-3) that are positioned either pre-, post-, or extra-synaptically. Unbound dopamine is taken back up into the presynaptic cell via reuptake by the dopamine transporter DAT-1.
HLH-17 is expressed in the glia-like cells surrounding the CEP dopaminergic neurons (McMiller and Johnson 2005) and in the sheath or socket cells of the inner labia and outer labia (Yoshimura et al. 2008). Our previous data revealed that HLH-17 affects dopamine signaling through the DOP-1, DOP-2, and DOP-3 receptors as shown by the impaired response of hlh-17(ns204) animals to endogenous and exogenous dopamine. The hlh-17(ns204) animals also have reduced levels of the dop-3 and dop-1 mRNAs and phenocopy dop-3 hypomorhs (Chase et al. 2004; Felton and Johnson 2011). Together, these data suggest that HLH-17 functions upstream of the dopamine receptor genes and that the loss of hlh-17 causes a reduction in dopamine receptor activity.
Here we continue our characterization of the role of HLH-17 in dopamine signaling. Our data suggest that HLH-17 influences dopamine-dependent behaviors by regulating genes that mediate levels of extracellular dopamine. The dat-1 mRNA levels are reduced, but not eliminated, in hlh-17(ns204) animals. Furthermore, hlh-17(ns204) animals display swimming-induced paralysis (SWIP) behavior in water that is an intermediate between the behavior in dat-1 animals and in wild-type animals and that is enhanced by treatment with the dopamine reuptake inhibitor, bupropion. We show that a null allele of dop-3 completely suppresses the SWIP phenotype of hlh-17 animals, supporting previous data that HLH-17 acts upstream of DOP-3. Surprisingly, the SWIP phenotype of hlh-17 animals is unaffected by treatment with the VMAT inhibitor reserpine or with the serotonin reuptake inhibitor, fluoxetine; however, this unresponsiveness is not due to reduced acetylcholine signaling. Taken together, our results suggest that HLH-17 influences extracellular dopamine levels in C. elegans, in part by its regulation of the dopamine receptors and the dopamine transporter.
Materials and Methods
Nematode strains and maintenance
The following strains were used in this study: wild-type: Bristol strain (N2); RM2702 [dat-1(ok157)]; OS2649: [hlh-17(ns204)]; and LX705 [dop-1 (vs100) dop-3 (vs106)]. OS2649 was a gift from Dr. S. Shaham. The strains CMJ2003 [hlh-17(ns204); dat-1(ok157)] and CMJ2004 [hlh-17(ns204); dop-1(vs100) dop-3(vs106)] were generated using traditional crossing techniques and the genotypes were confirmed by PCR. To generate CMJ2004, hlh-17(ns204) males were crossed with dop-1(vs100) dop-3(vs106) hermaphrodites, and the F1 males were backcrossed to dop-1(vs100) dop-3(vs106). F2 hermaphrodites were separately cloned, and their progeny were genotyped by PCR. The strain CMJ2005 [hlh-17(ns204); dat-1(ok157); dop-1(vs100) dop-3 (vs106)] was generated by crossing CMJ2003 males with CMJ2004. F1 hermaphrodites were separately cloned, and their progeny were genotyped by PCR for hlh-17 and dat-1. The progeny of hermaphrodites that were confirmed to be homozygous for both hlh-17 and dat-1 were then subcloned and their progeny were screened for homozygosity for dop-3 by PCR and for rescue of SWIP behavior.
The transgene, cmjEx22, is a 6.2-kb genomic fragment consisting of 2 kbp upstream of the hlh-17 translational start site, the entire hlh-17 coding region, the SV40 nuclear localization signal (NLS), and 850 bp of the sequences coding for green fluorescent protein (GFP). The GFP sequences were amplified from pPD95.67 (a gift from A. Fire) using serial overlap PCR. Transgenic lines to rescue loss of hlh-17 were produced by microinjecting the final PCR product (cmjEx22) into hlh-17(ns204) animals, along with the pCFJ90 [Pmyo-2::mCherry::unc-54utr] co-injection marker, using standard microinjection techniques(Rieckher et al. 2009), and is designated as CMJ2002 [hlh-17(ns204); cmjEX22, pCFJ90(Pmyo-2::mCherry::unc54utr)].
Three separate lines (15.1, 15.3, and 3.1) were tested for rescue of SWIP, basal slowing response, and dopamine paralysis. All three lines were able to at least partially rescue each of the phenotypes tested; however, the degree of rescue for each line was specific to the phenotype tested.
Unless otherwise noted, strains were cultured on solid nematode growth media (NGM) containing OP50 at 20° using standard methods and synchronized cultures were prepared by hypochlorite treatment of gravid adults, as previously described (Brenner 1974). The following primers were used for genotyping: HLH17F: 5′-TCTGGGGACCCTCTCCTCG-3′; HLH17R: 5′-CGATTTTTGCTGCTAATGGGCAACAC-3′; DAT1F: 5′-CTATTCGGATATCTTGCCAATGCTATACC-3′; DAT1R: 5′-CTATTCGGATATCTTGCCAATGCTATACC-3′; DOP3F: 5′-CTATTCGGATATCTTGCCAATGCTATACC-3′; and DOP3R: 5′-CTAACTCACCAGAAAATCAGAAACTGC-3′.
Gene expression analysis
Synchronized populations were collected at the L4 stage, pelleted, and frozen at −80°. Total RNA, cDNA synthesis, and real-time PCR were performed as previously described (Felton and Johnson 2011), except the cDNA was amplified from 1 µg of total RNA in 20 µL reactions. Real-time PCR was performed with Taqman Gene Expression Assays (Applied Biosystems/Invitrogen) using relative quantitation against glyceraldehyde 3-phosphate dehydrogenase (gpd-3) (Ce02616909_gH) as the endogenous control. The probe sets used were: hlh-17(Ce02616669_m1); dat-1(Ce02450896_g1); cat-1(Ce02495610_m1); mod-5 (Ce02415245_m1); dop-1 (Ce02494345_m1); dop-2 (Ce02479824_m1) dop-3(Ce02496462_m1); lev-8 (Ce02501240_g1); and unc-43 (Ce02458977_m1). Gene expression assays were performed in triplicate for at least three biological replicates.
Behavioral assays
Assays for dopamine paralysis and basal slowing response were as previously described (Felton and Johnson 2011), except animals were assayed at the late L4 stage. For SWIP, approximately 10 L4-stage animals were placed in 150 µL of water in a single well of 48-well tissue culture plate (Cat #677180; CELLSTAR). After 20 min, animals were categorized as paralyzed if they failed to exert the normal thrashing behavior within a 20-sec time frame (McDonald et al. 2007). For SWIP assay conducted with inhibitors, animals were grown on NGM plates containing the appropriate drug [reserpine (0.6 mM; Cat #S1601), fluoxetine (145 µM; Cat #S1333), and bupropion (10 μM; Cat #S2452)] and then analyzed in water. All inhibitors were obtained from Selleckchem. Aldicarb-induced paralysis and levamisole-induced paralysis assays were conducted using standard protocols (Lewis et al. 1980; Nguyen et al. 1995; Mahoney et al. 2006) with some modifications. Plates containing aldicarb (1.0 mM; FisherSci #US-PST-940) or levamisole (0.2 mM; FisherSci #ICN15522805) were prepared 1 hr before use. Drugs were prepared as 100-mM stocks in 70% ethanol, diluted in sterile M9 buffer, added to NGM plates already seeded with OP50, to the appropriate concentration, and allowed to diffuse into the media for 1 hr. L4-stage animals were manually selected to confirm their age and moved to plates using a platinum wire and were examined every hour for a 5-hr to 6-hr period. Animals were categorized as paralyzed if they failed to move after prodding with a platinum wire.
Results and Discussion
HLH-17 functions upstream of the D2-like dopamine receptor DOP-3 to regulate behavioral responses to dopamine
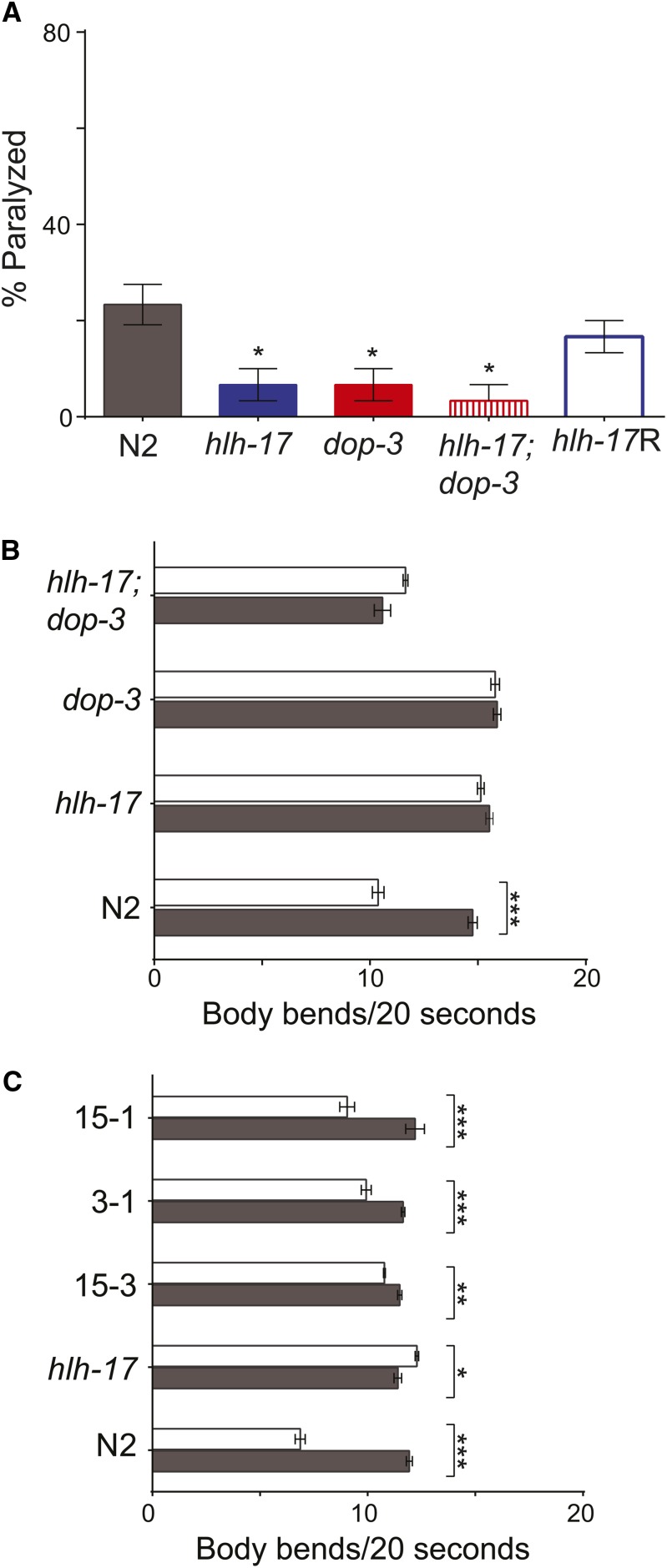
The effects of dopamine signaling in C. elegans are mediated by the three heterotrimeric G-protein receptors, DOP-1, DOP-2, and DOP-3 (Missale et al. 1998). Our previous studies demonstrated that mRNA levels of these three receptors are reduced in hlh-17(ns204) animals and that hlh-17(ns204) animals phenocopy those carrying loss-of-function alleles of dop-3 (Felton and Johnson 2011). As shown in Figure 1, and in our previous studies, fewer hlh-17(ns204) and dop-3(vs106) animals than wild-type animals were paralyzed after 40 min of exposure to 10 mM of exogenous DA, and both well-fed hlh-17(ns204) animals and well-fed dop-3(vs106) animals failed to exhibit the basal slowing response (BSR) when encountering a bacterial lawn. In this study, we used an extragenic, translational reporter for hlh-17 to rescue the dopamine paralysis and basal slowing phenotypes of transgenic hlh-17(ns204) animals. This reporter was able to restore dopamine sensitivity and to enhance BSR, showing that the previously reported phenotypes are indeed a result of loss of HLH-17 (Figure 1). We previously reported that a transcriptional reporter for hlh-17 drives expression in the glial-like, cephalic sheath cells of the dopaminergic neurons (McMiller and Johnson 2005), and others have detected weak hlh-17 expression in the sheath or socket cells of the inner labia and outer labia (Yoshimura et al. 2008). The translational reporter used in this study was driven by the same promoter sequences and was similarly expressed (data not shown). This expression pattern weakly correlates with expression of the dopamine receptors in neuronal support cells of the head (Chase et al. 2004); therefore, we looked for genetic interactions between hlh-17 and dop-3. As shown in Figure 1, the resistance to dopamine-induced paralysis and the BSR of hlh-17(ns204); dop-3(vs106) are not significantly different from the resistance and slowing response phenotypes of dop-3(vs106) and hlh-17(ns204) animals (Figure 1) and are consistent with a model in which HLH-17 is functioning in the same genetic pathway as DOP-3 to modulate these behaviors. Taken together, we conclude that the influence of HLH-17 on behaviors that are mediated by dopamine occurs through the transcriptional regulation of dop-3. Our existing data suggest that this regulation is indirect; however, it is possible that the transcriptional and translational constructs used in our studies do not fully report the wild-type expression pattern for hlh-17. In fact, recent gene expression profiles from FACS sorted cells point to overlapping expression of hlh-17 and dop-3 in the dopamine neurons of late embryos, in panneuronal cells and the glutamate receptor neurons of L2-stage animals, and in the cephalic sheath of young adult animals (Spencer et al. 2011; see WormViz at http://www.vanderbilt.edu/wormdoc/wormmap/WormViz.html). Additionally, dopamine receptor genes are expressed in mammalian glial cells (Biedermann et al. 1995; Kuric et al. 2013) and further support the possibility that HLH-17 directly regulates as dop-3 expression in the cephalic sheath and in selected neurons during C. elegans development.
Figure 1.
HLH-17 functions upstream of dop-3 to regulate dopamine signaling. (A) DA-induced paralysis: hlh-17(ns204), dop-3 (vs106), and hlh-17(ns204); dop-3 (vs106) animals are less sensitive than wild-type animals to 10 mM DA. Transgenic expression of HLH-17::GFP in hlh-17(ns204) animals rescues the DA-induced paralysis phenotype. The bar for hlh-17R represents the average measurements from three biological replicas of three independent lines. *Statistical significance when compared to wild-type, n = 10 animals/strain/rep for three biological replicas. (B) Basal slowing response: Well-fed wild-type animals, but not hlh-17(ns204), dop-3 (vs106), or hlh-17(ns204); dop-3 (vs106) animals, move significantly slower in the presence of food (white bars) than in the absence of food (gray bars). (C) Transgenic expression of HLH-17::GFP rescues the basal slowing response of hlh-17(ns204) animals. Three independent lines, 15.3, 15-1, and 3.1, were assayed. In (B) and (C), five animals/rep/strain for a total of three biological replicas were assayed. Each animal was analyzed for three separate 20-sec intervals, so that the total number of observations was 15 observations/rep/strain. *P < 0.05; **P < 0.005; ***P < 0.0005.
The hlh-17 mutants are defective in clearing dopamine from the synaptic cleft
In our previous studies, the mRNA levels of genes required for dopamine synthesis, those encoding tyrosine hydroxylase gene (cat-2) and the aromatic amino acid decarboxylase (bas-1), were not affected by loss of hlh-17. This suggested that the presynaptic synthesis of dopamine is not compromised in hlh-17(ns204) animals. Additionally, exogenous dopamine failed to repress egg-laying in naive hlh-17(ns204) animals; however, exogenous dopamine was able to repress the stimulation of egg-laying by the neurotransmitter, serotonin (Felton and Johnson 2011). Although we did not further address the serotonin responsiveness of hlh-17(ns204) animals, this result suggested that some ability of hlh-17(ns204) animals to respond to exogenous dopamine may be mediated by the binding of the neurotransmitter to other non-dopaminergic receptors. For example, dopamine can bind with low affinity to a number of the neurotransmitter receptors involved in serotonin-stimulated egg-laying, including MOD-1, SER-1, SER-2, and SER-7 (Chase and Koelle 2007; Dempsey et al. 2005).
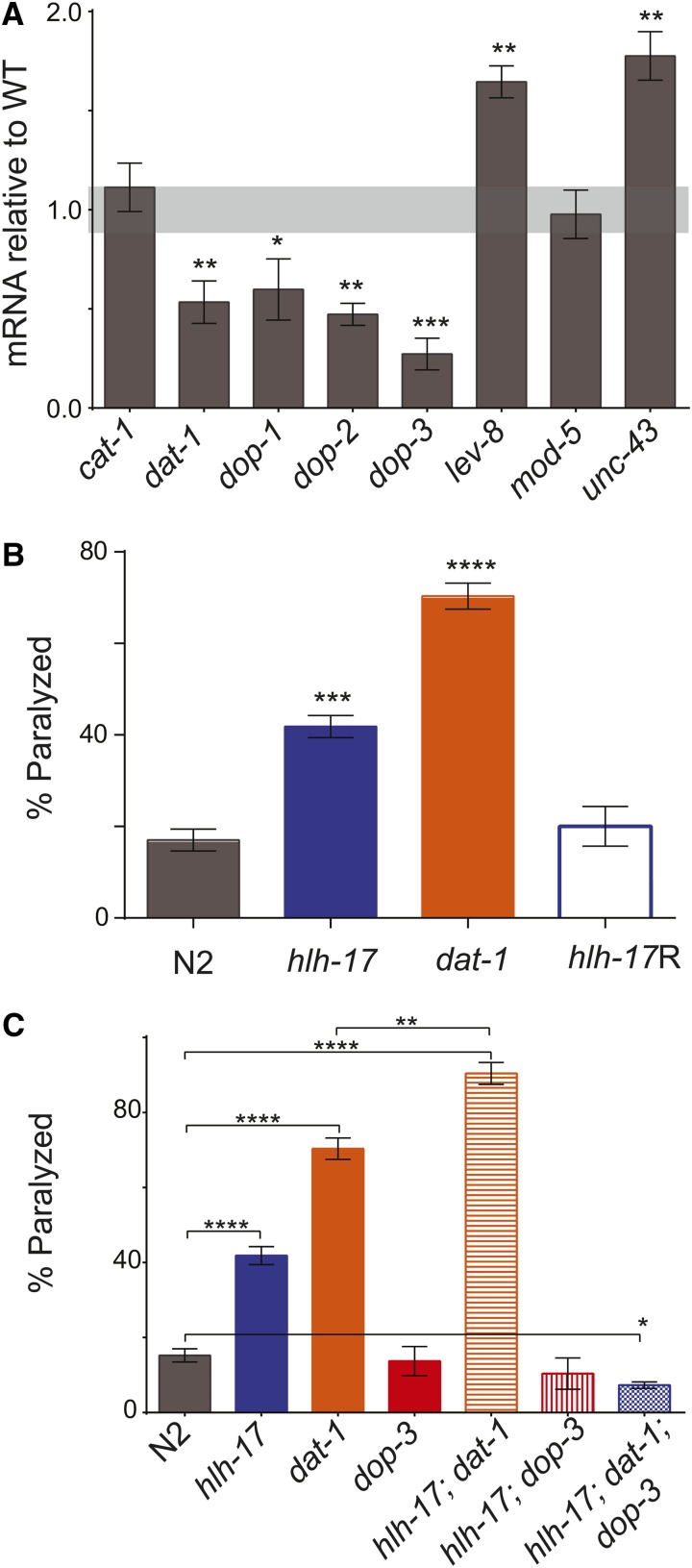
To further define the role of HLH-17 during dopamine signaling, we measured the mRNA levels of the genes encoding the vesicular monoamine transporter (VMAT), cat-1, and the dopamine reuptake transporter, dat-1, in hlh-17(ns204) animals. As shown in Figure 2, dat-1, but not cat-1, mRNA levels, are decreased in hlh-17(ns204) animals. We also found that the mRNA levels for the dopamine receptor genes, dop-1, dop-2, and dop-3, are downregulated in L4-stage animals, confirming that the decreased levels previously reported in L1-stage animals (Felton and Johnson 2011) remain low in animals at the stage used for our behavior assays.
Figure 2.
Loss-of hlh-17 affects extracellular DA levels. (A) mRNA levels in L4-stage hlh-17(ns204) animals when normalized against mRNA levels in age-matched wild-type animals. Light gray shading represents wild-type range of expression (1.0 ± 0.115). The levels of cat-1 and mod-5 mRNA are not significantly affected in hlh-17(ns204) animals. (B) hlh-17(ns204) animals demonstrate SWIP behavior that is an intermediate of the behavior in N2 and dat-1(ok157) animals, and that is rescued by transgenic expression of HLH-17::GFP. The bar for hlh-17R represents the average measurements from three biological replicas of three independent lines. For all strains except hlh-17R, n = 30 animals/rep/strain. For hlh-17R, n was equal to an average of at least 15 animals/line/biological rep (range, 12–26) because of differences in transmission frequency of the transgene. (C) SWIP phenotype in double mutant hlh-17(ns204); dat-1(ok157) and hlh-17(ns204); dop-3(vs106) animals is more similar to the phenotype in dat-1(ok157) and dop-3(vs106) animals, respectively, than in wild-type animals. The SWIP phenotype of hlh-17(ns204); dat-1(ok157); dop-3(vs106) animals is not significantly different from the SWIP phenotypes of dop-3 or hlh-17(ns204); dop-3(vs106) animals. n = 30 animals/rep/strain for three biological replicas. *P < 0.05; **P < 0.005; ***P < 0.0005; ****P < 0.0001.
Like the mammalian VMATs, CAT-1 mediates the packaging and transport of the biogenic amines into synaptic vesicles and is required for proper release of dopamine from presynaptic neurons in C. elegans (Duerr et al. 1999). The dopamine transporter, DAT-1, is localized to the synapses of all dopaminergic neurons of C. elegans males and hermaphrodites (McDonald et al. 2007) and is responsible for neurotransmitter clearance from the synaptic cleft (Carvelli et al. 2004; Torres et al. 2003). In otherwise wild-type animals, loss of dat-1 leads to increased activation of the DOP-3 receptors located on cholinergic motor neurons (Chase et al. 2004). Consequently, dat-1 animals are paralyzed in water as a result of DOP-3 hyperactivation; this behavior can be measured using a SWIP assay (Chase and Koelle 2007; McDonald et al. 2007). SWIP does not occur in cat-1 animals because dopamine is not efficiently packaged or subsequently released into the synaptic cleft. We reasoned that if hlh-17(ns204) animals synthesize and release normal levels of dopamine, but produce less DAT-1, then they would be less efficient than wild-type animals at clearing extrasynpatic dopamine from the synaptic cleft. To test this hypothesis, we conducted SWIP assays with wild-type, hlh-17(ns204), and dat-1(ok157) animals. As reported previously (McDonald et al. 2007), and as shown in Figure 2, dat-1(ok157) animals, but not wild-type animals, have a strong SWIP response after 20 min in water. The SWIP response of hlh-17(ns204) animals was an intermediate response, with approximately 40% of the animals becoming paralyzed after 20 min in water. This phenotype was rescued by transgenic expression of HLH-17. The result suggests that loss of HLH-17 compromises the ability of mutant animals to clear dopamine from the synaptic cleft and could be interpreted as representing a slight, rather than complete, loss of dat-1 activity.
The SWIP phenotype seen in dat-1 animals is completely rescued by loss of DOP-3 (Sugiura et al. 2005); hence, we reasoned that the reduced SWIP response of hlh-17(ns204) animals, which is an intermediate of the responses of wild-type and dat-1 animals, may be the result of having decreased levels of both dop-3 and dat-1. To test this hypothesis, we compared the SWIP phenotypes of hlh-17(ns204); dat-1(ok157) animals and of hlh-17(ns204); dop-3(vs106) animals with those of dat-1(ok157) and dop-3(vs106) animals, respectively. As shown in Figure 2, complete loss of dat-1 activity enhanced the SWIP response of hlh-17(ns204) animals, whereas complete loss of dop-3 activity significantly decreased the SWIP response. Furthermore, the SWIP phenotypes of hlh-17(ns204); dop-3(vs106) animals and hlh-17(ns204); dop-3(vs106); dat-1(ok157) animals were not significantly different from that of dop-3(vs106) animals (P = 0.574 and 0.265, respectively). Interestingly, loss of hlh-17 and dat-1 appears to be an additive effect: a comparison of the differences of the means shows that the difference for wild-type vs. hlh-17; dat-1 is equal to the sum of the differences for wild-type vs. hlh-17 and wild-type vs. dat-1. These results underscore the dependence of the SWIP phenotype on DOP-3. Furthermore, the results suggest that the SWIP response is not mediated solely through dat-1, and that hlh-17 may affect the SWIP phenotype through both dat-1-dependent and dat-1-independent mechanisms. A dat-1-independent, dop-3-dependent mechanism for the SWIP phenotype is consistent with the results of a previously reported forward genetics screen (Hardaway et al. 2012) and suggests that the HLH-17 transcriptional network may include genes that act in parallel to dat-1.
Our results from the dopamine paralysis assays and the egg-laying assays suggest that hlh-17(ns204) animals are less sensitive to exogenous dopamine, a result that is consistent with reduced dop-3 activity. The results from assays for BSR and SWIP, both of which rely on normal synthesis and release of endogenous dopamine from presynaptic neurons, suggest that hlh-17(ns204) animals produce normal amounts of dopamine but are deficient in the ability to transport the dopamine. This result is also consistent with reduced dop-3 activity. Likewise, a failure in the ability to transport dopamine from the synaptic cleft is consistent with reduced dat-1 activity, as is the SWIP phenotype of hlh-17(ns204) animals. From these results, we conclude that HLH-17 functions to control extrasynpatic dopamine levels, in part by its regulation of dop-3 and dat-1.
The hlh-17 mutants are responsive to reuptake inhibitors that are selective for dopamine, but not for serotonin
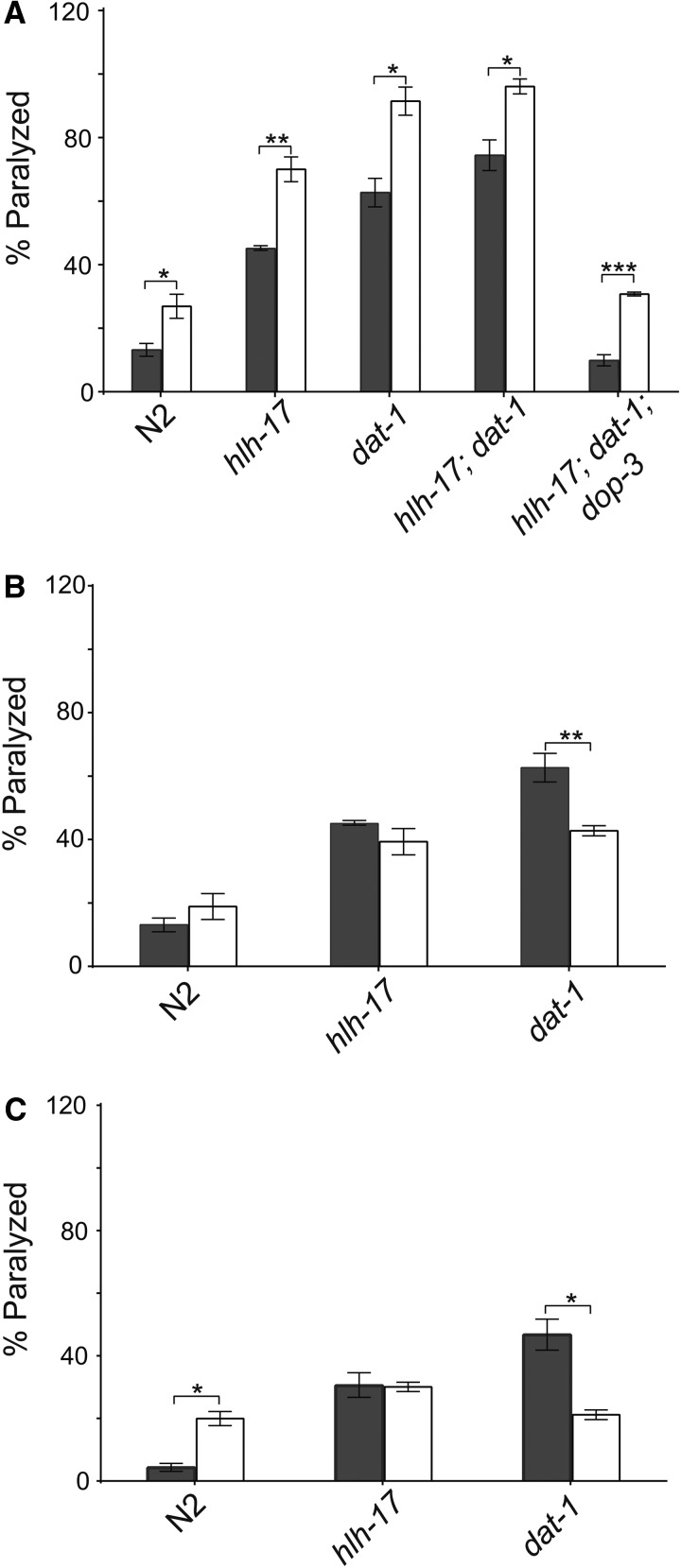
Bupropion is a selective norepinephrine and dopamine reuptake inhibitor commonly used in mice and human studies (Dellagioia et al. 2012; Roelands et al. 2012; Rosenberg et al. 2013) and in the treatment of ADHD (Cantwell 1998; Reimherr et al. 2005) and depression (Carlat 2012; Stahl et al. 2013). Reuptake inhibitors block the ability of a transporter to move a neurotransmitter from the synapse to the presynaptic neuron or the surrounding glial cells, thereby increasing extracellular concentrations that ultimately increase neurotransmission. We reasoned that the intermediate SWIP behavior of hlh-17(ns204) animals occurs because these animals still produce a small amount of functional DAT-1, and that treatment with bupropion would increase SWIP in hlh-17(ns204) animals. As expected, pretreatment of hlh-17(ns204) animals with bupropion increased their SWIP response to that of untreated dat-1(ok157) animals (Figure 3), supporting our mRNA studies showing that dat-1 expression is reduced but not completely eliminated in hlh-17(ns204) animals. It has been shown previously that SWIP can be rescued in dat-1(ok157) animals by pretreatment with the dopamine antagonist reserpine (McDonald et al. 2007), an antipsychotic drug that depletes vesicular dopamine stores by blocking the vesicular monoamine transporter (VMAT). As shown in Figure 3, pretreatment with reserpine reduced the SWIP responses of dat-1(ok157) animals but did not affect SWIP in wild-type animals or in hlh-17(ns204) animals. This result was unexpected because cat-1 mRNA levels are not affected in hlh-17(ns204) animals; however, others have reported reserpine insensitive mutants that show SWIP behavior in a dat-1-dependent manner (Hardaway et al. 2012). Bupropion pretreatment also increased SWIP in dat-1(ok157) animals, hlh-17(ns204); dat-1(ok157) animals, and hlh-17(ns204); dat-1(ok157); dop-3(vs106) animals (Figure 3). Together, these results further emphasize that SWIP behavior may not be mediated solely through dopamine reuptake by DAT-1. The ability to induce SWIP behavior in dop-3 animals suggests that the mechanism may occur through a dopamine-independent mechanism.
Figure 3.
The hlh-17 animals respond selectively to reuptake inhibitors. (A) Pretreatment with the DAT reuptake inhibitor, bupropion, increases the SWIP phenotype of N2, hlh-17(ns204), dat-1(ok157), and hlh-17(ns204); dop-3(vs106) animals. The ability of bupropion to enhance SWIP behavior is not dependent on DOP-3. The SWIP phenotype in hlh-17(ns204) animals is unaffected by pretreatment with reserpine (B) or fluoxetine (C). In all panels, n = 30 animals/rep/strain; dark bars = minus inhibitor; and light bars = plus inhibitor. *P < 0.05; **P < 0.005; ***P < 0.0005.
To test the possibility that the SWIP phenotype is also modulated through serotonin, although a role for 5HT during SWIP has not been reported to date, we measured the SWIP response of WT, hlh-17(ns204), and dat-1(ok157) animals after exposure to fluoxetine. Fluoxetine blocks the function of SERT/MOD-5, the serotonin (5HT) reuptake transporter (Keowkase et al. 2010; Kullyev et al. 2010). As seen in Figure 3, the SWIP response phenotype increased in wild-type animals that were pretreated with fluoxetine but decreased in similarly treated dat-1(ok157) animals. These results can be explained by the action of fluoxetine, which is known to increase extracellular concentrations of dopamine (Bymaster et al. 2002; Koch et al. 2002). The excess dopamine in treated wild-type animals would phenocopy mutants that have increased extracellular levels of dopamine and have an increased SWIP response. Fluoxetine can also aggressively inhibit any transport of dopamine by the serotonin transporters (Bymaster et al. 2002) so that treated dat-1 animals would show a reduced SWIP response, analogous to the reduced response of dat-1; dop-3 animals (McDonald et al., 2007). Interestingly, hlh-17(ns204) animals were insensitive to fluoxetine, although they have normal levels of mod-5 mRNA (see Figure 2) and respond to exogenous serotonin in egg-laying assays (Felton and Johnson 2011). Fluoxetine has previously been shown to act via both serotonin-dependent and serotonin-independent mechanisms in C. elegans (Kullyev et al. 2010; Ranganathan et al. 2001). In future studies we will further explore the role of HLH-17 in serotonin signaling, which may also address the mechanisms of fluoxetine resistance in hlh-17(ns204) animals.
The hlh-17(ns204) animals are not defective in acetylcholine release
It is possible that the SWIP response of hlh-17(ns204) animals is insensitive to both reserpine and fluoxetine because HLH-17 influences the activity of C. elegans biogenic amines in a manner that, with the exception of dopamine, does not involve the regulation of genes directly involved in neurotransmitter synthesis, packaging, or transport. A more attractive, alternative possibility is that HLH-17 influences acetylcholine release, as the phenotypic effects of both reserpine (Saharia et al. 2012) and fluoxetine (Bolanos et al. 2002; Chau et al. 2011) are dependent on acetylcholine. In support of this possibility, the inhibitory effect of fluoxetine on acetylcholine release in rats is dependent on activity of the dopaminergic D2 receptors (Bolanos et al. 2002). Furthermore, loss of dop-3 activity in C. elegans has recently been shown to increase acetylcholine release, whereas null alleles of genes required for acetylcholine release have been shown to rescue the SWIP phenotype in dat-1(ok157) animals (Allen et al. 2011).
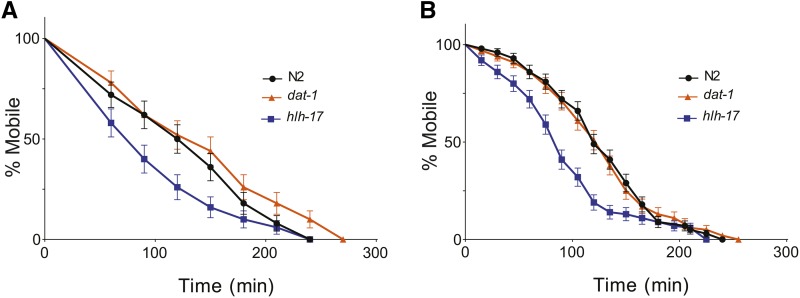
We used aldicarb and levamisole sensitivity assays to examine acetylcholine release and acetylcholine reception, respectively, in hlh-17(ns204) animals. Aldicarb is an acetylcholinesterase inhibitor and thereby increases the concentration of acetylcholine in the neuromuscular junction. Animals with reduced acetylcholine release are resistant to aldicarb-induced paralysis, whereas those with increased acetylcholine release are more sensitive (Allen et al. 2011; Rand 2007). As shown in Figure 4, hlh-17(ns204) animals are more sensitive to aldicarb than wild-type and dat-1(ok157) animals (P = 0.0428 and 0.132, respectively). This result is consistent with the weak effects of the dop-3(v106) mutation on aldicarb sensitivity that was previously reported, and suggests that acetylcholine release is otherwise normal in hlh-17(ns204) animals. We also found that hlh-17(ns204) animals are more sensitive to levamisole (P = 0.0002), a cholinergic agonist that binds selectively to acetylcholine receptors in body-wall muscles (Rand 2007). We are able to tentatively explain this increased sensitivity based on our unpublished microarray analysis that indicates that the activity of the nicotinic acetylcholine receptor gene, lev-8, is upregulated in hlh-17(ns204) animals. Interestingly, our microarray data indicated that the gene encoding the calcium/calmodulin-dependent protein kinase UNC-43 is also upregulated. Mutants carrying gain-of-function alleles of unc-43 have previously been reported to have increased resistance to fluoxetine. As shown in Figure 2, we were able to validate these results using RT-qPCR analysis. mRNA levels of unc-43 and lev-8 are increased in hlh-17 animals, whereas the level of mod-5, a gene that was not differentially affected in our microarray analysis, remained unaffected. To our knowledge, loss of dopamine receptor activity, in particular dop-3, has not been tested; however, animals that are defective in dopamine synthesis display normal sensitivity to levamisole (Suo and Ishiura 2013). Taken together, our results suggest that neither acetylcholine release nor acetylcholine reception is compromised in hlh-17(ns204) animals, and that the resistance to reserpine and fluoxetine may be mediated through other genes in the HLH-17 transcriptional network.
Figure 4.
The hlh-17 animals do not have reduced acetylcholine signaling. (A) hlh-17(ns204) animals are more susceptible to aldicarb-induced paralysis than wild-type (P = 0.0428) and dat-1(ok157) animals (P = 0.1319). (B) The hlh-17(ns204) animals are more susceptible to levamisole-induced paralysis than wild-type (P = 0.0002) and dat-1(ok157) animals (P = 0.0002). In all panels, n = 30 animals/rep/strain.
Conclusion
The Olig sub-family of bHLH transcription factors influences the specification of oligodendrocytes, myelin formation, and axon pathfinding of motor neurons in both invertebrates and vertebrates (Lu et al. 2002; Oyallon et al. 2012; Tiso et al. 2009; Zhou and Anderson 2002). In C. elegans, HLH-17 is an Olig homolog that is expressed in sheath cells of the dopaminergic neurons; however, this protein has no known role in glial cell specification, neurite extension, or axon guidance (Yoshimura et al. 2008). The work presented here and in previous studies points to a role for HLH-17 in controlling dopamine-dependent behaviors. Specifically, our work suggests that HLH-17 is needed to clear extracellular dopamine from the synaptic cleft. First, hlh-17(ns204) animals have reduced mRNA levels for dat-1, dop-3, dop-2, and dop-1 but maintain normal levels of cat-1 and cat-2. Second, the SWIP response of hlh-17(ns204) animals is consistent with reduced levels of dat-1 and dop-3 and is rescued when dop-3 activity is completely eliminated. Third, hlh-17(ns204) animals are not defective in acetylcholine release and, in fact, show an increased sensitivity to aldicarb that is consistent with the increased acetylcholine release that occurs in animals with reduced dop-3 activity.
The bHLH transcription factor family has well-established roles in neurogenesis and the specification and maintenance of neuronal identity. In Drosophila, for example, the bHLH gene, lethal of scute, is required for cell-specific transcription of the dopaminergic H-cell neuron of the ventral nerve cord and for specification of the non-midline dopaminergic neurons (Oyallon et al. 2012; Stagg et al. 2011) In zebrafish, Olig2 regulates expression of the gene encoding Sim1, a bHLH-PAS protein that drives specification of the diencephalic dopaminergic neurons (Borodovsky et al. 2009). Less clear, however, is whether HLH-17 plays a conserved role in the regulation of genes required for neurotransmitter signaling in general and dopamine signaling in particular. The gene encoding the human dopamine reuptake transporter is regulated by the hairy/enhancer of split-like bHLH protein, HesR1. HesR1 represses activity of the human DAT1 gene in cell culture by binding to sequences in the 3′ UTR(Fuke et al. 2005). HesR1 also affects dopamine receptor expression in mice, and hesr1 mutant mice show defects in dopamine-dependent behaviors (Fuke et al. 2006). Although both are basic helix-loop-helix proteins, HLH-17 shows no sequence similarity to HesR1 and is most similar to the human olig-related proteins, bHLHb5/Beta3 and bHLHb4. Neither of these proteins has been shown to directly regulate expression of the dopamine transporter or dopamine receptor genes in humans. However, both proteins are part of the bHLH transcriptional network that drives retina development (Feng et al. 2006; Pennesi et al. 2006; Skowronska-Krawczyk et al. 2004), and the dopamine receptors are critical for normal retinal function (He et al. 2013; Nguyen-Legros et al. 1999; Ogata et al. 2012; Reis et al. 2007; Yang et al. 2013). Our own transgenic expression data show strong expression of hlh-17 in the cephalic sheath cells of wild-type animals and, on its own, do not support the direct regulation of dat-1 and dop-3 by HLH-17. However, mRNA for both dop-3 and hlh-17 was recently detected in glutamate receptor neurons of L2-stage animals and in the cephalic sheath cells of young adult animals (Spencer et al. 2011; see also WormViz). Furthermore, mRNA for dop-3, dat-1, and hlh-17 was detected in the dopamine neurons and panneuronal neurons of late embryos and L2-stage animals, respectively. Taken together with the epistasis analysis presented in this study, the colocalization of these mRNAs supports the possibility that HLH-17 is a direct regulator of dop-3 and dat-1. However, further studies are in progress to confirm that prediction.
Interestingly, the SWIP response in hlh-17(ns204) animals is enhanced by pre-treatment with bupropion, an antidepressant and DAT inhibitor that is used to treat ADHD in adults and children (Faraone and Glatt 2010; Jafarinia et al. 2012) but is unaffected by the antidepressant fluoxetine and the dopamine antagonist, reserpine. This finding underscores the need to develop animal models of dopamine signaling that accurately reflect the effects of reduced expression of multiple neurotransmitter signaling pathway genes, rather than complete loss of function of a single gene. Our future studies are aimed at exploiting hlh-17(ns204) for this purpose.
Acknowledgments
We thank Dr. S. Shaham and the Caenorhabditis Genetics Center (CGC) for strains (supported by the National Institutes of Health, Office of Research Infrastructure Programs, grant number P40OD010440), and Dr. W. W. Walthall and members of the Johnson laboratory for comments on this manuscript. The work described here includes a portion of the dissertation research of C.M.F., who was supported by the Molecular Basis of Disease Area of Focus at GSU. C.M.J. was supported by National Science Foundation grant number MCB0919413.
Footnotes
Communicating editor: D. G. Moerman
Literature Cited
- Allen A. T., Maher K. N., Wani K. A., Betts K. E., Chase D. L., 2011. Coexpressed D1- and D2-like dopamine receptors antagonistically modulate acetylcholine release in Caenorhabditis elegans. Genetics 188: 579–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biedermann B., Frohlich E., Grosche J., Wagner H. J., Reichenbach A., 1995. Mammalian Muller (glial) cells express functional D2 dopamine receptors. Neuroreport 6: 609–612 [DOI] [PubMed] [Google Scholar]
- Bolanos C. A., Trksak G. H., Cohen O. S., Jackson D., 2002. Differential serotonergic inhibition of in vitro striatal [3H]acetylcholine release in prenatally cocaine-exposed male and female rats. Prog. Neuropsychopharmacol. Biol. Psychiatry 26: 1339–1348 [DOI] [PubMed] [Google Scholar]
- Borodovsky N., Ponomaryov T., Frenkel S., Levkowitz G., 2009. Neural protein Olig2 acts upstream of the transcriptional regulator Sim1 to specify diencephalic dopaminergic neurons. Dev. Dyn. 238: 826–834. [DOI] [PubMed] [Google Scholar]
- Brenner S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bymaster F. P., Zhang W., Carter P. A., Shaw J., Chernet E., et al. , 2002. Fluoxetine, but not other selective serotonin uptake inhibitors, increases norepinephrine and dopamine extracellular levels in prefrontal cortex. Psychopharmacology (Berl.) 160: 353–361 [DOI] [PubMed] [Google Scholar]
- Cantwell D. P., 1998. ADHD through the life span: the role of bupropion in treatment. J. Clin. Psychiatry 59(Suppl 4): 92–94 [PubMed] [Google Scholar]
- Carlat D., 2012. Evidence-based somatic treatment of depression in adults. Psychiatr. Clin. North Am. 35: 131–142 [DOI] [PubMed] [Google Scholar]
- Carvelli L., McDonald P. W., Blakely R. D., Defelice L. J., 2004. Dopamine transporters depolarize neurons by a channel mechanism. Proc. Natl. Acad. Sci. USA 101: 16046–16051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase D. L., Koelle M. R., 2007. Biogenic amine neurotransmitters in C. elegans. WormBook Feb 20: 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase D. L., Pepper J. S., Koelle M. R., 2004. Mechanism of extrasynaptic dopamine signaling in Caenorhabditis elegans. Nat. Neurosci. 7: 1096–1103 [DOI] [PubMed] [Google Scholar]
- Chau D. T., Rada P. V., Kim K., Kosloff R. A., Hoebel B. G., 2011. Fluoxetine alleviates behavioral depression while decreasing acetylcholine release in the nucleus accumbens shell. Neuropsychopharmacology 36: 1729–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Krause M., Sepanski M., Fire A., 1994. The Caenorhabditis elegans MYOD homologue HLH-1 is essential for proper muscle function and complete morphogenesis. Development 120: 1631–1641 [DOI] [PubMed] [Google Scholar]
- Choi Y. K., Tarazi F. I., 2010. Alterations in dopamine and glutamate neurotransmission in tetrahydrobiopterin deficient spr−/− mice: relevance to schizophrenia. BMB Rep 43: 593–598 [DOI] [PubMed] [Google Scholar]
- Dellagioia N., Devine L., Pittman B., Hannestad J., 2012. Bupropion pre-treatment of endotoxin-induced depressive symptoms. Brain Behav. Immun. 31: 197–204 [DOI] [PubMed] [Google Scholar]
- Dempsey C. M., Mackenzie S. M., Gargus A., Blanco G., Sze J. Y., 2005. Serotonin (5HT), fluoxetine, imipramine and dopamine target distinct 5HT receptor signaling to modulate Caenorhabditis elegans egg-laying behavior. Genetics 169: 1425–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doonan R., Hatzold J., Raut S., Conradt B., Alfonso A., 2008. HLH-3 is a C. elegans Achaete/Scute protein required for differentiation of the hermaphrodite-specific motor neurons. Mech. Dev. 125: 883–893 [DOI] [PubMed] [Google Scholar]
- Duerr J. S., Frisby D. L., Gaskin J., Duke A., Asermely K., et al. , 1999. The cat-1 gene of Caenorhabditis elegans encodes a vesicular monoamine transporter required for specific monoamine-dependent behaviors. J. Neurosci. 19: 72–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faraone S. V., Glatt S. J., 2010. A comparison of the efficacy of medications for adult attention-deficit/hyperactivity disorder using meta-analysis of effect sizes. J. Clin. Psychiatry 71: 754–763 [DOI] [PubMed] [Google Scholar]
- Felton C. M., Johnson C. M., 2011. Modulation of dopamine-dependent behaviors by the Caenorhabditis elegans Olig homolog HLH-17. J. Neurosci. Res. 89: 1627–1636 [DOI] [PubMed] [Google Scholar]
- Feng L., Xie X., Joshi P. S., Yang Z., Shibasaki K., et al. , 2006. Requirement for Bhlhb5 in the specification of amacrine and cone bipolar subtypes in mouse retina. Development 133: 4815–4825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank C. A., Baum P. D., Garriga G., 2003. HLH-14 is a C. elegans achaete-scute protein that promotes neurogenesis through asymmetric cell division. Development 130: 6507–6518 [DOI] [PubMed] [Google Scholar]
- Fuke S., Minami N., Kokubo H., Yoshikawa A., Yasumatsu H., et al. , 2006. Hesr1 knockout mice exhibit behavioral alterations through the dopaminergic nervous system. J. Neurosci. Res. 84: 1555–1563 [DOI] [PubMed] [Google Scholar]
- Fuke S., Sasagawa N., Ishiura S., 2005. Identification and characterization of the Hesr1/Hey1 as a candidate trans-acting factor on gene expression through the 3′ non-coding polymorphic region of the human dopamine transporter (DAT1) gene. J. Biochem. 137: 205–216 [DOI] [PubMed] [Google Scholar]
- Hallam S., Singer E., Waring D., Jin Y., 2000. The C. elegans NeuroD homolog cnd-1 functions in multiple aspects of motor neuron fate specification. Development 127: 4239–4252 [DOI] [PubMed] [Google Scholar]
- Hardaway J. A., Hardie S. L., Whitaker S. M., Baas S. R., Zhang B., et al. , 2012. Forward genetic analysis to identify determinants of dopamine signaling in Caenorhabditis elegans using swimming-induced paralysis. G3 (Bethesda) 2: 961–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harfe B. D., Vaz Gomes A., Kenyon C., Liu J., Krause M., et al. , 1998. Analysis of a Caenorhabditis elegans Twist homolog identifies conserved and divergent aspects of mesodermal patterning. Genes Dev. 12: 2623–2635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q., Xu H. P., Wang P., Tian N., 2013. Dopamine D1 receptors regulate the light dependent development of retinal synaptic responses. PLoS ONE 8: e79625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafarinia M., Mohammadi M. R., Modabbernia A., Ashrafi M., Khajavi D., et al. , 2012. Bupropion vs. methylphenidate in the treatment of children with attention-deficit/hyperactivity disorder: randomized double-blind study. Hum. Psychopharmacol. 27: 411–418 [DOI] [PubMed] [Google Scholar]
- Keowkase R., Aboukhatwa M., Luo Y., 2010. Fluoxetine protects against amyloid-beta toxicity, in part via daf-16 mediated cell signaling pathway, in Caenorhabditis elegans. Neuropharmacology 59: 358–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch S., Perry K. W., Nelson D. L., Conway R. G., Threlkeld P. G., et al. , 2002. R-fluoxetine increases extracellular DA, NE, as well as 5-HT in rat prefrontal cortex and hypothalamus: an in vivo microdialysis and receptor binding study. Neuropsychopharmacology 27: 949–959. [DOI] [PubMed] [Google Scholar]
- Krause M., Park M., Zhang J. M., Yuan J., Harfe B., et al. , 1997. A C. elegans E/Daughterless bHLH protein marks neuronal but not striated muscle development. Development 124: 2179–2189 [DOI] [PubMed] [Google Scholar]
- Kullyev A., Dempsey C. M., Miller S., Kuan C. J., Hapiak V. M., et al. , 2010. A genetic survey of fluoxetine action on synaptic transmission in Caenorhabditis elegans. Genetics 186: 929–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuric E., Wieloch T., Ruscher K., 2013. Dopamine receptor activation increases glial cell line-derived neurotrophic factor in experimental stroke. Exp. Neurol. 247: 202–208 [DOI] [PubMed] [Google Scholar]
- Lewis J. A., Wu C. H., Berg H., Levine J. H., 1980. The genetics of levamisole resistance in the nematode Caenorhabditis elegans. Genetics 95: 905–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligon K. L., Fancy S. P., Franklin R. J., Rowitch D. H., 2006. Olig gene function in CNS development and disease. Glia 54: 1–10 [DOI] [PubMed] [Google Scholar]
- Lu Q. R., Sun T., Zhu Z., Ma N., Garcia M., et al. , 2002. Common developmental requirement for Olig function indicates a motor neuron/oligodendrocyte connection. Cell 109: 75–86 [DOI] [PubMed] [Google Scholar]
- Mahoney T. R., Luo S., Monet M. L., 2006. Analysis of synaptic transmission in Caenorhabditis elegans using an alicarb-sensitivity assay. Nat Protoc. 1: 1772–1777 [DOI] [PubMed] [Google Scholar]
- McDonald P. W., Hardie S. L., Jessen T. N., Carvelli L., Matthies D. S., et al. , 2007. Vigorous motor activity in Caenorhabditis elegans requires efficient clearance of dopamine mediated by synaptic localization of the dopamine transporter DAT-1. J. Neurosci. 27: 14216–14227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMiller T. L., Johnson C. M., 2005. Molecular characterization of HLH-17, a C. elegans bHLH protein required for normal larval development. Gene 356: 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton L. S., Apparsundaram S., King-Pospisil K. A., Dwoskin L. P., 2007. Nicotine increases dopamine transporter function in rat striatum through a trafficking-independent mechanism. Eur. J. Pharmacol. 554: 128–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missale C., Nash S. R., Robinson S. W., Jaber M., Caron M. G., 1998. Dopamine receptors: from structure to function. Physiol. Rev. 78: 189–225 [DOI] [PubMed] [Google Scholar]
- Nguyen M., Alfonso A., Johnson C. D., Rand J. B., 1995. Caenorhabditis elegans mutants resistant to inhibitors of acetylcholinesterase. Genetics 140: 527–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen-Legros J., Versaux-Botteri C., Vernier P., 1999. Dopamine receptor localization in the mammalian retina. Mol. Neurobiol. 19: 181–204 [DOI] [PubMed] [Google Scholar]
- Ogata G., Stradleigh T. W., Partida G. J., Ishida A. T., 2012. Dopamine and full-field illumination activate D1 and D2–D5-type receptors in adult rat retinal ganglion cells. J. Comp. Neurol. 520: 4032–4049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyallon J., Apitz H., Miguel-Aliaga I., Timofeev K., Ferreira L., et al. , 2012. Regulation of locomotion and motoneuron trajectory selection and targeting by the Drosophila homolog of Olig family transcription factors. Dev. Biol. 369: 261–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennesi M. E., Bramblett D. E., Cho J. H., Tsai M. J., Wu S. M., 2006. A role for bHLH transcription factors in retinal degeneration and dysfunction. Adv. Exp. Med. Biol. 572: 155–161 [DOI] [PubMed] [Google Scholar]
- Poole R. J., Bashllari E., Cochella L., Flowers E. B., Hobert O., 2011. A genome-wide RNAi screen for factors involved in neuronal specification in Caenorhabditis elegans. PLoS Genet. 7: e1002109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand, J. B., 2007 Acetylcholine in C. elegans WormBook Jan 30: 1–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranganathan R., Sawin E. R., Trent C., Horvitz H. R., 2001. Mutations in the Caenorhabditis elegans serotonin reuptake transporter MOD-5 reveal serotonin-dependent and -independent activities of fluoxetine. J. Neurosci. 21: 5871–5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimherr F. W., Hedges D. W., Strong R. E., Marchant B. K., Williams E. D., 2005. Bupropion SR in adults with ADHD: a short-term, placebo-controlled trial. Neuropsychiatr. Dis. Treat. 1: 245–251 [PMC free article] [PubMed] [Google Scholar]
- Reis R. A., Ventura A. L., Kubrusly R. C., de Mello M. C., de Mello F. G., 2007. Dopaminergic signaling in the developing retina. Brain Res. Brain Res. Rev. 54: 181–188 [DOI] [PubMed] [Google Scholar]
- Rieckher M., Kourtis N., Pasparaki A., Tavernarakis N., 2009. Transgenesis in Caenorhabditis elegans. Methods Mol. Biol. 561: 21–39 [DOI] [PubMed] [Google Scholar]
- Roelands B., Watson P., Cordery P., Decoster S., Debaste E., et al. , 2012. A dopamine/noradrenaline reuptake inhibitor improves performance in the heat, but only at the maximum therapeutic dose. Scand. J. Med. Sci. Sports 22: e93–e98 [DOI] [PubMed] [Google Scholar]
- Rosenberg M. B., Carroll F. I., Negus S. S., 2013. Effects of monoamine reuptake inhibitors in assays of acute pain-stimulated and pain-depressed behavior in rats. J. Pain 14: 246–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saharia K., Arya U., Kumar R., Sahu R., Das C. K., et al. , 2012. Reserpine modulates neurotransmitter release to extend lifespan and alleviate age-dependent Abeta proteotoxicity in Caenorhabditis elegans. Exp. Gerontol. 47: 188–197 [DOI] [PubMed] [Google Scholar]
- Skowronska-Krawczyk D., Ballivet M., Dynlacht B. D., Matter J. M., 2004. Highly specific interactions between bHLH transcription factors and chromatin during retina development. Development 131: 4447–4454 [DOI] [PubMed] [Google Scholar]
- Spencer W. C., Zeller G., Watson J. D., Henz S. R., Watkins K. L., et al. , 2011. A spatial and temporal map of C. elegans gene expression. Genome Res. 21: 325–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stagg S. B., Guardiola A. R., Crews S. T., 2011. Dual role for Drosophila lethal of scute in CNS midline precursor formation and dopaminergic neuron and motoneuron cell fate. Development 138: 2171–2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl S. M., Lee-Zimmerman C., Cartwright S., Morrissette D. A., 2013. Serotonergic drugs for depression and beyond. Curr. Drug Targets 14: 578–585 [DOI] [PubMed] [Google Scholar]
- Sugiura M., Fuke S., Suo S., Sasagawa N., Van Tol H. H., et al. , 2005. Characterization of a novel D2-like dopamine receptor with a truncated splice variant and a D1-like dopamine receptor unique to invertebrates from Caenorhabditis elegans. J. Neurochem. 94: 1146–1157 [DOI] [PubMed] [Google Scholar]
- Suo S., Ishiura S., 2013. Dopamine modulates acetylcholine release via octopamine and CREB signaling in Caenorhabditis elegans. PLoS ONE 8: e72578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamai K. K., Nishiwaki K., 2007. bHLH transcription factors regulate organ morphogenesis via activation of an ADAMTS protease in C. elegans. Dev. Biol. 308: 562–571 [DOI] [PubMed] [Google Scholar]
- Tiso N., Filippi A., Benato F., Negrisolo E., Modena N., et al. , 2009. Differential expression and regulation of olig genes in zebrafish. J. Comp. Neurol. 515: 378–396 [DOI] [PubMed] [Google Scholar]
- Torres G. E., Carneiro A., Seamans K., Fiorentini C., Sweeney A., et al. , 2003. Oligomerization and trafficking of the human dopamine transporter. Mutational analysis identifies critical domains important for the functional expression of the transporter. J. Biol. Chem. 278: 2731–2739 [DOI] [PubMed] [Google Scholar]
- Xie W., Li X., Li C., Zhu W., Jankovic J., et al. , 2010. Proteasome inhibition modeling nigral neuron degeneration in Parkinson’s disease. J. Neurochem. 115: 188–199 [DOI] [PubMed] [Google Scholar]
- Yang J., Pahng J., Wang G. Y., 2013. Dopamine modulates the off pathway in light-adapted mouse retina. J. Neurosci. Res. 91: 138–150 [DOI] [PubMed] [Google Scholar]
- Yoshimura S., Murray J. I., Lu Y., Waterston R. H., Shaham S., 2008. mls-2 and vab-3 Control glia development, hlh-17/Olig expression and glia-dependent neurite extension in C. elegans. Development 135: 2263–2275 [DOI] [PubMed] [Google Scholar]
- Zhou Q., Anderson D. J., 2002. The bHLH transcription factors OLIG2 and OLIG1 couple neuronal and glial subtype specification. Cell 109: 61–73 [DOI] [PubMed] [Google Scholar]